Martín-Gayo et al. report that human early thymic progenitors can undergo a GATA2-dependent myeloid developmental program leading to resident dendritic cells (DCs) upon JAG1-Notch activation. The identification of JAG1+ DC-permissive intrathymic niches validates the human thymus as a DC-poietic organ.
Abstract
A key unsolved question regarding the developmental origin of conventional and plasmacytoid dendritic cells (cDCs and pDCs, respectively) resident in the steady-state thymus is whether early thymic progenitors (ETPs) could escape T cell fate constraints imposed normally by a Notch-inductive microenvironment and undergo DC development. By modeling DC generation in bulk and clonal cultures, we show here that Jagged1 (JAG1)-mediated Notch signaling allows human ETPs to undertake a myeloid transcriptional program, resulting in GATA2-dependent generation of CD34+ CD123+ progenitors with restricted pDC, cDC, and monocyte potential, whereas Delta-like1 signaling down-regulates GATA2 and impairs myeloid development. Progressive commitment to the DC lineage also occurs intrathymically, as myeloid-primed CD123+ monocyte/DC and common DC progenitors, equivalent to those previously identified in the bone marrow, are resident in the normal human thymus. The identification of a discrete JAG1+ thymic medullary niche enriched for DC-lineage cells expressing Notch receptors further validates the human thymus as a DC-poietic organ, which provides selective microenvironments permissive for DC development.
Introduction
DCs are specialized antigen-presenting cells that are essential mediators of immunity and tolerance (Banchereau and Steinman, 1998). DCs include two major subtypes conserved in humans and mice: conventional DCs (cDC) and plasmacytoid DCs (pDC), which are characterized on the basis of their surface marker expression, morphology and ability to respond to different pathogens (Shortman and Liu, 2002; Reizis et al., 2011; Steinman, 2012; Merad et al., 2013). Despite increasing knowledge of the physiological relevance and functional heterogeneity of DCs, their developmental origin has been a matter of intense debate in recent years, especially in humans, and only recently has a general consensus been reached about the developmental link of DCs with the myeloid lineage (Liu et al., 2009; Doulatov et al., 2010; Geissmann et al., 2010; Liu and Nussenzweig, 2010; Satpathy et al., 2012; Haniffa et al., 2013; Merad et al., 2013). In mice, both pDCs and cDCs arise from a BM-resident monocyte/DC progenitor (MDP; Fogg et al., 2006) via a Flt3+ (CD135+) common DC progenitor (CDP) with restricted DC potential, that branches off from the myeloid lineage in the BM and gives rise in situ to separate precursors of cDCs or pDCs, which then migrate to peripheral organs (del Hoyo et al., 2002; Naik et al., 2007; Onai et al., 2007; Liu et al., 2009). This progressively restricted DC differentiation pathway is conserved in humans, where MDPs and CDPs derived from CD34+ hematopoietic stem/progenitor cells (HSPCs) have been identified in the BM and also in cord blood, but not in peripheral blood or lymphoid tissues, which instead contain more mature pre-DCs (Breton et al., 2015a; Lee et al., 2015).
Besides their location in the periphery, cDCs and pDCs also reside in the steady-state thymus (Bendriss-Vermare et al., 2001; Vandenabeele et al., 2001; Okada et al., 2003; Wu and Shortman, 2005), where they play a key role in the establishment of central tolerance by inducing autoreactive T cell deletion and regulatory T cell generation (Gao et al., 1990; Brocker et al., 1997; Watanabe et al., 2005; Martín-Gayo et al., 2010), but the developmental origin of thymic DCs is far less well understood than the origin of peripheral DCs. The initial view that thymic DCs were derived in situ from intrathymic T/DC lymphoid progenitors (Ardavin et al., 1993; Shortman and Wu, 1996) was later challenged by cell-fate mapping experiments supporting separate lymphoid and myeloid origins for T cells and DCs, respectively (Schlenner and Rodewald, 2010). In addition, extrathymic myeloid-restricted progenitors were shown to generate all intrathymic DC subtypes (Li et al., 2009; Luche et al., 2011), supporting their extrathymic origin. However, these results did not formally exclude early thymic progenitors (ETPs) as precursors of myeloid-associated intrathymic DCs (discussed by von Boehmer, 2009; Krueger, 2011). Indeed, ETPs in both mice and humans display myeloid potential and behave as lymphomyeloid precursors able to generate monocytes and DCs at significant frequencies within the thymus (Márquez et al., 1995; Weijer et al., 2002; Bell and Bhandoola, 2008; Wada et al., 2008; Moore et al., 2012). Although these data indicate that commitment of ETPs to the DC lineage could in principle occur within the thymus, the physiological relevance of such developmental choice is difficult to reconcile with the unique function of the thymic microenvironment as an effective inducer of signals that impose T cell development and impair non–T cell fates of ETPs, two effects that rely on activation of the evolutionary conserved Notch signaling pathway (Pui et al., 1999; Radtke et al., 1999; Wilson et al., 2001).
Studies showing that Notch inactivation impairs T cell generation from ETPs and simultaneously diverts them to DC development (Feyerabend et al., 2009) provided evidence that ETPs have a latent non–T cell potential that is repressed in the steady-state thymus by a microenvironment supporting continuous Notch signaling (Schmitt et al., 2004). In mice, this repression occurs through interaction of Notch1 with the Notch ligand Delta-like ligand 4 (DLL4), which is the essential nonredundant axis responsible for keeping ETPs on track to T cell differentiation (Hozumi et al., 2008; Koch et al., 2008; Feyerabend et al., 2009). But Notch ligands other than DLL4, such as DLL1 and Jagged1 and 2 (JAG1 and JAG2), are presented as well by thymic epithelial cells to Notch receptors (Notch1–4) expressed on thymocytes, and some of them can repress DC development (De Smedt et al., 2005; Dontje et al., 2006; García-Peydró et al., 2006; Mohtashami et al., 2010), whereas others contribute to DC differentiation in distinct locations (Cheng et al., 2007). Although this controversy may reflect nonredundant functions of distinct Notch receptors and ligands, the key question is whether the normal thymus provides discrete niches expressing particular Notch ligands that are permissive for the development of non–T-lineage cells and whether thymus-seeding precursors with lymphomyeloid potential could escape T cell fate constraints imposed by a DLL4–Notch1 inductive microenvironment to undergo DC development. In this paper, we show that selective expression of JAG1 in the outer medulla of human thymus may provide permissive niches for DC development. The identification of thymus-resident MDPs and CDPs further supports the existence of a progressively restricted DC differentiation pathway in the human thymus.
Results
ETPs resident in the human postnatal thymus generate pDCs and cDCs in vitro and in vivo
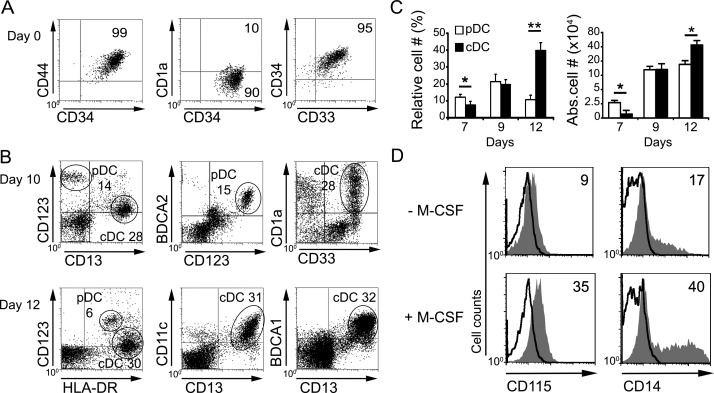
Independent studies have shown the intrinsic potential of human ETPs to generate either pDCs (Schotte et al., 2003) or cDCs (García-Peydró et al., 2006) when cocultured with OP9 stromal cells, suggesting that both DC subtypes could develop in situ within the thymus. However, the developmental relationship of intrathymic pDCs and cDCs remains unknown. To investigate this issue, we initially assessed the capability of the OP9 coculture assay for supporting the simultaneous development of pDC and cDC from CD34hi CD44hi CD1a− CD33lo ETPs isolated from the human postnatal thymus (>95% pure; Fig. 1 A). We found that ETPs cocultured with OP9 cells expanded gradually in the presence of FMS-related tyrosine kinase 3 ligand (FLT3L) and IL-7 up to day 9 (12.7 ± 2.6-fold; not depicted). Cellular proliferation paralleled the generation of significant proportions of cells with a characteristic pDC phenotype, which expressed high levels of IL-3R (CD123hi) and BDCA2 and lacked CD13 (Fig. 1 B). ETPs also gave rise to significant numbers of CD123lo CD13+ cells with high CD33 expression and variable CD1a levels (Fig. 1 B), consistent with intrathymic cDCs (Martín-Gayo et al., 2010). Absolute numbers of both DC subtypes increased in parallel up to day 9, but cDCs overgrew pDCs thereafter and represented the major DC subset by day 12 (Fig. 1 C), when most CD13+ cells coexpressed high levels of human leukocyte antigen–antigen D related (HLA-DR), CD11c, and BDCA1 cDC markers (Fig. 1 B). Detailed FACS analyses showed that, in addition to pDCs and cDCs, the ETP progeny arising along the OP9 culture comprised low numbers of CD14+ CD115+ (M-CSFR+) monocytes specifically included within the CD123lo CD13+ subset, which increased significantly in cultures supplemented with M-CSF (Fig. 1 D). Therefore, the OP9 culture system revealed a latent monocyte/DC potential of human ETPs.
Figure 1.
Human ETPs display monocyte/DC potential and generate in vitro pDCs and cDCs that resemble their intrathymic counterparts. (A) Flow cytometry phenotype of ETPs (CD34hi CD44hi CD33+ CD1a−) isolated from the human postnatal thymus. Numbers indicate percentages of positive cells for the indicated markers (n = 20). (B) Flow cytometry of cells generated from human ETPs cultured on OP9 cell monolayers in the presence of 100 IU/ml rhFLT3L and 200 IU/ml rhIL-7 (OP9 assay) at the indicated days. Numbers correspond to percentages of cells that display either a pDC (BDCA2+ CD123hi CD13−) or a cDC (BDCA2− CD123lo CD13hi) phenotype. Expression of CD1a, CD33, CD11c, and BDCA1 on cDCs and HLA-DR expression on cDCs and pDCs are also shown (n = 12). (C) Kinetics of pDC and cDC differentiation from ETPs cultured as in B. Data are shown as mean ± SEM of percentages (left) and absolute numbers (right) of pDCs and cDCs recovered at the indicated days of culture and normalized to 105 initial ETPs (n = 12). *, P < 0.05; **, P < 0.01. (D) Generation of monocytes from human ETPs cultured in the OP9 assay either as in B, or in the presence of M-CSF, for 7 d. Flow cytometry plots show the expression of CD115 and CD14 on electronically gated ETP-derived CD123lo CD13+ progenitor cells (shaded histograms). Background staining was determined with irrelevant isotype-matched Abs (empty histograms). Data are representative of one out of three experiments.
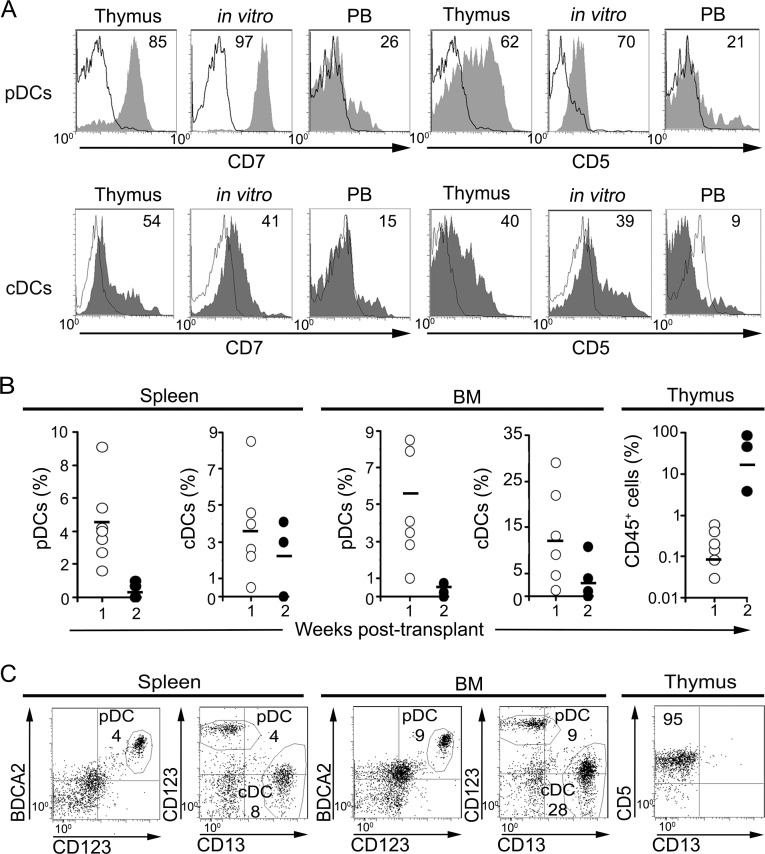
Thorough phenotypic characterization of pDCs derived in vitro from ETPs revealed that they expressed CD7 and CD5 molecules, similarly to intrathymic pDCs, whereas peripheral pDCs lack both markers (Fig. 2 A; Strobl et al., 1998; Bendriss-Vermare et al., 2001; de Yébenes et al., 2002). Likewise, ETP-derived cDCs were more similar to intrathymic cDCs in terms of CD7 and CD5 expression than to their peripheral counterparts (Fig. 2 A). These data support the possibility that at least part of intrathymic pDCs and cDCs, with a phenotype different from that of their peripheral counterparts, could arise in vivo from ETPs. Accordingly, we found that human ETPs infused into Rag2−/− × γc−/− immunodeficient mice were capable of generating pDCs and cDCs phenotypically equivalent to DCs resident in the human thymus (Fig. 2, B and C). Such DCs were already found in the BM and spleen of transplanted mice at 1 wk after transplantation, when human CD45+ cells were essentially undetectable in the thymus. Thereafter, ETP-derived pDCs and cDCs increased in mouse peripheral organs, but intrathymic DC production was hardly detectable at different times after transplantation, as human cells engrafting the host thymus typically developed into CD5+ CD13− T-lineage cells (Fig. 2, B and C). Although our xenotransplantation assay was not efficient enough to reveal the potential of human ETPs to generate DCs in the mouse thymus, our results confirmed that ETPs can generate in vivo both pDCs and cDCs with a thymus-related phenotype.
Figure 2.
ETPs resident in the human thymus generate pDCs and cDCs in vivo. (A) Flow cytometry plots show expression of CD7 (left) and CD5 (right) either on primary pDCs (top) and cDCs (bottom) resident in the human thymus and in peripheral blood (PB) or on their DC counterparts generated in vitro from ETPs in the OP9 assay. Data are representative of one of three experiments. (B) Percentages of pDCs and cDCs among human CD45+ cells recovered from the spleen and BM of Rag 2−/− × γc−/− mice transplanted by intrahepatic injection with 2–5 × 105 human ETPs. Percentages of total human CD45+ cells arising in the thymus are shown in the far-right graph. Mean values at 1 wk (n = 6) and 2 wk (n = 3) after transplantation are shown. (C) Flow cytometry analysis of human cells generated in mice transplanted with human ETPs at 1 wk after transplant. Numbers indicate percentages of CD45+ cells that display either a pDC (BDCA2+ CD123hiCD13−) or a cDC (BDCA2− CD123lo CD13hi) phenotype in the spleen and BM or percentages of CD5+ CD13− T-lineage cells developing in the thymus.
Human ETPs generate pDCs and cDCs through CD123+ progenitors transcriptionally primed for a myeloid fate
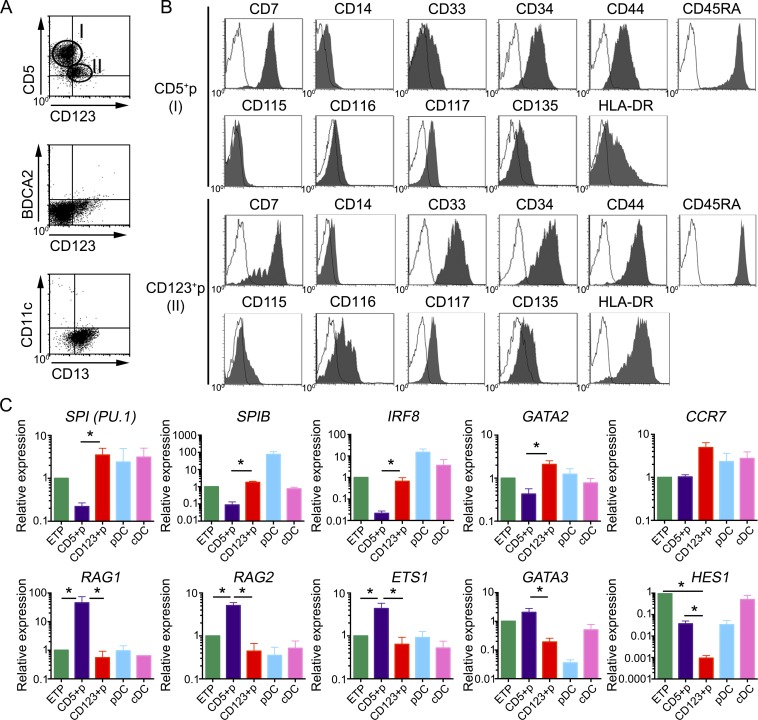
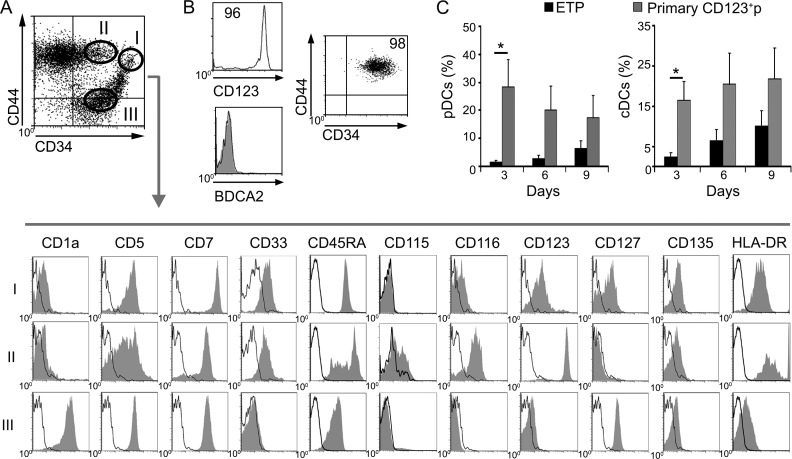
The ability of the OP9 culture to support the simultaneous generation of human pDC and cDC from ETPs prompted us to use this assay for modeling the putative DC differentiation pathways that ETPs would undergo in the human thymus. We found that cultured ETPs had diverged by day 3 into two discrete cell populations with a reciprocal expression of the lymphoid marker CD5 and the myeloid molecule CD123: a major (78 ± 2%) CD5hi CD123− population and a minor (22 ± 2%) CD5lo CD123+ subset, which lacked lineage-specific molecules (Lin−), including BDCA2 and CD11c markers of pDCs and cDCs, respectively (Fig. 3 A and not depicted). Supporting their progenitor nature, both cell subsets kept a significant expression of CD34 together with low but homogeneous levels of CD117 (c-kit) and CD135 receptors (Fig. 3 B), and they are thus hereafter referred to as CD5+ and CD123+ progenitors (CD5+p and CD123+p, respectively). According to their thymic origin, both progenitor subsets expressed high levels of CD45RA and were positive for CD7, but they displayed opposite expression levels of CD44 and HLA-DR and showed a differential distribution of CD33 and CD116 (GM-CSFRα-chain) myeloid markers, whose expression was specific of CD123+p cells. Expression of molecules associated to macrophages such as CD14 was negative, whereas CD115 expression was weak but detectable in a fraction of CD123+p cells (Fig. 3 B), suggesting that they include MDPs equivalent to MDPs identified in the human BM (Lee et al., 2015). Collectively, the divergent phenotypic features displayed by CD5+p and CD123+p suggest that they represent separate T- and myeloid-lineage progenitors, respectively.
Figure 3.
Human ETPs differentiate in vitro into separate progenitors transcriptionally primed for either a lymphoid/T or a myeloid cell fate. (A and B) Phenotype of cells generated from human ETPs cultured in the OP9 assay for 3 d. (A) Flow cytometry plots show two exclusive cell subsets defined by reciprocal expression of CD5 and CD123 as CD5+ CD123− (CD5+p) and CD5lo CD123+ (CD123+p), which lack pDC (BDCA2) and cDC (CD11c) markers but express low CD13. (B) Flow cytometry plots of electronically gated CD5+p (gate I) and CD123+p (gate II) subsets in A show expression of CD117 and CD135 cytokine receptors together with CD34 and CD45RA, a phenotype of hematopoietic progenitors. Expression of CD115 on CD123+p identifies progenitors of monocytes and DCs (MDPs), while CD116+ CD115− CD123+p correspond to CDPs with the capacity to give rise to all DCs but not to monocytes (Breton et al., 2015b). Background was defined using isotype-matched irrelevant Abs (empty histograms). Representative results from one experiment are shown (n = 9). (C) Quantitative PCR expression analysis of the indicated genes in primary ETPs, pDCs, and cDCs purified from human postnatal thymus samples and in FACS-sorted CD5+p and CD123+p ETP-derived progenitors shown in A. Data were normalized to GAPDH expression. All results are shown relative to those of ETP as mean ± SEM (n = 3). *, P < 0.05.
To characterize their lineage fate at the genetic level, FACS-sorted CD5+p and CD123+p cells derived from ETPs were compared with primary ETPs for expression of lineage-specific genes (Fig. 3 C). This study confirmed that CD5+p and CD123+p displayed divergent T versus myeloid transcriptional profiles, respectively, with a differential expression of 48 of 96 genes analyzed (not depicted). Of them, genes encoding transcription factors critical for myeloid development and pDCs and/or cDCs differentiation such as SPI1 (PU.1), SPIB, and IRF8 (reviewed by Miller et al., 2012), as well as the transcription factor GATA2 (Rodrigues et al., 2008), were highly expressed in CD123+p but were significantly lower in CD5+p (Fig. 3 C). This myeloid molecular signature was shared by primary pDCs and cDCs isolated from the human thymus and correlated with high expression of CCR7, which is also expressed on murine thymic DCs (Moore et al., 2012). Conversely, genes associated with lymphoid commitment and T cell development, including ETS1, GATA3, RAG1, RAG2, and the Notch target HES1 (Yui and Rothenberg, 2014), were overrepresented in CD5+p compared with CD123+p cells (Fig. 3 C). As a whole, these results confirm at the genetic level that CD5+p and CD123+p represent intermediate progenitors that have undertaken divergent developmental programs and indicate that human ETPs can be transcriptionally primed toward either a lymphoid/T or a myeloid fate.
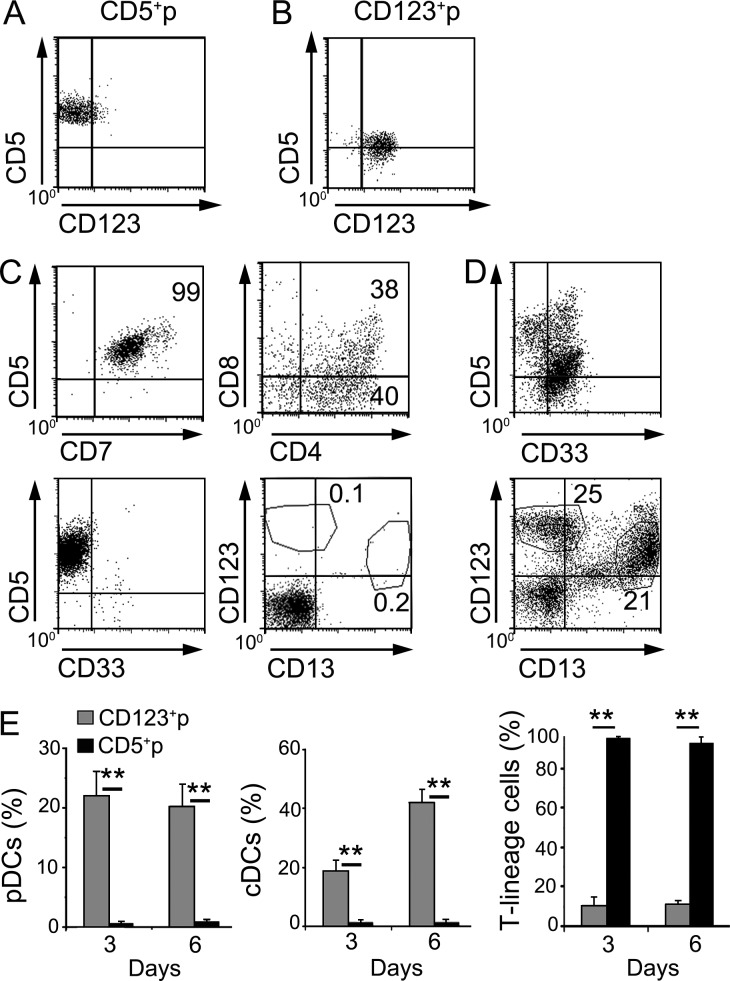
Differentiation analyses of FACS-sorted CD5+p and CD123+p subsets (Fig. 4, A and B) using the OP9 assay further confirmed at the functional level that CD5+p were lymphoid-primed progenitors that progressed efficiently along a CD5+ CD7+ T cell lineage and generated CD4+ CD8+ double-positive cells but were unable to generate CD33+ myeloid cells and were devoid of pDC and cDC potential (Fig. 4, C and E). In contrast, CD123+p progressed along a CD33+ myeloid pathway that efficiently generated large numbers of cDCs and pDCs, but they essentially lacked T cell potential (Fig. 4, D and E). Collectively, our results provide evidence that both pDCs and cDCs are generated in vitro from human ETPs through a CD123+ myeloid-primed progenitor that has lost the potential to generate T cells and may thus include CDPs. In contrast, ETPs generated T-lineage cells only through CD5+p intermediates (Fig. 4 E).
Figure 4.
Human ETPs generate pDCs and cDCs through myeloid-primed CD123+ CDPs. (A and B) Flow cytometry analysis of surface CD5 versus CD123 expression on FACS-sorted CD5+p (A) and CD123+p (B) progenitor subsets generated from human ETPs cultured for 3 d in the OP9 assay, as shown in Fig. 3 A. (C and D) Phenotype of cells generated from FACS-sorted CD5+p (C) and CD123+p (D) progenitors upon culture for 3 additional days in the OP9 assay. CD123+p progenitors generated pDCs (CD123+ CD13−) and cDCs (CD123− CD13+), whereas CD5+p progenitors exclusively generated T-lineage cells (CD5+ CD7+ CD4+ CD8+). (E) Differentiation kinetics of pDCs (top), cDCs (middle), and T-lineage cells (bottom), derived from either CD123+p or CD5+p sorted progenitors upon culture as in A for 3 and 6 additional days, respectively. Mean percentages ± SEM are shown (n = 8). **, P < 0.01.
Identification of myeloid-primed MDPs and CDPs resident in the human thymus
To determine the physiological relevance of the progenitor-progeny DC developmental pathway revealed in the OP9 assay, we next examined the postnatal human thymus for the presence of CD123+ DC progenitors. To this end, we analyzed thymus cell suspensions that were depleted of mature thymocytes and were thus enriched for CD34hi ETPs and more mature CD34+ intermediate progenitors. The latter included a subset of T cell committed precursors with down-regulated CD44 expression (CD44lo) and a population of non–T cell progenitors, which keep high CD44 expression (CD44hi; Márquez et al., 1995; de Yébenes et al., 2002). Accordingly, we found that primary CD44lo CD34+ progenitors lacked CD123 and other myeloid markers characteristic of myeloid-primed CD123+p generated in culture, but, like in vitro–derived CD5+p, they expressed CD1a, CD5, and CD127 (IL-7R) T-lineage molecules, in addition to CD7 and CD45RA (Fig. 5 A). In contrast, primary CD44hi CD34+ progenitors expressed CD123 like in vitro–derived CD123+p, although at higher levels, and also displayed weak levels of the CD135 progenitor molecule, and a significant expression of several myeloid-related markers, including CD33, HLA-DR, and CD116, but lacked the BDCA2 pDC molecule (Fig. 5, A and B). As expected given their intrathymic origin, CD123+ progenitors maintained significant CD7 and CD45RA and intermediate CD5 expression levels, despite their myeloid phenotype. Notably, some of them also expressed CD115, a hallmark of MDPs (Lee et al., 2015), although cells with a CD115− DC progenitor phenotype represented a major subset. Thus, we concluded that Lin− CD123+ progenitors resident in the human thymus include both CD115+ and mostly CD115− myeloid-primed DC progenitors, which may represent the intrathymic counterparts of MDPs and CDPs, respectively, identified in BM and cord blood (Lee et al., 2015). Confirming the immediate DC potential of primary CD123+ thymocytes (Fig. 5 B), we found that they generated both pDC and cDC subsets in the OP9 assay with kinetics similar to ETPs, but, as expected of more restricted precursors, they showed significantly higher DC generation efficiencies than ETPs (Fig. 5 C). Collectively, these results provide evidence that the human thymus includes CD123+ progenitors placed downstream of ETPs, which mostly contain CD115− DC-restricted precursors with high pDC and cDC potential and a CDP-like phenotype (hereafter referred to as thymic CDPs).
Figure 5.
Characterization of CD123+ MDPs and CDPs resident in the human thymus. (A) Flow cytometry of CD44 versus CD34 expression in human thymocytes depleted of T-lineage cells, as described in Materials and methods, defines three progenitor populations: (I) CD44hi CD34hi ETPs, (II) CD34lo CD44hi myeloid-like intrathymic progenitors, and (III) CD34lo CD44lo lymphoid-like intrathymic progenitors. Shaded histograms show expression of the indicated markers on electronically gated subsets I, II, and III (bottom). Isotype-matched irrelevant Abs were used to define background staining (empty histograms; n = 3). According to their phenotype, cells in gate II resemble CDPs and also include some CD115+ MDPs. (B) Flow cytometry plots show the phenotype of myeloid progenitors (gate II) that were magnetically sorted on the basis of CD123 expression. Data from a representative experiment are shown (n = 4). (C) Kinetics of pDC and cDC differentiation from ETPs and primary CD123+ myeloid progenitors shown in B, isolated from the same thymus sample and cultured in the OP9 assay as in Fig. 1 B. Data are shown as mean percentages ± SEM (n = 4). *, P < 0.05.
The human thymus contains a JAG1+ medullary niche enriched for pDCs and cDCs expressing Notch receptors
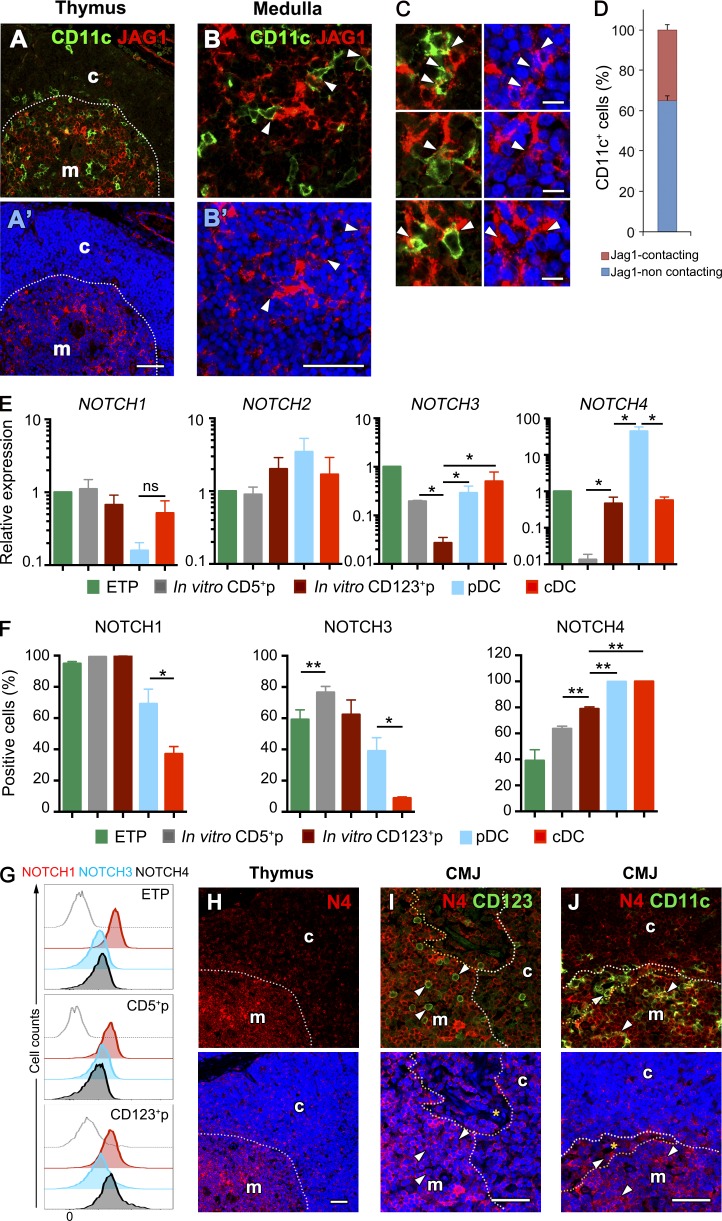
The identification of CDPs among primary CD123+ myeloid-primed thymocytes suggests that the thymus microenvironment might support their generation in situ from ETPs. Therefore, permissive niches for myeloid/DC development may exist in the human thymus. To analyze this possibility, we assessed the spatial distribution of intrathymic Notch ligands, with special attention to the outer medulla, where cDCs and pDCs are mostly located (Kurobe et al., 2006; Martín-Gayo et al., 2010). Immunohistochemistry and confocal microscopy showed that human CD11c+ cDCs were confined mostly to a particular region in the thymus outer medulla, close to the corticomedullary junction, which was rich in JAG1 expression (Fig. 6, A and B) but devoid of other Notch ligands (not depicted), whereas JAG1+ cells were essentially absent in the cortex. Quantitative analysis revealed high proportions (up to 25%) of CD11c+ cells establishing adjacent contacts with JAG1-expressing medullary cells (Fig. 6, C and D). Because JAG1-mediated signaling is known to impair T cell generation, at least in humans, but supports non–T cell development (Van de Walle et al., 2011), it is conceivable that human ETPs could develop along the DC lineage in the outer medulla upon interaction with JAG1 through surface Notch receptors. Quantitative PCR analyses of NOTCH(1–4) gene expression confirmed that ETPs transcribed all four Notch receptors (Fig. 6 E). Of them, expression of NOTCH1 and NOTCH2 remained essentially unchanged in both CD123+p and CD5+p derived from ETPs. In contrast, NOTCH3 was significantly down-regulated in CD123+p, whereas NOTCH4 was specifically down-regulated in CD5+p, but it was distinctively maintained in CD123+p and was also transcribed in intrathymic cDCs and pDCs (Fig. 6 E). FACS analyses confirmed that NOTCH4 was preferentially expressed at the protein level on myeloid-primed CD123+ progenitors, as well as on intrathymic pDCs and cDCs, whereas CD5+ T-lineage progenitors were enriched in NOTCH1- and NOTCH3-expressing cells (Fig. 6 F). According with a preferential expression of NOTCH4 in myeloid-lineage thymocytes, we found that primary CD123+ CDPs expressed increased levels of NOTCH4, compared with their CD5+p counterparts and with ETPs (Fig. 6 G), and NOTCH4 expression was observed as well in situ on thymic medullary DCs that coexpressed either CD123 or CD11c and thus include pDCs or cDCs, respectively (Fig. 6, H–J).
Figure 6.
The human thymus contains a JAG1+ medullary niche enriched for pDCs and cDCs expressing NOTCH4. (A–C) Immunohistochemistry and confocal microscopy analysis of JAG1 (red) and CD11c (green) expression in the human postnatal thymus. Topro labeling of cellular nuclei is shown in blue. Bars: 50 μm (A and B); 10 μm (C). (A) Thymus cortex (c) and medulla (m) are separated by the corticomedullary junction (dotted line). (B) Cell contacts between CD11c+ DCs and JAG1+ stromal cells confined to the thymus medulla (arrowheads). (C) Enlarged details of cell contacts between CD11c+ and JAG1+ cells in the thymus medulla marked by arrowheads. (D) Quantification of CD11c+ cells contacting JAG1+ cells in C. Data show mean percentages ± SEM per 63× field of three independent thymus samples (n = 30). (E) Quantitative PCR expression analysis of NOTCH (1–4) transcription in primary ETPs, pDCs, and cDCs purified from human postnatal thymus samples and in FACS-sorted CD5+p and CD123+p ETP-derived progenitors shown in Fig. 4 (A and B), respectively. Data were normalized to GAPDH expression. All results are shown relative to those of ETP as mean ± SEM (n = 3). *, P < 0.05. (F) Percentages of cells in E positive by flow cytometry for the expression of NOTCH1, 3, and 4. Data are shown as mean ± SEM of cell percentages (n = 3). *, P < 0.05; **, P < 0.01. (G) Flow cytometry analysis of cell surface NOTCH1, 3, and 4 expression on primary ETPs, CD5+p and CD123+p cells isolated from the human thymus (shaded histograms). Gray dotted histograms represent background staining obtained with irrelevant isotype-matched Abs. (H–J) Immunohistochemistry and confocal microscopy analysis of NOTCH4 protein expression (red) in combination with either CD123 or CD11c (green) in the human postnatal thymus. (H′–J′) Topro labeling of cellular nuclei is shown in blue. Bars, 50 μm. (H) NOTCH4 expression is confined to the human thymus medulla. Thymus cortex (c) and medulla (m) are separated by the corticomedullary junction (dotted line). (I and J) Enlarged details of the corticomedullary junction (CMJ) area of human thymus showing expression of NOTCH4 on CD123+ (I) and CD11c (J) DCs (arrowheads) confined to the medulla, close to the perivascular space (dotted lines).
JAG1- but not DLL1-induced Notch signaling is permissive for the generation of thymic CDPs from bipotent T/DC ETPs
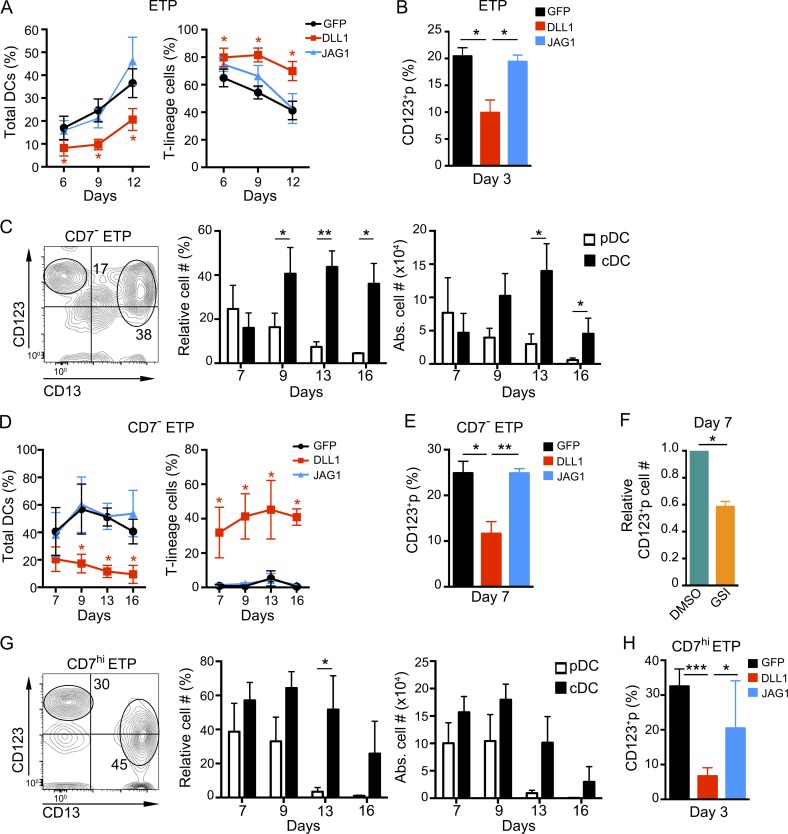
The restricted spatial distribution of JAG1 in a DC-rich thymus niche is compatible with a role for JAG1-mediated Notch signaling in intrathymic myeloid/DC priming and development. This possibility concurs with the fact that the OP9 cells used to model DC development of ETPs constitutively express low levels of surface JAG1, which may be sufficient to trigger a myeloid/DC developmental program in vitro (de Pooter et al., 2006), but they lack surface expression of other Notch ligands including JAG2, although they transcribe it (not depicted; Schmitt and Zúñiga-Pflücker, 2002; Van de Walle et al., 2011). To investigate the contribution of JAG1 to DC development, we compared the outcome of ETPs cultured with OP9 cells that were transduced with a retroviral bicistronic vector encoding either human JAG1 and GFP as reporter (OP9–JAG1), or GFP alone as control (OP9–GFP). OP9 cells transduced with DLL1 and GFP (OP9–DLL1), were used in parallel cultures to model T cell development of ETPs receiving high Notch signaling, which impairs diversion to the myeloid/DC lineage (De Smedt et al., 2005; García Peydró et al., 2006). As shown in Fig. 7 A, kinetic analyses revealed that production of total DCs, including pDCs and cDCs, was induced with similar efficiencies in OP9–JAG1 and OP9–GFP cultures but was significantly impaired in DLL1 cultures. Conversely, OP9–DLL1 cultures efficiently supported the development and long-term maintenance of T-lineage cells (Fig. 7 A), which acquire CD1a, CD4, and CD8 markers and finally express the mature TCR (not depicted; De Smedt et al., 2005; García Peydró et al., 2006). Therefore, JAG1 and DLL1 seem to prime ETPs toward either myeloid/DC or T cell development, respectively, a possibility supported by the fact that generation of CD123+p from ETPs was significantly impaired in OP9–DLL1 cultures compared with OP9–JAG1 and OP9–GFP cultures (Fig. 7 B). However, both OP9–JAG1 and OP9–GFP seemed permissive for the initial development of CD5+p and CD1a+ preT cells from ETPs, although such T cell progenitors decreased progressively thereafter in both conditions and never generated TCR+ thymocytes (Fig. 7 A and not depicted). According to previous results (Van de Walle et al., 2011), these data suggest that isolated ETPs may include some progenitors committed to the T cell lineage in vivo, which can progress in vitro along early T cell development in the absence of DLL1.
Figure 7.
JAG1- but not DLL1-mediated Notch signaling supports the generation of CD123+ thymic CDPs from human CD7−, CD7hi, and total ETPs. (A and D) Percentages of either total DCs, including pDCs and cDCs (left), or CD5+ T-lineage cells (right), derived from total ETPs (A) or CD7− ETPs (D), cultured onto OP9–GFP, OP9–DLL1, or OP9–JAG1 stromas, in the presence of FLT3L and IL-7 (n = 3). (B, E, and H) Percentages of CD123+ myeloid progenitors, mostly including CDPs as shown in Fig. 3, arising from human total (B), CD7− (E), or CD7hi ETPs (H), cocultured for the indicated days with OP9–DLL1, OP9–JAG1, or OP9–GFP stromal cells as in A (n = 3). (C and G) Flow cytometry phenotype (left) and relative (middle) and absolute (right) numbers of pDCs and cDCs derived from 105 FACS-sorted CD7− (C) or CD7hi (G) ETPs in the OP9 assay, at the indicated days. Numbers in quadrants correspond to percentages of gated cells that display either a pDC (CD123hi CD13−) or a cDC (CD123lo CD13hi) phenotype after 9 d of culture (n = 3 or 4). (F) Numbers of CD123+ myeloid progenitors generated from human CD7− ETPs in OP9–JAG1 cultures supplemented with either 100 nM of the gamma-secretase inhibitor (GSI) CompE or DMSO as a vehicle. Cell numbers in GSI cultures were normalized to those recovered in DMSO cultures (n = 3). Data in A–H are shown as mean ± SEM values. *, P < 0.05; ** P < 0.01; ***, P < 0.001.
ETPs defined as shown in Fig. 1 A are heterogeneous regarding CD7 expression and comprise mostly cells with high CD7 levels (CD7hi), including the more mature ETPs, and also a minor subset of earlier CD7− thymocytes (<2%), which have been identified as primitive lymphomyeloid thymic progenitors (Hao et al., 2008). Thus, we ought to assess the impact of Notch ligand-specific signaling on the DC/T developmental potential of both ETP populations. To this end, CD7− and CD7hi thymocytes were FACS-sorted from CD34hi ETPs, which were simultaneously depleted of putative CD123+ DC-lineage contaminants (Fig. S1), and both subsets were then assayed in OP9 cocultures. We first confirmed that as total ETPs, CD7− ETPs were able to generate CD123hi CD13− pDCs and CD123lo CD13+ cDCs with similar efficiencies early in OP9 cocultures (day 7), but cDCs increased progressively thereafter and overgrew pDCs (Fig. 7 C). The involvement of JAG1 in myeloid/DC priming and development of CD7− ETPs was assessed in OP9–GFP and OP9–JAG1 cultures, which were shown to support total DC generation with identical efficiencies, whereas both cultures completely prevented the emergence of T-lineage cells (Fig. 7 D). Conversely, OP9–DLL1-derived signals efficiently induced T cell commitment of CD7− ETPs and supported the efficient generation of CD7+ CD5+ cells but markedly impaired the development of DCs (Fig. 7 D). Therefore, our results suggest that levels of JAG1 ligand expressed on OP9–GFP and OP9–JAG1 stromal cells efficiently prime the most immature CD7− ETPs toward a myeloid/DC fate, whereas DLL1-mediated signaling selectively induces T cell priming at the expense of myeloid development. Accordingly, we found that DLL1 expression impaired the production of DCs by preventing the generation of myeloid-primed CDPs, whereas progression of CD7− ETPs along a myeloid/DC-lineage program and CDP generation seemed to be JAG1 dependent (Fig. 7 E). Supporting such an inductive role of JAG1 in myeloid/DC priming, we found that generation of CDPs from CD7− ETPs was significantly reduced when Notch activation was impaired by treatment with γ-secretase inhibitors (GSIs) in OP9–JAG1 cocultures (Fig. 7 F).
Besides CD7− ETPs, sorted CD7hi ETPs also displayed DC potential in OP9 cultures and generated both pDCs and cDCs with kinetics similar to those of total ETPs (Fig. 7 G). Notably, in both OP9 and OP9–JAG1 cocultures, CD7hi ETPs developed along the myeloid/DC pathway through CD123+ CDP progenitors, but CDP generation was significantly inhibited in OP9–DLL1 cultures (Fig. 7 H). Therefore, CD7hi ETPs, although enriched in T-lineage potential compared with their CD7− counterparts (not depicted; Hao et al., 2008), still retain the capability to generate pDCs and cDCs.
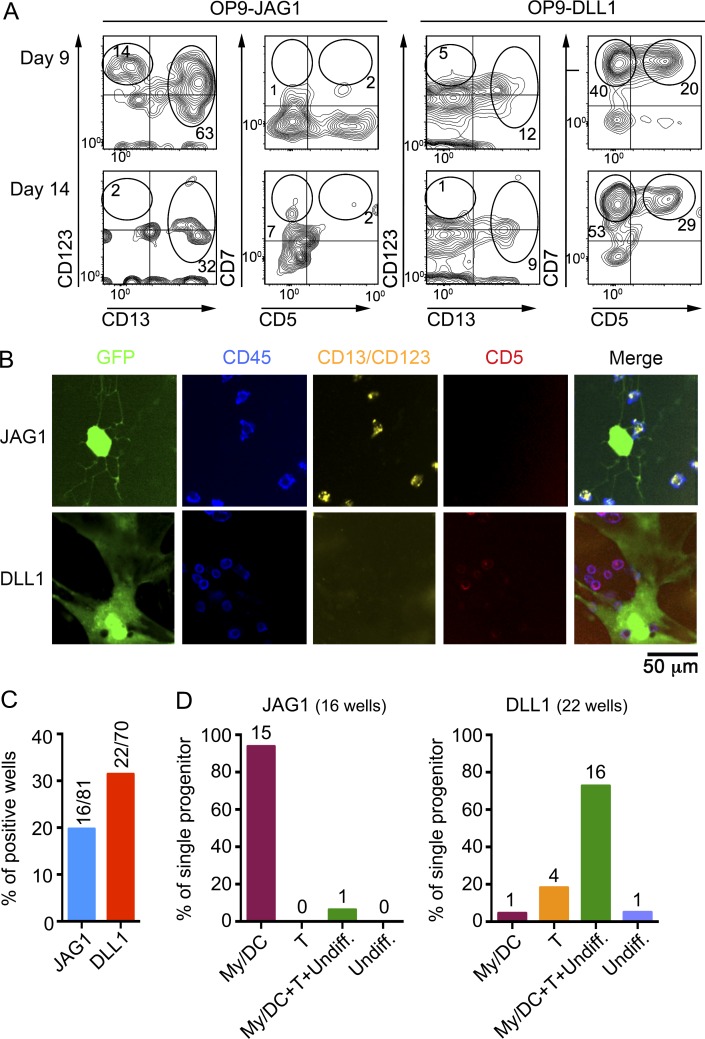
Collectively, our results suggest that the human thymus is seeded by CD7− T/DC bipotent progenitors that would develop along either the T cell or the myeloid/DC lineage in response to specific Notch ligands and propose that the latter is selectively dependent on JAG1. Formal support for this notion was obtained at the clonal level using a single cell progenitor/product coculture assay. To this end, FACS-sorted individual CD7− ETPs (Fig. S1) were deposited in 384-well culture plates seeded with either OP9–JAG1 or OP9–DLL1 cells and analyzed for the generation of DC- or T-lineage cells using appropriate marker combinations established in control CD7− ETP bulk cultures set up in parallel (Fig. 8 A). According to bulk culture developmental kinetics, the clonal progeny of CD7− ETPs cultured onto OP9–JAG1 or OP9–DLL1 cells was analyzed at day 9 or 14, respectively, after labeling with mAbs against CD45, CD5, and CD13 plus CD123. Images of clonal cultures were acquired and analyzed in an Opera confocal microscope (Fig. 8 B). The relative clonal efficiency of CD7− ETPs, as measured by their ability to give rise to CD45+ cells, was 20% (16 of 81 wells) and 33% (22 of 70 wells) in OP9–JAG1 and OP9–DLL1 cocultures, respectively (Fig. 8 C). All positive wells were further evaluated for clonal potential to generate T-lineage cells (CD5+ CD13− CD123−) or myeloid/DC-lineage cells (CD13+ and/or CD123+) cells. Of the positive wells obtained in OP9–JAG1-seeded plates, 94% (15 of 16) contained exclusively myeloid/DC cells, some of which expressed CD5 (9.0 ± 3.8%). No clones producing exclusively T-lineage cells were recorded, but one clone produced a multilineage progeny consisting of myeloid/DC, T-lineage cells, and undifferentiated CD45+ cells lacking CD5, CD13, and CD123 (Fig. 8 D). Of the 22 positive wells obtained in plates seeded with OP9–DLL1, one clone (4.5%) generated exclusively DCs, one contained only undifferentiated cells, 18% (4 of 22) only T-lineage cells, and 70% were multilineage clones that yielded T-lineage cells (32 ± 4.9%) in combination with myeloid/DC cells (23 ± 4.3%) and undifferentiated cells (41 ± 3.6%; Fig. 8 D). Therefore, these results formally prove the existence of a clonal thymic progenitor that possesses both myeloid/DC and T cell potential, and reveal the robust myeloid/DC potential of CD7− ETPs at the clonal level in response to JAG1-mediated Notch signaling.
Figure 8.
Single CD7− ETPs generate both dendritic cells and T-lineage cells in vitro. (A) Phenotype of myeloid/DC- and T-lineage cells generated at the indicated days in bulk cultures from human CD7− ETPs (105 cells) seeded onto OP9–JAG1 or OP9–DLL1 cells in the presence of FL3TL and IL-7. Numbers in quadrants represent percentages of pDCs (CD123hi CD13−), cDCs (CD13hiCD123lo), T-lineage (CD7+CD5+), and CD7+CD5− progenitors. (B) Representative confocal microscopy images of the clonal progeny obtained from culturing individual CD7− ETPs in OP9–JAG1 and OP9–DLL1 cultures at day 9 or 14, respectively. T-lineage cells were identified among CD45+ hematopoietic cells (blue) by exclusive expression of CD5 (red). Myeloid/DC-lineage cells were detected by coexpression of CD45 and CD13 and/or CD123 (yellow). OP9 cells expressing GFP are shown in green. Original magnification 20×. Scale bar is shown. (C) Clonal efficiency calculated on the basis of the number of positive wells is indicated. (D) Graphs represent the cellular output of all positive single cell cultures of CD7− ETPs. The number of wells per category is indicated at the top of each bar. My/DC, myeloid/DC-lineage (CD45+ CD13/CD123+); T, T-lineage (CD45+ CD13/CD123− CD5+); Undiff, undifferentiated cells (CD45+ CD13/CD123− CD5−).
Strong Notch signaling inhibits the generation of CDPs by repressing GATA2
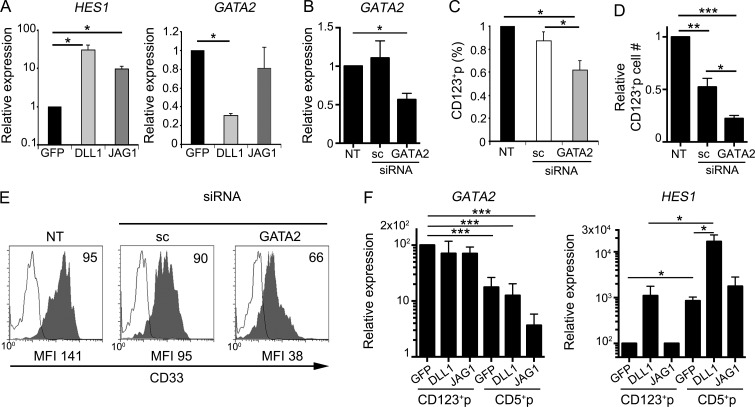
To investigate the molecular mechanisms underlying the differential lineage outcomes of ETPs receiving either DLL1- or JAG1-mediated Notch signaling, we next assessed the impact of each ligand on the expression of several lineage-associated genes. Because of cell number limitations, total ETPs were used in these experiments, using HES1 expression as readout of Notch signaling strength. We found that Notch activation induced by OP9–JAG1 after a 24 h resulted in a 10-fold up-regulation of HES1 in ETPs compared with control OP9–GFP cultures. As expected, DLL1 was more efficient than JAG1 in terms of Notch activation strength (Van de Walle et al., 2013) and induced a higher (about threefold) HES1 expression (Fig. 9 A). Notch activation levels were then correlated with expression of genes shown in Fig. 3 C to associate with the generation of CD123+ CDP-like progenitors. Of them, IRF8 and SPIB were expressed at similar levels in the ETP progeny recovered from OP9–JAG1, OP9–DLL1 and control OP9–GFP cocultures (not depicted). However, GATA2 transcriptional levels were significantly reduced (69.6 ± 2.5%) after coculture with OP9–DLL1 but not OP9–JAG1 cells compared with OP9–GFP controls (Fig. 9 A). Collectively, our data indicate that specific GATA2 suppression induced by DLL1-mediated strong Notch signaling correlates with an impaired generation of CD123+p progenitors, whereas high GATA2 expression supported by JAG1-mediated weaker Notch activation might be required for ETPs to acquire a myeloid/DC fate.
Figure 9.
JAG1- but not DLL1-mediated Notch signaling supports a GATA2-dependent generation of CD123+ thymic CDPs from human ETPs. (A) Quantitative PCR analysis of HES1 (left) and GATA2 (right) gene expression in total ETPs cultured for 24 h with OP9–DLL1, OP9–JAG1, or OP9–GFP cells, in the presence of FLT3L and IL-7. Data were normalized to GAPDH expression. Results are shown as mean ± SEM expression values normalized to those of ETPs cultured onto OP9–GFP controls (n = 3). *, P < 0.05. (B) Relative expression of GATA2 in human ETPs that were either nucleofected with GATA2-specific or scramble (sc) siRNAs or nontransfected (NT) and then cultured onto OP9–GFP cells for 3 d in the presence of FLT3L and IL-7. Data were normalized to GAPDH expression and are shown as mean ± SEM values normalized to those of nontransfected (NT) ETPs (n = 3). *, P < 0.05. (C and D) Numbers of CD123+ CDPs generated as shown in B from human ETPs nucleofected with GATA2 or sc siRNAs. Data are represented as mean ± SEM percentages (C) or absolute numbers (D) of cells normalized to those of nontransfected ETPs (NT; n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) Flow cytometry histograms show CD33 expression levels (shaded) of CD123+ CDPs in C and D. Background was determined using isotype-matched irrelevant Abs (empty histograms). Numbers indicate percentages of positive cells. Mean fluorescence intensity values are indicated at the bottom. (F) Quantitative PCR analysis of relative expression of GATA2 and HES1 transcripts in cells derived from FACS-sorted CD123+p or CD5+p subsets shown in Fig. 4 B that were cultured for 48 additional hours onto OP9–DLL1 or OP9–JAG1. Data are normalized to GAPDH expression and are shown as mean ± SEM values normalized to those of CD123+p cultured on OP9–GFP control stromal cells (n = 3). *, P < 0.05; ***, P < 0.001.
To directly investigate the role of GATA2 in myeloid/DC commitment, we assessed the lineage outcome of ETPs in which GATA2 expression was knocked down by specific siRNAs. GATA2 silencing (up to 65% inhibition; Fig. 9 B) resulted in a significant decrease, in both relative and absolute numbers, of CD123+ CDPs generated after 3 d of culture in the OP9 assay, compared with control ETPs expressing scramble siRNA (Fig. 9, C and D). Therefore, Notch-mediated control of GATA2 expression is at least partially involved in regulating the myeloid developmental program that triggers the generation of CD123+ CDPs from human ETPs. Moreover, the few GATA2-silenced CD123+ cells that managed to arise in culture consistently showed lower expression levels of myeloid-lineage markers, such as CD33, than controls (Fig. 9 E), suggesting that further maturation of CDPs along the DC lineage may likewise be GATA2 dependent. To investigate this possibility, we quantified GATA2 expression levels in the progeny of FACS-sorted CD123+p cells derived from ETPs in the OP9 assay as shown in Fig. 4 B, upon culture for 48 additional hours with either OP9–GFP, OP9–JAG1, or OP9–DLL1. Supporting the involvement of GATA2 in the generation of DCs from CDPs, we found that GATA2 expression was preserved in all culture conditions in the progeny of CD123+p cells, and relative expression was higher than that observed in their corresponding T-lineage counterparts derived from CD5+p (Fig. 9 F). More important, levels of GATA2 expression in CD123+p descendants were equivalent regardless of the Notch ligand expressed by OP9 cells (Fig. 9 F). Therefore, although weak JAG1- and strong DLL1-induced Notch signaling either support or inhibit, respectively, GATA2 expression in ETPs (Fig. 9 A), once ETPs have undergone the myeloid/DC developmental program, GATA2 transcription is preserved in committed CDPs, even upon strong Notch signaling provided by DLL1.
Both DLL1 and JAG1 support the survival of thymic CDPs and their development into DCs
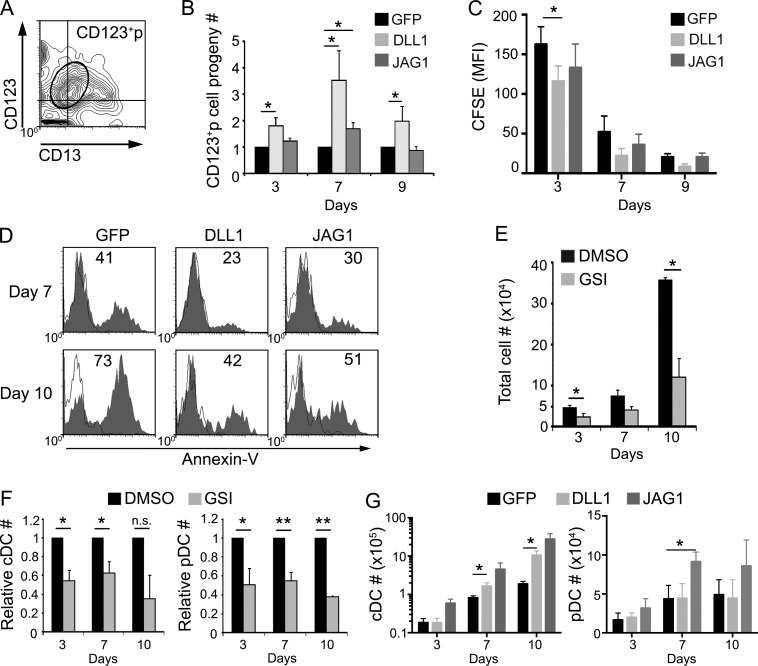
The aforementioned results could suggest that developmental progression of CD123+ CDPs is independent of ligand-specific Notch signaling. On the contrary, kinetic analysis of cell cultures set up from CD123+p as above (Fig. 9 F) showed that numbers of cells keeping the CD123+ CD13lo CDP phenotype (Fig. 10 A) increased significantly along time in OP9–DLL1 cultures compared with OP9–GFP controls but less so in OP9–JAG1 cultures, suggesting that Notch activation induced by Notch ligands, particularly DLL1, support the expansion of developing CDPs (Fig. 10 B). However, CFSE labeling experiments showed similar cell proliferation rates in all culture conditions, indicating that specific Notch signaling does not obviously impact CDP proliferation in vitro (Fig. 10 C). In contrast, Annexin V labeling revealed that numbers of apoptotic cells decreased in OP9–DLL1 and OP9–JAG1 cultures compared with OP9–GFP controls, especially at late culture time points (Fig. 10 D). Therefore, Notch signaling induced by DLL1, and to a lesser extent by JAG1, supports the survival of CD123+ CDPs generated in vitro from ETPs.
Figure 10.
Both DLL1- and JAG1-mediated Notch signaling support the survival of intrathymic CD123+ CDPs and their development into pDCs and cDCs. (A) Flow cytometry analysis of CD123 versus CD13 expression on the cell progeny of FACS-sorted CD123+ CDPs derived from ETPs as shown in Fig. 4 B (CD123+p), which were cultured for 3 additional days onto OP9–GFP cells. The electronic gate defines the original CD123+ CD13lo/- phenotype of the CD123+p cultured population. (B) Relative cell numbers of the CD123+p cell progeny gated as in A, recovered at the indicated days upon coculture with OP9–DLL1, OP9–JAG1, or OP9–GFP stroma. Data are shown as mean ± SEM of cell numbers normalized to those of OP9–GFP cultures (n = 7). *, P < 0.05. (C) Proliferation kinetics analyzed by CFSE labeling and flow cytometry of FACS-sorted CD123+p shown in Fig. 4 B, and cultured as in B. Bars and error bars are MFI ± SEM,(n = 3). *, P < 0.05. (D) Flow cytometry analysis of apoptotic cells (shaded histograms) recovered at the indicated days from cultures in B. Numbers show percentages of Annexin V+ apoptotic cells. Empty histograms show background staining. Data correspond to a representative experiment (n = 3). (E) Absolute numbers of cells derived from primary CD123+ CDPs, isolated from the human thymus as shown in Fig. 5, which were cultured for the indicated days on OP9 stroma in the presence of either 100 nM of CompE (GSI) or DMSO as control. Bars and error bars are mean ± SEM (n = 3). *, P < 0.05. (F) Relative numbers of cDCs (left) and pDCs (right) generated from intrathymic CD123+ CDPs in cultures shown in E. Data are shown as mean ± SEM of cell numbers in GSI cultures normalized to those in DMSO cultures (n = 3). *, P < 0.05; **, P < 0.01. (G) Absolute numbers of cDCs (left) and pDCs (right) generated from primary thymic CD123+ CDPs, cultured on OP9–DLL1, OP9–JAG1, or OP9–GFP stromas for the indicated days. Data are shown as mean ± SEM cell numbers normalized to 105 input CDPs (n = 7). *, P < 0.05.
Confirming the physiological relevance of the aforementioned results, we found that numbers of primary CD123+ CDPs isolated from the human postnatal thymus dropped markedly in OP9–GFP cultures when Notch activation was inhibited by treatment with GSI (Fig. 10 E), and generation of both pDCs and cDCs was concurrently reduced (Fig. 10 F). Therefore, survival of CD123+ CDPs within the human thymus may be dependent on a Notch signaling supportive microenvironment. Accordingly, development of intrathymic CDPs along the pDC and cDC pathways was improved in vitro in the presence of Notch ligands. Notably, ligand-specific signals had a differential impact on cDC versus pDC differentiation from thymic CDPs, as cDC production was significantly increased by DLL1, but pDC generation was more efficient in the presence of JAG1 (Fig. 10 G). As a whole, our data indicate that once intrathymic CDPs have been generated upon weak Notch activation, Notch signaling provided by distinct ligands may further regulate their differentiation into pDCs and cDCs in the human thymus.
Discussion
The presence of DCs in the steady-state thymus has been widely documented in mice and humans, but their developmental origin has been a matter of intense debate in recent years. In particular, whether intrathymic DCs are circulating DCs recruited to the thymus or are generated in situ remains an elusive but especially relevant issue, as peptide-presenting thymic DCs play a key role in the establishment of central tolerance (Banchereau and Steinman, 1998). Recent advances on the characterization of circulating DCs in mice suggest that at least some intrathymic DCs do not belong to the circulating subtype but may instead be generated in situ (Li et al., 2009). However, no DC-lineage–restricted progenitors have so far been identified in the thymus. In this study, we demonstrate that myeloid-primed progenitors for pDCs and cDCs and also macrophages are normally resident in the steady-state human postnatal thymus and show that these progenitors include the intrathymic counterparts of MDPs and CDPs, which develop from CD34+ HSPCs in the BM (Lee et al., 2015). Notably, modeling intrathymic DC development from ETPs in the OP9 in vitro assay allowed us to define the developmental pathway that may lead to DC production in the human thymus and to recognize that this pathway faithfully reproduces the DC hematopoiesis process described before in the marrow (Lee et al., 2015). As it was reported that MDPs and CDPs resident in the BM do not circulate through the blood and are unable to reach lymphoid tissues (Lee et al., 2015), we concluded that their intrathymic counterparts identified in this study must be generated in situ, supporting a role for the thymus as a DC-poietic organ.
Intrathymic DC hematopoiesis is difficult to reconcile with the conventional view that the primary function of the thymus is to provide a specific microenvironment that instructs BM-derived progenitors to adopt a T cell fate by inducing strong Notch signaling, while hampering non–T cell development (Pui et al., 1999; Radtke et al., 1999; Wilson et al., 2001; Feyerabend et al., 2009). Whereas DLL4 is the essential Notch ligand involved in this function, at least in mice, distinct Notch ligands are expressed in the thymus (Hozumi et al., 2008; Koch et al., 2008; Van de Walle et al., 2011), which induce different levels of Notch signal strength that affect hematopoietic lineage decisions very differently (Jaleco et al., 2001; Lehar et al., 2005; Van de Walle et al., 2011, 2013). We thus hypothesized that the differential distribution of Notch ligands at discrete intrathymic niches could be a mechanism operating in the thymus to provide permissive microenvironments for the generation of non–T cells, specifically myeloid-primed CDPs and DCs. In fact, multipotent progenitors that seed the thymus must migrate through distinct functional environments during their differentiation, first from the perimedullary region to the capsule through the cortex and then back to the medulla, which suggests that differentiation signals may be stratified in specific thymic regions (reviewed by Petrie and Zúñiga-Pflücker, 2007). Supporting our hypothesis, we show here that the outer region of human thymus medulla, a niche devoid of DLL1 and DLL4 ligands (Hozumi et al., 2008; Koch et al., 2008; unpublished data) and characterized by a high density of cDCs and pDCs, selectively express JAG1, the ligand that induces the lowest levels of Notch activation (Jaleco et al., 2001; Lehar et al., 2005; Van de Walle et al., 2011). Because intrathymic DCs are shown here to establish adjacent contacts with JAG1+ stromal cells in the medullary niche, we have proposed that JAG1 could support progression of ETPs along the myeloid/DC program and production of DCs, a possibility that concurs with our functional results in vitro.
Besides Notch ligands, specific Notch receptors can control the developmental fate of human thymocytes by inducing differential Notch activation levels (Van de Walle et al., 2013). Although DC-lineage thymocytes are found to express all NOTCH(1–4) receptors, thymic DCs, at least in mice, are absolutely independent of Notch1, which is, however, essential for T cell development (Radtke et al., 2000). Moreover, we found that NOTCH3, which like NOTCH1, is a strong inducer of Notch activation (Van de Walle et al., 2011), is down-regulated in CDPs but up-regulated in CD5+p, whereas the opposite expression pattern is found for NOTCH4, which is preferentially expressed in intrathymic myeloid/DC-lineage cells and also in ETPs. Although NOTCH4 could be the specific Notch receptor that interacts with JAG1 to induce myeloid/DC commitment of thymus-seeding progenitors during or immediately after their entry through the corticomedullary junction, formal proof of this notion requires future functional studies. Other receptors highly expressed on ETPs, such as NOTCH1, could participate in the process as well. Independent of the particular receptor involved, Notch signaling blocking experiments shown here provide evidence that Notch activation plays a key instructive role in ETP progression to CDPs, which seems to be specifically induced by JAG1. In fact JAG1, but no other Notch ligand, is expressed on the surface of OP9 stromal cells (Schmitt and Zúñiga-Pflücker, 2002; Van de Walle et al., 2011; unpublished data), and JAG1 has a unique role in preventing T-lineage differentiation of primitive cord blood HSPCs (Van de Walle et al., 2011), a finding that concurs with our clonal results derived from CD7− ETPs and with previous results in mice (de Pooter et al., 2006). However, JAG1 supports T cell development from downstream CD7+ intrathymic progenitors (Van de Walle et al., 2013; our results), which suggests a differential impact of this ligand on T cell differentiation at distinct developmental stages. Therefore, it is highly likely that CD7+ ETPs, although still able to generate CDPs and to develop along the DC pathway, may include progenitors that have already been primed for a T cell fate in vivo and can progress along early T cell development in response to JAG1, whereas JAG1 itself is unable to induce T cell priming of uncommitted precursors, as proposed before (Van de Walle et al., 2011). In this scenario, it can be proposed that only those thymus-seeding progenitors that scape strong Notch signaling induced by ligands such as DLL1 and DLL4, and encounter JAG1, would be myeloid primed, losing T cell potential, whereas those that experience DLL4-mediated strong Notch activation would undergo T-lineage priming. Regarding the nature of such primitive multipotent thymus seeding progenitors, available data indicate that they are confined to the most immature CD7− fraction of ETPs (Blom and Spits, 2006; Six et al., 2007; Hao et al., 2008), and we provide clonal evidence that individual CD7− ETPs are in fact bipotent myeloid/T progenitors. Nonetheless, as CD7+ ETPs also display myeloid potential at least at the population level in our in vitro assay, intrathymic precursors may display a certain degree of developmental plasticity, which highlights the importance of deciphering the molecular mechanisms underlying the divergent lymphoid vs myeloid developmental fates of ETPs.
By modeling strong Notch activation in the OP9–DLL1 assay, we have shown that lymphoid/T cell priming of ETPs occurs at the expense of myeloid/DC priming and results actually in an impaired production of CD123+ progenitors and DCs, whereas the reverse situation was observed in OP9–JAG1 cultures. Regarding the nature of the specific ligand-induced signals that regulate the myeloid/lymphoid developmental decision of ETPs, we found a tight regulation at the transcriptional level of divergent gene expression programs that results in a marked up-regulation of myeloid-related transcription factors and the simultaneous down-regulation of factors involved in T-lineage fate, or vice versa. Particularly relevant was the finding that GATA2 is selectively expressed in progenitor cells undergoing myeloid/DC differentiation, as well as in thymic pDCs and cDCs, but is markedly down-regulated in T cell progenitors. Accordingly, strong DLL1-mediated signaling leads to GATA2 down-regulation in ETPs developing in culture, whereas GATA2 expression is maintained in ETPs receiving JAG1-mediated Notch signaling. More importantly, GATA2 expression was found dissociated from expression of GATA3, which is the critical transcription factor involved in T cell commitment (Van de Walle et al., 2016) that was found to be expressed by CD5+p cells derived from ETPs. The finding that ETPs down-regulate GATA3 in JAG1 cultures indicates that this loss may be part of the transcriptional program selectively induced by JAG1 to prevent T-lineage differentiation while priming DC development. In addition because GATA2 silencing impairs the development of CDPs and the generation of DCs from ETPs, JAG1-supported GATA2 expression may be a key transcriptional mechanism required for triggering the intrathymic myeloid/DC program in the human thymus. Accordingly, GATA2 has recently emerged as a key regulator essential for DC differentiation in mice (Onodera et al., 2016). Therefore, ligand-specific Notch signaling may play a crucial regulatory role to balance GATA2/GATA3 expression and hence myeloid/DC versus T cell development.
Our proposal is at odds with the finding that DC development from CDPs is supported rather than impaired in OP9–DLL1 cultures. This apparent discrepancy could be explained by the fact that GATA2 expression persisted in developing CDPs regardless of the Notch ligand recognized. Thus, once ETPs are committed to the myeloid/DC lineage and CDPs are generated, regulation of GATA2 expression becomes independent of a particular Notch ligand. However, CDPs remain responsive to Notch signals, as shown by GSI blocking experiments, indicating that, as previously reported for T cell development (Schmitt et al., 2004), DC differentiation of ETPs requires recurrent Notch receptor-ligand interactions. Collectively, these data suggest that regulation of GATA2 transcription by selective Notch ligands during human thymic DC development is stage specific, a concept that may also explain previous results on Gata2 up-regulation by DLL1 in mouse HSPCs developing along the myeloid lineage (Robert-Moreno et al., 2005; de Pooter et al., 2006). The proposed stage-specific Notch ligand function also explains the finding that survival of committed intrathymic CDPs can be more efficiently supported by DLL1 than by JAG1, whereas preferential differentiation of CDPs toward either the pDC or the cDC pathway is differentially regulated by JAG1 and DLL1, respectively. Therefore, as previously reported for differentiation of BM DCs in mice, different Notch ligands have opposite effects on differentiation of thymic DC at distinct developmental stages (Cheng et al., 2007). Collectively, our results highlight the crucial regulatory role that ligand-specific Notch signaling provided by particular thymic microenvironments may play to balance expression and function of GATA2 in the context of other cooperative or interfering transcription factors and hence to control myeloid/DC versus lymphoid/T commitment during human thymopoiesis.
Materials and methods
Flow cytometry and antibodies
Phenotypic analyses were performed in FACSCalibur and FACSCanto II (BD Biosciences) flow cytometers using the following mAbs: PE-labeled anti-CD1a, anti-CD34, anti-CD127, and anti-TCRαβ (Beckman Coulter); anti-CD3, anti-CD13, anti-CD44, anti-CD45RA, anti-CD123, anti-CD135, anti-HLA-DR, and anti-Notch4 (BD Biosciences); anti-CD116 (Immunotech); anti-Notch1 and anti-Notch3 (BioLegend); PC5-labeled anti-CD7 (Caltag); anti-CD4, anti-CD5, anti-CD13, anti-CD33, and anti-CD34 (Beckman Coulter); BB515-labeled anti-CD115 (BD Biosciences); FITC-labeled anti-BDCA1 and anti-BDCA2 (Miltenyi Biotec); anti-CD11c (Life Technologies) and anti-CD5, anti-CD7, anti-CD8, anti-CD14, and anti-CD44 (BD Biosciences); V450-labeled anti-CD45; and APC-labeled anti-CD34 (Beckman Coulter) and anti-CD117 (BD Biosciences). Irrelevant isotype-matched Igs (Caltag) were used as controls. Staining with biotin-coupled Annexin V (Roche) plus streptavidin-PE (Invitrogen) and 7-AAD (BD Biosciences) was used for apoptosis analysis. Cell proliferation was quantified by flow cytometry using CFSE labeling, following the manufacturer’s instructions (BD Biosciences).
Isolation of human thymus cell populations
Experiments were performed in accordance with procedures approved by the Spanish Research Council Bioethics Committee. Human postnatal thymocytes were isolated from thymus fragments removed during corrective cardiac surgery of patients aged 1 mo to 4 yr, after providing informed consent in accordance with the Declaration of Helsinki. Thymocyte cell suspensions obtained after Ficoll-Hypaque (Lymphoprep; ATOM) centrifugation were depleted of T-lineage cells by sheep erythrocyte rosseting, as previously described (García-Peydró et al., 2006), and CD34+ thymocytes were positively selected from the resulting cell fraction using CD34 Dynabeads (Dynal Progenitor Cell Selection System; Invitrogen). ETPs and myeloid-primed intrathymic progenitors were isolated from CD34+ thymocytes either by magnetic depletion of CD1a+ cells with anti-CD1a Microbeads (Miltenyi Biotec) or by positive selection of CD123+ cells (>99% CD123hi CD34lo) after sequential incubation with anti-CD123PE mAbs (BD Biosciences) and anti–PE Microbeads (Miltenyi Biotec), respectively, followed by magnetic cell sorting (AutoMACS; Miltenyi Biotec). CD7− and CD7+ ETPs were independently isolated from total ETPs and simultaneously depleted of CD123hi cells using a FACSAria Fusion cell sorter (BD Biosciences). Thymic pDCs and cDCs were isolated from the CD34− cell fraction by AutoMACS depletion of CD14+ monocytes with CD14 Microbeads (Miltenyi Biotec) and subsequent positive selection of either CD123+ or CD13+ cells using PE-labeled anti-CD123 or anti-CD13 mAbs (BD Biosciences), respectively, and anti–PE Microbeads. Peripheral pDCs and cDCs were isolated from mononuclear cell suspensions obtained from peripheral blood of healthy donors by Percoll density gradients (1.068 density; GE Healthcare), after positive selection using the protocol described above for thymic DCs.
Isolation of CD123+p and CD5+p progenitor subsets derived in vitro from human ETPs was performed on the basis of reciprocal CD123 and CD5 expression levels, as CD123+ CD5lo and CD123− CD5hi cells, respectively, after labeling with PE-coupled anti-CD123 and FITC-coupled anti-CD5 mAbs, and further FACS sorting.
Differentiation of thymic pDCs and cDCs in vitro
For in vitro generation of pDCs and cDCs from human intrathymic progenitors, we used the OP9 coculture assay described by Schotte et al. (2003). In brief, either ETPs or primary CD123+ myeloid progenitors, isolated from human thymus samples as described above, were seeded (105 cells/ml) on semiconfluent monolayers of OP9 cells obtained from the American Type Culture Collection (ATCC CRL-2749) and then cultured for 10 d in P24-well plates (Falcon) with RPMI-1640 medium (BioWhitaker) supplemented with 10% FBS (Invitrogen Life Technologies), 100 IU/ml rhFLT3L (Preprotech), and 200 IU/ml rhIL-7 (National Institute of Biological Standards and Controls [NIBSC]; OP9 cultures herein). For simultaneous generation of monocytes and DCs, OP9 cultures were supplemented with 250 IU/ml rhM-CSF (NIBSC). To avoid OP9 cell overgrowth, cultured cells were transferred to fresh OP9 monolayers every 3 d, after filtering out OP9 cells through 70-µm Cup Filcons (BD Biosciences). CD5+p and CD123+p intermediate progenitors were derived from ETPs in these cultures by day 3. When indicated, in vitro–derived CD5+p and CD123+p isolated by cell sorting as described above were cultured for 10 additional days under the same conditions. For inhibition of Notch1 signaling, the OP9 cultures were supplemented with the GSI CompE (Enzo Biochem) at a final concentration of 100 nM. DMSO vehicle was used as control.
In some experiments, the generation of either pDCs and cDCs or CD5+p and CD123+p progenitors was assessed upon culture of ETPs on either OP9–DLL1 or OP9–JAG1 stromal monolayers, or their respective OP9–GFP controls, in the presence of rhFLT3L and rhIL-7. OP9 cells transduced with a bicistronic MigR1 retroviral vector expressing either murine Delta-like 1 and GFP (OP9–DLL1) or only GFP (OP9-MigR1), were provided by J.C. Zúñiga-Pflücker (University of Toronto, Toronto, Canada; Schmitt and Zúñiga-Pflücker, 2002). To obtain OP9–JAG1 cells, parental OP9 cells (ATCC CRL-2749) were transduced with a bicistronic pLZRS retroviral vector encoding human JAG1 and GFP, provided by L. Parreira (Faculdade de Medicina de Lisboa, Lisboa, Portugal; Neves et al., 2006). OP9 cells transduced with pLZRS encoding only GFP were used as controls (OP9–pLRZS). OP9–JAG1 and OP9–pLRZS cells expressing high GFP levels were selected by cell sorting using a FACSAria Fusion. Identical results were obtained using either OP9-MigR1 or OP9–pLRZS control cells; therefore, only results obtained with the former are presented (OP9–GFP).
Generation of ETP-derived pDCs and cDCs in vivo
To assess the pDC and cDC potential of human ETPs in vivo, 2–5 × 105 magnetically sorted ETPs were preincubated for 5–8 h in culture medium containing rhFLT3L and rhIL-7 and transplanted by intrahepatic injection of into sublethally irradiated (3.5 Gy) Rag2−/− × γc −/− immunodeficient mice aged 1–4 d, provided by K. Weijer (University of Amsterdam, Amsterdam, Netherlands; van Rijn et al., 2003). Generation of CD45+ human cells in the thymus, BM and spleen of transplanted animals was analyzed by flow cytometry.
Clonal assays and confocal microscopy analysis
Individual CD34+ CD7− ETPs depleted of CD123hi cells were deposited by cell sorting (FACSAria Fusion; BD Biosciences) directly into gelatin coated (0.1%; Sigma-Aldrich) 384-well plates (Cell Carrier; PerkinElmer) containing confluent monolayers of either OP9–JAG1 or OP9–DLL1 cells previously treated with mitomycin-C (10 mg/ml; Sigma-Aldrich) for 2 h at 37°C. Cells were kept in culture in RPMI plus 10% FBS supplemented with 100 IU/ml rhFLT3L and 200 IU/ml rhIL-7 for 9 d (OP9–JAG1) or 14 d (OP9–DLL1) and were then stained with anti-CD5-Dylight650 (Novus Biologicals), anti-CD13 biotin (ExBio), anti-CD123 biotin, and anti-CD45 V450 (BD Biosciences). Anti-CD13– and anti–CD123-biotin–coupled antibodies were simultaneously detected after labeling with Streptavidin-Alexa Fluor 546 (Thermo Fisher Scientific). Cells were imaged with an Opera (PerkinElmer) confocal microscope, and images of clonal cultures that contained at least 10 human CD45+ cells were then analyzed using ImageJ software. The quantification was done for a maximum of 100 cells/well. The lineage output potential of positive clones was defined by scoring when >70% of CD45+ cells were exclusively positive for CD5 (T-lineage) or were positive for CD13 and/or CD123 and either expressed or lacked CD5 (myeloid/DC lineage) or were negative for both markers (undifferentiated).
Quantitative PCR
Real-time PCR quantification of cDNA synthesized with oligo (dT) primers (Roche) from TRIzol-extracted (Invitrogen Life Technologies) total RNA was performed as described (González-García et al., 2009), using TaqMan Gene Expression Assays (Applied Biosystems), according to the manufacturer’s instructions, in an ABI PRISM 7900 HT Sequence Detection system (Applied Biosystems). Data were normalized to GAPDH mRNA expression (Hs99999905_m1; Applied Biosystems). All results are shown relative to those of ETPs.
Immunohistochemistry and confocal microscopy
Human thymus samples were dehydrated in graded ethanol series, fixed overnight on PFA 4% and paraffin included. Serial 8-µm sections were prepared and mounted on poly-lysine–treated slides. After deparaffinization and rehydration in ethanol series, antigen was retrieved by boiling during 10 min in sodium citrate (10 mM, pH 6.0), and, after cooling at room temperature, slides were washed in distilled water and PBS. For blocking, samples were incubated for 1 h in histoblock solution (3% BSA, 20 mM MgCl2, 0.3% Tween-20, and 5% FBS in PBS) before overnight incubation with the following antibodies: rabbit anti–human NOTCH4 (ab33163; Abcam), mouse anti–human CD123 (BioLegend), and mouse anti–human CD11c (Novocastra). Background staining was determined using isotype-matched irrelevant Igs. Tissue autofluorescence and endogenous biotin were quenched before addition of secondary antibodies by incubation for 1 h with quenching solution (3% Sudan Black, 70% ethanol) and avidin/biotin blocking solution (Vector Laboratories). NOTCH4 expression was detected by incubation for 1 h with an Alexa-555-coupled goat anti–rabbit antibody (Invitrogen). For CD123 and CD11c detection, incubation with a biotin-conjugated goat anti–mouse antibody (Vector Laboratories) was followed by amplification for 1 h with avidin/biotin complex (Elite Vectastain ABComplex kit; Vector Labs), and incubation with Alexa-488-conjugated streptavidin for 20 min. Finally, slides were counterstained with Topro-3 (Invitrogen) and mounted with Fluoromount-G (Southern Biotech). Images were acquired using an Axio Imager Z1 M (Zeiss) confocal microscope using 25× (scan zoom 0.7, NA 0.8) and 40× (scan zoom 1.0, NA 1.3) magnifications and were subsequently processed using ImageJ and Adobe Photoshop CS4 software.
GATA2 silencing by siRNA nucleofection
Human ETPs (0.5–106) preincubated for 8 h with rhFLT3L and rhIL-7 were washed in PBS and nucleofected with 2–5 µg of either GATA2-specific or scrambled siRNAs as control (ON-TARGETplus SMARTpool; Thermo Fisher Scientific), using the Human CD34+ Cell Nucleofector kit (Amaxa) and the U-008 protocol (Nucleofector II; Amaxa). To evaluate nucleofection efficiencies, ETPs were also nucleofected with 2.5 µg pmaxGFP Vector (Amaxa). Percentages of GFP+ cells were >70% in all experiments. GATA2 silencing was confirmed by quantitative PCR in nucleofected cells cultured during 48 h.
Statistics
Statistical significance was determined with Prism software (GraphPad), using two-tailed, unpaired, or paired Student’s t test, with the α level set at 0.05. For Fig. 7, the Wilcoxon paired test was used.
Online supplemental material
Fig. S1 shows the cell-sorting strategy used to isolate CD7− and CD7hi ETPs depleted of CD123hi cells.
Supplementary Material
Acknowledgments
We thank Dr. Berta Raposo and Mario Alía for cell sorting support; Drs. María Montoya, Laura Fernández de Manuel, and Irene Palacios for technical support with Opera image analysis; the Cellomics Unit and the ReDIB (Distributed Biomedical Imaging Network) facilities at Fundación Centro Nacional de Investigaciones Cardiovasculares Carlos III for image analysis; the Pediatric Cardiosurgery Units from Hospital Universitario La Paz and Hospital Universitario 12 de Octubre (Madrid, Spain) for thymus samples; and the Genomics Facility at Parque Científico de Madrid for quantitative PCR analyses.
This work was supported by funds from Plan Nacional, Ministerio de Ciencia e Innovación grants SAF2013-44857-R and SAF2016-75442-R (Agencia Estatal de Investigación/European Regional Development Fund, European Union), Instituto de Salud Carlos III (RTICC RD06/0014/1012), and the Seventh Framework Programme of the European Union (FP7-HEALTH-2013-INNOVATION-1-602587) to M.L. Toribio; by grant SAF2015-70880-R (Agencia Estatal de Investigación/European Regional Development Fund, European Union) to M.L. Gaspar; and by an Institutional Grant from Fundación Ramón Areces. M.J. García-León and A. Murcia-Ceballos were supported by Ministerio de Ciencia e Innovación.
The authors declare no competing financial interests.
Author contributions: E. Martín-Gayo, S. González-Garcia, and M.L. Toribio designed the study. E. Martín-Gayo and S. González-Garcia performed most of the experiments. A. Murcia-Ceballos performed single-cell progenitor cultures and labeling. M. García-Peydró, J. Alcain, and A. Murcia-Ceballos contributed to processing of thymus samples and to Opera image analyses. M.J. García-León performed immunohistochemistry analyses. B. de Andrés and M.L. Gaspar contributed to cell sorting studies. L. Allende provided thymus samples. M.L. Toribio supervised research and wrote the manuscript with E. Martín-Gayo and S. González-Garcia.
Footnotes
Abbreviations used:
- cDC
- conventional DC
- CDP
- common DC progenitor
- ETP
- early thymic progenitor
- FLT3L
- FMS-related tyrosine kinase 3 ligand
- GSI
- γ-secretase inhibitor
- HLA-DR
- human leukocyte antigen–antigen D related
- HSPC
- hematopoietic stem/progenitor cell
- MDP
- monocyte/DC progenitor
- pDC
- plasmacytoid DC
References
- Ardavin C., Wu L., Li C.L., and Shortman K.. 1993. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 362:761–763. 10.1038/362761a0 [DOI] [PubMed] [Google Scholar]
- Banchereau J., and Steinman R.M.. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252. 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- Bell J.J., and Bhandoola A.. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 452:764–767. 10.1038/nature06840 [DOI] [PubMed] [Google Scholar]
- Bendriss-Vermare N., Barthélémy C., Durand I., Bruand C., Dezutter-Dambuyant C., Moulian N., Berrih-Aknin S., Caux C., Trinchieri G., and Brière F.. 2001. Human thymus contains IFN-alpha-producing CD11c(-), myeloid CD11c(+), and mature interdigitating dendritic cells. J. Clin. Invest. 107:835–844. 10.1172/JCI11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom B., and Spits H.. 2006. Development of human lymphoid cells. Annu. Rev. Immunol. 24:287–320. 10.1146/annurev.immunol.24.021605.090612 [DOI] [PubMed] [Google Scholar]
- Breton G., Lee J., Zhou Y.J., Schreiber J.J., Keler T., Puhr S., Anandasabapathy N., Schlesinger S., Caskey M., Liu K., and Nussenzweig M.C.. 2015a Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 212:401–413. 10.1084/jem.20141441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Lee J., Liu K., and Nussenzweig M.C.. 2015b Defining human dendritic cell progenitors by multiparametric flow cytometry. Nat. Protoc. 10:1407–1422. 10.1038/nprot.2015.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker T., Riedinger M., and Karjalainen K.. 1997. Targeted expression of major histocompatibility complex (MHC) class II molecules demonstrates that dendritic cells can induce negative but not positive selection of thymocytes in vivo. J. Exp. Med. 185:541–550. 10.1084/jem.185.3.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Nefedova Y., Corzo C.A., and Gabrilovich D.I.. 2007. Regulation of dendritic-cell differentiation by bone marrow stroma via different Notch ligands. Blood. 109:507–515. 10.1182/blood-2006-05-025601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Hoyo G.M., Martín P., Vargas H.H., Ruiz S., Arias C.F., and Ardavín C.. 2002. Characterization of a common precursor population for dendritic cells. Nature. 415:1043–1047. 10.1038/4151043a [DOI] [PubMed] [Google Scholar]
- de Pooter R.F., Schmitt T.M., de la Pompa J.L., Fujiwara Y., Orkin S.H., and Zúñiga-Pflücker J.C.. 2006. Notch signaling requires GATA-2 to inhibit myelopoiesis from embryonic stem cells and primary hemopoietic progenitors. J. Immunol. 176:5267–5275. 10.4049/jimmunol.176.9.5267 [DOI] [PubMed] [Google Scholar]
- De Smedt M., Hoebeke I., Reynvoet K., Leclercq G., and Plum J.. 2005. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 106:3498–3506. 10.1182/blood-2005-02-0496 [DOI] [PubMed] [Google Scholar]
- de Yébenes V.G., Carrasco Y.R., Ramiro A.R., and Toribio M.L.. 2002. Identification of a myeloid intrathymic pathway of dendritic cell development marked by expression of the granulocyte macrophage-colony-stimulating factor receptor. Blood. 99:2948–2956. 10.1182/blood.V99.8.2948 [DOI] [PubMed] [Google Scholar]
- Dontje W., Schotte R., Cupedo T., Nagasawa M., Scheeren F., Gimeno R., Spits H., and Blom B.. 2006. Delta-like1-induced Notch1 signaling regulates the human plasmacytoid dendritic cell versus T-cell lineage decision through control of GATA-3 and Spi-B. Blood. 107:2446–2452. 10.1182/blood-2005-05-2090 [DOI] [PubMed] [Google Scholar]
- Doulatov S., Notta F., Eppert K., Nguyen L.T., Ohashi P.S., and Dick J.E.. 2010. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11:585–593. 10.1038/ni.1889 [DOI] [PubMed] [Google Scholar]
- Feyerabend T.B., Terszowski G., Tietz A., Blum C., Luche H., Gossler A., Gale N.W., Radtke F., Fehling H.J., and Rodewald H.R.. 2009. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 30:67–79. 10.1016/j.immuni.2008.10.016 [DOI] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., and Geissmann F.. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87. 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Gao E.K., Lo D., and Sprent J.. 1990. Strong T cell tolerance in parent----F1 bone marrow chimeras prepared with supralethal irradiation. Evidence for clonal deletion and anergy. J. Exp. Med. 171:1101–1121. 10.1084/jem.171.4.1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Peydró M., de Yébenes V.G., and Toribio M.L.. 2006. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J. Immunol. 177:3711–3720. 10.4049/jimmunol.177.6.3711 [DOI] [PubMed] [Google Scholar]
- Geissmann F., Manz M.G., Jung S., Sieweke M.H., Merad M., and Ley K.. 2010. Development of monocytes, macrophages, and dendritic cells. Science. 327:656–661. 10.1126/science.1178331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-García S., García-Peydró M., Martín-Gayo E., Ballestar E., Esteller M., Bornstein R., de la Pompa J.L., Ferrando A.A., and Toribio M.L.. 2009. CSL-MAML-dependent Notch1 signaling controls T lineage-specific IL-7Rα gene expression in early human thymopoiesis and leukemia. J. Exp. Med. 206:779–791. 10.1084/jem.20081922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Collin M., and Ginhoux F.. 2013. Ontogeny and functional specialization of dendritic cells in human and mouse. Adv. Immunol. 120:1–49. 10.1016/B978-0-12-417028-5.00001-6 [DOI] [PubMed] [Google Scholar]
- Hao Q.L., George A.A., Zhu J., Barsky L., Zielinska E., Wang X., Price M., Ge S., and Crooks G.M.. 2008. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7- lympho-myeloid thymic progenitors. Blood. 111:1318–1326. 10.1182/blood-2007-08-106294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi K., Mailhos C., Negishi N., Hirano K., Yahata T., Ando K., Zuklys S., Holländer G.A., Shima D.T., and Habu S.. 2008. Delta-like 4 is indispensable in thymic environment specific for T cell development. J. Exp. Med. 205:2507–2513. 10.1084/jem.20080134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaleco A.C., Neves H., Hooijberg E., Gameiro P., Clode N., Haury M., Henrique D., and Parreira L.. 2001. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J. Exp. Med. 194:991–1002. 10.1084/jem.194.7.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U., Fiorini E., Benedito R., Besseyrias V., Schuster-Gossler K., Pierres M., Manley N.R., Duarte A., Macdonald H.R., and Radtke F.. 2008. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med. 205:2515–2523. 10.1084/jem.20080829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger A. 2011. A missing link in thymic dendritic cell development. Eur. J. Immunol. 41:2145–2147. 10.1002/eji.201141850 [DOI] [PubMed] [Google Scholar]
- Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N., Arakaki R., Hayashi Y., Kitagawa T., Lipp M., et al. 2006. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 24:165–177. 10.1016/j.immuni.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Lee J., Breton G., Oliveira T.Y., Zhou Y.J., Aljoufi A., Puhr S., Cameron M.J., Sékaly R.P., Nussenzweig M.C., and Liu K.. 2015. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 212:385–399. 10.1084/jem.20141442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar S.M., Dooley J., Farr A.G., and Bevan M.J.. 2005. Notch ligands Delta 1 and Jagged1 transmit distinct signals to T-cell precursors. Blood. 105:1440–1447. 10.1182/blood-2004-08-3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Park J., Foss D., and Goldschneider I.. 2009. Thymus-homing peripheral dendritic cells constitute two of the three major subsets of dendritic cells in the steady-state thymus. J. Exp. Med. 206:607–622. 10.1084/jem.20082232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., and Nussenzweig M.C.. 2010. Origin and development of dendritic cells. Immunol. Rev. 234:45–54. 10.1111/j.0105-2896.2009.00879.x [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., and Nussenzweig M.. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luche H., Ardouin L., Teo P., See P., Henri S., Merad M., Ginhoux F., and Malissen B.. 2011. The earliest intrathymic precursors of CD8α(+) thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur. J. Immunol. 41:2165–2175. 10.1002/eji.201141728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez C., Trigueros C., Fernández E., and Toribio M.L.. 1995. The development of T and non-T cell lineages from CD34+ human thymic precursors can be traced by the differential expression of CD44. J. Exp. Med. 181:475–483. 10.1084/jem.181.2.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Gayo E., Sierra-Filardi E., Corbí A.L., and Toribio M.L.. 2010. Plasmacytoid dendritic cells resident in human thymus drive natural Treg cell development. Blood. 115:5366–5375. 10.1182/blood-2009-10-248260 [DOI] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium . 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohtashami M., Shah D.K., Nakase H., Kianizad K., Petrie H.T., and Zúñiga-Pflücker J.C.. 2010. Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. J. Immunol. 185:867–876. 10.4049/jimmunol.1000782 [DOI] [PubMed] [Google Scholar]
- Moore A.J., Sarmiento J., Mohtashami M., Braunstein M., Zúñiga-Pflücker J.C., and Anderson M.K.. 2012. Transcriptional priming of intrathymic precursors for dendritic cell development. Development. 139:373–384. 10.1242/dev.069344 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226. 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Neves H., Weerkamp F., Gomes A.C., Naber B.A.E., Gameiro P., Becker J.D., Lúcio P., Clode N., van Dongen J.J.M., Staal F.J.T., and Parreira L.. 2006. Effects of Delta1 and Jagged1 on early human hematopoiesis: correlation with expression of notch signaling-related genes in CD34+ cells. Stem Cells. 24:1328–1337. 10.1634/stemcells.2005-0207 [DOI] [PubMed] [Google Scholar]
- Okada T., Lian Z.X., Naiki M., Ansari A.A., Ikehara S., and Gershwin M.E.. 2003. Murine thymic plasmacytoid dendritic cells. Eur. J. Immunol. 33:1012–1019. 10.1002/eji.200323616 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., and Manz M.G.. 2007. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216. 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Onodera K., Fujiwara T., Onishi Y., Itoh-Nakadai A., Okitsu Y., Fukuhara N., Ishizawa K., Shimizu R., Yamamoto M., and Harigae H.. 2016. GATA2 regulates dendritic cell differentiation. Blood. 128:508–518. 10.1182/blood-2016-02-698118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie H.T., and Zúñiga-Pflücker J.C.. 2007. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 25:649–679. 10.1146/annurev.immunol.23.021704.115715 [DOI] [PubMed] [Google Scholar]
- Pui J.C., Allman D., Xu L., DeRocco S., Karnell F.G., Bakkour S., Lee J.Y., Kadesch T., Hardy R.R., Aster J.C., and Pear W.S.. 1999. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 11:299–308. 10.1016/S1074-7613(00)80105-3 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., and Aguet M.. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558. 10.1016/S1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Radtke F., Ferrero I., Wilson A., Lees R., Aguet M., and MacDonald H.R.. 2000. Notch1 deficiency dissociates the intrathymic development of dendritic cells and T cells. J. Exp. Med. 191:1085–1094. 10.1084/jem.191.7.1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B., Bunin A., Ghosh H.S., Lewis K.L., and Sisirak V.. 2011. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 29:163–183. 10.1146/annurev-immunol-031210-101345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., de la Pompa J.L., and Bigas A.. 2005. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 132:1117–1126. 10.1242/dev.01660 [DOI] [PubMed] [Google Scholar]
- Rodrigues N.P., Boyd A.S., Fugazza C., May G.E., Guo Y., Tipping A.J., Scadden D.T., Vyas P., and Enver T.. 2008. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood. 112:4862–4873. 10.1182/blood-2008-01-136564 [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Wu X., Albring J.C., and Murphy K.M.. 2012. Re(de)fining the dendritic cell lineage. Nat. Immunol. 13:1145–1154. 10.1038/ni.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner S.M., and Rodewald H.R.. 2010. Early T cell development and the pitfalls of potential. Trends Immunol. 31:303–310. 10.1016/j.it.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., and Zúñiga-Pflücker J.C.. 2002. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 17:749–756. 10.1016/S1074-7613(02)00474-0 [DOI] [PubMed] [Google Scholar]
- Schmitt T.M., Ciofani M., Petrie H.T., and Zúñiga-Pflücker J.C.. 2004. Maintenance of T cell specification and differentiation requires recurrent notch receptor-ligand interactions. J. Exp. Med. 200:469–479. 10.1084/jem.20040394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R., Rissoan M.C., Bendriss-Vermare N., Bridon J.M., Duhen T., Weijer K., Brière F., and Spits H.. 2003. The transcription factor Spi-B is expressed in plasmacytoid DC precursors and inhibits T-, B-, and NK-cell development. Blood. 101:1015–1023. 10.1182/blood-2002-02-0438 [DOI] [PubMed] [Google Scholar]
- Shortman K., and Liu Y.J.. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161. 10.1038/nri746 [DOI] [PubMed] [Google Scholar]
- Shortman K., and Wu L.. 1996. Early T lymphocyte progenitors. Annu. Rev. Immunol. 14:29–47. 10.1146/annurev.immunol.14.1.29 [DOI] [PubMed] [Google Scholar]
- Six E.M., Bonhomme D., Monteiro M., Beldjord K., Jurkowska M., Cordier-Garcia C., Garrigue A., Dal Cortivo L., Rocha B., Fischer A., et al. 2007. A human postnatal lymphoid progenitor capable of circulating and seeding the thymus. J. Exp. Med. 204:3085–3093. 10.1084/jem.20071003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M. 2012. Decisions about dendritic cells: past, present, and future. Annu. Rev. Immunol. 30:1–22. 10.1146/annurev-immunol-100311-102839 [DOI] [PubMed] [Google Scholar]
- Strobl H., Scheinecker C., Riedl E., Csmarits B., Bello-Fernandez C., Pickl W.F., Majdic O., and Knapp W.. 1998. Identification of CD68+lin- peripheral blood cells with dendritic precursor characteristics. J. Immunol. 161:740–748. [PubMed] [Google Scholar]
- Vandenabeele S., Hochrein H., Mavaddat N., Winkel K., and Shortman K.. 2001. Human thymus contains 2 distinct dendritic cell populations. Blood. 97:1733–1741. 10.1182/blood.V97.6.1733 [DOI] [PubMed] [Google Scholar]
- Van de Walle I., De Smet G., Gärtner M., De Smedt M., Waegemans E., Vandekerckhove B., Leclercq G., Plum J., Aster J.C., Bernstein I.D., et al. 2011. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood. 117:4449–4459. 10.1182/blood-2010-06-290049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle I., Waegemans E., De Medts J., De Smet G., De Smedt M., Snauwaert S., Vandekerckhove B., Kerre T., Leclercq G., Plum J., et al. 2013. Specific Notch receptor-ligand interactions control human TCR-αβ/γδ development by inducing differential Notch signal strength. J. Exp. Med. 210:683–697. 10.1084/jem.20121798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle I., Dolens A.C., Durinck K., De Mulder K., Van Loocke W., Damle S., Waegemans E., De Medts J., Velghe I., De Smedt M., et al. 2016. GATA3 induces human T-cell commitment by restraining Notch activity and repressing NK-cell fate. Nat. Commun. 7:11171–11185. 10.1038/ncomms11171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijn R.S., Simonetti E.R., Hagenbeek A., Hogenes M.C., de Weger R.A., Canninga-van Dijk M.R., Weijer K., Spits H., Storm G., van Bloois L., et al. 2003. A new xenograft model for graft-versus-host disease by intravenous transfer of human peripheral blood mononuclear cells in RAG2-/- gammac-/- double-mutant mice. Blood. 102:2522–2531. 10.1182/blood-2002-10-3241 [DOI] [PubMed] [Google Scholar]
- von Boehmer H. 2009. Notch 1 keeps pro-T cells on track. Immunity. 30:5–7. 10.1016/j.immuni.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Wada H., Masuda K., Satoh R., Kakugawa K., Ikawa T., Katsura Y., and Kawamoto H.. 2008. Adult T-cell progenitors retain myeloid potential. Nature. 452:768–772. 10.1038/nature06839 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Wang Y.H., Lee H.K., Ito T., Wang Y.H., Cao W., and Liu Y.J.. 2005. Hassall’s corpuscles instruct dendritic cells to induce CD4+CD25+ regulatory T cells in human thymus. Nature. 436:1181–1185. 10.1038/nature03886 [DOI] [PubMed] [Google Scholar]
- Weijer K., Uittenbogaart C.H., Voordouw A., Couwenberg F., Seppen J., Blom B., Vyth-Dreese F.A., and Spits H.. 2002. Intrathymic and extrathymic development of human plasmacytoid dendritic cell precursors in vivo. Blood. 99:2752–2759. 10.1182/blood.V99.8.2752 [DOI] [PubMed] [Google Scholar]
- Wilson A., MacDonald H.R., and Radtke F.. 2001. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 194:1003–1012. 10.1084/jem.194.7.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., and Shortman K.. 2005. Heterogeneity of thymic dendritic cells. Semin. Immunol. 17:304–312. 10.1016/j.smim.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Yui M.A., and Rothenberg E.V.. 2014. Developmental gene networks: a triathlon on the course to T cell identity. Nat. Rev. Immunol. 14:529–545. 10.1038/nri3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.