Abstract
Bacillus subtilis is an important model bacterium for the study of developmental adaptations that enhance survival in the face of fluctuating environmental challenges. These adaptations include sporulation, biofilm formation, motility, cannibalism, and competence. Remarkably, not all the cells in a given population exhibit the same response. The choice of fate by individual cells is random but is also governed by complex signal transduction pathways and cross talk mechanisms that reinforce decisions once made. The interplay of stochastic and deterministic mechanisms governing the selection of developmental fate on the single-cell level is discussed in this article.
Everything existing in the universe is the fruit of chance and necessity.
democritus (as quoted by J. Monod)
Biologists often think of regulation as deterministic; causes have invariant consequences, and changes predictably beget further changes. However, with the advent of techniques for the study of individual cell phenotypes, it has become evident that random cell-to-cell variation in the amounts of mRNA and protein is prevalent and often entails significant consequences, particularly for developmental processes (1, 2). This review focuses on the developmental pathways of Bacillus subtilis, with attention to the roles of this variation and of stochastic reactions in competence, motility, sporulation, and biofilm formation. We do not comprehensively review the regulatory mechanisms that control these pathways but present only what is needed for our purposes. Figure 1 identifies the major players that are discussed below and illustrates the competence, motility, cannibalism, biofilm, and sporulation modules that represent the major known developmental pathways of B. subtilis.
Figure 1.
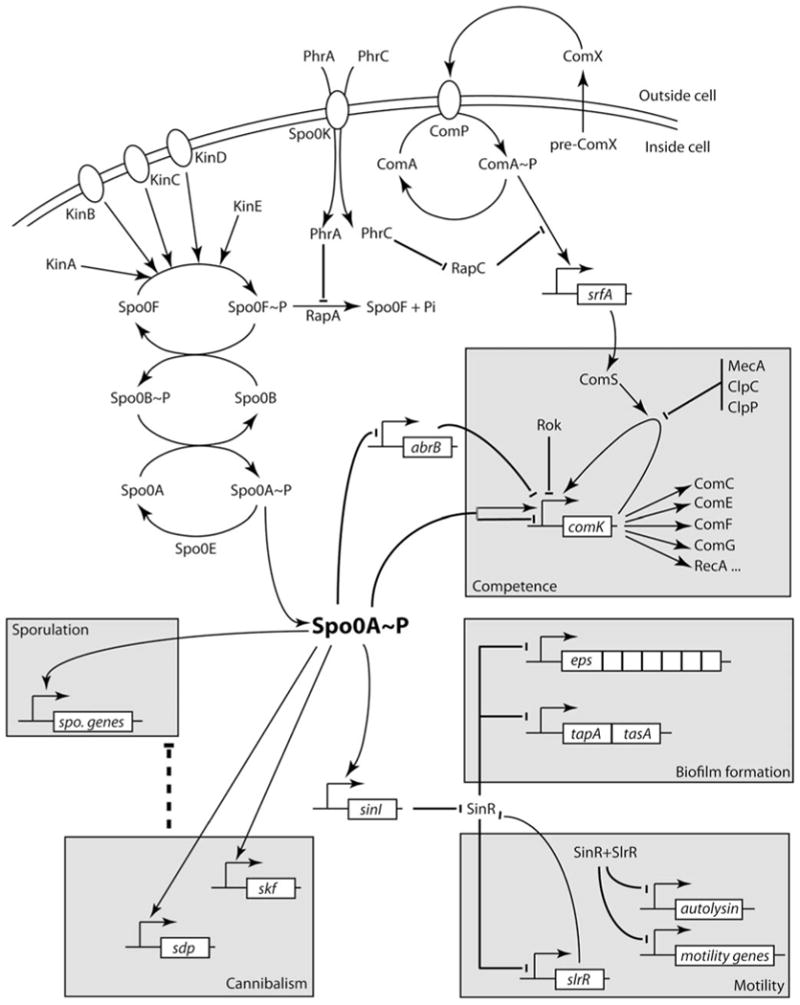
Developmental modules in B. subtilis and their major components. All of the indicated forms of development depend on 0A and on the phosphorelay that governs the phosphorylation of this transcription factor. This figure is intended to summarize many of the major interactions mentioned in the text that govern the developmental processes. It is not exhaustive, although it may be exhausting. Lines ending in perpendiculars and arrows denote negative and positive effects, respectively. Arrows associated with right-angled lines denote transcription initiation. The dotted line from the cannibalism module indicates that the release of nutrients from dead cells delays sporulation. Several kinases deliver phosphoryl groups to the phosphorelay, which results in the formation of 0A∼P. Under some conditions one or more kinase can dephosphorylate 0F∼P, draining phosphate from 0A∼P. RapA is one of several related proteins that can also dephosphorylate 0F∼P. RapC acts by preventing ComA∼P from interacting with its DNA target. These Rap proteins are inhibited by cognate secreted peptides (e.g., PhrA and PhrC), which are internalized by the oligopeptide permease Spo0K. ComX is a modified and secreted peptide which activates the autophosphorylation of ComP. ComP∼P donates a phosphate to ComA, and ComA∼P then activates the transcription of srfA. Embedded in the srfA operon is the gene for ComS. This small protein binds to the protease complex of MecA plus ComP plus ClpC, preventing the degradation of the transcription factor ComK. ComK is then free to activate its own expression by antagonizing the repressor Rok, activating a positive autoregulatory loop. When ComK accumulates, it, in turn, activates the transcription of many downstream genes, resulting in the induction of competence (the K-state). A low level of 0A∼P is also essential for competence due to its direct interaction with the comK promoter and its repression of abrB. A low to intermediate concentration of 0A∼P also activates the sinI promoter. SinI antagonizes SinR, lifting the repression of several transcription units that are essential for biofilm formation, as well as the repression of slrR. SlrR binds to SinR, further derepressing the biofilm operons. The SinR-SlrR hetero-complex represses the genes for motility as well as those that encode the autolysins that separate daughter cells following division. This results in the formation of chains of sessile cells. Low concentrations of 0A∼P also activate genes that encode toxins. Toxin-producing cells (cannibals) benefit by killing other cells, thus deriving nutrients. Finally, high concentrations of 0A∼P activate the sporulation genes. doi:10.1128/microbiolspectrum.TBS-0004-2012.f1
When a cell population bifurcates into two sub-populations that differ in their patterns of transcription, we use the term bimodal to describe the distribution of gene expression among the cells. The term bistable is reserved for a bimodal population in which the different cell types can be epigenetically inherited. Noise, or cell-to-cell variation in the abundance of gene products, results from the stochastic nature of chemical reactions and becomes important when small numbers of reactants are involved (3, 4). Noise in gene expression results in population heterogeneity, which may or may not be bimodal in nature. Transcription is the major contributor to noise because only a few copies of each promoter are present in a cell and because the engagement of RNA polymerase with a given promoter occurs randomly, with unpredictable delays between transcription events or between clusters of such events. Other processes, including mRNA decay, translation, and protein degradation, may also contribute to noise.
Distinction is often made between intrinsic and extrinsic noise (5, 6). The first is due to the inherent properties of a promoter and other gene expression sequences of a gene. Extrinsic noise results from cell-to-cell variation in transcription factors and other molecules that determine the rates of gene expression. Intuitively it would seem that as the average number of gene products increases, the importance of intrinsic noise will decrease and extrinsic noise will become dominant. This prediction has been confirmed, with the additional remarkable conclusion that at a given time, there is little correlation in a single cell between the number of transcripts from a gene and the number of its cognate protein molecules (7). This general result stems from the difference in mRNA and protein stability, so the number of mRNA molecules present at a given time does not predict the amount of accumulated protein.
B. subtilis provides a rich field for the investigator because of the wonderful variety of its adaptations. For example, cells can become motile and swim toward nutrients or away from repellents or may adopt a sessile lifestyle. As cultures approach stationary phase and the growth rate decreases while cell density increases, some cells express a large number of genes (in excess of 100) under the control of the transcription factor ComK (8–10). Among these so-called K-state genes are those that mediate transformation, the uptake and integration of environmental DNA. Under certain conditions, some cells elaborate products that form an extracellular matrix in which a multicellular community becomes embedded to form a biofilm (11). Within the biofilm, these matrix-producing cells produce toxic substances that cause the demise of certain of their sisters. This provides food for the aggressors, delaying their irreversible entry into the final, energy-expensive sporulation pathway. An important discovery is that the domesticated laboratory strains, derived from strain 168, have lost the ability to form robust biofilms and are modified in their motilesessile switch behavior compared to natural isolates (12). The full display of the biofilm-associated pheno-types is only revealed in natural isolates, notably in strain 3610, which has become the industry standard. It is worth mentioning that by adopting a standard, which valuably permits results in different laboratories to be compared, we also run the risk of ignoring interesting phenotypic diversity. For example, 3610 is much less transformable than other natural isolates.
Spo0A and the Phosphorelay as a Temporal Gatekeeper
This review will repeatedly return to Spo0A (OA) as a temporal gatekeeper. Although originally identified as the master regulator of spore formation, 0A in its phosphorylated form governs all of the developmental pathways mentioned above. 0A is phosphorylated via the famous phosphorelay (Fig. 1) (13). In the phosphorelay, one or more of five histidine kinases phosphorylates the response regulator protein Spo0F (OF), which donates its phosphoryl moiety to the phosphotransferase Spo0B (OB), which then passes the phosphoryl group to OA. The concentration of 0A∼P increases as cells approach and enter stationary phase, and the consequences of this increase are multiple and profound. Programmed changes in the average concentration of 0A∼P determine the frequencies at which cells enter the developmental pathways, while the choice of which cells do so is random. As described below, the phosphorelay, which is modulated by a large number of accessory proteins, has been called a “noise generator,” causing the level of 0A∼P to vary from cell to cell (14). Development in B. subtilis is thus both stochastic (chance) and deterministic (necessity). The mechanisms that regulate these changes in the concentration of 0A∼P are not completely understood, involve many genes, and exert their influence on the levels of transcription (including promoter switching), phosphorylation, and dephosphorylation. Only recently has light been shed on some of the upstream signal inputs that regulate this complex system, particularly with regard to biofilm formation.
Three important generalizations are basic to the various developmental processes. First, the average concentration of 0A∼P increases as cell division slows and cultures approach stationary phase. Second, promoters respond differently to 0A∼P depending on their affinities for this response regulator protein, and this has consequences for development (15, 16). Third, there are marked differences in the rates at which 0A∼P increases in the different cells of a single population (14, 17). This temporal heterogeneity has been assigned the useful term “heterochronicity” (17). In a number of cases, as described below, 0A∼P activates and then represses a given gene, allowing transient expression. In this sense, this master regulator acts as a temporal gatekeeper. We begin our review of developmental processes with competence, for which the role of noise has received considerable attention.
Competence: The K-State
An early response of B. subtilis to high cell density, as well as to other poorly understood signals, is competence for DNA uptake. The genes required for competence are transcribed only in the presence of the transcriptional regulator ComK (18). Although the incorporation of new genetic material potentially allows the bacteria to increase their fitness under challenging conditions, only some of the 100 or so genes under ComK control are devoted to transformation, and it is probably a mistake to discuss the evolution of the K-state only in terms of the fitness benefits that may be conferred by DNA uptake. (Despite this warning, here we use the terms competence and K-state interchangeably.) ComK not only activates transcription of the downstream K-state genes but also positively regulates its own promoter. In laboratory strains, this induction takes place in 10 to 20% of the cells. Induction in a given cell can be regarded as an all-or-none event that results in a bimodal distribution of competence gene expression. Natural isolates of B. subtilis vary widely in the fraction of cells that achieve the K-state, ranging from very few such cells to a frequency approaching that of the domesticated strain (10 to 20%), which has been selected in the laboratory for high transformation rates (unpublished data).
Temporal Control of Competence
During exponential growth, B. subtilis uses several mechanisms to prevent the development of competence (Fig. 1). First, the ComK concentration is kept low because it is targeted to the ClpC-ClpP protease complex by the MecA adaptor protein (19). In addition, comK transcription is inhibited by three repressors: Rok, AbrB, and CodY (20–22). This redundant inhibition of competence is necessary because K-state cells do not divide (23). As a culture approaches stationary phase and the cell density rises, a modified extracellular peptide (ComX) interacts with the ComP histidine kinase, which then donates a phosphoryl group to its cognate response regulator, ComA (24). As a result of this quorumsensing signal transduction pathway, comS transcription is activated. The anti-adaptor protein ComS competes for the binding of ComK to MecA, thereby lowering the rate of ComK degradation (25). The stabilization of ComK caused by ComS thus issues a license for transition to the K-state. Since ComS is produced in all the cells, and because the addition of ComX to cultures does not increase the percentage of the cells that become competent, the cellular decision point for competence must be sought outside the ComK stabilization pathway.
Two independent studies established that the positive autoregulation of comK is central to bimodal expression (26, 27). When comK was expressed from an inducible promoter as the only source of ComK protein in the cells, the response to increasing concentrations of inducer of a ComK-dependent reporter gene was unimodal and the expression of the reporter in the entire population increased continuously to a maximum value. In contrast, a bimodal response was observed when the same inducible construct was included in cells that retained the positive-feedback loop because they carried a wild-type copy of comK, which was still autoregulated. Based on these studies, it was suggested that noise in the basal expression of comK caused only a fraction of cells to exceed a threshold concentration of ComK needed to trigger the autoregulation of comK and drive these cells into the K-state. Since then, several studies have verified this hypothesis.
Two systems have been used to study the stochastic process that produces competent cells. In one, dispersed cultures are allowed to reach stationary phase, at which time about 15% of the cells convert to the competence ON state (28, 29). In the other, cells are deposited on agarose pads in nutrient-poor medium in which individual cells become competent, emerge from competence over an extended period, and eventually sporulate (30–32). We begin our discussion by addressing findings from the first system, in which programmed regulatory interactions temporally adjust the chances that a cell will become competent.
Changes in the Basal Expression of comK Establish a Window of Opportunity for Competence
The rate of transition to competence is high around the time of entry to stationary phase and then decreases, approaching zero 1.5 to 2 h later. This “window of opportunity” limits the bimodal expression of the K-state so that only 10 to 20% of the cells become competent. Leisner et al. (28) described this in detail and showed that the average basal level of comK transcription increases and then decreases, proposing that these changes establish the window of opportunity. Only those cells with a ComK level above a threshold become competent, and such cells exist transiently. Maamar et al. (29) also addressed the temporal regulation of the transition rate and used fluorescence in situ hybridization to count individual comK transcripts, confirming a transient rise in the basal average number of comK mRNA transcripts per cell (the “uptick”) that coincided with the timing of the window of opportunity described above. As the average mRNA content of the population increased, more cells crossed a threshold and became competent. Because this average number subsequently decreased, cells in which competence had not been induced during this period remained in the OFF state. Mathematical modeling and simulations confirmed the plausibility of this model. Following a method devised by Elowitz et al. (5), it was concluded that intrinsic noise in comK expression selects cells for competence. In agreement with this conclusion, when the promoter and start codon of comK were manipulated to decrease this noise without reducing the mean amount of ComK protein per cell, the proportion of K-state cells was predictably decreased. Using a different stratagem to reduce noise, Süel et al. (31) also demonstrated that noise in comK was responsible for the competence decision.
The Uptick Is Controlled by 0A∼P and by an Intrinsic Increase in Promoter Activity
The mechanism responsible for the increase and decrease in the basal rate of expression of comK (the uptick) is important to understand because these changes set the window of opportunity for the K-state. ComK and the positive autoregulatory loop play no role in the uptick, because the increase and decrease in the rate of transcription from the comK promoter (Pcomk) are unaffected in the absence of a functional comK gene (29).
All of the developmental adaptations of B. subtilis (e.g., biofilm formation, cannibalism, and sporulation) are tightly controlled by 0A∼P (Fig. 1), and competence is no exception, because a spo0A knockout does not express competence genes (33). It has been shown that as the concentration of 0A∼P increases during the approach to stationary phase, it first induces comK promoter activity and then represses it by direct binding (34). To accomplish the activation of basal expression, 0A∼P binds to three sites and represses by binding to two downstream sites, which likely have lower affinity. Interestingly, the activation is accomplished by anti-repression with respect to the Rok repressor (34), and this appears to be accomplished without the displacement of Rok from the DNA (unpublished data). Thus, 0A∼P regulates both the increasing and decreasing segments of the uptick. This is one more example of an emerging theme concerning 0A∼P; this molecule can exert both positive and negative effects on a given developmental gene as its concentration increases, establishing a temporal gate for gene expression (15, 35, 36). Interestingly, the details of how this so-called “bandpass” regulation (36) is achieved differ with each target gene, suggesting that evolution has repeatedly rediscovered the same use for 0A∼P. The mechanism for control of the comK uptick is illustrated in Fig. 2, in which additional details are provided.
Figure 2. Diagram of the uptick mechanism 34).
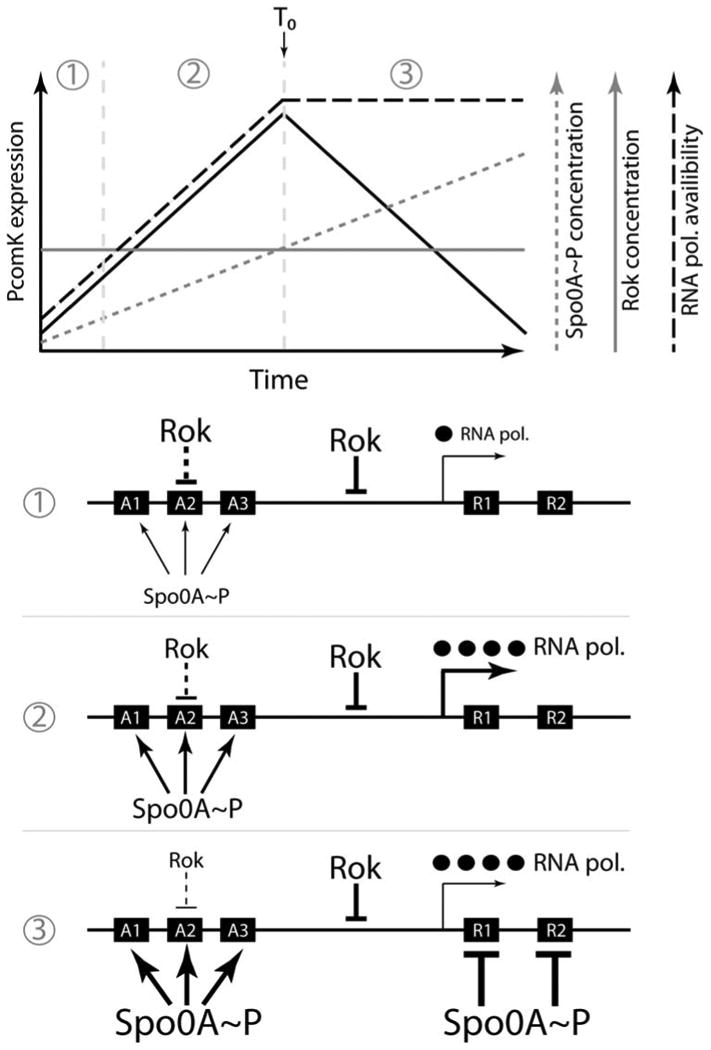
The top portion shows a graphical representation of the Rok and 0A∼P concentrations, as well as the availability of RNApol and the rate of comK basal transcription (solid black line) during the transition to stationary phase. (RNApol availability is used as a plausible stand-in for the cause of the global increase in transcription that was observed.) The peak rate of transcription coincides with T0, the time of departure from exponential growth. When the concentrations of available RNApol and of 0A∼P are low (1), Rok is dominant and the rate of comK transcription is also low. As the concentration of 0A∼P increases further, Rok is antagonized at sites A1, A2, and A3 and at the same time RNApol becomes more available. As a result, the rate of comK transcription increases (2). Finally, the 0A∼P concentration reaches a level that is able to repress at R1 and R2 and comK transcription slows (3). In reality, of course, three demarcated periods of time do not exist. Note that the concentration of Rok remains constant throughout and both RNApol and 0A∼P work to counteract its effects. Rok works at an unidentified site in addition to A1 to A3, shown here between A3 and R1. For simplicity, the availability of RNApol is shown as constant after T0, although the data would suggest that it varies somewhat (34). doi:10.1128/microbiolspectrum. TBS-0004-2012.f2
Surprisingly, in the absence of 0A∼P, an identical increase in comK basal activity takes place, but with a 10-fold-reduced amplitude (34). Thus, the roles of 0A∼P in regulation of the uptick are to amplify an inherent increase in transcription and then to repress transcription from PcomK. This inherent increase in the basal comK expression takes place during the approach to stationary phase and remarkably is also evident when several other promoters are studied, including completely synthetic σA- and σH-dependent promoters that consist of random sequences in which the canonical sequence motifs for these two promoter types are embedded. This inherent increase may be due to a passive mechanism in which RNA polymerase (RNApol) is released from stable RNA promoters as growth slows, thus activating repressed genes by competition with repressors as well as genes with promoters that have low RNApol affinities (Fig. 2) (37–39). Regardless of the mechanism, the inherent increase takes place as the culture approaches stationary phase and the growth rate drops, and so the competence transitions are likely to be influenced by some aspect of the metabolic state of the cells, perhaps indirectly by the same mechanisms as control transcription from stable RNA promoters as well as by the complex mechanisms that govern the formation of 0A∼P. Modeling of these interactions, including the roles of 0A∼P, of Rok as a repressor and of the inherent increase in comK activity reproduced the main features of the uptick (34).
If the amount of 0A∼P determines the time course of the window of opportunity for the K-state, competent cells examined at a fixed time should be restricted to those within a limited range of 0A∼P concentrations. This prediction was confirmed using spectrally distinct reporters fused to 0A∼P- and ComK-dependent promoters (Psdp and Pspo0A for 0A∼P and PcomK for ComK). Outside of this limited range the probability of competence decreased dramatically. However, within this range, there was no strong correlation between competence and the 0A∼P concentration. This result is consistent with the conclusion that the fate-determining noise is intrinsic to the comK promoter (29) and does not reflect variation in extrinsic factors such as the concentration of 0A∼P. Only the time course of competence development is dependent on 0A∼P (34).
Competence as an Excitable System
Süel et al. (31, 32) have exploited a different experimental system in which cells growing slowly in a nutrient-poor medium randomly enter and exit the competent state. Several observations made under these conditions strongly support the stochastic nature of the K-state; sister cells become competent with independent probabilities, and within a single cell lineage, cells enter and exit competence more than once, with no apparent “memory” effect. These observations led to the notion that competence is an “excitable” system. Older observations had shown that ComK negatively regulates the transcription of comS (40). Although the mechanism of this negative feedback is unknown, its existence suggested an explanation for the excitable behavior of the K-state, and indeed, a negative correlation between the activity of a ComK-dependent reporter and the comS promoter was documented by fluorescence microscopy (31). As a test of this idea, the feedback was bypassed by expressing comS from a ComK-dependent promoter, so that ComS remained abundant in the competent cells. As predicted, cells entered the K-state but exited about fivefold less frequently. These experiments suggest that ComS not only contributes to the ON state by preventing the degradation of ComK but also decays when a ComK-mediated negative-feedback loop prevents comS transcription, thus allowing cells to enter the OFF state.
This feedback model was further supported by expressing comK under the control of an inducible promoter in addition to the wild-type copy of comK (32). Within a certain inducer concentration range, the previously excitable behavior of the system was switched to oscillatory behavior, in which cells repeatedly entered and departed from the ON state. This would be expected (41) if induction raised the probability that the amount of ComK rapidly exceeded a threshold and then, with a delay, destabilized ComK by repressing comS. Expression of comS from an additional inducible promoter had little effect on the probability of competence initiation, suggesting that noise in comK expression remained the limiting factor. But increased expression of comS did have a positive effect on the average duration of competence expression. The model predicts this behavior, because extra ComS increases the time required for its decay to a level that permits the degradation of ComK. Taken together, the data from the two papers of Süel et al. strongly favor a model in which positive- and negative-feedback loops can facilitate the entry into and departure from the competence ON state and in which the noise in comK expression determines the probability of transitions.
To further understand the dynamics of this system, the ComS-mediated feedback loop was replaced by one in which MecA was placed under the control of a comK-dependent promoter (30). In this engineered feedback loop, a negative regulator (MecA) was regulated positively by ComK, whereas with the native circuitry, a positive regulator (ComS) is regulated negatively by ComK. The native and synthetic circuits differed in a number of interesting ways. For example, the time of exit from the ON state was less precisely defined with the native circuit and the mean duration of the ON state was longer. This difference can be appreciated intuitively. Since normally comS is repressed as ComK accumulates, causing a decrease in the number of ComS molecules per cell, noise in ComS increases in importance, thereby increasing the variability in the time of escape from competence. The opposite situation applies to the synthetic circuit. When ComK is high, so is MecA, and its noise decreases in importance. When the average amount of MecA per cell is high, ComK is sequestered and degraded and cells enter the OFF state. These experiments have at least two important and related biological implications. First, it appears that the circuit architecture can influence the dynamic properties of the system and may well have evolved to select these features. Second, the less precise timing of emergence from the K-state exhibited by the native circuit is suggested to increase fitness in the face of changing environments because for an extended time, some cells will be transformable, a bet-hedging strategy. This is certainly plausible, although it is important to recognize that the K-state involves more than transformability and the selective pressures that modulate its dynamics are therefore difficult to identify with certainty.
Two Experimental Systems for Studying Competence
Each of the two experimental systems used to analyze the regulation of competence has advantages. In the first and more classical approach, cells reach stationary phase, nutrients are depleted, and growth slows. This system seems to result in a relatively synchronous and global increase and decrease in the probability of switching to the K-state. Aside from its experimental simplicity, it displays competence as a programmed developmental system and has been used to uncover the role of the comK uptick as well as the importance of positive feedback and of stochastic decision making. The second system emphasizes the probabilistic nature of the transitions to both the ON and OFF states, facilitating the study of competence as an excitable system with a negative-feedback loop that governs the escape from the K-state. It is likely that both systems mimic situations that exist in nature: periods of near starvation and of transient abundance. These dual aspects, namely, programmed variation and stochastic transitions, are also evident for the sessile-motile switch, discussed below.
Living Together or Swimming Separately
Growing cultures of B. subtilis contain cells in two states with respect to motility (42). In swimmers, autolysin and motility genes are ON so that daughter cells detach from one another and can swim. In the sessile state, these genes are OFF and the cells remain in nonmotile chains. Because the cells are clonal, with identical genotypes, and are growing in a uniform environment, the choice between the ON and OFF states must be random. What is more, the two states can coexist and persist for several cell cycles, providing an example of epigenetic inheritance and of bistability in gene expression (43). Motile cells can exhibit chemotaxis, swimming toward attractants or away from repellents. Sessile cells can remain in place to exploit local riches, avoiding the risk of dispersal due to the random walk associated with motility even in the absence of attractants or repellents (44). They are also primed for biofilm formation. Two mechanisms have been proposed to explain the choice between the sessile and motile states: the epigenetic and σDswitch models.
Epigenetic Switch Model
The elegant epigenetic switch model (43) begins with the production of SinI in response to low levels of 0A∼P (Fig. 3A). In turn, SinI antagonizes the activity of the SinR repressor by direct protein-protein interaction (45). When SinI is absent, SinR homodimers repress the gene that encodes SlrR (46). When SinI is present, SlrR is produced and can form a heterocomplex with SinR, which directly represses the autolysin and motility genes, while SlrR antagonizes the activity of SinR homodimers (43). SinI and SlrR are both paralogs of SinR and bind to the latter, antagonizing its activity as a repressor of slrR and of the matrix genes (Fig. 3A). This unique situation creates a self-reinforcing, double negative-feedback loop, in which high SinR represses slrR and high SlrR inactivates SinR. As a result, two states can coexist in different cells within a population; high SinR corresponds to the motility ON state and high SlrR to the OFF state (Fig. 3A). Interestingly, a similar arrangement of mutually repressing proteins had been anticipated and analyzed in a synthetic-biology experiment in Escherichia coli (47), and this arrangement also resembles the Cro/CI double-negative lambda phage switch (48). SinR thus has two roles: as a partner with SlrR for repression of motility and autolysin genes and, as we shall see, as a direct repressor of genes for the expression of the biofilm matrix. The source of noise that triggers the switch is not known. Chai et al. (43) suggested that noise in sinI transcription is the primary stochastic event. This is plausible; sinI knockouts are permanently ON and overproducers are OFF. Of course the real “decider” may be variation in the level of 0A∼P. It has been shown recently that sinI transcription occurs within a window of 0A∼P concentration (35). Fluctuations in the level of 0A∼P during growth (39, 49) might thus cause transient spikes in the level of SinI, and decision making may be based on small, stochastic variations in the concentration of SinI or of 0A∼P.
Figure 3. Two proposed mechanisms controlling chaining and motility (A) The epigenetic switch (43).
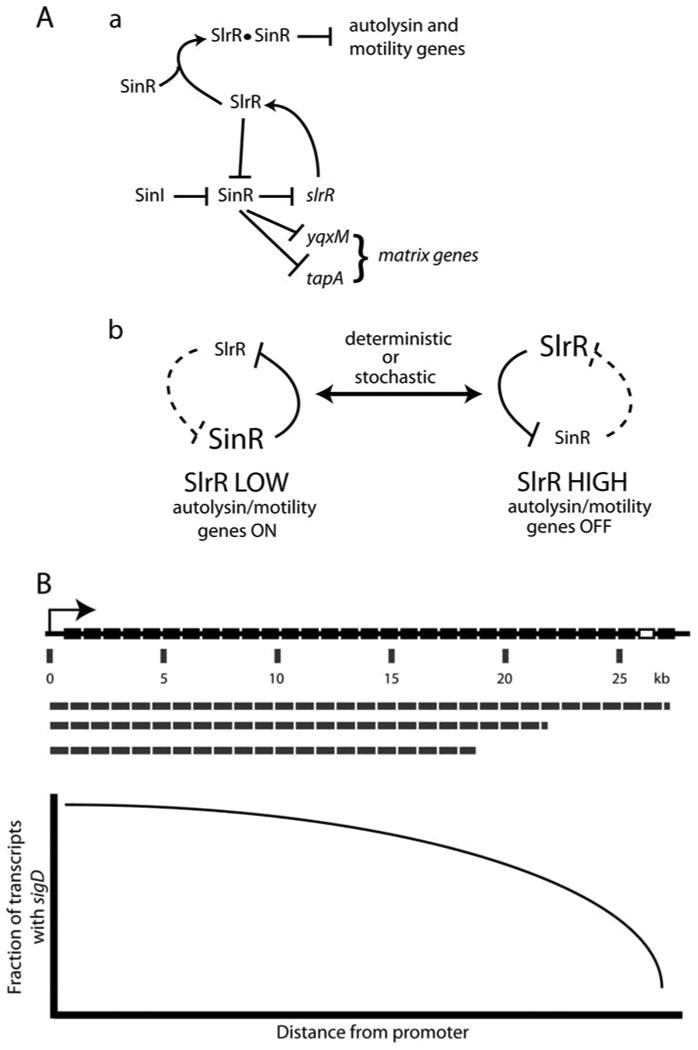
(Adapted with permission from the authors and the publisher from Fig. 1 in reference 43.) (a) SinI sequesters SinR, relieving repression of slrR. SlrR then binds to SinR, and the resulting complex represses the autolysin and motility genes and prevents repression of the matrix genes by SinR. (b) This circuitry allows for two metastable states. In one, when SlrR is low, the autolysin and motility genes are ON and the resulting cell is motile. In the other, when the SlrR concentration is high, these genes are OFF and the cells form chains and do not swim. The central feature of the circuitry that permits this bistable switch is the double-feedback mechanism involving repression of slrR by SinR and the inactivation of SinR for matrix gene repression by binding to SlrR. If SlrR is high, repression locks the cell in the motility OFF state, and vice versa. The transition between states can be stochastic, due to fluctuations in protein concentration (noise), or deterministic, in the sense that it is a programmed developmental switch. (B) Diagram of the gene position mechanism (50, 51). SigD is the penultimate gene in the 27-kb fla-che operon. For unknown reasons, the probability that promoter-distal genes are included in the operon transcript falls off with distance. Thus, if the mean number of transcripts per cell is low, some cells will have more SigD than others, and these cells will be motile. The distance-dependent fall-off in transcript abundance is reported to be due to the action of the SinR-SlrR heterocomplex (51). doi:10.1128/microbiolspectrum. TBS-0004-2012.f3
σD as the Switch Protein
A different mechanism has been suggested by Cozy and Kearns (50) (Fig. 3B). The motility and autolysin genes require an alternative sigma factor (σD) for their transcription, and σD is encoded near the end of the 27-kb flache operon. For unknown reasons, transcription of the 31 genes in this operon falls off with distance from a common promoter, implying that the amount of σD will vary from cell to cell if the transcription rate of this operon is low. Indeed, ON cells were shown to have about 10-fold more sigD transcript than OFF cells, and OFF cells had less σD protein. In this model, noise in the degree of processivity of transcription or in the degradation of fla-che mRNA from the 3′ end may determine whether a cell becomes motile. Similarly to comK expression, positive feedback may lock cells in the ON state, because an internal σD-dependent promoter is present within the fla-che operon and another is located upstream from the major SigA-dependent Pfla-che promoter (50).
What determines the fall-off in transcription of the fla-che operon? Cozy et al. (51) have recently shown that when a second copy of a gene known as slrA was introduced at a chromosomal ectopic site, σD-dependent gene expression was inhibited and the cells were completely locked into the OFF state. slrA had been identi-fied previously as a direct antagonist of SinR, acting like SinI in this respect (52, 53). As noted above (Fig. 3A), SinR is a repressor of slrR and an SlrR-SinR heterocomplex downregulates motility genes. Thus, it is reasonable to suggest that excess SlrA will titrate SinR, resulting in derepression of slrR (53). This will increase the amount of the SlrR-SinR complex and trigger the OFF state with respect to motility. Up to this point, the new finding with respect to SlrA is in accord with the epigenetic switch model and its circuitry. However, Cozy et al. (51) went on to show that the second slrA gene does not have its effect by repressing the fla-che promoter, although it does reduce the amount of σD protein and of σD-dependent gene expression. Instead, it dramatically decreases the amount of fla-che operon transcript distal to the promoter, apparently potentiating the phenomenon previously reported, in which transcript abundance falls off with distance from the initiation site (Fig. 3B). In other words, this evidence suggests that the SlrR-SinR heterocomplex acts post-initiation on the transcription of the fla-che operon. As predicted by this model, artificial expression of sigD bypassed the effect of extra SlrA, confirming that the amount of σD was limiting in the strain with two copies of slrA. These data do not agree with the epigenetic switch model, which is based partly on gel shifts and footprinting experiments demonstrating binding of SinR plus SlrR to the promoters of autolysin genes, implying direct repression by SinR plus SlrR. Cozy et al. (51) also used their system to demonstrate that the transition from the OFF to the ON state exhibits hysteresis and hyper-sensitivity with respect to the amount of σD, both hallmarks of a bistable system.
The epigenetic and σD switch models differ sharply in whether the autolysin and motility genes are regulated directly by the SlrR-SinR heterocomplex or indirectly via downstream effects on sigD. A problem with the σD switch model is that the dependence of the bistable behavior on σD is unexplained, although as noted above, this behavior may be due to an additional σD-dependent promoter believed to be embedded in the fla-che operon (50). It has been proposed that the epigenetic switch is responsible for the bistable regulation of motility but is biased by the amount of σD protein (43). In this view, the observation that motility ON cells exhibited enhanced sigD transcription, more σD-dependent gene expression, and more σD protein may reflect this bias. Nonetheless, the relationship between the two proposed mechanisms remains an important unresolved issue.
The roles of several other potentially important factors are not fully understood but hint at additional complexity (54). SwrA and SwrB bias the switch toward the ON position by increasing expression of the fla-che operon and by increasing the activity of σD, respectively (42, 55, 56). FlgM antagonizes σD, and DegU activates expression of flgM (57). Because SwrB is a membrane protein and DegU is a response regulator that is phosphorylated by DegS in response to unidentified signals, it is likely that these proteins are involved in signal transduction pathways that influence the switch bias. This would be reminiscent of the competence switch, where the probability of transition is modulated by signals that feed into the basal expression of comK. Also not fully understood is the role of YwcC in the control of biofilm formation (52).
Sporulation as a Response to Starvation
Spore formation, which results from the regulated expression of many genes, is the most-studied developmental adaptation of B. subtilis (58). The events that lead to the initiation of spore-specific gene expression are the focus of this discussion, rather than the later stages of spore formation. Classically, sporulation has been studied in poor growth media as a response to suboptimal conditions (59), although it apparently responds to different signals in the context of biofilms (60). In this section, we discuss the response to suboptimal conditions, which is typically studied in Difco Sporulation Medium (DSM) or resuspension media (61, 62).
As with competence and biofilm formation (see below), the initiation of spore formation is dependent on 0A∼P (Fig. 1), and indeed, spo0A was first identified as a spore gene. While some promoters, like PcomK and PsinI, respond to low levels of 0A∼P, the spore promoters require higher levels (15). Because the average concentration of 0A∼P increases during and after the transition to stationary phase, sporulation may be thought of as a later response than competence and the initiation of biofilm formation for the average cell.
Sporulation and the Phosphorelay
As noted above, the generation of 0A∼P depends on the phosphorelay (13), an extraordinary pathway that consists of only four proteins at its core (a kinase, 0F, 0B, and 0A), but is modulated by a number of 0F∼P phosphatases (63), several 0A∼P phosphatases (64), and other proteins, such as Sda (65), YaaT (66), YlbF (67), and YmcA (68). Also, at least some of the five kinases (KinA to -E) can dephosphorylate 0F∼P, and the entire pathway is reversible, so 0F can drain phosphoryl groups from 0B∼P and perhaps more slowly from 0A∼P. These proteins and chemical reactions represent points of information input, and the entire apparatus can be thought of as a signal integration device. In an early application of single-cell technology, the Grossman group reported that not all cells sporulate and that this difference was correlated with the low expression of early spore genes in the nonsporulating cells (69). Presciently, it was suggested that only cells with amounts of 0A∼P above a critical threshold would go on to sporulate, and this has been amply confirmed (14, 16, 17).
Because 0A∼P can activate the transcription of spo0A via its effect on σH, it was suggested that this positive-feedback loop generated heterogeneity and committed cells for spore formation, by analogy with other bimodal systems such as competence (70). This hypothesis has been refuted by two publications (14, 17). Both of these studies described the kinetics of expression and the roles of phosphorelay components on the single-cell level and have further sought the source of noise that determines which cells sporulate. The first important conclusion concerned the positive-feedback loop in which 0A∼P represses transcription of abrB, the resulting decrease in AbrB derepresses the gene encoding σH, and RNApol σH transcribes spo0A. This loop is not responsible for the major rise in the amount of 0A, because the rise precedes the observed decrease in the level of AbrB due to repression by 0A∼P (14). The same conclusion was reached by de Jong et al. (17). Chastanet et al. (14) also examined another loop, in which 0A∼P acts positively on the transcription of spo0F. Varying the expression of spo0F using an inducible promoter failed to increase the percentage of sporulating cells. Taken together, these and other results were interpreted as showing that the phosphorelay proteins are not rate limiting for spore formation. Instead, the synthesis of these proteins is adjusted to the need for 0A∼P by the action of feedback loops, resulting in the coordinated synthesis of the components and of 0A∼P. The feedback loops have been usefully described as comprising a “just-in-time” mechanism, which tunes the synthesis of the phosphorelay proteins (except 0B) to the needs of the system, ensuring that no component becomes rate limiting (14). An important contribution to this just-in-time mechanism is made by a complex arrangement of at least three 0A∼P binding sites upstream from the two spo0A promoters and also translational control, which mediate promoter switching and activation of 0A synthesis (71).
Further insight was obtained by examining the single-cell expression of 0A∼P activity, using a fusion of green fluorescent protein to the promoters of spoIIA and spoIIE, early spore genes that are transcribed only in the presence of 0A∼P (14, 17). The expression of both promoters was unimodal and quite noisy compared to that of a control promoter, leading to the conclusion that the activity of 0A∼P was likewise highly variable from cell to cell. Later in sporulation, expression from PspoIIA becomes bimodal, presumably because expression continues to increase in cells committed to sporulation (17). Similar experiments with fluorescent protein fusions to other relevant promoters showed that the expression of kinA, kinB, spo0F, and spo0A was unimodal and variable (17, 72). These results suggested an important conclusion. Whereas bimodal expression and the bifurcation of the cell population into sporulating and nonsporulating components were most likely dependent on heterogeneity in the level of 0A∼P, this heterogeneity was not rooted in bimodal expression of the phosphorelay components. Chastanet et al. (14) have proposed the appealing notion that the phosphorelay has evolved to be a “noise generator” and have proposed a computational model which was used to simulate the output of 0A∼P. Overexpression of KinA and inactivation of spo0E, which encodes a 0A∼P phosphatase, reduced cell-to-cell variation in 0A∼P, consistent with the idea that sporulation heterogeneity is rooted in the phosphorelay.
Remarkably, during the progression to sporulation, individual cells exhibit reversible bursts of spo0A transcription and corresponding bursts in Pspo0F activity, presumably due to activation of this promoter by 0A∼P (73). Thus, the noise generator acts stochastically to produce pulses of 0A∼P, which may be reversed by the action of Rap phosphatases acting on 0F, draining phosphate from 0A∼P, or by the direct action of Spo0E or other phosphatases on 0A∼P. A hallmark of spore formation is the localization of SpoIIE to the asymmetric septum (74, 75). This localization was also observed to be stochastic and reversible, so in about 2% of the cells localization of a SpoIIE fusion was seen to reverse, suggesting another layer of stochastic events on the level of protein localization (73). In contrast, the commitment to sporulation, measured by the activation of PspoIIR, was switch-like and not reversible; spores became apparent within a narrow window of time after PspoIIR was turned on. (The notion of irreversibility following activation of the σF-dependent spoIIR gene has been challenged recently [76], as explained below.) This “hybrid” model, which combines progression through reversible events with an irreversible commitment switch, was compared mathematically with two alternative models. In one, cells decide to sporulate in a single irreversible step. In the other, “reversible-only” model, even the decision to sporulate is reversible, distinguishing this model from the hybrid, or real-life, version. The three models were tested mathematically by subjecting them to stress of randomly varying durations, and the survival of the populations was determined. The hybrid model maximized survival in the face of unpredictable stress, with the reversible-only model doing well except with long stress durations. The latter result was explained by suggesting that if long periods of stress happen to be interrupted by short stress-free intervals, reversible-only cells run the risk of being caught sporeless, not able to turn on a dime and form spores. Perhaps an additional way to consider the bursts of 0A∼P is that it is a bet-hedging strategy that allows cells to repeatedly enjoy the possibility of entering alternative states during a period of stress. Thus, they continue to explore “probability space” and thereby enjoy the possibility of becoming cannibals, becoming competent, entering biofilms, or sporulating. This sampling would be precluded were they to move rapidly and irreversibly to spores. As noted above, when cells encounter more sustained stress, the hybrid mechanism will enjoy an advantage over a purely reversible one (73).
A recent study (76) has addressed the issue of commitment to sporulation, by utilizing an inducible kinA construct in which the level of transcription can be varied by adjusting the concentration of inducer (77). Remarkably, spore formation exhibited an ultrasensitive response, so the frequency of sporulation increased about 20-fold when the added inducer concentration was increased only 2.5-fold. This nonlinear response was analyzed by a combination of modeling and in vivo experiments, leading to a number of conclusions. First, it was shown that the production of 0A∼P exhibits a graded rather than an ultrasensitive response to kinA induction. Because an ultrasensitive response would be expected to generate a bimodal distribution, this is in accord with studies mentioned above, revealing that reporter gene expression for 0A∼P in a sporulating population is noisy but unimodal (14, 17). The source of ultrasensitivity was then sought in two downstream events, in which the alternative sigma factors σF and σE are activated in the forespore and mother cell, respectively. Both of these events exhibited an ultrasensitive response to the induction of kinA. In both cases, ultrasensitivity was ascribed to the existence of well-characterized regulatory pathways, in which 0A∼P directly activates the transcription of the sigma factor-encoding genes while indirectly activating each of the sigma factors posttranscriptionally. This arrangement establishes a pair of AND-gated coherent feed-forward loops (FFLs), which were shown by modeling to potentially lead to ultrasensitive responses (76) and are a common regulatory motif in both Saccharomyces cerevisiae and E. coli (78). Modeling and experimental data showed that the dynamic properties of the σF FFL cannot explain the ultrasensitive response of sporulation to kinA induction. In particular, σF is activated at a relatively low concentration of 0A∼P, not within the range of kinA inducer concentrations that elicits the sporulation response. In contrast, the properties of σE induction were found to be impressively in accord with the observed sporulation response to kinA. These results lead to an elegant picture of sporulation. The phosphorelay generates the unimodal but very noisy production of 0A∼P. At relatively low levels of 0A∼P, σF is activated. This is a necessary but insufficient condition for sporulation, and cells with activated σF in which σE is not yet activated can fail to sporulate (76). Cells with a higher concentration of 0A∼P cross a threshold that leads to a rapid (ultrasensitive) rise in activated σE, representing the point of irreversible commitment to sporulate (73, 79) and leading to the engulfment stage of spore formation. Coherent FFLs with AND-logic, operating exclusively on the level of transcription, have been described as “persistence detectors” (78). As pointed out previously (76), the analogous property in the present case may insulate cells from brief environmental fluctuations that might otherwise lead to a costly, unnecessary decision to sporulate.
The complexity of the phosphorelay, particularly with the intervention of numerous phosphatases and inputs from the cell cycle and cell-cell communication devices, offers many possibilities for generating variability while presenting a formidable challenge to deeper analysis. This complexity is underscored by the recent observation that 0A∼P formation varies during the cell cycle, mediated in part by a burst of Sda synthesis when DNA replication is initiated (80). Sda inhibits the activity of kinases that feed phosphate to the phosphorelay and thus downregulates sporulation (65, 81). Because the Sda concentration is minimal prior to the completion of a round of replication, 0A∼P presumably crosses the critical threshold for sporulation just before cell division. This timing mechanism tends to ensure that the developing spore will have the correct chromosome number. Also, in asynchronous cultures, only some cells will be in the appropriate stage of the division cycle for 0A∼P formation, and this may contribute to the observed heterogeneity in 0A∼P, in sporulation and perhaps in other forms of development.
Cross-Regulation Versus Temporal Competition Between Competence and Sporulation
Because several regulatory proteins have opposite effects on competence and sporulation, it has often been supposed that the pathways leading to these adaptations experience cross-regulation before commitment takes place (82–84). For example, SinI, which antagonizes SinR, is required for normal levels of spore formation, but inactivation of SinI has no effect on competence. In contrast, a null mutation of SinR depresses competence (40). It was reasonable to suppose that conditions leading to competence would require a low level of SinI and that prior to sporulation the SinI level would be elevated. However, the Süel group has posed a fundamental question concerning the relationship between competence and sporulation (85): does such cross-regulation take place prior to the decision point, or is there a “molecular race” in which the critical choice is made randomly, sending a cell toward one or another fate? According to this model, cross-regulation may indeed take place, but only after the decision point. This would serve to consolidate the choice, minimizing the appearance of dual-fate cells, which would be wasteful or even lethal.
To test this model, distinct fluorescent fusions of Pspo0A (a reporter of 0A∼P activity) and of PcomG (for competence) were coexpressed and single-cell measurements were carried out (85). It was concluded that the probability of competence initiation remained constant during the approach to sporulation. These data were consistent with the molecular race model and with the notion that the cell fate decisions take place independently of one another. As a further test, the expression of SinI and of AbrB (the latter being another potential cross-regulatory molecule) was measured using promoter fusions in cells that would eventually become competent or sporulate. No differences in these promoter activities were observed prior to the decision points, suggesting the absence of cross-regulation, at least involving these promoters, although it is still possible that the amounts or activities of the SinI and AbrB proteins differed in the cells with the two ultimate fates. Interestingly, the two promoter activities did differ after the decision point, during the execution of competence and sporulation. As might be expected, PabrB activity was lower in spores and PsinI was higher. A small number (∼0.1%) of “dual activity” cells which express comK also went on to sporulate. This number was predictable from the independent probabilities of the two cell fates. Smits et al. (86) have described another likely cross-regulation mechanism, which appears to prevent sporulation in competent cells. ComK induces the transcription of rapH. The RapH protein can dephosphorylate 0F∼P, thus preventing sporulation. Accordingly, inactivation of RapH led to the formation of dually expressing cells. Like the cases involving SinR and AbrB, this cross-regulation must take place after the decision point, because it depends on ComK. Since rapH must act after the decision point because it is produced as a product of ComK-dependent activation, it is possible to test its action as a cross-regulator by the use of a null mutant. This is not the case with sinR and abrB, because their elimination cannot distinguish between activity before or after the point of decision. Thus, although the data are consistent with the action of these molecules in cross-regulation after a decision has been made, there is no independent evidence that they do act in this manner.
Nevertheless, these findings are consistent with the following simple model for the relationship between sporulation and competence. The probabilities of competence and sporulation vary independently, partly in response to bursts in 0A∼P production (73) and to a gradual increase in the average concentration of this molecule and to excursions in the basal transcription of comK (34). Spore initiation requires more 0A∼P than competence and does, on the average, occur later. Once a cell enters one or the other pathway, cross-regulation precludes the alternative fate. If the choices to become competent and sporulate are made during a narrow interval, before the cross-regulation responses can be mounted, dual-activity cells result. Although there may be some cost to this infrequent outcome, it is presumably less than the cost of an elaborate cross-regulation mechanism that can operate before the decisions are made (85).
Biofilm Formation
Like many other microorganisms, B. subtilis forms complex communities known as biofilms (87). Indeed, the rich developmental potential of this model organism can be appreciated only in the context of biofilm development, because here the cell types discussed above appear with their temporal and spatial specificities revealed. Because biofilms contain motile, sessile, and sporulating cells, understanding their development depends on many of the interactions discussed above. The capacity to form robust biofilms has been lost by the descendants of laboratory strain 168 and is best studied with natural isolates, such as 3610, the probable parent of 168 (12). Not all media support the development of biofilms, and MsGG, in which glycerol and glutamate are the only organic molecules, is commonly used for biofilm investigations.
Biofilms Contain Several Cell Types
As a biofilm is initiated, cells become embedded in an insoluble matrix, consisting of protein fibers (encoded by the tapA [yqxM]-sipW-tasA operon) and an uncharacterized polysaccharide (the synthesis of which is encoded by the eps operon) (88). An important study used fluorescent reporters to the promoters of hag, tapA, and sspB as surrogates for the expression of motility, matrix, and spore genes, respectively (89). Competence is missing from this picture, only because strain 3610 is very poorly competent and other isolates have not been intensively studied. Early in biofilm formation at an agarair interface, hag was expressed in a majority of cells. At later times the number of motility-expressing cells decreased, and these were located at the edges and the base of the colony. Matrix-producing cells peaked in number after about 24 h and then declined. Sporulating cells increased steadily in number and were present preferentially in aerial projections from the biofilm surface, as shown previously (90). Time-lapse microscopy revealed that hag and tasA were not expressed in the same cells at the same time. Rather, cells expressing hag switched to tasA expression with time, consistent with the temporal progression noted above. Remarkably, sporulating cells arose mostly from matrix producers, resulting in a physical separation of motile and sporulating cells in different regions of the biofilm. Clearly, the various cell types tended to locate to different regions of the structure, and they showed specific temporal patterns of expression and distinct lineages. What is more, inactivation of tasA or eps resulted in a defect not only in biofilm architecture but also in the expression of a spore gene and in the regulation of the hag and tasA promoters.
The fact that different regions of a biofilm contain cells with different expression patterns tells us that geographically differentiated environmental signals influence the choice of cell fate. When studied in suspended, dispersed cultures, matrix genes are expressed in only a low percentage of the cells, and these cells are presumably selected randomly (91). The stochastic nature of decision making found for matrix production as well as for spore formation and for the expression of motility genes when dispersed cultures are studied suggests that chance as well as necessity probably plays a role in cell fate determination within a biofilm. Work emerging mainly from collaborations between R. Kolter and R. Losick and their coworkers has shed much light on the regulatory events that enable B. subtilis to produce biofilms.
SinR, 0A∼P, and Regulation in Biofilms
SinR regulates biofilm formation by at least two pathways and is the major proximal regulator of biofilm formation (Fig. 1). As described above, reduced SinR activity leads to a decrease in motility, driving cells into the sessile state, in which extensive chaining takes place. Additionally, SinR is a direct repressor of the biofilm-specific eps and tapA operons (92, 93). As noted above, SinI antagonizes SinR by forming SinI-SinR heterocomplexes (45). sinI transcription is driven by 0A∼P, and as 0A∼P increases in amount, the activity of SinR decreases and the repression of the eps and tapA operons is lifted. What is more, derepression of eps leads to the synthesis of EpsE, 1 of the 15 proteins encoded by this operon. In addition to participating in matrix synthesis, EpsE acts as a clutch protein, disconnecting the flagellar motor from its power source (94). Thus, a posttranslational mechanism contributes to the transition from the motile ON state to the OFF state accompanying biofilm formation.
In DSM starvation medium, as the amount of 0A∼P continues to increase, sinI transcription decreases, limiting the titration of SinR. It has been shown that this bandpass regulation of sinI transcription is due to the direct action of 0A∼P on activating and repressing sites near PsinI (35). On the other hand, in MsGG, as biofilms are formed, 0A∼P remains at an intermediate level and the amount of SinI continues to increase, leading to the formation of matrix-producing cells. The regulation of sinI by 0A∼P in DSM is analogous to the uptick in comK transcription, which is also dependent on direct binding of an increasing amount of 0A∼P (34). However, whereas the uptick in comK temporally adjusts the probability of competence, the uptick of sinI transcription in sporulation medium does not cause biofilm formation, and it appears to be an unavoidable consequence of a regulatory system that has evolved for dual use: sinI must be repressed by 0A∼P under sporulation conditions to avoid matrix production but must be expressed at an intermediate level to antagonize SinR for biofilm formation.
However, this simple description faces a potential dilemma within biofilms (35). The double negative loop proposed to regulate motility that was described above exhibits hysteresis, an inherent feature of such regulatory networks (43). In other words, when slrR was transiently overexpressed from an isopropyl-β-D-thiogalactopyranoside (IPTG)-inducible promoter, the system became epigenetically fixed in the motility OFF state. This is because SlrR titrates and inactivates SinR as a repressor of slrR, eliminating the need for the continued presence of an inducer to achieve high levels of SlrR (Fig. 2). In this motility OFF state, SinR is titrated not only by SinI but also by SlrR, and the matrix genes are derepressed. In a biofilm, most sporulating cells are derived from matrix producers. It is expected that if a matrix-producing cell goes on to sporulate, even if its elevated level of 0A∼P shuts down sinI expression, the previous production of SinI would have locked the cell into a matrix-producing state. But the evidence shows that under biofilm conditions, sporulating cells do not express matrix genes, although they derive from matrix producers (89). This problem is resolved by a beautifully simple proposed mechanism (35). SinR apparently binds cooperatively to its matrix gene targets but non-cooperatively to SinI. Thus, repression is hypersensitive to the concentration of SinR, but antirepression is not. Because sporulating cells contain two chromosomes, one destined for the forespore and the other for the mother cell, gene dosage of the sinI-sinR operon will ensure that the effect of repression by SinR will dominate over anti-repression by SinI and the hysteretic effect of prior high SinI will be overcome, shutting down matrix production. In support of this model, it was shown that doubling the copy number of the sinI-sinR operon shuts off matrix gene expression. Thus, two effects influence the transition from matrix production to sporulation. The first depends on the presence of activation and repression sites in the sinI promoter for 0A∼P that ensures an intermediate amount of SinI, and the second depends on gene dosage. This raises an interesting point. As noted above, in sporulating cells the activity of PsinI is elevated (85), likely serving to shut down competence after the decision to sporulate. For the transition from matrix production to sporulation to proceed in the biofilm, the concentration of SinI must be high enough to prevent SinR from inhibiting spore formation (95, 96), but not high enough to perpetuate matrix formation. This suggests that the reported binding of SinR to the promoters of spore genes must be relatively weak.
Why Spores in a Biofilm Come from Matrix-Producing Cells
Why are spores within a biofilm derived mainly from matrix producers, and why do mutant cells deficient in matrix formation fail to develop spores under biofilm conditions unless they are starved for nutrients (89)? Reporter fusions to 0A∼P-dependent promoters showed that in MsGG medium, matrix-deficient mutants contained decreased levels of 0A∼P, consistent with their failure to sporulate (60). Remarkably, inactivation of KinD, one of the five kinases that can feed phosphoryl groups to the phosphorelay, overcame the spore deficiency of a double eps tasA mutant. It was proposed that KinD, like other histidine kinases (97), can act as a phosphatase as well as a kinase and in this case was preventing spore formation by limiting the accumulation of 0A∼P. KinD is a membrane protein, and presumably, its phosphatase activity is downregulated when it senses the presence of matrix. This hypothesis is consistent with the observation that cocultivation of matrix mutants with matrix-producing sporulation mutants can complement sporulation in trans, demonstrating that matrix is sensed at the cell surface (60).
Quorum Sensing in Biofilm Development
It appears, then, that the decision of some cells in a biofilm to sporulate is at least in part deterministic, triggered by the production of matrix. But how are the matrix-producing cells determined? Lopez et al. (98) have discovered that a diverse set of molecules can trigger matrix production, even in a medium (LB broth) in which this does not usually occur. The common feature of these reagents is that they induce the leakage of potassium from the cell. Among the chemicals with this activity is surfactin, a natural product that is encoded by the srfA operon of B. subtilis (Fig. 4). KinC and KinD are both known to be required for biofilm formation (99), but only KinC, a membrane-localized protein, is needed for the response to surfactin (98). These and other observations strongly suggest that potassium leakage causes KinC to feed phosphate directly or via the phosphorelay to 0A, activating the transcription of eps and the tapA operon by the pathways described above.
Figure 4. Cell type determination in biofilms (60, 98, 104, 107).
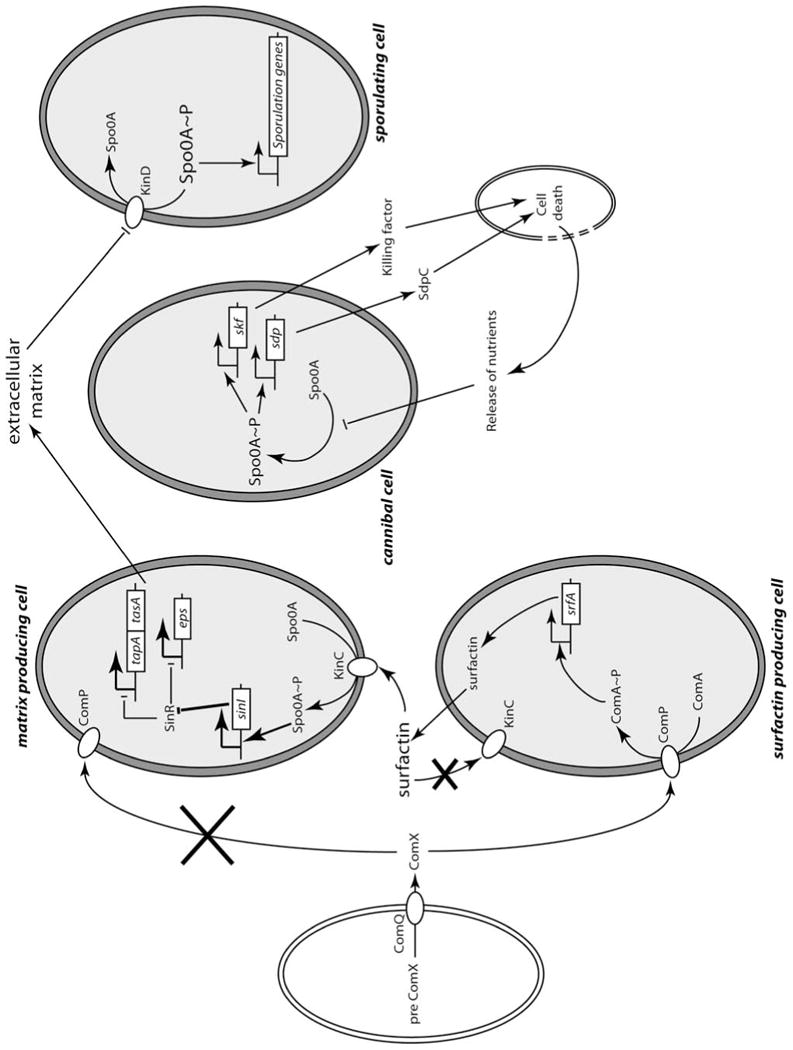
Pre-ComX is processed and ComX is secreted with the aid of ComQ. ComX interacts with ComP at the cell surface, resulting in the phosphorylation of ComA and the transcriptional activation of srfA. The surface-active SrfA molecule induces potassium flux in a susceptible cell, activating KinC and the formation of small amounts of 0A∼P. For unknown reasons, the surfactin-producing cell itself becomes refractory to activation by surfactin. In the susceptible cell, 0A∼P activates the transcription of sinI, which interacts with SinR, relieving repression of the matrix genes. For unknown reasons, matrix producers are not activated to produce surfactin. The presence of matrix downregulates the phosphatase activity of KinD, permitting the 0A∼P concentration to rise further, inducing sporulation. Matrix producers also become cannibals, because their intermediate 0A∼P concentration triggers toxin production. These toxins kill nonproducers, which release nutrients, delaying sporulation. As a result, matrix producers proliferate, increasing the population of eventual sporulating cells. doi:10.1128/microbiolspectrum.TBS-0004-2012.f4
These findings have several important implications. First, they establish the role of quorum-sensing systems for biofilm formation in B. subtilis. The two-component ComP-ComA proteins, which are activated by a quorum-sensing system, transcriptionally activate srfA by direct binding of ComA∼P to the srfA promoter. Surfactin then acts as a quorum-sensing signal to modulate KinC activity. Thus, a cascade of quorum-sensing and two-component regulatory proteins are involved in biofilm formation, emphasizing the importance of two-component regulators for Bacillus biology and the central role of cell-cell communication. Second, because diverse natural products produced by various species of soil-dwelling bacteria, including other bacilli, cause the release of potassium, it is likely that in nature, interstrain communication can cause biofilm formation. It is noteworthy that the identification of potassium leakage as a signal for KinC activation and of matrix as a signal for KinD are the only specific input signals so far identified leading to the phosphorylation of 0A via particular kinases. Although the molecular details of the signaling mechanisms are not understood, these are important findings.
It is also worth noting that the ComX pheromone that activates ComP exhibits a fascinating diversity, in that four specificity classes have been identified in natural isolates (100–103). Activation can occur within each class, but the ComX molecules produced by members of one class cannot activate the ComP receptors of another. A sequence relatedness tree constructed for the preComX proteins is congruent with that of the ComP receptors, and these sequences define the specificity classes. In contrast, the sequence relatedness of ComP and ComX is not congruent with that of housekeeping proteins. The evolutionary mechanisms that have led to this “pherotype” specificity are not understood, but somehow Bacillus has evolved to respond to the ComX pheromones produced by restricted classes of related bacteria, which are not necessarily the closest relatives. Whatever the selective pressures, this specificity potentially affects not only the K-state, which depends on the phosphorylation of ComA, but also the natural history of biofilms via their impact on surfactin production.
Cell Fate Determination
So far in our discussion, a linear signaling model has been described (Fig. 4). ComX activates ComP-ComA by phosphoryl transfer, and ComA∼P activates srfA. Then surfactin, a second quorum-sensing molecule, which is the product of srfA, activates KinC, triggering matrix production. Finally, matrix itself inactivates the phosphatase activity of KinD, acting extracellularly. While KinC allows sufficient 0A∼P formation to produce matrix, but not enough for sporulation, the inactivation of KinD-associated phosphatase activity permits sufficient 0A∼P to accumulate to trigger sporulation. How do these interactions generate cell type heterogeneity within the biofilm? By the use of promoter fusions to spectrally distinct fluorescent proteins, López et al. (104) observed that under conditions of biofilm development, only about 10% of the cells responded to the presence of the ComX pheromone (Fig. 4). This was not true for a domesticated 168 derivative, in which a more uniform response was observed. The explanation for this bimodal response is not known, but it was suggested that it may depend on a positive-feedback loop involving phrC, which is transcribed in the presence of ComA∼P (Fig. 1) (105). Thus, cells which initially respond more to ComX would produce more PhrC, preventing RapC from inhibiting the binding of ComA∼P to the srfA promoter, thus establishing a positive-feedback loop. Although ComA∼P activates transcription of the rapC phrC operon, a second promoter, dependent on σH, drives transcription of only phrC, suggesting that the synthesis of PhrC may exceed that of RapC. However, this scenario seems unlikely, because PhrC is secreted and its effects would presumably be distributed to non-producing cells. Also, if the cells that initially respond more to ComX are truly selected randomly, it is not clear why with time more and more cells do not become responders, unless some sort of additional regulation limits their number. Perhaps less-than-saturating amounts of ComX are produced by strain 3610, so only 10% of the cells cross the threshold for activation of ComP. Whatever the explanation, the stochastic selection of cells that respond to ComX provides a likely source of heterogeneity in cell fate.
Remarkably, the cells that produce surfactin in response to ComX are not the ones that produce matrix (104) (Fig. 4). This is because these responders cannot be activated by the surfactin that they produce. Also, once cells are producing matrix, they can no longer be activated by ComX to produce surfactin. Thus, the population bifurcates randomly into at least two populations with respect to matrix and surfactin production. In 168, a gene essential for competence (comS) is transcribed from the srfA promoter. Because 3610 is poorly competent, whether the surfactin producers have the potential to become competent cannot be readily studied in that strain. As noted above, matrix producers sporulate, and the bifurcation between matrix and surfactin producers has at least that additional consequence. It is interesting to note that some cells neither produce matrix nor become activated for surfactin production. Do these cells have an additional specialized role within the biofilm? Do they languish until the starvation pathway induces sporulation, do they lyse, or are they released from the biofilm to swim away (see below)?
Cannibalism in the Biofilm
In dispersed cultures grown under spore-forming conditions, some cells (cannibals) produce two toxins that kill sister cells (106). These martyrs release nutrients that are consumed by the cannibals, delaying their sporulation. The toxin-producing cells are immune to the lethal effects of the toxins. Cannibalism appears to be a “last chance” strategy that postpones the decision to sporulate. Because the toxin genes are controlled by 0A∼P, and because their promoters exhibit high affinity for this activator, cannibals are among the first cells with activated 0A∼P production. This is another example of the heterochronic production of 0A∼P underlying heterogeneity in B. subtilis development.
As noted above, small to moderate amounts of 0A∼P, produced in response to surfactin, trigger matrix synthesis in biofilms. It is therefore reasonable that surfactin can also activate toxin production when added under conditions (growth in LB broth) that do not ordinarily cause this to happen (107). These surfactin-responsive cells were shown to be the same ones that are triggered by surfactin to transcribe matrix genes, so the same cells in a biofilm produce toxins and matrix and only non-matrix-producing cells are killed. This has two consequences: sporulation is delayed, and the matrix toxin producers receive nutrients, which enables them to proliferate, and the proportion of matrix producers in the biofilm thus increases. It has been proposed that the production of matrix and toxins may be defensive and offensive weapons induced by other bacteria in the environment that secrete potentially damaging chemical agents causing potassium leakage.
Escape from the Biofilm
Although most cells in the biofilm eventually sporulate when nutrients are depleted, it appears that mechanisms exist that enable vegetative cells to swim away. This is perhaps one more example of bet hedging in which pioneers set out to explore the neighborhood for food or escape from toxins or predators, avoiding the costs of sporulation. Two dispersal mechanisms involve the secretion of D-amino acids, which interfere with the attachment of TasA fibers to the cell surface, facilitating breakdown of the matrix (108) and of norspermidine, which apparently attacks the exopolysaccharide matrix (109). It will be interesting to discover how the synthesis of these small molecules is regulated and whether they are expressed in only a subset of cells. An additional mechanism has been proposed to contribute to a return to motility, which aids in dispersal. As described above, SlrR maintains cells in the sessile, chaining state, and it has been shown that late in the life of biofilms, SlrR becomes unstable (110). SlrR degradation appears to be due in part to autocleavage and in part to a mechanism that requires ClpC.
Rare Versus Programmed Development: MecA as a Buffer
In the preceding discussion, we considered a number of developmental pathways that combine stochastic decision making and programmed regulatory events that modulate the probability that a given pathway is activated. Thus, the uptick in basal comK transcription or the programmed but noisy increase in 0A∼P production determines the chances that random cells will embark on competence or spore formation or form biofilms. However, because the critical events in these processes respond with thresholds for transcription factor binding and because the basal expression of determining molecules (ComK and 0A∼P) is nonzero and noisy, the probabilities of cells embarking on development is also not zero even during exponential growth in rich media. Thus, when reporter fusions are used, cells that express comK, spoIIG (a sporulation gene), or eps are readily detected in growing cells, albeit at very low frequencies (111). In fact, small numbers of mature spores and transformable cells also form in growing cultures. These considerations echo the dichotomy between excitable and programmed transitions in motility and competence.
It is plausible that the low frequency of development in exponential populations is a form of a priori bet hedging. Thus, a small investment may be made that increases the likelihood of survival in the face of an unexpected environmental catastrophe; a few cells will survive as spores, a few can receive new genes that may enhance fitness, and some may produce matrix, which primes them for biofilm formation. These stochastic switches of rare cells into developmental pathways during growth stand in contrast to the programmed transitions that occur when growth slows, and they are likely due to noise in the formation of 0A∼P or of a more downstream regulator like ComK or SinI. If these transitions serve to enhance fitness, as proposed, their rates have likely been adjusted by selection.
It has been observed that the frequency of these rare developing cells increases dramatically when mecA is inactivated (111). As described above, MecA has been studied as an adaptor protein that targets ComK for degradation unless the targeting is relieved by the synthesis of ComS late in growth in response to quorum sensing. In loss-of-function mecA mutants, the auto-stimulation of comK is uncontrolled, and massive production of competent cells takes place during growth. In addition, cells that express eps or spoIIG are more frequent, as are the heat-resistant spores produced during exponential growth. This effect appears to be due to the ability of MecA to bind 0A, somehow interfering with the ability of 0A∼P to activate transcription (111). This interference is not exerted via the phosphorelay, nor can MecA by itself dephosphorylate 0A∼P in vitro.
MecA may have evolved as a buffer that limits the chances that cells will enter developmental pathways until programmed mechanisms come into play and the buffer is inactivated (as by ComS) or overwhelmed by the activated phosphorelay. By adjusting the affinities of MecA for its binding partners, evolution can tinker with the rates of transitions for particular pathways.
Acknowledgments
We thank Jeanie Dubnau and Dan Kearns for helpful comments on the manuscript.
Work from our lab was supported by NIH grant GM057720.
Footnotes
The authors have no conflicts of interest regarding this manuscript.
References
- 1.Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 6.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30:5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- 11.Lemon KP, Earl AM, Vlamakis HC, Aguilar C, Kolter R. Biofilm development with an emphasis on Bacillus subtilis. Curr Top Microbiol Immunol. 2008;322:1–16. doi: 10.1007/978-3-540-75418-3_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 14.Chastanet A, Vitkup D, Yuan GC, Norman TM, Liu JS, Losick RM. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2010;107:8486–8491. doi: 10.1073/pnas.1002499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita M, Gonzalez-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong IG, Veening JW, Kuipers OP. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J Bacteriol. 2010;192:2053–2067. doi: 10.1128/JB.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 19.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamoen LW, Kausche D, Marahiel MA, van Sinderen D, Venema G, Serror P. The Bacillus subtilis transition state regulator AbrB binds to the −35 promoter region of comK. FEMS Microbiol Lett. 2003;218:299–304. doi: 10.1111/j.1574-6968.2003.tb11532.x. [DOI] [PubMed] [Google Scholar]
- 22.Hoa TT, Tortosa P, Albano M, Dubnau D. Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol Microbiol. 2002;43:15–26. doi: 10.1046/j.1365-2958.2002.02727.x. [DOI] [PubMed] [Google Scholar]
- 23.Haijema BJ, Hahn J, Haynes J, Dubnau D. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol Microbiol. 2001;40:52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- 24.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 25.Prepiak P, Dubnau D. A peptide signal for adapter protein-mediated degradation by the AAA+ protease ClpCP. Mol Cell. 2007;26:639–647. doi: 10.1016/j.molcel.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smits WK, Eschevins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol. 2005;56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 27.Maamar H, Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leisner M, Stingl K, Radler JO, Maier B. Basal expression rate of comK sets a ‘switching-window’ into the K-state of Bacillus subtilis. Mol Microbiol. 2007;63:1806–1816. doi: 10.1111/j.1365-2958.2007.05628.x. [DOI] [PubMed] [Google Scholar]
- 29.Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cagatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 31.Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 32.Süel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. Science. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 33.Albano M, Hahn J, Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987;169:3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirouze N, Desai Y, Raj A, Dubnau D. Spo0A∼P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet. 2012;8:e1002586. doi: 10.1371/journal.pgen.1002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen S, Garcia-Ojalvo J, Elowitz MB. Dynamical consequences of bandpass feedback loops in a bacterial phosphorelay. PLoS One. 2011;6:e25102. doi: 10.1371/journal.pone.0025102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 38.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci U S A. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirouze N, Prepiak P, Dubnau D. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 2011;7:e1002048. doi: 10.1371/journal.pgen.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn J, Kong L, Dubnau D. The regulation of competence transcription factor synthesis constitutes a critical control point in the regulation of competence in Bacillus subtilis. J Bacteriol. 1994;176:5753–5761. doi: 10.1128/jb.176.18.5753-5761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barkai N, Leibler S. Circadian clocks limited by noise. Nature. 2000;403:267–268. doi: 10.1038/35002258. [DOI] [PubMed] [Google Scholar]
- 42.Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berg HC. Random Walks in Biology. Princeton University Press; Princeton, NJ: 1983. [Google Scholar]
- 45.Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 46.Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 48.Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S. Switches in bacteriophage lambda development. Annu Rev Genet. 2005;39:409–429. doi: 10.1146/annurev.genet.39.073003.113656. [DOI] [PubMed] [Google Scholar]
- 49.Castilla-Llorente V, Salas M, Meijer WJ. kinC/D-mediated heterogeneous expression of spo0A during logarithmical growth in Bacillus subtilis is responsible for partial suppression of phi 29 development. Mol Microbiol. 2008;68:1406–1417. doi: 10.1111/j.1365-2958.2008.06234.x. [DOI] [PubMed] [Google Scholar]
- 50.Cozy LM, Kearns DB. Gene position in a long operon governs motility development in Bacillus subtilis. Mol Microbiol. 2010;76:273–285. doi: 10.1111/j.1365-2958.2010.07112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cozy LM, Phillips A, Calvo RA, Bate A, Hsueh Y-H, Bonneau R, Eichenberger P, Kearns DB. SlrA/SlrR/SinR inhibits motility gene expression upstream of a hypersensitive and hysteric switch at the level of sD in Bacillus subtilis. Mol Microbiol. 2012;83:1210–1228. doi: 10.1111/j.1365-2958.2012.08003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi K. SlrR/SlrA controls the initiation of biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;69:1399–1410. doi: 10.1111/j.1365-2958.2008.06369.x. [DOI] [PubMed] [Google Scholar]
- 53.Chai Y, Kolter R, Losick R. Paralogous antirepressors acting on the master regulator for biofilm formation in Bacillus subtilis. Mol Microbiol. 2009;74:876–887. doi: 10.1111/j.1365-2958.2009.06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patrick JE, Kearns DB. Swarming motility and the control of master regulators of flagellar biosynthesis. Mol Microbiol. 2012;83:14–23. doi: 10.1111/j.1365-2958.2011.07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Werhane H, Lopez P, Mendel M, Zimmer M, Ordal GW, Marquez-Magana LM. The last gene of the fla/che operon in Bacillus subtilis, ylxL, is required for maximal σD function. J Bacteriol. 2004;186:4025–4029. doi: 10.1128/JB.186.12.4025-4029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calvio C, Celandroni F, Ghelardi E, Amati G, Salvetti S, Ceciliani F, Galizzi A, Senesi S. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J Bacteriol. 2005;187:5356–5366. doi: 10.1128/JB.187.15.5356-5366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsueh YH, Cozy LM, Sham LT, Calvo RA, Gutu AD, Winkler ME, Kearns DB. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol Microbiol. 2011;81:1092–1108. doi: 10.1111/j.1365-2958.2011.07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins D, Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev. 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoch JA. spoO genes, the phosphorelay, and the initiation of sporulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and Other Gram-Positive Bacteria: Biochemistry, Physiology, and Molecular Genetics. American Society for Microbiology; Washington, DC: 1993. pp. 747–755. [Google Scholar]
- 60.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio. 2010;1(1):e00035–10. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaeffer P, Millet J, Aubert J-P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perego M, Hanstein C, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 64.Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol. 2001;42:133–143. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- 65.Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 66.Hosoya S, Asai K, Ogasawara N, Takeuchi M, Sato T. Mutation in yaaT leads to significant inhibition of phosphorelay during sporulation in Bacillus subtilis. J Bacteriol. 2002;184:5545–5553. doi: 10.1128/JB.184.20.5545-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tortosa P, Albano M, Dubnau D. Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol Microbiol. 2000;35:1110–1119. doi: 10.1046/j.1365-2958.2000.01779.x. [DOI] [PubMed] [Google Scholar]
- 68.Carabetta VJ, Tanner AW, Greco TM, Defrancesco M, Cristea IM, Dubnau D. A complex of YlbF, YmcA and YaaT regulates sporulation, competence and biofilm formation by accelerating the phosphorylation of Spo0A. Mol Microbiol. 2013;88:283–300. doi: 10.1111/mmi.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung JD, Stephanopoulos G, Ireton K, Grossman AD. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 71.Chastanet A, Losick R. Just-in-time control of Spo0A synthesis in Bacillus subtilis by multiple regulatory mechanisms. J Bacteriol. 2011;193:6366–6374. doi: 10.1128/JB.06057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eswaramoorthy P, Dinh J, Duan D, Igoshin OA, Fujita M. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology. 156:2294–2304. doi: 10.1099/mic.0.038497-0. [DOI] [PubMed] [Google Scholar]
- 73.Kuchina A, Espinar L, Garcia-Ojalvo J, Süel G. Reversible and noisy progression towards a commitment point enables adaptable and reliable cellular decision-making. PLoS Comp Biol. 2011;7:e1002273. doi: 10.1371/journal.pcbi.1002273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duncan L, Alper S, Arigoni F, Losick R, Stragier P. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science. 1995;270:641–644. doi: 10.1126/science.270.5236.641. [DOI] [PubMed] [Google Scholar]
- 75.Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 76.Narula J, Devi SN, Fujita M, Igoshin OA. Ultrasensitivity of the Bacillus subtilis sporulation decision. Proc Natl Acad Sci USA. 2012;109:E3513–E3522. doi: 10.1073/pnas.1213974109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eswaramoorthy P, Duan D, Dinh J, Dravis A, Devi SN, Fujita M. The threshold level of the sensor histidine kinase KinA governs entry into sporulation in Bacillus subtilis. J Bacteriol. 2010;192:3870–3882. doi: 10.1128/JB.00466-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alon U. An Introduction to Systems Biology Design Principles of Biological Circuits. Chapman & Hall/CRC; Boca Raton, FL: 2007. [Google Scholar]
- 79.Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 80.Veening JW, Murray H, Errington J. A mechanism for cell cycle regulation of sporulation initiation in Bacillus subtilis. Genes Dev. 2009;23:1959–1970. doi: 10.1101/gad.528209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cunningham KA, Burkholder WF. The histidine kinase inhibitor Sda binds near the site of autophosphorylation and may sterically hinder autophosphorylation and phosphotransfer to Spo0F. Mol Micro-biol. 2009;71:659–677. doi: 10.1111/j.1365-2958.2008.06554.x. [DOI] [PubMed] [Google Scholar]
- 82.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 83.Hahn J, Roggiani M, Dubnau D. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J Bacteriol. 1995;177:3601–3605. doi: 10.1128/jb.177.12.3601-3605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schultz D, Wolynes PG, Ben Jacob E, Onuchic JN. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106:21027–21034. doi: 10.1073/pnas.0912185106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuchina A, Espinar L, Cagatay T, Balbin AO, Zhang F, Alvarado A, Garcia-Ojalvo J, Süel GM. Temporal competition between differentiation programs determines cell fate choice. Mol Syst Biol. 2011;7:557. doi: 10.1038/msb.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 87.Lopez D, Vlamakis H, Kolter R. Biofilms. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 89.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008;22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- 93.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- 94.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 95.Mandic-Mulec I, Doukhan L, Smith I. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J Bacteriol. 1995;177:4619–4627. doi: 10.1128/jb.177.16.4619-4627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mandic-Mulec I, Gaur N, Bai U, Smith I. Sin, a stage-specific repressor of cellular differentiation. J Bacteriol. 1992;174:3561–3569. doi: 10.1128/jb.174.11.3561-3569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huynh TN, Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol. 2011;82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci USA. 2009;106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kobayashi K, Kuwana R, Takamatsu H. kinA mRNA is missing a stop codon in the undomesticated Bacillus subtilis strain ATCC 6051. Microbiology. 2008;154:54–63. doi: 10.1099/mic.0.2007/011783-0. [DOI] [PubMed] [Google Scholar]
- 100.Tran L-SP, Nagai T, Itoh Y. Divergent structure of the ComQXPA quorum sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol Microbiol. 2000;37:1159–1171. doi: 10.1046/j.1365-2958.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- 101.Tortosa P, Logsdon L, Kraigher B, Itoh Y, Mandic-Mulec I, Dubnau D. Specificity and genetic polymorphism of the Bacillus competence quorum-sensing system. J Bacteriol. 2001;183:451–460. doi: 10.1128/JB.183.2.451-460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ansaldi M, Marolt D, Stebe T, Mandic-Mulec I, Dubnau D. Specific activation of the Bacillus quorum-sensing systems by isoprenylated pheromone variants. Mol Microbiol. 2002;44:1561–1573. doi: 10.1046/j.1365-2958.2002.02977.x. [DOI] [PubMed] [Google Scholar]
- 103.Stefanic P, Mandic-Mulec I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J Bacteriol. 2009;191:1756–1764. doi: 10.1128/JB.01290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.López D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis. Mol Microbiol. 2003;49:1509–1522. doi: 10.1046/j.1365-2958.2003.03659.x. [DOI] [PubMed] [Google Scholar]
- 106.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 107.Lopez D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009;74:609–618. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kolodkin-Gal I, Romero D, Cao S, Clardy J, Kolter R, Losick R. D-Amino acids trigger biofilm disassembly. Science. 2010;328:627–629. doi: 10.1126/science.1188628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kolodkin-Gal I, Cao S, Chai L, Bottcher T, Kolter R, Clardy J, Losick R. A self-produced trigger for biofilm disassembly that targets exopolysaccharide. Cell. 2012;149:684–692. doi: 10.1016/j.cell.2012.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 110.Chai Y, Kolter R, Losick R. Reversal of an epigenetic switch governing cell chaining in Bacillus subtilis by protein instability. Mol Microbiol. 2010;78:218–229. doi: 10.1111/j.1365-2958.2010.07335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Prepiak P, Defrancesco M, Spadavecchia S, Mirouze N, Albano M, Persuh M, Fujita M, Dubnau D. MecA dampens transitions to spore, biofilm exopolysaccharide and competence expression by two different mechanisms. Mol Microbiol. 2011;80:1014–1030. doi: 10.1111/j.1365-2958.2011.07627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


