Abstract
Trypsin is the most important digestive enzyme produced in the pancreas, and is a useful biomarker for pancreatitis. In this work, a rapid and sensitive method for the quantitative determination of trypsin activity is developed by using a biological alpha-hemolysin protein nanopore. Due to its much larger molecular diameter than the narrow pore constriction, trypsin itself cannot transport through the alpha-hemolysin channel. Hence, an indirect trypsin detection method is developed by monitoring its proteolytic cleavage of a lysine-containing peptide substrate. Based on the current modulations produced by the translocation of the substrate degradation products in the nanopore, the activity levels of trypsin could be determined. The method is rapid and highly sensitive, with picomolar concentrations of trypsin detected in minutes. In addition, the effects of cation and temperature on the sensor sensitivity, trypsin inhibition, and serum sample analysis are also investigated.
Keywords: trypsin activity, nanopore, label-free, biomarker, serum
TOC image
Trypsin is a serine protease with 223 amino acid residues (M.W. = 23.8 kDa), which is the most important digestive enzyme produced in the pancreas as the inactive proenzyme trypsinogen1. Trypsin cleaves peptide bonds mainly at the carboxyl side of the lysine or arginine amino acid residue. Pancreatic trypsin production can be adversely affected by pathologies (e.g., pancreatitis), resulting in organ damage and release of enzyme into the blood. It is well known that the trypsin level is increased with some types of pancreatic diseases. For example, it has been reported that the mean trypsin concentration in a pancreas transplant patient’s urine was 84.4 μg/mL, while no trypsin was found in healthy person’s urine2; in healthy people, the serum trypsin concentration ranges from 140–400 ng/mL, while the mean serum trypsin concentration in acute pancreatitis patients was 1,400 ng/mL3. Therefore, rapid and sensitive methods for trypsin detection, activity assay, and inhibitor screening are highly desired for the efficient diagnosis and treatment of pancreatic diseases.
Thus far, two major approaches have been developed for protease detection. They are affinity-based methods where an antibody or a protease template is usually employed4, 5, and activity-based assay, which typically use a synthetic substrate consisting of a short peptide of known sequence to identify a specific protease and detect its activity6. While the conventional affinity-based methods allow for analysis of protease abundance, they are usually time-consuming and do not succeed in providing a measure of protease activity. Several such examples include radioimmunoassay, enzyme-linked immunosorbent assay, electrochemical immunoassay, molecular imprinting technology, etc4, 5, 7, 8. However, activities of protease are tightly regulated through post-translational modifications. These modifications typically occur in the absence of any detectable change in protease abundance and are thus invisible to the conventional protease assay, leading to significant difference between overall abundance levels and activity of proteases. In contrast, measurement of protease activities can reveal valuable information regarding the type of chemical entities accepted by the protease as its potential substrates, and thus helps to better understand the properties and the catalytic mechanism of the protease9. Due to its potential applications in inhibitor screening, disease diagnosis, and drug development, measurement of protease activities has attracted increasing attention in the recent years, and numerous methods have been developed. Examples of activity-based trypsin assays include gel electrophoresis10, gelatin-based film techniques11, colorimetry6, chemiluminescence12, fluorescence13, and electrochemical methods14. However, most of these methods require the use of labels or sophisticated instruments.
In this work, we report a novel, rapid, and label-free nanopore stochastic sensing method to detect trypsin and quantitatively determine its activity. Nanopore sensing is an emerging amplification-free technique for measuring single molecules15–20. When analyte molecules pass through a single nanopore, they would induce ionic current modulations. These modulation events can be utilized to provide valuable information regarding both the concentration and the identity of the analyte, the former from the event frequency or event counts, while the latter from the event residence time and the extent of current blockage (i.e., amplitude). Since analytes pass through the nanopore at one molecule at a time, simultaneous detection and quantification of multiple components in a solution mixture can be readily accomplished using a single nanopore as long as the nanopore itself can provide enough resolution21. During the past decade, nanopore technology has shown great potential applications in numerous fields, such as medical diagnosis22–24, homeland security and bio-defense25, 26, pharmaceutical screening27, 28, and environmental protection29, 30.
It should be noted that nanopore stochastic sensors have been utilized to detect enzymatic reactions31–34. By using a substrate (tethered or untethered) for a particular enzyme/protease and monitoring its translocation in the nanopore, we can readily identify whether the sample contains the target enzyme/protease or not. Specifically, the appearance of new types of events or the change in the event frequency in the nanopore indicates the presence of the target analyte. However, to the best of our knowledge, until now, there is no report of employing nanopore sensing technology for the quantitative determination of enzyme/protease activity.
Experimental Section
Materials and reagents
The trypsin substrate, β-amyloid (10–20) peptide (sequence: YEVHHQKLVFF; PI: 7.7), was synthesized from American Peptide Company (Sunnyvale, CA). The two substrate cleavage fragments (peptide sequence: LVFF and YEVHHQK) were purchased from WatsonBio (Houston, TX), and Biomatik Corporation (Wilmington, DE), respectively. All the other chemicals, including Trypsin (from bovine pancreas, its optimum pH ranging from 7 to 9), were obtained from Sigma-Aldrich (St. Louis, MO). All of the solutions were prepared with HPLC-grade water (ChromAR, Mallinchkrodt Baker). The concentrations of the stock solutions were 0.5 mM, 10 mM, 0.5 mM, and 100 μg/mL for the peptide substrate, its two breakdown products (peptides YEVHHQK and LVFF), and trypsin, respectively. All these solutions were kept at −20 °C before and immediately after use.
Bilayer experiment and data analysis
The procedure of electrical recording has been described previously (see Supporting Information, Fig. S-1 for a schematic illustration of the protein nanopore sensing system)30. Briefly, nanopore experiments were performed in a two-compartment chamber separated by a Teflon film (Goodfellow Malvern, PA) having a 150 μm diameter orifice, on which planar bilayer was formed using 1, 2-diphytanoylphosphatidylcholine (DPhPC, Avanti Polar Lipids, Alabaster, AL, USA) according to the Montal-Mueller method35. Single channel recordings were carried out at 24 ± 1 °C using engineered α-hemolysin (αHL) (M113F) 7 protein pores31 under symmetric buffer conditions, with each compartment containing a 2.0 mL electrolyte solution (1.0 M NaCl,10 mM Tris, pH 7.5). With the exception of the αHL protein nanopore (approximately 0.2–2.0 ng/mL), which was added to the grounded cis side of the chamber device, other substances were introduced to the trans chamber. Currents were recorded with an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA), filtered with a four-pole low-pass Bessel filter at 5 KHz, and then digitized with a Digidata 1440A converter (Molecular Devices) at a sampling frequency of 50 KHz. The signatures of current modulations (or events), including the mean values of residual currents, residence times, and amplitude histograms, were obtained using Clampfit 10.3 software (Molecular Devices).
Serum sample analysis
100 ng trypsin was spiked into 2 μL bovine serum (Thermo Fisher Scientific, Grand Island, NY) to create a mock serum sample. The simulated serum sample, 4.24 μL reaction buffer (containing 1 mM NaCl and 10 mM tris, pH = 7.5), and 20 μL 0.5 mM substrate β-amyloid (10–20) peptide were mixed and incubated for 30 minutes in water bath at 22 °C. Then, the mixture was introduced to the trans chamber compartment for single channel recording. The control sample was prepared using the same concentrations and volumes of trypsin and the substrate peptide but with HPLC water instead of the serum. Both experiments were carried out using the (M113F)7 α-hemolysin protein nanopore in an electrolyte buffer solution (1.0 M NaCl, 10 mM Tris, pH = 7.5).
Results and discussion
Detection of trypsin
The principle for nanopore detection of trypsin is shown in Scheme 1. When there is no presence of trypsin, the substrate peptide molecules pass through the nanopore giving one signal reading, i.e., a type of characteristic current modulation events with unique residence time and blockage amplitude. By contrast, if trypsin is present in the solution, acting like a scissors, cuts the peptide molecules, the cleavage products produce entirely different current modulations from those of the substrate. A comparison of these events quantifies the trypsin enzymatic activity.
Scheme 1.
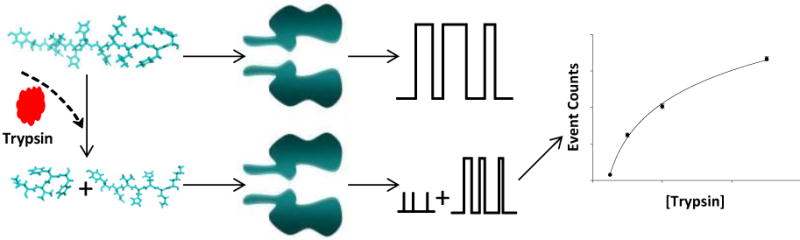
Schematic representation of the principle of nanopore detection of trypsin. The interaction of the substrate cleavage products with the nanopore produces current modulation events having significantly different signatures from those attributed to the translocation of the substrate peptide in the nanopore. A comparison of these events quantifies the trypsin activity.
As proof of concept, our effort in the development of trypsin nanopore sensor was initially conducted at −40 mV in an electrolyte buffer solution containing 1 M NaCl and 10 mM Tris (pH 7.5). The mutant αHL (M113F)7 protein nanopore was used as the stochastic sensing element, while β-amyloid (10–20) peptide (sequence: YEVHHQKLVFF, 5 μM) was employed as the substrate. It is well known that catalytic cleavage of peptides by trypsin occurs after positively charged lysine or arginine amino acid residues, while the αHL (M113F)7 protein pore has been shown to offer an improved sensor resolution/sensitivity (e.g., prolonged event residence time for the analytes) over the wild-type αHL pore36. The −40 mV voltage and 5 μM substrate were chosen based on our previous work37. Specifically, at this applied potential bias, peptide translocation in the αHL nanopore showed a largest signal-to-noise ratio (i.e., small-amplitude, transient, rarely-observed background spikes vs. large-amplitude, long-duration, and frequent peptide blockage events), and hence, the nanopore sensor offered the best resolution. On the other hand, 5 μM YEVHHQKLVFF could produce enough translocation events for statistical data analysis using a relatively short recording time and a relatively small amount of expensive peptide substrate. The experimental results were summarized in Fig. 1. It is apparent that, with the addition of β-amyloid (10–20) peptide alone in the trans chamber compartment, only one major type of current modulations was observed. These current blockage events had a mean residence time of ~0.31 s and a mean residual current of ~ −2.25 pA, suggesting the translocation of the peptide substrate through the nanopore. By contrast, after trypsin was additionally added to the trans chamber, two types of new events (blue and yellow marked ones in Figs. 1b and 1c) appeared. These new events presented much smaller residence times and amplitudes than those of the peptide substrate (Figs. 1a, 1b, and 1c). Although they themselves had similar residence time (~1.1 ms), the two types of new events could be well separated and differentiated based on their significantly different residual currents (~−5 pA vs. ~ −12 pA). Furthermore, our experiments showed that the frequencies of the new events increased with the recording time and with the increased concentration of added trypsin, a clear indication that they were attributed to the products formed by the trypsin’s cleavage of the β-amyloid (10–20) peptide, i.e., YEVHHQK and LVFF, respectively. It should be noted that, the possibility that these new events were attributed to trypsin was ruled out by the control experiment (Supporting Information, Fig. S-2), in which trypsin itself couldn’t produce translocation events in the nanopore as single-stranded nucleic acids and short peptides did19, 36. This result not only agreed very well with the fact that trypsin has a larger spherical molecular diameter than that of the αHL transmembrane domain (38Å vs. 20 Å) so that it can not enter the protein pore38, 39, but also suggested that trypsin would not be denatured and pulled through the nanopore under our experimental condition, i.e., at a small applied voltage bias of −40 mV.
Figure 1.
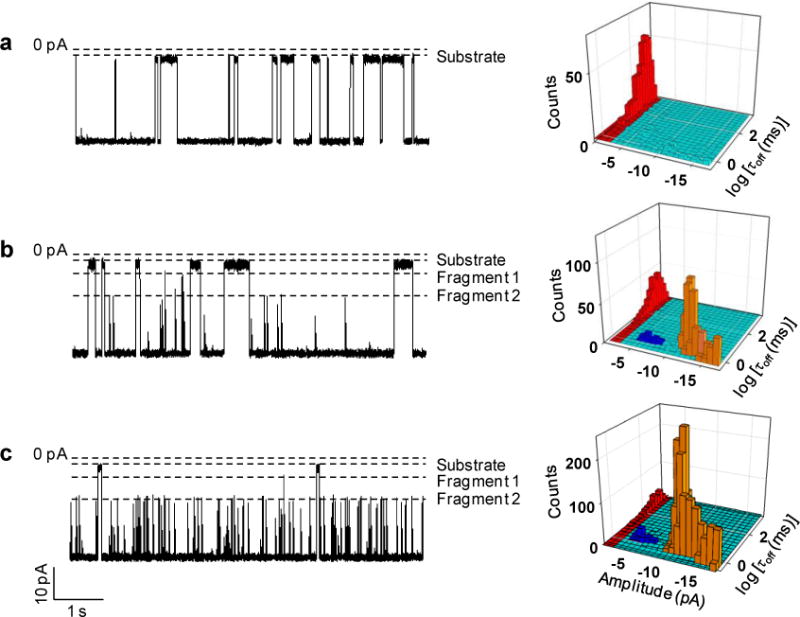
Nanopore measurement of trypsin activity: (a) 0 ng/mL; (b) 10 ng/mL; and (c) 100 ng/mL. (Left) Typical single-channel recording trace segments after 30-min proteolytic cleavage of peptide β-amyloid (10–20). Dashed lines represent the levels of zero current, and residual currents of the substrate (peptide sequence: YEVHHQKLVFF) and its two cleavage products (the sequence of fragment 1: YEVHHQK, and that of fragment 2: LVFF); (Right) the corresponding 3D plots of event count vs. residence time vs. blockage amplitude, showing the significant difference in the signatures of the blockage events attributed to the peptide substrate (red), fragment 1 (blue), and fragment 2 (yellow). The experiments were performed at −40 mV in 1 M NaCl solution buffered with 10 mM Tris (pH 7.5) using the mutant α-hemolysin protein (M113F)7 pore. The concentration of the substrate β-amyloid (10–20) was 5 μM.
Detection limit of the nanopore trypsin sensor
Under the current experimental conditions (i.e., with the α-hemolysin (M113F)7 pore, at an applied potential of −40 mV, in 1M NaCl buffer solution of pH 7.5), a series of experiments was carried out to investigate the proteolytic cleavage of the substrate peptide β-amyloid (10–20) by trypsin, where we kept the concentration of the substrate constant (5 μM), but varied the trypsin concentration (ranging from 2 to 100 ng/mL). Similar to the observation we made in the previous section, in addition to the substrate events, two new types of current modulations were identified in all of the different trypsin concentration situations, providing further evidence that these events were attributed to the substrate degradation products. It should be noted that, after trypsin digestion of the substrate, in principle, the two cleavage products should have identical concentrations. However, due to the length and structure difference in these two peptide fragments, the event frequencies for their translocation in the nanopore might vary greatly, which has been demonstrated in our previous study36. Since the LVFF events were significantly more frequent than the YEVHHQK events, for convenience, only LVFF events were included in the data analysis. It is worth mentioning that, one advantage of obtaining enzyme kinetic and thermodynamic information based on the substrate degradation product signal instead of the substrate signal (which is commonly used by various conventional enzyme assays) is that the substrate cleavage site can be utilized as a useful parameter to improve the selectivity and the accuracy of the protease assay. In the event that other potential interfering proteases and the target trypsin cleave the substrate peptide at different positions, their generated different peptide fragments allow the differentiation between the target analyte from false positives. The plot of the number of LVFF events collected in a 30-min recording versus the trypsin concentration is summarized in Fig. 2. It was found that the detection limit for trypsin (which is defined as the trypsin concentration corresponding to three times the standard deviation of a blank signal) in a 30-minute reaction period was 1.4 ng/mL (corresponding to 59 pM). Such a detection limit is as good as those (ranging from 100 ng/mL to 2 ng/mL) of other various sensitive trypsin detection methods developed thus far5,6,13,14, and more than good enough for analysis of trypsin in clinical samples (note that the trypsin concentration in healthy people’s serum ranges from 140–400 ng/mL).
Figure 2.
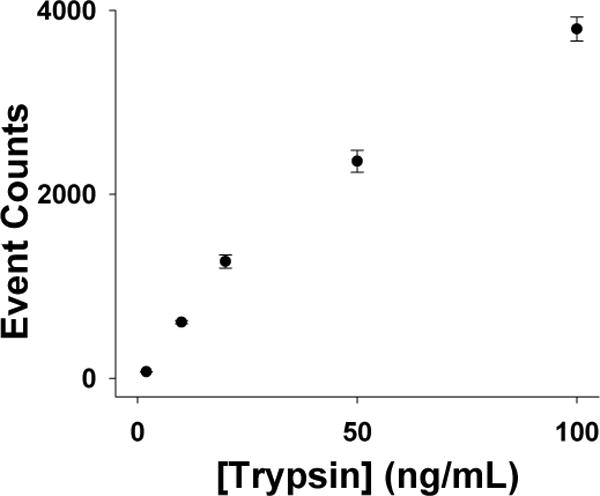
Dose-response curve for trypsin detection. The experiments were performed with the (M113F)7 α-hemolysin pore in a buffer solution comprising 1.0 M NaCl and 10 mM Tris (pH 7.5) at −40 mV in the presence of 5 μM β-amyloid (10–20). The data analysis was performed based on 30 min trace segments.
Effects of metal ions and temperature on the nanopore sensor sensitivity
It has been well-documented that the existence of Ca2+ ions in the solution would benefit the process of proteolytic reaction catalyzed by trypsin40. In fact, trypsin itself is a protein, it is capable of digesting itself. However, this autolysis process (more commonly known as self-digestion) can be prevented by Ca2+ ions (due to their binding to trypsin). In addition, Ca2+ ions can also promote the formation of active trypsin from trypsinogen, the protein produced in its inactive form within the pancrease of humans41–43. To examine whether the existence of Ca2+ ions in the electrolyte solution could improve the nanopore sensor sensitivity for trypsin analysis, the effect of Ca2+ ions on the trypsin cleavage of the β-amyloid (10–20) peptide was further investigated. Our experimental results showed that, compared with the electrolyte solution without Ca2+, the buffer solution containing additional 5 mM Ca2+ ions would result in a (19.5±0.5)% increase in the number of LVFF events (Fig. 3a), confirming that Ca2+ indeed enhanced the sensor sensitivity.
Figure 3.
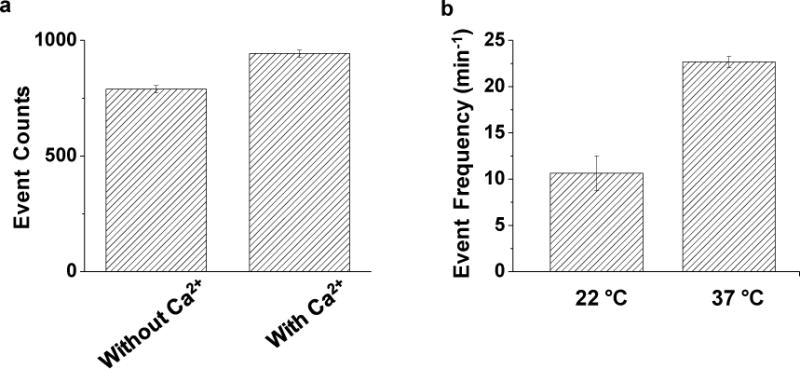
Effects of (a) Ca2+ ions and (b) temperature on the trypsin sensor sensitivity. The experiments were performed using the (M113F)7 α-hemolysin pore in a buffer solution comprising 1.0 M NaCl and 10 mM Tris (pH 7.5) at −40 mV in the absence and presence of 5 mM Ca2+, and at 22 °C or 37 °C, respectively. The concentrations of trypsin shown in Figs. 3a and 3b were 10 ng/mL and 50 ng/mL, respectively. Event counts shown in Fig. 3a were calculated based on the last 20 min trace segments of 30-min single channel recordings.
The influence of temperature on trypsin activity was further investigated since enzymatic activity is temperature dependent. Briefly, 1 μL of 100 μg/mL trypsin and 20 μL 5 mM β-amyloid (10–20) peptide was incubated for 30 min using a water bath at 22 ͦ C and 37 ͦ C, respectively. Then, the mixture was introduced to the trans compartment of the nanopore sensing chamber for single-channel recording. The final concentrations of trypsin and the substrate peptide in the nanopore sensor were 50 ng/mL and 5 μM, respectively. Our experiments (Fig. 3b) showed that the frequency of LVFF events increased by (113±20)% as the temperature of incubation increased from 22 °C to 37 °C.
Measurement of trypsin activity
In order to measure the activity of trypsin by nanopore analysis, a modified version of the universal protease activity assay44 was developed. Briefly, peptide LVFF, one of the cleavage products of the substrate β-amyloid (10–20) peptide, was custom synthesized, and the dose-response curve (event counts vs. peptide concentration) for its translocation in the nanopore was constructed (Supporting information, Fig. S-3). By comparing the number of events of LVFF generated by the trypsin cleavage of β-amyloid (10–20) with this calibration curve, we could obtain the concentration of LVFF in the enzymatic reaction mixture solution. Then, the activity of trypsin could be determined in terms of Units, which is the amount of micromoles of the peptide LVFF fragment released from the substrate per minute. The activities of trypsin at various concentrations were summarized in Table 1. From the table, we could see that, with the increase in the concentration of the trypsin (freshly bought from Sigma), its activity indeed increased. However, the percent increase in trypsin activity was not linearly related to that of trypsin concentration, but gradually decreased as the concentration of trypsin increased. One of the main reasons might be because trypsin cleaved the substrate so rapidly that the concentration of the substrate was low in our trypsin assay experiments, especially at high trypsin concentrations. This interpretation is supported by the time curve of substrate digestion (Supporting Information, Figure S-4), where we found that under our experimental condition (5 μM substrate and 100 ng/mL trypsin), ~70% of the substrate was digested in one hour. In principle, in order to determine the protease activity accurately, a large excess of substrate over enzyme should be used. In addition, to show the difference between trypsin abundance and trypsin activity, the same substrate digestion experiment was performed using 50 ng/mL trypsin, with its stock solution prepared one year ago and stored at −80 °C freezer. Our experimental results (data not shown) showed that the substrate cleavage events were rarely observed, thus confirming that abundance and activity were indeed different concepts.
Table 1.
Determination of trypsin activity using a nanopore single-molecule biosensor
| Trypsin Concentration (ng/mL) | Trypsin Activity (Units) | Relative deviation (%) |
|---|---|---|
| 2 | 9.8 × 10−6 | 1.01 |
| 10 | 8.5 × 10−5 | 2.83 |
| 20 | 1.8 × 10−4 | 5.60 |
| 50 | 3.6 × 10−4 | 13.3 |
| 100 | 5.3 × 10−4 | 3.47 |
The activity of trypsin was determined in terms of Units, which is the amount in micromoles of the LVFF fragment released from trypsin digestion of the peptide substrate β-amyloid (10–20) per minute. The data analysis was performed based on 30 min trace segments.
Trypsin inhibition and simulated serum sample analysis
Proteases are valuable diagnostic or prognostic markers for a variety of diseases and conditions, such as inflammation, cardiovascular diseases and cancer, and are becoming increasingly important targets for drug discovery. For example, inhibitors of well-established protease targets such as HCV protease and HIV protease have been widely used to treat hepatitis C and AIDS. To demonstrate the feasibility of utilizing our developed nanopore sensing platform as a screening tool for protease inhibitors, trypsin cleavage of the β-amyloid (10–20) peptide was further investigated in the presence of the trypsin inhibitor from bovine pancreas (TIBP), a small globular protein with 58 amino acid residues (M. W. = 6.5 kDa)45. Similar to the observation we made in the interaction between trypsin and the α-hemolysin nanopore, due to its large molecular size, low concentrations of TIBP (e.g., 0.125 nM, Supporting Information, Fig. S-2) would rarely produce translocation events with blockage amplitudes similar to those of the substrate degradation products, thus not interfering with our investigation of peptide translocation in the nanopore. In principle, TIBP would bind to trypsin, thus inhibiting trypsin cleavage of β-amyloid (10–20) peptide. Therefore, compared with the situation without trypsin inhibitor, the presence of TIBP in the solution would lead to a decrease in the event frequency of the substrate cleavage products. Our experiments (Fig. 4) showed that, in the absence of TIBP, 2075 LVFF events were observed in a 456-s single-channel recording. In contrast, in the presence of TIBP, we only identified 1030 LVFF events in the same time frame. The results confirmed that TIBP could indeed inhibit trypsin activity.
Figure 4.
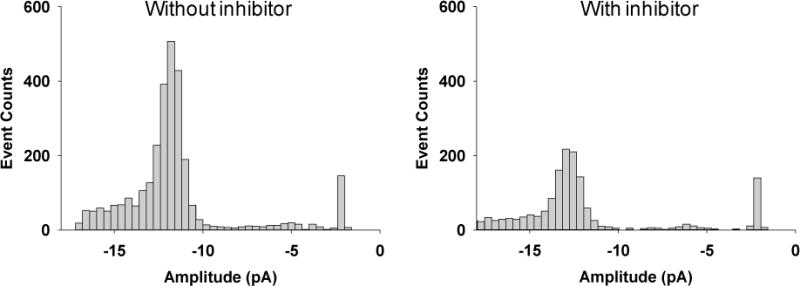
Effect of inhibitor on trypsin digestion of β-amyloid (10–20). The experiments were performed with the (M113F)7 α-hemolysin pore in a buffer solution comprising 1.0 M NaCl and 10 mM Tris (pH 7.5) at −40 mV in the presence of 5 μM β-amyloid (10–20) and 100 ng/mL trypsin. The concentration of the inhibitor was 0.125 nM. Data analysis was performed based on 456-s trace segments after 30-min proteolytic cleavage of β-amyloid (10–20) by trypsin and the trypsin/inhibitor mixture. In the latter case, trypsin and trypsin inhibitor were incubated at 37 °C for 10 min before performing the substrate digestion experiment.
To demonstrate the application of our developed nanopore sensor for potential clinical sample analysis, trypsin was spiked into the bovine serum to create a mock serum sample. It should be noted that, one of the challenges to analyze clinical samples using the protein nanopore sensor is that the amphiphilic proteins in serum samples might disturb the lipid bilayer34 using the vertical design used in this work, although it has been successfully used in other horizontal conducting chambers.24 One feasible approach to overcome this difficulty is to use spin-columns to remove interfering blood serum components46. In our previous work, we found that by incubating mock serum samples with peptide substrates, and then analyzing the mixture solution using the protein nanopore sensor, we could sensitively determine the protease activity in the serum within minutes, thus overcoming the lipid bilayer’s short life-time limitation31. This same approach was utilized to analyze the mock trypsin serum sample in this work. Our experimental results showed that the trypsin-spiked serum sample produced LVFF events with a similar frequency to that of the control sample (i.e., trypsin standard solution) (Supporting Information, Fig. S-5), supporting the feasibility of our developed nanopore sensor for potential clinical applications.
Conclusions
In summary, a new type of nanoscale-sized biosensor was developed to measure the activity of trypsin by monitoring the enzymatic reactions between the substrate and trypsin in an α-hemolysin nanopore. Compared with the time-consuming commercial assay kits, which typically use a two-antibody sandwich immunoassay approach, to detect trypsin, our developed label-free nanopore sensor is real time, and is more suitable for point-of-care applications. In particular, in our sensor design, we take advantage of the substrate cleavage site, and use the signal from the cleavage product instead of the substrate to measure the protease activity, thus achieving improved sensor selectivity and accuracy over the conventional activity-based protease assay. To the best of our knowledge, this is the first time for the nanopore sensing technology to be utilized for the quantitative determination of protease activity. Given the importance of proteases as valuable diagnostic or prognostic markers for a variety of diseases and as important targets for drug discovery, developing sensitive and selective sensors for other proteases, especially those of biological, pharmaceutical, and medical importance (e.g., prostate-specific antigen, beta-secretase, caspases, and so on), is highly desirable. The nanopore sensor design strategy developed in this work would serve as a generic approach for that purpose.
Supplementary Material
Acknowledgments
This work was financially supported by the National Institutes of Health (1R15GM110632).
Footnotes
Supporting Information Available: The following file is available free of charge on the ACS Publications website.
Additional figures, including the schematic illustration of trypsin nanopore sensor, typical trace segments for trypsin and TIBP, LVFF dose-response curve, time curve of substrate digestion, and serum sample analysis.
References
- 1.Stoytcheva M, Zlatev R, Cosnier S, Arredondo M. Square Wave Voltammetric Determination of Trypsin Activity. Electrochim Acta. 2012;76:43–47. [Google Scholar]
- 2.See WA, Smith JL. Urinary Levels of Activated Trypsin in Whole-Organ Pancreas Transplant Patients with Duodenocystostomies. Transplantation. 1991;52:630–633. doi: 10.1097/00007890-199110000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Artigas JMG, Garcia ME, Faure MRA, Gimeno AMB. Serum Trypsin Levels in Acute Pancreatic and Non-Pancreatic Abdominal Conditions. Postgrad Med J. 1981;57:219–222. doi: 10.1136/pgmj.57.666.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braatz JA, Elias C, Finny JG, Tran H, McCaman M. Quantitation of Residual Trypsin in Cell-Based Therapeutics Using Immobilized α-1-Antitrypsin or SBTI in an ELISA Format. J Immunol Methods. 2015;417:131–133. doi: 10.1016/j.jim.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Hayden O, Haderspöck C, Krassnig S, Chen X, Dickert FL. Surface Imprinting Strategies for the Detection of Trypsin. Analyst. 2006;131:1044–1050. doi: 10.1039/b608354b. [DOI] [PubMed] [Google Scholar]
- 6.Miao P, Liu T, Li X, Ning L, Yin J, Han K. Highly Sensitive, Label-Free Colorimetric Assay of Trypsin Using Silver Nanoparticles. Biosens Bioelectron. 2013;49:20–24. doi: 10.1016/j.bios.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 7.Lake-Bakaar G, McKavanagh S, Rubio CE, Epstein O, Summerfield JA. Measurement of Trypsin in Duodenal Juice by Radioimmunoassay. Gut. 1980;21:402–407. doi: 10.1136/gut.21.5.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yi Q, Liu Q, Gao F, Chen Q, Wang G. Application of an Electrochemical Immunosensor with a MWCNT/PDAA Modified Electrode for Detection of Serum Trypsin. Sensors. 2014;14:10203–10212. doi: 10.3390/s140610203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan R, Huang X, Ng SL, Yao SQ. Activity-Based Fingerprinting of Proteases. Chembiochem. 2006;7:32–36. doi: 10.1002/cbic.200500340. [DOI] [PubMed] [Google Scholar]
- 10.Johnston D, Hermans JM, Yellowlees D. Isolation and Characterization of a Trypsin from the Slipper Lobster, Thenus Orientalis (Lund) Arch Biochem Biophys. 1995;324:35–40. doi: 10.1006/abbi.1995.9933. [DOI] [PubMed] [Google Scholar]
- 11.Kersey AD, Berkoff TA, Morey WW. Multiplexed Fiber Bragg Grating Strain-Sensor System with a Fiber Fabry–Perot Wavelength Filter. Opt Lett. 1993;18:1370–1372. doi: 10.1364/ol.18.001370. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Yu D, Zhao Y, Fan A. Turn-on Chemiluminescent Sensing Platform for Label-Free Protease Detection Using Streptavidin-Modified Magnetic Beads. Biosens Bioelectron. 2014;61:45–50. doi: 10.1016/j.bios.2014.04.050. [DOI] [PubMed] [Google Scholar]
- 13.Hu L, Han S, Parveen S, Yuan Y, Zhang L, Xu G. Highly Sensitive Fluorescent Detection of Trypsin Based on BSA-Stabilized Gold Nanoclusters. Biosens Bioelectron. 2012;32:297–299. doi: 10.1016/j.bios.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Ionescu RE, Cosnier S, Marks RS. Protease Amperometric Sensor. Anal Chem. 2006;78:6327–6331. doi: 10.1021/ac060253w. [DOI] [PubMed] [Google Scholar]
- 15.Mereuta L, Asandei A, Seo CH, Park Y, Luchian T. Quantitative Understanding of pH- and Salt-Mediated Conformational Folding of Histidine-Containing, β-Hairpin-Like Peptides, through Single-Molecule Probing with Protein Nanopores. ACS Appl Mater Interfaces. 2014;6:13242–13256. doi: 10.1021/am5031177. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Gu J, Brandin E, Kim YR, Wang Q, Branton D. Probing Single DNA Molecule Transport Using Fabricated Nanopores. Nano Lett. 2004;4:2293–2298. doi: 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majd S, Yusko EC, Billeh YN, Macrae MX, Yang J, Mayer M. Applications of Biological Pores in Nanomedicine, Sensing, and Nanoelectronics. Curr Opin Biotechnol. 2010;21:439–476. doi: 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Movileanu L. Watching Single Proteins Using Engineered Nanopores. Protein Pept Lett. 2014;21:235–246. doi: 10.2174/09298665113209990078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haque F, Li J, Wu H, Liang X, Guo P. Solid-State and Biological Nanopore for Real-Time Sensing of Single Chemical and Sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shasha C, Henley RY, Stoloff DH, Rynearson KD, Hermann T, Wanunu M. Nanopore-Based Conformational Analysis of a Viral RNA Drug Target. ACS Nano. 2014;8:6425–6430. doi: 10.1021/nn501969r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, Wang L, Han Y, Zhou S, Guan X. Nanopore Stochastic Detection: Diversity, Sensitivity, and Beyond. Acc Chem Res. 2013;46:2867–2877. doi: 10.1021/ar400031x. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zheng D, Tan Q, Wang MX, Gu LQ. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat Nanotechnol. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kakish J, Tavassoly O, Lee JS. Rasagiline, a Suicide Inhibitor of Monoamineoxidases, Binds Reversibly to α-Synuclein. ACS Chem Neurosci. 2015;6:347–355. doi: 10.1021/cn5002914. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, Haque F, Rychahou PG, Evers M, Guo P. Engineered Nanopore of phi29 DNA-Packaging Motor for Real-Time Detection of Single Colon Cancer Specific Antibody in Serum. ACS Nano. 2013;7:9814–9822. doi: 10.1021/nn404435v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Y, Zhou S, Wang L, Guan X. Nanopore Back Titration Analysis of Dipicolinic Acid. Electrophoresis. 2014;36:467–470. doi: 10.1002/elps.201400255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu A, Zhao Q, Guan X. Stochastic Nanopore Sensors for the Detection of Terrorist Agents: Current Status and Challenges. Anal Chim Acta. 2010;675:106–115. doi: 10.1016/j.aca.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Steffensen MB, Rotem D, Bayley H. Single-Molecule Analysis of Chirality in a Multicomponent Reaction Network. Nat Chem. 2014;6:603–607. doi: 10.1038/nchem.1949. [DOI] [PubMed] [Google Scholar]
- 28.Kang XF, Cheley S, Guan X, Bayley H. Stochastic Detection of Enantiomers. J Am Chem Soc. 2006;128:10684–10685. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 29.Wang G, Zhao Q, Kang X, Guan X. Probing Mercury (II)-DNA Interactions by Nanopore Stochastic Sensing. J Phys Chem B. 2013;117:4763–4769. doi: 10.1021/jp309541h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Wang L, Han Y, Zhou S, Guan X. Nanopore Detection of Copper Ions Using a Polyhistidine Probe. Biosens Bioelectron. 2014;53:453–458. doi: 10.1016/j.bios.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Han Y, Zhou S, Guan X. Real-Time Label-Free Measurement of HIV-1 Protease Activity by Nanopore Analysis. Biosens Bioelectron. 2014;62:158–162. doi: 10.1016/j.bios.2014.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukwikila M, Howorka S. Electrically Sensing Protease Activity with Nanopores. J Phys Condens Matter. 2010;22:454103. doi: 10.1088/0953-8984/22/45/454103. [DOI] [PubMed] [Google Scholar]
- 33.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of Individual Polynucleotide Molecules Using a Membrane Channel. Proc Nat Acad Sci. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Montana V, Grubišić V, Stout RF, Jr, Parpura V, Gu LQ. Nanopore Sensing of Botulinum Toxin Type B by Discriminating an Enzymatically Cleaved Peptide from a Synaptic Protein Synaptobrevin 2 Derivative. ACS Appl Mater Interfaces. 2015;7:184–192. doi: 10.1021/am5056596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montal M, Mueller P. Formation of Bimolecular Membranes from Lipid Monolayers and a Study of Their Electrical Properties. Proc Nat Acad Sci. 1972;69:3561–3566. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Q, Jayawardhana DA, Wang D, Guan X. Study of Peptide Transport through Engineered Protein Channels. J Phys Chem B. 2009;113:3572–3578. doi: 10.1021/jp809842g. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Q, de Zoysa RS, Wang D, Jayawardhana DA, Guan X. Real-Time Monitoring of Peptide Cleavage Using a Nanopore Probe. J Am Chem Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- 38.Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of Staphylococcal α-Hemolysin, a Heptameric Transmembrane Pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 39.Diaz JF, Balkus KJ., Jr Enzyme Immobilization in MCM-41 Molecular Sieve. J Mol Catal B Enzym. 1996;2:115–126. [Google Scholar]
- 40.Morari D, Stepurina T, Rotari VI. Calcium Ions Make Phytohemagglutinin Resistant to Trypsin Proteolysis. J Agric Food Chem. 2008;56:3764–3771. doi: 10.1021/jf0734222. [DOI] [PubMed] [Google Scholar]
- 41.Sipos T, Merkel JR. Effect of Calcium Ions on the Activity, Heat Stability, and Structure of Trypsin. Biochem. 1970;9:2766–2775. doi: 10.1021/bi00816a003. [DOI] [PubMed] [Google Scholar]
- 42.Singh M, Krikorian AD. Inhibition of Trypsin Activity in vitro by Phytate. J Agric Food Chem. 1982;30:799–800. [Google Scholar]
- 43.Papaleo E, Fantucci P, de Gioia L. Effects of Calcium Binding on Structure and Autolysis Regulation in Trypsins. A Molecular Dynamics Investigation. J Chem Theory Comput. 2005;1:1286–1297. doi: 10.1021/ct050092o. [DOI] [PubMed] [Google Scholar]
- 44.Cupp-Enyard C. Sigma’s Non-specific Protease Activity Assay - Casein as a Substrate. J Vis Exp. 2008;19:e899. doi: 10.3791/899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laskowski M, Jr, Kato I. Protein Inhibitors of Proteinases. Annu Rev Biochem. 1980;49:593–526. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 46.Kukwikila M, Howorka S. Nanopore-Based Electrical and Label-Free Sensing of Enzyme Activity in Blood Serum. Anal Chem. 2015;87:9149–9154. doi: 10.1021/acs.analchem.5b01764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


