Abstract
Rabies is enzootic among dog populations in some parts of Cameroon and the risk of human rabies is thought to be steadily high in these regions. However, the molecular epidemiology of circulating Rabies Virus (RABV) has been hardly considered in Cameroon as well as in most neighboring central African countries. To address this fundamental gap, 76 nucleoprotein (N) gene sequences of dog-derived RABV were obtained from 100 brain specimens sampled in Cameroon from 2010 to 2016. Studied sequences were subjected to molecular and phylogenetic analyses with reference strains retrieved from databases. The 71 studied Africa-1 isolates displayed 93.5–100% nucleotide (nt) and 98.3–100% amino-acid (aa) identities to each other while, the 5 studied Africa-2 isolates shared 99.4–99.7% sequence similarities at nt and aa levels. Maximum Likelihood based phylogenies inferred from nucleotide sequences confirmed all studied RABV isolates as members of the dog-related species 1 of the Lyssavirus genus. Individual isolates could be unambiguously assigned as either the Africa-1 subclade of the Cosmopolitan clade or the Africa 2 clade. The Africa-1 subclade appeared to be more prevalent and diversified. Indeed, 70 studied isolates segregated into 3 distinct circulating variants within Africa-1a lineage while a unique isolate was strikingly related to the Africa-1b lineage known to be prevalent in the neighboring Central African Republic and eastern Africa. Interestingly, all five Africa-2 isolates fell into the group-E lineage even though they appeared to be loosely related to databases available reference RABV; including those previously documented in Cameroon. This study uncovered the co-circulation of several Africa-1 and Africa-2 lineages in the southern regions of Cameroon. Striking phylogenetic outcasts to the geographic differentiation of RABV variants indicated that importation from close regions or neighboring countries apparently contributes to the sustainment of the enzootic cycle of domestic rabies in Cameroon.
Author summary
Rabies has been repeatedly reported among dog populations in Cameroon, especially in Yaounde, its capital city. However, the relative rates and genetic variability of Rabies Virus (RABV) variants circulating among dog populations in Cameroon are still to be documented. This study aimed to estimate the frequency and genetic diversity of RABV isolates originating from rabid dogs in the southern regions of Cameroon from 2010 to 2016. Overall, 76 of the 100 dog-derived RABV isolates sampled in Cameroon from 2010 to 2016 were successfully characterized. Our findings revealed that studied isolates belonged to the dog-related species 1 of the Lyssavirus genus, specifically 70 Africa-1a, 1 Africa-1b and 5 Africa-2 group-E lineages. The general phylogenetic pattern suggested an in-country geographic differentiation of the circulating RABV variants. This apparent geographic differentiation was contradicted by striking outcasts indicating importation from close or distant regions. Overall, this study uncovered the co-circulation of several Africa-1 and Africa-2 lineages in some southern regions of Cameroon, thus providing base-line molecular data that would be of interest for future stages of implementation of the rabies surveillance and control plan that is being setup in Cameroon.
Introduction
Rabies is a neglected lethal neurological disease which has a case-fatality rate of almost 100% [1–3]. It causes an estimated 59,000 human deaths primarily in developing and low-income countries where the disease is endemic in animal populations [1,3]. Bites from rabid domestic dogs account for over 99% of the human rabies cases [3,4], most of which occur in Asia and Africa [5,6]. Post-exposure prophylaxis (PEP) efficiently prevents disease development in humans bitten by rabid animals when administrated immediately. Unfortunately, PEP is often unavailable in all settings or not affordable in many developing countries [3].
Canine rabies has been shown to be endemic in Cameroon and relatively higher frequencies of rabid dogs have been reported in urban settings compared to rural areas [7,8]. In the absence of a multiannual active national surveillance and control strategy, the actual burden of human and animal rabies in Cameroon is likely underestimated as in other African countries [3,9–12]. Some rabies control interventions, such as yearly discount of pet vaccination and irregular radio communication campaigns, are conducted in Cameroon but their actual impact remains unknown [8,9,13].
The etiological agent of rabies, the Rabies Virus (RABV), of the genus Lyssavirus and family Rhabdoviridae, has a single-stranded RNA genome of approximately 12 kb in length and of negative polarity [14]. The RABV genome consists of five genes encoding the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G) and the large protein which is the polymerase (L). These five genes N, P, M, G, and L are separated by intergenic regions of variable lengths [14,15]. Like other RNA viruses, RABV displays high rates of mutation due to the lack of proofreading activity of the L protein [16]. RABV is the only virus among the 16 known Lyssavirus species found worldwide in a wide range of mammalian reservoirs of the orders Chiroptera and Carnivora [17–20]. It has been recently demonstrated that individual gene or complete genome sequences of RABV isolates segregate into two major phylogenetic clusters gathering bat- and dog-derived RABV isolates respectively. Within these clades, isolates fall into several major clades [21].
Bat-derived RABV isolates have been shown to circulate specifically in the New World mainly among bats and, to a lesser extent, in some terrestrial carnivores such as skunks (Mephitis mephitis) and raccoons (Procyon lotor) [22–25]. Conversely, dog-specific RABV isolates have been documented worldwide mainly among domestic dogs, but also among wild-living carnivores comprising foxes and raccoon dogs in Europe [26], foxes in the Middle East [27], raccoon dogs and ferret-badgers in Asia [28–30], skunks, foxes, coyotes and mongooses in the Americas [23,24], African civet and mongooses in Africa [31,32]. Within the divergent dog-specific cluster, six major well-defined clades, respectively assigned as Africa-2, Africa-3, Arctic-related, Asian, Cosmopolitan and Indian clades, have been documented [21,33].
In particular, molecular studies in Africa uncovered the presence of Cosmopolitan, Africa-2, and Africa-3 clades. All these clades comprise classical RABV species that segregate into several subclades and lineages varying by geographic area, virus variability, and reservoir species in Africa [21,33–37]. Two major subclades are defined by field RABV isolates of the Cosmopolitan clade: Africa-1 and Africa-4. Africa-4 has been recently identified in northern Africa [21,34] while Africa-1 subclade has been shown to circulate in the northern, eastern and southern parts of Africa [33,38]. Africa-1a lineage has been suggested to have a very broad distribution across Africa. It is predominant in northern and eastern Africa [38–40] and has also been previously reported in Cameroon, Gabon, Equatorial Guinea, Ghana and Madagascar [38,41,42]. Africa-1b lineage circulates mainly in eastern and southern Africa [38,39,43,44].
Africa-2 lineages are uninterruptedly found across West and Central Africa [35–37,42] and has been shown to co-circulate with the Africa-1 lineages in Nigeria and Central African Republic [36,38,42,44–46]. Although Africa-1 and Africa-2 lineages have been documented in several domestic and wild carnivore species, domestic dogs are virtually the only population essential for maintaining canid variants in some parts of Africa [47]. Conversely, wild canids have been suggested to contribute to the sustainment of canine rabies cycles in specific geographic locations in South Africa, Namibia and Zimbabwe [32,48–50].
The third Africa-3 clade is well adapted to mongooses [31,51,52] and is sustained through an independent epidemiological cycle (distinct from that of dog RABV) within viverrid species in southern Africa [31,32,51,53].
Although RABV has been continuously reported in Cameroon [7,8,54], its molecular epidemiology among dog populations have not yet been documented. Less than five genomic sequences of dog-related Africa-1 and Africa-2 RABV originating from Cameroon have been described in previous studies [21,36,38,44]. Based on very limited sequence data available so far in Cameroon, it was thought that there may be an in-country geographic differentiation of dog-derived RABV; with Africa-1 isolates found in the Center region including the capital city, Yaounde, and Africa-2 isolates detected in the northern part of Cameroon.
The purpose of this study was to provide insight into the frequency, genetic variability and phylogenetic relatedness of the RABV isolates derived from rabid dog in Cameroon. Interestingly, this study uncovered the co-circulation of both Africa-1 and Africa-2 in two southern regions of Cameroon and indicated that they circulate in close proximity between neighboring administrative regions of Cameroon as well as between Cameroon and neighboring countries.
Results
Geographic and temporal distribution of the RABV-positive specimens analyzed
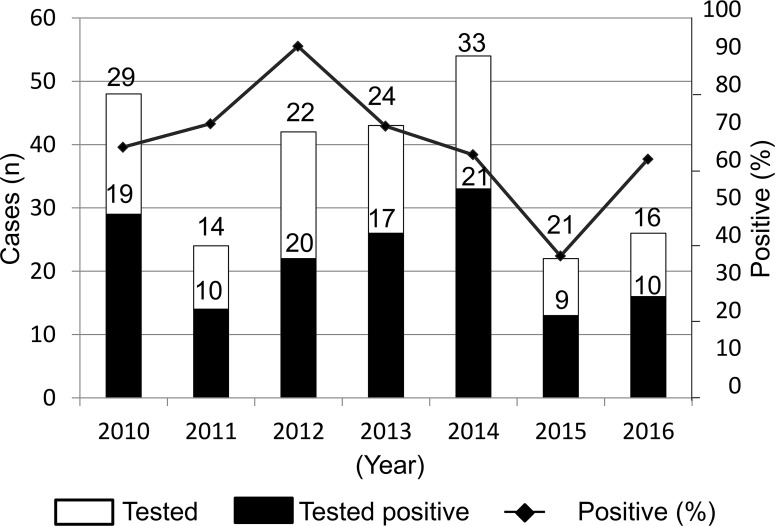
From January 2010 to December 2016, a total of 163 animal specimens were analyzed for rabies diagnosis at CPC, comprising 159 specimens originating from domestic dogs and 4 specimens from other animal species (1 cat, 1 cow, 1 monkey and 1 pig). Overall, 65.4% (104/159) of all dog specimens analyzed were from the Center region amongst which 61.5% (64/104) originated from the capital city, Yaounde where CPC is located. All specimens from cat, cow, monkey and pig were found rabies-negative whereas 66.0% (105/159) of dog specimens were confirmed rabies-positive. The number of dog specimens analyzed from 2010 to 2016 as well as annual rates of positive samples were variable as depicted in Fig 1. Overall, 100 of the 105 rabies positive specimens were available for molecular characterization in this study. They originated from the Center region (68 samples amongst which 46 from Yaounde), East region (1 sample), Littoral region (3 samples), North West region (8 samples), West region (14 samples), South West region (3 samples) and South region (3 samples) (Table 1).
Fig 1. Annual distribution of laboratory confirmed rabies cases among dogs originating from the southern regions of Cameroon, 2010–2016.
Table 1. Summary of the characteristics and genotyping results of the Rabies Virus isolates derived from brain specimens of rabid dogs enrolled in this study.
| Virus Name | Origina | Host species | Source | Phylogenetic clade (subclade)b | Sequence Accession number | ||||
|---|---|---|---|---|---|---|---|---|---|
| District codes | Regions of origin | Districts of origin | |||||||
| 14V-9183 | CEN-BOK | Centre | Bokito | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537529 | ||
| 13V-1970 | CEN-EKO | Centre | Ekoumtik | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537550 | ||
| 14V-1505 | CEN-MAT | Centre | Matomb | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-1507 | CEN-MAT | Centre | Matomb | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-2281 | CEN-MBA | Centre | Mbalmayo | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537549 | ||
| 14V-6062 | CEN-MBA | Centre | Mbalmayo | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537554 | ||
| 14V-7214 | CEN-MBA | Centre | Mbalmayo | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537551 | ||
| 11V-5200 | CEN-MON | Centre | Monatélé | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537541 | ||
| 11V-6266 | CEN-MON | Centre | Monatélé | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537536 | ||
| 11V-18499 | CEN-NTU | Centre | Ntui | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537542 | ||
| 11V-19185 | CEN-NTU | Centre | Ntui | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 12V-2731 | CEN-NTU | Centre | Ntui | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 16V-608 | CEN-NTU | Centre | Ntui | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537519 | ||
| 10V-5902 | CEN-OBA | Centre | Obala | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537511 | ||
| 11V-18800 | CEN-OBA | Centre | Obala | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537520 | ||
| 11V-7563 | CEN-OBA | Centre | Obala | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537509 | ||
| 12V-746 | CEN-OBA | Centre | Obala | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537539 | ||
| 13V-5439 | CEN-OBA | Centre | Obala | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-1546 | CEN-OKO | Centre | Okola | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537544 | ||
| 16V-2918 | CEN-OKO | Centre | Okola | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 12V-1395 | CEN-SAA | Centre | Sa'a | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537558 | ||
| 16V-1728 | CEN-SAA | Centre | Sa'a | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537547 | ||
| 10V-2182 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537538 | ||
| 10V-2240 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537553 | ||
| 10V-2374 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537543 | ||
| 10V-2510 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537540 | ||
| 10V-2713 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537534 | ||
| 10V-3801 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537537 | ||
| 11V-1662 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537513 | ||
| 11V-267 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 12V-1202 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537535 | ||
| 12V-1379 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537559 | ||
| 12V-3708 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537548 | ||
| 12V-3857 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537532 | ||
| 12V-3955 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537552 | ||
| 12V-4917 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 12V-5365 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537524 | ||
| 12V-5642 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537507 | ||
| 12V-5644 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537531 | ||
| 12V-5932 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537557 | ||
| 12V-6225 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537512 | ||
| 12V-6270 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537518 | ||
| 12V-6272 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 12V-6274 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537510 | ||
| 12V-805 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 13V-0091 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537514 | ||
| 13V-1784 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 13V-2178 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537505 | ||
| 13V-3676 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537528 | ||
| 13V-4531 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537533 | ||
| 13V-4610 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537556 | ||
| 13V-5495 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537517 | ||
| 13V-6289 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537515 | ||
| 13V-7152 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537516 | ||
| 13V-7292 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537508 | ||
| 13V-7409 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537525 | ||
| 14V-278 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-4391 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-4642 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537521 | ||
| 14V-5015 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537523 | ||
| 14V-5583 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 15V-3509 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 15V-3966 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537527 | ||
| 15V-5099 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537526 | ||
| 16V-1501 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 16V-2297 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537522 | ||
| 16V-470 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537546 | ||
| 16V-5173 | CEN-YAO | Centre | Yaounde | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537545 | ||
| 14V-4292 | EAS-GAR | East | Garoua Boulai | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1b) | MF537575 | ||
| 15V-1406 | LIT-DIB | Littoral | Dibombari | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-4199 | LIT-DOU | Littoral | Douala | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537555 | ||
| 14V-6269 | LIT-DOU | Littoral | Douala | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-7840 | NOW-BAL | North West | Balikumbat | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537568 | ||
| 12V-4015 | NOW-BAM | North West | Bamenda | Dog (Canis familiaris) | Original brain | Africa-2 (group-E) | MF537580 | ||
| 13V-6144 | NOW-FUN | North West | Fundong | Dog (Canis familiaris) | Original brain | Africa-2 (group-E) | MF537579 | ||
| 10V-3477 | NOW-KUM | North West | Kumbo | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537570 | ||
| 10V-3478 | NOW-KUM | North West | Kumbo | Dog (Canis familiaris) | Original brain | Africa-2 (group-E) | MF537577 | ||
| 14V-1960 | NOW-KUM | North West | Kumbo | Dog (Canis familiaris) | Original brain | Africa-2 (group-E) | MF537576 | ||
| 13V-5717 | NOW-MBE | North West | Mbengui | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 15V-2175 | NOW-BAT | North West | Batibo | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537560 | ||
| 16V-6323 | SOU-AMB | South | AMBAM | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-3979 | SOU-EBO | South | Ebolowa | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 15V-6614 | SOU-EBO | South | Ebolowa | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-6473 | SOW-FON | South West | Fontem | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
| 14V-6432 | SOW-KUM | South West | Kumba | Dog (Canis familiaris) | Original brain | Africa-2 (group-E) | MF537578 | ||
| 14V-6475 | SOW-KUM | South West | Kumba | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537566 | ||
| 12V-007 | WES-BAF | West | Bafoussam | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537506 | ||
| 14V-1636 | WES-BAF | West | Bafoussam | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537565 | ||
| 14V-4880 | WES-BAF | West | Bafoussam | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537567 | ||
| 13V-4215 | WES-BAT | West | Batié | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537530 | ||
| 10V-4586 | WES-DSC | West | Dschang | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537564 | ||
| 11V-18506 | WES-DSC | West | Dschang | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537563 | ||
| 13V-5885 | WES-KEK | West | Kekem | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537561 | ||
| 14V-8641 | WES-KEK | West | Kekem | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537562 | ||
| 10V-2375 | WES-PEN | West | Penka Michel | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537573 | ||
| 10V-2444 | WES-PEN | West | Penka Michel | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537571 | ||
| 10V-3476 | WES-PEN | West | Penka Michel | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537574 | ||
| 10V-3680 | WES-PEN | West | Penka Michel | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537569 | ||
| 10V-3834 | WES-PEN | West | Penka Michel | Dog (Canis familiaris) | Original brain | Cosmopolitan (AF1a) | MF537572 | ||
| 13V-0472 | WES-SAN | West | Santchou | Dog (Canis familiaris) | Original brain | Not applicable | / | ||
a District codes were derived from the respective regions and districts’ names.
b Rabies Virus molecular typing from 24 specimens was not applicable because of failure to efficiently amplify the nucleocapsid coding gene (n = 17) or because of unexploitable sequencing results (n = 7).
Partial sequencing and genetic features of the studied RABV isolates
Of the 100 rabies-positive cases whose brains specimens were analyzed, 76 were confirmed by the molecular analyses performed in this study. The remaining 24 cases without molecular confirmation included 7 cases whose sequences were unexploitable (because of chromatograms with superimposed peaks and/or short reads), 4 cases which showed very weak amplification signals (insufficient for sequencing), and 13 cases which did not amplify. Comparison of the newly determined sequences with homologous sequences obtained from databases identified all 76 RABV isolates as strains of the Lyssavirus species 1 and specifically as Africa-1 or Africa-2 lineages. While 71 isolates were identified as belonging to the Africa-1 lineage displaying 93.5–100% nt and 98.3–100% aa identities to each other, 5 RABVs were closely related to Africa-2 isolates sharing 99.4–99.7% sequence similarities at nt and aa levels. Comparison between the studied and few database available Africa-1 RABV sequences from Cameroon showed 93.3–95.2% nt and 93.6–95.4% aa identities. The same analysis showed less sequence divergence between the studied and reference Africa-2 isolates from Cameroon: 96.4–99.0% nt and 94.5–99.7% aa sequence identities.
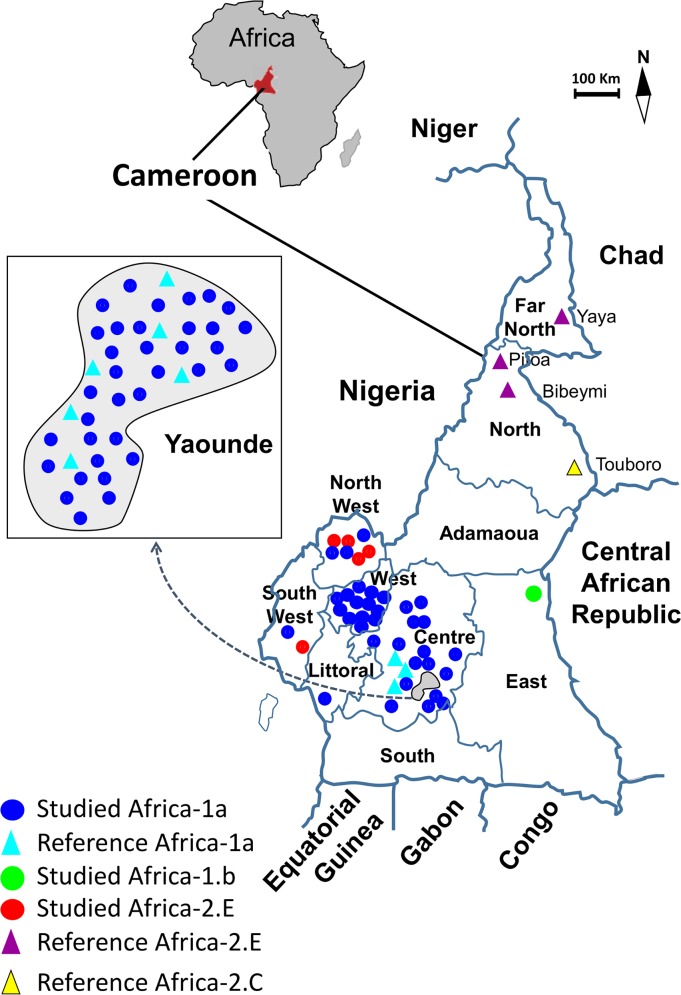
Africa-1 isolates, representing 71 of the 76 sequences obtained, were more prevalent and distributed across the southern regions of Cameroon (5 out of the 7 southern regions) (Fig 2). In contrast, Africa-2 isolates, detected from 2010 to 2014, were geographically restricted to the two neighboring South West and North West regions (Fig 2 and Table 1). This indicates that Africa-1 and Africa-2 RABV co-circulate in some regions of the southern part of Cameroon.
Fig 2. Geographic distribution of the major genetic lineages and variants of the Rabies Virus circulating in Cameroon.
Studied isolates are represented by circles color-coded according to their genetic variants: Africa-1a (blue), Africa-1b (green) and Africa-2 group E (red). Strains previously described in Cameroon are also represented and highlighted on the map, with triangles color-coded according to their genetic variants: Reference Africa-1a (blue), Africa-2 group-C (yellow) and Africa-2 group E (purple). The capital city, Yaounde, has been oversized in order to allow better visibility.
Phylogenetic relatedness of circulating RABV strains
We analyzed the phylogenetic relationships of the nucleoprotein gene sequences (1040 nt) of the studied RABV isolates with database available homologous sequences representing RABV lineages originating from a wide geographic range in Africa (S1 Table). The resulting Maximum Likelihood phylograms confirmed all newly sequenced RABV isolates from Cameroon as dog-related Lyssavirus species 1.
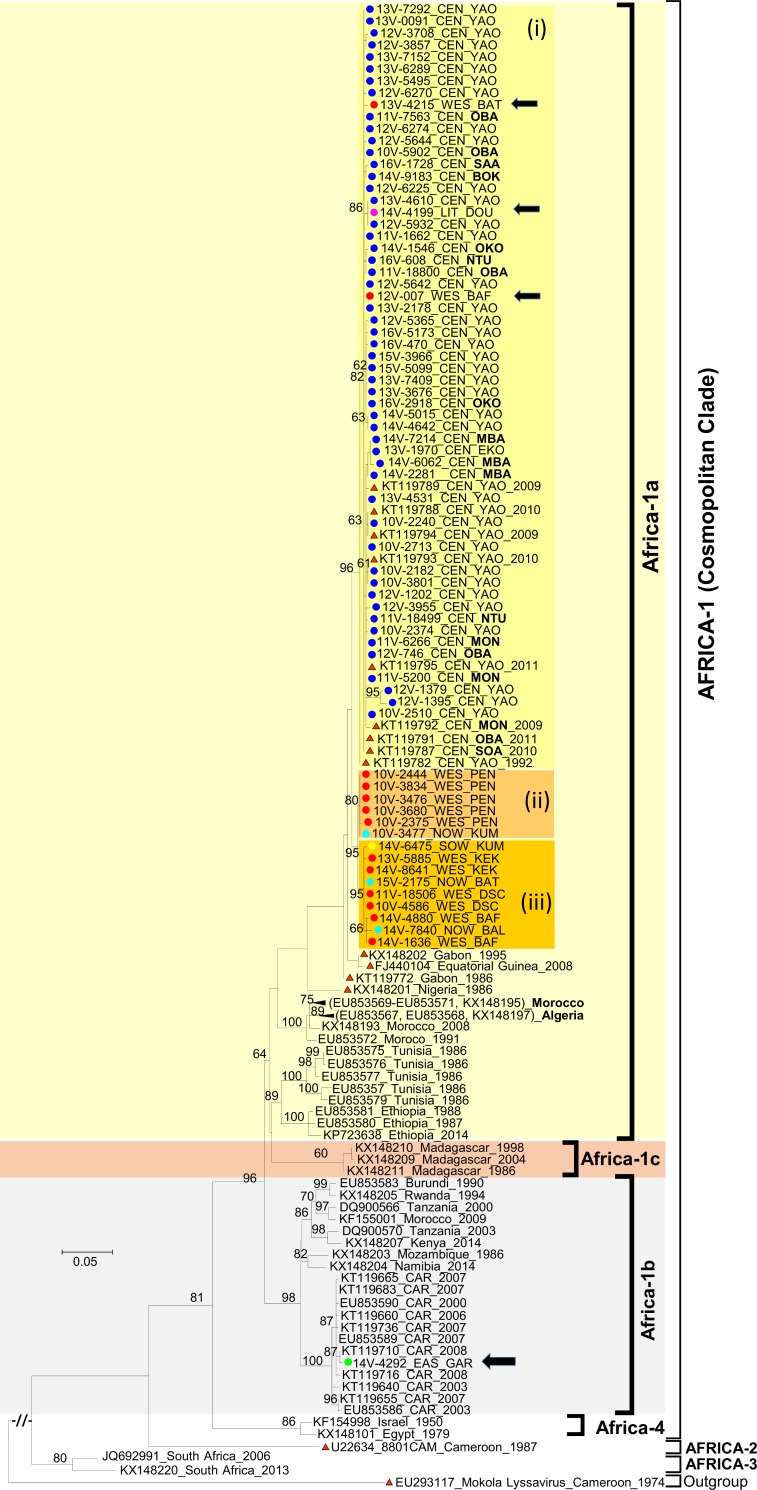
Within the divergent clade of Cosmopolitan RABV, all 71 studied isolates fell in the Africa-1 subclade (Fig 3). None of them grouped with the recently reported Africa-4 subclade defined by RABV isolates from Egypt (in North Africa) and Israel (in Middle East) [34]. Within the Africa-1 subclade, the studied isolates segregated into two distinct lineages with a remarkable association to the geographic origin. One isolate (14V-4292) detected in 2014 in Garoua Boulai (East region) fell within the Africa-1b lineage with previously described isolates from Central African Republic (Fig 3). This isolate displayed 99.9% nt and 100.0% aa sequence identities with its closest match (GenBank N° KT119710) detected in 2008 in the Western part of Central African Republic. Identification of Africa-1b lineage in the East region of Cameroon is consistent with its high prevalence in eastern Africa and in Central African Republic [21,41,44]. This indicates the potential role of intercountry circulation in the sustainment of the enzootic cycle of rabies in Africa.
Fig 3. Maximum-likelihood phylogenetic tree of nucleocapsid gene sequences depicting the phylogenetic relationships of Africa-1 Rabies Viruses originating from Cameroon with other Africa-1 from Africa.
The phylogenetic tree was estimated from the alignment of 1040 nucleotides (nt) long sequence alignment (positions 41–1080 nucleotides according to the genome of the Rabies Virus strain RRV_ON-99-2) using a maximum-likelihood (ML) method under the general time-reversible (GTR) model of nucleotide substitution, with the rate of each substitution type estimated from the dataset using PHYML 3.0 [55]. The ML base frequencies, the proportion of invariable sites (I) and a gamma distribution of rate variation among sites (Γ with four rate categories) were estimated from the dataset. Newly sequenced Rabies Viruses are indicated with circles color-coded according to their respective regions of origin: CEN, Centre; EAS, East; NOW, North West; SOW, South West and WES, West. Their districts of origin are coded as specified by district codes provided in Table 1. The districts of the Centre region, other than Yaounde, are specified in bold black. The years of origin of the studied viruses are provided by two digit numbers preceding the letter “V” in the virus name (10V-, 2010; 11V-, 2011; 12V-, 2012; 13V-, 2013; 14V-, 2014; 15V-, 2015, 16V-; 2016). Database available reference viruses are named with corresponding GenBank accession numbers followed by the country (CAR, Central African Republic) and year of origin, if known. Viruses from Central Africa are specifically highlighted with red-filed triangles; except those from Cameroon that are further distinguished by the indication of the regions (CEN, Centre) and district (OBA, Obala; MON, Monatélé; SAA, Sa’a; YAO, Yaounde) codes. Clades, subclades and lineages are designated as reported in a reference study based on the ML phylogeny of 321 RABV sequences from five concatenated genes [21]. The major clades, lineages and variants of the Rabies Virus commented in the main text are gathered in color-shaded boxes. Viruses displaying peculiar features, commented in the main text, are further highlighted by black arrows. ML bootstrap values (generated from 100 replicates) >60% are shown next to the nodes. Scale is shown at the left as substitutions per site.
The other 70 studied isolates of the Africa-1 subclade grouped in the Africa-1a lineage along with previously described isolates from diverse geographic origin in North Africa (Algeria, Ethiopia, Nigeria, and Morocco) and Central Africa (Cameroon, Gabon and Equatorial Guinea) (Fig 3). Within the Africa-1a lineage, studied isolates fell into three distinguishable groups. The first group, (i), was defined by all viruses originating from Yaounde and other districts of the Centre region, indicating some association between the phylogenetic pattern and the regional origin of the RABV isolates from Cameroon. This first reliable group (bootstrap value of 96%) included sequences of RABV previously documented in the Centre region of Cameroon from 1992 to 2010 (Fig 3). However, the apparent feature of in-country geographic differentiation of the RABV was contradicted by several isolates: 12V-007 and 13V-4215, originating respectively from Bafoussam and Batcham in the West region; and 14V-4199 from Douala in the Littoral region. Indeed, they were strikingly related (99.8 to 100% nt and 100.0% aa identity) to their counterparts originating from the Centre region (Fig 3). The second distinguishable group, (ii), gathered Africa-1a isolates originating from the West and North West regions while third group (iii) was defined by Africa-1a isolates from the West, South West and North West regions (Fig 3). These results indicate that RABV circulate in close proximity between geographically close regions.
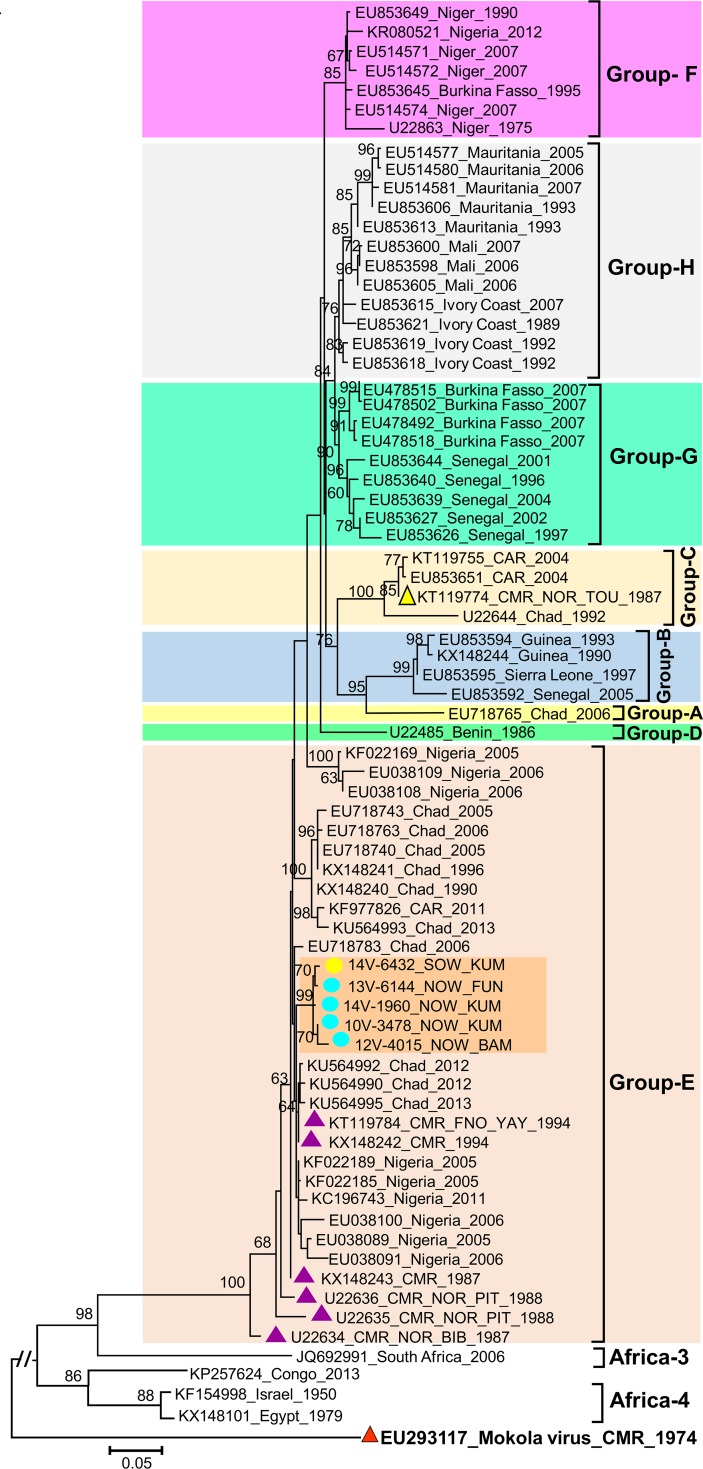
Interestingly all five newly sequenced Africa-2 RABV belonged specifically to the group-E (Fig 4). This Africa-2 group-E also comprised RABV strains previously reported in Cameroon and neighboring countries including Central African Republic, Chad, Niger and Nigeria [36,37]. Within the Africa-2 group-E, the newly described isolates were closely related to each other but were loosely related to databases available isolates, including those originating from the Northern regions of Cameroon from 1987 to 1994 (Fig 4). This study revealed that Africa-2 isolates circulate in two southern regions of Cameroon whereas the most prevalent Africa-1 lineage have been documented in five of the seven southern regions.
Fig 4. Maximum-likelihood phylogenetic tree of nucleocapsid gene sequences depicting the phylogenetic relationships of Africa-2 Rabies Viruses originating from Cameroon with other Africa-2 from Africa.
The phylogenetic tree was estimated from the alignment of 1040 nucleotides (nt) long sequence alignment (positions 41–1080 nucleotides according to the genome of the Rabies Virus strain RRV_ON-99-2) using a maximum-likelihood (ML) method under the K80 model of nucleotide substitution, with the rate of each substitution type estimated from the dataset using PHYML 3.0 [55]. The ML base frequencies, the proportion of invariable sites (I) and a gamma distribution of rate variation among sites (Γ with four rate categories), were estimated from the dataset. Newly sequenced Rabies Virus isolates are indicated with circles color-coded according to their respective regions of origin (NOW, North West and SOW, South West). Their districts of origin, in the North West (BAM; Bamenda; FUN, Fundong; KUM; Kumbo) and South West (KUM, Kumba), are also specified. The years of origin of the studied isolates are provided by two digit numbers preceding the letter “V” in the isolate names (10V-, 2010; 12V-, 2012; 13V-, 2013; 14V-, 2014). Database available reference viruses are named with corresponding GenBank accession numbers followed by the country (CMR, Cameroon; CAR, Central African Republic) and year of origin if known. Those from Cameroon are specifically highlighted with color-coded triangles as follows: yellow for Africa-2 group-C, purple for Africa-2 group-E and red for Mokola virus). Reference viruses from Cameroon are further distinguished by the specification of their respective districts of origin in the North region [(NOR, North): BIB, Bibemy; PIT, Pitoa and TOU, Touboro) and Far North region [(FNO, Far North): YAY, Yaya] if known. Clades, subclades and lineages are designated as reported in a reference study based on the Maximum clade credibility (MCC) of 134 RABV sequences from the nucleocapsid coding genes [36]. The major clades, lineages and variants of the Rabies Virus commented in the main text are gathered in color-shaded boxes. ML bootstrap values (generated from 100 replicates) >60% are shown next to nodes. Scale is shown at bottom as substitutions per site.
Discussion
This study confirms previous reports suggesting continuous circulation of the RABV in Cameroon; especially in the capital city, Yaounde [7,8,54]. In the absence of a multiannual national surveillance and control plan, data on the actual burden of human and animal rabies in Cameroon is certainly underestimated as previously reported in comparable settings in Africa [3,9–12]. This fundamental gap prevents substantial conclusion about the geographical and temporal variation of rabies incidence in Cameroon (Figs 1 and 2).
This study is the first to provide data from elaborate phylogenetic analysis of RABV from Cameroon. Three clades of RABV have been previously documented as being specific to Africa: Cosmopolitan (Africa-1 and Africa-4 subclades), Africa-2 and Africa-3 clades [56]. In this study no isolate of the Africa-3 and Africa-4 was identified. This finding is consistent with the facts that Africa-3 and Africa-4 variants have been shown to be specific to southern [31,51,52] and northern Africa [34], respectively.
Based on the few previous molecular data, it was hypothesized that Africa-2 was exclusively found in the northern part of Cameroon whereas Africa-1 isolates were reported only in the southern regions. This study uncovered RABV of the Africa-2 group-E lineage in two southern regions of Cameroon (North West and South West regions) (Figs 2 and 4). Surprisingly, Africa-2 was the less prevalent lineage in this study whereas it has been shown to be uninterruptedly widespread in western and central Africa, including neighboring countries of Cameroon (Niger, Nigeria, Chad and Central African Republic) [35–37,42,44,45].
This study revealed Africa-1 of the Cosmopolitan clade as the most prevalent RABV lineage circulating in the southern regions of Cameroon (Table 1 and Fig 3). This observation is substantially true for the Centre region of Cameroon where all 51 RABV isolates that could be identified were assigned as Africa-1a lineage. Concerning specifically the North West and South West regions, which share borders with Nigeria, it might be possible to find that Africa-1 and Africa-2 co-circulate with comparable rates if more RABV isolates from these regions were characterized. The Africa-1 sub-clade of the RABV is predominant in the northern, eastern and southern parts of Africa [39,40,57], and have been shown to co-circulate with Africa-2 in Central African Republic [44,45,58] and Nigeria [35,36]. Interestingly, this study provides substantial evidence of co-circulation of Africa-1 and Africa-2 isolates of RABV in at least two regions of the southern part of Cameroon.
In accordance with previous reports, individual RABV sequences fell into a variety of groups, in association with the geographic origin. There was an apparent in-country geographic differentiation of the RABV, however few odds were observed. Similar findings suggesting region-specific variants of the RABV have been documented in some African countries [40,44], thus confirming genomic sequence relatedness as useful marker of intra- and inter-countries RABV dissemination among domestic dogs’ populations. An outstanding application of that marker is provided by the recent finding by Bourhy et al., suggesting that the maintenance of the enzootic cycle of rabies at local geographic level in Bangui is more likely driven by human-mediated waves of spread rather than by continuous dispersion in a relatively large and homogenous dogs’ population [45].
A limitation to this study was the fact that no specimen originated from the three northern regions of Cameroon (Fig 2). Furthermore, restricted geographic range covered by the study was associated to the fact that only few specimens originated from the East, Littoral and South regions while as high as 68.0% (68/100) of all specimens were from Yaounde and its neighborhoods (Fig 2). These shortcomings prevent final conclusion to be drawn on the relative rates and genetic diversity of RABV variants co-circulating in Cameroon. In particular, it remains unknown whether the apparently most prevalent Africa-1 lineage of the Cosmopolitan clade circulates in the northern regions of Cameroon (Adamoua, North and Far North regions) (Fig 2). However, our findings suggest that the Cosmopolitan subclade, Africa-1, circulate extensively in the Centre region of Cameroon, and in Yaounde in particular (Fig 2). Meanwhile the Africa-1a lineage was remarkably more frequent, Africa-1b was represented by only one isolate (Fig 3). Given the relatively high rate of Africa-1b RABV reported in the neighboring Central African Republic [44,45,58], it could be hypothesized that the unique Africa-1b identified in this study was introduced from Central African Republic. Accordingly, it has been recently suggested that RABV of the Africa-1 and Africa-2 variants circulate along the trunk roads between Cameroon and the city of Bangui. However, it is not possible to rule out the direction of RABV dissemination across the border between Cameroon and Central African Republic without extensive sampling in both sides.
In contract to the unique Africa-1b isolate, that was strikingly related to their counterparts from Central Africa, Africa-2 viruses were reliably separate from Africa-2 group C previously found along the trunk roads linking the Bangui city to Cameroon [44]. Africa-2 isolates from this study fell within Africa-2 group E and were only loosely related to their counterparts previously reported in the northern part of Cameroon as well as in neighboring countries (Figs 2 and 4). No Africa-2 group-C was found in this study despite the fact that this lineage has been reported in the northern part of Cameroon; that was not covered by this study.
Although it is tempting to explain away failure to efficiently amplify some RABV isolates by low RABV load in 17 studied rabies-positive specimens, the presence of potentially divergent Lyssaviruses among domestic dogs cannot be wiped out. Remarkably, none of the studied isolates displayed close phylogenetic relationships either with the divergent shrew-derived Mokola Lyssavirus strain 86100CAM (GenBank N° EU293117) from Cameroon, or with the Africa-4 and Africa-3 clades which are specific to northern [34] and southern [32,43,52] Africa, respectively. One explanation for the failure to efficiently amplify potential divergent variants of the RABV in this study may be that they were so divergent from the more prevalent variants that they could be refractory to amplification with generic primers used in this study. More powerful experimental approaches (including the use of divergent primers’ systems, virus propagation in cell cultures or high throughput sequencing) will be helpful for the complete assessment of the genetic landscape of Lyssaviruses in the studied specimens’ collection. Whole genome sequencing will also help to differentiate genetically-related isolates; thus providing more insights into the spatial dynamics of RABV epidemics.
This study is the first to tackle the molecular epidemiology of RABV isolates in Cameroon. We uncovered the presence of diverse lineages and variants of RABV co-circulating among dog populations in Cameroon. Striking phylogenetic evidence of outcasts to the apparent in-country geographic differentiation of RABV variants provided further support to the idea that the movements of rabid animals may be involved in the spread of dog rabies at least in urban areas. Molecular data reported here constitute potential baseline that would be interesting for the design, optimization and evaluation of rabies surveillance, prevention and control during the future stages of rabies elimination in Central Africa.
Methods
Ethical considerations
Animal specimens were collected with the approval of the Cameroon Ministry of Livestock, Fisheries and Animal Industries within the framework of routine rabies surveillance in Cameroon. All studied brain specimens originated from naturally infected rabid dogs enrolled by the Cameroon’s government veterinary services. No specimens were obtained from an experimental procedure nor animals used for experimental purposes.
Study area
The Republic of Cameroon is a Central African country (Fig 2); sharing borders towards the east with Nigeria, towards the west with Central African Republic, towards the north with Chad and Niger, and towards the south with Equatorial Guinea, Gabon and Congo. Cameroon is characterized by diverse ecosystems that are correlated, and thus attributed to, the patterns of rainfall and geological topology. The highlands of the West and North West regions define an area rich in volcanic lands having an average altitude of ≥ 1,100 meters. The southern rain forest of Cameroon is located in the maritime and equatorial zones (Center, East, Littoral, South and South-West regions) while its northern regions (Adamaoua, North and Far North) are progressively dominated by the savannah and steppe. There is no geographical features that may represent barriers to rabies spread in Cameroon. According to the 2010 estimates, the population of the Cameroon is at 19,406,100 people with 10,091,172 and 9,314,928 inhabitants in urban and rural settings, respectively [59]. In particular, the Center region has approximately 2,638,648 inhabitants in urban settings as compared to 887,016 people in rural settings.
Specimens
This was a retrospective and transversal study based on the biological collection of brain specimens collected from rabid domestic dogs. Originally, heads of domestic dogs suspected of rabies were obtained from both private and public veterinary services. Domestic dogs were suspected of rabies if they displayed at least two of the following signs and symptoms: unprovoked aggression, foaming at the mouth, paralysis, incoordination, hoarse bark, hydrophobia, weakness, seizures, or loss of appetite. Brain specimens were collected during necropsy performed on dogs’ heads submitted for rabies diagnosis at the Centre Pasteur du Cameroun (CPC) located in the capital city, Yaounde. Laboratory confirmation of rabies at CPC was based on the detection of RABV nucleocapsid antigen in brain specimens by direct Fluorescent Antibody Test (dFAT) using rabbit IgG antibodies (Bio-Rad, Marnes-la-Coquette, France) [8]. Brain specimens negative for FAT were further confirmed by virus isolation on Murina neuroblastoma cell cultures as previously described [60]. After rabies diagnosis, remaining brain samples were kept frozen at -80°C. A total of 100 specimens derived from 100 rabid dogs were available and were thus considered in this study. All studied specimens originated from the 7 southern regions of Cameroon from 2010 to 2016 (Table 1 and Fig 2).
RNA extraction and amplification of RABV nucleocapsid gene
Each brain specimen was crushed in PBS (10% weight/volume) and 250 μL of supernatant resulting from clarified brain suspension was subjected to RNA extraction using TRIzol LS (Invitrogen, Paris, France), as recommended by the manufacturer's instructions. RNA samples were stored at -80°C prior to analysis.
Complementary DNA (cDNA) synthesis was performed in a final volume of 20 μL using pd(N)7 random primers and AMV reverse transcriptase (Promega). Briefly, 7 μL of purified RNA was incubated at 65°C for 10 minutes with 2 μL of RNase- and DNase-free water, 100 ng of random primers (1 μL) and 10 nmol. of each deoxynucleotide triphosphate (1 μL). Reaction tubes were then transferred on ice for at least 2 minutes and completed with 3 μL of RNase- and DNase-free water, 4 μL of AMV 5X Buffer, 40 U of RNAsin (1 μL) and 10 U of AMV reverse transcriptase (1 μL). Resulting 20 μL reaction mixtures were incubated 10 min at 25°C, 90 min at 42°C and 5 min at 95°C.
A 1485-base pairs DNA fragment encompassing the entire 1353 nucleotides (nt) of the N gene of RABV was amplified by reverse transcription-nested polymerase chain reaction (RT-nPCR) using previously described consensus oligonucleotide primers. First round PCR was performed with the primers pair RHN1 (5’-ACAGACAGCGTCAATTGCAAAGC-3’, nucleotides (nt) 28–52) and N8m (5’-CAGTCTCYTCNGCCATCTC-3’; nt 1584–1568) [21,26,61] in a final volume of 50 μL containing: 5 μL of cDNA, 34.5 μL of RNase- and DNase-free water, 5 μL of 10X PCR Buffer, 200 μM of each dNTP, 2 mM of MgCl2, 25 pmol of each primers and 2.5U of Taq DNA polymerase (Invitrogen, Cergy-Pontoise, France). The thermocycler profile was as follows: 5 min at 95°C followed by 35 cycles of 30 s at 95°C, 30 s at 56°C and 2 min at 72°C, and a final elongation at 72°C for 10 min. Second round PCR was carried out from 2 μL of the PCR product using the primers pair N127 (5’-ATGTAACACCTCTACAATGG-3’, nt 55–74) and 304 (5’-GAGTCACTCGAATATGTC-3’; nt 1539–1516) [21,62] under the same experimental conditions.
PCR products were analyzed by migration on Gelgreen-stained agarose gels and reveled on an ultraviolet transilluminator.
Sequencing and sequences analyses
Amplicons were purified using the QIAquick PCR Purification kit (Qiagen, Courtaboeuf, France) following the manufacturer’s protocol. Purified amplicons were subjected to direct double strands sequencing using nested PCR primers, the BigDye terminator v3.1 kit (Applied Biosystems) and the ABI Prism 3140 automated sequencer (Applied Biosystems).
Consensus sequence editing, multiple sequences alignments and pairwise sequence comparisons were carried out with the CLC Main Workbench 5.7.2 software (CLC bio, Aarhus, Denmark).
GenBank accession numbers for the nucleocapsid gene sequences of RABV determined in this study have been assigned as MF537505 to MF537580.
Phylogenetic analyses
To determine the phylogenetic relatedness of the newly sequenced RABV isolates, sequences were originally aligned with all relevant reference sequences available from online databases and originating from Cameroon and neighboring countries as well as representative sequences from other parts of Africa. Based on initial trees obtained, the alignment was downsized by removing duplicate sequences and by splitting the alignment into two datasets. We used Smart Model Selection [63] to determine the best-fit model of nucleotide substitution based on the Bayesian Information Criterion. This revealed that the General Time Reversible model with proportion of invariable sites plus gamma-distributed rate heterogeneity (GTR+I+Γ4) was optimal for the Africa 1 related dataset while the K80 model with proportion of invariable sites plus gamma-distributed rate heterogeneity (K80+I+Γ4) was the most suitable for the Africa 2 related dataset. Phylogenetic trees using individual datasets were then estimated by the maximum likelihood (ML) method available in PhyML 3.0 [55] using SPR branch-swapping. The reliability of individual nodes on the phylogenetic trees was estimated using 1,000 bootstrap pseudoreplicates.
Supporting information
(PDF)
Acknowledgments
We thank all field veterinarians/technicians for their valuable contributions to specimens’ collection and shipment. We also thank Mrs Gwladys Monamele for her assistance related to English language.
Data Availability
The nucleocapsid gene sequences of RABV determined in this study are available from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MF537505 to MF537580.
Funding Statement
This study was co-funded by the Centre Pasteur du Cameroun and the Cameroon’s Ministry of Livestock, Fisheries and Animal Industries. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fooks AR, Banyard AC, Horton DL, Johnson N, McElhinney LM, et al. (2014) Current status of rabies and prospects for elimination. Lancet 384: 1389–1399. doi: 10.1016/S0140-6736(13)62707-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hemachudha T, Ugolini G, Wacharapluesadee S, Sungkarat W, Shuangshoti S, et al. (2013) Human rabies: neuropathogenesis, diagnosis, and management. Lancet Neurol 12: 498–513. doi: 10.1016/S1474-4422(13)70038-3 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (2013) World Health Organization expert consultation on rabies. World Health Organization Technical report series 931: 1–88. [PubMed] [Google Scholar]
- 4.Hampson K, Coudeville L, Lembo T, Sambo M, Kieffer A, et al. (2015) Estimating the global burden of endemic canine rabies. PLoS Negl Trop Dis 9: e0003709 doi: 10.1371/journal.pntd.0003709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knobel DL, Cleaveland S, Coleman PG, Fevre EM, Meltzer MI, et al. (2005) Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ 83: 360–368. doi: /S0042-96862005000500012 [PMC free article] [PubMed] [Google Scholar]
- 6.Shwiff S, Hampson K, Anderson A (2013) Potential economic benefits of eliminating canine rabies. Antiviral Res 98: 352–356. doi: 10.1016/j.antiviral.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awah NJ, Tchoumboue J, Tong JC (2002) Canine and Human Rabies in Cameroon. Trop Vet 20: 162–168. [Google Scholar]
- 8.Sadeuh-Mba SA, Besong L, Demanou M, Loul S, Nchare A, et al. (2014) Laboratory data of dog rabies in southern Cameroon from 2010 to 2013. BMC Res Notes 7: 905 doi: 10.1186/1756-0500-7-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodet B (2009) The fight against rabies in Africa: From recognition to action. Vaccine 27: 5027–5032. doi: 10.1016/j.vaccine.2009.06.030 [DOI] [PubMed] [Google Scholar]
- 10.Dodet B, Adjogoua EV, Aguemon AR, Amadou OH, Atipo AL, et al. (2008) Fighting rabies in Africa: the Africa Rabies Expert Bureau (AfroREB). Vaccine 26: 6295–6298. doi: 10.1016/j.vaccine.2008.04.087 [DOI] [PubMed] [Google Scholar]
- 11.Dodet B, Tejiokem MC, Aguemon AR, Bourhy H (2015) Human rabies deaths in Africa: breaking the cycle of indifference. Int Health 7: 4–6. doi: 10.1093/inthealth/ihu071 [DOI] [PubMed] [Google Scholar]
- 12.Nel LH (2013) Discrepancies in data reporting for rabies, Africa. Emerg Infect Dis 19: 529–533. doi: 10.3201/eid1904.120185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ajoke E, Solomon A, Ikhide E (2014) The role of dog trading and slaughter for meat in rabies epidemiology with special reference to Nigeria—a review. J Exp Biol Agric Sci 2: 130–136. [Google Scholar]
- 14.Ribadeau-Dumas F, Dacheux L, Bourhy H (2013) [Rabies]. Med Sci (Paris) 29: 47–55. [DOI] [PubMed] [Google Scholar]
- 15.Wunner WH, Larson JK, Dietzschold B, Smith CL (1988) The molecular biology of rabies viruses. Rev Infect Dis 10 Suppl 4: S771–784. [DOI] [PubMed] [Google Scholar]
- 16.Voloch CM, Capellao RT, Mello B, Schrago CG (2014) Analysis of adaptive evolution in Lyssavirus genomes reveals pervasive diversifying selection during species diversification. Viruses 6: 4465–4478. doi: 10.3390/v6114465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayman DT, Fooks AR, Marston DA, Garcia RJ (2016) The Global Phylogeography of Lyssaviruses—Challenging the 'Out of Africa' Hypothesis. PLoS Negl Trop Dis 10: e0005266 doi: 10.1371/journal.pntd.0005266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nel LH, Markotter W (2007) Lyssaviruses. Crit Rev Microbiol 33: 301–324. doi: 10.1080/10408410701647602 [DOI] [PubMed] [Google Scholar]
- 19.Nel LH, Rupprecht CE (2007) Emergence of lyssaviruses in the Old World: the case of Africa. Curr Top Microbiol Immunol 315: 161–193. [DOI] [PubMed] [Google Scholar]
- 20.Gunawardena PS, Marston DA, Ellis RJ, Wise EL, Karawita AC, et al. (2016) Lyssavirus in Indian Flying Foxes, Sri Lanka. Emerg Infect Dis 22: 1456–1459. doi: 10.3201/eid2208.151986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troupin C, Dacheux L, Tanguy M, Sabeta C, Blanc H, et al. (2016) Large-Scale Phylogenomic Analysis Reveals the Complex Evolutionary History of Rabies Virus in Multiple Carnivore Hosts. PLoS Pathog 12: e1006041 doi: 10.1371/journal.ppat.1006041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA (2007) A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc Natl Acad Sci U S A 104: 7993–7998. doi: 10.1073/pnas.0700741104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis R, Nadin-Davis SA, Moore M, Hanlon C (2013) Genetic characterization and phylogenetic analysis of skunk-associated rabies viruses in North America with special emphasis on the central plains. Virus Res 174: 27–36. doi: 10.1016/j.virusres.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 24.Kuzmin IV, Shi M, Orciari LA, Yager PA, Velasco-Villa A, et al. (2012) Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001–2009. PLoS Pathog 8: e1002786 doi: 10.1371/journal.ppat.1002786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzmina NA, Lemey P, Kuzmin IV, Mayes BC, Ellison JA, et al. (2013) The phylogeography and spatiotemporal spread of south-central skunk rabies virus. PLoS One 8: e82348 doi: 10.1371/journal.pone.0082348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourhy H, Kissi B, Audry L, Smreczak M, Sadkowska-Todys M, et al. (1999) Ecology and evolution of rabies virus in Europe. J Gen Virol 80 (Pt 10): 2545–2557. [DOI] [PubMed] [Google Scholar]
- 27.Seimenis A (2008) The rabies situation in the Middle East. Dev Biol 131: 43–53. [PubMed] [Google Scholar]
- 28.Oem JK, Kim SH, Kim YH, Lee MH, Lee KK (2013) Complete genome sequences of three rabies viruses isolated from rabid raccoon dogs and a cow in Korea. Virus Genes 47: 563–568. doi: 10.1007/s11262-013-0923-1 [DOI] [PubMed] [Google Scholar]
- 29.Tsai KJ, Hsu WC, Chuang WC, Chang JC, Tu YC, et al. (2016) Emergence of a sylvatic enzootic formosan ferret badger-associated rabies in Taiwan and the geographical separation of two phylogenetic groups of rabies viruses. Vet Microbiol 182: 28–34. doi: 10.1016/j.vetmic.2015.10.030 [DOI] [PubMed] [Google Scholar]
- 30.Zhao J, Liu Y, Zhang S, Zhang F, Wang Y, et al. (2014) Molecular characterization of three ferret badger (Melogale moschata) rabies virus isolates from Jiangxi province, China. Arch Virol 159: 2059–2067. doi: 10.1007/s00705-014-2044-0 [DOI] [PubMed] [Google Scholar]
- 31.Nel LH, Sabeta CT, von Teichman B, Jaftha JB, Rupprecht CE, et al. (2005) Mongoose rabies in southern Africa: a re-evaluation based on molecular epidemiology. Virus Res 109: 165–173. doi: 10.1016/j.virusres.2004.12.003 [DOI] [PubMed] [Google Scholar]
- 32.Sabeta CT, Shumba W, Mohale DK, Miyen JM, Wandeler AI, et al. (2008) Mongoose rabies and the African civet in Zimbabwe. Vet Rec 163: 580 [DOI] [PubMed] [Google Scholar]
- 33.Bourhy H, Reynes JM, Dunham EJ, Dacheux L, Larrous F, et al. (2008) The origin and phylogeography of dog rabies virus. J Gen Virol 89: 2673–2681. doi: 10.1099/vir.0.2008/003913-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David D, Hughes GJ, Yakobson BA, Davidson I, Un H, et al. (2007) Identification of novel canine rabies virus clades in the Middle East and North Africa. J Gen Virol 88: 967–980. doi: 10.1099/vir.0.82352-0 [DOI] [PubMed] [Google Scholar]
- 35.De Benedictis P, Sow A, Fusaro A, Veggiato C, Talbi C, et al. (2010) Phylogenetic analysis of rabies viruses from Burkina Faso, 2007. Zoonoses Public Health 57: e42–46. doi: 10.1111/j.1863-2378.2009.01291.x [DOI] [PubMed] [Google Scholar]
- 36.Talbi C, Holmes EC, de Benedictis P, Faye O, Nakoune E, et al. (2009) Evolutionary history and dynamics of dog rabies virus in western and central Africa. J Gen Virol 90: 783–791. doi: 10.1099/vir.0.007765-0 [DOI] [PubMed] [Google Scholar]
- 37.Traore A, Picard-Meyer E, Mauti S, Biarnais M, Balmer O, et al. (2016) Molecular Characterization of Canine Rabies Virus, Mali, 2006–2013. Emerg Infect Dis 22: 866–870. doi: 10.3201/eid2205.150470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kissi B, Tordo N, Bourhy H (1995) Genetic polymorphism in the rabies virus nucleoprotein gene. Virology 209: 526–537. doi: 10.1006/viro.1995.1285 [DOI] [PubMed] [Google Scholar]
- 39.Brunker K, Marston DA, Horton DL, Cleaveland S, Fooks AR, et al. (2015) Elucidating the phylodynamics of endemic rabies virus in eastern Africa using whole-genome sequencing. Virus Evol 1: vev011 doi: 10.1093/ve/vev011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amouri IK, Kharmachi H, Djebbi A, Saadi M, Hogga N, et al. (2011) Molecular characterization of rabies virus isolated from dogs in Tunisia: evidence of two phylogenetic variants. Virus Res 158: 246–250. doi: 10.1016/j.virusres.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 41.Andriamandimby SF, Heraud JM, Ramiandrasoa R, Ratsitorahina M, Rasambainarivo JH, et al. (2013) Surveillance and control of rabies in La Reunion, Mayotte, and Madagascar. Vet Res 44: 77 doi: 10.1186/1297-9716-44-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayman DT, Johnson N, Horton DL, Hedge J, Wakeley PR, et al. (2011) Evolutionary history of rabies in Ghana. PLoS Negl Trop Dis 5: e1001 doi: 10.1371/journal.pntd.0001001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mollentze N, Weyer J, Markotter W, le Roux K, Nel LH (2013) Dog rabies in southern Africa: regional surveillance and phylogeographical analyses are an important component of control and elimination strategies. Virus Genes 47: 569–573. doi: 10.1007/s11262-013-0974-3 [DOI] [PubMed] [Google Scholar]
- 44.Tricou V, Bouscaillou J, Kamba Mebourou E, Koyanongo FD, Nakoune E, et al. (2016) Surveillance of Canine Rabies in the Central African Republic: Impact on Human Health and Molecular Epidemiology. PLoS Negl Trop Dis 10: e0004433 doi: 10.1371/journal.pntd.0004433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bourhy H, Nakoune E, Hall M, Nouvellet P, Lepelletier A, et al. (2016) Revealing the Micro-scale Signature of Endemic Zoonotic Disease Transmission in an African Urban Setting. PLoS Pathog 12: e1005525 doi: 10.1371/journal.ppat.1005525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tricou V, Berthet N, Nakoune E, Kazanji M (2014) Complete genome sequence of a rabies virus isolated from a human in central african republic. Genome Announc 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lembo T, Haydon DT, Velasco-Villa A, Rupprecht CE, Packer C, et al. (2007) Molecular epidemiology identifies only a single rabies virus variant circulating in complex carnivore communities of the Serengeti. Proc Biol Sci 274: 2123–2130. doi: 10.1098/rspb.2007.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nel L, Jacobs J, Jaftha J, Meredith C (1997) Natural spillover of a distinctly Canidae-associated biotype of rabies virus into an expanded wildlife host range in southern Africa. Virus Genes 15: 79–82. [DOI] [PubMed] [Google Scholar]
- 49.Pfukenyi DM, Pawandiwa D, Makaya PV, Ushewokunze-Obatolu U (2009) A retrospective study of wildlife rabies in Zimbabwe, between 1992 and 2003. Trop Anim Health Prod 41: 565–572. doi: 10.1007/s11250-008-9224-4 [DOI] [PubMed] [Google Scholar]
- 50.Bellan SE, Cizauskas CA, Miyen J, Ebersohn K, Kusters M, et al. (2012) Black-backed jackal exposure to rabies virus, canine distemper virus, and Bacillus anthracis in Etosha National Park, Namibia. J Wildl Dis 48: 371–381. doi: 10.7589/0090-3558-48.2.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis PL, Rambaut A, Bourhy H, Holmes EC (2007) The evolutionary dynamics of canid and mongoose rabies virus in Southern Africa. Arch Virol 152: 1251–1258. doi: 10.1007/s00705-007-0962-9 [DOI] [PubMed] [Google Scholar]
- 52.Van Zyl N, Markotter W, Nel LH (2010) Evolutionary history of African mongoose rabies. Virus Res 150: 93–102. doi: 10.1016/j.virusres.2010.02.018 [DOI] [PubMed] [Google Scholar]
- 53.Johnson N, Letshwenyo M, Baipoledi EK, Thobokwe G, Fooks AR (2004) Molecular epidemiology of rabies in Botswana: a comparison between antibody typing and nucleotide sequence phylogeny. Vet Microbiol 101: 31–38. doi: 10.1016/j.vetmic.2004.03.007 [DOI] [PubMed] [Google Scholar]
- 54.Ngangnou A (2011) Country report 2011: Cameroon. Rabies diagnostics and surveillance (SEARG).
- 55.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. doi: 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 56.Troupin A, Shirley D, Londono-Renteria B, Watson AM, McHale C, et al. (2016) A Role for Human Skin Mast Cells in Dengue Virus Infection and Systemic Spread. J Immunol 197: 4382–4391. doi: 10.4049/jimmunol.1600846 [DOI] [PubMed] [Google Scholar]
- 57.Johnson N, McElhinney LM, Ali YH, Saeed IK, Fooks AR (2004) Molecular epidemiology of canid rabies in Sudan: evidence for a common origin of rabies with Ethiopia. Virus Res 104: 201–205. doi: 10.1016/j.virusres.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 58.Nakoune E, Digol M, Konamna X, Selekon B, Le Faou A (2012) New introduction and spread of rabies among dog population in Bangui. Acta Trop 123: 107–110. doi: 10.1016/j.actatropica.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 59.INS (2010) The population of Cameroon in 2010; National Institute of Statistics; http://www.statistics-cameroon.org/news.php?id=18. [Google Scholar]
- 60.Bourhy H, Rollin PE, Vincent J, Sureau P (1989) Comparative field evaluation of the fluorescent-antibody test, virus isolation from tissue culture, and enzyme immunodiagnosis for rapid laboratory diagnosis of rabies. J Clin Microbiol 27: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ito M, Itou T, Sakai T, Santos MF, Arai YT, et al. (2001) Detection of rabies virus RNA isolated from several species of animals in Brazil by RT-PCR. J Vet Med Sci 63: 1309–1313. [DOI] [PubMed] [Google Scholar]
- 62.David D, Yakobson B, Smith JS, Stram Y (2000) Molecular epidemiology of rabies virus isolates from Israel and other middle- and Near-Eastern countries. J Clin Microbiol 38: 755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lefort V, Longueville JE, Gascuel O (2017) SMS: Smart Model Selection in PhyML. Mol Biol Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
The nucleocapsid gene sequences of RABV determined in this study are available from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession numbers MF537505 to MF537580.