Abstract
Background
Atrial fibrillation (AF) affects over 33 million individuals worldwide. Genome-wide association studies have identified at least 30 AF loci, but the mechanisms through which individual variants lead to altered disease risk have remained unclear for the majority of these loci. At the 1q24 locus, we hypothesized that the transcription factor PRRX1 could be a strong candidate gene as it is expressed in the pulmonary veins, a source of AF in many individuals. We sought to identify the molecular mechanism whereby variation at 1q24 may lead to AF susceptibility.
Methods and Results
We sequenced a ~158 kilobase (kb) region encompassing PRRX1 in 962 individuals with and without AF. We identified a broad region of association with AF at the 1q24 locus. Using in silico prediction and functional validation, we identified an enhancer that interacts with the promoter of PRRX1 in cells of cardiac lineage. Within this enhancer, we identified a single nucleotide polymorphism (SNP), rs577676, which alters enhancer activity in a mouse atrial cell line and in embryonic zebrafish and differentially regulates PRRX1 expression in human left atria. We found that suppression of PRRX1 in human embryonic stem cell derived cardiomyocytes and embryonic zebrafish resulted in shortening of the atrial action potential duration, a hallmark of AF.
Conclusions
We have identified a functional genetic variant that alters PRRX1 expression, ultimately resulting in electrophysiological alterations in atrial myocytes that may promote AF.
Keywords: atrial fibrillation, arrhythmia, genetics, genomics, Genome Wide Association Study, functional genomics
Journal Subject Terms: Arrhythmias; Atrial Fibrillation; Functional Genomics; Genetic, Association Studies; Translational Studies
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia, currently affecting over 3 million people in the United States1, and 33.5 million worldwide2. AF also is associated with substantial morbidity and mortality3. Although many clinical risk factors have been described, in recent years it has become increasingly clear that AF is heritable4. To date, at least 30 distinct genetic loci for AF have been identified in genome wide association studies (GWAS)5–12. However, as with most GWAS loci, the specific mechanism(s) how an AF-associated risk variant leads to the arrhythmia remains unknown.
The top variant at the AF locus on chromosome 1q24 was previously identified approximately 63kb upstream of the gene PRRX1 in both Europeans (rs3903239; odds ratio (OR) = 1.14, 95% confidence interval (CI) = 1.10–1.17; P = 8.4 ×10−14)8 and Japanese (rs639652, r2 of 0.935 with rs3903239). PRRX1 is a homeobox containing transcriptional co-activator13, 14 that is expressed throughout embryonic development, especially in mesenchymal tissues14, and has been implicated in regulation of epithelial to mesenchymal transition15. PRRX1 is an intriguing candidate gene for AF since it is known to be expressed in the heart and pulmonary veins16, 17, the source of AF triggers in many patients18.
We therefore hypothesized that the AF associated variants at the 1q24 locus may regulate the expression of PRRX1. We identified an AF associated SNP which modulates the activity of an enhancer upstream of PRRX1 and results in diminished PRRX1 expression in human left atrial tissue. Furthermore, we found that loss of PRRX1 expression results in shortening of the atrial APD in human cardiomyocytes and embryonic zebrafish myocardium. In sum, our findings implicate PRRX1 as the causative gene at this locus and provide a mechanism for the genetic association at the 1q24 locus for AF.
Methods
Enhancer/Promoter Activity Detection in Zebrafish
Zebrafish embryos were microinjected at the single cell stage with 0.5nL of the enhancer sequence upstream of the cfos minimal promoter driving eGFP expression (25ng/μl) along with transposase mRNA (10ng/μl). The PRRX1 promoter region was cloned in frame upstream of eGFP using the backbone vector lacking cfos minimal promoter. EGFP fluorescence was assessed at 72 hpf. mRNA was collected (Zymo Research, USA) and cDNA synthesized (BioRad Laboratories, USA) according to standard protocols. qRT-PCR analysis was conducted using SYBR green supermix on a BioRad CFX384 Real-Time System. ef-1α and β-actin were used as housekeeping genes. Primer sequences are listed in Supplemental Table S1.
Study Samples for Sequencing of the PRRX1 Locus
Written informed consent was obtained from all participants prior to study enrollment. The study was approved by the Institutional Review Board at Massachusetts General Hospital. We selected cases from the Massachusetts General Hospital Atrial Fibrillation Study with AF onset before 66 years of age, and no history of myocardial infarction, heart failure, or structural heart disease as assessed by echocardiography. As expected, there is a male predominance among younger patients with AF. We selected referents from the Framingham Heart Study with no personal or family history of AF. Referents were further matched to cases on the basis of age, sex, and hypertension status. Principal components analysis on previously genotyped samples was performed to exclude ancestral outliers (Supplemental Figure 1). Both cases and referents were self-identified to be of European descent and both studies were from the same geographical region.
Sequencing and Bioinformatics Analysis of the PRRX1 Locus
A custom NimbleGen capture kit was created for chromosome 1: 170,555,163–170,713,376 and sequencing was performed on the Illumina HiSeq 2500 platform. We used BWA19 to align the short reads to the reference human genome (NCBI Build 37, hg19), which were then converted into BAM files by SAMtools 20. We also performed local realignment to minimize mismatching of bases across all the reads, and base quality recalibration to correct base qualities using the GATK software package. Duplicate reads were marked by MarkDuplicates using the Picard software package. Only the first read pair in each duplicate cluster was kept for downstream analysis. Single nucleotide variations were then called by the UnifiedGenotyper function using the GATK software package21. All samples were called simultaneously, and a project VCF file was created that included the genotype calls for all samples.
Common variants were defined as variants with a minor allele frequency (MAF) of ≥1% and rare variants with a MAF of <1.0%. Common variants were tested for association with AF using logistic regression and adjusting for age at enrollment, sex and hypertension status. We employed a conservative Bonferroni correction for the common variant analysis (0.05 / 333 common SNPs in the region sequenced = 1.5 × 10−4). For the 1,285 rare variants, we examined the associations of SNP sets with AF, using sequence kernel association test (SKAT) with the default (Wu) weight 22. We tested groups of common and rare variants sequenced at the PRRX1 gene region for association with AF using a rolling window approach (window size of 10,000 base pairs and an overlap of 5,000 base pairs). We also tested coding variants within the PRRX1 gene itself for association with AF. We then stratified analyses by allele frequency classes (MAF<0.01; MAF>0.01 and MAF<0.05; MAF<0.05) to specifically assess whether rare or low frequency variation associated with AF. All analyses were adjusted for age, sex, and hypertension status. SKAT testing was performed using the R version 3.1 seqMeta package (https://www.r-project.org/). The effect of variants on protein coding was predicted using the RefGene gene definition (https://genome.ucsc.edu/). Analysis of evolutionary conservation of the amino acids was performed using the PhyloP tool. PolyPhen-2 was used to predict the effect of the amino acid substitution in protein function.
Identification of Putative Enhancers
To identify potential PRRX1 enhancers, we analyzed both mammalian conservation and defined genomic markers of transcriptional enhancers (H3K4Me1 and DNAse hypersensitivity from the ENCODE database23). Region chr1:170560110–170642899, the region with a R2>0.3 (SNAP, 1000 Genomes Pilot 1) with the most significant AF SNP from GWAS, rs3903239, was screened for potential enhancer regions based on UCSC browser tracks(hg19) for mammalian conservation, H3K4Me1, and DNAseI hypersensitivity in human cardiac fibroblasts and human cardiac myoblasts. For the H3K4Me1 track, human skeletal muscle myoblasts (HSMM) or normal human lung fibroblasts (NHLF) were selected for further analysis. Potential enhancer regions were selected based on the fulfillment of 2 out of the 3 of the following criteria: 1) mammalian conservation score by PhyloP more than 1, 2) DNAseI hypersensitivity by ENCODE more than 50, and presence of DNAseI hypersensitivity peak (FDR=0.5%) in human cardiac fibroblasts or human cardiac myocytes, 3) ENCODE enhancer and promoter-associated histone mark (H3K4Me1) greater than 10 in (HSMM) or (NHLF).
Cell Culture and Luciferase Assays
HL-1 cells were grown on gelatin-fibronectin coated plates under standard conditions24. For luciferase assays, cells were transfected with reporter vectors (pGL4.23 [luc2/minP]) containing the various enhancer regions upstream of the CMV minimal promoter. Renilla luciferase driven by a SV40 promoter (pGL4.73[luc2/SV40]) was co-transfected as a transfection control. Transfection was performed using Lipofectamine LTX with Plus reagent (Invitrogen) according to manufacturer’s instructions. Luciferase activity was assayed using the Dual-Luciferase Reporter (DLR) Assay System according to manufacturer’s instructions in six biological replicates. Enhancer activity was calculated by normalization of Firefly luminescence to Renilla luminescence and the DNA concentration. To ensure that sufficient nucleotide context was provided for enhancer activity measurement, all final constructs were determined to have activity greater than an empty vector control with at least one allele.
Chromatin Conformation Capture
A human cardiac fibroblast (HCF) cell line was purchased from Promo Cell (Heidelberg, Germany) and maintained according to manufacturer’s instructions. The H7 hESC line (WA07; WiCell) was used as a reference cell line. Cells were maintained in feeder-free culture in Essential 8 medium25 on Geltrex coated plates (Invitrogen).
HCF and hESC were dissociated in 0.04% and 0.25% Trypsin EDTA, respectively, with subsequent neutralization with 0.05% trypsin inhibitor (for HCF) or DMEM medium containing 10% of fetal bovine serum (for hESC). 5×106 cells per experiment were used. The 3C library was generated as described by Naumova et al26 using HindIII as restriction enzyme. BAC clone R11-907L24 was utilized as random ligation control. Interaction frequency was measured by TaqMan qPCR. Quality of primer pairs was assessed with mock controls, defined as samples in which one of the crucial steps: fixation, restriction, or ligation, was omitted. Primer pairs that showed amplification in any of the mock controls were excluded from the analysis. Supplemental Table S1 contains the primer sequences used with respective distance between the restriction site and the probe. A separate primer was designed to assess the rate of self-ligation (self-loop) to control for ligation efficiency. Serial dilutions of BAC clone random ligation DNA were used to generate a standard curve (lg[DNA] to cycle number) for each primer pair. The linear regression derived from the standard curve for each primer pair was used to calculate arbitrary concentration of the ligation product in the sample. This number was then divided by the number obtained for the self-loop in this sample to normalize for the ligation efficiency. For each cell type, values were normalized to the average value for the Pr6 and Pr4 primers (on each side of the enhancer) as internal control to normalize for the efficiency of the ligation of the enhancer with the nearby regions. The average value for different biological regions; enhancer region, PRRX1 promoter region, PRRX1 gene and distant region was used for comparison between the two cell types.
Hi-C Data Analysis
Hi-C data from Dixon et al., Jin et al. and Naumova et al.27–29 were mapped and processed using hiclib software30 at 10kb, 20kb and 40kb resolutions. All hESC Hi-C datasets from Dixon et al.27 and Jinet et al.28 were pooled together; all wild type IMR90 Hi-C datasets and were also pooled together. Hi-C interaction maps at 10kb and 20kb were processed as in Naumova et al.29; directionality ratios were obtained as in Naumova et al29, with 20kb resolution and 400kb distances.
Expression Quantitative Trait Loci Analysis from Human Atrial Samples
A total of 121 human left atrial samples were used for the expression quantitative trait loci (eQTL) analysis. Surgical samples were obtained at Massachusetts General Hospital during cardiac surgery for valvular heart disease (n=83) or cardiac transplantation (n=27). Normal left atrial tissue was obtained from the National Disease Research Interchange repository (n=11). The sample collection was approved by the Institutional Review Board at Massachusetts General Hospital. Atrial tissue was homogenized in 1 ml of TRIZOL® Reagent (Applied Biosystems, Foster City, CA) and mRNA was extracted according to the manufacturer’s instructions. Reverse transcription reaction was performed using iScript (BioRad). qRT-PCR analysis was conducted using SYBR green supermix (BioRad) on a BioRad CFX384 Real-Time System. TBP and HPRT were used as housekeeping genes. Sequences of the qRT-PCR primers used are listed in Supplemental Table S1.
Optical Mapping in Human Embryonic Stem Cell Derived Cardiomyocytes
H7 embryonic stem cells (WA07; WiCell) were electroporated with PX458 construct (Addgene, 48138) carrying both spCas9-GFP nuclease and single guide RNA (sgRNA) targeting exon 1 of the PRRX1 gene (sgRNA sequence: GAGTCGCCGGGACTCACCAG, hg19 chr4:170633555–170633574). Cells positive for GFP signal were selected by flow cytometry and expanded as single colonies in 96-well plates. Sanger-sequencing was performed to identify the colonies with an insertion or deletion that led to a frameshift in the PRRX1 transcript. A single clone with a bi-allelic single base pair insertion after base 211 of the coding region induced a frameshift that resulted in a protein truncation at amino acid 81. Wild-type and PRRX1-knockout stem cells were differentiated towards cardiac lineage using previously published protocols31. Arclight, a genetically encoded fluorescent voltage sensor32, was transduced into the differentiated cardiomyocytes (ESC-CMs) using a lentiviral vector. At day 33 of differentiation, ESC-CMs were imaged 96 h post infection with Tyrode’s solution (136mM NaCl, 5.4mM KCl, 10mM Dextrose, 1mM MgCl, 1.8mM CaCl2, 0.33mM NaH2PO4, 5mM HEPES, pH 7.35) perfusion at 37 °C. A CardioCCD-SMQ camera (RedShirt Imaging) and a Nikon Eclipse Ti-U inverted microscope equipped with a FITC filter set were used for ArcLight fluorescence recording at 500Hz. ESC-CMs were rate-controlled by field pacing using a S48 Stimulator (Grass Products). APD80 was calculated for both wild-type and PRRX1-null ESC-CMs using a method described previously32.
Morpholino Design and Zebrafish Embryo Microinjections
Zebrafish of the Tübingen/AB strain were maintained according to standard methods. Morpholino oligonucleotides (MOs) designed to disrupt the proper splicing of zebrafish genes prrx1a (Exon1/Intron1 (1), Exon3/Intron3 (2)), prrx1b (Exon1/Intron1 (1), Exon2/Intron2 (2)), were obtained from Gene Tools LLC. Semi-q RT-PCR was used to assess knock-down efficiency. For control of non-specific toxicity, a non-targeting MO of equal length but differing nucleotide concentration was used. MOs were diluted in injection buffer (0.4 mM MgSO4, 0.6 mM CaCl2, 0.7 mM KCl, 58 mM NaCl, 25 mM HEPES pH 7.1) and microinjected into the yolks of single cell stage embryos at a final concentration of 0.2 mM (for prrx1 E1I1 and prrx1b E1I1 MOs), 0.1mM (for prrx1a E3I3 MO), and 0.3 mM (for prrx1b E2I2) in a standardized volume. At 72 hours post fertilization (hpf) mRNA was extracted using TRIZOL® Reagent according to manufacturer’s instructions; cDNA was synthesized using iScript (BioRad). Effective knockdown was confirmed by RT-PCR at 27 cycles. ef-1α was utilized as a reference gene. Densitometry analysis was performed using ImageJ version 1.47. All MOs and percent of calculated knock-down are listed in Supplemental Figure S2. All primers used for analysis of knock-down efficiency are contained with Supplemental Table S1.
Color brightfield images of morpholino-injected embryos were obtained on an Olympus SZX16 microscope at 72 hpf. Heart rate was measured at 72 hpf by phase contrast imaging on a Nikon Eclipse Ti equipped with a Hamamatsu Orca-Flashcam 2.8 at an effective framerate of acquisition 88fps. Heart rate was determined manually using ImageJ version 1.47. Measurements of cardiac contractility were conducted at 72 hpf. Ventricular fractional shortening was assessed by measuring the change in inner chamber diameter when measured perpendicular to the direction of blood flow. Atrial area was measured in ImageJ version 1.47 at the end of diastole. Optical mapping to determine atrial action potential durations at 72hpf was performed as previously described33.
To examine the presence of fibrosis in morpholino-injected embryonic zebrafish hearts (72 hpf), excised hearts were subjected to aniline blue stain for 5 min (Trichrome Stain; Abcam; ab150686), and mounted in synthetic resin (DPX Mountant for histology; Sigma; 06522). Color micrographs were generated using an Olympus SZX16 microscope.
Statistical Analyses
Results are presented as mean ± standard error of the mean. Statistical analysis was performed using Prism 6 (GraphPad Software, La Jolla, CA, USA). Statistical analysis of differences was determined by Student’s two-tailed t-test for comparison of two groups, or one-way ANOVA with Dunnett’s post hoc test for data with greater than two groups.
Results
Targeted resequencing of the PRRX1 locus reveals a broad region of association with AF
The association between PRRX1/1q24 locus and AF had previously been established in a discovery sample consisting of 6,707 individuals with and 52,426 without AF, and replicated in an additional 5,381 individuals with and 10,030 without AF.8 The identified SNP, rs3903239, was significantly associated with AF and resides upstream of PRRX1 (RR 1.14, CI 1.10–1.17, P= 8.4 × 10−14).8
To comprehensively identify the common and rare genetic variation at the PRRX1 locus, we sequenced an approximately 158kb region in 462 AF cases from the Massachusetts General Hospital (MGH) AF Study and 464 referents from the Framingham Heart Study (Table 1 and Figure 1). The sequencing was performed with >200X coverage. Raw reads were aligned to the reference human genome (NCBI Build 37, 2009) after which we applied stringent filters to exclude variants with low quality, as defined by those with a calling quality less than 30, a missing call rate higher than 5%, or a strand bias >−0.1, or depth of coverage <10× or higher than 300x.
Table 1.
Baseline characteristics of study samples used for sequencing of the PRRX1 locus.
| MGH AFStudy | Framingham Heart Study | P value | |
|---|---|---|---|
| Number of individuals | 462 | 464 | - |
| Phenotype | AF | Referents | - |
| Sex, male (%) | 363 (79) | 366 (79) | - |
| Mean age of AF onset (MGH) orDNA blood draw (FHS), years | 47±11 | 55±11 | <0.0001 |
| Height, cm | 177±10 | 173±8 | <0.0001 |
| Weight, kg | 91±19 | 85±17 | <0.0001 |
| BMI, kg/m2 | 28.1±6.6 | 28.3±4.8 | 0.59 |
| Systolic blood pressure, mmHg | 125±16 | 128±18 | 0.008 |
| Diastolic blood pressure, mmHg | 76±10 | 78±9 | 0.001 |
| Family history of AF, percent | 28 | 0 |
AF – atrial fibrillation; BMI – body mass index; MGH – Massachusetts General Hospital; Data are presented as mean ± SD or percentage
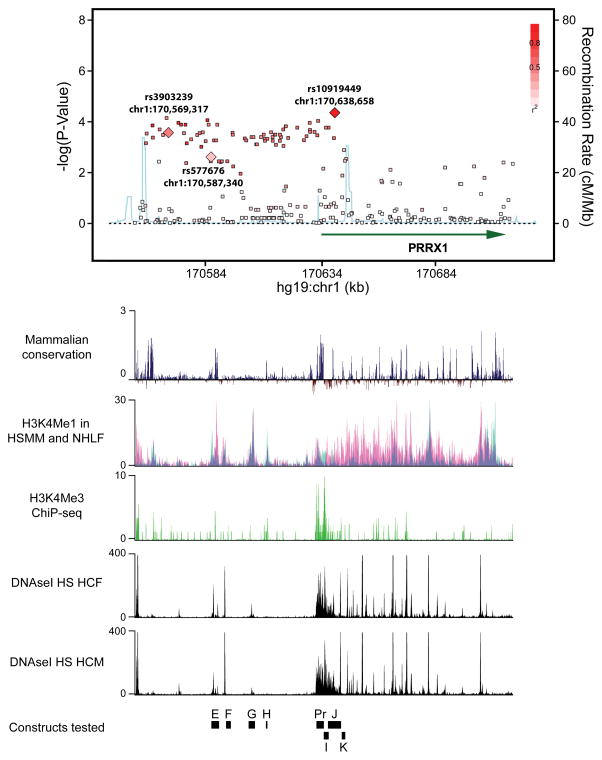
Figure 1. Plot association of the region upstream of PRRX1, with corresponding ENCODE and mammalian conservation tracks.
Regional plot displaying the association of common SNPs with AF (upper panel). Each dot represents an individual SNP plotted according to genomic location (x-axis) and strength of association (y-axis). The gradation of red color indicates the degree of linkage disequilibrium with the most significantly AF associated SNP, rs10919449. The light blue line indicates the rate of recombination.
Lower panels indicate corresponding UCSC browser tracks (from ENCODE) detailing mammalian conservation, and measures of H3K4Me1 in myoblast and fibroblast cell lines (HSMM, NHLF respectively), H3K4Me3 ChiP-seq raw signal and DNaseI Hypersensitivity(HS) in human cardiac fibroblasts and human cardiac myocytes. E, F, G, H, Pr, I, J, K indicate the locations of constructs tested in the enhancer/promoter screen.
We identified 333 common SNPs with a minor allele frequency of >1%, and 1,285 rare or singleton SNPs (Supplemental Table S2). Included within this set were 6 missense variants (Supplemental Table S3), 2 synonymous variants, and 1 proximal to a splice site, all of which were contained within the PRRX1 gene. We then tested the association of each common variant with AF status by logistic regression, and adjusted the analysis by age, sex, and hypertension. We identified a broad, approximately 82 kb, region that was associated with AF (Figure 1). Within this region, the SNP most significantly associated with AF, rs10919449, is intronic to PRRX1 (OR 1.48, 95% CI 1.22–1.79; P = 4.4 × 10−5) (Supplemental Table S2) and is located approximately 69kb away from the most significant SNP identified in a prior AF GWAS, rs3903239 (OR 1.41, 95%CI 1.17–1.71, P=2.8×10−4)8. We did not find any association between AF and rare variants (MAF<1%), indels or coding variants at this locus, although given our sample size the power for these analyses was limited (Supplemental Table S2, S3). Similarly, we did not observe associations between specific rolling windows comprised of both common and rare variants and AF, or coding variation in PRRX1 with AF.
Identification of functional regulatory elements at the PRRX1 locus
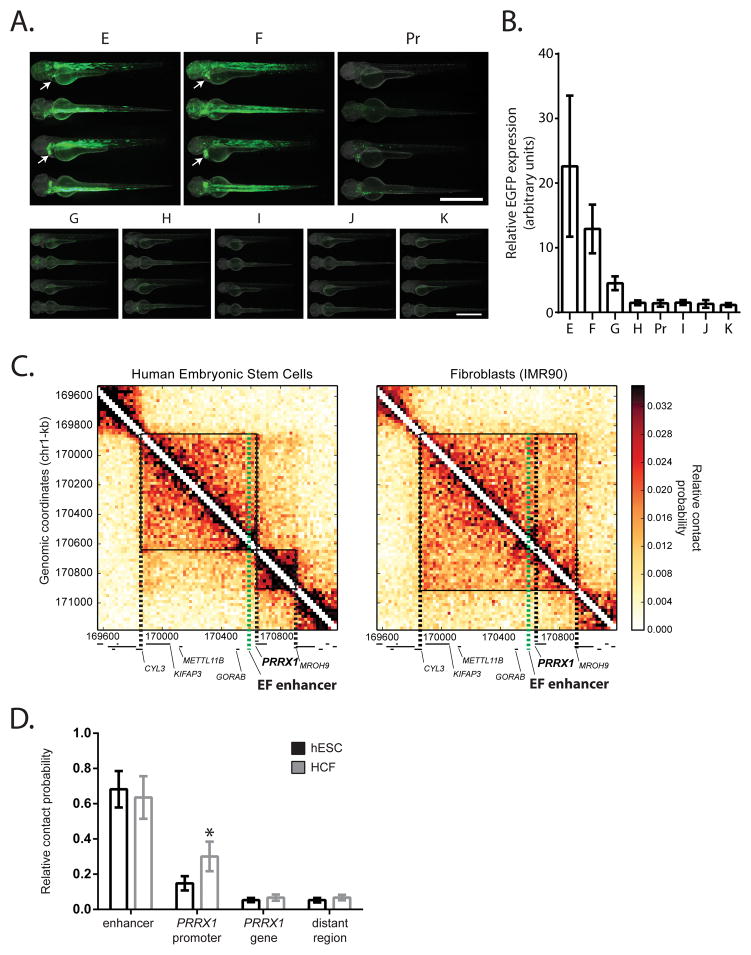
Given the broad and largely intergenic region that was associated with AF, we hypothesized that the causative variant for AF was regulating the expression of the adjacent PRRX1 gene. To identify potential PRRX1 enhancers, we analyzed both mammalian conservation and defined genomic markers of transcriptional enhancers (H3K4Me1 and DNAse hypersensitivity from the ENCODE database23) within this region. Potential enhancer regions were selected from the genomic region (chr1:170560110–170642899 as defined by the region with R2 >0.3 with the most significant AF SNP, rs3903239) based on fulfillment of two out of the three criteria: 1) mammalian conservation score by PhyloP34 more than 0.5; 2) ENCODE DNAseI hypersensitivity raw signal of more than 50 in human cardiac fibroblasts or human cardiac myocytes; and 3)ENCODE enhancer and promoter-associated histone mark (H3K4Me1) of more than 10 in skeletal myoblasts (HSMM) or neonatal fibroblasts (NHLF). As a result, seven putative enhancer regions (E–K) and PRRX1 promoter region (Pr) were prioritized for further analysis. Genomic coordinates for these regions are contained within Supplementary Table S4. Microinjection of eGFP reporter constructs in embryonic zebrafish revealed that two putative enhancer regions, E and F, displayed strong eGFP expression in cardiac and skeletal muscle tissue while the PRRX1 promoter (Pr) exhibited a similar pattern of activity at notably lower levels (Figure 2A,B).
Figure 2. Identification of the PRRX1 enhancer underlying the 1q24 AF-association locus.
A. The representative zebrafish for the initial enhancer (E–K) / promoter(Pr) screen displaying the tissue distribution and intensity of the enhancer and promoter activity. Arrows indicate the cardiac region. Scale bar indicates 1mm. B. qRT-PCR results measuring relative eGFP expression levels in zebrafish enhancer/promoter screen (n=3). C. Hi-C data showing organization of topologically associated domains around E and F enhancers in fibroblast cell lines (IMR90) as compared to human embryonic stem cells (hESC). D. Results of chromatin conformation capture (3C) experiment displaying increased frequency of interaction between the E and F enhancers and PRRX1 promoter in human cardiac fibroblasts (HCF)(n=5) as compared to hESC (n=4). *Represents p<0.05.
Chromatin conformation capture links active regulatory elements to PRRX1 promoter
To determine the interaction between the E and F enhancer regions and nearby promoters, we utilized a tiered approach, using both Hi-C and 3C (chromatin conformation capture) to further analyze the target regions. In silico analysis revealed that DNAse hypersensitivity around the E and F enhancer regions in human cardiac myocytes (HCM) and human cardiac fibroblasts (HCF) was lacking in human embryonic stem cells (hESC), establishing the hESC as a negative reference for further interaction analysis (Supplemental Figure S3). First, to determine which promoters may be in physical contact with the E and F enhancer regions at a lower resolution but with a wider genomic context we explored the Hi-C map in hESC and IMR90 fibroblasts. In hESC cells, the E and F enhancer regions are embedded in a topological domain that contains the PRRX1 promoter at the margin (Figure 2C). In Hi-C data for a fibroblast cell line (IMR90)27, 29, this region undergoes a major structural reorganization during differentiation, characterized by a merging of two adjacent topological domains (Figure 2C, Supplemental Figure S4) and significant alterations in the patterns of interactions between the E and F enhancer regions and the PRRX1-containing domain (Supplemental Figure S4). Next, to identify a potential increase in interaction frequency between the enhancer region and PRRX1 promoter also occurs in cardiac cells, we employed a targeted approach using 3C on hESC and human cardiac fibroblast cell lines. The E and F enhancer region had an increased interaction frequency with the PRRX1 promoter in cardiac fibroblasts as compared to hESC (103 ± 57%, P=0.03; Figure 2D). No difference in relative contact probability was detected between the cell lines when examining regions proximal to the enhancer, the PRRX1 coding region, or a distant genomic region. In sum, these data demonstrate a physical link between the E and F enhancer regions and the PRRX1 promoter in relevant tissues, supporting the hypothesis that alterations in PRRX1 expression via the E and F enhancer regions may underlie the AF association signal.
Identification of a Functional SNP at 1q24 Locus
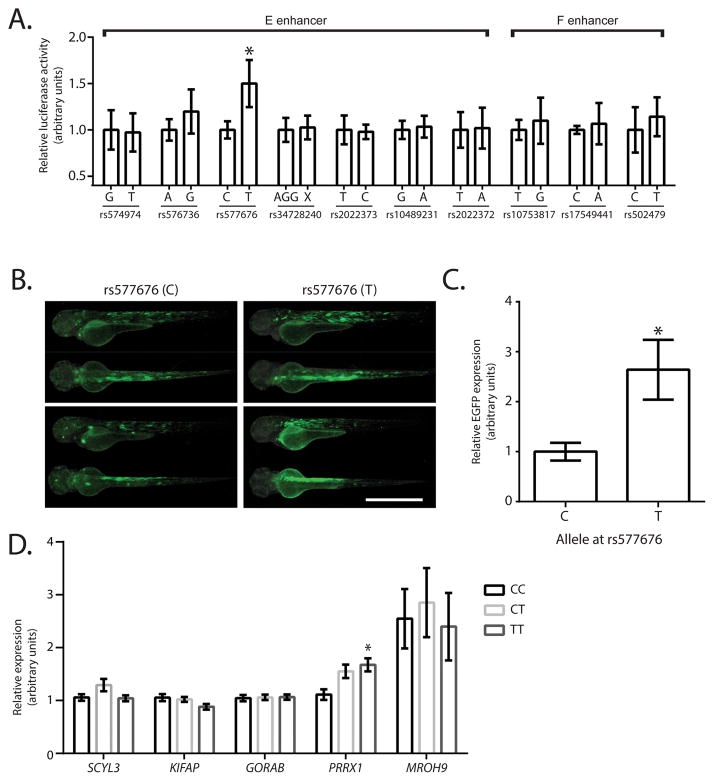
Next, we sought to identify any functional SNPs within the E and F enhancers and PRRX1 promoter. We cloned 100–300bp regions around the 21 common SNPs (MAF>5%) and compared the enhancer activity of risk vs non-risk alleles by luciferase assays in a murine atrial myocyte (HL-1) cell line. Among SNPs within E and F enhancer and PRRX1 promoter regions, only the region surrounding SNP rs577676 exhibited altered enhancer activity, with the AF risk allele displaying an approximately 1.5-fold decrease in luciferase activity (Figure 3A, Supplemental Figure S5). Importantly, the immediate region surrounding rs577676 has a positive PhyloP conservation score of 0.99, suggesting the evolutionary stability expected for putative enhancers. Further, the enhancer activity of the rs577676 region in zebrafish embryos was localized to the heart and skeletal muscle, with consistently higher expression in non-risk allele shown by epifluorescence imaging and qRT-PCR (Figure 3B,C). Interestingly, although rs577676 has moderate association with AF in the targeted sequencing analysis (P = 2.7×10−4, Risk Allele Frequency (RAF) = 0.57, OR = 1.33, 95% CI 1.10–1.38), it is only in modest linkage disequilibrium with adjacent SNPs at the 1q24 locus (r2=0.605 with rs3903239). Importantly, although rs577676 does not reach the genome wide significance threshold in the present study due to the limited sample size used for sequencing, it was significantly associated with AF in a previous GWAS (P = 1.3×10−10, RAF = 0.56, OR = 1.14, 95% CI 1.10–1.19). Importantly, after conditioning on the top SNP from previous GWAS (rs3903239), the association of rs577676 with AF is abrogated (p = 0.67). We also performed a sensitivity analysis to ensure that observed associations were not due to population sub-structure. After adjusting for principle components derived from previously genotyped samples, rs577676 remained significantly associated with AF, although the association was attenuated.
Figure 3. Identification of the functional SNP at PRRX1 locus for AF.
A. Identification of the functional SNP at E and F enhancer regions. Luciferase assay results (n=6) showing the alteration of the enhancer activity depending on the SNP allele within E and F enhancer regions from major to minor allele. Regions ranging 100–300bp in length containing a single SNP are labeled as SNP number. *Represents p<0.05 as compared to the major allele. B. Representative fluorescent micrographs of 72 hours post fertilization zebrafish displaying the tissue distribution and intensity of the enhancer of the 237bp region containing different alleles of rs577676. Scale bar represents 1mm. C. qRT-PCR results showing relative eGFP expression levels in zebrafish injected with a construct containing rs577676(C) or rs577676(T) (n=11). * Represents p<0.05 as compared to the major (risk) allele. D. Expression quantitative trait loci analysis showing the comparison of PRRX1 mRNA levels depending on the genotype at rs577676 (n(CC)=28,n(CT)=66, n(TT)=27). *Represents p<0.05 when compared to homozygous C (risk) allele.
To define the necessity of the rs577676 to drive enhancer activity, we performed serial deletions within this region and found that removal of 20 base pairs surrounding rs577676 led to almost complete loss of the enhancer activity (Supplemental Figure S6). In silico analyses of potential transcription factor binding sites which may be disrupted by genotype at rs577676 were performed using JASPAR. 33 potential enhancers were predicted to be generated by the minor allele while 3 potential repressors were identified exclusively with the major allele (Supplemental Table S5).
eQTL analyses link AF associated SNP to expression of PRRX1
Chromatin conformation capture experiments demonstrate that the E enhancer is in contact with the PRRX1 promoter, but did not rule out the possibility of effects on other promoters within the topologically associated domain. Given the differential activity of the rs577676 enhancer by reporter assays, we then sought to directly link this altered enhancer activity to altered expression of nearby genes in human left atrial tissue samples. To ensure our approach was unbiased, we determined whether the genotype of rs577676 was associated with the expression of any genes in same topologically associated domain as defined by the Hi-C analysis. We performed expression quantitative trait loci (eQTL) analysis in 121 human left atrial tissue samples (Figure 3D). Among the 5 genes within the topologically associated domain, only PRRX1 transcript levels correlated with genotype at rs577676, with a 1.7 fold decreased expression in those homozygous for the risk allele. Thus, PRRX1 expression is regulated by the PRRX1 enhancer in the heart and its expression is dependent on the genotype at rs577676.
PRRX1 suppression results in action potential shortening
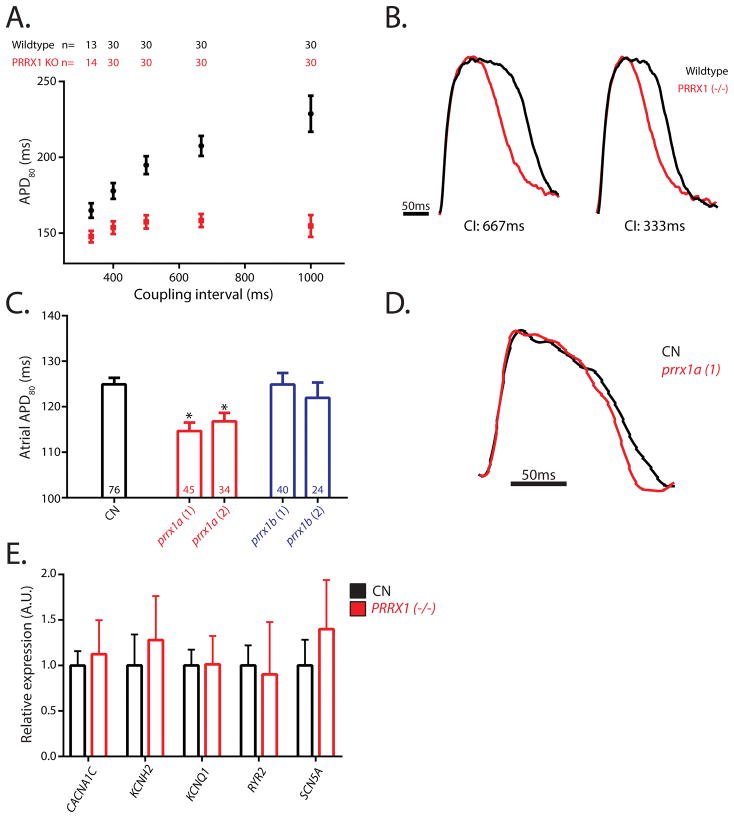
Since shortening of the atrial APD is a well described in the pathophysiology of AF,35 we determined the role of PRRX1 in cardiac electrophysiology in both stem cell derived cardiomyocytes (Figure 4A,B,E) and embryonic zebrafish (Figure 4C,D). For the former, we utilized CRISPR-Cas9 mediated knockout of PRRX1 in human embryonic stem cells and morpholino-mediated knockdown of the putative PRRX1 orthologues in the zebrafish (prrx1a and prrx1b). PRRX1-null human ES cells were differentiated into cardiomyocytes by standard protocols, infected with the fluorescent voltage reporter, Arclight, and then assayed for action potential duration (APD80) by optical mapping. PRRX1 knockout cardiomyocytes displayed significantly shortened action potentials at all pacing rates when compared to controls (Figure 4A,B). Interestingly, PRRX1 cells also maintained a relatively stable action potential duration regardless of pacing rate, a finding in contrast to the typical use-dependent action potential modulation observed in wild-type cardiomyocytes (p<0.001). We then attempted to identify potential mechanisms by which PRRX1 modulation may affect the action potential duration. We assayed the expression levels of 5 major ion channels known to affect the action potential duration in cardiomyocytes (CACNA1C, KCNH2, KCNQ1, RYR2, and SCN5A). No significant differences were observed between control and PRRX1 null myocytes (Figure 4E).
Figure 4. The effect of PRRX1 suppression on the action potential in ES-derived cardiomyocytes and zebrafish.
A. Action potential duration at 80% repolarization (APD80) assayed by Arclight fluorescence in field-paced ES-derived cardiomyocytes. n’s for each respective measurement are displayed above in the corresponding color text. B. Representative traces of ES-derived cardiomyocyte action potentials for wild-type and PRRX1 knockout cells. CI represents the coupling interval between stimulations. C. APD80 as assayed by optical mapping with 2hz field pacing in 72 hours post fertilization zebrafish hearts. prrx1a(1) and prrx1a(2) represent the two distinct prrx1a targeting morpholinos used to ensure the specificity of the effects observed. Numbers of replicates are displayed within the bars. D. Representative traces of atrial action potentials for prrx1a(1) and control morpholino injected zebrafish hearts. *Represents p<0.05 when compared to control. CN-control. Error bars in all panels represent the SEM. E. Relative expression by qRT-PCR of major ion channels in control and PRRX1 (−/−) stem cell derived cardiomyocytes. Error bars represent the SEM derived from 4 control and 3 PRRX1 (−/−) biological replicates.
For the zebrafish, minimal effective doses of morpholino were evaluated by RT-PCR (Supplemental Figure S2). At 72 hours post fertilization, a time point when distinct cardiac chambers are formed and both prrx1a and prrx1b are robustly expressed within the cardiac regions (Supplemental Figure S2), the gross morphology of prrx1a and prrx1b knockdown embryos was indistinguishable from controls (Supplemental Figure S2). Similarly to the human myocytes, significant shortening of atrial action potential (114.7±1.9ms and 116.8±1.9ms vs 124.9±1.4ms in controls, P=0.0002 and P=0.006, respectively) was observed in prrx1a knockdown embryos (Figure 4C,D). s rs577676-mediated modulation of PRRX1 expression in human left atrium is more modest than the severe diminishment utilized in our models, we next sought to examine action potential durations with various doses of prrx1a morpholinos. This action potential duration shortening was reduced in a dose-dependent manner when progressively lower amounts of prrx1a morpholinos were injected, suggesting that even small modulations in PRRX1 expression may be sufficient to modulate the action potential duration (Supplemental Figure S2). In contrast, knockdown of prrx1b had no effect on action potential duration when compared to controls (124.9±2.5ms and 122±3.4ms). 1 Neither prrx1a nor prrx1b knockdown had significant effect on resting heart rate or ventricular contractility, nor were any ectopic foci of contractile initiation observed (Supplemental Figure S2).
Discussion
Over the past decade, thousands of genetic loci have been identified for a wide variety of diseases and traits using GWAS. However, despite the rapid pace of discovery, relatively few studies have directly linked a genetic variant at a locus to plausible biological mechanism for disease. 36–42 For AF, at least 30 loci have been identified to date, yet the underlying molecular mechanism at most of these loci remains incompletely understood. In the present work, we have used an integrative functional genomics approach that combines resequencing of the candidate locus, phenotypic modeling in both zebrafish and stem cell derived cardiomyocytes, chromatin conformation analyses, the identification of gene regulatory elements, and eQTL mapping to elucidate a functional variant at the 1q24 locus for AF.
PRRX1 is the AF-associated gene at the 1q24 locus
Although genome wide studies have identified many genomic regions associated with AF, determining which nearby gene or genes are related to the association signal is a critical step for understanding the biological pathways underlying disease. In the present study, we provide three independent analyses that strongly suggest that PRRX1 is the causative gene at the 1q24 locus for AF. First, diminished PRRX1 expression in ES-derived cardiomyocytes and zebrafish resulted in action potential shortening, a hallmark of AF in humans.35 Second, eQTL analyses directly linked the genotype of rs577676 to the expression of PRRX1 in human left atrial samples. Finally, chromatin conformation studies demonstrated that an enhancer modified by rs577676 is in close proximity to the PRRX1 promoter in three-dimensional space. Additionally, in recent work Lin and colleagues found an increased frequency of rare, nonsynonymous variants in the coding region of PRRX1 in individuals with AF. In sum, these findings strongly suggest that PRRX1 is the gene related to AF at this locus.43 Ideally, an RNA-seq based eQTL dataset from human left atrial tissue could be used to not only identify the cis-QTL, which is PRRX1 for the 1q24 locus, but also trans-QTLs which may give insight into downstream function of this gene alteration. Generation of these publically available datasets should be the focus of future effort.
In addition to the present data detailing a role for PRRX1 in atrial myocyte electrophysiology, previous work on PRRX1 also suggests plausible biological pathways whereby altered PRRX1 expression would result in an increased risk of AF. Among these, PRRX1 is highly expressed in mesenchymal tissues14 including the heart and pulmonary veins, 16, 17 and has been shown to regulate epithelial to mesenchymal transition15, a critical feature of human cardiac development. Moreover, Prrx1-deficient mice showed abnormalities of great vessels, such as abnormal positioning and awkward curvature of the aortic arch, misdirected and elongated ductus arteriosus and an anomalous retro-esophageal right subclavian artery44. In future studies, it will be interesting to examine function of atrial or pulmonary venous restricted knockouts of PRRX1 in mice to further delineate the role of this gene in atrial structure and function.
Identification of rs577676 as a functional variant for AF at the 1q24 locus
The disease variants identified by a GWAS are typically a proxy rather than the functional variant at a locus. It remains challenging to move from a SNP identified by GWAS to a functional variant, particularly when an association signal can tag a large haplotype block associated with disease. In our current work, we resequenced a large genomic region at 1q24, encompassing the entire PRRX1 gene as well as nearly 80kb upstream from the transcriptional start site. In order to be inclusive for potential functional variants which may be somewhat distant either in genomic distance or in modest linkage disequilibrium with a sentinel SNP, our sequencing efforts the sequencing included the genomic location bounded by and r2 of 0.3, as well as the additional 75kb extending through the PRRX1 gene region. This sequencing effort was performed to identify not only common risk variants, but also rare genetic variation, structural, and copy number variation at the 1q24 locus. The latter were of particular interest as they are potential sources of AF risk that are not readily identified by traditional GWAS methodologies. Although our power was modest, we found a broad area of association with AF similar to our initial GWAS.
We therefore employed an in silico prioritization strategy for identifying functionally active regulatory regions at this locus. We took advantage of existing ENCODE datasets on histone post-translational modifications and DNase hypersensitivity in cardiac cells as a surrogate for active enhancers and tested their activity in a vertebrate model. We identified two enhancers and the PRRX1 proximal promoter as regions with tissue-specific activity in the heart. In reporter assays in a mouse atrial cell line, we found a single AF-associated variant, rs577676 that significantly alters enhancer activity. Conditional analyses definitively linked rs577676 to the top signal at this locus, although the possibility remains that other SNPs within the risk haplotype may also contribute to altered enhancer activity and gene regulation. Although our approach provided both qualitative and quantitative assessments of PRRX1 enhancer function, it is clear that future analyses of GWAS loci can be improved, particularly in light of the recent data releases from the NIH Roadmap Epigenomics Mapping Consortium.45
Summary and Limitations
In summary, PRRX1 expression is associated with action potential shortening in two independent model systems. We have identified an enhancer upstream of PRRX1 that is physically associated with the PRRX1 promoter, and is modulated by SNP rs577676. This SNP in turn is associated with reduced PRRX1 expression. Thus, the AF risk allele (C) of rs577676 is associated with reduced PRRX1 expression and an electrophysiological phenotype that would be expected to promote AF. Based on the multiple lines of functional evidence, we believe that the rs577676 risk allele most likely acts through modulating PRRX1 expression.
Our study was subject to a number of potential limitations. First, despite our extensive analysis of functional variation at the PRRX1 locus, it is possible that there is more than a single functional variant at a locus. Second, it would be ideal to identify the specific transcription factor that is modulated by rs577676. Unfortunately, the in silico prediction of transcription factor binding remains imperfect and CHIP-seq databases are currently incomplete. Third, future studies directed at defining the spatiotemporal expression of the enhancer modulated by rs577676 would be interesting. Finally, while our study identifies the major effector of the AF risk, reduced PRRX1 expression, and the cellular phenotype which creates a pro-arrhythmic substrate, action potential shortening, the downstream transcriptional pathway through which this action potential shortening occurs remains unclear. We attempted a targeted candidate gene analysis in our stem cell derived cardiomyocyte model, but observed no significant alterations. Further work examining the entire transcriptome, ideally in a single cell manner due to the heterogeneity of stem cell-derived cardiomyocyte cultures, will be necessary to further elaborate on these transcriptional pathways. It is also possible that additional pathogenic mechanisms for AF may not be sufficiently captured by our models, including fibrotic deposition, myolysis and inflammation. Determining whether these factors also result from altered PRRX1 activity would be of interest in future work. Ultimately, the best platform for studying these possibilities will be primary human atrial tissue, where the cardiovascular effects of the PRRX1 enhancer and gene may be studied in a more native context.
Conclusion
In conclusion, we have implicated PRRX1 as the causative gene at the 1q24 locus for AF, and we have identified a SNP that functionally modulates a PRRX1 enhancer. Our approach provides a potentially generalizable path for identifying the functional variants at other cardiovascular GWAS loci.
Supplementary Material
Clinical Perspective.
Atrial fibrillation (AF) is a common and morbid arrhythmia. In addition to the well-known clinical risk factors, AF has a considerable genetic risk component and nearly 30 genetic loci have been identified in genome-wide association studies. At the 1q24 locus for AF we found that a common genetic variant leads to altered expression of the transcription factor PRRX1 in human left atria. In turn, suppression of PRRX1 expression in zebrafish or stem cell models revealed significant shortening of the action potential duration. Our findings provide a link between the genetic association at this locus and a potential mechanistic link between PRRX1 and AF. Further study of this pathway will ultimately provide greater insight into the pathogenesis of AF and may provide new potential targets for therapeutic intervention.
Acknowledgments
Sources of Funding
This study was supported by NIH grants K23HL114724 (Lubitz), an institutional T32 award, T32HL007208 (Tucker), 2R01HL092577 (Ellinor, Lunetta, and Benjamin), R01HL104156, and K24HL105780 (Ellinor), an American Heart Association Established Investigator Award 13EIA14220013 (Ellinor), a Doris Duke Charitable Foundation Clinical Scientist Development Award 2014105 (Lubitz), and from the Fondation Leducq (14CVD01). This study was also supported by the George L. Nardi, MD fund at MGH.
Resequencing services were provided by the Northwest Genomics Center at the University of Washington, Department of Genome Sciences, under U.S. Federal Government contract number HHSN268201100037C from the National Heart, Lung, and Blood Institute.
Drs. Tucker, Dolmatova and Ellinor had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Disclosures
Dr. Ellinor is the principal investigator on a grant from Bayer HealthCare to the Broad Institute regarding the genetics and therapeutics of atrial fibrillation.
References
- 1.Miyasaka Y, Barnes ME, Gersh BJ, Cha SS, Bailey KR, Abhayaratna WP, et al. Secular trends in incidence of atrial fibrillation in Olmsted County, Minnesota, 1980 to 2000, and implications on the projections for future prevalence. Circulation. 2006;114:119–25. doi: 10.1161/CIRCULATIONAHA.105.595140. [DOI] [PubMed] [Google Scholar]
- 2.Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, et al. Worldwide epidemiology of atrial fibrillation: a Global Burden of Disease 2010 Study. Circulation. 2014;129:837–47. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.January CT, Wann LS, Alpert JS, Calkins H, Cleveland JC, Jr, Cigarroa JE, et al. 2014 AHA/ACC/HRS Guideline for the Management of Patients With Atrial Fibrillation: Executive Summary: A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the Heart Rhythm Society. Circulation. 2014 doi: 10.1161/CIR.0000000000000040. [DOI] [PubMed] [Google Scholar]
- 4.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. Jama. 2010;304:2263–9. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubitz SA, Sinner MF, Lunetta KL, Makino S, Pfeufer A, Rahman R, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–84. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–5. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res. 2014;114:1469–82. doi: 10.1161/CIRCRESAHA.114.302225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christophersen IE, Rienstra M, Roselli C, Yin X, Geelhoed B, Barnard J, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49:946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Low SK, Takahashi A, Ebana Y, Ozaki K, Christophersen IE, Ellinor PT, et al. Identification of six new genetic loci associated with atrial fibrillation in the Japanese population. Nat Genet. 2017;49:953–958. doi: 10.1038/ng.3842. [DOI] [PubMed] [Google Scholar]
- 13.Grueneberg DA, Natesan S, Alexandre C, Gilman MZ. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257:1089–95. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 14.Libório TN, Acquafreda T, Matizonkas-Antonio LF, Silva-Valenzuela MG, Ferraz AR, FDN In situ hybridization detection of homeobox genes reveals distinct expression patterns in oral squamous cell carcinomas. Histopathology. 2011;58:225–33. doi: 10.1111/j.1365-2559.2011.03751.x. [DOI] [PubMed] [Google Scholar]
- 15.Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–24. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 16.Bergwerff M, Gittenberger-de Groot AC, DeRuiter MC, van Iperen L, Meijlink F, REP Patterns of paired-related homeobox genes PRX1 and PRX2 suggest involvement in matrix modulation in the developing chick vascular system. developmental dynamics. 1998;213:59–70. doi: 10.1002/(SICI)1097-0177(199809)213:1<59::AID-AJA6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 17.Leussink B, Brouwer A, el Khattabi M, Poelmann RE, Gittenberger-de Groot AC, FM Expression patterns of the paired-related homeobox genes MHox/Prx1 and S8/Prx2 suggest roles in development of the heart and the forebrain. Mechanisms of development. 1995;52:51–64. doi: 10.1016/0925-4773(95)00389-i. [DOI] [PubMed] [Google Scholar]
- 18.Haissaguerre M, Jais P, Shah DC, Takahashi A, Hocini M, Quiniou G, et al. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–66. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas DJ, Rosenbloom KR, Clawson H, Hinrichs AS, Trumbower H, Raney BJ, et al. The ENCODE Project at UC Santa Cruz. Nucleic Acids Res. 2007;35:D663–7. doi: 10.1093/nar/gkl1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2979–84. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naumova N, Smith EM, Zhan Y, Dekker J. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012;58:192–203. doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–4. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, et al. Organization of the mitotic chromosome. Science. 2013;342:948–53. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat Protoc. 2013;8:162–75. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leyton-Mange JS, Mills RW, Macri VS, Jang MY, Butte FN, Ellinor PT, et al. Rapid Cellular Phenotyping of Human Pluripotent Stem Cell-Derived Cardiomyocytes using a Genetically Encoded Fluorescent Voltage Sensor. Stem Cell Reports. 2014;2:163–70. doi: 10.1016/j.stemcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, et al. Drug-sensitized zebrafish screen identifies multiple genes, including GINS3, as regulators of myocardial repolarization. Circulation. 2009;120:553–9. doi: 10.1161/CIRCULATIONAHA.108.821082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heijman J, Voigt N, Nattel S, Dobrev D. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ Res. 2014;114:1483–99. doi: 10.1161/CIRCRESAHA.114.302226. [DOI] [PubMed] [Google Scholar]
- 36.Guenther CA, Tasic B, Luo L, Bedell MA, Kingsley DM. A molecular basis for classic blond hair color in Europeans. Nat Genet. 2014;46:748–52. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–4. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–9. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Cowper-Sal lari R, Bailey SD, Moore JH, Lupien M. Integrative functional genomics identifies an enhancer looping to the SOX9 gene disrupted by the 17q24.3 prostate cancer risk locus. Genome Res. 2012;22:1437–46. doi: 10.1101/gr.135665.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linsel-Nitschke P, Heeren J, Aherrahrou Z, Bruse P, Gieger C, Illig T, et al. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–9. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- 41.Duan J, Shi J, Fiorentino A, Leites C, Chen X, Moy W, et al. A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am J Hum Genet. 2014;95:744–53. doi: 10.1016/j.ajhg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kapoor A, Sekar RB, Hansen NF, Fox-Talbot K, Morley M, Pihur V, et al. An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am J Hum Genet. 2014;94:854–69. doi: 10.1016/j.ajhg.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin H, Sinner MF, Brody JA, Arking DE, Lunetta KL, Rienstra M, et al. Targeted sequencing in candidate genes for atrial fibrillation: the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Targeted Sequencing Study. Heart Rhythm. 2014;11:452–7. doi: 10.1016/j.hrthm.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, et al. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Archiv. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- 45.Roadmap Epigenomics C. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.