Abstract
Cells are capable of rapid replication and performing tasks adaptively and ultra-sensitively and can be considered as cheap “biological-robots”. Here we propose to engineer cells for screening biomolecules in parallel and with high sensitivity. Specifically, we place the biomolecule variants (library) on the bacterial phage M13. We then design cells to screen the library based on cell-phage interactions mediated by a specific intracellular signal change caused by the biomolecule of interest. For proof of concept, we used intracellular lysine concentration in E. coli as a signal to successfully screen variants of functional aspartate kinase III (AK-III) under in vivo conditions, a key enzyme in L-lysine biosynthesis which is strictly inhibited by L-lysine. Comparative studies with flow cytometry method failed to distinguish the wild-type from lysine resistance variants of AK-III, confirming a higher sensitivity of the method. It opens up a new and effective way of in vivo high-throughput screening for functional molecules and can be easily implemented at low costs.
Introduction
High-throughput screening (HTS) technologies are powerful tools with many successful applications, especially in directed evolution of biomolecules such as enzymes. They are primarily based on chemical or physical readouts such as fluorescence and assisted with miniaturized and/or parallel devices such as microfluidics and microchip, increasingly in an automated manner with the help of robotics1–4. These systems require expensive infrastructure and special expertise. The major focus was put on speeding up the screening process. For example, the state-of-the-art HTS technology based on fluorescence activated cell sorting (FACS) can reach 18,000–20,000 events per second5. However, signal detection with fast moving cells is a challenge which can result in noisy signals as shown by previous studies6–9. Furthermore, single cell variations are another source of signal noise which cannot be avoided by FACS based methods10. These represent some of the shortcomings of presently used HTS technologies when the molecules to be evolved and optimized are to be used for regulation and improvement of metabolic pathways in the context of metabolic engineering or for creating new synthetic pathways and regulation tools.
Similar to the electric robots, microbial cells can be considered as a kind of “biological robots” that can sense the information of fast changing environment, compute and make decisions for survival. Cells are highly programmable as proved by recent developments in synthetic biology. Programming cells to perform specific tasks have been achieved successfully in many cases. For example, cells have been programmed to produce pharmaceuticals, fuels, amino acids, fine and bulk chemicals and even metal nanoparticles11–18. Cells also have been programmed to sense toxic compounds in environments19, to record the environment signal in human gut20 and to eradicate human pathogen21. Although the capability of a single cell is limited, cells can reproduce themselves exponentially and work simultaneously to solve complicated tasks or accomplish sophisticated tasks in principle. However, these capabilities of cells have not yet been well exploited, especially for HTS purpose.
Recently, concentrations of intracellular molecules have been used as signals for overexpression of fluorescence for screening purposes in the context of strain improvement8. For example, Binder et al. successfully used the intracellular concentration of lysine, a natural lysine-responsive transcriptional activator LysG and fused expression of eYFP to screen high lysine producer from C. glutamicum 7. Later, by using the same sensor for in vivo detection of the desired end-product in single cells, they established a screening method with FACS to screen for enzymes without allosteric inhibition. However, due to the complexity of metabolic pathways, one enhanced enzyme usually has limited effects on productivity of the end-product. Genetic modifications are required to enhance the signal of the end-product in their studies8. Esvelt et al. (2011) presented an interesting phage-assisted method for continuous evolution of a specific gene-coded biomolecule that is linked to the infectivity of the phage mediated by the expression of a specific protein in host cells22. Specifically, M13 filamentous bacteriophage carrying the molecule of interest was used to infect E. coli cells in a lagoon with continuous inflow and outflow of the host cells, where the evolving gene is transferred from host cell to host cell in a manner that is dependent on the activity of the molecule of interest. The method was demonstrated with the evolution of a T7 RNA polymerase for new binding properties. It was later on used to successfully evolve proteases with significantly increased drug resistance to protease inhibitor23–25.
Here, we propose to use the cell-phage interactions mediated by the intracellular concentration of a specific metabolite for parallel and highly sensitive screening of biomolecules for metabolic pathway optimization under in vivo conditions. The basic idea is to program the cells to perform a certain screening task which is linked to the desired property or activity of the molecule of interest. The latter is in turn linked to the infectivity of the phage. Compared to physical robots the biological robots have the decisive advantage of fast replication, resulting in a large pool for simultaneously screening under in vivo conditions. Thus, the screening throughput can be expanded simply by using a larger population of cells, indicating a massively parallel screening manner potentially far beyond the current HTS technologies. It is also worth to mention that the cost for such an approach is almost zero compared to methods based on expensive FACS or microcapillary arrays, making it applicable in almost all biological labs.
We demonstrated the concept by screening mutants of a protein with reduced allosteric inhibition. Allosteric regulation is one of the fundamental mechanisms that control almost all cellular metabolism and gene regulation26. Deregulation of allosteric inhibition is essential in designing and optimizing metabolic pathways for the production of target metabolites such as amino acids27. Aspartate kinase III (AK-III), encoded by lysC, catalyzes the phosphorylation of aspartate and controls the biosynthesis of several industrially important amino acids such as lysine, threonine, and methionine in E. coli 28. AK-III is allosterically inhibited by L-lysine strictly. AK-III was chosen in this work as a model enzyme because of our extensive previous work on the rational design of it27,29. The new approach is shown to be more sensitive than the widely used flow cytometry method owing to a novel way of signal capturing.
Results
Principle of programming cells as robots for screening
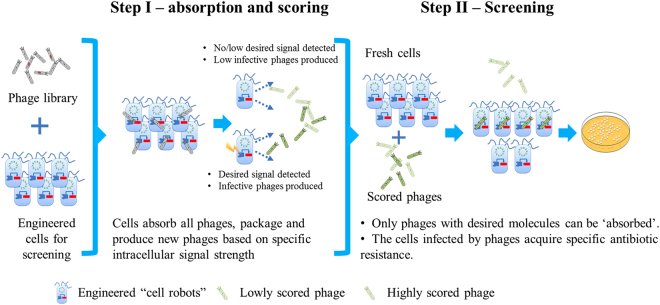
The workflow of programming cells as robots for the screening of molecule of interest (target) is shown in Fig. 1. Briefly, instead of placing the screening targets inside of the host cells as in most of the traditional screening methods, we place the targets to be screened on M13 phages. We then engineer the host cells so that they can screen for phages carrying the targets with desired properties. Specifically, we use E. coli XL1-Blue cells as the host cells for this purpose. To enable the host cells to control the infectivity of packaged phages, we transfer an essential gene for phage infectivity from the M13 phage to the host cells. The essential gene applied in this study is gene III encoding the attachment protein pIII which mediates adsorption of the phage to its primary receptor, the tip of E. coli F-pilus30. We then design an intracellular biological circuit to control the infectivity of packaged phages by controlling the expression level of gene III based on a specific intracellular signal that is related to the performance of the biomolecules to be screened, such as the concentration of an end product or an intermediate metabolite of a metabolic pathway. The targets are then cloned into VCSM13 by replacing the original gene III. A helper plasmid pJ175-Str which can supply the gene III product is used for preparing infective phage library at the first step of screening (see below). Elimination of gene III does not affect the phage secretion. However, the infectivity of the produced phages is very low. Thus, to enable an effective screening, we design a two-step strategy as illustrated in Fig. 1. In the first step, the phage library with the variants is ‘absorbed’ and ‘scored’ by the host cells based on the strength of the specific signal representing the performance of the target molecule. High-performance targets will produce more infectious phages than the low-performance ones. In the second step, the ‘scored’ phages are collected and screened in another run of cell–phage interaction. In this step, only infectious phages carrying the molecule with desired property can be ‘absorbed’. Since a kanamycin resistance gene (aph) is placed on the M13 phage, the cells capturing phages with desired properties can be selected easily by incubation under antibiotic pressure. In such a way, the target with the best performance under in vivo conditions can be effectively identified.
Figure 1.
Work flow of cell robot based screening. A two-step screening strategy is suggested. First, phages are absorbed by engineered cells and packaged (scored) based on the performance of the molecules carried by the phages. Only the phages carrying molecules with desired properties are packaged in an effective way. Second, the ‘scored’ phages are absorbed by fresh host cells. In this step, only the infective phages, i.e. phages carrying molecules with the desired properties, are ‘absorbed’ by the host cells. The cells infected by the phages with desired molecules/targets acquire kanamycin resistance and can be easily identified by cultivation under kanamycin stress.
Proof of concept of the method
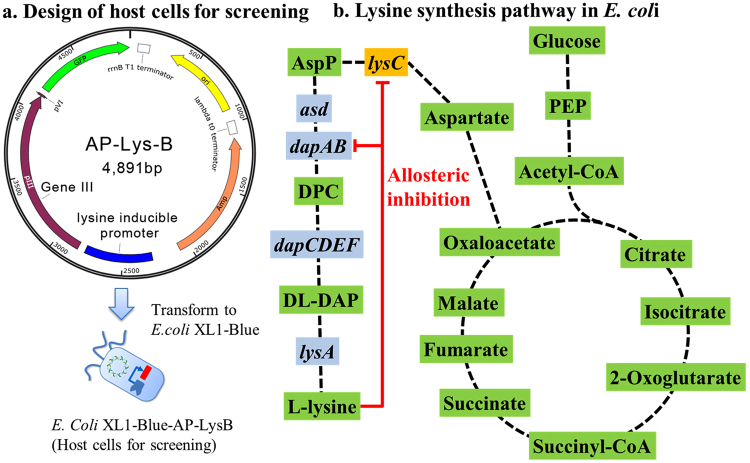
To use intracellular lysine concentration as a signal for the screening of an L-lysine-resistant aspartate kinase (AK-III, encoded by lysC gene in E. coli) based on the cell-phage interaction, a lysine inducible promoter was cloned from Corynebacterium glutamicum ATCC13032 as the lysine sensor31. The gene III from M13 phage was cloned into the plasmid AP-Lys-B and placed under the control of the lysine inducible promoter (Fig. 2a). The screening “cell robots”, namely E. coli XL-Blue-AP-Lys-B, were then obtained by transforming E. coli XL-Blue cells with the plasmid AP-Lys-B. In such a way, if a phage carrying a target gene that can increase the intracellular lysine concentration is absorbed by the E. coli cells, it will increase the gene III expression level, which will result in the production of infective phages. Thus, the desired targets which can increase intracellular lysine concentration can be identified by using the two-step screening strategy.
Figure 2.
Illustration of designed “cell robots” screening for targets which can increase intracellular lysine concentration. (a) Plasmid map of the biological device controlling the scoring process based on intracellular lysine concentration. A lysine inducible promoter was cloned from C. glutamicum ATCC13032 as lysine sensor. The gene III of M13 phage and a green fluorescence protein (GFP) encoding gene were placed under the control of the lysine inducible promoter. The GFP-encoding gene was used for comparing the sensitivity of the cell-phage based screening with the flow cytometry-based screening. It is not required for the cell robot based screening. (b) Biosynthesis pathway of lysine in E. coli. Green rectangles represent metabolites, light blue and yellow rectangles the names of related genes. In principle, the engineered host cells can be used to screen any enzymes in the lysine biosynthesis pathway for enhanced enzyme performance. In the proof of concept study, we focused on screening mutants of AK-III (encoded by the lysC gene) with reduced allosteric inhibition by lysine. PEP – phosphoenolpyruvate; AspP – L-aspartyl-4-phosphate; DPC – Tetrahydrodipicolinate; DL-DAP – D,L-diaminopimelate.
To experimentally demonstrate the functioning of the method, the wild-type lysC gene was first cloned from E. coli MG1655 to VCSM13 phage by replacing the Gene III. The obtained phagemid is named as M13-lysC. We then constructed another phagemid M13-lysC-V339A by introducing a site mutation to the lysC gene of M13-lysC. The V339A mutant of AK-III has been previously proven to be well resistant to allosteric inhibition by lysine27. We mixed roughly equal amount phage particles of M13-lysC and M13-lysC-V339A and screened the mixture of the phages using the designed host cells. If the screening robots function as expected, the phages of M13-lysC-V339A should be screened out. We repeated the experiments three times with the designed host cells using lysine as signals for screening. One time we used cells cultivated with LB medium and twice with cells cultivated with M9 medium, representing differential expression levels of lysine synthesis pathway genes under various conditions. To verify the genotypes of the resulting phages, plasmids extracted from six individual colonies were sequenced for each experiment. All colonies were verified to be M13-lysC-V339A in all three experiments, confirming a robust screening function of the designed “cell robots”.
In a further experiment, we screened a variant pool containing the wild-type AK-III, six AK-III mutants from previous study27 and a new mutant (R300C) obtained from screening a relatively small size of library with the present method (data not shown). In the first run of screening, we selected 12 colonies for sequencing which gave the following results: 5 for the variant V339A, 3 for the variant I337P, 2 for the variant S338L, and 1 for each of the variant T253R and H320A. In the second run using the phage mix from the 1st screening, we obtained only one colony of V339A. As shown in Fig. 3, the variant V339A has the highest activity and resistance against lysine, confirming the effectiveness of the two-step approach.
Figure 3.
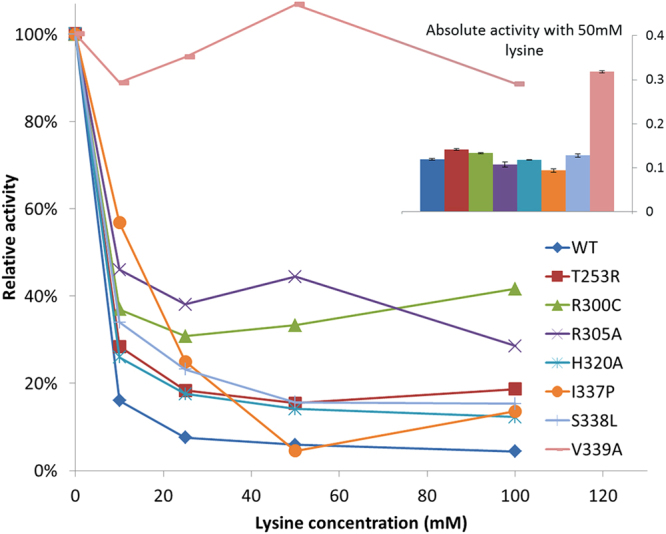
Inhibition profiles of wild-type and muteins of AK-III. In vitro enzyme assays were performed to characterize the inhibition profiles of wild-type and mutants of AK-III by lysine. The activities were displayed as relative activities normalized by the specific activities without lysine inhibition. The specific activities with 50 mM lysine presented by normalized absorbance by protein concentration are shown by the small histogram top-right. Data represent mean values and standard deviation from three assays.
Screening with a higher sensitivity than fluorescence-based method
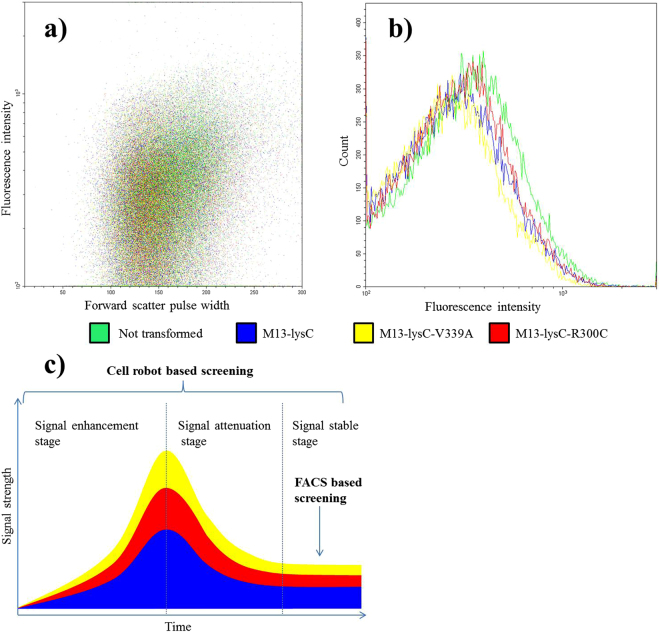
To compare the sensitivity of the current screening system with methods based on fluorescence and flow cytometry, the state-of-the-art screening technology, we transformed E. coli XL1-Blue/AP-Lys-B cells with M13-lysC, M13-lysC-V339A, and M13-lysC-R300C individually to obtain cells of XL1-Blue/AP-Lys-B/M13-lysC-WT, XL1-Blue/AP-Lys-B/M13-lysC-V339A, and XL1-Blue/AP-Lys-B/M13-lysC-R300C. A GFP-encoding gene was placed under the control of the same lysine sensing sensor in E. coli XL1-Blue/AP-Lys-B. Over-night cultivated cells of XL1-Blue/AP-Lys-B, XL1-Blue/AP-Lys-B/M13-lysC-WT, XL1-Blue/AP-Lys-B/M13-lysC-V339A and XL1-Blue/AP-Lys-B/M13-lysC-R300C were harvested and washed twice with 50 mM, ice-cooled PBS buffer. The fluorescence activities of the four different cell populations were measured using flow cytometry. As shown in Fig. 4b, although slight differences could be observed, it was not possible to set up a gain setting to select the mutants. In other words, the different cell populations cannot be distinguished by the flow cytometry method. On the other hand, our method based on cell-phage interaction can successfully screen out V339A as the best mutant of AK-III as shown in Fig. 3, confirming a higher sensitivity of the cell robot based screening method.
Figure 4.
Flow Cytometry assays of cell populations harboring wild-type AK-III and AK-III muteins. (a) Dot plot of flow cytometry assay results. (b) Statistic analysis of the total cell numbers at different fluorescence intensities. (c) Illustration of differences in signal capture of the cell-robot based and the FACS-based screening methods. Phagemids M13-lysC, M13-lysC-V339A and M13-lysC-R300C were transformed to E. coli XL1-Blue/AP-Lys-B cells individually. Flow Cytometry assays were performed on the obtained cell populations by measuring green fluorescence intensity. Although slight differences could be observed in figure (b), the flow cytometry method failed to distinguish the mutants despite varying the gain setting. Introduction of molecular variants into cells can be regarded as perturbations to the cells. Figure c illustrates possible change of signal after introduction of variants (perturbations): enhancement, attenuation and stabilization.
Biological systems are complex and highly adaptive, meaning that the cells always try to reduce the perturbations introduced. Introducing molecular variants into cells can be regarded as perturbations to the cells. As illustrated in Fig. 4c, after the introduction of molecular variants, the cells may undergo three stages of signal change: enhancement, attenuation and stabilization. The signal enhancement stage is the direct consequence of the perturbations induced by the introduced molecular variants. The signal attenuation stage is caused by adaptive response of cells to the perturbation. Finally, the signal reaches a stable state which might be slightly different from the state before the perturbation. The time interval for these changes may be relatively short. The curves in Fig. 4c are theoretical response patterns of cells to the disturbance by over-expression of the different AK-III variants respectively. For the FACS based method, the cells to be measured may have already reached the stable stage where the signal strength may not be significantly different in the cell populations with different variants. However, our method captures signals during the whole response and adaptation processes which correspond to the area below the curves and can be therefore more sensitive.
Furthermore, the “cell robots” based screening works in principle like an autocatalytic process of signal amplification: the target molecule with desired performance will increase the intracellular concentration of the signal molecule (in this case lysine) in the cell, the increased concentration of the signal molecule will amplify the population of phage carrying the target molecule. The amplified phages can infect other cells to further enhance the signal. In such a way the screening process is highly effective and sensitive compared to the other presently used methods, such as those based on single cells using fluorescence as readout signal8.
Discussion
In this study, we demonstrated that the biological ‘robots’, i.e. the cells, can be engineered to perform screening tasks in protein engineering. By capturing the signals during the whole response and adaptation processes, which cannot be achieved by screening based on electric machines, the cell-phage based screening system has an inherent higher sensitivity. The current proof of concept study shows that cell-phage interaction system does not require any genetic modifications of the host cells to enhance the signal for screening. In a recent similar work which used FACS as the screening method, genetic modifications are required to enhance the signal8. Furthermore, FACS based screening suffers often signal noise caused by single cell variations and signal detection under conditions of fast moving cells7,8,10. By equally accessing all cells, the cell-phage interaction system can avoid the problem of single cell variation in principle. Cells as biological ‘robots’ have a unique feature of reproducing themselves to generate a vast population exponentially and cheaply. Thus, the screening throughput can be expanded simply by using a larger population of cells with minimal additional costs, indicating a massively parallel screening manner beyond the current electric based machines. As proved by the power of parallel computing in computational science32, parallelization is a great solution for speeding up the process of parallel tasks. The sensitivity and throughput are key factors determining the success of a screening experiment. The cell-phage screening system shows clear advantages in both sensitivity and throughput. Furthermore, the cost of cell robots is almost zero compared to that of expensive electric machines/robots. It should be mentioned that, while electric machines can utilize various types of signals for screening, screening based on the cell robots uses a “biological signal and sensor”, which might represent a limitation in some cases. However, many natural or purposefully designed biological elements or sensors such as promoters and riboswitches can be used for this purpose33,34 and the signal molecules can be intermediates of metabolic pathways.
Materials and Methods
Strains, phages and plasmids
The M13 phage (VCSM13) was purchased from Agilent Technology (5301 Stevens Creek Blvd. Santa Clara, CA 95051, USA). The wild lysC gene encoding AK-III was amplified by PCR from the genomic DNA of E. coli K12 MG1655. For over-expression and purification of the wild-type AK-III and relevant muteins, the wild-type lysC gene was cloned to pET-22b(+) with the introduction of an additional His-tag at the C-terminal to generate the plasmid pET22-lysC. Site-mutagenesis was performed on pET22-lysC to generate over-expression plasmids for AK-III muteins. The lysC gene was also cloned to VCSM13 by replacing the original gene III to generate a phagemid M13-lysC. Similarly, site-mutagenesis was also performed on M13-lysC to generate phagemid derivations carrying different AK-III muteins.
For construction of plasmid AP-Lys-B, i.e. the device harnessed by the host cells to control the phage packaging process based on intracellular lysince concentration, we ultilized a lysine inducible promoter from Corynebacterium glutamicum ATCC13032 as a lysine sensor31. The lysine inducible promoter, gene III from M13 phage and a GFP-encoding gene were cloned into the plasmid pZE21MCS to obtain AP-Lys-B as shown in Fig. 2a. The transcriptional levels of gene III and GFP encoding gene are controlled by the lysine inducible promoter. The antibiotic resistance type of AP-Lys-B was changed to ampicillin resistance by replacing the kanamycin resistance gene with an ampicillin resistance gene. All used strains, plasmids in current study are listed in Table 1.
Table 1.
Strains, plasmids and primers used in this study.
| Strains/phages/plasmids/primers | Description/Genotype | Note |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | Agilent | |
| E. coli XL1-Blue/pJ175e | E. coli XL1-Blue harboring pJ175e | |
| E. coli XL1-Blue/AP-Lys-B | E. coli XL1-Blue harboring AP-Lys-B | |
| E. coli XL1-Blue/AP-Lys-B /M13-lysC | E. coli XL1-Blue harboring AP-Lys-B and M13-lysC | |
| E. coli XL1-Blue/AP-Lys-B /M13-lysC-V339A | E. coli XL1-Blue harboring AP-Lys-B and M13-lysC-V339A | |
| E. coli XL1-Blue/AP-Lys-B /M13-lysC-R300C | E. coli XL1-Blue harboring AP-Lys-B and M13-lysC-R300C | |
| Phages | ||
| VCSM13 | Kan | Agilent |
| M13-lysC | Derived from VCSM13 by replacing gene III with wild lysC from E. coli K12 | Plasmid map and full sequence are detailed in Supplementary information. |
| M13-lysC-T253R | Derived from M13-lysC by site mutagenesis | |
| M13-lysC-R300C | Obtained by screening with a library of M13-lysC generated by in vivo random mutagenesis | |
| M13-lysC-R305A | Derived from M13-lysC by site mutagenesis | |
| M13-lysC-H320A | Derived from M13-lysC by site mutagenesis | |
| M13-lysC-I337P | Derived from M13-lysC by site mutagenesis | |
| M13-lysC-S338L | Derived from M13-lysC by site mutagenesis | |
| M13-lysC-V339A | Derived from M13-lysC by site mutagenesis | |
| Plasmids | ||
| pJ175e | Amp, obtained from David Group | |
| AP-Lys-B | Amp, Derived from pZE21 plasmid; | Plasmid map and full sequence are detailed in Supplementary information. |
| pET22-lysC | Amp, Expression plasmid for wild-type AK-III | |
| pET22-lysC-T253R | Expression plasmid for T253R mutant of AK-III | |
| pET22-lysC-R300C | Expression plasmid for R300C mutant of AK-III | |
| pET22-lysC-R305A | Expression plasmid for R305A mutant of AK-III | |
| pET22-lysC-H320A | Expression plasmid for H320A mutant of AK-III | |
| pET22-lysC-I337P | Expression plasmid for I337P mutant of AK-III | |
| pET22-lysC-S338L | Expression plasmid for S338L mutant of AK-III | |
| pET22-lysC-V339A | Expression plasmid for V339A mutant of AK-III | |
| Primers | Description | Sequence (5′−3′) |
| M13Seq-G3-P1 | Sequencing primer | TCTGTAGCCGTTGCTACCCTCGTT |
| M13Seq-G3-P2 | Sequencing primer | AAGAAACAATGAAATAGCAATA |
| M13-ln4Genes-P1 | Primer for linearization of VCSM13 | CTAGTATTTCTCCTCTTTCTCTAGTATAATTGTATCGGTTTATCAGCTTGCT |
| M13-ln4Genes-P2 | Primer for linearization of VCSM13 | CTCCCTCAATCGGTTGAATGT |
| LysC-4M13-P1 | For cloning of lysC | GAGGAGAAATACTAGATGTCTGAAATTGTTGTCTCC |
| LysC-4M13-P2 | For cloning of lysC | AACCGATTGAGGGAGTTACTCAAACAAATTACTATG |
| V339A-P1 | Site-directed mutagenesis of lysC to generate V339A mutant | GCAGACTTAATCACCACGTCAGAA G |
| V339A-P2 | Site-directed mutagenesis of lysC to generate V339A mutant | CGAAATATTATGCCGCGCGAGGAT G |
| T253R-P1 | Site-directed mutagenesis of lysC to generate T253R mutant | CGTTTTGGTGCAAAAGTACTGC |
| T253R-P2 | Site-directed mutagenesis of lysC to generate T253R mutant | TGCCATCTCTGCCGCTTCGGCA |
| R305A-P1 | Site-directed mutagenesis of lysC to generate R305A mutant | TGCTCGCAATCAGACTCTGCTC |
| R305A-P2 | Site-directed mutagenesis of lysC to generate R305A mutant | AGCGCCAGAGCGCGGAACAGCG |
| H320A-P1 | Site-directed mutagenesis of lysC to generate H320A mutant | TTCTCGCGGTTTCCTCGCGGAA |
| H320A-P2 | Site-directed mutagenesis of lysC to generate H320A mutant | GCCAGCATATTCAGGCTGTGCA |
| I337P-P1 | Site-directed mutagenesis of lysC to generate I337P mutant | CTTCGGTAGACTTAATCACCAC |
| I337P-P2 | Site-directed mutagenesis of lysC to generate I337P mutant | GATTATGCCGCGCGAGGATGCC |
| S338L-P1 | Site-directed mutagenesis of lysC to generate S338L mutant | TGGTAGACTTAATCACCACGTC |
| S338L-P2 | Site-directed mutagenesis of lysC to generate S338L mutant | AAATATTATGCCGCGCGAGGAT |
| R300C-P1 | Site-directed mutagenesis of lysC to generate R300C mutant | TGCGCTCTGGCGCTTCGTCGCAATC |
| R300C-P2 | Site-directed mutagenesis of lysC to generate R300C mutant | GAACAGCGGCGGATTTTCAGTTTTA |
Molecular cloning
All PCR experiments for cloning were performed using Thermo Scientific PCR Master Mix or CloneAmp HiFi PCR Premix. Cloning experiments were performed using In-Fusion HD Cloning Plus. Site-mutagenesis was performed using a protocol similar to the NEB Q5® Site-Directed Mutagenesis Kit. Briefly, none overlap primers were designed and synthesized which contain the desired mutations. Then PCR amplification was performed with the designed primers using the original plasmid as templates to generate linear plasmids. Template DNA was eliminated by enzymatic digestion with DpnI. Finally, phosphorylation and ligation using T4 Polynucleotide Kinase and T4 Ligase were carried out to obtain circular DNA before transformation.
Mutagenesis
Random in vivo mutagenesis was enabled by using the plasmid pJ184-Str harboring genes which can increase intracellular DNA replication error rates. The plasmid pJ184-Str was derived from pJ184 by replacing the chloramphenicol acetyltransferase encoding gene with a streptomycin resistance gene. The pJ184 plasmid which has been described previously was obtained from David R. Liu’s group of Harvard Medical School22.
Preparation of infective phages for screening
Since the engineered phages lack gene III, the helper plasmid pJ175e was harnessed by the host cells to supply gene III products intracellularly to obtain infective phages. The plasmid pJ175e was obtained from David R. Liu’s group. Specifically, engineered phages were co-transformed with pJ175e into XL1-Blue cells. Overnight cultures were deposited for centrifuge and the supernatant containing the packaged infective phages was collected.
Screening based on cell robots
XL1-Blue/AP-Lys-B cells, i.e. the “cell robots”, were incubated in LB medium to an OD600 value around 1.0. Roughly 200ul XL1-Blue/AP-Lys-B cells were mixed with 2ul proper diluted phages (Cells to phage number ratio above 10:1 to make sure that all phages could be captured and evaluated by host cells. Different types of phages in a total number of roughly 10,000 were used as inputs in the present study). The mixture was incubated at 37 °C for 15 minutes without shaking to allow the phages to attach to the cells, following by incubation at 37 °C with shaking for 1 to 2 hours. Inactivate the host cells at 65 °C for 15 min. The cell debris were spinned down and the supernatant containing the “scored” phages was transferred to a fresh tube. A proper amount of “scored” phages were mixed with fresh XL1-Blue/AP-Lys-B cells and incubated at 37 °C for 15 minutes without shaking to allow the “cell robots” to absorb the highly “scored” phages. A proper amount of the culture was then sprayed on LB agar plates with kanamycin (50 mg/ml) for screening.
Protein purification and enzyme activity assay
Enzymes were expressed in E. coli BL21 (DE3) cells (New England Biolabs, Germany) using pET derived plasmids. The recombinant cells were first cultivated in LB media supplemented with 50 μg/mL kanamycin at 37 °C to reach an OD600 of 0.6 and then protein expression was induced by adding 0.1 mM isopropyl β-D-thiogalactopyranoside (IPTG) for overnight at 30 °C. The harvested cells were washed twice with 20 mM Tris-HCl buffer (pH 7.0) and suspended in a buffer of 50 mM Na2HPO4 (pH 7.0), 0.2 mM EDTA and 0.1 mM dithiothreitol. Suspended cells were disrupted and centrifuged at 100,000 g for 1 h. The crude enzymes, e.g. the supernatant, was purified using a Ni2 +-NTA column (GE Healthcare Bio-Sciences, Piscataway, NJ) to obtain samples for activity assay.
The aspartokinase activities of AK-III and muteins were assayed using the hydroxamate method35. In details, 1 ml reaction mixture contained 200 mM Tris–HCl (pH 7.5), 10 mM MgSO4·6H2O, 10 mM aspartate, 10 mM ATP, 160 mM NH2OH·HCl and appropriate amounts of enzyme. After incubation at 30 °C for 30 min, the reaction was stopped by mixing with 1 ml 5% (w/v) FeCl3 solution and the absorbance at 540 nm was monitored.
Flow cytometry analysis
Overnight cultured cells were washed and re-suspended in ice-cooled 50 mM PBS buffer and diluted to a cell density of approximately 3 × 106 cells/ml. Then, cellular GFP was analyzed by using flow cytometry (Beckman Coulter CytoFlex) with excitation at 488 nm and detecting fluorescence at 529 ± 14 nm. For each sample, 50,000 events were recorded. Data were analyzed using the CytExpert software.
Data availability statement
All data are provided in full in the results section of this paper.
Electronic supplementary material
Plasmid map and full sequences of M13-lysC and AP-Lys-B
Author Contributions
L.F.S. proposed the original idea, performed the studies and drafted the manuscript. A.P.Z. supervised the studies and improved the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15621-0.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bornscheuer UT. Protein engineering: Beating the odds. Nature chemical biology. 2016;12:54–55. doi: 10.1038/nchembio.1989. [DOI] [PubMed] [Google Scholar]
- 2.Chen B, et al. High-throughput analysis and protein engineering using microcapillary arrays. Nature chemical biology. 2016;12:76–81. doi: 10.1038/nchembio.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colin P-Y, et al. Ultrahigh-throughput discovery of promiscuous enzymes by picodroplet functional metagenomics. Nature communications. 2015;6:10008. doi: 10.1038/ncomms10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colin P-Y, Zinchenko A, Hollfelder F. Enzyme engineering in biomimetic compartments. Current opinion in structural biology. 2015;33:42–51. doi: 10.1016/j.sbi.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Aghaeepour N, et al. Critical assessment of automated flow cytometry data analysis techniques. Nature methods. 2013;10:228–238. doi: 10.1038/nmeth.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold, L. W. & Lannigan, J. Practical issues in high-speed cell sorting. Current protocols in cytometry/editorial board, J. Paul Robinson, managing editor[et al.] Chapter 1, Unit 1.24.1–30 (2010). [DOI] [PubMed]
- 7.Binder S, et al. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome biology. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schendzielorz G, et al. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS synthetic biology. 2014;3:21–29. doi: 10.1021/sb400059y. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Y. et al. Evolving the L-lysine high-producing strain of Escherichia coli using a newly developed high-throughput screening method. Journal of industrial microbiology & biotechnology (2016). [DOI] [PMC free article] [PubMed]
- 10.Young JW, et al. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. Nature protocols. 2011;7:80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ro D-K, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 12.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 13.Ahn JH, Jang Y-S, Lee SY. Production of succinic acid by metabolically engineered microorganisms. Current opinion in biotechnology. 2016;42:54–66. doi: 10.1016/j.copbio.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Choi SY, et al. One-step fermentative production of poly(lactate-co-glycolate) from carbohydrates in Escherichia coli. Nature biotechnology. 2016;34:435–440. doi: 10.1038/nbt.3485. [DOI] [PubMed] [Google Scholar]
- 15.Park TJ, Lee KG, Lee SY. Advances in microbial biosynthesis of metal nanoparticles. Applied microbiology and biotechnology. 2016;100:521–534. doi: 10.1007/s00253-015-6904-7. [DOI] [PubMed] [Google Scholar]
- 16.Park SH, et al. Metabolic engineering of Corynebacterium glutamicum for L-arginine production. Nature communications. 2014;5:4618. doi: 10.1038/ncomms5618. [DOI] [PubMed] [Google Scholar]
- 17.Liao JC, Mi L, Pontrelli S, Luo S. Fuelling the future: microbial engineering for the production of sustainable biofuels. Nature reviews. Microbiology. 2016;14:288–304. doi: 10.1038/nrmicro.2016.32. [DOI] [PubMed] [Google Scholar]
- 18.Bommareddy RR, Chen Z, Rappert S, Zeng A-P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metabolic engineering. 2014;25:30–37. doi: 10.1016/j.ymben.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Lee JH, Mitchell RJ, Kim BC, Cullen DC, Gu MB. A cell array biosensor for environmental toxicity analysis. Biosensors & bioelectronics. 2005;21:500–507. doi: 10.1016/j.bios.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeidi N, et al. Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular systems biology. 2011;7:521. doi: 10.1038/msb.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472:499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickinson BC, Packer MS, Badran AH, Liu DR. A system for the continuous directed evolution of proteases rapidly reveals drug-resistance mutations. Nature communications. 2014;5:5352. doi: 10.1038/ncomms6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badran AH, et al. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature. 2016;533:58–63. doi: 10.1038/nature17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson JC, Badran AH, Guggiana-Nilo DA, Liu DR. Negative selection and stringency modulation in phage-assisted continuous evolution. Nature chemical biology. 2014;10:216–222. doi: 10.1038/nchembio.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai C-J, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Molecular bioSystems. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, Rappert S, Sun J, Zeng A-P. Integrating molecular dynamics and co-evolutionary analysis for reliable target prediction and deregulation of the allosteric inhibition of aspartokinase for amino acid production. Journal of biotechnology. 2011;154:248–254. doi: 10.1016/j.jbiotec.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Theze J, Margarita D, Cohen GN, Borne F, Patte JC. Mapping of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. Journal of bacteriology. 1974;117:133–143. doi: 10.1128/jb.117.1.133-143.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma C-W, Xiu Z-L, Zeng A-P. A new concept to reveal protein dynamics based on energy dissipation. PloS one. 2011;6:e26453. doi: 10.1371/journal.pone.0026453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gailus V, Ramsperger U, Johner C, Kramer H, Rasched I. The role of the adsorption complex in the termination of filamentous phage assembly. Research in microbiology. 1994;145:699–709. doi: 10.1016/0923-2508(94)90042-6. [DOI] [PubMed] [Google Scholar]
- 31.Park, Y. H. et al. Nouveau Promoteur Inductible Par L-Lysine. Kr20040117104 (2005), C12n15/31.
- 32.Owens JD, et al. GPU Computing. Proceedings of the IEEE. 2008;96:879–899. doi: 10.1109/JPROC.2008.917757. [DOI] [Google Scholar]
- 33.Deaner, M. & Alper, H. S. Promoter and Terminator Discovery and Engineering. Advances in biochemical engineering/biotechnology (2016). [DOI] [PubMed]
- 34.Ma, C.-W., Zhou, L.-B. & Zeng, A.-P. Engineering Biomolecular Switches for Dynamic MetabolicControl. Advances in biochemical engineering/biotechnology (2016). (In print). [DOI] [PubMed]
- 35.BLACK S, WRIGHT NG. Beta-Aspartokinase and beta-aspartyl phosphate. The Journal of biological chemistry. 1955;213:27–38. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plasmid map and full sequences of M13-lysC and AP-Lys-B
Data Availability Statement
All data are provided in full in the results section of this paper.