ToC figure
Keywords: graphene oxide, circulating tumor cell, thermoresponsive polymer, breast cancer, pancreatic cancer
With over 1600 people dying of cancer in the United States every day,[1] the prevention of the second leading cause of death is a clear area of research interest in the medical community. The spread of tumor cells to distant locations in the body, or metastasis, is the cause of 90% of cancer related deaths,[2] presenting an impetus for the study of those cells most responsible for cancer mortality. Circulating tumor cells (CTCs) are those cells shed from the primary tumor into the blood circulation, potentially en route to forming a secondary tumor, and are present at the incredibly low frequency of on the order of one in one billion normal blood cells in the peripheral blood of cancer patients.[3] CTCs can not only provide biological insight into primary and metastatic tumors but also have the potential to serve as real time biomarkers for making treatment decisions and monitoring drug efficacy.[4] Indeed, over 270 clinical trials have now been proposed using CTCs as surrogate biomarkers.[5] However, to date, CTCs have not been incorporated into clinical practice for management of patients with cancer. The main challenges to this field include: (i) reaching the sensitivity needed to isolate these extremely rare cells from the surrounding blood cells (1 in 1 billion), (ii) minimizing processing to preserve the viability of cells, and (iii) achieving the specificity necessary to acquire pure population to enable meaningful genomic and functional analysis.
Microfluidic technologies have emerged as a solution to isolate live CTCs from small amounts of blood collected from cancer patients. A common separation technique involves immunocapture, the tethering of an antibody against a CTC-specific marker to a surface or structure to bind CTCs but not the normal blood cells. Functionalized microposts have been used in a number of CTC isolation devices.[6–9] Antibody-functionalized silicon microposts for CTC capture were used in the first microfluidic device designed for this purpose, the CTC Chip.[6] Subsequent microfluidic CTC capture devices also featured microfeatures coated with antibodies, such as the geometrically enhanced differential immunocapture (GEDI) chip,[7] chaotic micromixer HB CTC Chip,[10] high throughput microsampling unit (HTMSU)[11] and the HD-CTC module of an integrated system.[12] These immunocapture devices included features fabricated from polymers such as polydimethylsiloxane (PDMS), poly(methyl methacrylate) (PMMA), and cyclic olefin copolymer (COC). In order to push the field to realize the opportunities afforded by these cells, which may be captured at early stage as well as mid-metastasis, orthogonal techniques and materials would be necessary to enhance the sensitivity. Nanomaterials provide one such avenue, with advantageous properties such as a high surface area to volume ratio and a length scale on the order of magnitude of extracellular features. Many different classes of nanomaterials have been incorporated into CTC research.[8, 13] One example, graphene oxide (GO) has a number of proven biomedical applications.[14] We have recently developed a graphene oxide (GO) based device, the GO Chip, that took advantage of the increased surface area afforded by graphene oxide for highly sensitive and selective cell capture.[15] Using this method, we were able to demonstrate the capture of CTCs from peripheral blood samples from pancreatic, breast, and early stage lung cancer patients with low white blood cell contamination. However, this device shares the common drawback across most immunoaffinity based technologies reliant on antibodies attached to a surface: the limitation of post-capture analysis because of difficulty in releasing viable cells from the capture substrate.
Thermoresponsive polymers, a class of stimuli-responsive polymers that respond to temperature changes by undergoing conformational changes, have found wide applications in drug delivery,[16] tissue engineering,[17] controlling cell adhesion[18] and bacterial growth,[19] protein encapsulation,[20] and the release of captured CTCs from the surface of such capturing devices.[21, 22] Alternative CTC release techniques take advantage of alginate hydrogel[23, 24] or layer-by-layer assembled[25] degradable capture substrates. However, these approaches all feature performance limitations in throughput,[24] purity requiring additional processing,[26] ability to process blood collected by standard conditions,[23] immense fabrication facility requirements,[21, 26] time-consuming chemistry,[27] and inconvenient experimental temperature conditions.[27]
Graphene- and graphene oxide (GO)-based polymer composites are a new class of materials which combine the excellent properties of graphene, such as high surface-to-volume ratio, high Young’s modulus, and high thermal and electrical characteristics,[28] with the easy processability of polymers. Such composites have found uses in fields ranging from energy storage[29] and electronic devices,[30] to biomedical applications such as drug and gene delivery,[31, 32] cancer therapy,[33] cell differentiation,[34] coating of biomedical implants,[35] and bio-imaging.[32]
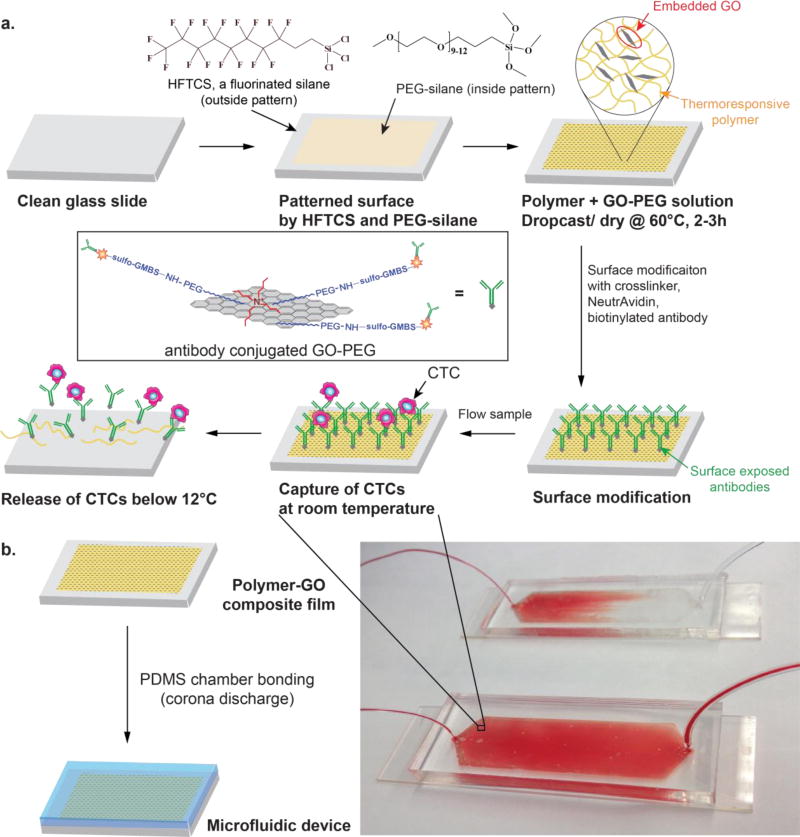
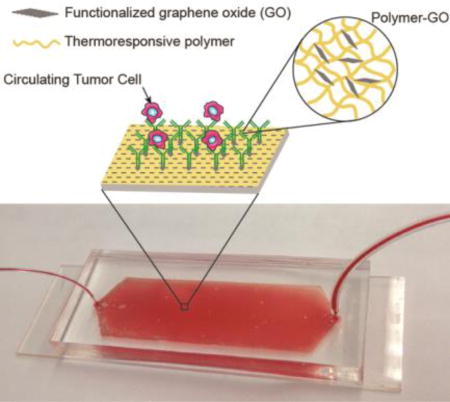
We hypothesized that the combined advantages of a biocompatible functionalized nanomaterial with a thermoresponsive polymer that promotes effective cell release could address the challenge of sensitive capture while simultaneously allowing viable cell release. This could lead to improvement in downstream analysis such as fluorescence in situ hybridization (FISH), molecular analysis, and single cell analysis. We present a new tunable thermal-sensitive polymer-GO Chip for highly efficient capture and subsequent release of CTCs incorporated into a microfluidic device (Figure 1a).
Figure 1.
a) Schematic concept of a polymer-GO microfluidic device for the capture/release of CTCs. b) Enclosure within polydimethylsiloxane chamber and photograph of patient blood samples being processed by the polymer-GO devices.
In the current work, the microfluidic device bottom substrate was coated with a composite film of functionalized GO dispersed in a matrix of thermoresponsive polymer with a lower critical solution temperature (LCST) of 13°C. Surface available functionalized GO (described below) provided anchors for attaching the CTC capture antibody while the polymer matrix provided temperature dependent modulation of capture or release functionality. The microfluidic assembly facilitated the processing of patient blood samples within a simple planar device (Figure 1b). Drop-casting the polymer-GO blend on a patterned and surface modified substrate made such a device cheap and easy to fabricate. Moreover, the LCST of around 13°C for the polymer matrix made it possible to use the device at room temperature as opposed to higher temperatures, [26] such that there are no concerns about inadvertently releasing the cells during the capture step. Additionally, cell release occurred under gentle conditions, maximizing the viability of released cells. The consolidation of the advantageous properties of GO-based capture with superior release functionality of the chosen polymer yielded a device that enables the study of these clinically interesting cells without many of the shortcomings of past technologies (Supplementary Table 3), while simultaneously presenting an easy, scalable fabrication method.
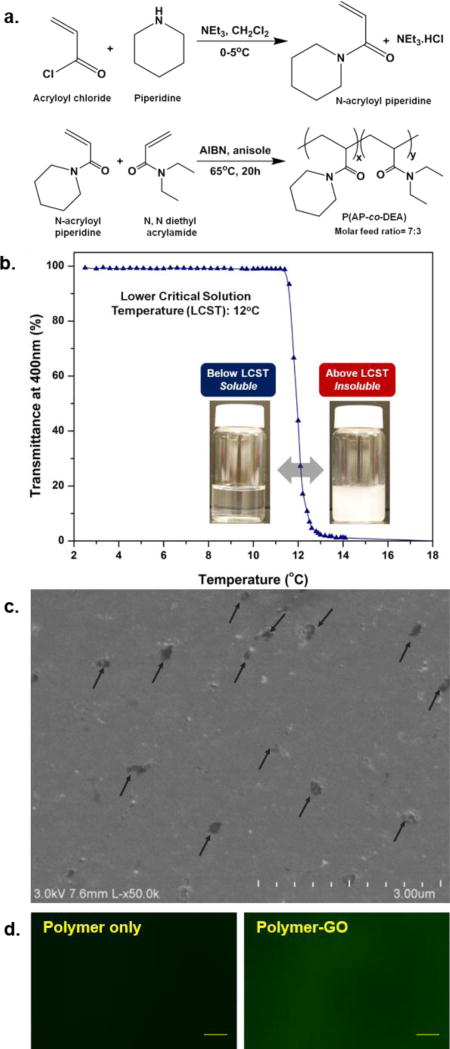
To create a tunable thermal responsive polymer, copolymer poly(N-acryloyl piperidine-co-N,N-diethyl acrylamide) was synthesized via free radical polymerization using AIBN as an initiator and was characterized for its molecular weight and LCST (Figure 2a). LCST was modulated by employing a copolymerization technique using two acrylamide monomers with different degrees of hydrophobicity: N-acryloyl piperidine (AP) and N,N-diethyl acrylamide (DEA). The homopolymers poly(N-acryloyl piperidine) (PAP) and poly(N,N-diethyl acrylamide) (PDEA) have LCSTs of 4°C and 25°C respectively.[36] The required capture/release modulation temperature for the CTC device can be achieved by changing the ratio of the two monomers in the copolymer. For example, a copolymer synthesized with 7:3 molar ratio of AP:DEA showed a critical temperature of around 12–13°C, which was used in this study (Figure 2b). The detailed procedure of polymer synthesis and characterization can be found in the Supporting Information. GO nanosheets were functionalized with phospholipid-polyethylene-glyco-amine according to an earlier reported method.[15] The polymer-GO nanocomposite films were prepared through drop-casting a DMF solution of polymer and functionalized GO. The drop-cast films were dried at 60°C in oven for 2–3 hours to yield 3–4 µm thick composite film. Figure 2c shows the SEM image of polymer-GO composite surface. The polymer-GO microfluidic devices for cell capture and release were fabricated in two steps (Figure S1). Poly(ethylene glycol) (PEG) is well known to render surfaces non-fouling.[37] The PEG monolayer was necessitated to avoid recapturing of the released CTCs on the glass substrate. In the first step, the polymer-GO composite film was deposited on a patterned and surface modified glass substrate followed by assembly with a PDMS chamber to form a microfluidic device. In the second step, the device was functionalized by immobilizing anti-EpCAM on the surface available GO through a cross-linker (N-γ-maleimidobutyryl-oxysuccinimide ester, sulfo-GMBS) and avidin-biotin mediated bio-conjugation, providing cell capture/release functionality (Figure S2).
Figure 2.
a) Synthetic scheme for copolymer. b) UV-vis transmittance vs temperature plots for the copolymer showing LCST of ~12°C. c) SEM image of polymer-GO composite surface. Arrows indicate suspended GO present on the surface of the film. d) Fluorescence images of polymer-only and polymer-GO films. The films were incubated with an amine-reactive dye (FSE, 0.25mM aq. solution) for 30 minutes at 40°C. Scale bar: 20.0 µm.
To show the surface availability of the amine groups from the GO-PEG in polymer-GO composite films, the drop-cast films were incubated with 0.25mM aqueous solution of an amine reactive dye, FSE (5-(and-6)-carboxyfluorescein, succinimidyl ester; Life Technologies (C1311)) for 30 minutes at 40°C and then washed with copious amount of DI water. The dye treated films were then imaged with fluorescence microscope (Olympus BX51 coupled with Olympus DP71 camera and EXFO X-cite Series 120 light source). While polymer-GO composite films showed bright green fluorescence from the surface tethered dye, polymer-only films showed very low to no fluorescence (Figure 2d). Though the possibility of physically adsorbed dye molecules cannot be completely ruled out, it is most likely that the dye molecules were primarily tethered to the surface through covalent bonding between the amine groups on film surface and succinimidyl ester groups on the dye, as suggested by large contrast in fluorescence intensity from polymer-GO and polymer-only films. Time dependence of dissolution of polymer-GO composite films in cold water was also determined. Dye treated films were dipped in cold water for different lengths of time and the fluorescence images before and after dipping were compared. Films were dipped in cold water (5°C) for 5, 10, 20, and 30 minutes, and in room temperature water (20°C) for 30 minutes. The beakers with the dipped films were kept on an orbital shaker to weakly simulate conditions in microfluidic devices where the films are subjected to shearing by the flowing fluids. Figure S3 shows the fluorescent images of the films before and after dipping in water at 5°C for different time durations. While the film was completely dissolved and washed off in 20 to 30 minutes under cold condition as evident from gradual disappearance of green fluorescence, it remains stable and intact at room temperature even after 30 minutes. It is to be noted that in the actual device, the dissolution time is much shorter at 10 minutes, most likely due to the shear of the constant flow rate.
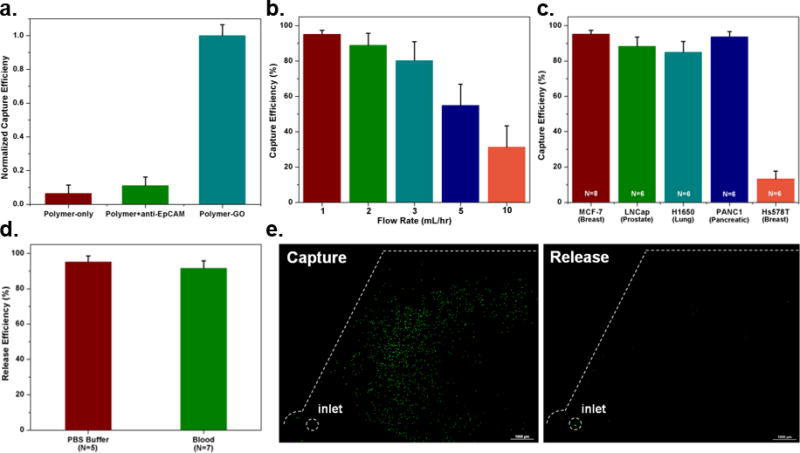
To verify the steps of the conjugation chemistry, experiments were performed to compare capture by (1) a polymer film lacking GO alone; (2) a polymer film lacking GO with the addition of anti-EpCAM; and (3) the polymer-GO film with full conjugation chemistry. The two control films showed significantly lower levels of capture with the polymer film and the polymer film with antibody capturing at 6.4% and 11.0% the level of the full chemistry, respectively (Figure 3a), with the increase in capture of the polymer with antibody condition a likely a result of physically adsorbed anti-EpCAM. This also suggests that very little of the capture antibody on the fully functional device is non-specifically bound.
Figure 3.
a) Capture efficiency of microfluidic devices featuring only the thermo-sensitive polymer, the thermo-sensitive polymer and non-specifically bound anti-EpCAM, and the polymer-GO film with specific conjugation chemistry as normalized by this last condition. b) Cell capture efficiency of the microfluidic polymer-GO device at various flow rates evaluated using a breast cancer cell line (MCF-7). Error bars show standard deviations (n=6). c) Capture efficiency of cell lines of varying origin and EpCAM expression levels. MCF-7 (n=8), PANC1, H1650, LNCaP, Hs578T (n=6). d) Release efficiency of the microfluidic polymer-GO device (MCF-7 cells were spiked into 1 mL of buffer or blood). e) Fluorescence microscope images of devices after capture and release of fluorescently-labeled MCF-7 cells.
To test the performance of the GO-polymer device for CTC capture, fluorescence labeled human breast cancer cell lines MCF-7 cells (1,000 cells/mL) were spiked into buffer and flowed through the GO-polymer device at different flow rates (1–10 mL/hr). The captured cells in the device and the non-captured cells collected in the waste were then counted. As expected, the capture efficiency decreased with flow rate. We observed that the efficiency rapidly decreased at flow rates ≥ 5mL/hr. In the 1–3 mL/hr range, the average capture efficiency was over 88.2% (n = 6 at each flow rate) (Figure 3b) with the highest capture of 95.21% at 1mL/hr. To further investigate the effect of tumor type and EpCAM expression on capture efficiency, three high EpCAM expressing cell lines for various cancer types (MCF-7 breast cancer cells, LNCaP prostate cancer cells, and H1650 lung cancer cells), one low EpCAM expressing cancer-cell line (Panc-1 pancreatic cancer cells), and one EpCAM negative cancer cell lines (Hs578T breast cancer cells) were selected for capture experiments at the flow rate of 1 mL/hr. The cells were fluorescently labeled and spiked into buffer at a concentration of 1000 cells/ml. The results in Figure 3c indicate that the anti-EpCAM-coated GO-polymer device achieved high capture efficiency (84.93–95.21%) for EpCAM-positive cancer cells. In contrast, a relatively low number of EpCAM-negative cells (Hs578T) were captured. Furthermore, the device is comparably effective in capturing different tumor cells, indicating the robust sensitivity of the device. After capturing cells on the devices, cell release experiments were carried out by flowing 1 mL PBS through the devices in a room maintained at 5°C at 100 µL/min (Figure 3e). Quantification of the cells in the devices before and after release showed an average cell release of 95.21% and 91.56% in buffer and blood experiments, respectively (Figure 3d). Furthermore, we tested the viability of the released cells by live dead assay. 91.68% of cells remained viable after release (Table S2).
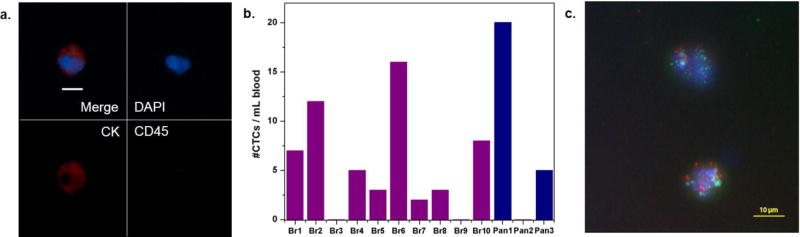
To demonstrate the CTC capture and release in clincial samples using the tunable polymer-GO composite film based device, we processed blood samples obtained from 10 metastatic breast cancer patients and 3 pancreatic cancer patients. Whole blood samples collected into EDTA tubes were processed at a flow rate of 1 mL/hr. Following a washing step, cells were released from the chip and deposited/spun onto glass slides by a cytospin centrifuge. CTCs in these samples were identified as DAPI-positive (shown in blue) nucleated cells staining positive for tumor markers (cytokeratin 7/8, visualized with a secondary antibody tagged with Alexa Fluor 546, shown in red) and negative for leukocyte markers (CD45, visualized with a secondary antibody tagged with Alexa Fluor 488, shown in green) (Figure 4a, b). CTCs were successfully recovered from 8 breast cancer patient samples and 2 pancreatic cancer patients (ranging from 2 to 20 CTCs/mL) (Figure 4c). The average number of CTCs recovered from breast samples was 5.6 CTCs/mL and from pancreatic samples was 8.3 CTCs/mL.
Figure 4.
a) Fluorescence images of CTCs from breast cancer patient sample. Nucleated cells (blue) staining positive for cytokeratin 7/8 (red) and negative for the white blood cell marker CD45 (green) were enumerated as CTCs. Scale bar = 10 µm. b) CTC enumeration results from 10 breast cancer patients and 3 pancreatic cancer patients. c) Fluorescence in situ hybridization (FISH) image of CTCs of breast cancer patient sample Br10. HER2(green)/centromere 17 probe(red).
Released CTCs were viable and structurally intact, and hence could be readily investigated via standard clinical cytopathological and genetic testing. Here we examined the feasibility of detecting HER2 amplification by fluorescence in situ hybridization (FISH). CTCs released from the chip were subsequently made into “cell blocks” by first fixing them with ethanol and then embedding them in Histogel (Thermo Scientific). Blocks were then formalin fixed and stored in 70% ethanol until slide preparation. Blocks were paraffin embedded and sectioned at the University of Michigan Histology Core. FISH was conducted using probes for HER2 (BAC clone RP11-94L15) and chromosome 17 control probe (BAC clone RP11-100E5), revealing HER2 amplification in one breast cancer patient (Figure 4c). FISH hybridization and image capture were performed essentially as previously described.[38] One green signal indicates the presence of one copy of HER2, while one red signals indicates one copy of centromere 17 probe; the multiple green signals in the figure imply HER2 amplification.
The downstream analysis facilitated by the efficient release of captured cells highlights the potential for this device’s use in basic and clinical cancer investigation. Through the incorporation of a composite that combines the advantages of a temperature-sensitive modality and sensitive nanomaterial-enabled capture, the polymer-GO film that serves as the basis of this technology overcomes some of the key shortcomings of previous CTC capture technologies (Table S3). As evidenced by data obtained from physiologic solutions containing spiked labelled cancer cells from multiple cancers and the processing of primary breast and pancreatic cancer patient blood samples, isolation of these rare cells with this device is highly feasible, completing the first step to unlocking the research opportunities presented by CTCs.
Compared with other CTC isolation strategies, immunoaffinity based technologies harvest CTCs with high sensitivity and purity,[39] but has the drawback of tethering cells within the device. Overcoming this limitation, we are able to collect viable and intact CTCs in suspension after immunocapture, making it ideal for various downstream analyses that require the high integrity and purity of the targeted cell population, such as genotyping and single cell profiling. This advanced analysis of CTCs could become a ‘real-time’ indicator to develop personalized therapy, as well as to bring valuable insights into the mechanism underlying cancer metastasis. Due to the low cost and ease of fabrication, this technology is scalable for commercialization. Future study will optimize it for large-scale clinical study and investigate its clinical utility for therapeutic marker discovery, treatment selection, and management.
Supplementary Material
Acknowledgments
The authors thank Mary Owczarczak and G Su Park for technical assistance in experiments. This work was supported by the National Institutes of Health (NIH) Director’s New Innovator Award (1DP2OD006672-01) to S. Nagrath. Financial support from Converging Research Center Program funded by the Ministry of Science, ICT and Future Planning (Project No. NRF-2014M3C1A8048791) was awarded to J. Kim. This material is based in part upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE 1256260 to M. Kozminsky. The work was performed in part at the Lurie Nanofabrication Facility, a member of the National Nanotechnology Infrastructure Network, which is supported by the National Science Foundation.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Prof. Hyeun Joong Yoon, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Department of Electrical Engineering and Computer Science, South Dakota State University Brookings, SD 57007, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA; Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
Apoorv Shanker, Macromolecular Science and Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA.
Yang Wang, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA; Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
Molly Kozminsky, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA; Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
Qu Jin, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA.
Dr. Nallasivam Palanisamy, Henry Ford Health System, 1 Ford Place, Detroit, MI 48202, USA
Monika L. Burness, Department of Internal Medicine, University of Michigan Comprehensive Cancer Center, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA
Dr. Ebrahim Azizi, Department of Internal Medicine, University of Michigan Comprehensive Cancer Center, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA
Prof. Diane M. Simeone, Department of Internal Medicine, University of Michigan Comprehensive Cancer Center, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
Prof. Max S. Wicha, Department of Internal Medicine, University of Michigan Comprehensive Cancer Center, 1500 E. Medical Center Drive, Ann Arbor, MI 48109, USA Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA; Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
Prof. Diane M. Kim, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Macromolecular Science and Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Department of Material Science and Engineering, Department of Biomedical Engineering, Department of Chemistry, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA.
Prof. Sunitha Nagrath, Department of Chemical Engineering, University of Michigan, 2300 Hayward Street, Ann Arbor, MI 48109, USA; Biointerfaces Institute, University of Michigan, 2800 Plymouth Road, Ann Arbor, MI 48109, USA; Translational Oncology Program, University of Michigan Health System, 1600 Huron Pkwy, Ann Arbor, MI 48109, USA.
References
- 1.Brongersma ML, Halas NJ, Nordlander P. Nat Nano. 2015;10:25. doi: 10.1038/nnano.2014.311. [DOI] [PubMed] [Google Scholar]
- 2.Gupta GP, Massagué J. Cell. 2006;127:679. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Maheswaran S, Haber DA. Current opinion in genetics & development. 2010;20:96. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paterlini-Brechot P, Benali NL. Cancer Letters. 2007;253:180. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 5.Alix-Panabieres C, Pantel K. Nat Rev Cancer. 2014;14:623. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 6.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleghorn JP, Pratt ED, Denning D, Liu H, Bander NH, Tagawa ST, Nanus DM, Giannakakou PA, Kirby BJ. Lab on a chip. 2010;10:27. doi: 10.1039/b917959c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon HJ, Kozminsky M, Nagrath S. ACS nano. 2014;8:1995. doi: 10.1021/nn5004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Nagrath S. Biomedical microdevices. 2013;15:595. doi: 10.1007/s10544-012-9734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, Fouladdel S, Chang AC, Lin L, Jiang H, Waghray M, Luker G, Simeone DM, Wicha MS, Beer DG, Ramnath N, Nagrath S. Oncotarget. 2014;5:12383. doi: 10.18632/oncotarget.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stott SL, Hsu C, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Gilman AJ, Lord JBWD, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. PNAS. 2010;107:18392. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Göttert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. Journal of the American Chemical Society. 2008;130:8633. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamande JW, Hupert ML, Witek MA, Wang H, Torphy RJ, Dharmasiri U, Njoroge SK, Jackson JM, Aufforth RD, Snavely A, Yeh JJ, Soper SA. Analytical chemistry. 2013;85:9092. doi: 10.1021/ac401720k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Asghar W, Demirci U, Wan Y. Nano Today. 2013;8:374. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun X, Liu Z, Welsher K, Robinson JT, Goodwin A, Zaric S, Dai H. Nano research. 2008;1:203. doi: 10.1007/s12274-008-8021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jung JH, Cheon DS, Liu F, Lee KB, Seo TS. Angewandte Chemie International Edition. 2010;49:5708. doi: 10.1002/anie.201001428. [DOI] [PubMed] [Google Scholar]; Feng L, Chen Y, Ren J, Qu X. Biomaterials. 2011;32:2930. doi: 10.1016/j.biomaterials.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nature nanotechnology. 2013;8:735. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura K, Maitani Y, Lowman AM, Takayama K, Peppas NA, Nagai T. Journal of Controlled Release. 1999;61:329. doi: 10.1016/s0168-3659(99)00150-9. [DOI] [PubMed] [Google Scholar]
- 17.Nitschke M, Gramm S, Götze T, Valtink M, Drichel J, Voit B, Engelmann K, Werner C. Journal of Biomedical Materials Research Part A. 2007;80A:1003. doi: 10.1002/jbm.a.31098. [DOI] [PubMed] [Google Scholar]
- 18.Stile RA, Healy KE. Biomacromolecules. 2001;2:185. doi: 10.1021/bm0000945. [DOI] [PubMed] [Google Scholar]
- 19.Cunliffe D, Smart CA, Tsibouklis J, Young S, Alexander C, Vulfson EN. Biotechnology Letters. 2000;22:141. [Google Scholar]
- 20.Huber DL, Manginell RP, Samara MA, Kim B-I, Bunker BC. Science. 2003;301:352. doi: 10.1126/science.1080759. [DOI] [PubMed] [Google Scholar]
- 21.Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo SC, Tseng HR, Yu HH. Adv Mater. 2013;25:1547. doi: 10.1002/adma.201203185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S, Jiang L. Adv Mater. 2013;25:922. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 23.Hatch A, Hansmann G, Murthy SK. Langmuir. 2011;27:4257. doi: 10.1021/la105016a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah AM, Yu M, Nakamura Z, Ciciliano J, Ulman M, Kotz K, Stott SL, Maheswaran S, Haber DA, Toner M. Analytical chemistry. 2012;84:3682. doi: 10.1021/ac300190j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reategui E, Aceto N, Lim EJ, Sullivan JP, Jensen AE, Zeinali M, Martel JM, Aranyosi AJ, Li W, Castleberry S, Bardia A, Sequist LV, Haber DA, Maheswaran S, Hammond PT, Toner M, Stott SL. Adv Mater. 2015;27:1593. doi: 10.1002/adma.201404677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR. ACS Nano. 2015;9:62. doi: 10.1021/nn5056282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Liu X, Meng J, Zhang P, Yang G, Su B, Sun K, Chen L, Han D, Wang S. Advanced Materials. 2013;25:922. doi: 10.1002/adma.201203826. [DOI] [PubMed] [Google Scholar]
- 28.Das TK, Prusty S. Polymer-Plastics Technology and Engineering. 2013;52:319. [Google Scholar]
- 29.Wu Q, Xu Y, Yao Z, Liu A, Shi G. ACS nano. 2010;4:1963. doi: 10.1021/nn1000035. [DOI] [PubMed] [Google Scholar]
- 30.Zhuang XD, Chen Y, Liu G, Li PP, Zhu CX, Kang ET, Noeh KG, Zhang B, Zhu JH, Li YX. Adv Mater. 2010;22:1731. doi: 10.1002/adma.200903469. [DOI] [PubMed] [Google Scholar]; Li GL, Liu G, Li M, Wan D, Neoh KG, Kang ET. The Journal of Physical Chemistry C. 2010;114:12742. [Google Scholar]
- 31.Sahoo NG, Bao H, Pan Y, Pal M, Kakran M, Cheng HKF, Li L, Tan LP. Chemical Communications. 2011;47:5235. doi: 10.1039/c1cc00075f. [DOI] [PubMed] [Google Scholar]
- 32.Kim H, Namgung R, Singha K, Oh I-K, Kim WJ. Bioconjugate Chemistry. 2011;22:2558. doi: 10.1021/bc200397j. [DOI] [PubMed] [Google Scholar]
- 33.Hu S-H, Chen Y-W, Hung W-T, Chen IW, Chen S-Y. Advanced Materials. 2012;24:1748. doi: 10.1002/adma.201104070. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Raj S, Kolanthai E, Sood AK, Sampath S, Chatterjee K. ACS applied materials & interfaces. 2015;7:3237. doi: 10.1021/am5079732. [DOI] [PubMed] [Google Scholar]; Chaudhuri B, Bhadra D, Moroni L, Pramanik K. Biofabrication. 2015;7:015009. doi: 10.1088/1758-5090/7/1/015009. [DOI] [PubMed] [Google Scholar]
- 35.Thampi S, Muthuvijayan V, Parameswaran R. Journal of Applied Polymer Science. 2015;132 [Google Scholar]; Pant HR, Pokharel P, Joshi MK, Adhikari S, Kim HJ, Park CH, Kim CS. Chemical Engineering Journal. 2015;270:336. [Google Scholar]
- 36.Hoshino K, Taniguchi M, Kitao T, Morohashi S, Sasakura T. Biotechnol Bioeng. 1998;60:568. [PubMed] [Google Scholar]
- 37.Cecchet F, De Meersman B, Demoustier-Champagne S, Nysten B, Jonas AM. Langmuir : the ACS journal of surfaces and colloids. 2006;22:1173. doi: 10.1021/la052507z. [DOI] [PubMed] [Google Scholar]
- 38.Ithimakin S, Day KC, Malik F, Zen Q, Dawsey SJ, Bersano-Begey TF, Quraishi AA, Ignatoski KW, Daignault S, Davis A, Hall CL, Palanisamy N, Heath AN, Tawakkol N, Luther TK, Clouthier SG, Chadwick WA, Day ML, Kleer CG, Thomas DG, Hayes DF, Korkaya H, Wicha MS. Cancer research. 2013;73:1635. doi: 10.1158/0008-5472.CAN-12-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arya SK, Lim B, Rahman ARA. Lab on a Chip. 2013;13:1995. doi: 10.1039/c3lc00009e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.