Abstract
Children who grow up in traditional farm environments are protected from developing asthma and allergy. This “farm effect” can be largely explained by the child’s early life contact with farm animals, in particular cows, and their microbes. Our studies in Amish and Hutterite school children living on farms in the U.S. have further demonstrated that this protection is mediated through innate immune pathways. Although very similar with respect to ancestry and many lifestyle factors that are associated with asthma risk, Amish and Hutterites follow farming practices that are associated with profound differences in the levels of house dust endotoxin, in the prevalence of asthma and atopy among school children, and in the proportions, phenotypes, and functions of immune cells from these children. In this review, we will consider our studies in Amish and Hutterites children in the context of the many previous studies in European farm children and discuss how these studies have advanced our understanding of the asthma-protective “farm effect”.
Graphical abstract
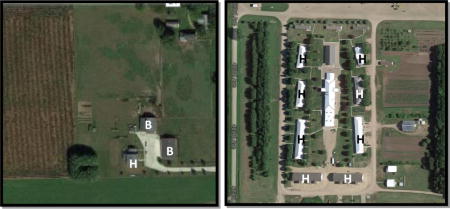
Aerial photographs of an Amish single-family farm (left) and a Hutterite communal farm (right), shown at the same scale. Note the close proximity of the Amish barns (B) to their homes (H), and that there are no barns within the area shown in the photograph of the eight Hutterite homes (each including 2–4 single family homes).
Introduction
Childhood asthma is a complex disease of the airways characterized by inflammation, remodeling and hyperresponsiveness to stimuli [1,2]. Asthma affects 30 million people in the U.S. and is the most common chronic disease in childhood [3]. Wheezing symptoms develop in the first years of life, but most children with wheezing illnesses in infancy do not go on to develop asthma (transient wheeze). Because clinical manifestations of transient wheeze and asthma (persistent wheeze) are indistinguishable in early life, childhood asthma cannot be diagnosed with certainty before the age of 5, even though most childhood asthma likely begins in the first 3 years of life [4]. The development of atopic sensitization to food and inhalant allergens in the first years of life significantly contributes to the development of asthma [5–7], but epidemiologic patterns of asthma and atopy differ: the prevalence of atopy rises from infancy to preschool age and then it reaches a plateau [7]. Infections with rhinovirus (RV) and respiratory syncytial virus (RSV) early in life are likewise strong determinants of subsequent asthma development [8,9].
Asthma and the Farm Effect
Asthma and allergic diseases have a strong environmental component, eloquently illustrated by the rapid rise of their prevalence in the Western world [10]. While, as mentioned above, some environmental exposures (for instance, respiratory viruses) are associated with asthma risk, others have been consistently found to confer protection [11]. Traditional farming in particular appears to have the most potent and consistent protective effects, especially when exposure occurs in early life or even prenatally [12,13]. Data from the PASTURE (Protection against Allergy STUdy in Rural Environments) birth cohort enrolled in rural areas of Austria, Finland, France, Germany, and Switzerland demonstrated that early life exposure to certain farm characteristics, such as animal barns (particularly cows) and consumption of unprocessed cow’s milk, provided stronger protection than exposures occurring later in life [14]. Moreover, not only asthma, but also rhinitis, respiratory tract infections, otitis, fever and C-reactive protein levels at 12 months were reduced by about 30% following raw milk consumption in early life [12], a finding that significantly extends the scope of the protection provided by farm exposure. Interestingly, whereas the overall farm effect can be explained by specific exposures (cows, straw, and unprocessed farm milk for asthma, and fodder storage rooms and manure for atopic dermatitis), the link between the farm effect and hay fever and/or atopic sensitization remains incompletely understood.
Traditional, asthma-protective farms are rich in microbes that modulate immune responses. Consistent with this notion, several recent studies have focused on the connections among the farm effect, the microbiome and immune maturation, and their impact on asthma development. Work from the Netherlands [15–18] and Australia [18] suggests that delayed maturation of immune responses and airway microbial communities contribute to the early development of asthma. The microbiome more generally has been linked to asthma development, with observations of dysbiosis in the gut and airways of asthma patients [19–23], higher relative abundance of Proteobacteria such as Haemophilus influenza, Moraxella catarrhalis and Streptococcus pneumoniae [20–22], greater diversity [20–22], and other alterations of microbial community structure and composition [23]. However, it remains unknown whether dysbiosis is a primary determinant of asthma development or is secondary to airway inflammation.
European farm studies have linked both the environmental and the host microbiome to asthma [24,25]. Asthma was associated with an altered nasal, but not throat, microbiota that was characterized by lower diversity and an abundance of Moraxella [24]. Farm exposure in turn was positively associated with bacterial diversity in mattress dust samples as determined by richness (P = 8.1×10−6); asthma was inversely associated with richness (aOR= 0.48 [0.22–1.02]) in mattress dust and to a lower extent to richness in nasal samples (richness aOR 0.63 [0.38–1.06]) [25].
Asthma Genetics and the Farm Effect
Asthma risk is influenced not only by environmental exposures but also by genetic factors. To date, over 20 loci have been associated with asthma in large genome-wide association studies (GWAS) [26–36]. Many of these loci map to genes that encode molecules involved in immune responses (e.g., IL33, IL1RL1, IL13, TSLP, HLA) or transcription factors that mediate immune responses (e.g., SMAD3, GATA3, RORA, STAT6). However, the most significant and highly replicated associations are with single nucleotide polymorphisms (SNPs) at a novel locus on chromosome 17q21 that is characterized by extensive linkage disequilibrium (LD) spanning ~200 kb and encoding six genes. Two of these genes (ORMDL3 and GSDMB) have emerged as the most likely candidates because SNPs associated with asthma in GWAS are expression quantitative trait loci (eQTLs) for these two co-regulated genes, primarily in blood cells [37–43]. Moreover, SNPs at this locus are robustly associated with childhood-onset asthma and asthma in children exposed to environmental tobacco smoke, with respiratory infections and hospitalizations, and with wheezing illnesses in early life [26,44–48], but not with allergic phenotypes [26,37,45,49,50] (with two exceptions [51,52]). In fact, the risk for asthma associated with genotype at the 17q21 locus is confined to children who wheeze in early life: the 17q21 genotype is not associated with asthma among children who do not wheeze in early life [37,53]. Surprisingly, however, the 17q21 genotype is also associated with protection from wheezing in the first year of life and subsequent asthma among children exposed to barn animals [53]. While neither the specific 17q21 gene(s) involved in risk or protection nor the role of ORMDL3 or GSDMB in asthma pathogenesis is currently known, it is clear that this asthma locus modulates risk or protection associated with two of the most important environmental exposures associated with asthma: virus-induced wheezing illness in early life and exposure to farm animals. These observations support the possibility that the 17q21 locus indirectly impacts risk of childhood onset asthma through its direct effect on early life wheezing illnesses.
Immune Responses and the Farm Effect
Mechanistically, the farm effect likely reflects the ability of farm exposures to modify both immune profiles at birth and immune maturation in early life. Indeed, differences between farm and non-farm children are already detectable in cord blood. For example, expression of the pattern recognition receptors TLR7 and TLR8 was already higher in cord blood of farm neonates [12], and enhanced expression of TLR5 and TLR9 was associated with a lower risk of developing atopic dermatitis later in life [54]. Furthermore, significantly higher levels of IFN-γ and TNF-α were found in farm compared to non-farm children after stimulation of cord blood mononuclear cells [55], and cord blood levels of fetal IgE correlated negatively with IFN-γ production [56]. Moreover, increased numbers and improved function of regulatory T cells were found in neonates of farming mothers compared to neonates born to non-farm mothers [57].
Innate immunity appears to represent a key target of the farm effect. Studies in European farm children have revealed increased expression of Toll-like receptor genes and their downstream signaling molecules in leukocytes from farm compared to non-farm children [12,58,59]. Increased expression of IRAK4 and RIPK1 partially explained the protective farm effect on asthma in the PARSIFAL Study [59]. Moreover, expression of the ubiquitin-modifying enzyme A20 in the airway epithelium was required to support the ability of dust extracts from European animal (cow) sheds to protect mice against experimental allergic asthma, and A20 mRNA and protein expression was significantly reduced in air liquid interface-cultured bronchial epithelial cells from mild and severe asthmatics compared with healthy controls [60]. While Treg function and numbers are improved in farm neonates [57], how the altered innate immune response affects the development and function of adaptive immunity in farm children remains largely unexplored.
A Tale of Two U.S. Farming Populations
The latest chapter in the evolving story of the “farm effect” is based on data gathered by comparing and contrasting two U.S. farming populations characterized by striking similarities in their histories and lifestyles and striking differences in asthma prevalence. Amish farm children from Indiana have an even lower prevalence of asthma (5.2%) and allergic sensitization (7.2%) than Swiss farm children (6.8% and 25.2%, respectively)[61], while Hutterite farm children from South Dakota have a higher prevalence of asthma (21.3%) and allergic sensitization (33.3%) [62]. These observations were unexpected because of the similarities between these two populations (Box 1). However, the Amish live on single-family dairy farms and use traditional farming methods that rely on horses for field work and transportation, whereas the Hutterites live on large, communal farms of approximately 15 to 25 families and utilize modern, highly industrialized farming technologies. These observations led us to ask why the “farm effect” in Amish children was so pronounced, while Hutterite children lacked the asthma- and allergy-protection typically conferred by being raised on a farm [14,63–65]. By addressing this question, our study provided many important insights into the mechanistic pathways through which protection may be mediated.
Box 1. The Amish and Hutterite Populations.
The Amish and Hutterite populations arose during the Anabaptist movement in 16th century Switzerland (Amish) and the South Tyrol (Hutterites). Due to religious persecution, members of these groups migrated to the United States. The Amish emigrated in the 18th and 19th centuries and settled on single family farms in Pennsylvania, Ohio and Indiana. The Hutterites emigrated in the 19th century and settled on three communal farms in South Dakota. The sizes of both populations have grown rapidly due to high rates of fertility and strong values for large families. As of 2015, there were more than 270,000 Amish living in 24 U.S. states, although 65% still live in Pennsylvania, Ohio and Indiana. In 2010, there were over 45,000 Hutterites living in 475 communal farms (called colonies) in the U.S. (South Dakota, Montana, eastern Washington, western Minnesota, North Dakota) and Canada (Manitoba, Alberta, Saskatchewan).
Both populations have retained traditional lifestyles based on their strict interpretation of the bible. Their primary language is their original Swiss/German (Amish) or Tyrolean/German (Hutterites) dialect. Children learn English when they start Amish-only or Hutterite-only schools in the 1st grade and graduate after the 8th grade. In addition to both having large sibship sizes, the Amish and Hutterite populations are very similar with respect to many of the known asthma risk or protective factors. For example, they both have diets rich in fat, salt, and raw milk, low rates of childhood obesity, long durations of breast-feeding, minimal exposure to tobacco smoke and air pollution, and taboos against indoor pets, TV, and internet. However, these two populations differ in one very important respect. The Amish avoid all modern technology and, as a result, they practice traditional single family dairy farming and use horses for fieldwork and transportation. A variety of farm animals are kept in addition to cows, such as horses, poultry, rabbits, and other birds to which all children of both sexes are exposed from an early age. In contrast, the Hutterites live on large communal farms and embrace modern technology. Their industrial-sized farms can house up to 100,000 turkeys, 20,000 hogs, and 600 cows, although the number and types of animals on each colony vary considerably. Because of the size of the Hutterite barns and their distance from their homes, young Hutterite children are not exposed to farm animals or barns. Moreover, due to their strict division of labor, girls and women have little exposure to the animals and barns throughout their lives.
We focused our studies on 30 Amish and 30 Hutterite school children who were balanced for age and gender, and sampled in November and December of 2012. Our primary objective was to assess the prevalence of asthma (by questionnaire) and atopy (by the presence of serum allergen-specific IgE) in these children, and determine whether differences existed in their environment (as assessed by measuring levels of endotoxin and allergens in house dust) and immune response profiles (characterized as whole blood gene expression, cytokine responses, and immune cell phenotypes) [66].
After ruling out the possibility that the large disparity in the prevalence of asthma and allergy observed between Amish and Hutterite children was rooted in genetic differences (Fig. 1a), we turned our attention to differences in environmental exposures. To this end, we collected airborne dust from 10 Amish and 10 Hutterite homes using electrostatic dust collectors (EDCs), as in the GABRIEL Advanced Study [64]. Allergen levels were not different among these homes, but median levels of endotoxin were 6.7-fold higher in the Amish homes (Fig. 1b). Moreover, 16S rRNA sequencing in a pooled sample of mattress dust from each population revealed a greater relative abundance of the bacterial phylum Proteobacteria in the Amish dust and greater relative abundance of Firmicutes and Bacteroidetes in Hutterite dust. Collectively, these data showed that microbial burden and composition differ between the Amish and Hutterite home environments.
Figure 1.
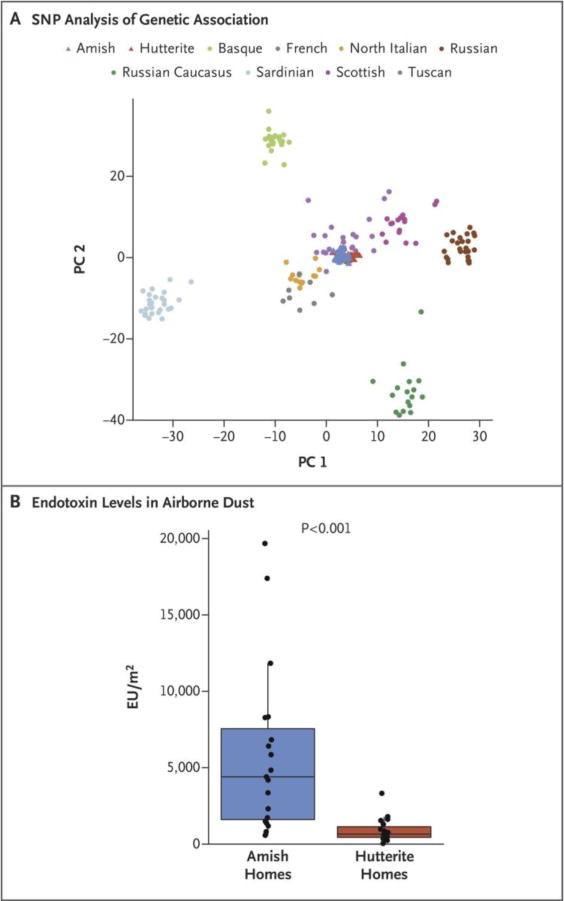
Ancestries and Environments of Amish and Hutterite Children. A) Amish and Hutterite children have similar genetic ancestries. Principal components plot of the first two principal components (PC1 and PC2) of 72,034 SNPs. Amish and Hutterite genotypes are shown relative to European individuals from the Human Genome Diversity Project (HGDP). B) Box-and-whisker plots of endotoxin levels in airborne dust from 10 Amish and 10 Hutterite homes. The horizontal lines show median values, the box represents the interquartile range, and whiskers show the 95% confidence interval. The P value was calculated from the Wilcoxon rank-sum test. EU, endotoxin units.
From New England Journal of Medicine: M.M. Stein, C.L. Hrusch, J. Gozdz, C. Igartua, V. Pivniouk, S.E. Murray, J.G. Ledford, M. Marques dos Santos, R.L. Anderson, N. Metwali, J.W. Neilson, R.M. Maier, J.A. Gilbert, M. Holbreich, P.S. Thorne, F.D. Martinez, E. von Mutius, D. Vercelli, C. Ober, A.I. Sperling. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. Volume 375, pages 411–421. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission.
To explore the impact of these distinct environments on immune profiles, we assessed clinical phenotypes, cell proportions and phenotypes, and gene expression in peripheral blood leukocytes (PBLs) from the Amish and Hutterite children. None of the Amish children and six (20%) of the Hutterite children had asthma, and total serum IgE levels and numbers of children with high (>3.5 kUA/L) levels of IgE against common allergens were lower in Amish than in Hutterite children, similar to our earlier studies [61,62].
Notably, circulating innate immune cells from Amish children had increased proportions of neutrophils and decreased proportions of eosinophils, but similar proportions of monocytes compared to Hutterite children (Fig. 2a). When we phenotyped PBLs from Amish and Hutterite children by flow cytometry, we found that the expression of the chemokine receptor CXCR4 and the adhesion molecules CD11b and CD11c on the surface of neutrophils was lower in Amish children, suggesting that these cells represented a less mature developmental stage. Although monocyte proportions were similar in Amish and Hutterite children, monocytes from Amish children expressed HLA-DR at lower levels and the inhibitory molecule ILT3 at higher levels, a pattern compatible with a suppressive phenotype [67,68]. Collectively, these analyses revealed profound quantitative and qualitative differences in the development and functional properties of innate immune cells in Amish and Hutterite children, likely reflecting the distinct environments to which these children are exposed in early life. In contrast to previous studies in European farm children [57,69], however, Amish and Hutterite children did not differ in the percentage of T regulatory cells, defined as CD3+CD4+FoxP3+CD127− (0.056±0.054% vs. 0.079±0.081% of PBLs, respectively, P = 0.29).
Figure 2.
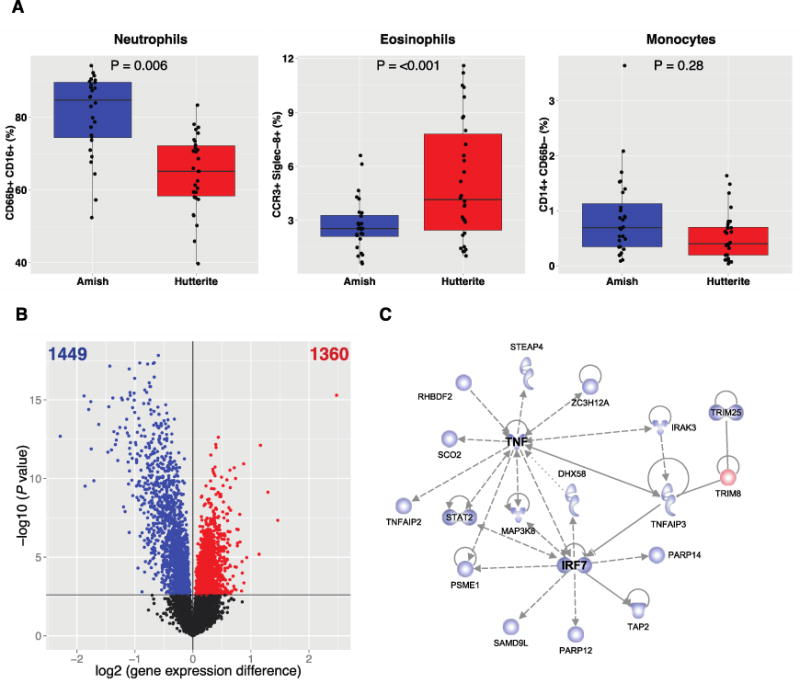
Amish and Hutterite peripheral blood cells differ with respect to composition and gene expression patterns. A) The percentages of total peripheral-blood leukocytes were determined with flow cytometry for neutrophils (defined as CD66b+CD16+), eosinophils (defined as CCR3+Siglec-8+), and monocytes (defined as CD14+CD66b−). Box-and-whisker plots show a line indicating median value, with the box showing the interquartile range and whiskers showing the 95% confidence interval. B) A volcano plot showing the differences in gene expression in peripheral blood leukocytes from Amish and Hutterite children. The x axis shows the log2 differences in gene-expression level between groups; larger positive values are genes with higher expression in the Hutterites compared to the Amish (1360 genes, shown as red points) and larger negative values are genes with higher expression in the Amish compared to the Hutterites (1449 genes, shown as blue points). The y axis shows the –log10 of the P values for each gene. The black horizontal line shows the 1% false discovery rate. C) The most significant network of differentially expressed genes. Genes shown in blue had increased expression in Amish children; the gene shown in red had increased expression in Hutterite children. The gene shapes represent the type of each gene’s protein product (spirals = enzymes, v-shape = cytokines, conjoined circles = transcription regulators, hollow upside-down triangles = kinases, cups = transporters, circles = other products). Solid lines indicate direct interaction and dashed lines indirect interaction. Arrows indicate the direction of activation, arrows with a horizontal line direction of activation and inhibition, and lines without arrows indicate binding only.
Modified from New England Journal of Medicine: M.M. Stein, C.L. Hrusch, J. Gozdz, C. Igartua, V. Pivniouk, S.E. Murray, J.G. Ledford, M. Marques dos Santos, R.L. Anderson, N. Metwali, J.W. Neilson, R.M. Maier, J.A. Gilbert, M. Holbreich, P.S. Thorne, F.D. Martinez, E. von Mutius, D. Vercelli, C. Ober, A.I. Sperling. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. Volume 375, pages 411–421. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission.
Differences in PBL proportions in Amish and Hutterite children were also reflected in their gene expression profiles: the expression of 1,449 genes was higher in Amish PBLs and 1,360 genes were higher in Hutterite PBLs (false discovery rate [FDR] of 1%), (Fig. 2b). These differentially expressed genes formed 15 co-expression modules [70]. When we used Ingenuity Pathway Analysis (IPA) to construct unsupervised protein-protein interaction networks, the most significant network was within a module that was associated with Amish and Hutterite membership (P=7.1×10−9), as well as with neutrophil (P=1.5×10−6) and eosinophil (P=1.0×10−3) proportions. Major hubs in this network were TNF and IRF7, key proteins in the innate immune response to microbial stimuli (Fig. 2c). Seventeen of the 18 network genes were more highly expressed in PBLs from Amish children, and among them was TNFAIP3, which encodes A20, a ubiquitin-editing enzyme critical to limit the activity of multiple NF-κB-dependent inflammatory pathways [71] and previously implicated in the asthma protective effect of European farm dust in a murine model [60]. The one gene that was more highly expressed in PBLs from Hutterite children was TRIM8, a positive regulator of TNFα- and IL-1β-dependent NF-κB activation [72]. Thus, our integrated analysis of the proportions and transcriptional activity of peripheral blood immune cells in Amish and Hutterite children highlighted as a feature prominent in Amish children an enrichment in innate immunity genes involved in the response to microbes, both bacteria and viruses.
To place the immune profiles detected in Amish and Hutterite children in a more mechanistic context and further understand the role of innate immunity in asthma protection, we compared the activity of Amish and Hutterite house dust extracts in a classic ovalbumin (OVA)-driven mouse model of asthma. Consistent with the lack of protection observed in Hutterite children, mice treated with OVA and Hutterite dust extracts developed substantial broncho-alveolar lavage (BAL) eosinophilia and airway hyperresponsiveness (AHR) at levels even higher than those measured in mice that received OVA alone (Fig. 3A). In contrast, intra-nasal administration of Amish dust extracts was sufficient to significantly inhibit OVA-induced AHR, BAL eosinophilia, and OVA-specific IgE (Figs. 3a–b). All of these protective effects required innate immunity because they were strongly reduced in MyD88-deficient mice (Fig. 3c) and completely abrogated in mice lacking both MyD88 and Trif, molecules critical for multiple innate immune signaling pathways (Fig. 3d). That administration to mice of airborne dust collected in Amish and Hutterite homes was sufficient to protect from or enhance experimental asthma, respectively, and result in immune profiles consistent with those seen in Amish and Hutterite children, supports the critical contribution of the environment to the disparate rates of asthma in these two farming populations.
Figure 3.
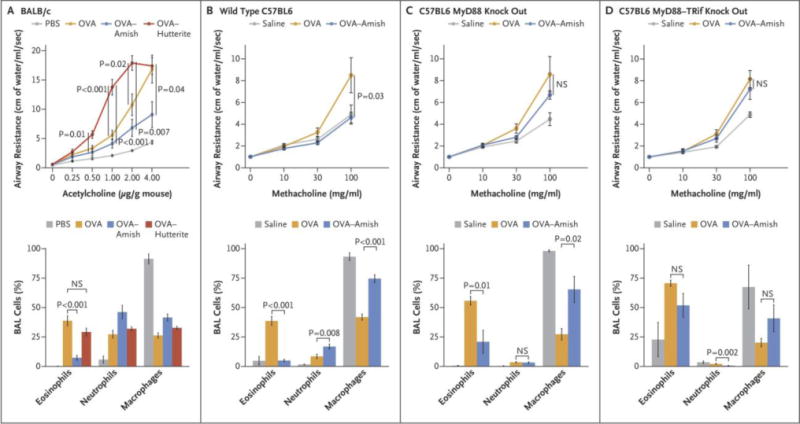
Effects of Amish and Hutterite House-Dust Extracts on Airway Responses in Mouse Models of Allergic Asthma. Panel A shows the effects of the intranasal instillation of 50 μl of Amish or Hutterite dust extract in 7-week-old mice (BALB/c strain) every 2 to 3 days for a total of 14 times beginning at day 0. The mice were sensitized with ovalbumin (OVA) intraperitoneally on days 0 and 14 and challenged with ovalbumin intranasally on days 28 and 38. Airway resistance (shown as centimeters of water per milliliter per second and stimulated in response to increasing doses of acetylcholine administered intravenously) and bronchoalveolar-lavage (BAL) cellularity were measured on day 39 (4 to 6 mice per group). The total amount of Amish and Hutterite dust extract administered over the course of the experiment represented the total load of airborne dust deposited on electrostatic dust collectors placed in Amish or Hutterite homes for 1 month. Statistical differences in experimental measures were assessed with the use of Student’s t-test. Amish house-dust extracts (7.5 mg of dust equivalent in 50 μl) were instilled intranasally every 2 to 3 days for a total of 14 times beginning 5 days before day 0 into 7-week old wild-type mice (Panel B), mice deficient in MyD88 (Panel C), and mice deficient in MyD88 and Trif (Panel D) (all C57BL6 strains). These mice were sensitized intraperitoneally with 20 μg of ovalbumin on days 0 and 14 and were challenged intranasally with 75 μg of ovalbumin on days 26, 27, and 28. Airway resistance (shown as an increase from baseline in response to increasing doses of nebulized methacholine) and bronchoalveolar-lavage cellularity were measured on day 30 (12 mice per group for wild-type mice and 6 mice per group for those deficient in MyD88 or MyD88 and Trif). Statistical differences in experimental measures were assessed with the use of Student’s t-test. I bars represent the standard errors of the data. NS denotes not significant and PBS phosphate-buffered saline.
From New England Journal of Medicine: M.M. Stein, C.L. Hrusch, J. Gozdz, C. Igartua, V. Pivniouk, S.E. Murray, J.G. Ledford, M. Marques dos Santos, R.L. Anderson, N. Metwali, J.W. Neilson, R.M. Maier, J.A. Gilbert, M. Holbreich, P.S. Thorne, F.D. Martinez, E. von Mutius, D. Vercelli, C. Ober, A.I. Sperling. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. Volume 375, pages 411–421. Copyright © 2016 Massachusetts Medical Society. Reprinted with permission.
However, a robust asthma-protective potential exists not only in the Amish but also within the Hutterite farm environment. Indeed, in a subsequent study we demonstrated that inhalation of dust extracts from Hutterite barns could inhibit allergen-induced AHR, BAL eosinophilia and specific IgE as effectively as inhalation of Amish barn dust extract [73]. These results suggest to us that lack of protection among Hutterite children (21.3% of whom are asthmatic) may be due not to the absence of protective exposures from their farms, but to a lack of access to these protective exposures. In fact, before the age of 6 Hutterite children do not spend time near farm animals or in the barns, in part because the latter are located at much greater distances from their homes than are Amish barns (Box 1).
Ongoing Questions and Future Directions
The remarkable genetic similarities between Amish and Hutterite children, and the opposite effects their house dust has on airway responses and inflammation in mouse models, suggest that environmental exposures confer strong protection from asthma and allergies among the Amish by engaging innate immune responses, whereas the lack of such exposures and/or the presence of unidentified risk exposures promotes asthma risk among the Hutterites. However, a number of critical questions remain. First, our studies were performed at school age, when protection from childhood-onset asthma and atopy is already established. As a result, we were unable to investigate the impact of the environment on immune maturation in different groups of children. Second, because we examined only 30 Amish and 30 Hutterite children, we could not evaluate differences between children with asthma or allergies. Third, our immune profiling studies were limited by both technological constraints and small amounts of blood cells that prevented functional studies. Fourth, the design of our study did not allow us to investigate how the airway mucosa – a plausible and even likely target of environmental effects [74] – contributes to asthma protection. And last, but not least, although we have strong supportive evidence that the protective exposures are microbial in nature, we do not know which bacteria, fungi and/or archaea are involved, which metabolites play a protective role, and the sources of these microorganisms and metabolites.
Closing Remarks
Taken together, our studies on farm and non-farm children across Europe, and Amish and Hutterite farm children from U.S. communities that utilize distinct farming practices indicate that early life exposure to airborne substances present in animal barns protects against allergic asthma by engaging and shaping innate immune pathways. Many fundamental questions remain unanswered, but intensive investigation in research laboratories around the world will likely provide some answers over the next several years.
Highlights.
Studies of European and U.S. farm and nonfarm children show that early life exposure to airborne substances protects against allergic asthma by activating innate immune pathways
School age Amish children, who are exposed to a protective farm environment, have distinct immune cell proportions, phenotypes, and gene expression profiles compared to Hutterite children who are not exposed to farm products
Genotype at the 17q21 asthma locus is associated with both protection from asthma among European farm children and with risk for asthma among non-exposed children.
Acknowledgments
The authors gratefully acknowledge their collaborators who contributed toward their studies in the Amish and Hutterite children, with special thanks to Michelle Stein, Cara Hrusch, Justyna Gozdz, Mark Holbreich, Peter Thorne, Jack Gilbert, and Fernando Martinez.
Funding: C.O. is supported in part by NIH grants R01 HL085197, U19 AI095230, U19 AI106683, P01 HL070831, R01 HL122712, R01 HL129735; A.I.S is supported in part by NIH grants R01 HL118758, R01 AI125644, R21 AI126031, and U19 AI095230. E.v.M. received support from the German Research Foundation (DFG) and the German Federal Ministry of Education and Research (BMBF); and D.V. is supported by NIH grants R01 HL129735 and by research grants from Johnson and Johnson and OM Pharma.
References
- 1.Raedler D, Schaub B. Immune mechanisms and development of childhood asthma. Lancet Respir Med. 2014;2:647–656. doi: 10.1016/S2213-2600(14)70129-8. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 3.http://www.aafa.org/page/asthma-facts.aspx.
- 4.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332:133–138. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 5.Hose AJ, Depner M, Illi S, Lau S, Keil T, Wahn U, Fuchs O, Pfefferle PI, Schmausser-Hechfellner E, Genuneit J, et al. Latent class analysis reveals clinically relevant atopy phenotypes in 2 birth cohorts. J Allergy Clin Immunol. 2016 doi: 10.1016/j.jaci.2016.08.046. [DOI] [PubMed] [Google Scholar]
- 6*.Jackson DJ, Evans MD, Gangnon RE, Tisler CJ, Pappas TE, Lee WM, Gern JE, Lemanske RF., Jr Evidence for a causal relationship between allergic sensitization and rhinovirus wheezing in early life. Am J Respir Crit Care Med. 2012;185:281–285. doi: 10.1164/rccm.201104-0660OC. This review considers the interplay between allergic sensitization, viruses and bacteria in early life on the pathogenesis of asthma and allergic diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simpson A, Tan VY, Winn J, Svensen M, Bishop CM, Heckerman DE, Buchan I, Custovic A. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181:1200–1206. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- 8.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson DJ, Gern JE, Lemanske RF., Jr The contributions of allergic sensitization and respiratory pathogens to asthma inception. J Allergy Clin Immunol. 2016;137:659–665. doi: 10.1016/j.jaci.2016.01.002. quiz 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 11.Schaub B, Vercelli D. Environmental protection from allergic diseases: From humans to mice and back. Curr Opin Immunol. 2015;36:88–93. doi: 10.1016/j.coi.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Loss G, Bitter S, Wohlgensinger J, Frei R, Roduit C, Genuneit J, Pekkanen J, Roponen M, Hirvonen MR, Dalphin JC, et al. Prenatal and early-life exposures alter expression of innate immunity genes: the PASTURE cohort study. J Allergy Clin Immunol. 2012;130:523–530 e529. doi: 10.1016/j.jaci.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 13.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 14.Riedler J, Braun-Fahrlander C, Eder W, Schreuer M, Waser M, Maisch S, Carr D, Schierl R, Nowak D, von Mutius E. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 15.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, Bogaert D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 16.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, Chaussabel D, Cohen DM, Sanders EA, Ramilo O, et al. Nasopharyngeal Microbiota, Host Transcriptome, and Disease Severity in Children with Respiratory Syncytial Virus Infection. Am J Respir Crit Care Med. 2016;194:1104–1115. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017 doi: 10.1038/nrmicro.2017.14. This paper reviews the epidemiological, biological and functional evidence that supports the physiological role of the respiratory microbiota in the maintenance of human health. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, Holt BJ, Hales BJ, Walker ML, Hollams E, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–715. doi: 10.1016/j.chom.2015.03.008. This study describes comprehensive prospective data on changes in the composition and strutcture of the nasopharygeal microbiome and its role for the development of aculte upper and lower respiratry tract infections and wheeze. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiemsma LT, Turvey SE. Asthma and the microbiome: defining the critical window in early life. Allergy Asthma Clin Immunol. 2017;13:3. doi: 10.1186/s13223-016-0173-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, Woyke T, Allgaier M, Bristow J, Wiener-Kronish JP, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. e371–373. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–352. e341–343. doi: 10.1016/j.jaci.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Woerden HC, Gregory C, Brown R, Marchesi JR, Hoogendoorn B, Matthews IP. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Depner M, Ege MJ, Cox MJ, Dwyer S, Walker AW, Birzele LT, Genuneit J, Horak E, Braun-Fahrlander C, Danielewicz H, et al. Bacterial microbiota of the upper respiratory tract and childhood asthma. J Allergy Clin Immunol. 2017;139:826–834 e813. doi: 10.1016/j.jaci.2016.05.050. This study investigates the role of the upper respiratory tract microbiome on asthma in the cross-sectional GABRIEL study. The composition of the nasal, but not throat, microbiome was associated with asthma risk. [DOI] [PubMed] [Google Scholar]
- 25*.Birzele LT, Depner M, Ege MJ, Engel M, Kublik S, Bernau C, Loss GJ, Genuneit J, Horak E, Schloter M, et al. Environmental and mucosal microbiota and their role in childhood asthma. Allergy. 2017;72:109–119. doi: 10.1111/all.13002. This paper described the interrelationship between the environmental microbiome, the nasal microbiome and asthma in the cross-sectional GABRIEL study. They show that the composition of both environmental and nasal microbiomes are related to childhood asthma. [DOI] [PubMed] [Google Scholar]
- 26.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bønnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, den Dekker HT, Husby A, Sevelsted A, Faura-Tellez G, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 29.Yan Q, Brehm J, Pino-Yanes M, Forno E, Lin J, Oh SS, Acosta-Perez E, Laurie CC, Cloutier MM, Raby BA, et al. A meta-analysis of genome-wide association studies of asthma in Puerto Ricans. Eur Respir J. 2017;49 doi: 10.1183/13993003.01505-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sleiman PM, Flory J, Imielinski M, Bradfield JP, Annaiah K, Willis-Owen SA, Wang K, Rafaels NM, Michel S, Bonnelykke K, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 31.Ramasamy A, Curjuric I, Coin LJ, Kumar A, McArdle WL, Imboden M, Leynaert B, Kogevinas M, Schmid-Grendelmeier P, Pekkanen J, et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 32.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, Bush A, Chung KF, Cookson WO, Strachan DP, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67:762–768. doi: 10.1136/thoraxjnl-2011-201262. [DOI] [PubMed] [Google Scholar]
- 33.Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, Danoy P, Baltic S, Nyholt DR, Jenkins M, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noguchi E, Sakamoto H, Hirota T, Ochiai K, Imoto Y, Sakashita M, Kurosaka F, Akasawa A, Yoshihara S, Kanno N, et al. Genome-wide association study identifies HLA-DP as a susceptibility gene for pediatric asthma in Asian populations. PLoS Genet. 2011;7:e1002170. doi: 10.1371/journal.pgen.1002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, Fujita K, Miyatake A, Enomoto T, Miyagawa T, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Çalışkan M, Bochkov YA, Kreiner-Moller E, Bønnelykke K, Stein MM, Du G, Bisgaard H, Jackson DJ, Gern JE, Lemanske RF, Jr, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 39.Murphy A, Chu JH, Xu M, Carey VJ, Lazarus R, Liu A, Szefler SJ, Strunk R, Demuth K, Castro M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Acevedo N, Reinius LE, Greco D, Gref A, Orsmark-Pietras C, Persson H, Pershagen G, Hedlin G, Melen E, Scheynius A, et al. Risk of childhood asthma is associated with CpG-site polymorphisms, regional DNA methylation and mRNA levels at the GSDMB/ORMDL3 locus. Hum Mol Genet. 2015;24:875–890. doi: 10.1093/hmg/ddu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halapi E, Gudbjartsson DF, Jonsdottir GM, Bjornsdottir US, Thorleifsson G, Helgadottir H, Williams C, Koppelman GH, Heinzmann A, Boezen HM, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–908. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, Grundberg E, Kwan T, Ouimet M, Ge B, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–393. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E, Kabesch M, Schaub B. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol. 2011;127:1587–1594 e1586. doi: 10.1016/j.jaci.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Bonnelykke K, Vissing NH, Sevelsted A, Johnston SL, Bisgaard H. Association between respiratory infections in early life and later asthma is independent of virus type. J Allergy Clin Immunol. 2015;136:81–86 e84. doi: 10.1016/j.jaci.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, Chateigner N, Gormand F, Just J, Le Moual N, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 46.Flory JH, Sleiman PM, Christie JD, Annaiah K, Bradfield J, Kim CE, Glessner J, Imielinski M, Li H, Frackelton EC, et al. 17q12–21 variants interact with smoke exposure as a risk factor for pediatric asthma but are equally associated with early-onset versus late-onset asthma in North Americans of European ancestry. J Allergy Clin Immunol. 2009;124:605–607. doi: 10.1016/j.jaci.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 47.Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, Bousquet J, Gormand F, Just J, Le Moual N, et al. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 48.van der Valk RJ, Duijts L, Kerkhof M, Willemsen SP, Hofman A, Moll HA, Smit HA, Brunekreef B, Postma DS, Jaddoe VW, et al. Interaction of a 17q12 variant with both fetal and infant smoke exposure in the development of childhood asthma-like symptoms. Allergy. 2012;67:767–774. doi: 10.1111/j.1398-9995.2012.02819.x. [DOI] [PubMed] [Google Scholar]
- 49.Bisgaard H, Bonnelykke K, Sleiman PM, Brasholt M, Chawes B, Kreiner-Moller E, Stage M, Kim C, Tavendale R, Baty F, et al. Chromosome 17q21 gene variants are associated with asthma and exacerbations but not atopy in early childhood. Am J Respir Crit Care Med. 2009;179:179–185. doi: 10.1164/rccm.200809-1436OC. [DOI] [PubMed] [Google Scholar]
- 50.Wu H, Romieu I, Sienra-Monge JJ, Li H, del Rio-Navarro BE, London SJ. Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood asthma. Allergy. 2009;64:629–635. doi: 10.1111/j.1398-9995.2008.01912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomita K, Sakashita M, Hirota T, Tanaka S, Masuyama K, Yamada T, Fujieda S, Miyatake A, Hizawa N, Kubo M, et al. Variants in the 17q21 asthma susceptibility locus are associated with allergic rhinitis in the Japanese population. Allergy. 2013;68:92–100. doi: 10.1111/all.12066. [DOI] [PubMed] [Google Scholar]
- 52.Fuertes E, Soderhall C, Acevedo N, Becker A, Brauer M, Chan-Yeung M, Dijk FN, Heinrich J, de Jongste J, Koppelman GH, et al. Associations between the 17q21 region and allergic rhinitis in 5 birth cohorts. J Allergy Clin Immunol. 2015;135:573–576. doi: 10.1016/j.jaci.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 53**.Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvarinen A, Roduit C, Kabesch M, Lauener R, Pfefferle PI, et al. The Early Development of Wheeze. Environmental Determinants and Genetic Susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193:889–897. doi: 10.1164/rccm.201507-1493OC. This study reports for the first time that the protection against asthma among children exposed to farm animals in the first year of life is restricted to children carrying the allele at the 17q21 asthma locus that is associated with asthma risk in non-farm children. Thus, they made the important observation that the same alleles associated with risk in genome wide association studies of asthma is also associated with protection among children exposed to barn animals. [DOI] [PubMed] [Google Scholar]
- 54.Roduit C, Wohlgensinger J, Frei R, Bitter S, Bieli C, Loeliger S, Buchele G, Riedler J, Dalphin JC, Remes S, et al. Prenatal animal contact and gene expression of innate immunity receptors at birth are associated with atopic dermatitis. J Allergy Clin Immunol. 2011;127:179–185. 185 e171. doi: 10.1016/j.jaci.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 55.Pfefferle PI, Buchele G, Blumer N, Roponen M, Ege MJ, Krauss-Etschmann S, Genuneit J, Hyvarinen A, Hirvonen MR, Lauener R, et al. Cord blood cytokines are modulated by maternal farming activities and consumption of farm dairy products during pregnancy: the PASTURE Study. J Allergy Clin Immunol. 2010;125:108–115. e101–103. doi: 10.1016/j.jaci.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 56.Pfefferle PI, Sel S, Ege MJ, Buchele G, Blumer N, Krauss-Etschmann S, Herzum I, Albers CE, Lauener RP, Roponen M, et al. Cord blood allergen-specific IgE is associated with reduced IFN-gamma production by cord blood cells: the Protection against Allergy-Study in Rural Environments (PASTURE) Study. J Allergy Clin Immunol. 2008;122:711–716. doi: 10.1016/j.jaci.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 57.Schaub B, Liu J, Hoppler S, Schleich I, Huehn J, Olek S, Wieczorek G, Illi S, von Mutius E. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123:774–782 e775. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 58.Lauener RP, Birchler T, Adamski J, Braun-Fahrlander C, Bufe A, Herz U, von Mutius E, Nowak D, Riedler J, Waser M, et al. Expression of CD14 and Toll-like receptor 2 in farmers’ and non-farmers’ children. Lancet. 2002;360:465–466. doi: 10.1016/S0140-6736(02)09641-1. [DOI] [PubMed] [Google Scholar]
- 59.Frei R, Roduit C, Bieli C, Loeliger S, Waser M, Scheynius A, van Hage M, Pershagen G, Doekes G, Riedler J, et al. Expression of genes related to anti-inflammatory pathways are modified among farmers’ children. PLoS One. 2014;9:e91097. doi: 10.1371/journal.pone.0091097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60**.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, Madeira FB, Beyaert R, van Loo G, Bracher F, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349:1106–1110. doi: 10.1126/science.aac6623. This paper is the first to show a potential role of the ubiquitin-modifying enzyme A20 in lung epithelium. Deletion of A20 in murine airway epithelium resulted in the loss of the protective effect of farm dust extracts on experimental allergic asthma. [DOI] [PubMed] [Google Scholar]
- 61.Holbreich M, Genuneit J, Weber J, Braun-Fahrlaender C, Waser M, von Mutius E. Amish children living in Northern Indiana have a very low prevalence of allergic sensitization. J Allergy Clin Immunol. 2012;129:1671–1673. doi: 10.1016/j.jaci.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Motika CA, Papachristou C, Abney M, Lester LA, Ober C. Rising prevalence of asthma is sex-specific in a US farming population. J Allergy Clin Immunol. 2011;128:774–779. doi: 10.1016/j.jaci.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, Maisch S, Carr D, Gerlach F, Bufe A, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347:869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 64.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, Heederik D, Piarroux R, von Mutius E. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 65.Illi S, Depner M, Genuneit J, Horak E, Loss G, Strunz-Lehner C, Buchele G, Boznanski A, Danielewicz H, Cullinan P, et al. Protection from childhood asthma and allergy in Alpine farm environments-the GABRIEL Advanced Studies. J Allergy Clin Immunol. 2012;129:1470–1477 e1476. doi: 10.1016/j.jaci.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 66**.Stein MM, Hrusch CL, Gozdz J, Igartua C, Pivniouk V, Murray SE, Ledford JG, Marques dos Santos M, Anderson RL, Metwali N, et al. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N Engl J Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. This study of Amish and Hutterite children demonstrates that the lower prevalence of asthma and allergic sensitization among Amish children is associated with profound differences in immune cell composition and function, and that inhalation of dust extracts from Amish and Hutterite homes is sufficient to recapitulate these differences in a murine model of allergic asthma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim-Schulze S, Scotto L, Vlad G, Piazza F, Lin H, Liu Z, Cortesini R, Suciu-Foca N. Recombinant Ig-like transcript 3-Fc modulates T cell responses via induction of Th anergy and differentiation of CD8+ T suppressor cells. J Immunol. 2006;176:2790–2798. doi: 10.4049/jimmunol.176.5.2790. [DOI] [PubMed] [Google Scholar]
- 68.Velten FW, Duperrier K, Bohlender J, Metharom P, Goerdt S. A gene signature of inhibitory MHC receptors identifies a BDCA3(+) subset of IL-10-induced dendritic cells with reduced allostimulatory capacity in vitro. Eur J Immunol. 2004;34:2800–2811. doi: 10.1002/eji.200324732. [DOI] [PubMed] [Google Scholar]
- 69.Lluis A, Depner M, Gaugler B, Saas P, Casaca VI, Raedler D, Michel S, Tost J, Liu J, Genuneit J, et al. Increased regulatory T-cell numbers are associated with farm milk exposure and lower atopic sensitization and asthma in childhood. J Allergy Clin Immunol. 2014;133:551–559. doi: 10.1016/j.jaci.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 70.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. doi: 10.1371/journal.pcbi.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Q, Yan J, Mao AP, Li C, Ran Y, Shu HB, Wang YY. Tripartite motif 8 (TRIM8) modulates TNFalpha- and IL-1beta-triggered NF-kappaB activation by targeting TAK1 for K63-linked polyubiquitination. Proc Natl Acad Sci U S A. 2011;108:19341–19346. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gozdz J, Ober C, Vercelli D. Innate immunity and asthma risk. N Engl J Med. 2016;375:1897–1899. doi: 10.1056/NEJMc1611699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, Galanter JM, Gignoux CR, Roth LA, Kumar R, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol. 2014;133:670–678 e612. doi: 10.1016/j.jaci.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]


