Abstract
The periplasmic conversion of ferribactin to pyoverdine is essential for siderophore biogenesis in fluorescent pseudomonads, such as pathogenic Pseudomonas aeruginosa or plant growth-promoting Pseudomonas fluorescens. The non-ribosomal peptide ferribactin undergoes cyclizations and oxidations that result in the fluorophore, and a strictly conserved fluorophore-bound glutamic acid residue is converted to a range of variants, including succinamide, succinic acid, and α-ketoglutaric acid residues. We recently discovered that the pyridoxal phosphate-containing enzyme PvdN is responsible for the generation of the succinamide, which can be hydrolyzed to succinic acid. Based on this, a distinct unknown enzyme was postulated to be responsible for the conversion of the glutamic acid to α-ketoglutaric acid. Here we report the identification and characterization of this enzyme in P. fluorescens strain A506. In silico analyses indicated a periplasmic transaminase in fluorescent pseudomonads and other proteobacteria that we termed PtaA for “periplasmic transaminase A.” An in-frame-deleted ptaA mutant selectively lacked the α-ketoglutaric acid form of pyoverdine, and recombinant PtaA complemented this phenotype. The ptaA/pvdN double mutant produced exclusively the glutamic acid form of pyoverdine. PtaA is homodimeric and contains a pyridoxal phosphate cofactor. Mutation of the active-site lysine abolished PtaA activity and affected folding as well as Tat-dependent transport of the enzyme. In pseudomonads, the occurrence of ptaA correlates with the occurrence of α-ketoglutaric acid forms of pyoverdines. As this enzyme is not restricted to pyoverdine-producing bacteria, its catalysis of periplasmic transaminations is most likely a general tool for specific biosynthetic pathways.
Keywords: enzyme, iron, Pseudomonas, pyridoxal phosphate, siderophore, periplasmic tailoring, pyoverdines, transaminase
Introduction
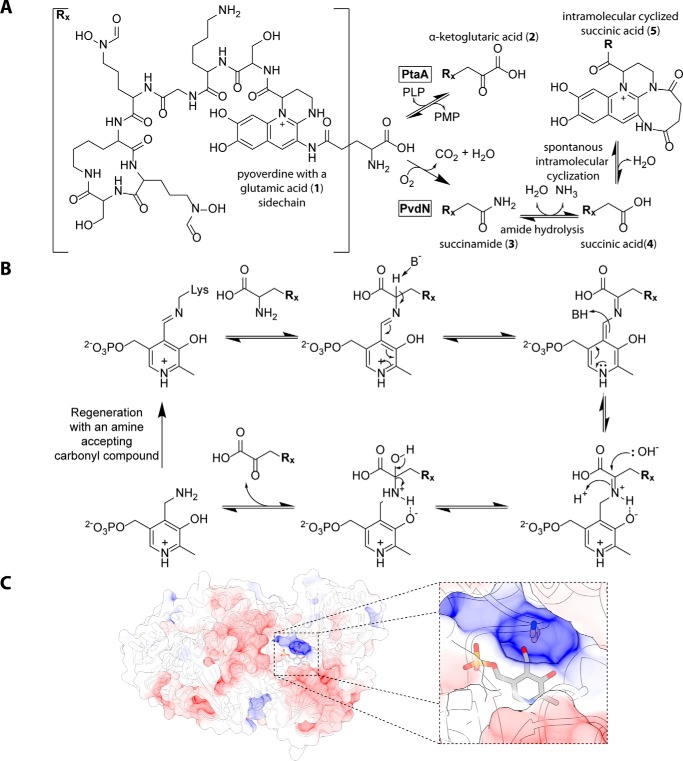
Iron ions play important roles in all living organisms, but in many habitats their availability is limited due to low solubility of Fe(III) oxide hydrates (1). Pyoverdines are siderophores that permit growth of many pseudomonads under iron limitation. Examples include pathogenic Pseudomonas aeruginosa as well as non-pathogenic or even plant-growth promoting Pseudomonas fluorescens strains (2). All pyoverdines originate from non-ribosomally synthesized peptides that are translocated into the periplasm where a quinoline fluorophore is formed (3, 4). The many known “fluorescent pseudomonads” produce a multitude of distinct pyoverdine variants that differ generally in the sequence of the peptide chain and the nature of a fluorophore-attached residue that is synthesized as a glutamic acid and usually converted to succinamide, succinic acid, or α-ketoglutaric acid and in rare cases to malamide and malic acid (4). While the formation of the fluorophore is known to depend on the activity of the periplasmic enzyme PvdP (5), the chemical modification of the glutamic acid that is amide-bonded to the fluorophore via its γ-carboxylic acid has long been puzzling.
We recently revealed that the periplasmic enzyme PvdN is responsible for the direct conversion of the glutamic acid to succinamide by a novel PLP2-catalyzed reaction (6). From these findings, it was concluded that there had to exist a yet unknown transaminase in the periplasm that should, in competition with the PvdN-catalyzed succinamide formation, convert the glutamic acid residue to α-ketoglutaric acid (6). Here we demonstrate that this enzyme indeed exists. Initially, we identified in silico a unique candidate that was predicted to possess all required characteristics. The cofactor content, transport, and quaternary structure of this enzyme were experimentally confirmed. We inactivated the corresponding gene by scarless in-frame deletion and showed that the α-ketoglutaric acid variant was indeed absent in the mutant strain, a phenotype that was fully complemented in trans. Further analyses indicate that in pseudomonads the occurrence of this transaminase correlates with α-ketoglutaric acid-containing pyoverdines. However, there is strong evidence for additional functions of this transaminase in the periplasm, which is why we term this enzyme “periplasmic transaminase A” (PtaA).
Results
In silico prediction of a pyoverdine-modifying periplasmic transaminase in P. fluorescens A506
Recent genetic and biochemical analyses of our model strain P. fluorescens A506 indicated the existence of a yet unknown periplasmic enzyme that converts the fluorophore-attached glutamic acid residue of its pyoverdine (PVDA506) into α-ketoglutaric acid (6). We approached the identification of this enzyme initially by combining in silico tools, anticipating that the conversion should be carried out by a PLP-containing transaminase, and like the PLP-containing PvdN (6), this enzyme should be Tat-dependently transported into the periplasm.
A TATFIND v.1.4 analysis (7) predicted the presence of 29 putative Tat substrates in the proteome of P. fluorescens A506 of which three are most likely false positives. The functional annotation based on the InterPro web service (8, 9) indicated that two of these predicted Tat substrates were sequence-related to transaminases, namely PvdN and PflA506_4424 (Table 1). PvdN has been structurally and functionally characterized (6, 10). It contains a PLP cofactor and catalyzes the unusual amine-retaining oxidative decarboxylation of the fluorophore-attached glutamic acid residue of pyoverdine to succinamide. PvdN thus has already been demonstrated not to be involved in the searched-for transamination reaction. Therefore, the only remaining candidate was PflA506_4424, which we will hereafter term PtaA to avoid confusion and to facilitate reading.
Table 1.
Potential Tat substrates in P. fluorescens A506
Potential Tat substrates were identified with TATFIND v.1.4 (7) and classified using the InterPro web service (8, 9). ABC, ATP-binding cassette.
| Locus tag | Classification |
|---|---|
| PflA506_0002a | DNA polymerase III, β-chain |
| PflA506_0050 | Gluconate 2-dehydrogenase subunit 3 |
| PflA506_0159 | Pectin lyase/hemagglutinin |
| PflA506_0418 | Flavin-dependent halogenase |
| PflA506_0574 | Leu/Ile/Val-binding protein/urea ABC transporter, substrate-binding protein UrtA-like |
| PflA506_0673 | Acid phosphatase, AcpA |
| PflA506_0796 | Alkaline phosphatase d-related/metallo-dependent phosphatase-like |
| PflA506_0832 | Rieske iron--sulphur protein |
| PflA506_0959 | Mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase |
| PflA506_1047 | Multicopper oxidase (CumA) |
| PflA506_1182 | Glucan biosynthesis, MdoD |
| PflA506_1897 | Alkaline phosphatase d-related/metallo-dependent phosphatase-like |
| PflA506_2032a | Hypothetical protein/no classification possible |
| PflA506_2213 | Oxidoreductase molybdopterin-binding subunit, LorB-related |
| PflA506_2582 | Deferrochelatase/peroxidase EfeB |
| PflA506_2836 | Periplasmic binding protein-like II/Aliphatic sulfonate-binding protein-related |
| PflA506_2902 | Dienelactone hydrolase |
| PflA506_3071 | Copper chaperone SCO1/SenC |
| PflA506_3083 | PvdP (uncharacterized domain, dicopper center, tyrosinase family protein) |
| PflA506_3085 | PvdN (aminotransferase class V domain) |
| PflA506_3359 | Leucine-binding protein domain |
| PflA506_3490 | Aldehyde oxidase/xanthine dehydrogenase, molybdopterin-binding |
| PflA506_3831 | Copper-resistance protein CopA |
| PflA506_4010a | 3-Oxoacyl-[acyl-carrier-protein] synthase 2 |
| PflA506_4424 | Aminotransferase, class I/class II |
| PflA506_4509 | Oxidoreductase, molybdopterin-binding domain (YedY) |
| PflA506_4922 | Six-bladed β-propeller, TolB-like/protein of unknown function, DUF839 |
| PflA506_5086 | Hypothetical protein/no classification possible |
| PflA506_5390 | N-Acetylmuramoyl-l-alanine amidase |
a This hit might represent a false positive in the opinion of the authors.
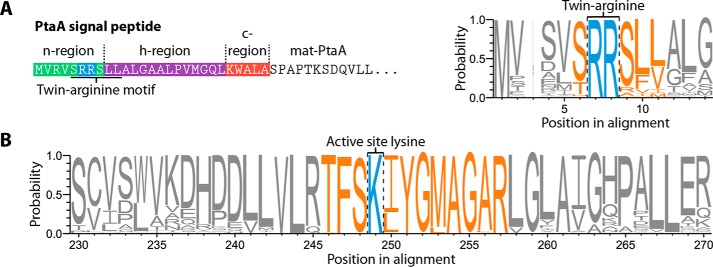
To gain a better understanding of PtaA's properties, we searched all genomes contained in the Pseudomonas Genome Database (11) for homologs of PtaA. To reduce the bias by overrepresented sequences within the identified homologs, we performed a redundancy reduction with an identity threshold of 0.95 utilizing the CD-HIT web service (12). Thereafter, the sequences were aligned using the T-Coffee web service (13), and the multiple sequence alignment was visualized as sequence logo with WebLogo 3 (14). From the results of these analyses, it can be inferred that Tat signal peptides are common in PtaA homologs (Fig. 1A). Additionally, the initial classification of PtaA by the InterPro web service (8, 9) allowed us to identify the active-site lysine residue Lys-224 in a highly conserved sequence pattern (TFSK(I/L)YG(M/L)AGAR), which is highlighted in the sequence logo (Fig. 1B). Furthermore, the InterPro predictions suggested that PtaA likely forms a homodimer.
Figure 1.
Analysis of the N-terminal Tat signal peptide of PtaA and its conserved PLP-binding site, the lysine residue Lys-224. A, depiction of the N-terminal Tat signal peptide of PtaA containing the eponymous twin-arginine motif (ZRRXΦΦ; Z denotes a polar amino acid, R is an l-arginine residue, X is any amino acid, and Φ is a hydrophobic amino acid; Ref. 30). Furthermore, a subsection of the sequence logo of the signal peptide region demonstrates the conserved twin-arginine motif. B, sequence logo of the active-site lysine, Lys-224, and the sequence in its vicinity. The process to generate the sequence logo is detailed under “Experimental procedures.”
PtaA is a periplasmic homodimeric enzyme that requires cytoplasmic cofactor assembly for folding and transport
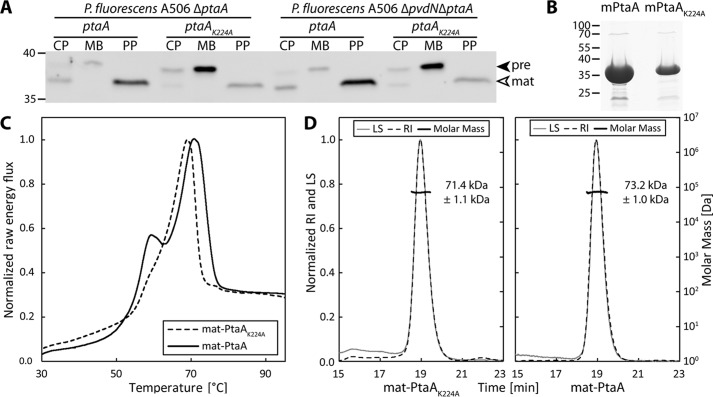
To identify the predicted PLP cofactor, PtaA was overproduced in its mature form in Escherichia coli and purified as described under “Experimental procedures.” By means of a PLP binding assay, we removed the PLP cofactor from PtaA and detected the PLP oxime via its fluorescence (Fig. 2). The predicted periplasmic localization was addressed in complemented P. fluorescens mutant strains that were constructed for subsequent functional analyses (described below). We carried out an in-frame deletion of the gene encoding PtaA in the wild-type P. fluorescens A506 strain as well as in a ΔpvdN deletion strain that was generated previously (6). In P. fluorescens A506, the ptaA gene is flanked by genes encoding a predicted DNA-binding protein upstream in the opposite direction and a putative autoinducer-binding transcriptional regulator downstream in the same direction. The in-frame deletion removed the sequence from codons 5 to 357 of the coding region to avoid any potential polar effects.
Figure 2.
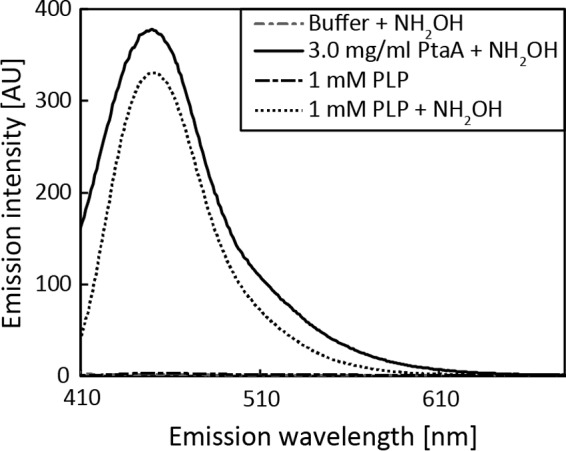
PLP binding assay. Shown are the fluorescence spectrum of the PLP oxime as generated by transimination with hydroxylamine with mature PtaA purified from E. coli (continuous line), the reference spectrum of the PLP oxime (dotted line), and buffer as well as PLP negative control spectra (dashed lines), indicating that PtaA binds PLP. Due to different sensitivity modes, spectra are to be qualitatively compared. AU, arbitrary units.
With the ΔptaA, ΔpvdN, and ΔptaA/ΔpvdN strains in our hands, we constructed complementation vectors for recombinant production of functional PtaA or a PtaA(K224A) variant that is expected to abolish the PLP-dependent activity. The constructs were C-terminally tagged with a Strep-tag II to facilitate detection and purification.
Western blot detection in subcellular fractions proved that PtaA is indeed localized in the periplasmic fraction. Interestingly, the PtaA(K224A) variant accumulated in the membrane fraction, suggesting folding defects that are known to influence the efficiency and accuracy of Tat-dependent translocation (Ref. 15 and Fig. 3A). The precursor form still carrying the N-terminal signal peptide could be distinguished from the mature form in which the signal peptide was cleaved off. These observations were made with both the single- and the double-mutant strains. To examine whether folding defects were caused by the K224A exchange that could influence transport, the protein was analyzed by differential scanning calorimetry (DSC) (Fig. 3C). The proteins used for this study were purified via affinity chromatography and subsequent size-exclusion chromatography (SEC). A Coomassie-stained SDS-PAGE gel of the purified proteins is depicted in Fig. 3B. The DSC data indicated that covalent PLP binding stabilized the overall protein structure to some extent since the unfolding transition temperature of the PtaA(K224A) variant was lowered from 70.8 to 69.0 °C in comparison with the wild-type PtaA. More importantly, a folded subdomain that gives rise to a minor peak at approximately 60 °C was almost absent in the PtaA(K224A) variant, which can be explained by a stabilization of the binding pocket in wild-type PtaA by the covalently attached PLP cofactor. To assess whether PtaA forms a homodimer, we analyzed the molecular size via SEC coupled with static multiangle light scattering/refractive index (MALS-RI) detectors as detailed under “Experimental procedures.” The experiment allowed us to calculate the approximate molecular mass from the data obtained (Fig. 3D). Since the size of mature monomeric PtaA and PtaA(K224A) (with the Strep-tag II) amounts to 37.3 and 37.2 kDa, respectively, the measured molecular masses of 73.2 ± 1.0 kDa for PtaA and 71.4 ± 1.1 kDa for PtaA(K224A) correlate well with the calculated molecular masses of the dimers when the influence of protein shape on MALS-derived values is considered. From these data, it can be concluded that both the wild-type PtaA and the PtaA(K224A) variant form homodimeric structures.
Figure 3.
PtaA is periplasmically localized and forms a homodimer that is stabilized by covalent PLP binding. A, SDS-PAGE/Western blot analysis of subcellular fractions of P. fluorescens A506 deletion strains carrying plasmids for the production of the indicated C-terminally Strep-tagged PtaA variants. PtaA was detected using tag-specific antibodies. CP, cytoplasm; MB, membrane; PP, periplasm; pre, PtaA precursor; mat, mature PtaA. B, Coomassie stained SDS-PAGE gel of the purified mature PtaA (mPtaA) variants used for the DSC experiment depicted in C. C, DSC analysis of PtaA and PtaA(K224A). D, SEC-MALS-RI molecular mass determination of the indicated PtaA variants, indicating the dimeric structure of the enzyme. LS, light scattering; RI, refractive index. Data in C and D are normalized to the global maximum of the respective data set. In A and B, masses (in kDa) of standard proteins are indicated on the left.
PtaA is not essential for the overall pyoverdine production and physiological function
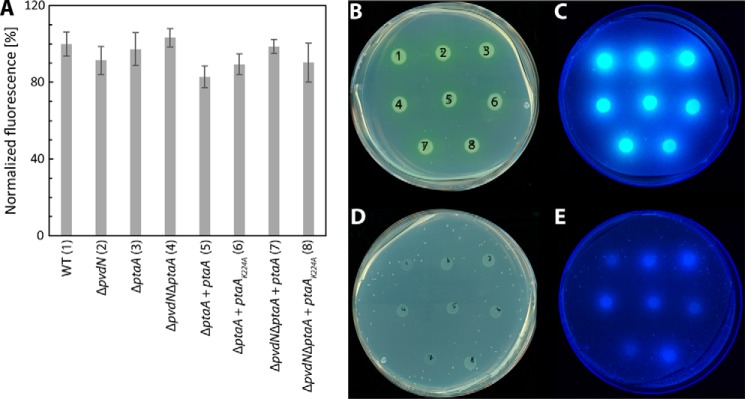
The influence of the ΔptaA deletion on formation/export of PVDA506 was assessed by means of a semiquantitative pyoverdine production assay (Fig. 4A). Furthermore, in the same assay, we included the analysis of ΔpvdN single- and ΔptaA/ΔpvdN double-deletion strains and the above mentioned in trans complementation systems in the analyses. The results obtained (Fig. 4A) demonstrate that the deletions do not affect the overall pyoverdine production. Certainly, this type of assay did not differentiate between predicted differences in the side chain at the chromophore. To exclude that potential side-chain modifications impair iron affinity or pyoverdine import, iron binding and import were examined qualitatively by growth experiments (Fig. 4, B–E). In agreement with the formation of functional siderophores, the mutant strains were able to grow similarly well on pyoverdine-inducing casamino acid (CAA) medium (Fig. 4, B and C), and even in the presence of 0.5 g/liter iron-depleting chelator ethylenediamine di(o-hydroxy)phenylacetic acid (EDDHA) (Fig. 4, D and E), growth was normal, indicating physiological siderophore functionality of the pyoverdines produced in the mutant strains.
Figure 4.
Semiquantitative pyoverdine production and iron binding assay for all P. fluorescens A506 strains used in this study. A, relative quantification of pyoverdine production in liquid culture, normalized to P. fluorescens A506 (WT) levels. The error bars indicate the standard deviation calculated from triplicates. B–E, droplet assays of all P. fluorescens A506 strains used in this study as indicated by numeric notation (1–8) in A. The incubation was performed either in the absence (B and C) or presence of EDDHA (D and E). The same plates were scanned (B and D), or fluorescence was detected on a UV table (C and E).
PtaA is the searched-for transaminase
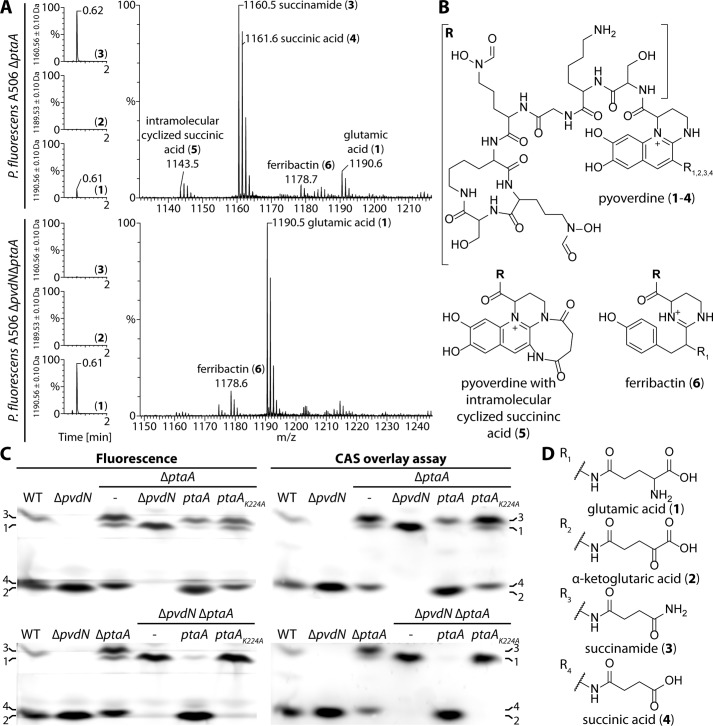
To examine whether PtaA is responsible for the predicted side-chain modification, we isolated the pyoverdines from all strains and analyzed the molecular composition via ultraperformance liquid chromatography (UPLC)-coupled high-resolution mass spectrometry (HR-MS) and isoelectric focusing (IEF) gels in combination with the chrome azurol S (CAS) overlay assay. The results verified our hypothesis about the nature of the modification (Fig. 5, A–D). For convenience, the structures of all pyoverdine variants identified in this study are denoted in Fig. 5, B and D, and the assigned numbers 1-6 will be used to unambiguously attribute the compounds discussed. The masses of compounds 1-6 were determined by HR-MS and compared with the calculated molecular masses (denoted in parentheses) of the respective molecules: 1, 1190.544 atomic mass units (a.m.u.) (1190.544 a.m.u.); 2, 1189.510 a.m.u. (1189.513 a.m.u.); 3, 1160.534 a.m.u. (1160.534 a.m.u.); 4, 1161.520 a.m.u. (1161.518 a.m.u.); 5, 1143.507 a.m.u. (1143.507 a.m.u.); 6, 1178.581 a.m.u. (1178.581 a.m.u.).
Figure 5.
The enzyme PtaA is the periplasmic transaminase that converts the glutamic acid side chain of pyoverdine into the α-ketoglutaric acid variant. A, mass spectrometry results showing the UPLC chromatograms filtered for the masses of the succinamide, α-ketoglutaric acid, and glutamic acid variants on the left. On the right, the corresponding mass spectra of the peak at 0.6 min are depicted. The mass peaks were assigned to the structures shown in B and D. B, structure of the pyoverdine backbone, the intramolecular cyclized succinic acid variant, and the non-cyclized pyoverdine precursor ferribactin. The residues R1–4 correspond to the substructures shown in D. C, IEF gels visualized by pyoverdine fluorescence (left) and a CAS overlay assay (right). Numbers denote the identified pyoverdine variants, whose structures are represented in B in conjunction with D.
To analyze whether the identified structures were truly distinct molecules and not artifacts of MS (e.g. dehydrations), we used a shallower gradient for elution and could thereby prove that all compounds have a distinct elution profile (supplemental Fig. S1). From the MS results, it was obvious that, in the absence of PtaA, no α-ketoglutaric acid variant (2) of PVDA506 was produced (Fig. 5A). The equilibrium between the side-chain modifications was clearly shifted in favor of the succinamide variant (3) of PVDA506. Additionally, the unmodified glutamic acid precursor (1) of PVDA506 was detectable, indicating an incomplete turnover by PvdN. It should be noted that further pyoverdine species could by detected by MS, including the hydrolysis product of the succinamide variant, namely the succinic acid variant (4); traces of the intramolecular cyclization product thereof (5); and the non-cyclized pyoverdine precursor ferribactin (6). As expected from our previously published analysis of PvdN (6), the ΔptaA/ΔpvdN double-deletion strain produced neither the succinamide variant (3) or further derivatives thereof nor the α-ketoglutaric acid variant (2) of PVDA506. Consequently, the only detectable variants were the fluorophore-containing precursor with the original glutamic acid residue (1) and its non-cyclized precursor ferribactin (6). To examine the validity of our MS data interpretation by an independent method, we performed IEF experiments in combination with the CAS overlay assay (Fig. 5C). The assignments of the bands are denoted on the sides of the images using the definitions given in Fig. 5D. The results obtained by IEF correlate well with the data obtained by MS. Noticeably, the wild-type strain produced an excess of the α-ketoglutaric acid variant (2), whereas the succinamide variant (3) was a minor side product. The ΔpvdN deletion strain did not produce the succinamide variant (3) anymore, and all pyoverdine was converted into the α-ketoglutaric acid variant (2) by PtaA. The ΔptaA deletion strain in turn produced at least three variants of pyoverdine, namely the unmodified glutamic acid precursor (1), the succinamide variant (3), and the hydrolysis product thereof, the succinic acid variant (4). It should be noted that the lower band in the ΔptaA strain could easily be mistaken for the α-ketoglutaric acid variant (2) due to its nearly identical pI, but the succinic acid variant (4) migrated slightly higher. Furthermore, in the ΔptaA/ΔpvdN double-deletion strain, no succinic acid variant (4) was present due to the lack of its precursor, the succinamide variant (3). This lane unambiguously clarifies the assignment of the closely migrating α-ketoglutaric acid (2) and succinic acid variants (4) of PVDA506.
The ΔptaA phenotype was entirely complemented in trans by the ptaA complementation strain, whereas the PtaA(K224A) variant did not complement the phenotype and was therefore inactive. This conclusion can be deduced from the comparison of the IEF pyoverdine band patterns of these strains with the patterns obtained for the wild-type and the ΔptaA deletion strains. The same holds true for the complementation of the ΔptaA/ΔpvdN double-deletion strains as the in trans complementation with ptaA resulted in the quantitative turnover of the glutamic acid precursor (1) to the α-ketoglutaric acid variant (2), whereas the strain with PtaA(K224A) produced only the unmodified glutamic acid precursor (1). Moreover, the CAS overlay assay indicated that the side-chain modifications did not alter the iron affinity of the pyoverdine variants as can be inferred from the identical band patterns in comparison with fluorescence imaging of the IEF gels (Fig. 4C).
Discussion
More than 30 years ago, an α-ketoglutaric acid was identified for the first time as an amide-bonded residue at the fluorophore of a pyoverdine in strains of P. aeruginosa and P. fluorescens (16, 17). Also the at that time already known succinamide (18) and succinic acid residues were detected at this position in those studies. Interestingly, pyoverdines of P. aeruginosa strains greatly differ in their peptide moiety but generally have the three above mentioned residues in common that are amidically attached to their fluorophore (19). Also a glutamic acid residue bound to the fluorophore via its γ-carboxylic acid was discovered at this position (20) as well as malamide (21), malic acid (22), and an intramolecular cyclized succinate (23). These variations of the pyoverdine are made in a species- and strain-specific manner (24). Based on the large variability of amino acid sequences and fluorophore-attached side chains, IEF-based siderotyping methods have been developed that use the strain-specific characteristics for systematic purposes (25).
The glutamic acid residue is always the first residue in ferribactins, initially discovered by Hohlneicher et al. (26). This glutamic acid residue is myristoylated for transport into the periplasm where it is deacylated by PvdQ (27–29). After formation of the fluorophore, it thus must be the glutamic acid residue that is initially present at this position, and the found succinamide, succinic acid, α-ketoglutaric acid, malamide, and malic acid residues must result from glutamic acid modifications.
Biochemistry of PtaA
It was clear from our recent study on PvdN (6) that an unknown enzyme had to be responsible for the formation of the α-ketoglutaric acid. Using P. fluorescens A506 as model organism, we have now identified PtaA as the periplasmic transaminase responsible for the formation of the α-ketoglutaric acid residue in PVDA506 (2). PtaA is a PLP-containing enzyme (Fig. 2) that is transported together with its covalently bound cofactor via the Tat pathway in a folded conformation (Fig. 3). The folded state is influenced by cofactor binding to position Lys-224 (Fig. 3C), which is most likely the reason for a partial mislocalization of the mutated protein in the membrane (Fig. 3A). A similar effect had been already observed with PvdN, which also requires PLP binding for Tat-dependent transport (6). In the case of PvdN, the exchange of the active-site Lys resulted even in a complete mislocalization of the enzyme in the membrane fraction (6). PvdN and PtaA are both homodimeric enzymes (Ref. 6 and Fig. 3D) with their PLP most likely bound close to the dimer interface (Ref. 6 and Fig. 6C). This class of enzymes includes therefore good examples for typical Tat substrates that need to be folded prior to translocation (30). They possess twin-arginine signal peptides that can be used for identification purposes in addition to transaminase sequence specifications (Fig. 1).
Figure 6.
Role and mechanism of PtaA in periplasmic pyoverdine tailoring. A, beginning from the glutamic acid variant (1) of PVDA506, two competing tailoring pathways are present. Transamination by PtaA results in the α-ketoglutaric acid variant (2) PVDA506, whereas the PvdN modification results in the succinamide variant (3), which can be partially hydrolyzed to the succinic acid (4) and then intramolecularly cyclized (6). B, proposed mechanism of transamination as catalyzed by PtaA in the periplasm, postulating a carbonyl compound for regeneration of PLP. C, structure of PtaA as calculated by homology modeling, highlighting the Lys-224 residue and the PLP cofactor. The model was visualized with UCSF Chimera.
Fig. 6A summarizes the now established modifications of the fluorophore-attached glutamic acid residue of pyoverdines. The transamination probably proceeds via a standard transaminase mechanism, which is interesting to exist in the periplasm as it requires a yet unknown periplasmic carbonyl compound that we postulate to exist (Fig. 6B). As the Tat transport and its PLP requirement indicate that PtaA assembles PLP inside the cytoplasm to fold to an active enzyme, PLP must be regenerated from PtaA-bound pyridoxamine inside the periplasm. The enzymes PvdN and PtaA are responsible for the initial conversions of the glutamic acid pyoverdine variant. The existence of these competing enzyme reactions explains the reported regulatory differences for the formation of succinamide and α-ketoglutaric acid forms of pyoverdine (31). As a side aspect, the glutamic acid variant of pyoverdine that is produced by the ΔptaA/ΔpvdN double mutant can be very helpful for drug delivery by “Trojan horse” antibiotics (32) because bulky side chains may be attached to this residue without affecting uptake (33). Malamide is likely formed by a hydroxylating enzyme acting on succinamide. The acids succinic acid and malic acid are hydrolysis products of their respective amides. A non-enzymatic hydrolysis has been observed and postulated to be responsible for the formation of these acids in cultures (34). It certainly might be that amidases contribute to the rates of these reactions.
PtaA, a transaminase for pyoverdines and other periplasmic compounds
In P. aeruginosa PAO1, the homolog of PtaA (53% sequence identity) is encoded by the locus PA2531, which is PvdS-dependently up-regulated in response to iron (35); hence this gene belongs to the regulon of PvdS, the σ factor that mediates pyoverdine production (36). An “iron starvation box” for PvdS binding (37) has been identified in the promoter region of PA2531 (35). We found the consensus box as described for P. aeruginosa PAO1 by Ochsner et al. (35) also in the ptaA promoter region of P. fluorescens A506, namely “TAAATN16CGT.” However, the genetic context of ptaA is not conserved between P. aeruginosa PAO1 and P. fluorescens A506. While the iron-responsive regulation of ptaA correlates with its function in pyoverdine modification, its usually pvd-unrelated genomic environment is suggestive for additional functions. This interpretation is further strengthened by a previous study on chorismate mutases that included the detection of transaminase activity in the periplasm of P. aeruginosa PAO1 (38). In that study, the periplasmic transaminase activity could be assigned to PA2531, the above mentioned PtaA homolog. PA2531 is dimeric just like PtaA from P. fluorescens in our study (Fig. 3D). At that time, the authors could not know about its role in pyoverdine modification, which is now clarified by our study. They used α-ketoglutaric acid and phenylalanine as substrates for the transaminase assay. Therefore, PtaA homologs cannot be highly specific for pyoverdine substrates and are likely to be involved in other conversions as well. In support of this conclusion, ptaA homologs are also present in some acidobacteria and α-proteobacteria, such as Zymomonas mobilis (supplemental Fig. S2), that do not produce any pyoverdines. Therefore, this periplasmic transaminase must be able to exert its function for distinct physiological purposes, possibly in conjunction with the generation of distinct secondary metabolites.
Distribution of PtaA among pseudomonads
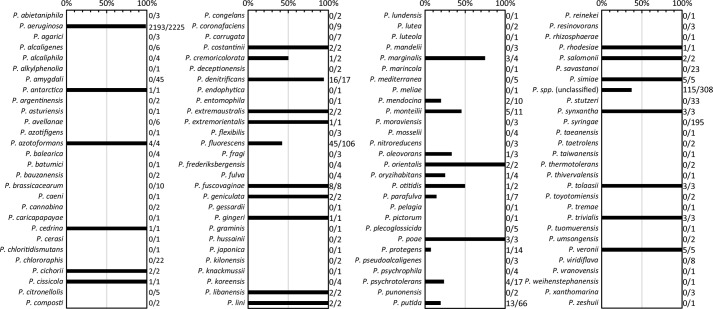
As summarized in Fig. 7, we searched for ptaA homologs in all Pseudomonas genomes available in the Pseudomonas Genome Database (11) (see “Experimental procedures” for details). In agreement with the known occurrence of α-ketoglutaric acid forms of pyoverdines, we found this gene in almost all sequenced P. aeruginosa strains (2193 out of 2225 genomes; ∼99%). The absence of hits in the ∼1% of strains without this gene might be due to the inclusion of draft genomes in the analysis.
Figure 7.
Distribution of PtaA homologs within the genus Pseudomonas. Shown is an evaluation of the distribution of PtaA within the genus Pseudomonas. Numbers to the right of the bar graph indicate the ratio between genomes positive for the presence of PtaA and the total number of analyzed genomes of the respective species.
In the more diverse groups of P. fluorescens and Pseudomonas putida strains, only 45 out of 106 genomes and 13 out of 66 genomes, respectively, encode PtaA, suggesting that the presence of the α-ketoglutaric acid variant of pyoverdine is not strictly conserved in these species. In agreement with this prediction, P. fluorescens Pf0-1 has no PtaA and, as expected, has been shown not to contain the α-ketoglutaric acid (24). On the contrary, there are strains that form α-ketoglutaric acid pyoverdine variants but not any succinamide/succinate variants (24), such as the genome-sequenced P. putida strain H8234. We found that this strain has a pvdMO-ptaA operon instead of the commonly found pvdMNO operon. Therefore, in the rarely found strains that do not possess the succinamide/succinic acid modification pathway, ptaA can be associated with the pvd gene cluster that encodes the periplasmic pyoverdine maturation enzymes. In conclusion, some Pseudomonas species with at least the exception of P. aeruginosa show the interesting characteristic of containing either only the PtaA pathway or only the PvdN pathway (39). Strains with both pathways acting in parallel also exist. We did not find a strain that produces pyoverdine without modifying its glutamic acid. The reason for the apparent requirement for such modifications is unknown and must be unrelated to iron affinity and uptake (Ref. 40 and Fig. 4). Future studies will have to clarify the exact functions of these modifications as well as the additional roles that PtaA can have in the periplasm of proteobacteria.
Experimental procedures
Strains and growth conditions
For physiological studies, P. fluorescens A506 was used. For cloning E. coli DH5α λ pir+ and for expression E. coli Rosetta 2 (DE3) pLysSRARE2 were utilized. P. fluorescens A506 was cultivated at 30 °C, whereas E. coli strains were cultivated at 37 °C unless noted otherwise. The standard cultivation medium was LB (1% (w/v) tryptone, 1% (w/v) NaCl, 0.5% (w/v) yeast extract). If necessary, the appropriate antibiotics were added to the cultivation media at the following final concentrations: 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 20 μg/ml tetracycline.
The production of pyoverdine and the pyoverdine plate assay, pertaining to P. fluorescens A506, were carried out as described previously (6). For relative pyoverdine quantification, 5-ml LB overnight cultures (ONCs) of P. fluorescens A506 were inoculated from cryocultures and incubated at 30 °C at 180 rpm overnight. Thereafter, 50 of ml CAA medium (41) (5 g/liter casamino acids, 5 mm K2HPO4, 1 mm MgSO4) precultures in 100-ml Erlenmeyer flasks with one baffle were inoculated with 50 μl of the respective ONC and incubated for ∼16 h at 30 °C and 180 rpm. 2 ml of the respective preculture were sedimented at 16,000 × g for 2 min at room temperature and subsequently washed twice with 1.5 ml of CAA medium. Afterward, the OD600 of the cell suspensions was adjusted to 1.0 with CAA medium, and for each sample three 4.5-ml CAA cultures (without antibiotics) were inoculated with 0.5 ml of the respective adjusted cell suspension. The cultures were then incubated overnight at 30 °C and 180 rpm. These cultures were sedimented by centrifugation at 3,260 × g and 4 °C for 10 min in 15-ml screw-top plastic tubes. 2.7 ml of the supernatant were mixed with 300 μl of a 1 m HEPES buffer, pH 8.0, and transferred into 1-cm acrylic cuvettes. The samples were measured with a Jasco FP-6500 spectrofluorometer using the following settings for acquisition: excitation at 405 nm, emission at 460 nm, bandwidth of 3 nm for both excitation and emission, and 0.5 s response time in “low-sensitivity” mode.
Genetic methods and plasmids
The construction of all scarless and markerless deletions in P. fluorescens A506 were performed as described previously (6). Furthermore, complementation and production vectors as well as constructs with single-point mutations (pME6010 and pEXH5 derivatives) were generated according to our previously published procedure (6). All primers used for cloning are listed in Table 2. Constructs were verified by restriction analysis and sequencing. E. coli DH5α λ pir+ cells were rendered competent and transformed as described previously (42).
Table 2.
Primers used in this study
oePCR, overlap extension PCR.
| Name | Sequence | Restriction site | Purpose |
|---|---|---|---|
| PfA506-4424-F1-MR | ATAGCCGGATCCTAGACAGGTAGCGCCAAATCACG | BamHI | Forward primer for ptaA left flanking region |
| PfA506-4424-R1-MR | AGGAGTAGTCACCATGGTGCGTGTCACGCAGGTGGTCTGATCAGGCGGCGCTATAGCTG | Reverse primer for ptaA left flanking region | |
| PfA506-4424-F2-MR | GACACGCACCATGGTGACTACTCCTTG | Forward primer for ptaA right flanking region | |
| PfA506-4424-R2-MR | GGCCGCGAATTCTTTGTACAGATTCTTGATTTCCATCTTG | EcoRI | Reverse primer for ptaA right flanking region |
| PfA506-4424-DF-MR | TTTGAGTGCTGCGCCTATTG | ptaA genomic deletion control primer | |
| PfA506-4424-DR-MR | CGCGGCAATGCTGGTGAAGATATAG | ptaA genomic deletion control primer | |
| PfA506-4424-F-MR | TCATCGCATATGGTGCGTGTCAGTCGTCGATCC | NdeI | Forward primer for cloning PtaA-coding region into pEXH5 |
| PfA506-4424-strep-R-MR | GGCCGCAAGCTTTTACTTTTCGAACTGCGGGTGGCTCCAGACCACCTGCGTCGCAAAGGCCTCGC | HindIII | Reverse primer for cloning PtaA-coding region into pEXH5 |
| PfA506-4424_K224A-F-MR | CTGGTGCTGCGCACCTTCTCCGCCATCTAC | Primer for K224A exchange by oePCR | |
| PfA506-4424_K224A-R-MR | CCGGCCATGCCGTAGATGGCGGAGAAGGTG | Primer for K224A exchange by oePCR | |
| PfA506-mat4424-F-MR | GATATACATATGAGCCCAGCGCCGACAAAATCTGACC | NdeI | Forward primer for cloning mature PtaA-coding region into pEXH5 |
| pEXH5-RBS-F-MR | GGCGCGGGATCCGTTTAACTTTAAGAAGGAGATATAC | BamHI | Forward primer for subcloning from pEXH5 into pME6010 |
| pEXH5-strep-term-HindIII-R-MR | CCCCTTAAGCTTAAAAAAAACCCCGCCCTGTCAGGGGCGGGGTTTTTTTTTTTACTTTTCGAACTGCGGGTGGCTCC | HindIII | Reverse primer for subcloning from pEXH5 into pME6010 |
Biochemical methods
For analysis of subcellular fractionations and protein overexpression by SDS-PAGE, successive Western blotting, or in-gel colloidal Coomassie staining, standard protocols were used (43–46). Western blots were developed according to the manufacturer's instructions using StrepMAB-Classic (IBA, Göttingen, Germany) as primary antibody and anti-mouse-HRP conjugate (Carl Roth, Karlsruhe, Germany) as secondary antibody. Images were acquired with the MF-ChemiBIS 4.2 imaging system (DNR Bio-Imaging Systems, Jerusalem, Israel).
The overproduction of mature PtaA and mature PtaA(K224A) was performed using E. coli Rosetta 2 (DE3) pLysSRARE2 as described previously (47) with minor modifications. Briefly, the respective construct was transformed into E. coli Rosetta 2 (DE3) pLysSRARE2 by the transformation and storage solution method (48). After recovery, the cells were spread on MDAG-11 plates (47) and incubated at 37 °C overnight. A 5-ml MDAG-135 (47) ONC was then inoculated with a single colony and incubated at 180 rpm and 37 °C. For overproduction, two 0.5-liter ZYM-5052 preheated (37 °C) autoinducer medium cultures (47) in 3-liter Erlenmeyer flasks with four baffles were each inoculated with 0.5 ml of the ONC. The cultures were incubated for 3 h at 200 rpm and 37 °C. Subsequently, the temperature was decreased to 30 °C, and the cultures were incubated for approximately 19 h at 200 rpm. The cells were harvested, and the respective protein was purified by means of four 1.5-ml gravity-flow Strep-Tactin®-Sepharose columns (IBA) as described previously (6). The purified proteins were concentrated using Vivaspin® 6 concentrators with a cutoff of 10 kDa (Sartorius, Göttingen, Germany). For molecular weight determination, a Pharmacia FPLC (LKB Pump P-500, V-7 valve, 50-μl sample loop) was connected to a size-exclusion chromatography column (Superdex 200 Increase 10/300 GL, GE Healthcare) coupled to MALS (miniDAWN TREOS, Wyatt Technology Europe GmbH, Dernbach, Germany) and refractive index detectors (Shodex RI-101, Showa Denko Europe GmbH, Munich, Germany). The flow rate of the mobile phase (PBS; 10 mm phosphate buffer, pH 7.4, 140 mm NaCl) was set to 0.75 ml/min. The chromatograms were recorded and analyzed with ASTRA 6.1 software (Wyatt Technology). For DSC, a NANO DSC in conjunction with a degassing station from TA Instruments (Lindon, UT) was used. For preparation of DSC samples, the protein elution peak (19-min retention time) from the SEC-MALS-RI system was collected by hand and subsequently concentrated with Vivaspin 6 concentrators (cutoff, 10 kDa). The protein concentration was determined using Roti®-Nanoquant according to the instruction manual (Carl Roth) using a SpectraMax M3 spectrophotometer (Molecular Devices, Biberach an der Riss, Germany). The protein of interest was diluted with degassed PBS to a concentration of ∼1.5 mg/ml prior to analysis. The DSC reference cell was filled with degassed PBS. Analyses covered the temperature range from 20 to 110 °C at a heating rate of 1 °C/min with a 900-s pre-equilibration time at a constant pressure of 3 atm.
The subcellular fractionation of P. fluorescens A506 cultures was performed as described previously (49) with slight modifications. 50-ml cultures were grown to an OD600 of 1 and fractionated into 1-ml fractions that were further analyzed without precipitation.
For PLP cofactor detection, a method modified from that of Ojha et al. (50) was used. The purified mature PtaA enzyme in 100 mm potassium phosphate buffer, pH 7.2, was treated with 5 mm hydroxylamine and incubated for ∼72 h at 4 °C. The sample was loaded onto a Vivaspin 6 concentrator with 5-kDa cutoff, and the PLP oxime in the flow-through was detected at 446-nm emission (scan from 400 to 700 nm) with an excitation of 353 nm, 3-nm bandwidth for both excitation and emission, 0.2-s response time, and three accumulations in “high-sensitivity” mode using a Jasco FP-6500 spectrofluorometer. Reference solutions with the buffer only or with 1 mm PLP in the same buffer were treated identically. PLP- or PLP oxime-containing reference spectra were recorded with the same settings but in “medium-sensitivity” mode.
The extraction of pyoverdine, IEF analysis in conjunction with the CAS overlay assay, and UPLC-MS analysis were performed as described previously (6). To prove that the identified compounds are not artifacts of the MS, an adjusted linear gradient profile was utilized as follows: solvent A (double-distilled water with 0.1% (v/v) formic acid) and solvent B (acetonitrile with 0.1% (v/v) formic acid) at a flow rate of 0.6 ml/min; 2% B (0.0 min), 10% B (10.0 min), 90% B (10.5 min), 90% B (13.0 min), 2% B (13.5 min), and 2% B (20.0 min). The electrospray ionization voltage was set to 100 V, and the injection volume was 5 μl. All other instrument parameters and all instrument hardware were identical to the method described previously (6) except that an ACQUITY UPLC Column Manager (Waters) was additionally installed on the instrument.
Bioinformatics methods
To perform an initial distribution analysis of PtaA within the genus Pseudomonas, we downloaded all amino acid sequences of all complete and draft genomic sequences from the Pseudomonas Genome Database (11) and used TATFIND v.1.4 (7) to search for potential Tat substrates. Subsequently, phmmer v3.1b (51) was used to search for PtaA homologs (cutoff, 1e−45) within all potential Tat substrates. Then a taxonomy table reduced to species level was constructed by means of in-house developed software. Thereafter, the ratio between the total number of genomes of one species and the number of genomes of the same species carrying a potential PtaA homolog was calculated.
To construct the sequence logos, the sequence redundancy of all PtaA homologs was reduced by means of the CD-HIT algorithm contained in the CD-HIT Suite (12) using a similarity threshold of 0.95. Then the resulting sequences were aligned with the T-Coffee web server (13). The sequence alignment was subsequently visualized with WebLogo 3 (14) by plotting the probability of each amino acid at each alignment position. For further investigations into the distribution of PtaA homologs across other bacterial species, a profile hidden Markov model (pHMM) was generated from the multiple sequence alignment with hmmbuild v.3.1b (51). Then all available reference sequences were searched with the generated model (E-value cutoff, 1e−100) using the HMMER web server (52, 53). The resulting sequences were then analyzed regarding their phylogenetic relationship using the Phylogeny.fr3 web service (54) in conjunction with the ETE toolkit (55) for visualization.
To initially predict the enzyme class and active-site lysine residue, Lys-224, the InterPro web service (8, 9) was used. The SWISS-MODEL web service (56) was used to model the structure of PtaA (template, Protein Data Bank code 3LY1), and UCSF Chimera (57) in conjunction with APBS (58) was used to visualize the structure of PtaA.
Author contributions
M. T. R. performed the experiments, prepared the figures, and analyzed the data together with T. B. G. D. performed the MS analyses. T. B. conceived and coordinated the study. T. B. and M. T. R. wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgment
We thank Sybille Traupe for technical support.
This work was supported by the German Science Foundation (Deutsche Forschungsgemeinschaft) GRK1798 “Signaling at the Plant-Soil Interface” and Project BR 2285/7-1. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- PLP
- pyridoxal phosphate
- CAS
- chrome azurol S
- EDDHA
- ethylenediamine di(o-hydroxy)phenylacetic acid
- IEF
- isoelectric focusing
- ONC
- overnight culture
- Tat
- twin-arginine translocation
- UPLC
- ultraperformance liquid chromatography
- PVD
- pyoverdine
- DSC
- differential scanning calorimetry
- SEC
- size-exclusion chromatography
- MALS-RI
- multiangle light scattering/refractive index
- CAA
- casamino acid
- a.m.u.
- atomic mass units.
References
- 1. Andrews S. C., Robinson A. K., and Rodríguez-Quiñones F. (2003) Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237 [DOI] [PubMed] [Google Scholar]
- 2. Cézard C., Farvacques N., and Sonnet P. (2015) Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem. 22, 165–186 [DOI] [PubMed] [Google Scholar]
- 3. Gulick A. M. (2017) Nonribosomal peptide synthetase biosynthetic clusters of ESKAPE pathogens. Nat. Prod. Rep. 34, 981–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schalk I. J., and Guillon L. (2013) Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ. Microbiol. 15, 1661–1673 [DOI] [PubMed] [Google Scholar]
- 5. Nadal-Jimenez P., Koch G., Reis C. R., Muntendam R., Raj H., Jeronimus-Stratingh C. M., Cool R. H., and Quax W. J. (2014) PvdP is a tyrosinase that drives maturation of the pyoverdine chromophore in Pseudomonas aeruginosa. J. Bacteriol. 196, 2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ringel M. T., Dräger G., and Brüser T. (2016) PvdN enzyme catalyzes a periplasmic pyoverdine modification. J. Biol. Chem. 291, 23929–23938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rose R. W., Brüser T., Kissinger J. C., and Pohlschröder M. (2002) Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45, 943–950 [DOI] [PubMed] [Google Scholar]
- 8. Jones P., Binns D., Chang H.-Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S.-Y., et al. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., Bridge A. J., Chang H.-Y., Dosztányi Z., El-Gebali S., Fraser M., Gough J., Haft D., Holliday G. L., Huang H., Huang X., et al. (2017) InterPro in 2017—beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drake E. J., and Gulick A. M. (2016) 1.2 Å resolution crystal structure of the periplasmic aminotransferase PvdN from Pseudomonas aeruginosa. Acta Crystallogr. F Struct. Biol. Commun. 72, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winsor G. L., Griffiths E. J., Lo R., Dhillon B. K., Shay J. A., and Brinkman F. S. (2016) Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang Y., Niu B., Gao Y., Fu L., and Li W. (2010) CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.-M., Taly J.-F., and Notredame C. (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crooks G. E., Hon G., Chandonia J.-M., and Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halbig D., Wiegert T., Blaudeck N., Freudl R., and Sprenger G. A. (1999) The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263, 543–551 [DOI] [PubMed] [Google Scholar]
- 16. Briskot G., Taraz K., and Budzikiewicz H. (1986) Pyoverdine type siderophores from Pseudomonas aeruginosa. Z. Naturforsch. C 41, 497–506 [PubMed] [Google Scholar]
- 17. Poppe K., Taraz K., and Budzikiewicz H. (1987) Pyoverdine type siderophores from Pseudomonas fluorescens. Tetrahedron 43, 2261–2272 [Google Scholar]
- 18. Teintze M., Hossain M. B., Barnes C. L., Leong J., and van der Helm D. (1981) Structure of ferric pseudobactin: a siderophore from a plant growth promoting Pseudomonas. Biochemistry 20, 6446–6457 [DOI] [PubMed] [Google Scholar]
- 19. Meyer J. M., Stintzi A., De Vos D., Cornelis P., Tappe R., Taraz K., and Budzikiewicz H. (1997) Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143, 35–43 [DOI] [PubMed] [Google Scholar]
- 20. Geisen K., Taraz K., and Budzikiewicz H. (1992) New pyoverdin type siderophores from Pseudomonas fluorescens. Monatsh. Chem. 123, 151–178 [Google Scholar]
- 21. Yang C. C., and Leong J. (1984) Structure of pseudobactin 7SR1, a siderophore from a plant-deleterious Pseudomonas. Biochemistry 23, 3534–3540 [DOI] [PubMed] [Google Scholar]
- 22. Demange P., Bateman A., Mertz C., Dell A., Piémont Y., and Abdallah M. A. (1990) Bacterial siderophores: structures of pyoverdins Pt, siderophores of Pseudomonas tolaasii NCPPB 2192, and pyoverdins Pf, siderophores of Pseudomonas fluorescens CCM 2798. Identification of an unusual natural amino acid. Biochemistry 29, 11041–11051 [DOI] [PubMed] [Google Scholar]
- 23. Lenz C., Amann C., Briskot G., Taraz K., and Budzikiewicz H. (2000) Succinopyoverdins—a new variety of the pyoverdin chromophore. Z. Naturforsch. C 55, 146–152 [DOI] [PubMed] [Google Scholar]
- 24. Meyer J.-M., Gruffaz C., Raharinosy V., Bezverbnaya I., Schäfer M., and Budzikiewicz H. (2008) Siderotyping of fluorescent Pseudomonas: molecular mass determination by mass spectrometry as a powerful pyoverdine siderotyping method. Biometals 21, 259–271 [DOI] [PubMed] [Google Scholar]
- 25. Koedam N., Wittouck E., Gaballa A., Gillis A., Höfte M., and Cornelis P. (1994) Detection and differentiation of microbial siderophores by isoelectric focusing and chrome azurol S overlay. Biometals 7, 287–291 [DOI] [PubMed] [Google Scholar]
- 26. Hohlneicher U., Hartmann R., Taraz K., and Budzikiewicz H. (1992) The structure of ferribactin from Pseudomonas fluorescens ATCC 13525. Z. Naturforsch. B 47, 1633–1638 [Google Scholar]
- 27. Yeterian E., Martin L. W., Guillon L., Journet L., Lamont I. L., and Schalk I. J. (2010) Synthesis of the siderophore pyoverdine in Pseudomonas aeruginosa involves a periplasmic maturation. Amino Acids 38, 1447–1459 [DOI] [PubMed] [Google Scholar]
- 28. Drake E. J., and Gulick A. M. (2011) Structural characterization and high-throughput screening of inhibitors of PvdQ, an NTN hydrolase involved in pyoverdine synthesis. ACS Chem. Biol. 6, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hannauer M., Schäfer M., Hoegy F., Gizzi P., Wehrung P., Mislin G. L., Budzikiewicz H., and Schalk I. J. (2012) Biosynthesis of the pyoverdine siderophore of Pseudomonas aeruginosa involves precursors with a myristic or a myristoleic acid chain. FEBS Lett. 586, 96–101 [DOI] [PubMed] [Google Scholar]
- 30. Hou B., and Brüser T. (2011) The Tat-dependent protein translocation pathway. Biomol. Concepts 2, 507–523 [DOI] [PubMed] [Google Scholar]
- 31. Schäfer H., Taraz K., and Budzikiewicz H. (1991) On the genesis of the dicarboxylic acids bound amidically to the chromophore of the pyoverdins. Z. Naturforsch. C 46, 398–406 [Google Scholar]
- 32. Schalk I. J., and Mislin G. L. A. (2017) Bacterial iron uptake pathways: gates for the import of bactericide compounds. J. Med. Chem. 60, 4573–4576 [DOI] [PubMed] [Google Scholar]
- 33. Schons V., Atkinson R. A., Dugave C., Graff R., Mislin G. L., Rochet L., Hennard C., Kieffer B., Abdallah M. A., and Schalk I. J. (2005) The structure-activity relationship of ferric pyoverdine bound to its outer membrane transporter: implications for the mechanism of iron uptake. Biochemistry 44, 14069–14079 [DOI] [PubMed] [Google Scholar]
- 34. Budzikiewicz H. (2004) Siderophores of the Pseudomonadaceae sensu stricto (fluorescent and non-fluorescent Pseudomonas spp.), in Progress in the Chemistry of Organic Natural Products (Falk H., and Kirby G. W., eds), pp. 81–237, Springer, Vienna, Austria: [DOI] [PubMed] [Google Scholar]
- 35. Ochsner U. A., Wilderman P. J., Vasil A. I., and Vasil M. L. (2002) GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45, 1277–1287 [DOI] [PubMed] [Google Scholar]
- 36. Cunliffe H. E., Merriman T. R., and Lamont I. L. (1995) Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative σ factor. J. Bacteriol. 177, 2744–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson M. J., McMorran B. J., and Lamont I. L. (2001) Analysis of promoters recognized by PvdS, an extracytoplasmic-function σ factor protein from Pseudomonas aeruginosa. J. Bacteriol. 183, 2151–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Calhoun D. H., Bonner C. A., Gu W., Xie G., and Jensen R. A. (2001) The emerging periplasm-localized subclass of AroQ chorismate mutases, exemplified by those from Salmonella typhimurium and Pseudomonas aeruginosa. Genome Biol. 2, RESEARCH0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gwose I., and Taraz K. (1992) Pyoverdine from Pseudomonas putida. Z. Naturforsch. B 47, 487–502 [PubMed] [Google Scholar]
- 40. Meyer J. M. (2000) Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 174, 135–142 [DOI] [PubMed] [Google Scholar]
- 41. Ochsner U. A., Snyder A., Vasil A. I., and Vasil M. L. (2002) Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 8312–8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inoue H., Nojima H., and Okayama H. (1990) High efficiency transformation of Escherichia coli with plasmids. Gene. 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 43. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 44. Burnette W. N. (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 45. Towbin H., Staehelin T., and Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neuhoff V., Arold N., Taube D., and Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9, 255–262 [DOI] [PubMed] [Google Scholar]
- 47. Studier F. W. (2014) Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol. 1091, 17–32 [DOI] [PubMed] [Google Scholar]
- 48. Chung C. T., Niemela S. L., and Miller R. H. (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U.S.A. 86, 2172–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ize B., Viarre V., and Voulhoux R. (2014) Cell fractionation. Methods Mol. Biol. 1149, 185–191 [DOI] [PubMed] [Google Scholar]
- 50. Ojha S., Wu J., LoBrutto R., and Banerjee R. (2002) Effects of heme ligand mutations including a pathogenic variant, H65R, on the properties of human cystathionine β-synthase. Biochemistry 41, 4649–4654 [DOI] [PubMed] [Google Scholar]
- 51. Eddy S. R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Finn R. D., Clements J., and Eddy S. R. (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Finn R. D., Clements J., Arndt W., Miller B. L., Wheeler T. J., Schreiber F., Bateman A., and Eddy S. R. (2015) HMMER web server: 2015 update. Nucleic Acids Res. 43, W30–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.-F., Guindon S., Lefort V., Lescot M., Claverie J.-M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huerta-Cepas J., Serra F., and Bork P. (2016) ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 33, 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 58. Baker N. A., Sept D., Joseph S., Holst M. J., and McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.