Abstract
West Nile virus (WNV) is an important agent of human encephalitis that has quickly become endemic across much of the United States since its identification in North America in 1999. While the majority (~75%) of infections are subclinical, neurologic disease can occur in a subset of cases, with outcomes including permanent neurologic damage and death. Currently, there are no WNV vaccines approved for use in humans. This study introduces a novel vaccine platform for WNV to reduce viral replication in the central nervous system while maintaining peripheral replication to elicit strong neutralizing antibody titers. Vaccine candidates were engineered to incorporate microRNA (miRNA) target sequences for a cognate miRNA expressed only in neurons, allowing the host miRNAs to target viral transcription through endogenous RNA silencing. To maintain stability, these targets were incorporated in multiple locations within the 3′-untranslated region, flanking sequences essential for viral replication without affecting the viral open reading frame. All candidates replicated comparably to wild type WNV in vitro within cells that did not express the cognate miRNA. Insertional control viruses were also capable of neuroinvasion and neurovirulence in vivo in CD-1 mice. Vaccine viruses were safe at all doses tested and did not demonstrate mutations associated with a reversion to virulence when serially passaged in mice. All vaccine constructs were protective from lethal challenge in mice, producing 93–100% protection at the highest dose tested. Overall, this is a safe and effective attenuation strategy with broad potential application for vaccine development.
Keywords: miRNA, Immunogenicity, WNV, Neurovirulence, Attenuation, Neuroinvasive
1. Introduction
Since first identified in North America in 1999, West Nile virus (WNV) has become endemic and the leading cause of arboviral encephalitis in the United States [1,2]. Approximately 70–80% of WNV infections are asymptomatic but can cause acute encephalitis resulting in permanent neurological damage or death in approximately 0.1% of infections [3–9]. WNV has been responsible for ~40,000 cases of human disease and ~1700 deaths in the U.S. (http://www.cdc.gov/westnile/statsmaps/index.html). While the mechanism(s) of viral entry into the central nervous system (CNS) are not completely understood [10], replication in neurons of the CNS is critical for the virus to elicit neuropathology. Despite the high prevalence of the WNV in endemic areas, there are currently no vaccines or targeted therapeutics for humans.
A member of the family Flaviviridae, genus Flavivirus, WNV is an enveloped virus with a positive-sense single-stranded RNA genome of ~11 kb encoding a single open reading frame flanked by 5′ and 3′-untranslated regions (UTRs). The genome strongly resembles a mammalian messenger RNA (mRNA) including a 5′ capped RNA with a 3′-UTR lacking 3′-polyadenylation. Both UTRs contain conserved structural elements that are essential for viral replication and viral RNA synthesis [11–14].
MicroRNAs (miRNAs) are small (~23 nt) RNAs that bind to target sequences in the 3′-UTR of eukaryote mRNAs, thus serving an important role in regulating post-transcriptional gene expression [15]. miRNAs with perfect or near-perfect complementarity for their targets, especially in the 5′ “seed” sequence, facilitate direct cleavage of target mRNAs [15] and miRNA targets spaced in short intervals within the mRNA (13–35 nt apart) have been shown to promote optimal silencing of protein expression. By using strategically placed target sequences for the neuron-specific mature miRNA (miR)-124a within the 3′-UTR of WNV, this study investigated the capacity for using miRNA-mediated WNV silencing in neurons as a strategy for WNV vaccination [16]. This strategy has been previously described for other viruses but has not been applied to WNV [17,18].
2. Materials and methods
2.1. Plasmids, viruses, and cells
Stocks of recombinant WNVs containing miRNA target sequences were derived as previously described from cDNA clones, pWN-AB and pWN-CG, containing the 5′ and 3′ portions of the NY99 strain (WT WNV), respectively [19]. A schematic and descriptions are presented in Fig. 1 and Table 1. For construction of vaccine (WNVΔVax-1, -2a, -2b, and -3) and control (WNVΔCtrl-1, -2a, -2b, and -3) viruses, cassettes containing miRNA target sequences specific for mammalian neurons (miR-124a) and for insect cell cycle regulation (miR-14) were synthesized and inserted in the 3′-UTR in several combinations and expressing multiple copies [20,21]. Control viruses were generated using the miR-1175 target sequence, a highly expressed miRNA identified in the mosquito midgut [22]. Unlike miR-14, miR-1175 is not expressed in C6/36 cells, therefore WNVΔCtrl viruses served as insertional controls for C6/36 replication. All miR-1175 targets were inserted in identical locations in control viruses vs. vaccine constructs (miR-124a/miR-14 targets). Finally, to assess specificity of miR-124a on peripheral replication and neuroinvasiveness, viruses were generated using 3 copies of only either miR-14 and miR-124a in a cassette in the same position as the WNVΔCtrl/Vax-1 constructs. Transcription of viral RNA from ligated, linearized cDNA template was performed as previously described [19]. Transcribed RNA was then electroporated into BHK-21 cells [19].
Fig. 1.
Schematic of WNV recombinant constructs. cDNA copies of WNV NY99 were genetically modified to include neuron-specific (miR124a) and mosquito-specific (miR14) miRNA target sequences (WNVΔVax), or mosquito midgut epithelium-specific (miR1175) target sequences (WNVΔCtrl). (A) Multiple targets for each miRNA were inserted in cassettes at locations indicated by dark grey boxes in the 3′-untranslated region. Cassettes were placed specifically to avoid known secondary structures. (B) Description and sequences of miRNA targets used. (C) Serial passage studies (5 passages per virus) were performed assessing the stability of viral sequence in vivo. Mutations were described as either “mixed” (both parental and mutated viruses were detected) or “complete” (only mutated viruses were detected).
Table 1.
WNVΔCtrl virus neuroinvasiveness and neurovirulence in mice.
| Dose (IU) | WT WNV Survivors/Total[AST (days) of decedents]
|
WNVΔCtrl-1 Survivors/Total [AST (days) of decedents]
|
WNVΔCtrl-2a Survivors/Total [AST (days) of decedents]
|
WNVΔCtrl-2b Survivors/Total [AST (days) of decedents]
|
WNVΔCtrl-3 Survivors/Total [AST (days) of decedents]
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Route | IC | IP | IC | IP | IC | IP | IC | IP | IC | IP |
| 10−1 | 2/5 (7.3) | ND | 3/5 (7) | ND | 5/5 | ND | 5/5 | ND | 5/5 | ND |
| 100 | 1/5 (7.5) | 3/5 (7) | 1/5 (7.8) | 5/5 | 3/5 (10) | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 |
| 101 | 0/5 (6.6) | 2/5 (7) | 1/5 (7) | 3/5 (8) | 5/5 | 4/5 (9) | 3/5 (10.5) | 4/5 (8) | 4/5 (8) | 4/5 (9) |
| 102 | ND | 1/5 (7) | 0/5 (6.4) | 3/5 (7) | 1/5 (8.5) | 4/5 (9) | 0/5 (7.8) | 2/5 (8) | 1/6 (6.8) | 2/5 (9) |
| 103 | ND | ND | ND | 0/5 (6.4) | ND | 2/5 (8) | ND | 2/5 (10.7) | ND | 2/5 (8) |
| LD50 as log10 (IU) (95% CI) | −1.18 (−2.43,0.08) | 0.48 (−0.90,1.87) | −0.75 (−1.90,0.40) | 1.71 (1.04,2.39) | 1.49 (0.44,2.54) | 2.75 (1.60,3.90) | 1.02 (−9.30,11.34) | 2.23 (1.27,3.18) | 1.48 (0.99,1.97) | 2.23 (1.27,3.18) |
Vero and BHK-21 cells were maintained in Dulbecco’s minimal essential medium (DMEM) with 5% FBS, 100 units/mL penicillin, and 100 lg/mL streptomycin at 37 °C and 5% CO2 (Life Technologies). SH-SY5Y neuroblastoma cells (gift of D. Beckham) were maintained in DMEM/F12 with 10% FBS and antibiotics, temperature, and CO2 as above [23]. Aedes albopictus mosquito (C6/36) cells were maintained as Vero and BHK-21 cells with 10% FBS at 28 °C.
2.2. Viral growth profiles and quantification assays
Vero and C6/36 cells were plated in 6-well plates and inoculated with viruses in triplicate (3 wells/virus) at a multiplicity of infection of 0.1. Viral adsorption was performed for 1 h at 37 °C (Vero) or 28 °C (C6/36), then cells were washed three times with PBS and 2 mL of media added to the culture. At 0, 24, 48, and 72 h post-inoculation (h.p.i.), 50 lL supernatant was removed from each well, diluted 1:10 in media containing 20% FBS, and stored at −80 °C. Viral titers were determined by plaque assay or 50% tissue culture infectious dose (TCID50) on Vero cells as described [24]. No difference in titer was observed for viruses utilized between plaque [25] or TCID50 [26] assay in this cell line; thus, all titers presented herein are reported as infectious units (IU). Plaque size was determined by measuring the size in mm of 10 well-isolated plaques. All plaque size measurements were made during the same experiment to preclude any intra-experimental variation in agarose concentration and cell density. SH-SY5Y cells were plated in 12-well plates and inoculated with virus and titrated as above.
2.3. Mouse viremia, neurovirulence, and neuroinvasiveness assessments
All animal procedures were reviewed and approved by the CDC IACUC.
2.3.1. Viremia study
Groups of 21 5-week-old female CD-1 outbred mice (Charles River Laboratories) were inoculated intraperitoneally (IP) with [100 μL] of viral inoculum (1000 IU) in PBS using WT, miR-14, and miR-124a viruses. Three animals were bled by cardiac puncture and euthanized daily through dpi 7 for serum viremia titers. Blood was spun in serum separator tubes, and viruses from sera were titrated by plaque assay as described above on Vero cells. An additional 15 3-week-old CD-1 females were inoculated IP with 1000 IU using the same three viruses in order to independently assess induction of morbidity/mortality.
2.3.2. Neuroinvasiveness/neurovirulence study
Groups of 5 3-week-old female CD-1 outbred mice (Charles River Laboratories) were inoculated intracranially (IC) or by the IP route with 10 or 100 μL, respectively, of viral inoculum in PBS. Doses of 0.1, 1, 10, and 100 IU (IC) or 1, 10, 100, and 1000 IU (IP) were used for WT and miRNA insertional mutants. IC inoculations were performed with a 29-gauge needle with an affixed stopper to limit penetration to 2 mm while IP inoculations were performed with a 27-gauge needle. Mice were monitored for clinical signs of encephalitis, including hunched posture, ataxia, hind limb paralysis, or recumbency, and euthanized when signs of encephalitis or paralysis were apparent. Groups of 9 additional 3-week-old CD-1 mice were inoculated by the IC route and three processed daily at dpi 1, 3 and 5 for histology and immunohistochemistry (IHC) (see below).
2.4. Histology and immunohistochemistry
Mice were euthanized as described above and brains were immediately fixed in 10% buffered formalin for at least 48 h and then embedded in paraffin. 4-μm sections were processed by standard histologic techniques, and side-by-side sections were stained with either hematoxylin and eosin (H&E) or were processed for IHC. Slides for IHC were stained with polyclonal rabbit serum primed for the WNV NS5 protein, using Dako Invision + system-HRP labeled polymer anti-rabbit (Dako) and NovaRED peroxidase substrate (Vector Laboratories), and counterstained with Mayer’s hematoxylin.
2.5. Assay of viral stability in mice
Groups of 3 5-week-old CD-1 mice were inoculated with 1000 IU IP of each WNVΔVax with a 27-gauge needle. Mice were bled by cardiac puncture and euthanized at 3 days post infection (d.p.i.). Virus from sera was titrated, and new cohorts of 3 5-week-old mice inoculated with passaged virus at the same dose. This was repeated for five total passages. Viruses were sequenced retrospectively until the first passage where a mutation arose was determined.
2.6. Mouse protection studies
Groups of 15 5-week-old CD-1 mice were immunized IP with 100 μL of each WNVΔVax virus at 100/1000 IU doses with 16 PBS-inoculated mice serving as a control group. No morbidity or mortality was observed for 21 days post-immunization. At this point, whole blood was collected in a serum separator tube, spun at 5000g for 5 min, and sera collected for quantification of neutralizing antibody by 90% plaque reduction neutralization test (PRNT90) (see below). Following blood collection, mice were challenged with an estimated 100,000 LD50 doses of WNV strain NY99 (10,000 IU) IP and monitored for clinical signs of encephalitis as above. At 21 days post-challenge, blood was collected for PRNT90 as described above.
2.7. Measurement of antibody responses
Sera were collected as described above from immunized mice at 21 d.p.i., prior to challenge, and from all surviving mice at 21 days post-challenge. Sera were diluted 1:5, heat inactivated at 56 °C for 30 min, and incubated with an equal volume of WT WNV to a concentration of 100 IU/0.1 mL. Serum samples were co-incubated at 37 °C for 1 h with the virus suspension and 0.2 mL of the serum/virus mixture was inoculated onto Vero cells in 6-well plates and plaques enumerated as described above. A 90% reduction in IU, as compared to the serum-negative control, was used as the determinant of neutralization and titers were represented as the reciprocal of the highest final sample dilution demonstrating ≥90% neutralization.
2.8. Statistical analyses
Peak titers of recombinant viruses were compared using a linear model to contrast recombinant viruses with wild type virus. Peak titers and plaque sizes were compared between vaccine/control viruses by pairwise t tests with Welch’s correction for unequal variance. Significance cutoffs were set a priori at α = 0.05, and p values for t tests were adjusted for multiple comparisons using the Holm method. log2(PRNT90) titers were compared using a linear model to estimate the change in PRNT90 titer from post-vaccination to subsequent challenge. For these statistical analyses, data were analyzed using R version 3.1.2 (R Core Team 2014). Differences in mean peak growth kinetic and mouse serum titers for miR-14 and miR-124a virus studies were assessed by ANOVA.
3. Results
3.1. Generation of recombinant WNV
A schematic and descriptions of the recombinant viruses generated for this study are presented in Fig. 1A and B (modified from Funk et al. [14]). The mature miRNA (miR) target sequences were inserted in cassettes into 1–3 different locations per construct within 4 different regions in the 3′-UTR of the parental WNV strain using an infectious clone strategy previously described [19]. Insertional points were arranged to avoid interrupting known secondary RNA structures referred to in this study following the nomenclature of Pijlman et al. [13]. miRNA target sequence insertion points were incorporated at WNV genomic (Genbank Accession AF196835) positions 10,437/10,438 (1), 10,657/10,658 (2), 10,749/10,750 (3) and 10,833/10,834 (4) (see Fig. 1). All viruses were engineered to contain multiple miR targets placed consecutively within cassettes. Vaccine cassettes contained either three (WNVΔVax-1) or two (WNVΔVax-2a, 2b, 3) copies of each of the mammalian neuron-specific miR-124a target and the mosquito-specific miR-14 target. Cassettes were placed in combinations between SL1 and SL2 structures (all), between SL3 and SL4 (WNVΔVax-2a, 3), between SL4 and dumbbell (DB) 1 (WNVΔVax-2b) and DB1-DB2 (WNVΔVax-3). Matching control viruses were generated with miR-1175 insect-specific miRNA targets (WNVΔCtrl-1, 2a, 2b, 3), not known to be expressed in C6/36 cells (miR-14 was expressed in C6/36 cells) (data not shown). Thus, control viruses served as insertional controls for C6/36 cells. Supplemental sequence files contain the exact sequences of the constructed viruses.
3.2. Recombinant WNV characterization in vitro and in vivo
MicroRNA target-containing viruses were designed to balance immunogenicity and safety. To maximize the immunogenicity against reversion to virulence, insertion sites and combinations were chosen based on known 3′-UTR structures. However, tertiary structural interactions could not be predicted with certainty. Therefore, plaque morphology and in vitro replication phenotypes were assessed for all rescued viruses.
Vero cells do not express miR-124a, verified by transcript screening on microarray chips; as such, alterations in viral replication in Vero cells were interpreted as being due to 3′-UTR modifications (A. M. Lewis, personal communication, database from [27]). All recombinant viruses replicated similarly to WT WNV (Fig. 2A and C), and none was attenuated by >10-fold IU at peak titer as compared to WT WNV (average 0.5 log10 IU at peak titer, Fig. 2E, Supp. Table 1). For all viruses except one (WNVΔCtrl-2b), the mild attenuation was statistically significant (Supp. Table 1). Attenuation was most pronounced for WNVΔVax-3 and WNVΔCtrl-3, which contained the largest number of insertion sites (attenuation of 0.9 and 0.8 log10 IU, respectively).
Fig. 2.
Replication profiles in vitro. Vero (panels A, C, E) and C6/36 (panels B, D, F) cells were inoculated with wild type (WT) and recombinant WNV strains at a multiplicity of infection of 0.1. Supernatants were harvested at 0, 24, 48, and 72 h.p.i. and titrated by plaque assay (reported in IU). (A–D) All infections were performed in triplicate, and growth profiles plotted represent the mean titer and standard deviations. (E and F) Peak titer represents the mean of the three highest values for each virus, and error bars represent standard error. (G) 10 well-isolated plaques were measured in mm at 72 hpi. Sizes are plotted as mean and standard error.
In C6/36 cells, control but not vaccine viruses, were able to replicate with titers and kinetics similar to that of WT WNV (Fig. 2B and D). Vaccine viruses showed partial miRNA-mediated attenuation, particularly at 24–48 h.p.i. Control viruses exhibited peak titers no>0.5 log10-fold lower than WT WNV (Fig. 2F), and this attenuation was not statistically significantly different from WT WNV (Supp. Table 1). Vaccine viruses, in contrast, demonstrated at least 3 log10-fold lower peak titers compared to WT WNV, and the only vaccine viruses able to replicate with any efficiency were WNVΔVax-1 and -2a. However, these viruses were still attenuated significantly as compared to WT.
Additionally, peak titers for each pair of vaccine and control viruses were compared in each cell line (e.g., WNVΔCtrl-1 vs. WNVΔVax-1). These pairs would be expected to replicate similarly in Vero cells but differentially in C6/36 cells. Mean peak titers in Vero cells did not differ significantly between vaccine and control viruses (Fig. 2E, Supp. Table 2). However, as expected in C6/36 cells, all pairs were statistically significantly different (WNVΔVax < WNVΔCtrl) (Fig. 2F, Supp. Table 2).
Finally, plaque sizes were compared between WT and recombinant viruses in Vero cells (Fig. 2G). All but WNVΔCtrl-1 had significantly smaller average plaques than WT WNV (Supp. Tables 3 and 4), despite the similarities in the viruses’ replication profiles described above. Comparing WNVΔCtrl and WNVΔVax pairs of viruses, mean plaque sizes were not significantly different for the WNVΔVax/Ctrl-1 pair or for the WNVΔVax/Ctrl-3 pair (Supp. Table 4). For both the -2a and -2b pairs, WNVΔCtrl had significantly smaller average plaques than their respective WNVΔVax viruses.
After completion of in vitro profiling, in vivo characterization of recombinant viruses was performed. Viruses were generated to contain 3 copies of each miR-14 or miR-124a at the position used for WNVΔCtrl/Vax-1; these viruses are referred to herein as miR-14 and miR-124a. Groups of 21 3-week-old CD1 mice were inoculated with 1000 IU intraperitoneally (IP) of each WT WNV, miR-14, and miR-124a. Three mice were euthanized daily to collect serum for viral titration (Fig. 3A). No differences in mean peak serum titer were observed between the three viruses (p < 0.05). Survival analysis was performed on 15 3-week-old mice per group inoculated with the same three viruses at 1000 IU IP (Fig. 3B). While all mice inoculated with miR-124a survived, there was 93–100% mortality seen in WT and mir-14 virus-inoculated groups. Additionally, histologic characterization and immunohistochemistry for the NS5 protein (to indicate active infection) was performed at day 5 post-infection with 1000 IU inoculated IC for these same 3 viruses in 3-week-old CD-1 mice (Fig. 3C). Inflammation and neuronal cytoplasmic virus were both evident in animals infected with WT WNV and miR-14, but these were both absent in mice infected with miR-124a. Finally, SH-SY5Y neuroblastoma cells were inoculated with each of these 3 viruses at an MOI of 0.1 and supernatant was titrated daily for WNV. Mean peak titer of miR-124a was significantly lower than that of either WT WNV or miR-14 (p < 0.01) (Fig. 3D).
Fig. 3.
Neuronal miR insertion exerts specific effects on peripheral replication and neuropathogenesis. Mice were inoculated IP at 1000 IU with WT WNV, miR-14, and miR-124a. Mice were either euthanized for serum viremia (A, n = 3 per virus per time point) or held for survival analysis for 21 days (B, n = 15). IC-inoculated mice were assayed for neuronal infection by histologic characterization (C). Top row: H&E; bottom row: immunohistochemistry. Human neuronal SH-SY5Y cells were inoculated with these viruses at an MOI of 0.1 and assayed for viral titer daily through 6 days post infection (D).
3.3. Neuroinvasiveness and neurovirulence in vivo
To test the in vivo attenuation of recombinant WNV from insertion of sequence in the 3′-UTR alone, all WNVΔCtrl strains were compared to WT WNV for the ability to elicit neurologic disease in groups of 5 3-week-old CD-1 mice (clinical signs of ataxia, hunching, ruffled fur, paresis/paralysis). Both neuroinvasiveness (by IP inoculation) and neurovirulence (by IC inoculation) were assessed. All insertion sites were attenuating compared to WT WNV as demonstrated by increased LD50 values (Table 1). WNVΔCtrl-1 was the least attenuated by both routes compared to the WT WNV. All viruses were able to elicit encephalitic disease by the IP route at a dose of 10 IU, and by both routes at 100 IU.
3.4. Stability assessed by serial passage in vivo
To address safety concerns about reversion to virulence, serial passage studies were performed. While the likelihood of each target being individually inactivated is very low [28], a deletion event eradicating all target sequence cassettes could occur. However, WNVΔVax-2a, 2b, and 3 flank highly conserved regions of the 3′−UTR encoding secondary RNA structures, so this type of deletion would likely render the virus avirulent. To test this, each virus was serially passaged 5 times in groups of 3 5-week-old CD-1 mice.
Overall only 3 total point mutations were discovered; no deletion events were observed. WNVΔVax-1 showed a single point mutation downstream of the miR target cassette but upstream of the SL2 structure in 2 mice at passage 4 (p4), which was maintained through p5 (Fig. 1A and C). In one mouse, both parental and mutant viruses were identified (“mixed”), and in the other, only mutant viruses were identified (“complete”). Additionally, a single complete point mutation in WNVΔVax-2b was detected at nt10 within the first cassette at p5. No mutations were detected for either WNVΔVax-2a or -3.
3.5. Protection from challenge and serum neutralization profiles of vaccinated mice
Mice were vaccinated at 5 weeks of age with 100 and 1000 IU IP for each vaccine construct. No mortality or adverse events were observed for 21 days post vaccination, at which point mice were challenged with 10,000 IU of WT WNV. An age-matched PBS (mock-vaccinated) control group was included. PRNT90 assays were performed on mice at 21 days post vaccination and again 21 days after challenge in surviving mice (Figs. 4 and 5). All PRNT90 values reported here represent the reciprocal of serum dilution values.
Fig. 4.
WNVΔVax constructs elicit high antibody titers in mice. Mice were vaccinated IP at doses of 100 and 1000 IU. At 21 days post-vaccination, serum was collected and PRNT90 titers were determined for each animal. PRNT90 values indicated represent the reciprocal of serum titers. Grey bars represent geometric mean titers for each vaccine at each dose, and error bars represent standard error.
Fig. 5.
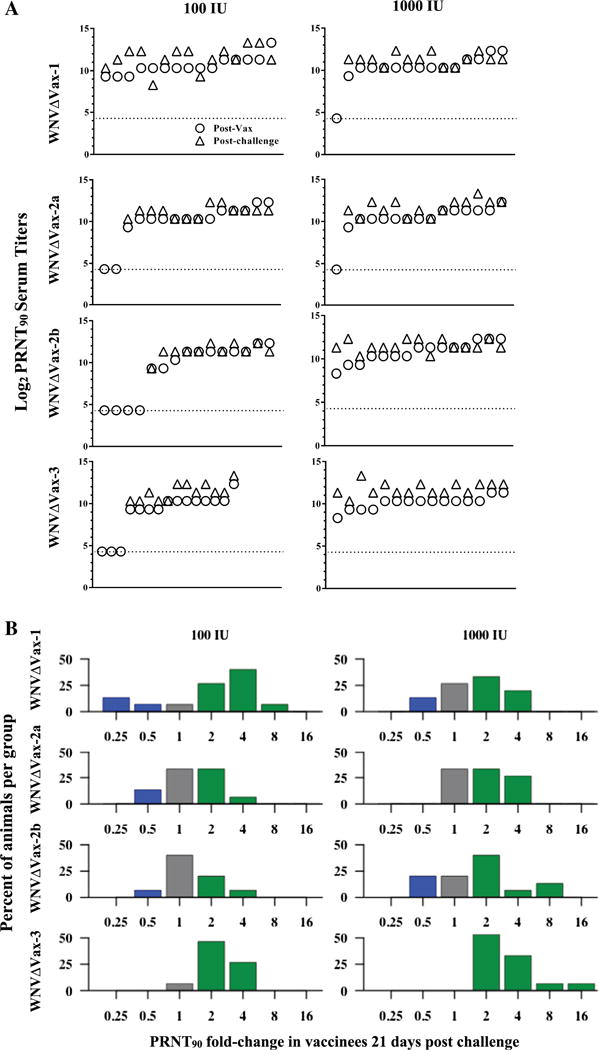
PRNT90 titers in mice after vaccination and after challenge. (A) Mouse sera were tested by PRNT90 assay at 21 days post vaccination (circles) then challenged, and serum was tested subsequently by PRNT90 at 21 days post challenge (triangles). Each stacked pair represents an individual mouse and the x-axis distribution for each graph represents individual mice from 1 (far left) to 15 (far right). Mice whose titers fell below the limit of detection (20 PRNT90, ~4.5 log2 PRNT90) are reported here at the limit of detection, indicated by the dotted line. (B) Fold-change titers from Fig. 4A from 21 days post-vaccination to 21 days post-challenge were compared. Green bars represent groups of animals whose titers increased (>1-fold-change), grey bars represent groups of animals with no change in titer (=1-fold-change), and blue bars represent groups of animals with a decrease in titer (<1-fold-change) after challenge. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Overall, 3 weeks after vaccination, reciprocal serum titers varied from undetectable (<20 PRNT90) to 10,240 PRNT90 (Fig. 5A, circles). The lowest measured detectable titer (>20) was 320. PRNT90 titers varied by construct and by dose, and no statistically significant differences were found overall (data not shown). Importantly, any detectable neutralizing titer, regardless of magnitude, was found to be protective against challenge. Mortality after challenge was low in vaccinated mice (Fig. 6), and corresponded directly with mice that had undetectable PRNT90 titers post-vaccination. Only 1 of the 16 mock-vaccinated mice survived challenge (6%). Survivor-ship in challenged vaccine group mice ranged from 73% to 100% (mean 85%) in the low-dose vaccinees and from 93% to 100% (mean 96.5%) in the high-dose vaccinees. For only WNVΔVax-1, survivor-ship was higher for the lower dose of vaccination as compared to the higher vaccine dose, mimicking the geometric mean titer pattern for that virus. However, this difference was not statistically significant. Both WNVΔVax-2b and -3 showed 100% protection at the higher dose (and correspondingly 100% seroconversion). Of the 3 multiple insertional viruses, the matched control viruses of these two insertion points demonstrated lower LD50 values of 162 by the IP route (compared to 533 for WNVΔCtr-2a), indicating that those contained the least attenuating insertion sites in vivo (Table 1).
Fig. 6.
WNVΔVax constructs protect mice from lethal challenge. Groups of 15 mice were vaccinated with 100 or 1000 IU of each recombinant vaccine with PBS controls. At 3 weeks post vaccination, mice were challenged with 10,000 IU of WT WNV, and survival was assayed for 21 days post-challenge. Data points in the 1000 IU vaccination groups were offset slightly on the y-axis for visualization purposes.
To assess the presence of an anamnestic response, the neutralizing antibody titers three weeks after vaccination were compared with the neutralizing responses observed 3 weeks following challenge (Fig. 5A and B). No statistically significant differences were observed for the likelihood of increasing, decreasing, or maintaining titer post-challenge by vaccine construct or by dose. Regardless, if a titer was detectable after vaccination, it was again detectable (and protective) after challenge.
4. Discussion
This study describes an attenuation strategy for WNV with potential application as a safe and effective vaccine platform. miRNA silencing holds several improvements over current methods of virus attenuation as an approach to vaccine development. MicroRNA silencing has been used for other viruses previously, including picornavirus vaccination strategies, as oncolytic cancer therapeutics, and other flavivirus vaccines [17,29–31]. The study described herein has demonstrated that WNV vaccine constructs were stable and safe through serial passage in vivo. Viral replication was significantly reduced in neurons, which is critical for preventing potentially fatal encephalitis. Immunogenicity was retained via peripheral, non-neuronal viral replication, and was sufficient to prevent morbidity/mortality following robust lethal challenge.
The safety and stability of these vaccine viruses were addressed in this study in several ways, including: (1) using miRNA targets with perfect complementarity for miR-124a, (2) placing multiple target sequences in cassettes with ideal spacing for multiplicative silencing, and (3) incorporating these cassettes in locations flanking regions of the 3′-UTR that are necessary for efficient viral replication. Despite the finding that several passaged WNVΔVax viruses demonstrated single nucleotide substitutions within the miRNA target sequences, there were at least two intact target sequences still present in those viruses. If warranted, future vaccine constructs could incorporate additional CNS-specific miRNA targets such as miR129, miR154, or miR433 to decrease selection pressure on miR-124a [32]. However, the viruses used here had significant nonspecific attenuation (delayed average survival time in the neuroinvasiveness/neurovirulence study, higher LD50 of control compared to WT WNV). This attenuation provides an additional level of safety, reducing the potential for selection of mutants in peripheral (non-neuronal) cells.
These vaccines elicited protective antibody responses. All vaccine constructs were capable of eliciting a neutralizing antibody response in mice, and two constructs (WNVΔVax-2b and WNVΔVax-3) provided 100% protection while retaining complete safety at a high dose of vaccination (1000 IU). Regardless of magnitude, any detectable neutralizing antibody titer was completely protective against challenge.
Although these viruses provided strong overall protection, not all mice seroconverted, likely due to nonspecific attenuation. While all mice included in the time course viremia study demonstrated comparable titers between the WT, miR-14, and miR-124a constructs, the sample sizes were small (n = 3 per virus per day). It is possible that some mice in the larger vaccination cohorts (n = 15 per vaccine per dose) simply failed to generate a viremia sufficient for seroconversion. Modifications including administering a higher vaccine dose and modifying vaccines with multiple inserts exhibiting the best neutralization response (WNVΔVax-2b and WNVΔVax-3) could be explored. Additionally, a prime-boost strategy could be used to increase the percentage of animals that seroconvert. Furthermore, because a transient viremic phase would be expected in vivo in vaccinees, this vaccine could be transmitted to other individuals via mosquito vectors. Vector-specific miRNA targets could be incorporated as a further level of safety. This study provided in vitro evidence for the potential that insect-specific targets could be employed to block transmissibility by mosquitoes.
Overall, this platform for WNV vaccination demonstrates high efficacy and a strong safety profile. This miRNA-mediated attenuation strategy could also be manipulated for further study of viral pathogenesis and development of targeted therapeutics. Once optimized, these vaccines have the potential to induce precise and specific immunity in their host while incorporating several targeted mechanisms to prevent the generation of viral attenuation escape mutants.
Supplementary Material
Acknowledgments
This work was supported by a developmental proposal from the Pacific Southwest Regional Center for Excellence (PSWRCE) U54 AI065359. Funding for AMBL, JLK and EAD was provided American Society for Microbiology postdoctoral fellowships. CMB acknowledges financial support from the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate, Department of Homeland Security and Fogarty International Center, National Institutes of Health.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2016.08.063.
References
- 1.Reimann CA, Hayes EB, DiGuiseppi C, Hoffman R, Lehman JA, Lindsey NP, et al. Epidemiology of neuroinvasive arboviral disease in the United States, 1999–2007. Am J Trop Med Hyg. 2008;79:974–9. [PubMed] [Google Scholar]
- 2.Hayes EB, Komar N, Nasci RS, Montgomery SP, O’Leary DR, Campbell GL. Epidemiology and transmission dynamics of West Nile virus disease. Emerg Infect Dis. 2005;11:1167–73. doi: 10.3201/eid1108.050289a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen LR, Marfin AA. West Nile virus: a primer for the clinician. Ann Intern Med. 2002;137:173–9. doi: 10.7326/0003-4819-137-3-200208060-00009. [DOI] [PubMed] [Google Scholar]
- 4.Watson JT, Pertel PE, Jones RC, Siston AM, Paul WS, Austin CC, et al. Clinical characteristics and functional outcomes of West Nile Fever. Ann Intern Med. 2004;141:360–5. doi: 10.7326/0003-4819-141-5-200409070-00010. [DOI] [PubMed] [Google Scholar]
- 5.Hayes EB, Sejvar JJ, Zaki SR, Lanciotti RS, Bode AV, Campbell GL. Virology, pathology, and clinical manifestations of West Nile virus disease. Emerg Infect Dis. 2005;11:1174–9. doi: 10.3201/eid1108.050289b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi SL, Ross TM, Evans JD. West Nile virus. Clin Lab Med. 2010;30:47–65. doi: 10.1016/j.cll.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou S, Foster GA, Dodd RY, Petersen LR, Stramer SL. West Nile fever characteristics among viremic persons identified through blood donor screening. J Infect Dis. 2010;202:1354–61. doi: 10.1086/656602. [DOI] [PubMed] [Google Scholar]
- 8.Petersen LR, Brault AC, Nasci RS. West Nile virus: review of the literature. JAMA, J Am Med Assoc. 2013;310:308–15. doi: 10.1001/jama.2013.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mostashari F, Bunning ML, Kitsutani PT, Singer DA, Nash D, Cooper MJ, et al. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet. 2001;358:261–4. doi: 10.1016/S0140-6736(01)05480-0. [DOI] [PubMed] [Google Scholar]
- 10.Clark DC, Brault AC, Hunsperger E. The contribution of rodent models to the pathological assessment of flaviviral infections of the central nervous system. Arch Virol. 2012;157:1423–40. doi: 10.1007/s00705-012-1337-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinton MA, Dispoto JH. Sequence and secondary structure analysis of the 5′−terminal region of flavivirus genome RNA. Virology. 1988;162:290–9. doi: 10.1016/0042-6822(88)90468-0. [DOI] [PubMed] [Google Scholar]
- 12.Brinton MA, Fernandez AV, Dispoto JH. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology. 1986;153:113–21. doi: 10.1016/0042-6822(86)90012-7. [DOI] [PubMed] [Google Scholar]
- 13.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008;4:579–91. doi: 10.1016/j.chom.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, et al. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010;84:11407–17. doi: 10.1128/JVI.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–48. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–48. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiss BL, Maximova OA, Thach DC, Speicher JM, Pletnev AG. MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J Virol. 2012;86:5647–59. doi: 10.1128/JVI.07125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinney RM, Huang CY, Whiteman MC, Bowen RA, Langevin SA, Miller BR, et al. Avian virulence and thermostable replication of the North American strain of West Nile virus. J Gen Virol. 2006;87:3611–22. doi: 10.1099/vir.0.82299-0. [DOI] [PubMed] [Google Scholar]
- 20.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol: CB. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 21.Fagegaltier D, Konig A, Gordon A, Lai EC, Gingeras TR, Hannon GJ, et al. A genome-wide survey of sexually dimorphic expression of drosophila mirnas identifies the steroid hormone-induced miRNA let-7 as a regulator of sexual identity. Genetics. 2014;198(2):647–68. doi: 10.1534/genetics.114.169268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–62. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forster JI, Koglsberger S, Trefois C, Boyd O, Baumuratov AS, Buck L, et al. Characterization of differentiated SH-SY5Y as neuronal screening model reveals increased oxidative vulnerability. J Biomol Screen. 2016;21:496–509. doi: 10.1177/1087057115625190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brault AC, Langevin SA, Bowen RA, Panella NA, Biggerstaff BJ, Miller BR, et al. Differential virulence of West Nile strains for American crows. Emerg Infect Dis. 2004;10:2161–8. doi: 10.3201/eid1012.040486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang CY, Butrapet S, Pierro DJ, Chang GJ, Hunt AR, Bhamarapravati N, et al. Chimeric dengue type 2 (vaccine strain PDK-53)/dengue type 1 virus as a potential candidate dengue type 1 virus vaccine. J Virol. 2000;74:3020–8. doi: 10.1128/jvi.74.7.3020-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed LJ, Muench H. A Simple method of estimating fifty per cent endpoints. Am J Epidemiol. 1938;27:493–7. [Google Scholar]
- 27.Teferedegne B, Murata H, Quinones M, Peden K, Lewis AM. Patterns of microRNA expression in non-human primate cells correlate with neoplastic development in vitro. PLoS ONE. 2010;5:e14416. doi: 10.1371/journal.pone.0014416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–3. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278–83. doi: 10.1038/nm.1776. [DOI] [PubMed] [Google Scholar]
- 30.Teterina NL, Liu G, Maximova OA, Pletnev AG. Silencing of neurotropic flavivirus replication in the central nervous system by combining multiple microRNA target insertions in two distinct viral genome regions. Virology. 2014:456–457. 247–58. doi: 10.1016/j.virol.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.tenOever BR. RNA viruses and the host microRNA machinery. Nat Rev Microbiol. 2013;11:169–80. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- 32.Jovicic A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci: Off J Soc Neurosci. 2013;33:5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


