Abstract
A templated assembly approach for Hepatitis B virus-like particles was employed to determine how the T = 3 and T = 4 polymorphs of the Hepatitis B virus (HBV) icosahedral cores respond to a systematic, gradual change in the encapsulated cargo size. It was found that assembly into complete virus-like particles occurs cooperatively around a variety of core diameters, albeit the degree of cooperativity varies. Among these virus-like particles, it was found that those of an outer diameter similar to T = 4 are able to accommodate the widest range of cargo sizes.
Keywords: hepatitis B virus, physical virology, directed assembly, encapsulation, nanomedicine
The most generic representation of a virus is embodied by a protein cage encapsulating macromolecules necessary for replication of the entire complex by a host. Though many viruses are more complex, even in this generic view the protein cage is more than a passive container of genomic cargo. Its outstanding capacity of responding to environment changes, crucial to virus replication, relies on structural and compositional properties that have just recently begun to be understood.1
In addition to an external environment, virus shells also have to adapt to the presence and state of their cargo. Understanding the specifics of cargo/protein interaction in viruses is central not only for biological function but also for virus-based nanotechnological applications. More specifically, virus-like particles hold promise as therapeutic or diagnostic delivery agents.2–5 Moreover, DNA is among the most dense and stable information storage media known, and has been proposed as a viable solution for high latency future information storage needs.6 As a consequence, bioinspired strategies for packaging DNA could take advantage of and even enhance information storage density.
One of the most striking examples of efficient genome packaging is the case of dsDNA bacteriophages, which spool their long nucleic acid molecules into a pre-assembled icosahedral capsule by means of an ATP-driven packaging motor.7, 8 In this case too, specific virus shell properties enable DNA to be packaged at close to crystalline densities. Thus, significant hydration forces, electrostatic repulsion and bending of the rigid dsDNA have to be overcome in the process.9 As a consequence, the internal pressure that the capsid has to withstand was estimated at ~ 30 atm, to take the Φ29 phage for example.10 Such great differences in internal pressure between empty and filled capsids are often accommodated by maturation processes involving global morphological and compositional changes, as in the case of HK9711 or T712 phages.
The most plentiful viruses on the planet are (+) strand RNA viruses,13 whose genomes can be directly translated into proteins by host ribosomes. Single stranded RNA is much more flexible than double stranded DNA and its packaging generally occurs by spontaneous assembly of capsid proteins and nucleic acids without help required from powerful ATP-driven motors (such as in the case of certain bacteriophages), and even could be achieved in vitro, in total absence of a cellular environment.14 For genome presentation, capsids of some RNA viruses disassemble inside the cell, while in others, e.g. picornaviruses, the genome appears to be injected into the cytoplasm via a channel through the capsid.15 In either case, the switch between stable and unstable states occurs in response to chemical cues. Because in their life cycle ssRNA viruses take advantage of this purposefully marginal stability, RNA virus capsids are orders of magnitude weaker, by nanoindentation probing, than their bacteriophage counterparts.16 Therefore, from a nanotechnology perspective, RNA virus capsids are expected to offer a different set of challenges and opportunities than bacteriophages do.
An intriguing case situated between the two extremes of dsDNA phages and ssRNA viruses, is the Hepatitis B virus (HBV), an important pathogen responsible for hepatitis and liver cancer in millions of people.17 In HBV, the genome is initially incorporated into the assembling capsid as a single-stranded RNA molecule.18 The result of this process can be reproduced in vitro, by spontaneous assembly similar to that of many other ssRNA viruses.19 However, in this case, the RNA pregenome is subsequently retrotranscribed into circular dsDNA inside the capsid by the viral reverse transcriptase.20 The stiffness of the packaged dsDNA adds strain to the capsid, decreasing its stability.21 In the light of the above comparison between dsDNA and ssRNA virus mechanics, it is conceivable that the passage from a ssRNA to a dsDNA-loaded nucleocapsid is facilitated by specific but currently unknown capsid adaptation features.
However, interpreting data on capsid stability is complicated by the fact that HBV has the unusual property of packaging its genome in particles of two different sizes corresponding to T numbers of 3 (180 capsid proteins) and 4 (240 capsid proteins).22 While single-particle nanoindentation studies performed by atomic force microscopy indicated that both polymorphs have similar stabilities to mechanical compression (within current detection limit of atomic force microscopy),23 subunit exchange rates measured by native mass spectrometry showed marked differences between T = 3 and T = 4 capsids.24
The goal of the work reported here is to determine by alternative techniques whether the two morphologies respond differently to changes in the physical properties of the cargo. By using the nanoparticle template method,25–30 we have been able to show that T = 4 capsids are more resilient than T = 3 capsids to internal constraints in the form of changes in the size of the cargo. These results align with Uetrecht et al. findings by mass spectrometry24 of a T = 3 particle which more readily exchanges protein subunits (dimers) with the surrounding solution than a T = 4 particle, which has undetectable exchange rates, and also with the observation by Bourne et al. that T = 4 particles can undergo symmetry-preserving structural changes in response to small-molecule drugs, suggesting increased T = 4 flexibility.31 Furthermore, protease sensitivity experiments suggest that HBV capsids are highly dynamic.20
Mature HBV is an enveloped virus consisting of an outer lipid envelope studded with glycoproteins that surrounds an inner icosahedral capsid containing a gaped, 3.2 kb dsDNA genome. The HBV capsid protein (CP) is a polypeptide of 183 residues, composed of two major domains: the N-terminus domain - or the “assembly domain” (residues 1–149) - and a highly basic C-terminus domain - or “protamine” domain (residues 150–183).32, 33 The N-terminus domain embodies the minimal component required for in vitro self-assembly.34 The basic C-terminal domain is flexible, is on the interior of the capsid, and is believed to interact electrostatically with the genome.30, 35–39 The T = 3 and T = 4 capsid polymorphs have been observed with same relative abundances in both in vivo and in vitro samples.18 RNA-filled recombinant HBV virus-like particles, which share the same morphological properties as their natural counterparts, can be expressed in large quantity in E. coli.40 Furthermore, previous experimental evidence has suggested that the specific encapsidation of the virus genome inside its native host is achieved by the virus polymerase and a highly conserved stem loop in the 5′ sequence of pre-genomic RNA.41, 42 In E. coli, which lacks the polymerase and the HBV genome, the capsid packages random RNAs, such as the E. coli RNAs and also can package genomic RNAs of other viruses, suggesting the involvement of a nonspecific, electrostatic component in the interaction between HBV capsid and its cargo.19
This non-specific character of the native interaction between the positively charged carboxy-terminal protamine-like domain of the CP (CTD) and the viral RNA was exploited here to encapsulate functionalized Au nanoparticles (NPs) of various sizes instead of the native pre-genomic RNA (pgRNA) and examine the effect of increasing the diameter of encapsulated NP on the outer diameter of the virus-like particle (VLP), Fig. 1. The main question is therefore whether the virus shell is able to accommodate in a T number-dependent manner an increase in the core diameter as a consequence of the same capsid properties that allow the shell to accommodate internal retro-translation.
Figure 1.
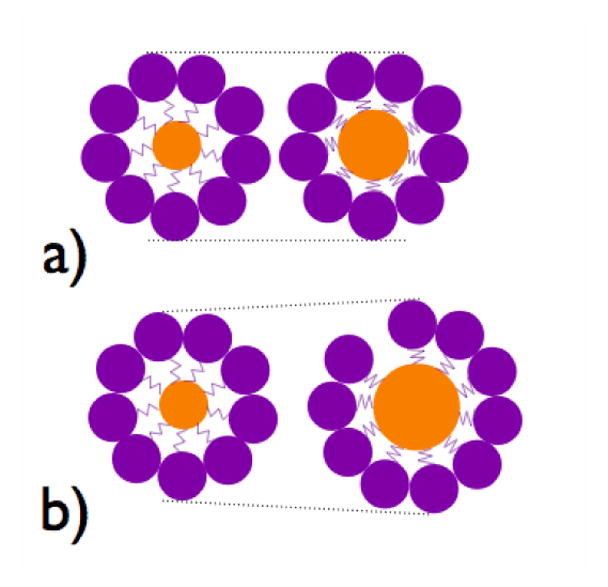
Schematic principle of testing the possibility for differential response of capsid polymorphs to an increase in the cargo size. a) closed capsid resisting to an increase in cargo diameter. b) CP shell adapting to increase in cargo size by an increase in the outer diameter facilitated by packing defects (opening).
RESULTS AND DISCUSSION
Testing capsid stability by template assembly is based upon the assumption that the energetics of assembling a CP shell around a cargo is similar to that of keeping a cargo contained within a capsid. Thus, in the experiments described here we are measuring resilience to an increase in cargo size by the magnitude of change by which the outer diameter yields to changes in the NP core diameter. A less stable capsid will more readily yield to internal pressure and change its outer diameter than a more stable capsid, even if this means going away from the preferred curvature of the coat proteins themselves.43 Note that, with respect to previous work by AFM nanoindentation,23 in which stress was applied from the outside, in the template assembly approach, stress is applied by the NP template from the inside of the forming CP shell.
Two main experimental goals were pursued: First, we determined whether assembly of Au NP encapsulating VLPs exhibit similar cooperativity and selectivity with the HBV capsid assembly19 and similar structures with those encountered in wild-type viruses, and second, whether the T = 3 and T = 4 polymorphs exhibit different responses to cargo size increase.
HBV VLP assembly is cooperative
Pre-genomic RNA interacts strongly with the CTD,19 which is thought to carry the role of a nucleic acid chaperone.44, 45 In unphosphorylated capsids, pgRNA forms an icosahedral cage indicative of a partially double-stranded state, while in a capsid constructed of a phosphorylation mimic, the RNA distribution is less condensed (based on cryo-em data) and more consistent with a single-stranded state. At the same time, the assembly domain structure remains relatively unperturbed.45 Thus, because differences in the RNA structure do not appear to greatly influence the assembly domain, we surmise that despite the fact that the HBV CP is different from the canonical coat-protein model the interaction between cargo and CP can be modeled in terms of an internal, flexible domain responsible for the nucleic acid/coat protein interface, and a structural domain, principally responsible for icosahedral assembly. In this prototypical view, valid for some other simple icosahedral viruses, the carboxy-terminal and assembly domains act as parts mechanically decoupled by a hinge region.46 This organization explains why Au NP VLPs of the Brome mosaic virus could tolerate small variations in the diameter of their encapsulated colloidal nanoparticle cargo, without significant changes of their outer surface structure, Fig. 1 (a).47
HBV architecture is unlike that of Brome Mosaic virus in that subunit-subunit interactions are much stronger, they are based on hydrophobic interactions rather than hydrogen bonds, and HBV forms T=4 as well as T=3 icosahedra. Therefore it was unclear whether HBV would behave similarly with a prototypical icosahedral virus such as BMV in terms of templated assembly. Thus, we first tested the ability of HBV CP to form closed shells around NPs. HBV capsids have a lumen diameter of ~ 19 nm for T = 3 and ~ 23 nm for T = 4.48 Assuming that one of the principal driving forces for in vitro assembly is electrostatic attraction between RNA and the C-termini of the CP,19 a negative surface charge density was supplied in the form of a carboxyl-terminated polyethylene glycol brush covalently bound to the Au NP surface by a thiol bond.25 PEG prevents the NP surface from perturbing CP function, while the charge density of ~ 5e/nm2 (presuming dissociation of all the carboxyl moieties the surface) drives the assembly.49 The PEG brush thickness was ~2 nm by negative stain transmission electron microscopy (EM). Thus, particle cores of 15 nm nominal diameter had a 19 nm diameter after pegylation (commensurate with the T = 3 lumen) while the 19 nm NP cores achieved a diameter of ~23 nm (commensurate with the T = 4 lumen). Upon mixing of CPs with NPs, assembly was induced by dialysis into assembly buffer.
In vitro, and in absence of a polyanionic cargo, the CP-CP interaction (plausibly involving hydrophobic interactions, and hydrogen bonding) will support formation of empty capsids. In presence of RNA, empty particles, aggregates, and filled particles will form.45 With respect to empty capsids, the role of a polyanionic core is both kinetic and thermodynamic,28, 30, 37 because it decreases the kinetic barrier to VLP assembly with respect to that for empty capsid formation by increasing, through long-range non-specific electrostatic interactions, the local CP concentration around the NP core, it may promote VLP assembly by activating the CP into an assembly-active state, thus reducing the energy barrier required for VLP assembly, and it shifts the minimum Gibbs free energy of the final structure as result of contributions from shell reorganization by cooperative CP-CP interactions under core-subunit binding constraints.50
We observed similar outcomes for HBV VLP assembly, but also differences. Thus, negative stain TEM data provide evidence for encapsulation of Au NPs and protein shell assembly exhibiting regular capsomer array signatures are suggestive of capsid formation, Fig. 2 a)–b).
Figure 2.
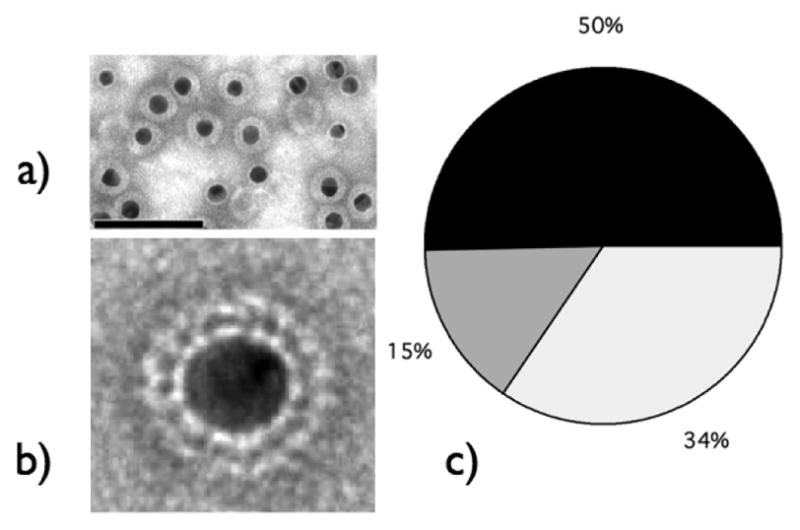
a) Completely formed VLPs and bare Au NPs are the two main states in which a Au NP can be found at CP concentrations below NP surface saturation value. Scale bar: 100 nm. b) TEM picture of VLP encapsulating a 15 nm NP core is suggestive of a regular array of CPs. c) Pie-chart showing relative frequencies of complete VLPs (black), incomplete VLPs (dark gray), and bare NPs for an initial CP:NP molar ratio of 50.
A hallmark of wt HBV assembly is the cooperative character of protein subunit interactions.19 Cooperativity of assembly arises in part from the multivalent character of CP. It has been noted that HBV assembly appears to be allosterically propagated by an induced fit mechanism, which will also contribute to cooperativity. Preservation of this feature in HBV VLP assembly is important because, if the subunit interaction energy were much weaker than the driving force for subunit adsorption onto the NP core, nonspecific association of subunits with the nanoparticle core may result in protein shells ridden with defects in jammed conformations.51 In NP-template assembly, the strength of CP/NP interaction can be adjusted by chemical means,49 an approach adopted here, too.
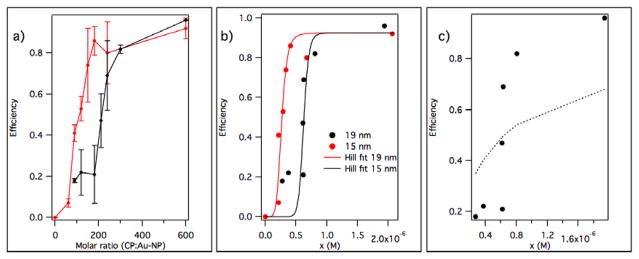
To investigate cooperativity, efficiency of assembly measured by TEM was determined through titration experiments of NP solutions by CP in assembly buffer. Efficiency of assembly was defined here as the ratio between the number of apparently complete VLPs to the total number of NPs. The shape of the titration curve, Fig. 3, provides a test for self-assembly cooperativity as discussed in the following.
Figure 3.
a) Assembly efficiency as a function of CP:NP molar ratio for the two core sizes which correspond to the T=3 and T=4 HBV capsid lumen diameters, respectively (error bars from replicates). b) Same data but shown against estimated free CP molar concentration, x. Lines represent Hill plots (eq. 1). c) A Langmuir model (dotted line) cannot fit the 19 nm data.
As expected for the formation of a spherical-templated monolayer, efficiencies of assembly plateaued at close to 90% for both diameters. In Fig. 3 a sharp rise is easily noticeable only above a certain pseudo-critical concentration.52 This is a feature suggestive of cooperativity.49, 53 For 15 nm NPs, efficiencies higher than ~ 80% are attained for ~ 180 CPs per NP. However, for 19 nm NPs, the same elevated efficiency values are attained only after ~ 290 CPs per NP. The ratio of the two saturation values is 290/180 = 1.61, which is close to the surface area ratio of the two NPs: (19/15)2 = 1.604. We infer that, at least initially, the NP surface area available strongly influences CP binding to the gold particle However, a simple Langmuir non-cooperative binding model to the surface (Fig. 3 c) would predict a rise from 0. If, on the other hand, based upon the small fraction of incomplete VLPs, we assume that binding occurs in such a way that the number of CPs bound may be either zero or q, according to the reaction:
Where A1 represents CP subunits in monomeric form (actually dimers), B is the Au NP, and AqB represents the complete VLP, then we can plot the encapsulation efficiency η vs. the free protein concentration, x, Fig. 3 b):
and
| (1) |
with r being the molar fraction CP : Au NP, [B]0 the concentration of Au NPs (kept constant at 4 nM), β is a fitting factor of 0.8–0.9 accounting for the fact that some of the protein is lost in empty capsids and aggregates, and η representing the average number of filled capsids per nanoparticle (efficiency). The Hill coefficient values for the plots in Fig. 3 b are q = 6±2 (K = 2.8±0.9 × 1039) for 15 nm NPs, and q = 15±4 (K = 1.2±0.5 × 1093) for 19 nm NPs. These are rough estimates limited by statistical data quality and validity of the original assumption of a binary mixture of CP states, but they do suggest that: (1) CP:NP binding is cooperative for both T=4 and T=3 VLPs, (2) binding constants appear to be different between T=3 and T=4. It also is interesting to note that, for BMV which has weaker CP-CP interactions than HBV, a much stronger cooperative character (as measured by the magnitude of q) was observed in similar in vitro experiments.49
To further test for selective assembly, a competition assay with a mixture of different particle diameters was performed, at a molar ratio (CP: Au NP) of 50, that is in conditions of limited CP supply, Fig. 4. Particle diameters constituting the extremes and the mean of the range for which assembly was observed were chosen. In a purely nonselective adsorption scenario, competition between different particle diameters for CP would lead to a monotonic decrease of the fraction of complete particles with core radius (red dots in Fig. 4). However, experimental evidence shows the trend is clearly non-monotonic, peaking at ~17 nm, (Fig. 4). A similar effect was recently observed in in vitro encapsidation experiments of RNA of different length by CCMV.54 Presence of selectivity and cooperativity, two characteristics of the native subunit-subunit interactions, is important because it suggests that native interactions are preserved in VLPs and electrostatic association between cores and CP does not purely dominate the assembly process.
Figure 4.
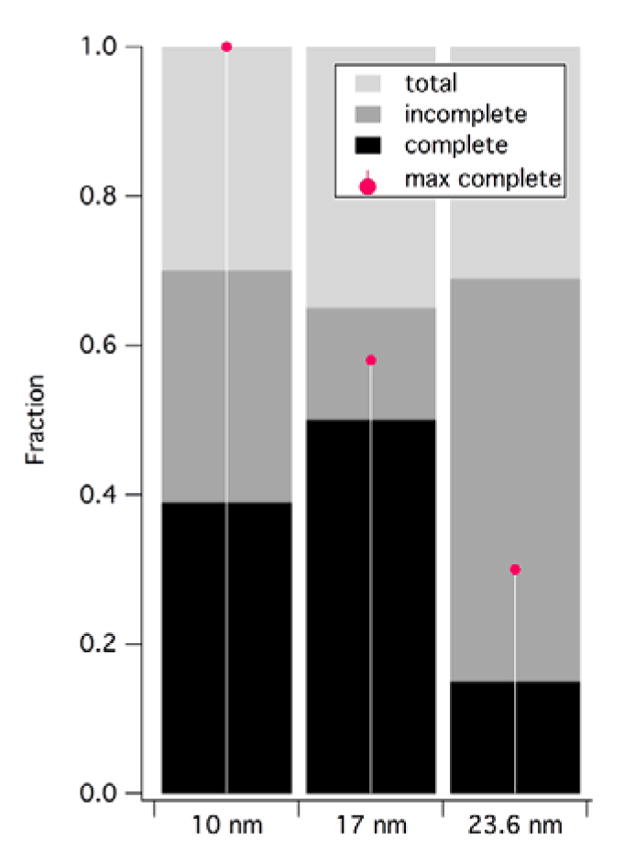
Fractions of complete and incomplete VLPs from a competition experiment in conditions of limited CP supply. Red dots: expected from non-cooperative adsorption.
We have mentioned that, in principle, the magnitude of the electrostatic driving force can be adjusted by mixing charged and neutral ligands on the core NP surface. Efficiency of encapsulation of 15 nm particles is represented as a function of both the NP surface charge and the number of net positive charges on the CP C-termini in Fig. 5. In these experiments, the surface charge for four different 15 nm NP preparations was adjusted by ligand exchange with different mixtures of PEG-COOH and PEG-OH, which yielded 100%, 80%, 60% and 33% PEG-COOH coverage, respectively, as determined by gel electrophoresis and dynamic light scattering (DLS).55 Besides wt CP, two HBV mutants, denoted as Cp183EEE and Cp154, were used to decrease the protein charge and thus compensate for the decreased core charge. With respect to the wt protein, Cp154 is a truncated mutant that has only 4 positive charges of the C-terminus. Cp183EEE is a phosphorylation-mimicking mutant used in previous studies.19 It has the same length as Cp183, but three serines were mutated to glutamic acids, making the net charge on its tail drop from +15 to +12. Therefore, decreasing the net surface charge and compensating for it by decreasing the charge on the CP tails does not restore efficiency. This is not surprising because reduction in the number of charges also reduces the free energy of binding.30, 36 This changes the critical association concentration, and hence the efficiency.28 This hypothesis has been tested for the 15 nm particles by reconstituting VLPs at higher CP:AuNP molar ratios and measuring assembly efficiency (Table 1)
Figure 5.
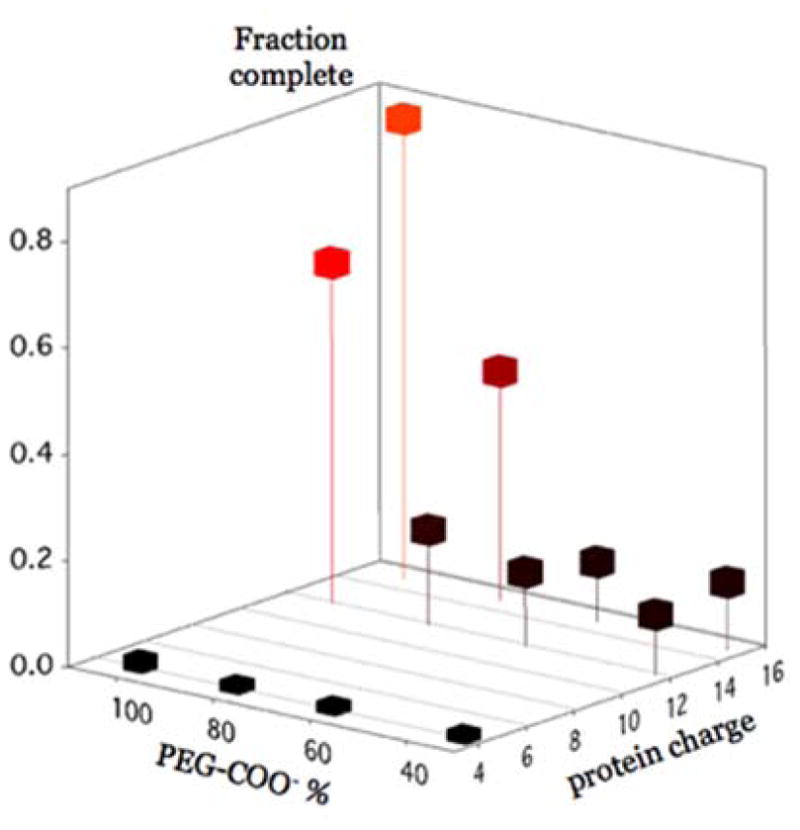
Efficiency of encapsulation of 15 nm diameter NPs as a function of PEG-COOH coverage and number of positive charges per CP tail.
Table 1.
| Protein:Core Ratio | Cp183wt + 15nm Au-60%PEG-COOH | Cp183wt + 15nm Au-80%PEG-COOH |
|---|---|---|
| 180 | 0.113 | 0.431 |
| 300 | 0.116 | 0.747 |
| 600 | 0.708 | 0.808 |
Mapping stability by templated assembly
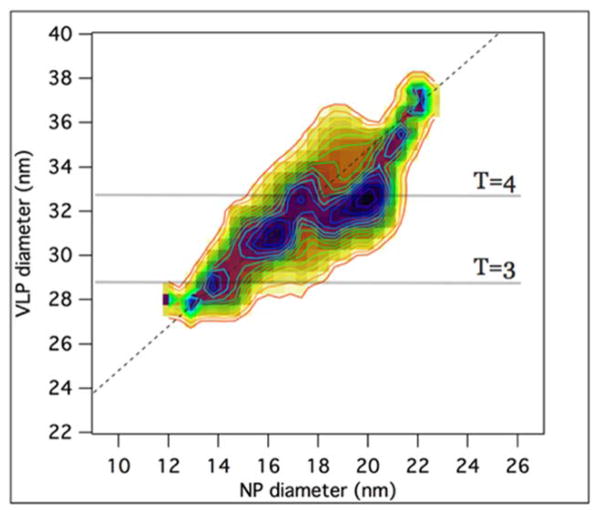
Finally, to establish whether the CP shells corresponding to T = 3 and T = 4 numbers adapt differently to an increase in the size of the NP core, we measured the NP diameter and the VLP diameter for ~ 700 VLPs formed from NP sizes with a distribution spanning 12–23 nm, Fig. 6. Since we are interested in the correlation between the NP core diameter and the VLP outer diameter, a joined histogram was constructed, which provides a map of probability density for the NP diameter-VLP diameter pair. The 2D probability density map was normalized to the starting NP core diameter probability density to account for the fact that the latter was not flat. Clearly, the NP core diameter affects the VLP size distribution. The NP cargo strongly biases the assembly towards the T number that best matches the NP, thus reducing capsid polymorphism.51
Figure 6.
Probability density map of VLP diameters resulting from certain NP core diameters. The dotted line represents the increase in VLP diameter with NP core diameter in conditions of a hypotethical constant protein layer thickness of 7.5 nm.
The distribution exhibits statistically relevant local maxima corresponding to 32.7 nm, 30.8 nm, and 28.3 nm VLP diameters. The most prominent, 32.7 nm maximum is close to the value reported for T = 4 diameter (~ 33 nm).48 Note that this outer diameter value also corresponds to a relatively broad range of core sizes (17 to 20 nm diameter). Moreover, at constant concentration of all species, mass action would work in favor of the smaller species.50
The 28.5 nm maximum is close to the value expected for a T = 3 particle diameter. With respect to the T = 4 VLP, this maximum corresponds to a much narrower range of core diameters, suggesting cores outside this range are tolerated less and lead the outer diameter to change.
A third local maximum can be observed with a VLP diameter of 30.8 nm (core diameter 16.5 nm). However, this local maximum could not be assigned to a T number. Assuming same packing density as a T = 4, it would correspond to ~ 210 subunits. Its morphology is unknown. It may represent a kinetic intermediate or a non-quasi-equivalent structure of lower stability relative to T = 4.
The dotted diagonal in Fig. 6 represents the hypothetical increase in VLP diameter as a function of core diameter assuming a constant protein layer thickness (of ~ 7.5 nm). We see that the data deviates significantly from the straight line at ~ T = 4 diameter values, then it gradually regains it. In the vicinity of the T = 4 VLP size on the Fig. 6 map, the outer diameter remains more or less constant. This means that the inner portion of the CP has to adapt, probably by a conformational change, to the increase in cargo diameter, a behavior not seen yet for the putative T = 3 particles or for other viruses that have been tested against NP encapsulation.27, 56 Outside of the narrow range where geometry specifies a Caspar-Klug approved diameter, we suggest that the cores acquire a CP coat with numerous disclination defects, arguably not dissimilar to what is observed in colloidosomes in which colloidal particles decorate the surface of a spherical droplet.57 Disclinations result in a higher energy for the complex relative to defect-free shells, decreasing the efficiency of complex formation and also its stability. The weaker association of defective capsids in turn is also expected to favor increased formation of kinetically accessible empty capsids.
CONCLUSION
It was found that the coat protein of HBV has high affinity for nanoparticle cores functionalized with a polyanionic coat which they bind to in a cooperative manner. Encapsulation efficiencies peak at diameters commensurate with Caspar-Klug T numbers. However, we have also observed a relatively frequent occurrence of VLP diameters which cannot be assigned to icosahedral structures.
When the diameter of the NP core is increased, for most core diameters, the HBV VLP diameter follows the cargo size increase linearly. However, once the VLP diameter reaches the value expected for T = 4, it remains unchanged upon further increase of the core diameter of up to 18%. In contrast, T = 3 VLPs will react immediately to change in the cargo diameter, presumably by accommodating new proteins and defects in the shell. The difference suggests that the T=4 HBV capsids may have a built-in mechanism of adaptation to changes in the cargo size which is absent in T = 3 capsids. This could be important for nanomaterials applications since it provides an intriguing example of two cage architectures built by self-assembly from the same subunits, but having different stabilities.
MATERIALS AND METHODS
Synthesis of Functionalized Gold Nanoparticles
GNPs of different sizes, including 10.0nm (10 nm), 15.5nm (15 nm), 16.8nm (17nm), 19.1nm (19 nm), 23.6nm (23 nm) were prepared by citrate reduction of HAuCl4.58 Ligand exchange reactions were carried out by adding a large excess of thiol ligand (10 equiv/particle) to Au-citrate NPs solution.59 The amount of the ligand was estimated by assuming that the surface area occupied by a single thiol molecule is 0.16 nm.2 The mixture was then stirred at room temperature for 48 h and then subjected to centrifugation at 14,000 rpm for 15 min. The supernatant was removed and the pellet was resuspended in water and subjected to sonication for 10 min. Centrifugation was carried out under the same conditions for two more times in order to completely eliminate unreacted ligands.
Two ligands were used for varying the anionic surface charge density as described in previous work:55 thiolated carboxylated tetraethylene glycol (PEG) ligand (HS-C11-TEGCH2-COOH) and hydroxylated TEG ligand (HS-C11-TEG-OH). Functionalization of gold nanoparticles was performed by exchanging the initial citrate molecules with a mixture composed of different molar ratios of PEG-COOH and PEG-OH ligands.
The PEG-COOH coverage was verified by dynamic light scattering (DLS) and gel electrophoresis as described previously.55 In short, dynamic light scattering (DLS) measurements were carried out with a Zetasizer NanoS (Malvern Instruments). Gold NPs functionalized with different amount of PEG-COOH were first diluted, then sonicated for 10–20 min, and filtered through a 0.22 μm syringe filter. Measurement duration was set to be determined automatically, and data were averaged from at least three runs. Intensity and volume distributions of the particle sizes were recorded. For gel electrophoresis experiments, 8μL of each sample were loaded on 0.6% agarose gel together with 2μL of pure glycerol and run in TAE buffer (40 mM Tris, 1 mM EDTA, pH 7.4).
Preparation of HBV-VNPs
HBV capsid protein sequences (Cp183, Cp183-EEE, and Cp154) with a C-terminal domain codons optimized for expression in E. coli were used for HBV capsid protein expression.19 HBV capsids from E coli lysate were purified by ammonium sulfate precipitation and size exclusion chromatography and then stored in −80 °C. Before the experiments, the HBV capsids were dissociated and separated from E coli RNA by dialysis into 1.5M GnCl, 0.5M LiCl, 50mM Tris-base pH 9.5 and 5mM DTT overnight followed by separation through a Sucrose 6 10/300 column. The mutant protein Cp154 is a deletion mutant with the last 29 C-terminus amino acids of CP183 truncated. Cp154 retains the ability to form virus-like particles both in vitro and in the cell expression system. Another mutant protein (Cp183-EEE) contains three Ser to Glu mutations corresponding to the significant phosphorylation sites S155, S162, and S170. Previous experiments have demonstrated that Cp183-EEE successfully packages genomic RNA in cell culture60 as well as random RNA in vitro.19
For assembly, protein dimers were mixed with gold nanoparticles of different sizes and functionalizations, and the mixtures dialyzed against assembly buffer: with 0.1 M NaCl, 50 mM Tris-base pH 7.5 (low ionic strength buffer) or 1 M NaCl, 50 mM Tris-base pH 7.5 (high ionic strength buffer) overnight. Typical initial concentrations of protein and NP in these reactions were 0.5–1 μM protein dimer and 3–5 nM NP.
VLP Analysis
HBV VLPs were first centrifuged at 14,000 rpm in 10% sucrose cushion for 10 min and then sonicated for 10min. The step was repeated for up to 6 times in order to eliminate the protein aggregates and empty capsids. 10μL of the purified sample was put on a glow-discharged carbon-coated copper grid. After 10 min, the excess solution on the grid was removed with filter paper. A 10μL portion of 2% uranyl acetate was used to stain for 10s. Excess solution was removed by blotting with filter paper. The sample was then left to dry for at least 30 min.
The assembly efficiency of VLPs was determined from the TEM images by dividing the number of complete VLPs over the total number of gold particles (involved in complete VLPs, incomplete VLPs or bare gold NPs). Images were taken from at least two independent TEM grids. Average diameters of VLPs were obtained from a horizontal and a vertical measurement per particle using ImageJ software. 300–800 particles were analyzed for each sample.
Acknowledgments
The authors gratefully acknowledge funding from the International Human Frontier Science Program Organization (B.D.), the National Science Foundation (B.D., grant #1014947), the National Institutes for Health (A.Z., grant R01 AI077688), and helpful discussions with Dr. Irina Tsvetkova.
References
- 1.Veesler D, Johnson JE. Virus Maturation. Annu Rev Biophys. 2012;41:473–496. doi: 10.1146/annurev-biophys-042910-155407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douglas T, Young M. Viruses: Making Friends with Old Foes. Science. 2006;312:873–875. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- 3.Manchester M, Steinmetz FN. Viruses and Nanotechnology. Preface. Curr Top Microbiol Immunol. 2009;327:v–vi. [PubMed] [Google Scholar]
- 4.Irvine DJ. Drug Delivery: One Nanoparticle, One Kill. Nat Mater. 2011 Jun 01;:342–343. doi: 10.1038/nmat3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minten IJ, Claessen VI, Blank K, Rowan AE, Nolte RJM, Cornelissen JJLM. Catalytic Capsids: the Art of Confinement. Chem Sci. 2011;2:358–362. [Google Scholar]
- 6.Church GM, Gao Y, Kosuri S. Next-Generation Digital Information Storage in DNA. Science. 2012;337:1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 7.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The Bacteriophage Straight Phi29 Portal Motor Can Package DNA Against a Large Internal Force. Nature (London, U K) 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 8.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic Pressure Inhibition of DNA Ejection From Phage. Proc Natl Acad Sci U S A. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purohit PK, Kondev J, Phillips R. Mechanics of DNA Packaging in Viruses. Proc Natl Acad Sci U S A. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelbart WM, Knobler CM. VIROLOGY: Pressurized Viruses. Science. 2009;323:1682–1683. doi: 10.1126/science.1170645. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix RW, Johnson JE. Bacteriophage HK97 Capsid Assembly and Maturation. Adv Exp Med Biol. 2012;726:351–363. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- 12.Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular Mechanisms in Bacteriophage T7 Procapsid Assembly, Maturation, and DNA Containment. Adv Protein Chem. 2003;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- 13.Flint SJ, Enquist LW, Racaniello VR, Skalka AM. Principles of Virology. John Wiley & Sons. 2009:1. [Google Scholar]
- 14.Fraenkel-Conrat H, Singer B. Virus Reconstitution and The Proof of The Existence of Genomic RNA. Philos Trans R Soc, B. 1999;354:583–586. doi: 10.1098/rstb.1999.0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyth MS, Martin JH. Picornavirus Uncoating. Mol Pathol. 2002;55:214–219. doi: 10.1136/mp.55.4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roos WH, Bruinsma R, Wuite GJL. Physical Virology. Nat Phys. 2010;6:733–743. [Google Scholar]
- 17.Ganem D, Prince AM. Hepatitis B Virus Infection--Natural History and Clinical Consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 18.Steven A, Conway J, Cheng N, Watts N, Belnap D, Harris A, Stahl S, Wingfield P. Structure, Assembly, and Antigenicity of Hepatitis B Virus Capsid Proteins. Adv Virus Res. 2005;64:125–164. doi: 10.1016/S0065-3527(05)64005-5. [DOI] [PubMed] [Google Scholar]
- 19.Porterfield JZ, Dhason MS, Loeb DD, Nassal M, Stray SJ, Zlotnick A. Full-Length Hepatitis B Virus Core Protein Packages Viral and Heterologous RNA with Similarly High Levels of Cooperativity. J Virol. 2010;84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassal M. Hepatitis B Viruses: Reverse Transcription a Different Way. Virus Res. 2008;134:235–249. doi: 10.1016/j.virusres.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Dhason MS, Wang JC-Y, Hagan MF, Zlotnick A. Differential Assembly of Hepatitis B Virus Core Protein on Single- and Double-Stranded Nucleic Acid Suggest the dsDNA-Filled Core is Spring-Loaded. Virology. 2012;430:20–29. doi: 10.1016/j.virol.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caspar DLD, Klug A. Physical Principles in the Construction of Regular Viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 23.Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, Steven AC, Heck AJR. High-Resolution Mass Spectrometry of Viral Assemblies: Molecular Composition and Stability of Dimorphic Hepatitis B Virus Capsids. Proc Natl Acad Sci U S A. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uetrecht C, Watts NR, Stahl SJ, Wingfield PT, Steven AC, Heck AJR. Subunit Exchange Rates in Hepatitis B Virus Capsids are Geometry- and Temperature-Dependent. Phys Chem Chem Phys. 2010;12:13368–13371. doi: 10.1039/c0cp00692k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Daniel M-C, Quinkert ZT, De M, Stein B, Bowman VD, Chipman PR, Rotello VM, Kao CC, Dragnea B. Nanoparticle-Templated Assembly of Viral Protein Cages. Nano Lett. 2006;6:611–615. doi: 10.1021/nl0600878. [DOI] [PubMed] [Google Scholar]
- 26.Loo L, Guenther RH, Lommel SA, Franzen S. Encapsidation of Nanoparticles by Red Clover Necrotic Mosaic Virus. J Am Chem Soc. 2007;129:11111–11117. doi: 10.1021/ja071896b. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, DuFort C, Daniel M-C, Murali A, Chen C, Gopinath K, Stein B, De M, Rotello VM, Holzenburg A, Kao CC, Dragnea B. Core-Controlled Polymorphism in Virus-Like Particles. Proc Natl Acad Sci U S A. 2007;104:1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin H-K, van der Schoot P, Zandi R. Impact of Charge Variation on the Encapsulation of Nanoparticles by Virus Coat Proteins. Phys Biol. 2012;9:066004. doi: 10.1088/1478-3975/9/6/066004. [DOI] [PubMed] [Google Scholar]
- 29.Siber A, Zandi R, Podgornik R. Thermodynamics of Nanospheres Encapsulated in Virus Capsids. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2010;81:051919. doi: 10.1103/PhysRevE.81.051919. [DOI] [PubMed] [Google Scholar]
- 30.Hagan MF. A Theory for Viral Capsid Assembly Around Electrostatic Cores. J Chem Phys. 2009;130:114902. doi: 10.1063/1.3086041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, Zlotnick A. Small-Molecule Effectors of Hepatitis B Virus Capsid Assembly Give Insight into Virus Life Cycle. J Virol. 2008;82:10262–10270. doi: 10.1128/JVI.01360-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birnbaum F, Nassal M. Hepatitis B Virus Nucleocapsid Assembly: Primary Structure Requirements in the Core Protein. J Virol. 1990;64:3319–3330. doi: 10.1128/jvi.64.7.3319-3330.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nassal M. The Arginine-Rich Domain of The Hepatitis B Virus Core Protein is Required for Pregenome Encapsidation and Productive Viral Positive-Strand DNA Synthesis but not for Virus Assembly. J Virol. 1992;66:4107–4116. doi: 10.1128/jvi.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceres P, Zlotnick A. Weak Protein-Protein Interactions Are Sufficient to Drive Assembly of Hepatitis B Virus Capsids. Biochemistry. 2002;41:11525–11531. doi: 10.1021/bi0261645. [DOI] [PubMed] [Google Scholar]
- 35.Belyi VA, Muthukumar M. Electrostatic Origin of the Genome Packing in Viruses. Proc Natl Acad Sci U S A. 2006;103:17174–17178. doi: 10.1073/pnas.0608311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Schoot P, Bruinsma R. Electrostatics and the Assembly of an RNA Virus. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2005;71:061928. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- 37.Hu Y, Zandi R, Anavitarte A, Knobler CM, Gelbart WM. Packaging of a Polymer by a Viral Capsid: The Interplay Between Polymer Length and Capsid Size. Biophys J. 2008;94:1428–1436. doi: 10.1529/biophysj.107.117473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Šiber A, Podgornik R. Nonspecific Interactions in Spontaneous Assembly of Empty versus Functional Single-Stranded RNA Viruses. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2008;78:051915. doi: 10.1103/PhysRevE.78.051915. [DOI] [PubMed] [Google Scholar]
- 39.Ting CL, Wu J, Wang Z-G. Thermodynamic Basis for the Genome to Capsid Charge Relationship in Viral Encapsidation. Proc Natl Acad Sci U S A. 2011;108:16986–16991. doi: 10.1073/pnas.1109307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen BJ, Richmond JE. Electron Microscopy Of Hepatitis B Core Antigen Synthesized in. E Coli Nature (London, U K) 1982;296:677–678. doi: 10.1038/296677a0. [DOI] [PubMed] [Google Scholar]
- 41.Hirsch RC, Lavine JE, Chang LJ, Varmus HE, Ganem D. Polymerase Gene Products of Hepatitis B Viruses are Required for Genomic RNA Packaging as well as for Reverse Transcription. Nature (London, U K) 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 42.Junker-Niepmann M, Bartenschlager R, Schaller H. A Short cis-acting Sequence is Required for Hepatitis B Virus Pregenome Encapsidation and Sufficient for Packaging of Foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prinsen P, van der Schoot P, Gelbart WM, Knobler CM. Multishell Structures of Virus Coat Proteins. J Phys Chem B. 2010;114:5522–5533. doi: 10.1021/jp911040z. [DOI] [PubMed] [Google Scholar]
- 44.Lewellyn EB, Loeb DD. The Arginine Clusters of the Carboxy-Terminal Domain of the Core Protein of Hepatitis B Virus Make Pleiotropic Contributions to Genome Replication. J Virol. 2011;85:1298–1309. doi: 10.1128/JVI.01957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang JC-Y, Dhason MS, Zlotnick A. Structural Organization of Pregenomic RNA and the Carboxy-Terminal Domain of the Capsid Protein of Hepatitis B Virus. PLoS Pathog. 2012;8:e1002919. doi: 10.1371/journal.ppat.1002919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson J, Speir J. Quasi-Equivalent Viruses: A Paradigm for Protein Assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068. [DOI] [PubMed] [Google Scholar]
- 47.DuFort CC, Dragnea B. Bio-Enabled Synthesis of Metamaterials. Annu Rev Phys Chem. 2010;61:323–344. doi: 10.1146/annurev.physchem.012809.103300. [DOI] [PubMed] [Google Scholar]
- 48.Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, Ose V, Pumpens P. Three-Dimensional Structure of Hepatitis B Virus Core Particles Determined by Electron Cryomicroscopy. Cell (Cambridge, MA, U S) 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 49.Tsvetkova I, Chen C, Rana S, Kao CC, Rotello VM, Dragnea B. Pathway Switching in Templated Virus-Like Particle Assembly. Soft Matter. 2012;8:4571–4577. [Google Scholar]
- 50.Zandi R, van der Schoot P. Size Regulation of ss-RNA Viruses. Biophys J. 2009;96:9–20. doi: 10.1529/biophysj.108.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elrad OM, Hagan MF. Mechanisms of Size Control and Polymorphism in Viral Capsid Assembly. Nano Lett. 2008;8:3850–3857. doi: 10.1021/nl802269a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zlotnick A. To Build a Virus Capsid. An Equilibrium Model of the Self Assembly of Polyhedral Protein Complexes. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
- 53.Dill KA, Bromberg S. Molecular Driving Forces: Statistical Thermodynamics in Chemistry, Physics, Biology, and Nanoscience. Garland Pub; 2003. [Google Scholar]
- 54.Comas-Garcia M, Cadena-Nava RD, Rao ALN, Knobler CM, Gelbart WM. In Vitro Quantification of The Relative Packaging Efficiencies of Single-Stranded RNA Molecules by Viral Capsid Protein. J Virol. 2012;86:12271–12282. doi: 10.1128/JVI.01695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel M-C, Tsvetkova IB, Quinkert ZT, Murali A, De M, Rotello VM, Kao CC, Dragnea B. Role of Surface Charge Density in Nanoparticle-Templated Assembly of Bromovirus Protein Cages. ACS Nano. 2010;4:3853–3860. doi: 10.1021/nn1005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang CB, Knobler CM, Gelbart WM, Mason TG. Curvature Dependence of Viral Protein Structures on Encapsidated Nanoemulsion Droplets. ACS Nano. 2008;2:281–286. doi: 10.1021/nn700385z. [DOI] [PubMed] [Google Scholar]
- 57.Bausch AR, Bowick MJ, Cacciuto A, Dinsmore AD, Hsu MF, Nelson DR, Nikolaides MG, Travesset A, Weitz DA. Grain Boundary Scars and Spherical Crystallography. Science. 2003;299:1716–1718. doi: 10.1126/science.1081160. [DOI] [PubMed] [Google Scholar]
- 58.Slot JW, Geuze HJ. A New Method of Preparing Gold Probes for Multiple-Labeling Cytochemistry. Eur J Cell Biol. 1985;38:87–93. [PubMed] [Google Scholar]
- 59.Zhu T, Vasilev K, Kreiter M, Mittler S, Knoll W. Surface Modification of Citrate-Reduced Colloidal Gold Nanoparticles with 2-Mercaptosuccinic Acid. Langmuir. 2003;19:9518–9525. [Google Scholar]
- 60.Lan YT, Li J, Liao W-y, Ou J-h. Roles of the Three Major Phosphorylation Sites of Hepatitis B Virus Core Protein in Viral Replication. Virology. 1999;259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]