Abstract
The DREAM (Dp/Retinoblastoma(Rb)-like/E2F/MuvB) transcriptional repressor complex acts as a gatekeeper of the mammalian cell cycle by establishing and maintaining cellular quiescence. How DREAM’s three functional components, the E2F-DP heterodimer, the Rb-like pocket protein, and the MuvB subcomplex, form and function at target gene promoters remains unknown. The current model invokes that the pocket protein links E2F-DP and MuvB and is essential for gene repression. We tested this model by assessing how the conserved yet less redundant DREAM system in Caenorhabditis elegans is affected by absence of the sole C. elegans pocket protein LIN-35. Using a LIN-35 protein null mutant, we analyzed the assembly of E2F-DP and MuvB at promoters that are bound by DREAM and the level of expression of those "DREAM target genes" in embryos. We report that LIN-35 indeed mediates the association of E2F-DP and MuvB, a function that stabilizes DREAM subunit occupancy at target genes. In the absence of LIN-35, the occupancy of E2F-DP and MuvB at most DREAM target genes decreases dramatically and many of those genes become upregulated. The retention of E2F-DP and MuvB at some target gene promoters in lin-35 null embryos allowed us to test their contribution to DREAM target gene repression. Depletion of MuvB, but not E2F-DP, in the sensitized lin-35 null background caused further upregulation of DREAM target genes. We conclude that the pocket protein functions primarily to support MuvB-mediated repression of DREAM targets and that transcriptional repression is the innate function of the evolutionarily conserved MuvB complex. Our findings provide important insights into how mammalian DREAM assembly and disassembly may regulate gene expression and the cell cycle.
Author summary
The 8-subunit DREAM transcriptional repressor complex contains 3 functional components that together control expression of cell cycle and developmental genes. How the E2F-DP transcription factor heterodimer, the pocket protein, and the highly conserved MuvB complex coalesce on chromatin and repress DREAM target genes has yet to be determined. We directly tested the prevailing model that the DREAM pocket protein links E2F-DP to MuvB and is required for gene repression. Using a protein null mutant of the sole C. elegans pocket protein LIN-35, we demonstrate that the pocket protein indeed links E2F-DP and MuvB, which aids in the stable occupancy of DREAM components near target genes. Depletion of additional DREAM components in lin-35 null worms revealed that the remaining chromatin-bound MuvB represses target genes. We conclude that the MuvB subcomplex mediates DREAM’s critical repressive function. Our functional genomics approach in the simplified C. elegans system reveals that the ancestral function of the pocket protein is to stabilize the innate repressive activity of MuvB, ensuring proper regulation of DREAM target genes through development.
Introduction
As embryonic cells develop and differentiate, a conserved transcriptional program establishes their ordered exit from the cell cycle into a resting phase, also called G0 or quiescence. The highly conserved DREAM (for Dp, Retinoblastoma (Rb)-like, E2F, and MuvB) transcriptional repressor complex mediates this cell cycle quiescence program [1–3]. DREAM contains 3 components: a “repressive” E2F-DP heterodimer (in mammals, E2F4/5-DP1/2), a retinoblastoma-like pocket protein (in mammals, p130 or p107), and a 5-subunit subcomplex called MuvB (in mammals, LIN9, LIN37, LIN52, LIN54, and RBAP48) [1, 4, 5]. Together, the 8-subunit DREAM complex negatively regulates cell cycle reentry by directly repressing key cell cycle genes [1, 2]. How DREAM assembly culminates in gene repression is not understood.
The prevailing model for DREAM complex activity is that DREAM assembly at promoters, driven by p130/p107 linkage of E2F-DP and MuvB, mediates gene repression. Biochemical analyses in mammalian cells have revealed that DREAM assembly is triggered by DYRK1A phosphorylation of the MuvB subunit LIN52, directing MuvB association with the pocket protein [6, 7]. The p130/p107 pocket domain simultaneously interacts with MuvB and the transactivation domain of repressive E2F-DP through separate binding interfaces, completing assembly of the complex [8]. To repress transcription, DREAM localizes to chromatin through E2F-DP and the MuvB subunit LIN54 binding to DNA sequence motifs called cell cycle-dependent elements (CDEs) and cell cycle genes homology regions (CHRs), respectively [9–13]. Loss-of-function analyses in mice support a central role for the pocket protein in DREAM complex activity, since both p130/p107 double knockout mice and E2F4/5 double knockout mice display neonatal lethality, which has been attributed to defects in chondrocyte proliferation [14–16]. However, no direct evidence has demonstrated how E2F-DP, the pocket protein, and MuvB coordinate to repress target genes.
Because of the transcriptional dynamics observed during the cell cycle, how each component, especially MuvB, contributes to gene repression remains obscure. Upon progression from G0 into the cell cycle, p130 is phosphorylated by CDK4/6-cyclin D and dissociates from DREAM [2, 7, 17]. The transcription factor B-Myb then binds MuvB, forming the Myb-MuvB (MMB) complex, which activates late cell cycle genes [18–22]. This dual role for MuvB in repressing and activating genes, depending on the cell cycle context, complicates studies that have attempted to address its role in DREAM gene repression. For example, in contrast to E2F-DP and pocket protein loss-of-function, loss of the MuvB subunit LIN9 in mice causes early embryonic lethality [23]. This phenotype is likely due to MuvB’s activating role in MMB in the late cell cycle and not its role in DREAM [23]. Therefore, how MuvB’s inclusion in the DREAM complex contributes to target gene repression remains an outstanding question [2].
In C. elegans, the homologous complex, called DRM, acts similarly to the mammalian DREAM complex on chromatin (Fig 1A). C. elegans possesses only one pocket protein, LIN-35, which functionally resembles p130/p107 [24]. C. elegans E2F-DP (EFL-1-DPL-1) resembles the repressive E2F4-DP1 heterodimer [25]. Importantly, C. elegans MuvB likely acts predominantly or solely in a repressive capacity, because C. elegans LIN-52 lacks the phosphorylation switch present in mammalian LIN52 and no C. elegans B-Myb homolog has been identified [7, 26]. A key role of the C. elegans DRM complex is to keep germline genes repressed in developing somatic cells, as mutations in 7 of the 8 subunits cause ectopic expression of germline genes in somatic cells [27, 28]. Thus, loss-of-function analyses of C. elegans DRM subunits are well suited to address how E2F-DP, the pocket protein, and MuvB coordinate on chromatin to repress DRM target genes.
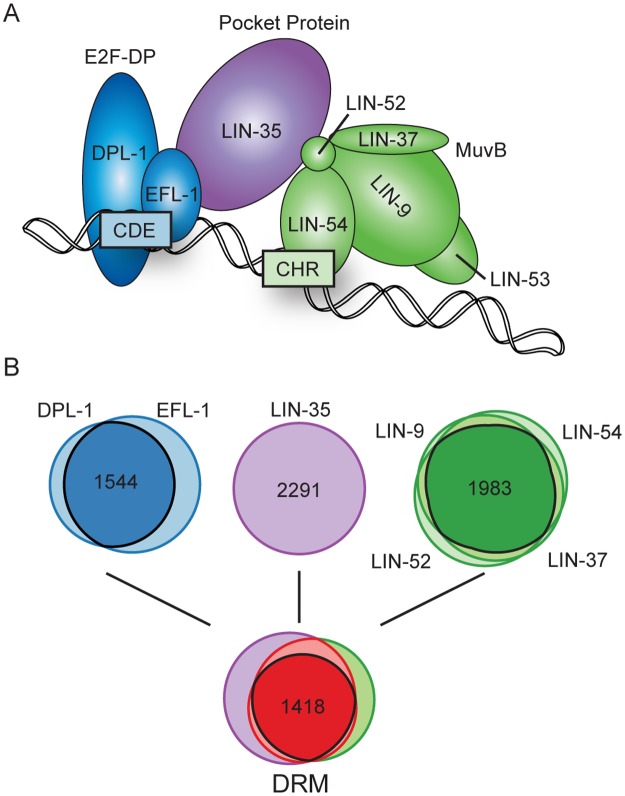
Fig 1. Chromatin association of the DRM complex in late C. elegans embryos.
(A) Model of the C. elegans DRM complex bound to DNA: E2F-DP (blue), the pocket protein LIN-35 (purple), and the 5-subunit MuvB core (green). Subunit interactions and binding of E2F-DP and LIN-54 to CDE and CHR DNA motifs, respectively, are based on [7, 11]. (B) Tier 1: Venn diagrams showing ChIP-seq peak overlaps in wild-type late embryos for E2F-DP (1771 DPL-1 peaks, 2119 EFL-1 peaks), LIN-35 (2291 peaks), and 4 of 5 subunits of the MuvB core (2067 LIN-9 peaks, 2111 LIN-37 peaks, 2284 LIN-52 peaks, 2290 LIN-54 peaks). Tier 2: Venn diagram showing the total peak overlaps between E2F-DP, LIN-35, and MuvB.
Here we utilize a protein null mutant of the sole C. elegans pocket protein LIN-35 to test how each DRM component contributes to target gene repression. Using co-immunoprecipitation, we demonstrate that LIN-35 mediates E2F-DP and MuvB association. Using chromatin immunoprecipitation linked to high throughput sequencing (ChIP-seq), we demonstrate that E2F-DP and MuvB continue to localize to chromatin in lin-35 null embryos, but their occupancy is reduced globally. Thus, DRM components can assemble on chromatin in the absence of LIN-35 bridging E2F-DP and MuvB association, but LIN-35 is required for maximal DRM chromatin occupancy on all regulatory elements. We also determine that misregulation of many genes with promoter-bound DRM, or “DRM target genes,” begins in lin-35 null late-stage embryos. DRM target genes that retain some DRM chromatin occupancy in the absence of LIN-35 remain at least partially repressed; repression is mediated by MuvB, as revealed by upregulation of those target genes when MuvB components are depleted from lin-35 null mutants. Loss of MuvB activity in lin-35 null worms also results in sterility. Our findings highlight that MuvB's innate function is as a transcriptional repressor of DRM target genes and provide evidence for how this function is facilitated by LIN-35. These results shed light on how DRM components coordinate gene repression in worms, which has important implications for the current model of DREAM-mediated gene repression in mammals.
Results
E2F-DP and MuvB act primarily within the repressive DRM complex in C. elegans embryos
Chromatin association of the 8-subunit DRM complex occurs through the sequence-specific DNA-binding activities of E2F-DP and LIN-54 (Fig 1A) [11]. To begin to address how E2F-DP, LIN-35, and MuvB contribute to transcriptional repression, we re-evaluated each DRM subunit’s in vivo chromatin-bound landscape in C. elegans late embryos. Expanding upon our previously published 2 replicates of ChIP-seq data [29], we performed an additional biological replicate of each E2F-DP and MuvB subunit in wild-type (WT) late embryo extracts. Late embryos represent a predominantly somatic cell sample, as only 2 germ cells (Z2/Z3) are present in each 200- to 550-cell late embryo. LIN-53 was not included, because its chromatin localization was not as robust as the other MuvB subunits [29]. We applied a 1% Irreproducible Discovery Rate (IDR) threshold on peaks called from the 3 biological replicates to identify the high-confidence chromatin binding sites for each individual DRM subunit (S1 Table). The overlap of individual subunit peaks defined 1544 E2F-DP peaks (EFL-1 and DPL-1 overlap) and 1983 MuvB peaks (at least 3 of 4 subunit overlap). The overlap between E2F-DP, MuvB, and 2291 LIN-35 peaks defined 1418 high-confidence DRM binding sites that were used for all subsequent analyses (Fig 1B). We did not observe any appreciable chromatin association of individual subunits or sub-complexes independent of DRM in late embryos (S1 Fig). To determine whether DRM subunits preferentially assemble with other transcription factors on high occupancy target (HOT) regions, we compared our DRM peak regions with HOT regions observed in C. elegans embryos [30]. We observed that 20–30% of individual subunit peaks and 425 of the 1418 high-confidence DRM binding regions coincide with embryonic HOT regions (S1 Table). Overall, we conclude that E2F-DP and MuvB operate primarily within the context of the repressive DRM complex during late C. elegans embryogenesis, which represents an ideal stage for evaluating the mechanism of DRM action.
The C. elegans pocket protein LIN-35 mediates the association of E2F-DP and MuvB
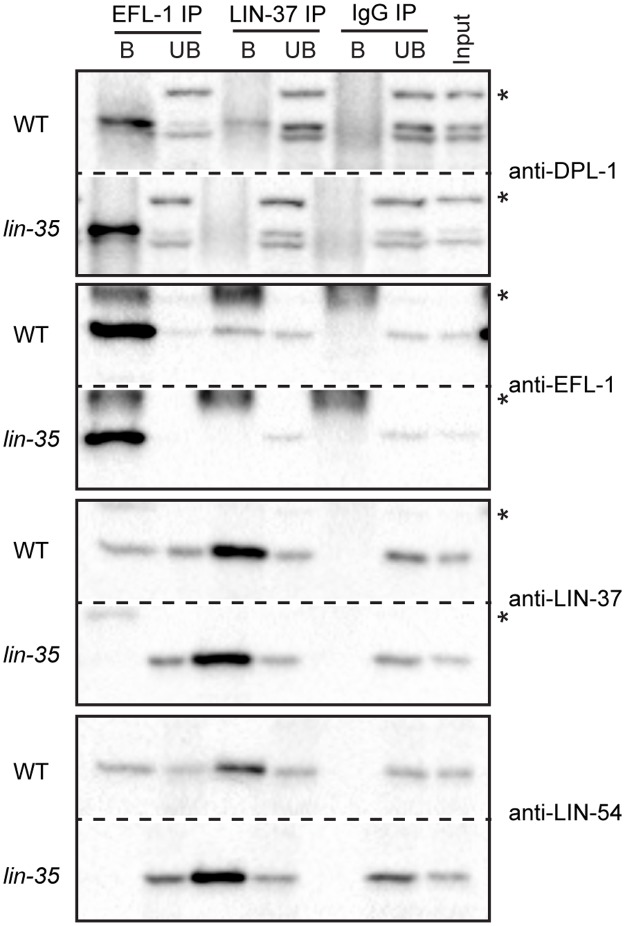
According to the current model of DRM complex assembly (Fig 1A), LIN-35 acts as a scaffold through concurrent interactions with EFL-1 and LIN-52 on opposite faces of the pocket domain [7, 16]. To address how LIN-35 contributes to DRM complex formation in C. elegans, we performed co-immunoprecipitation (co-IP) experiments from WT and lin-35(n745) late embryo protein extracts (Fig 2). The n745 allele introduces an early stop codon and is a protein-null allele [24]. EFL-1 immunoprecipitation successfully pulled down the MuvB components LIN-37 and LIN-54 from WT extracts but not from lin-35 null extracts. In reciprocal pull-downs, LIN-37 immunoprecipitation successfully pulled down EFL-1 and DPL-1 from WT extracts but not from lin-35 null extracts. As controls, the EFL-1 and LIN-37 IPs successfully pulled down their respective complex partners DPL-1 or LIN-54 from both extracts. These results were observed in 2 biological replicate experiments (S2 Fig) and demonstrate that E2F-DP and MuvB association requires LIN-35.
Fig 2. Co-immunoprecipitation analysis of DRM components from wild-type and lin-35 null late embryo cell lysates.
Late embryo extracts from wild-type (WT) and a lin-35 null mutant were immunoprecipitated with anti-EFL-1, anti-LIN-37, or control IgG antibodies. Proteins bound (B) and unbound (UB) were separated by SDS/PAGE, and western blot analysis was performed using the antibodies indicated on the right. 5% of Input was included. Asterisks indicate non-specific bands. Full blots are shown in S2 Fig.
Loss of LIN-35 impairs but does not eliminate E2F-DP and MuvB chromatin association
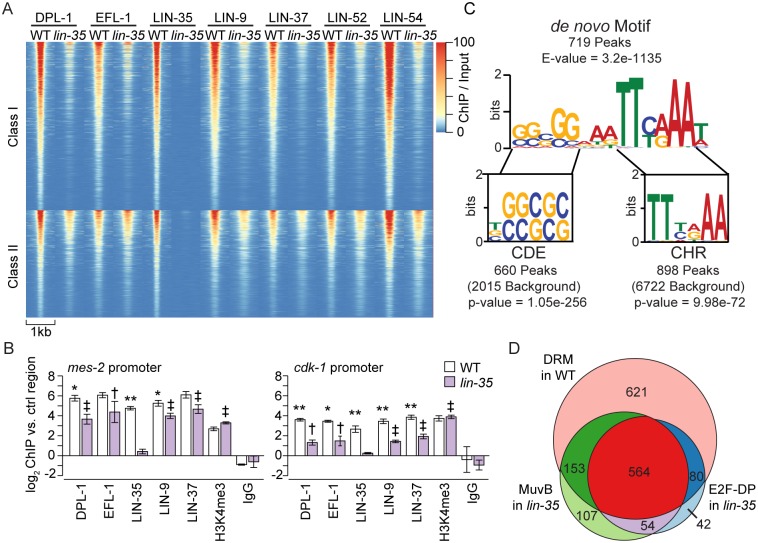
To test if loss of LIN-35 affects E2F-DP and/or MuvB chromatin localization, we performed ChIP-seq of E2F-DP (DPL-1 and EFL-1) and MuvB (LIN-9, LIN-37, LIN-52, and LIN-54) on 3 biological replicates of lin-35 null late embryos. LIN-35 ChIP-seq was performed on 2 lin-35 null biological replicates as a negative control. We observed a genome-wide decrease in E2F-DP and MuvB subunit chromatin occupancy in lin-35 null embryo extracts (Fig 3A, S3A Fig). To define the reproducible differences between lin-35 null and WT DRM subunit ChIP-seq, we used DEseq2 differential binding analysis [31]. Of the 1418 high-confidence DRM peaks identified in WT (Fig 1B, S1 Table), 866 peaks displayed significantly decreased chromatin occupancy by at least 1 E2F-DP/MuvB subunit in lin-35 null (Class I, FDR < 0.05) (Fig 3A). 71% of Class I peaks showed a significant decrease in chromatin occupancy of at least 3 of the 6 analyzed subunits in lin-35 null compared to WT, and 27% showed a significant decrease for all 6 subunits. Of the remaining 552 peaks that were not significantly decreased (Class II), 6 showed a significant increase in chromatin occupancy of at least one subunit; this corresponds to 0.4% of all high-confidence DRM peaks. Our data demonstrate that in the absence of LIN-35, the chromatin association of E2F-DP and MuvB is significantly reduced (Fig 3A, S3A Fig).
Fig 3. Comparison of DRM subunit chromatin localization in wild-type and lin-35 null late embryos.
(A) Heatmap of normalized ChIP-seq profiles of pooled replicates for each DRM subunit sorted from highest to lowest signal in the following classes. Class I: DRM peak occupancy significantly decreased in lin-35 compared to wild type (WT) for at least 1 subunit (866 peaks). Class II: DRM peak occupancy not significantly decreased in lin-35 compared to WT (552 peaks). Significance was determined using DEseq2 with an FDR < 0.05. (B) ChIP-qPCR of 5 DRM subunits at 2 Class II peaks in the mes-2 and cdk-1 promoters. H3K4me3 ChIP was included as a positive control. Signals are presented as the log2 fold enrichment of the ChIP signal for each region vs. a negative control region (a non-coding region of chromosome IV). Error bars indicate standard error of the mean. Significance was determined by a student’s T test between subunit ChIP values in WT versus lin-35 (* p-value < 0.05, ** p-value < 0.01) or between subunit versus IgG ChIP values in lin-35 († p-value < 0.05, ‡ p-value < 0.01). (C) Sequence logo of the DRM motif observed de novo in 719 DRM peaks using MEME. Aligned below are the sequence logos of the conserved CDE and CHR motifs observed in 660 DRM peaks and 898 DRM peaks, respectively. The p-value for the conserved motif enrichment over background was calculated using a hypergeometric distribution. Additional motif enrichment analyses are shown in S6 Fig. (D) Venn diagram showing ChIP-seq peak overlaps in late embryos for DRM in WT (pink, 1418 peaks), MuvB in lin-35 (green, 878 peaks), and E2F-DP in lin-35 (blue, 740 peaks).
Although the 552 Class II peaks did not meet our significance cut-off for being reduced in lin-35 null compared to WT, these peaks appeared reduced in lin-35 (Fig 3A). In fact, the signal from 2 of the 3 lin-35 null ChIP-seq replicates suggested that LIN-35 loss greatly affected DRM subunit occupancy at all sites (S4 Fig). When we removed the remaining lin-35 null ChIP-seq replicate from the differential binding analysis, then 93% of all DRM peaks displayed significantly decreased chromatin occupancy by at least one DRM subunit, and 64% showed a significant decrease for all 6 subunits, in lin-35 null compared to WT. Taken together, our genome-wide analysis revealed that E2F-DP and MuvB chromatin occupancy is impaired globally in lin-35 null embryos.
We validated our ChIP-seq findings by ChIP-qPCR analysis of 8 Class I DRM peaks and 6 Class II peaks. In lin-35 null embryos, we observed a significant decrease in chromatin occupancy of the majority of DRM subunits at all 14 sites tested (Fig 3B, S3B Fig). Two representative Class II peaks within the mes-2 and cdk-1 promoters are shown in Fig 3B. Histone H3 lysine 4 trimethylation (H3K4me3) ChIP was used as a positive control; it confirmed that the observed reduction of DRM subunit chromatin occupancy in lin-35 null compared to WT late embryos was specific to DRM subunits (Fig 3B, S3B Fig). Similar to our ChIP-seq analysis, in lin-35 null embryos, the chromatin occupancy of the DRM subunits DPL-1, EFL-1, LIN-9, and LIN-37 was significantly enriched over the IgG negative control near 11 of the 14 promoter regions tested, including mes-2 and cdk-1 (Fig 3B, S3B Fig). Interestingly, the DRM complex occupies the promoter region of most of the genes that encode DRM subunits (S5A Fig), suggesting that the DRM complex represses its own expression. Indeed, the majority of DRM subunit transcripts were elevated in the 2 DRM mutants we tested (S5B Fig); protein levels were similar or slightly elevated in lin-35 null compared to WT late embryos, based on western blot analysis (S5C Fig). These results indicate that the observed decrease in DRM subunit chromatin occupancy was not driven by reduced protein levels. To extend this analysis beyond embryos, we also performed ChIP-qPCR to test DPL-1 and LIN-37 occupancy in L1 larvae. As in embryos, we observed a decrease in DPL-1 and LIN-37 occupancy in lin-35 null L1 larvae compared to WT L1 larvae (S5D Fig). Together, our ChIP analyses of DRM subunits in the lin-35 null indicate that E2F-DP and MuvB chromatin occupancy is significantly impaired but not eliminated at most genomic sites in the absence of the pocket protein.
We tested whether the DRM DNA binding motif is enriched in Class I targets and/or Class II targets. We performed de novo DNA motif discovery using MEME [32] on the 1418 high-confidence DRM peak regions and identified the characteristic hybrid CDE/CHR motif similar to what has been observed previously (Fig 3C) [33]. Additionally, we performed a search for the known E2F-DP binding motif (CDE, IUPAC code: BSSSSS) and LIN-54 binding motif (CHR, IUPAC code: TTYRAA), restricting our search to phylogenetically conserved motifs similar to an analysis performed on the mammalian genome (S6 Fig) [34]. The analyses together identified 719 peaks with the de novo CDE/CHR motif, 660 peaks with a conserved CDE motif, and 898 peaks with a conserved CHR motif (Fig 3C, S1 Table). However, the motif content of a DRM peak does not predict whether E2F-DP or MuvB remain more or less stably bound in the absence of LIN-35 (S6 Fig).
E2F-DP and MuvB remain confined to WT peak regions in lin-35 null embryos
In mammalian cells, phosphorylation of the pocket protein triggers DREAM disassembly and reentry into the cell cycle [2, 7]. Activating E2F-DPs replace repressive E2F-DP, and MuvB forms the activating MMB complex to drive cell cycle gene expression [22, 35]. Since C. elegans E2F-DP alone can activate genes in the germline [36], we assessed whether the chromatin-bound landscape of the DRM components changes in lin-35 null embryos. We performed peak calling analysis on the lin-35 null ChIP-seq replicates, as described previously (S2 Table). For each subunit, roughly half of the WT peaks were lost in lin-35 null embryos (S7 Fig). After overlapping WT high-confidence DRM peaks with E2F-DP and MuvB peaks detected in lin-35 null embryos, we observed that 621 of the 1418 high-confidence DRM peaks in WT lose detectable E2F-DP and MuvB occupancy in lin-35 null embryos (Fig 3D). 564 of the 1418 high-confidence DRM peaks in WT retain appreciable E2F-DP and MuvB occupancy in lin-35 null embryos, underscoring that loss of LIN-35 impairs but does not eliminate DRM complex chromatin localization (Fig 3A). We did not observe any appreciable or consistent appearance of E2F-DP or MuvB at new sites (S7 Fig). Thus, loss of LIN-35 does not appear to result in de novo binding of E2F-DP or MuvB to new sites but rather destabilizes binding to their native sites, resulting in elimination of DRM occupancy from about half of the native sites.
DRM subunit destabilization affects target gene expression in late stage embryos
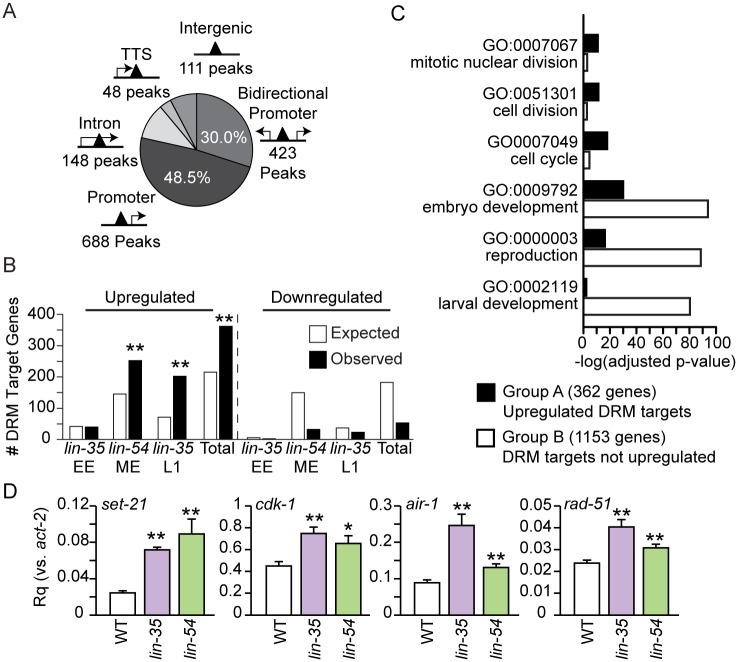
To address the effects of LIN-35 loss on target gene transcription, we mapped the 1418 high-confidence DRM binding sites to likely gene targets and then analyzed transcript levels from those genes in lin-35 null mutants. 78% of all DRM binding sites map to within at least one gene’s promoter region, which we defined as 1000 base pairs upstream to 100 base pairs downstream (-1000 bp, +100 bp) of a transcriptional start site (Fig 4A, S3 Table). Of these promoter peaks, 688 DRM binding sites were observed within 677 single gene promoters, and 423 DRM binding sites were observed within bidirectional promoters for 844 genes. Of the remaining DRM binding sites, 148 were observed within an intron of 137 genes, 48 were observed within 1000 bp of 46 transcriptional termination sites, and 111 were observed in intergenic regions (Fig 4A, S3 Table).
Fig 4. Locations of DRM peaks and misregulation of DRM target genes.
(A) Pie chart showing the number of peaks within a promoter (-1000 bp to +100 bp) of bidirectional gene pairs (423 peaks) or single genes (688 peaks); within an intron (148 peaks); at the transcriptional termination site (TTS, 48 peaks); or intergenic (> 1000 bp from a gene, 111 peaks). The percentage of total peaks is indicated for promoter peaks only. (B) Enrichment analysis of DRM target genes expected and observed among genes up- or downregulated in microarray expression analyses reported in [37] (lin-35 early embryos (EE) and L1s) and [33] (lin-54 mixed-stage embryos (ME)) and a combination of all 3 experiments (Total). The significance of enrichment observed over expected was determined using a hypergeometric distribution (** p-value < 1x10-5). (C) Bar chart plotting the –log10 Benjamini adjusted p-value of selected GO term enrichment observed for 362 upregulated DRM target genes (Group A, black) compared to GO term enrichment for 1153 DRM target genes not upregulated (Group B, white). (D) RT-qPCR analysis comparing transcript levels of 4 Group A genes (set-21, cdk-1, air-1, and rad-51) in lin-35(n745) (purple) and lin-54(n2231) (green) late embryos to the level in wild-type (WT, white) late embryos. Expression values from 2 independent experiments each consisting of 4 biological replicates were averaged and are presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean, and significance was determined by a student’s T test between transcript levels in mutant vs WT (* p-value < 0.05, ** p-value < 0.01). Additional genes are shown in S5 Table.
To generate a list of high-confidence DRM targets, we assessed whether genes with DRM bound in their promoter region in WT (DRM target genes) were misregulated in previously published expression analyses that compared lin-35 null to WT in early embryos and L1 larvae [37] and compared lin-54(n2990) mutant to WT in mixed-stage embryos [33] (S3 Table) using microarray analysis. The lin-54 allele produces protein with impaired DNA-binding ability, which reduces its chromatin occupancy by ~50% [33]. Upregulated genes in both lin-35 null L1s and lin-54 mutant mixed embryos were significantly enriched for DRM target genes (Fig 4B). Our analysis defined 362 "high-confidence DRM targets" (bound by DRM and upregulated in DRM mutants), which we refer to as Group A DRM targets (Fig 4B, S3 Table, S8A Fig). The remaining "lower-confidence DRM targets" (1153 genes) we refer to as Group B DRM targets, because we do not have evidence that DRM represses these genes.
To investigate why the expression of many DRM target genes appears to be unaffected by disruption of DRM activity, we performed a Gene Ontology (GO) enrichment analysis of the 362 Group A genes and the 1153 Group B genes using DAVID (Fig 4C, S4 Table) [38, 39]. REViGO was applied to remove redundant GO terms (S9A Fig) [40]. Group A includes genes for cell cycle, while Group B lacks those genes; both groups include genes for development and reproduction (Fig 4C). DRM target genes marked by Class I peaks, Class II peaks, or HOT regions were not enriched for any particular GO term (S9 Fig). We speculate that the genes in Group A and Group B have distinct requirements for transcriptional activation: Group A genes, which include cell cycle genes that are likely expressed in all cell types, turn on when DRM activity is lost, while Group B genes do not turn on when DRM activity is lost, perhaps because they require additional transcriptional activators that are present only at specific developmental stages and/or in specific cell types.
Since developmental stage appears to greatly affect whether loss of DRM function causes target gene misregulation (Fig 4B, S8B Fig), we performed RT-qPCR analysis on lin-35 null and lin-54(n2231) mutant late embryos to match our ChIP-seq analysis. The lin-54(n2231) allele contains the same point mutation as lin-54(n2990), which impairs its DNA binding [33]. We included lin-54 late embryos to address whether the difference in number of genes significantly upregulated in lin-35 null early embryos and lin-54 mutant mixed-stage embryos (Fig 4B) is due to their different stages. We tested 4 candidates from Group A DRM target genes, which we expected to be upregulated in late embryos: set-21, cdk-1, air-1 and rad-51. All 4 candidates were significantly upregulated in both lin-35 null and lin-54 mutant late embryos as compared to WT late embryos (Fig 4D). Out of a total of 25 Group A genes tested, 22 were significantly upregulated in lin-35 null and/or lin-54 mutant late embryos (S5 Table). We also tested 5 Group B genes, genes not upregulated in the microarray analyses; 2 genes were significantly upregulated (S5 Table). Observing similar upregulation of genes in lin-35 null and lin-54 mutant late embryos suggests that the differences in number of genes upregulated in the microarray analyses are due to stage differences and that DRM target gene misregulation becomes detectable by late embryogenesis. Together with our ChIP-seq analysis, these results allow us to consider 2 possible mechanisms for how LIN-35 may function in DRM: 1) LIN-35 is essential for DRM-mediated repression, and loss of LIN-35 causes target gene upregulation or 2) LIN-35 is not essential for DRM-mediated repression, and loss of LIN-35 causes reduced chromatin occupancy by E2F-DP and MuvB, which in turn causes target gene upregulation.
MuvB mediates DRM target gene repression
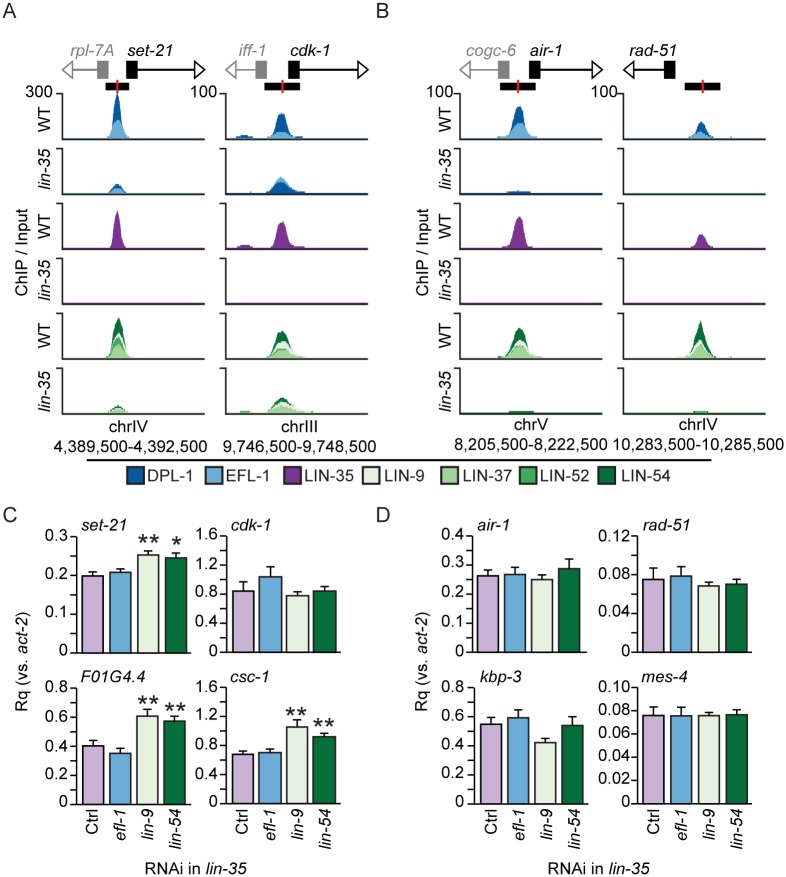
We reasoned that if LIN-35 is essential for repression of DRM target genes, as the current model suggests, then further disruption of E2F-DP or MuvB should not cause further upregulation of DRM target genes. Conversely, if the reduced chromatin occupancy of E2F-DP or MuvB causes upregulation of DRM target genes, then further disruption of E2F-DP or MuvB should cause further upregulation of DRM target genes whose promoters retained those components. We tested these predictions on 4 DRM target genes that retained some E2F-DP and MuvB in lin-35 null late embryos (set-21, cdk-1, F01G4.4, and csc-1), and 4 DRM target genes that had no detectable E2F-DP or MuvB in lin-35 null late embryos (air-1, rad-51, kbp-3, and mes-4) (Fig 5). We confirmed that DRM subunit binding was retained or eliminated at 4 of these respective sites by ChIP-qPCR (S3B Fig). We performed RT-qPCR analysis on lin-35 null late embryos after two generations of RNA interference (RNAi) knockdown of efl-1, lin-9, or lin-54. The transcript levels of each targeted subunit were knocked down by at least 50% compared to empty vector control (S10A Fig). Knockdown of efl-1 had no effect on transcript levels from the 8 genes tested (Fig 5C and 5D); we also confirmed a previous report [41] that in the absence of LIN-35, E2F-DP does not activate germline genes in late embryos (S10 Fig). In contrast to efl-1, knockdown of lin-9 and lin-54 caused a significant increase in transcript levels from 3 of the 4 genes where MuvB retains some observable chromatin occupancy in lin-35 null late embryos and 0 of the 4 genes where MuvB chromatin occupancy is abolished in lin-35 null late embryos (Fig 5C and 5D). Testing of additional genes confirmed the above findings (S5 Table). Of 24 total genes to which MuvB remains bound in lin-35 null late embryos, 10 genes were further upregulated following knockdown of MuvB but not following knockdown of efl-1 (S5 Table). Of 6 total genes where MuvB occupancy is lost in lin-35 null embryos, 0 genes were further upregulated following knockdown of MuvB or efl-1. These results indicate that MuvB continues to repress DRM target genes in lin-35 null embryos, strongly supporting a role for MuvB as the mediator of DRM target gene repression.
Fig 5. Effects of depletion of additional DRM subunits in lin-35 null mutants.
(A,B) Genomic profiles of each DRM component in wild-type (WT) and lin-35 late embryos near (A) set-21 and cdk-1 and (B) air-1 and rad-51. E2F-DP subunits (blues), LIN-35 (purple), and MuvB subunits (greens) were overlaid on the same track. Normalized ChIP-seq enrichment values for each data track are indicated on the y-axis, and the chromosomal coordinates of the areas shown are indicated below the x-axis. Gene transcriptional start sites are indicated by black boxes, and coding strand directions are indicated by white arrowheads. Other genes in the region are similarly indicated in grey. Each DRM peak location is indicated by a black rectangle with the peak center highlighted in red. (C,D) RT-qPCR analysis of (C) set-21, cdk-1, F01G4.4, and csc-1 and (D) air-1, rad-51, kbp-3, and mes-4 following efl-1 (blue), lin-9 (light green), or lin-54 (dark green) RNAi in lin-35 null late embryos compared to empty-vector RNAi (Ctrl, purple). Expression values from 2 independent experiments each consisting of 4 biological replicates for each RNAi condition were averaged and are presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean, and significance was determined by a student’s T test comparing DRM subunit RNAi to Control RNAi (Ctrl) (* p-value < 0.05, ** p-value < 0.01). Additional genes are shown in S5 Table.
Loss of MuvB activity in lin-35 null worms has a catastrophic effect on fertility
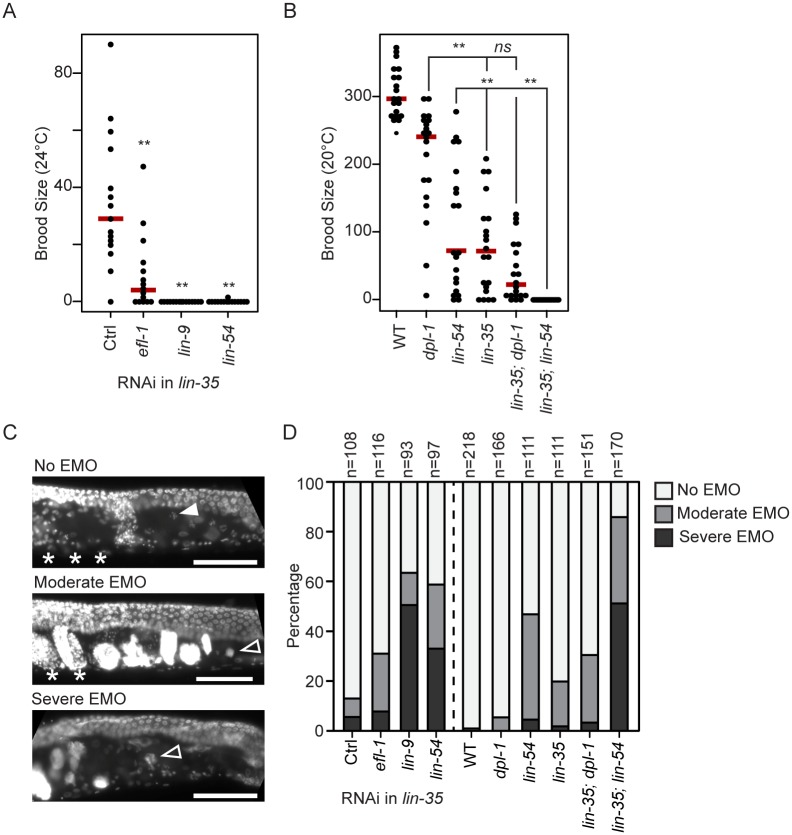
The analyses described above show that E2F-DP and MuvB chromatin occupancy is reduced but not eliminated in the absence of LIN-35, even though E2F-DP and MuvB protein association is completely decoupled. In our RNAi RT-qPCR analysis, we observed that knockdown of MuvB in lin-35 null embryos resulted in further upregulation of DRM targets where MuvB retains some chromatin occupancy. While performing this experiment, we observed far fewer collected embryos in MuvB RNAi conditions compared to efl-1 and control RNAi. Indeed, the brood size of individual lin-35 null worms following knockdown of other DRM subunits was significantly reduced in efl-1 RNAi and severely reduced in lin-9 and lin-54 RNAi compared to empty vector controls (Fig 6A). Reduction in brood size was also seen after we introduced hypomorphic alleles dpl-1(n3643), which produces a truncated DPL-1 protein product [25], and lin-54(n2231) into the sensitized lin-35 null background. We observed a significant decrease in brood size in lin-35 null, dpl-1(n3643) hypomorphic, and lin-54(n2231) hypomorphic single mutants compared to WT (Fig 6B). We did not observe a significant reduction of brood size in lin-35(n745); dpl-1(n3643) double mutants compared to the lin-35(n745) single mutants (Fig 6B). In contrast, in lin-35(n745); lin-54(n2231) double mutants, we observed complete sterility. Together, these results indicate that further disruption of MuvB activity in lin-35 null worms causes a severe reduction in the production of viable progeny.
Fig 6. Phenotypic effects of depletion of MuvB or E2F-DP in lin-35 null worms.
(A,B) Strip chart of the brood size of (A) lin-35 null worms following efl-1, lin-9, or lin-54 RNAi at 24°C and (B) wild-type (WT), single DRM mutants, and double DRM mutants at 20°C. Each dot indicates the total brood of an individual worm, and a red bar indicates the median for each genotype. Significance (** p-value < 0.01) was determined by a Wilcoxon-Mann-Whitney test comparing RNAi to empty vector control (Ctrl, A) or double mutants to their respective single mutants (B). (C) DAPI staining of young adults. The filled arrowhead indicates a representative healthy oocyte, open arrowheads indicate representative endomitotic oocytes (EMO), and asterisks indicate normal-appearing embryos. No EMO: no DAPI-stained DNA masses in the oviduct, and presence of normal-appearing embryos in the uterus. Moderate EMO: presence of some DAPI-stained DNA masses in the oviduct and some normal-appearing embryos in the uterus. Severe EMO: presence of DAPI-stained DNA masses in the oviduct and no normal-appearing embryos in the uterus. Scale bar, 50 μm. (D) Normalized stacked bar chart of the EMO phenotype of Day 1 adult lin-35 null worms with DRM subunit RNAi or double DRM mutants compared to single mutants and WT.
Importantly, null mutations of individual E2F-DP and MuvB subunits cause sterility due to a severe EMO phenotype (production of endomitotic oocytes) [33, 36, 42], whereas the brood size reduction observed in lin-35 null worms is associated with a moderate EMO phenotype (Fig 6C) [33]. E2F-DP null sterility is linked to its activating role in the germline, which is distinct from its role in DRM [36, 43]. However, MuvB null sterility is due to a defect in somatic sheath development [42]. We tested for EMO in lin-35 null worms depleted of other DRM subunits. In lin-35 null worms, RNAi knockdown of lin-9 or lin-54, but not of efl-1, caused an increase in severe EMO (Fig 6D). Similarly, lin-35; lin-54(n2231) double mutants, but not lin-35; dpl-1(n3643) double mutants, displayed an increase in severe EMO (Fig 6D). We conclude that further loss of MuvB repression in lin-35 null worms is catastrophic, reproducing the severe EMO phenotype previously observed only in null alleles that likely completely eliminate DRM activity.
Discussion
Using genomic, genetic, and cell biological experiments on C. elegans lacking the sole pocket protein, we report evidence that MuvB mediates DRM target gene repression and that MuvB does not require LIN-35 to act as a transcriptional repressor. We demonstrate that loss of the sole C. elegans pocket protein LIN-35, which abolishes E2F-DP and MuvB association, results in a dramatic decrease in chromatin occupancy by both E2F-DP and MuvB and upregulation of many DRM target genes in late embryos. To determine how LIN-35 contributes to DRM gene repression, we depleted E2F-DP or MuvB subunits in lin-35 null animals. Depletion of MuvB, but not E2F-DP, causes further target gene upregulation and enhances the lin-35 null phenotype, indicating that MuvB directs repression of DRM target genes. Our study demonstrates that LIN-35 bridges E2F-DP and MuvB association, supporting their chromatin occupancy at target genes, and that MuvB functions innately as a transcriptional repressor of DRM target genes.
Studies of the mammalian DREAM DNA-binding motifs support the notion that MuvB mediates target gene repression. Most mammalian genes that are repressed in G0/G1 and activated in the later cell cycle contain a CDE, the binding motif for repressive E2F-DP, and a CHR, the binding motif for the LIN-54 subunit of MuvB [11]. Promoter analyses have shown that the CDE is not essential for DREAM binding; however, the presence of a CDE can enhance DREAM binding [12]. In contrast, the CHR is fully required for gene repression and activation at appropriate cell cycle stages [12]. Although our experimental approach does not eliminate the possibility that E2F-DP mediates repression of some target genes, taken together with the above motif analysis, our data support a model in which MuvB chromatin localization mediates DRM’s core repressive function.
Our assessment of the endomitotic oocytes (EMO) phenotype in the sensitized lin-35 null background may provide a clue as to how C. elegans E2F-DP contributes to DRM activity. EMO can result from germline dysfunction, e.g. defects in meiosis or fertilization, or somatic dysfunction, e.g. defects in somatic sheath cell formation or function [44, 45]. EMO and sterility have previously been observed in null mutants of dpl-1, efl-1, lin-9, and lin-54 [33, 36, 42, 43]. The dpl-1 and efl-1 null phenotype results from loss of E2F-DP’s transcriptional activation role in the germline, although some unknown somatic component is also involved [36, 43]. In contrast, the lin-9 null phenotype results from defects in somatic sheath development [42]. With E2F-DP decoupled from MuvB in lin-35 null worms, efl-1 knockdown resulted in significant reduction in fertility but no enhancement of the EMO phenotype. In E2F-DP null worms, we suspect that E2F-DP loss affects MuvB function in somatic cells but only when LIN-35 is present, and that effect is overshadowed by loss of E2F-DP activating function in the germline. Thus, we speculate that E2F-DP function in DRM is to support the association of MuvB with gene promoters, with the pocket protein acting as the intermediary.
In C. elegans, loss of DRM activity leads to ectopic expression of germline genes in somatic cells [27, 28]. Interestingly, in wild-type worms LIN-35 chromatin localization is dramatically reduced in the germline compared to somatic tissues [41]. Additionally, EFL-1 and DPL-1 are known activators of the germline oogenic program [36], and by ChIP-seq they localize to more genomic regions in the adult germline than in late embryos [41]. However, a previous study found no evidence for E2F-DP adopting an activating function in the absence of LIN-35 and directly activating germline gene expression in somatic cells [41]. Our analysis also found no evidence for C. elegans E2F-DP occupying new genomic sites in lin-35 null embryos that are normally only observed in the adult germline. Perhaps an additional event following loss of LIN-35 must occur before E2F-DP can localize to and activate a germline program. Our study further suggests that some DRM target genes require secondary regulatory factors in addition to loss of DRM for their activation. As an example, E2F-DP was recently shown to co-regulate some developmental genes with heat-shock factor (HSF) [46]. It is likely that additional regulatory networks overlap DRM functionality at certain DRM targets and coordinate their activation in specific tissues and times during development.
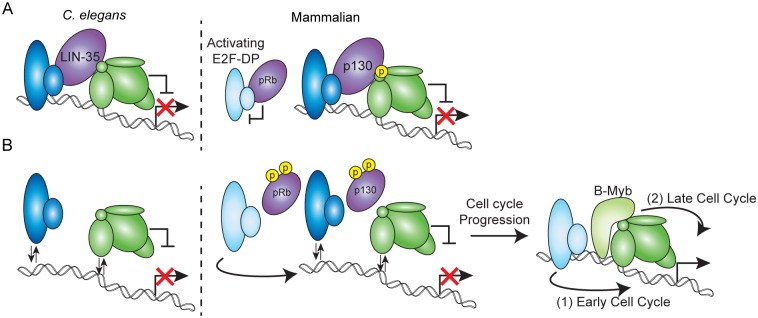
Our analysis of C. elegans DRM provides insight into how the mammalian DREAM complex components maintain cellular quiescence and promote the transition into the cell cycle. Upon exit from the cell cycle, p130/p107 promotes E2F-DP and MuvB association on chromatin, stabilizing MuvB-mediated transcriptional repression of important cell cycle gene targets (Fig 7A). We speculate that the effects on DRM activity observed in lin-35 null embryos mimic the events that immediately follow phosphorylation-mediated release of p130 from the mammalian DREAM complex, an event triggered by progression into the cell cycle (Fig 7B) [2, 7]. Loss of the pocket protein destabilizes the chromatin association of E2F-DP and MuvB, but remaining MuvB continues to repress some target genes. In mammalian cells, MuvB-mediated repression is likely overwhelmed by 1) the release of activating E2F-DP heterodimers from the retinoblastoma protein (pRb), which promotes progression into the early cell cycle [47] and 2) incorporation of MuvB into a transcriptional activation complex with B-Myb in the late cell cycle [20, 21]. Future work on how MuvB’s regulatory role is altered from repression to activation by B-Myb will help answer how precise temporal activation of cell cycle genes is achieved.
Fig 7. Model of DRM/DREAM function in cell cycle progression.
(A) Model of the C. elegans DRM complex (left) and mammalian DREAM complex (right) activity with an intact pocket protein: E2F-DP (blue), the pocket proteins LIN-35/p130 and pRb (purple), and the 5-subunit MuvB core (green). Mammalian activating E2F-DP heterodimers (light blue) are indicated bound to pRb. Phosphorylation of LIN-52 in mammalian DREAM is based on [6]. (B) Model of the C. elegans DRM complex (left) and mammalian DREAM complex (right) activity in the absence of a pocket protein. Phosphorylation of mammalian pRb and p130 is based on [2, 7, 8]. MuvB-mediated repression of target genes is released as the cell cycle progresses, which leads to 1) activating E2F-DP heterodimers activating early cell cycle genes and 2) B-Myb association with MuvB activating late cell cycle genes [3].
The MuvB complex’s dual transcriptional role in mammalian cells has obscured how it may contribute to mammalian DREAM-mediated gene repression. Recently, it was hypothesized that DREAM stably positions a nucleosome at the transcriptional start site of mammalian DREAM target gene promoters to prevent transcriptional activation [13]. A likely candidate for mediating DREAM-nucleosome interactions is the MuvB component RBAP48 (in C. elegans, LIN-53), a WD-repeat family protein known to bind histones when present in other complexes [24]. The RBAP48 ortholog in Drosophila has been shown to be required for dREAM-mediated gene repression [48]. Additionally, in C. elegans, LIN-53’s role may include facilitating deposition of the histone variant HTZ-1/H2A.Z within the body of target genes [29]. If MuvB positions nucleosomes through LIN-53/RBAP48, this would represent a novel mechanism for transcriptional repression that employs targeted non-enzymatic inhibition of transcriptional initiation.
Our demonstration of MuvB’s innate repressive role may provide clues as to how the DREAM complex components evolved to meet diverse biological needs. Phylogenetic analyses suggest that the activating E2F-DPs and pRb coevolved, diverging from their respective common ancestors, which are more similar to the repressive E2F-DPs and p130/p107 DREAM components [49, 50]. Our findings in C. elegans indicate that MuvB mediates the repressive functions of the worm DREAM complex, and can perform this activity in the absence of p130/p107 and a link to E2F-DP. Thus, if DREAM-associated gene repression represents the ancestral function of E2F-DP and pocket proteins, MuvB may have emerged prior to or coincident with E2F-DP and a pocket protein. Moreover, since C. elegans does not have a B-Myb homolog, MuvB’s repressive mechanism likely represents its ancestral function, with its activating role emerging more recently [26].
Compared to the strong tumor suppressor activity of pRb, functional redundancy in p130/p107 has obscured if and how they act as tumor suppressors [51]. Interestingly, all 3 mammalian pocket proteins are specifically targeted for degradation by the E7 oncoprotein in high-risk human papillovirus (HPV) [52, 53]. Concurrently, HPV16 E7 stimulates MMB assembly and gene activation [54]. These findings illustrate how cancer cells may drive cell cycle progression by promoting disassembly of DREAM through inactivation of p130/p107 while also coaxing MuvB into its activating function. Our results describe how the pocket protein stabilizes MuvB-mediated repression, revealing that loss of the pocket protein in cancer cells may not immediately or completely relieve MuvB repression. Cancer cell retention of the MuvB complex could allow for both oncogenic transformation through MMB-mediated activation of cell cycle genes [55] and escape from cytotoxic chemotherapy by induced reentry into quiescence through MuvB’s innate repressive activity [56]. We expect that future work on how MuvB both activates and represses its target genes will provide much-needed insights into how cancers hijack cell cycle control.
Materials and methods
Worm strains, culture conditions, and late embryo harvesting
All strains were cultured at 20°C using standard methods, unless otherwise noted. N2 (Bristol) was used as wild type (WT). Mutant alleles that were used in this study are listed in S6 Table. For CoIP and ChIP experiments, late stage embryos were collected by bleaching gravid worms and aging embryos for up to 3.5 hours before freezing them in liquid nitrogen.
Co-immunoprecipitation (CoIP)
Following embryo collection, extracts were prepared and CoIP performed based on [5]. Frozen late embryos were ground using a mortar and pestle, resuspended in lysis buffer (25 mM HEPES pH 7.6, 150 mM NaCl, 1mM DTT, 1mM EDTA, 0.5 mM EGTA, 0.1% Nonidet P-40, 10% glycerol) with Complete EDTA-free protease inhibitors (Roche), and sonicated twice for 30 seconds. Lysates were clarified and precleared using Protein A Dynabeads (ThermoFisher, Waltham, MA). Protein concentrations of lysates were determined using a Qubit fluorometer (ThermoFisher). 15 μg of antibody was crosslinked to 100 μL Dynabeads using dimethyl pimelimidate in 0.2 M trimethylamine pH 8.2. Crosslinking was stopped using 0.1M Tris pH 8.0, and beads were washed with 0.1 M glycine pH 2.8 before being stored in phosphate buffered saline pH 7.2 with 0.05% Tween-20. 5 mg of protein lysate and 20 μL antibody-conjugated Dynabeads were incubated for 2 hours at 4°C. Each IP was washed with lysis buffer, eluted with 50 μL 2x SDS gel-loading sample buffer for 5 minutes at 98°C, and separated by SDS/PAGE. Antibodies used in CoIP and ChIP experiments are listed in S7 Table. Western blot analysis was performed using a 1:5,000 dilution of primary antibody and a 1:2,000 dilution of an appropriate HRP-conjugated secondary antibody. Serial western blot analysis was performed by stripping the blot with buffer containing 0.2M glycine (pH 2.2), 0.1% SDS, and 1% Tween-20 between antibody probings.
Chromatin immunoprecipitation (ChIP)
Following embryo collection, extracts were prepared and ChIP performed based on [29]. Frozen late embryos were ground, crosslinked for 10 minutes in 1% formaldehyde, and sonicated to an average size of 250 base pairs in FA buffer (50 mM HEPES/KOH pH 7.5, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 150 mM NaCl). Protein concentrations of lysates were determined using a Qubit fluorometer. Lysates were precleared with Protein A Dynabeads. ChIPs were performed with 1–2 mg of extract, and 2% of the extract was set aside for an input reference control. 1–5 μg of antibody were used for ChIP-seq analysis, and 0.5 μg of antibody were used for ChIP-qPCR analysis. ChIPs were incubated overnight at 4°C with 1% sarkosyl. Protein A Dynabeads equilibrated in 20 μL FA buffer were added and incubated for 2 hours at 4°C. ChIPs were washed with the following buffers: once with FA buffer containing 1 M NaCl, once with FA buffer containing 0.5 M NaCl, once with TEL buffer (10 mM Tris-HCl pH 8.0, 0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA), and twice with TE buffer (10 mM Tris-HCl pH 8.0 and 1 mM EDTA). 2 elutions of 50 μL elution buffer containing TE plus 1% SDS and 250 mM NaCl were incubated at 55°C. Eluted ChIP and input samples were incubated with proteinase K for 1 hour at 55°C. Crosslinks were reversed overnight at 65°C. DNA was purified by phenol-chloroform extraction and ethanol precipitation using glycogen as a carrier. Quantitative PCR was performed using SYBR green reagents on a LightCycler 480 (Roche, Basel, Switzerland) or an Applied Biosystems ViiA 7 Real-Time PCR System (ThermoFisher) using primers specific to select promoter regions, which are provided in S8 Table, and normalized against signal from a negative control region on chromosome IV.
ChIP-seq preparation and analysis
Sequencing libraries were prepared from genomic DNA fragments (input) or those obtained after ChIP using the TruSeq ChIP Sample Prep Kit (Illumina, San Diego, CA). Amplified libraries were size selected to obtain 200–500 bp fragments using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). After verifying library fragment sizes using a 2100 Bioanalyzer (Agilent, Santa Clara, CA), sequencing was performed using the Illumina HiSeq 2000/4000 platforms. Libraries were sequenced at the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley. Sequencing reads were mapped to the ce10 reference genome using bowtie-1.1.2 [57], allowing a maximum of 2 reported alignments. ChIP-seq data were normalized to input using the signal extraction scaling (SES) method using deepTools and visualized using the UCSC genome browser [58–60]. Peak calling and data reproducibility checks were performed using the SPP peak caller and Irreproducible Discovery Rate (IDR) pipeline established by the ENCODE project [61, 62]. Peak overlaps and gene mapping were processed using HOMER [63], with gene transcriptional start sites (TSS) and transcriptional terminal sites (TTS) based on RefSeq annotations. Differential binding analysis was performed using the DiffBind package in Bioconductor (www.bioconductor.org) and the R statistical programming language [64]. Motif analysis was performed de novo using the MEME suite [32], or overlapping known motifs using HOMER and phastCons [65]. Motif sequence logos were generated using WebLogo [66]. Important quality control metrics are provided in S9 Table.
RNA interference and transcript analysis
Bacteria from the Ahringer RNAi feeding library [67] expressing dsRNA against efl-1, lin-9, and lin-54 were sequence-verified and fed to lin-35(n745) worms. Progeny from gravid worms grown in liquid media were synchronized as L1 larvae and fed RNAi on NGM plates containing 1 mM IPTG and ampicillin. RNAi feeding was administered for 2 generations at 24°C, after which late embryos from 2nd generation gravid adults were collected in Trizol for RNA isolation and transcript analysis. A total of 1.5 μg RNA was DNAse treated and reverse transcribed using the High Capacity cDNA Kit (Applied Biosystems, Foster City, CA). qPCR was performed using SYBR green reagents on a LightCycler 480 (Roche) or an Applied Biosystems ViiA 7 Real-Time PCR System (ThermoFisher) using primers specific to a select gene set, which are provided in S10 Table. The relative quantity of experimental transcripts was calculated with act-2 as the control gene using the ΔCt method with efficiency correction. Statistical analysis of genome-wide expression data was performed using R, using the Quantile normalization and Robust Multichip Average (RMA) algorithm in the affy package from Bioconductor [68, 69]. To determine differential expression, moderated T Statistics were applied using the limma package, using q-value < 0.05 and fold change > 1.5 as the significance thresholds [70]. For EMO and brood size scoring, 2nd generation RNAi-fed L4 larvae were aged overnight and scored.
C. elegans phenotype scoring
For endomitotic oocyte (EMO) scoring, L4 larvae were aged overnight, fixed using Carnoy’s solution, stained with the DNA intercalator DAPI, and scored blind. The definitions of severe EMO, moderate EMO, and no Emo are in the Fig 6 legend. Images were acquired using a Zeiss Axioskop (Oberkochen, Germany) and processed using Image J [71]. For brood size analyses, individuals were cloned to fresh plates every 24 hours starting at the L4 larval stage and all progeny were counted.
Supporting information
(A) UpSet visualization of DRM subunit peak overlaps. Black filled circles indicate the factor(s) present within an intersection category. Intersection categories consisting of less than 20 represented peaks were omitted. (B) Heatmap of normalized ChIP-seq profiles of pooled replicates for each DRM subunit across all identified peaks. High confidence DRM peaks (identified in Fig 1) are separated from the peaks that did not pass the overlap requirements (remaining DRM peaks). ChIP-seq signal appears consistently stronger in the high confidence DRM peaks when compared to the remaining DRM peaks. Even though many peaks do not pass the overlap requirements, the occupancy observed for each DRM subunit appears similar at all peak regions.
(TIF)
Full western blots from 2 co-immunoprecipitation experiments performed on biological replicate late embryo lysates. Proteins bound (B) and unbound (UB) by EFL-1, LIN-37, or control IgG immunoprecipitation were separated by SDS/PAGE, and western blot analysis was performed using the antibodies indicated in the bottom right corner. 5% of Input was included. Arrows indicate protein bands presented in main Fig 2. Asterisks indicate bands that carried over from a previous blot.
(TIF)
(A) Scatter plot of normalized ChIP-seq reads in lin-35 vs. wild type (WT) for each DRM subunit for Class I (dark) and Class II (light) peaks, as described in the Fig 3 legend. Each point indicates the WT (x-axis) and lin-35 null (y-axis) read density of pooled replicates within a single wild-type DRM peak from E2F/DP (blue), LIN-35 (purple), and MuvB (green) subunit ChIP-seq. Dotted lines indicate the slope expected if DRM occupancy is equivalent in WT and lin-35 ChIP-seq. (B) ChIP-qPCR of 5 DRM subunits at 8 Class I peaks and 4 Class II peaks. H3K4me3 ChIP was included as a positive control. Signals are presented as the log2 fold enrichment of the ChIP signal for each region vs. a negative control region (a non-coding region of chromosome IV). Error bars indicate standard error of the mean. Significance was determined by a student’s T test between subunit ChIP values in WT versus lin-35 (* p-value < 0.05, ** p-value < 0.01) or between subunit versus IgG ChIP values in lin-35 († p-value < 0.05, ‡ p-value < 0.01). The chromatin occupancy of the majority of DRM subunits significantly decreased at all sites tested. Additionally, the majority of DRM subunits retained significant chromatin enrichment over IgG in lin-35 null embryos at 9 of the 12 sites tested.
(TIF)
Box plots of normalized read counts in DRM peaks in wild-type (WT) versus lin-35 null replicates for each DRM subunit. Box plots show the median (black bar) with each box extending from the 25th to 75th percentile. Extended whiskers indicate the 2.5th and 97.5th percentile, and wedges indicate the 95% confidence interval for the medians. Outliers were removed from the graphs. Signal from the 2nd lin-35 null ChIP-seq replicate appears to be abnormally elevated compared to the 1st and 3rd replicates.
(TIF)
(A) Genomic profiles of each DRM component in wild-type (WT) and lin-35 late embryos near DRM subunit genes. E2F-DP subunits (blues), LIN-35 (purple), and MuvB subunits (greens) were overlaid on the same track. Normalized ChIP-seq enrichment values for each data track are indicated on the y-axis, and the chromosomal coordinates of the areas shown are indicated below the x-axis. Gene transcriptional start sites are indicated by black boxes, coding strand directions are indicated by white arrowheads, and gene transcriptional termination sites are indicated by black arrowheads. Other genes in the region are similarly indicated in grey. Each DRM peak location is indicated by a black rectangle with the peak center highlighted in red. Of the 8 DRM genes, only lin-37 and lin-52 are not targeted by a promoter-bound DRM peak. (B) RT-qPCR analysis comparing transcript levels of DRM subunit genes in lin-35(n745) (purple) and lin-54(n2231) (green) late embryos to the level in WT (white) late embryos, presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean, and significance was determined by a student’s T test between transcript levels in mutant vs WT (* p-value < 0.05, ** p-value < 0.01). 7 of the 8 DRM genes were upregulated in lin-35 and/or lin-54 mutants. lin-35 transcripts in lin-35 null embryos were significantly decreased compared to WT, consistent with the W151Stop* mutation leading to nonsense-mediated decay of the transcript. (C) Western blot analysis of 2 biological replicates of WT and lin-35 lysates performed using the antibodies indicated on the right. Alpha-tubulin served as a loading control. (D) ChIP-qPCR of DRM subunits at the mes-2, cdk-1, and pcn-1 gene promoters in L1 larvae. Signals are presented as the log2 fold enrichment of the ChIP signal for each region vs. a negative control region (a non-coding region of chromosome IV). Error bars indicate standard error of the mean. Significance was determined by a student’s T test between subunit ChIP values in WT versus lin-35 (* p-value < 0.05, ** p-value < 0.01) or between subunit versus IgG ChIP values in lin-35 († p-value < 0.05, ‡ p-value < 0.01).
(TIF)
(A,B) Enrichment of phylogenetically conserved (phasCons > 0.7) CDE-like sequences (IUPAC code: BSSSSS, allowing 1 mismatch) and CHR-like sequences (IUPAC code: TTYRAA) in DRM peak regions (1418 regions) compared to background promoters of all non-overlapping and non-DRM target genes (16,471 background regions). Black bars and bolded sequences indicate significant enrichment over background using a hypergeometric distribution (p-value < 1e-5). Not all potential CDE sequences are shown. (C,D) Motif enrichment in Class I versus Class II DRM peaks and in HOT versus not HOT DRM peaks using a chi-squared test (p < 0.05). Although we observed enrichment of the de novo motif in Class I peaks compared to Class II peaks, the enrichment was lost when we tested conserved CDE and/or CHR motifs. This result suggests that the presence of a DRM motif does not distinguish the two peak classes. Conserved CDE and CHR motifs were observed in HOT DRM peaks, suggesting that many DRM peaks in HOT regions are true DRM binding sites.
(TIF)
(A,B) Venn diagrams showing total peak overlaps for each E2F-DP subunit (A, blue) and each MuvB subunit (B, green) in lin-35 compared to wild-type (WT) ChIP-seq. The number of subunit peaks observed only in WT (i), in both WT and lin-35 (ii), and only in lin-35 (iii) are indicated. Total overlaps of subunit peaks observed only in lin-35 (iii) for E2F-DP (A) or MuvB (B) are indicated in the 2nd venn diagram (grey). (C) Heatmap of normalized ChIP-seq profiles of pooled replicates for each DRM subunit. Plotted regions for each subunit are separated based on whether the peak was observed only in wild type (WT) (i), observed in both WT and lin-35 (ii), or observed only in lin-35 (iii). Many "lin-35 only" peaks reside near DRM binding sites already observed in WT.
(TIF)
(A) Venn diagrams showing DRM target genes that are upregulated based on microarray analysis from [37] (lin-35 early embryo (EE) and L1, light and dark purple) and [33] (lin-54 mixed-stage embryo (ME), green) (Group A genes). (B) Box plots of wild-type (WT) or mutant (lin-35 or lin-54) log2 average expression values of Upregulated DRM targets (Group A), Not Upregulated DRM Targets (Group B), and All Genes. Staging of the samples for each experimental group is indicated: early embryos (EE), mixed-stage embryos (ME), and L1 larvae (L1). Box plots show the median (black bar) with each box extending from the 25th to 75th percentile. Extended whiskers indicate the 2.5th and 97.5th percentile, and wedges indicate the 95% confidence interval for the medians. Outliers were removed from the graphs. Significance (* p-value < 0.05, ** p-value < 0.01) was determined by a Wilcoxon-Mann-Whitney test. These results indicate that detectable upregulation reflects a drop in the average expression of DRM target genes in WT over developmental time, not an increase in the average expression of DRM target genes in mutants. This effect is also observed in Group B genes but the repression is not as dramatic. We speculate that Group A genes are progressively more repressed by DRM over developmental time; DRM dysfunction manifests as a failure in this repression and hence gene upregulation as development progresses. In contrast, Group B genes may include genes that 1) DRM does not fully repress or 2) are activated intermittently as development progresses. These findings suggest that detection of DRM target gene upregulation is dependent on developmental stage because wild-type DRM requires developmental time to establish repression of gene targets.
(TIF)
(A) REViGO gene ontology (GO) visualization of terms, which helped identify informative GO terms, enriched in Group A targets (left) vs. Group B targets (right), as described in Fig 5. GO terms are clustered on the x-y plane based on semantic similarity measures, with the size of each bubble indicating the GO term frequency and the color indicating the Benjamini adjusted p-value. Representative GO terms are highlighted and labeled. (B,C) Bar chart plotting the –log10 Benjamini adjusted p-value of selected GO term enrichment observed in (B) DRM targets marked by Class I peaks vs. Class II peaks, and (C) DRM targets marked by HOT regions vs. not HOT regions.
(TIF)
(A) RT-qPCR quantification of the effectiveness of RNAi depletion of efl-1 (blue), lin-9 (light green), and lin-54 (dark green) transcript levels in lin-35 null (lin-35(n745)) late embryos compared to empty-vector (Ctrl, purple) RNAi. (B) Genomic profiles of each DRM component in wild-type (WT) and lin-35 null late embryos at rme-2 and mex-5. E2F/DP subunits (blues), LIN-35 (purple) and MuvB subunits (greens) were overlaid on the same track. Normalized ChIP-seq enrichment values for each data track are indicated on the y-axis, and the chromosomal coordinates of the areas shown are indicated below the x-axis. Gene transcriptional start sites are indicated by black boxes, and coding strand directions are indicated by white arrowheads. (C) RT-qPCR analysis of rme-2 and mex-5 following efl-1 (blue), lin-9 (light green), and lin-54 (dark green) RNAi in lin-35 null late embryos as compared to empty-vector (Ctrl, purple). Expression values from 2 independent experiments each consisting of 4 biological replicates for each RNAi condition were averaged and presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We thank Seth Rubin, Joseph Lipsick, Andreas Rechtsteiner, and members of the Rubin and Strome labs for helpful discussions. We thank Robert Horvitz for antibodies. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 Instrumentation Grants S10RR029668, S10RR027303, and S10OD018174. Some strains used in this study were provided the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Data Availability
The ChIP-seq and expression data used in this study are available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo). ChIP-seq data used in this study are accessible through GEO Series accession number GSE95071. Microarray expression analysis data used in this study are accessible through GEO Series accession number GSE6547 [37] and GSE28853 [33].
Funding Statement
This work was supported by California Institute for Regenerative Medicine Postdoctoral Fellowship TG-11057 (https://www.cirm.ca.gov/) and American Cancer Society Postdoctoral Fellowship PF-16-106-01-DDC (https://www.cancer.org/) to PDG and NIH R01 grant GM34059 (https://www.nih.gov/) to SS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Molecular Cell. 2007;26(4):539–51. doi: 10.1016/j.molcel.2007.04.015 [DOI] [PubMed] [Google Scholar]
- 2.Pilkinton M, Sandoval R, Colamonici OR. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26(54):7535–43. doi: 10.1038/sj.onc.1210562 . [DOI] [PubMed] [Google Scholar]
- 3.Sadasivam S, DeCaprio JA. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer. 2013;13(8):585–95. doi: 10.1038/nrc3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119(2):181–93. doi: 10.1016/j.cell.2004.09.034 [DOI] [PubMed] [Google Scholar]
- 5.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci U S A. 2006;103(45):16782–7. doi: 10.1073/pnas.0608461103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litovchick L, Florens LA, Swanson SK, Washburn MP, DeCaprio JA. DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly. Genes Dev. 2011;25(8):801–13. doi: 10.1101/gad.2034211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guiley KZ, Liban TJ, Felthousen JG, Ramanan P, Litovchick L, Rubin SM. Structural mechanisms of DREAM complex assembly and regulation. Genes Dev. 2015;29(9):961–74. doi: 10.1101/gad.257568.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14(7):804–16. [PMC free article] [PubMed] [Google Scholar]
- 9.Zwicker J, Lucibello FC, Wolfraim LA, Gross C, Truss M, Engeland K, et al. Cell cycle regulation of the cyclin A, cdc25C and cdc2 genes is based on a common mechanism of transcriptional repression. EMBO J. 1995;14(18):4514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276(19):5703–16. doi: 10.1111/j.1742-4658.2009.07261.x . [DOI] [PubMed] [Google Scholar]
- 11.Müller GA, Engeland K. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 2010;277(4):877–93. doi: 10.1111/j.1742-4658.2009.07508.x . [DOI] [PubMed] [Google Scholar]
- 12.Müller GA, Quaas M, Schümann M, Krause E, Padi M, Fischer M, et al. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 2012;40(4):1561–78. doi: 10.1093/nar/gkr793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marceau AH, Felthousen JG, Goetsch PD, Iness AN, Lee HW, Tripathi SM, et al. Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters. Nat Commun. 2016;7:12301 doi: 10.1038/ncomms12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10(13):1633–44. . [DOI] [PubMed] [Google Scholar]
- 15.Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6(3):729–35. . [DOI] [PubMed] [Google Scholar]
- 16.Forristal C, Henley SA, MacDonald JI, Bush JR, Ort C, Passos DT, et al. Loss of the mammalian DREAM complex deregulates chondrocyte proliferation. Mol Cell Biol. 2014;34(12):2221–34. Epub 2014/04/07. doi: 10.1128/MCB.01523-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tedesco D, Lukas J, Reed SI. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2). Genes Dev. 2002;16(22):2946–57. doi: 10.1101/gad.1011202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18(23):2929–40. doi: 10.1101/gad.1255204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B, et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. The EMBO Journal. 2006;26(1):144–57. doi: 10.1038/sj.emboj.7601478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6(15):1903–13. doi: 10.4161/cc.6.15.4512 . [DOI] [PubMed] [Google Scholar]
- 21.Knight AS, Notaridou M, Watson RJ. A Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28(15):1737–47. doi: 10.1038/onc.2009.22 . [DOI] [PubMed] [Google Scholar]
- 22.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26(5):474–89. doi: 10.1101/gad.181933.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichert N, Wurster S, Ulrich T, Schmitt K, Hauser S, Probst L, et al. Lin9, a subunit of the mammalian DREAM complex, is essential for embryonic development, for survival of adult mice, and for tumor suppression. Mol Cell Biol. 2010;30(12):2896–908. doi: 10.1128/MCB.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95(7):981–91. . [DOI] [PubMed] [Google Scholar]
- 25.Ceol CJ, Horvitz HR. dpl-1 DP and efl-1 E2F act with lin-35 Rb to antagonize Ras signaling in C. elegans vulval development. Mol Cell. 2001;7(3):461–73. . [DOI] [PubMed] [Google Scholar]
- 26.Davidson CJ, Guthrie EE, Lipsick JS. Duplication and maintenance of the Myb genes of vertebrate animals. Biol Open. 2013;2(2):101–10. doi: 10.1242/bio.20123152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang D, Kennedy S, Conte D, Kim JK, Gabel HW, Kamath RS, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436(7050):593–7. doi: 10.1038/nature04010 [DOI] [PubMed] [Google Scholar]
- 28.Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development. 2011;138(6):1069–79. doi: 10.1242/dev.059501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latorre I, Chesney MA, Garrigues JM, Stempor P, Appert A, Francesconi M, et al. The DREAM complex promotes gene body H2A.Z for target repression. Genes Dev. 2015;29(5):495–500. doi: 10.1101/gad.255810.114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Araya CL, Kawli T, Kundaje A, Jiang L, Wu B, Vafeados D, et al. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature. 2014;512(7515):400–5. doi: 10.1038/nature13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550 doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. Epub 2009/05/20. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabuchi TM, Deplancke B, Osato N, Zhu LJ, Barrasa MI, Harrison MM, et al. Chromosome-biased binding and gene regulation by the Caenorhabditis elegans DRM complex. PLoS Genet. 2011;7(5):e1002074 doi: 10.1371/journal.pgen.1002074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller GA, Wintsche A, Stangner K, Prohaska SJ, Stadler PF, Engeland K. The CHR site: definition and genome-wide identification of a cell cycle transcriptional element. Nucleic Acids Res. 2014;42(16):10331–50. doi: 10.1093/nar/gku696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, Scime A, et al. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25(18):8166–78. doi: 10.1128/MCB.25.18.8166-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi W, Reinke V. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development. 2006;133(16):3147–57. doi: 10.1242/dev.02490 . [DOI] [PubMed] [Google Scholar]
- 37.Kirienko NV, Fay DS. Transcriptome profiling of the C. elegans Rb ortholog reveals diverse developmental roles. Dev Biol. 2007;305(2):674–84. doi: 10.1016/j.ydbio.2007.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 39.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. Epub 2008/11/25. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800 Epub 2011/07/18. doi: 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kudron M, Niu W, Lu Z, Wang G, Gerstein M, Snyder M, et al. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol. 2013;14(1):R5 doi: 10.1186/gb-2013-14-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beitel GJ, Lambie EJ, Horvitz HR. The C. elegans gene lin-9,which acts in an Rb-related pathway, is required for gonadal sheath cell development and encodes a novel protein. Gene. 2000;254(1–2):253–63. . [DOI] [PubMed] [Google Scholar]
- 43.Chi W, Reinke V. DPL-1 (DP) acts in the germ line to coordinate ovulation and fertilization in C. elegans. Mech Dev. 2009;126(5–6):406–16. doi: 10.1016/j.mod.2009.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasaki K, McCarter J, Francis R, Schedl T. emo-1, a Caenorhabditis elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J Cell Biol. 1996;134(3):699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCarter J, Bartlett B, Dang T, Schedl T. Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev Biol. 1997;181(2):121–43. doi: 10.1006/dbio.1996.8429 . [DOI] [PubMed] [Google Scholar]
- 46.Li J, Chauve L, Phelps G, Brielmann RM, Morimoto RI. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev. 2016;30(18):2062–75. doi: 10.1101/gad.283317.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burke JR, Deshong AJ, Pelton JG, Rubin SM. Phosphorylation-induced conformational changes in the retinoblastoma protein inhibit E2F transactivation domain binding. J Biol Chem. 2010;285(21):16286–93. doi: 10.1074/jbc.M110.108167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor-Harding B, Binné UK, Korenjak M, Brehm A, Dyson NJ. p55, the Drosophila ortholog of RbAp46/RbAp48, is required for the repression of dE2F2/RBF-regulated genes. Mol Cell Biol. 2004;24(20):9124–36. doi: 10.1128/MCB.24.20.9124-9136.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao L, Peng B, Yao L, Zhang X, Sun K, Yang X, et al. The ancient function of RB-E2F pathway: insights from its evolutionary history. Biol Direct. 2010;5:55 Epub 2010/09/20. doi: 10.1186/1745-6150-5-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liban TJ, Medina EM, Tripathi S, Sengupta S, Henry RW, Buchler NE, et al. Conservation and divergence of C-terminal domain structure in the retinoblastoma protein family. Proc Natl Acad Sci U S A. 2017. Epub 2017/04/24. doi: 10.1073/pnas.1619170114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev. 2004;18(23):2952–62. doi: 10.1101/gad.322004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation. Proc Natl Acad Sci U S A. 2006;103(2):437–42. doi: 10.1073/pnas.0510012103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huh K, Zhou X, Hayakawa H, Cho JY, Libermann TA, Jin J, et al. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J Virol. 2007;81(18):9737–47. doi: 10.1128/JVI.00881-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pang CL, Toh SY, He P, Teissier S, Ben Khalifa Y, Xue Y, et al. A functional interaction of E7 with B-Myb-MuvB complex promotes acute cooperative transcriptional activation of both S- and M-phase genes. (129 c). Oncogene. 2014;33(31):4039–49. doi: 10.1038/onc.2013.426 . [DOI] [PubMed] [Google Scholar]
- 55.Iltzsche F, Simon K, Stopp S, Pattschull G, Francke S, Wolter P, et al. An important role for Myb-MuvB and its target gene KIF23 in a mouse model of lung adenocarcinoma. Oncogene. 2017;36(1):110–21. Epub 2016/05/23. doi: 10.1038/onc.2016.181 . [DOI] [PubMed] [Google Scholar]
- 56.Boichuk S, Parry JA, Makielski KR, Litovchick L, Baron JL, Zewe JP, et al. The DREAM complex mediates GIST cell quiescence and is a novel therapeutic target to enhance imatinib-induced apoptosis. Cancer Res. 2013;73(16):5120–9. doi: 10.1158/0008-5472.CAN-13-0579 . [DOI] [PubMed] [Google Scholar]
- 57.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. Article published online before print in May 2002. doi: 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diaz A, Park K, Lim DA, Song JS. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol. 2012;11(3):Article 9. doi: 10.1515/1544-6115.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–91. doi: 10.1093/nar/gku365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26(12):1351–9. doi: 10.1038/nbt.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22(9):1813–31. doi: 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–93. doi: 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. Epub 2005/07/15. doi: 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421(6920):231–7. doi: 10.1038/nature01278 . [DOI] [PubMed] [Google Scholar]
- 68.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. . [DOI] [PubMed] [Google Scholar]
- 69.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249 . [DOI] [PubMed] [Google Scholar]
- 70.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47 Epub 2015/01/20. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–5. . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) UpSet visualization of DRM subunit peak overlaps. Black filled circles indicate the factor(s) present within an intersection category. Intersection categories consisting of less than 20 represented peaks were omitted. (B) Heatmap of normalized ChIP-seq profiles of pooled replicates for each DRM subunit across all identified peaks. High confidence DRM peaks (identified in Fig 1) are separated from the peaks that did not pass the overlap requirements (remaining DRM peaks). ChIP-seq signal appears consistently stronger in the high confidence DRM peaks when compared to the remaining DRM peaks. Even though many peaks do not pass the overlap requirements, the occupancy observed for each DRM subunit appears similar at all peak regions.
(TIF)
Full western blots from 2 co-immunoprecipitation experiments performed on biological replicate late embryo lysates. Proteins bound (B) and unbound (UB) by EFL-1, LIN-37, or control IgG immunoprecipitation were separated by SDS/PAGE, and western blot analysis was performed using the antibodies indicated in the bottom right corner. 5% of Input was included. Arrows indicate protein bands presented in main Fig 2. Asterisks indicate bands that carried over from a previous blot.
(TIF)
(A) Scatter plot of normalized ChIP-seq reads in lin-35 vs. wild type (WT) for each DRM subunit for Class I (dark) and Class II (light) peaks, as described in the Fig 3 legend. Each point indicates the WT (x-axis) and lin-35 null (y-axis) read density of pooled replicates within a single wild-type DRM peak from E2F/DP (blue), LIN-35 (purple), and MuvB (green) subunit ChIP-seq. Dotted lines indicate the slope expected if DRM occupancy is equivalent in WT and lin-35 ChIP-seq. (B) ChIP-qPCR of 5 DRM subunits at 8 Class I peaks and 4 Class II peaks. H3K4me3 ChIP was included as a positive control. Signals are presented as the log2 fold enrichment of the ChIP signal for each region vs. a negative control region (a non-coding region of chromosome IV). Error bars indicate standard error of the mean. Significance was determined by a student’s T test between subunit ChIP values in WT versus lin-35 (* p-value < 0.05, ** p-value < 0.01) or between subunit versus IgG ChIP values in lin-35 († p-value < 0.05, ‡ p-value < 0.01). The chromatin occupancy of the majority of DRM subunits significantly decreased at all sites tested. Additionally, the majority of DRM subunits retained significant chromatin enrichment over IgG in lin-35 null embryos at 9 of the 12 sites tested.
(TIF)
Box plots of normalized read counts in DRM peaks in wild-type (WT) versus lin-35 null replicates for each DRM subunit. Box plots show the median (black bar) with each box extending from the 25th to 75th percentile. Extended whiskers indicate the 2.5th and 97.5th percentile, and wedges indicate the 95% confidence interval for the medians. Outliers were removed from the graphs. Signal from the 2nd lin-35 null ChIP-seq replicate appears to be abnormally elevated compared to the 1st and 3rd replicates.
(TIF)
(A) Genomic profiles of each DRM component in wild-type (WT) and lin-35 late embryos near DRM subunit genes. E2F-DP subunits (blues), LIN-35 (purple), and MuvB subunits (greens) were overlaid on the same track. Normalized ChIP-seq enrichment values for each data track are indicated on the y-axis, and the chromosomal coordinates of the areas shown are indicated below the x-axis. Gene transcriptional start sites are indicated by black boxes, coding strand directions are indicated by white arrowheads, and gene transcriptional termination sites are indicated by black arrowheads. Other genes in the region are similarly indicated in grey. Each DRM peak location is indicated by a black rectangle with the peak center highlighted in red. Of the 8 DRM genes, only lin-37 and lin-52 are not targeted by a promoter-bound DRM peak. (B) RT-qPCR analysis comparing transcript levels of DRM subunit genes in lin-35(n745) (purple) and lin-54(n2231) (green) late embryos to the level in WT (white) late embryos, presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean, and significance was determined by a student’s T test between transcript levels in mutant vs WT (* p-value < 0.05, ** p-value < 0.01). 7 of the 8 DRM genes were upregulated in lin-35 and/or lin-54 mutants. lin-35 transcripts in lin-35 null embryos were significantly decreased compared to WT, consistent with the W151Stop* mutation leading to nonsense-mediated decay of the transcript. (C) Western blot analysis of 2 biological replicates of WT and lin-35 lysates performed using the antibodies indicated on the right. Alpha-tubulin served as a loading control. (D) ChIP-qPCR of DRM subunits at the mes-2, cdk-1, and pcn-1 gene promoters in L1 larvae. Signals are presented as the log2 fold enrichment of the ChIP signal for each region vs. a negative control region (a non-coding region of chromosome IV). Error bars indicate standard error of the mean. Significance was determined by a student’s T test between subunit ChIP values in WT versus lin-35 (* p-value < 0.05, ** p-value < 0.01) or between subunit versus IgG ChIP values in lin-35 († p-value < 0.05, ‡ p-value < 0.01).
(TIF)
(A,B) Enrichment of phylogenetically conserved (phasCons > 0.7) CDE-like sequences (IUPAC code: BSSSSS, allowing 1 mismatch) and CHR-like sequences (IUPAC code: TTYRAA) in DRM peak regions (1418 regions) compared to background promoters of all non-overlapping and non-DRM target genes (16,471 background regions). Black bars and bolded sequences indicate significant enrichment over background using a hypergeometric distribution (p-value < 1e-5). Not all potential CDE sequences are shown. (C,D) Motif enrichment in Class I versus Class II DRM peaks and in HOT versus not HOT DRM peaks using a chi-squared test (p < 0.05). Although we observed enrichment of the de novo motif in Class I peaks compared to Class II peaks, the enrichment was lost when we tested conserved CDE and/or CHR motifs. This result suggests that the presence of a DRM motif does not distinguish the two peak classes. Conserved CDE and CHR motifs were observed in HOT DRM peaks, suggesting that many DRM peaks in HOT regions are true DRM binding sites.
(TIF)
(A,B) Venn diagrams showing total peak overlaps for each E2F-DP subunit (A, blue) and each MuvB subunit (B, green) in lin-35 compared to wild-type (WT) ChIP-seq. The number of subunit peaks observed only in WT (i), in both WT and lin-35 (ii), and only in lin-35 (iii) are indicated. Total overlaps of subunit peaks observed only in lin-35 (iii) for E2F-DP (A) or MuvB (B) are indicated in the 2nd venn diagram (grey). (C) Heatmap of normalized ChIP-seq profiles of pooled replicates for each DRM subunit. Plotted regions for each subunit are separated based on whether the peak was observed only in wild type (WT) (i), observed in both WT and lin-35 (ii), or observed only in lin-35 (iii). Many "lin-35 only" peaks reside near DRM binding sites already observed in WT.
(TIF)
(A) Venn diagrams showing DRM target genes that are upregulated based on microarray analysis from [37] (lin-35 early embryo (EE) and L1, light and dark purple) and [33] (lin-54 mixed-stage embryo (ME), green) (Group A genes). (B) Box plots of wild-type (WT) or mutant (lin-35 or lin-54) log2 average expression values of Upregulated DRM targets (Group A), Not Upregulated DRM Targets (Group B), and All Genes. Staging of the samples for each experimental group is indicated: early embryos (EE), mixed-stage embryos (ME), and L1 larvae (L1). Box plots show the median (black bar) with each box extending from the 25th to 75th percentile. Extended whiskers indicate the 2.5th and 97.5th percentile, and wedges indicate the 95% confidence interval for the medians. Outliers were removed from the graphs. Significance (* p-value < 0.05, ** p-value < 0.01) was determined by a Wilcoxon-Mann-Whitney test. These results indicate that detectable upregulation reflects a drop in the average expression of DRM target genes in WT over developmental time, not an increase in the average expression of DRM target genes in mutants. This effect is also observed in Group B genes but the repression is not as dramatic. We speculate that Group A genes are progressively more repressed by DRM over developmental time; DRM dysfunction manifests as a failure in this repression and hence gene upregulation as development progresses. In contrast, Group B genes may include genes that 1) DRM does not fully repress or 2) are activated intermittently as development progresses. These findings suggest that detection of DRM target gene upregulation is dependent on developmental stage because wild-type DRM requires developmental time to establish repression of gene targets.
(TIF)
(A) REViGO gene ontology (GO) visualization of terms, which helped identify informative GO terms, enriched in Group A targets (left) vs. Group B targets (right), as described in Fig 5. GO terms are clustered on the x-y plane based on semantic similarity measures, with the size of each bubble indicating the GO term frequency and the color indicating the Benjamini adjusted p-value. Representative GO terms are highlighted and labeled. (B,C) Bar chart plotting the –log10 Benjamini adjusted p-value of selected GO term enrichment observed in (B) DRM targets marked by Class I peaks vs. Class II peaks, and (C) DRM targets marked by HOT regions vs. not HOT regions.
(TIF)
(A) RT-qPCR quantification of the effectiveness of RNAi depletion of efl-1 (blue), lin-9 (light green), and lin-54 (dark green) transcript levels in lin-35 null (lin-35(n745)) late embryos compared to empty-vector (Ctrl, purple) RNAi. (B) Genomic profiles of each DRM component in wild-type (WT) and lin-35 null late embryos at rme-2 and mex-5. E2F/DP subunits (blues), LIN-35 (purple) and MuvB subunits (greens) were overlaid on the same track. Normalized ChIP-seq enrichment values for each data track are indicated on the y-axis, and the chromosomal coordinates of the areas shown are indicated below the x-axis. Gene transcriptional start sites are indicated by black boxes, and coding strand directions are indicated by white arrowheads. (C) RT-qPCR analysis of rme-2 and mex-5 following efl-1 (blue), lin-9 (light green), and lin-54 (dark green) RNAi in lin-35 null late embryos as compared to empty-vector (Ctrl, purple). Expression values from 2 independent experiments each consisting of 4 biological replicates for each RNAi condition were averaged and presented as the relative quantity (Rq) compared to act-2. Error bars indicate standard error of the mean.
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
The ChIP-seq and expression data used in this study are available from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo). ChIP-seq data used in this study are accessible through GEO Series accession number GSE95071. Microarray expression analysis data used in this study are accessible through GEO Series accession number GSE6547 [37] and GSE28853 [33].