Mitochondria are crucial organelles for providing energy for a cell. It is known that MIC60/mitofilin is important for maintaining mitochondrial structure in dissociated cells; however, its physiological roles in an intact animal are less clear. In this study, we unravel the functional consequences of deleting MIC60/mitofilin in fruit flies.
Abstract
MIC60/mitofilin constitutes a hetero-oligomeric complex on the inner mitochondrial membranes to maintain crista structure. However, little is known about its physiological functions. Here, by characterizing Drosophila MIC60 mutants, we define its roles in vivo. We discover that MIC60 performs dual functions to maintain mitochondrial homeostasis. In addition to its canonical role in crista membrane structure, MIC60 regulates mitochondrial motility, likely by influencing protein levels of the outer mitochondrial membrane protein Miro that anchors mitochondria to the microtubule motors. Loss of MIC60 causes loss of Miro and mitochondrial arrest. At a cellular level, loss of MIC60 disrupts synaptic structure and function at the neuromuscular junctions. The dual roles of MIC60 in both mitochondrial crista structure and motility position it as a crucial player for cellular integrity and survival.
INTRODUCTION
The membrane architecture of a mitochondrion is intricately shaped and strictly maintained to allow efficient biochemical reactions within mitochondrial compartments. Crista membranes are protrusions of inner mitochondrial membranes (IMM) into the matrix and house the electron transport chain (ETC) complexes and ATP synthase, key enzymes for ATP production (Mannella et al., 2013). The crista space is exceptionally slim and elongated and joins the intermembrane space through crista junctions. Inner boundary membranes are the part of the IMM that are closely apposed to the outer mitochondrial membranes (OMM) and connect to the OMM at contact sites to facilitate mitochondrial protein import and fission-and-fusion (Zerbes et al., 2012). Yeast and cell culture work has shown that MIC60 (also known as mitofilin/IMMT/fcj1), a crucial component of the mitochondrial contact site and cristae organizing system (MICOS) complex on the IMM, maintains both crista junctions and contact sites (John et al., 2005; Xie et al., 2007; Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Zerbes et al., 2012; Ding et al., 2015; Friedman et al., 2015). However, the physiological roles of MIC60 in multicellular organisms in vivo are not well understood.
Mitochondrial movement and distribution are tightly regulated on a spatial and temporal scale to balance energy homeostasis. Microtubule-based mitochondrial motility is mediated by Miro, an OMM GTPase that bridges mitochondria to the microtubule motors (Guo et al., 2005; Fransson et al., 2006; Glater et al., 2006; Wang and Schwarz, 2009b; Wang et al., 2011; Koutsopoulos et al., 2010). Multiple cellular signals target Miro to influence mitochondrial motility and distribution (Lovas and Wang, 2013; Course and Wang, 2016). Several signals trigger removal of Miro from the OMM and thus dissociate mitochondria from the microtubule motors and arrest mitochondrial motility (Wang et al., 2011; Hsieh et al., 2016). In this study, we discover that loss of MIC60 causes mitochondrial arrest and down-regulates Miro protein. Furthermore, we reveal novel physiological and developmental roles of MIC60 in vivo, linking it to synaptic structure and function.
RESULTS AND DISCUSSION
Characterization of dMIC60 mutants
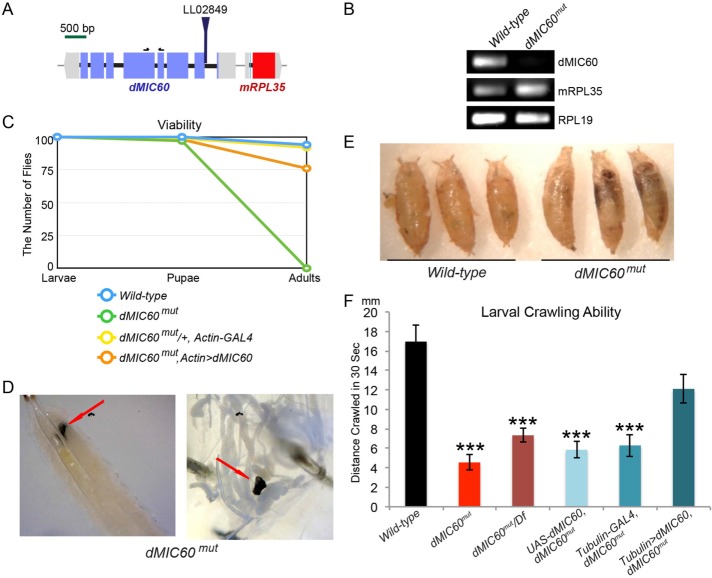
We identified a mutant allele of Drosophila MIC60 (dMIC60) containing a PiggyBac insertion (LL02849) in the dMIC60 gene named dMIC60mut (Figure 1A) (Schuldiner et al., 2008), which eliminated dMIC60 expression detected by reverse transcription PCR (RT-PCR) (Figure 1, A and B). dMIC60mut flies were viable only until the late pupal stage, suggesting that dMIC60 is essential for development and viability in Drosophila. The lethality of dMIC60mut was rescued by ubiquitous expression of a UAS-dMIC60 transgene (Figure 1C). dMIC60mut larvae displayed melanotic masses inside their bodies (Figure 1D), and their pupal cases showed an abnormal “banana”-like shape (Figure 1E). dMIC60mut third instar larvae were sluggish. Their crawling ability was significantly compromised compared with controls, which was rescued by ubiquitous expression of dMIC60 (Figure 1F). Transheterozygous dMIC60mut over a deficiency locus that covers dMIC60 showed impaired crawling ability equivalent to that of homozygous mutants (Figure 1F), indicating this dMIC60mut allele is a null or strong hypomorph mutation.
FIGURE 1:
Characterization of a mutant allele of dMIC60. (A) Depiction of the genomic site of the PiggyBac insertion (LL02849). Black arrows indicate the locations of the primers used in B. (B) RT–PCR on third instar larvae of wild-type flies (w1118) or flies with LL02849 (dMIC60mut), using primers amplifying genes as indicated. (C) Viability analysis. One hundred late third instar larvae of each genotype were collected and the pupae and adults were counted subsequently. dMIC60mut,Actin>dMIC60: UAS-dMIC60-Myc;Actin-GAL4, dMIC60mut. (D) Images of melanotic masses (red arrows) in third instar larvae of dMIC60mut. The left image shows the lower half of a larva, dorsal side up; the right image shows the mass amongst the guts after dissection of a larva. (E) Images of pupal cases as indicated. (F) Quantification of the crawling ability of third instar larvae. Df: Deficiency. Comparisons with “Wild-type.” n = 20–21 flies. * p < 0.05, ** p < 0.01, *** p < 0.001, and error bars represent mean ± SEM here and for all figures unless otherwise stated.
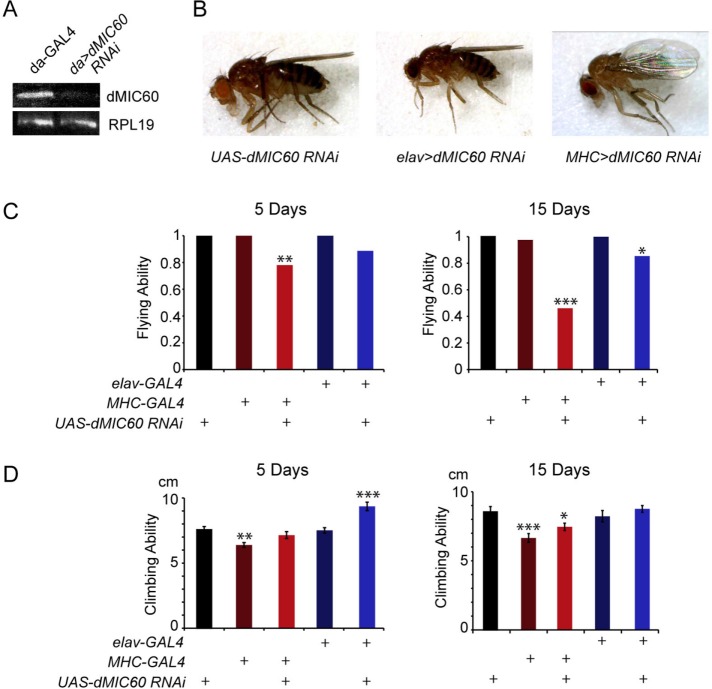
We also obtained UAS-dMIC60 RNA interference (RNAi) lines from Vienna Drosophila RNAi Center (VDRC). Using RT–PCR and quantitative real-time PCR (qPCR) we confirmed significant reduction of dMIC60 expression by ubiquitous dMIC60 RNAi (Figure 2A and Supplemental Figure S1A). Knockdown of dMIC60 allowed adult survivors using the ubiquitous driver da-GAL4 or tissue-specific drivers. We found that dMIC60 RNAi in muscles by MHC-GAL4, but not in neurons by elav-GAL4, caused a drooped wing posture (Figure 2B). dMIC60 RNAi in both muscles and neurons impaired the flying ability of adult flies in an age-dependent manner (Figure 2C). dMIC60 RNAi in neither neurons nor muscles disrupted climbing ability, and, surprisingly, dMIC60 RNAi in neurons of 5-d-old flies improved climbing ability (Figure 2D). Thus, dMIC60 is essential for maintaining flying ability and wing posture in adult flies.
FIGURE 2:
RNAi Knockdown of dMIC60 causes behavioral defects. (A) RT–PCR on third instar larvae as indicated. (B) Images of wing postures. An average of 13% of “MHC>dMIC60 RNAi” flies showed the drooped-wing posture. (C, D) Quantifications of flying (C) and climbing (D) abilities of adult flies. Comparisons with “UAS-dMIC60 RNAi.” The mean is shown and the chi-square test with Yates correction is used for C due to categorical data. n = 20–68 flies.
dMIC60 is required for maintaining mitochondrial crista structure
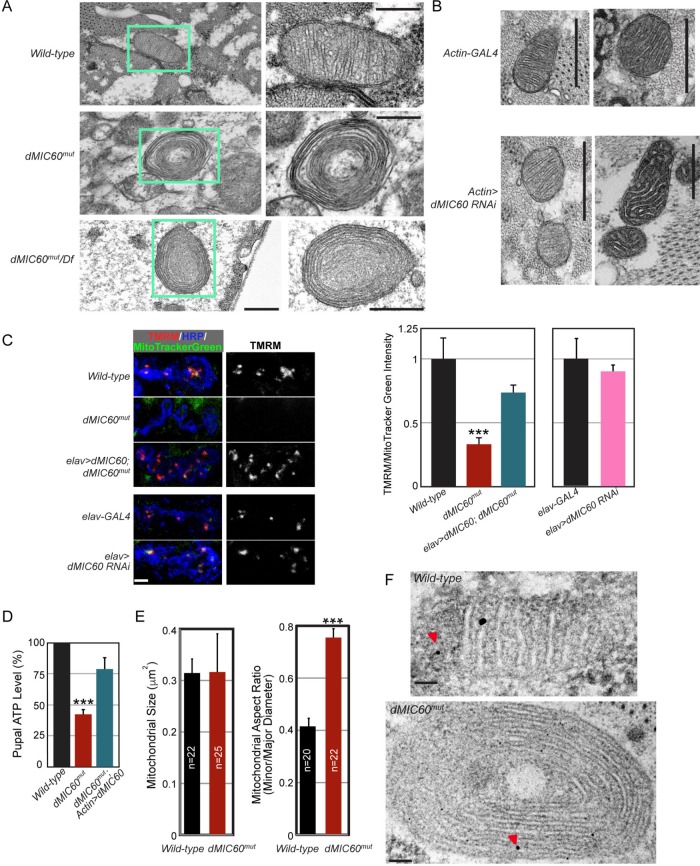
It has been reported that in yeast, Caenorhabditis elegans, and HeLa cells lacking MIC60, mitochondria display a unique “onion”-like structure (John et al., 2005; Rabl et al., 2009; Mun et al., 2010; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Zerbes et al., 2012). By examining mitochondrial ultrastructure using transmission electron microscopy (TEM), we also found the “onion”-like mitochondria in body wall muscles of dMIC60mut third instar larvae (Figure 3A). Mitochondrial crista membranes formed multiple concentric layers and crista junctions were lost. Transheterozygous dMIC60mut over the deficiency locus caused the same “onion”-like mitochondria as the homozygous mutant (Figure 3A). This phenotype was fully penetrant as 100% of the mitochondria exhibited this “onion”-like morphology in homozygous dMIC60mut or transheterozygous dMIC60mut over the deficiency (n = 272–303 mitochondria from six larvae). These results suggest that MIC60 has an evolutionarily conserved role in maintaining crista structure. However, ubiquitous knockdown of dMIC60 by RNAi was insufficient to impair crista structure in body wall muscles of third instar larvae (n = 912–1147 mitochondria from 10 larvae) (Figure 3B). We next detected the mitochondrial membrane potential (Δψm), which is highly dependent on an intact crista membrane structure (Mannella et al., 2013). The Δψm was significantly reduced in dMIC60mut but unaffected in dMIC60 RNAi flies in synaptic boutons stained by the Δψm-dependent tetramethylrhodamine (TMRM) (Gerencser et al., 2012; Devireddy et al., 2014) (Figure 3C), consistent with the result showing that crista structure is impaired in dMIC60mut but normal in dMIC60 RNAi flies (Figure 3, A and B). The total ATP level was significantly decreased in dMIC60mut pupae, which was rescued by ubiquitous expression of dMIC60 (Figure 3D). In addition, the mitochondrial shape appeared round despite no alteration in the mitochondrial size in dMIC60mut larval muscles (Figure 3E). Distribution of mitochondrial DNA as detected by immunogold staining under TEM (Cameron et al., 2011) appeared normal (the number of immunogold particles per mitochondrion for wild-type: 2.57 ± 0.19; for dMIC60mut: 2.28 ± 0.22; p > 0.05, n = 33–38 mitochondria from 21–25 images from 3 larvae) (Figure 3F). Taken together, dMIC60 is essential for maintaining mitochondrial crista structure and function in Drosophila.
FIGURE 3:
Mitochondrial crista structure and function are impaired in dMIC60mut. (A, B) TEM images of thin sections, performed on the body wall muscles of late third instar larvae. (A) The right panel is a higher magnification image of the mitochondrion marked by the green-lined box in the left. Scale bars: 500 nm. (C) Representative confocal images of TMRM staining in NMJ boutons at muscle 4 of third instar larvae. Red is TMRM staining in polarized mitochondria, green is MitoTracker Green staining in all mitochondria, and blue is horseradish peroxidase (HRP) on bouton membranes. To the right is quantification of the TMRM/MitoTracker Green fluorescence intensity in boutons as shown. The values are normalized to the mean of “Wild-type” or “elav-GAL4.” n = 11–14 NMJs from five to six larvae. Scale bar: 10 μm. (D) Quantification of the total ATP level in 48-h APF pupae, expressed as a percentage of the wild-type values. Comparisons with “Wild-type.” n = 5 pupae for each experiment and total six independent experiments. (E) The mitochondrial size is quantified between genotypes by measuring the mitochondrial area, and the mitochondrial shape is quantified by measuring the mitochondrial aspect ratio (dividing the minor diameter by the major diameter) of a mitochondrion, using Image J from the 10,000 × TEM images. n = 20–25 mitochondria from 12–15 images from four to five larvae. (F) Immunogold TEM images of thin sections, performed on the body wall muscles of late third instar larvae, show mitochondrial DNA clusters exampled by red arrowheads. Scale bars: 100 nm.
dMIC60 regulates mitochondrial motility and miro levels
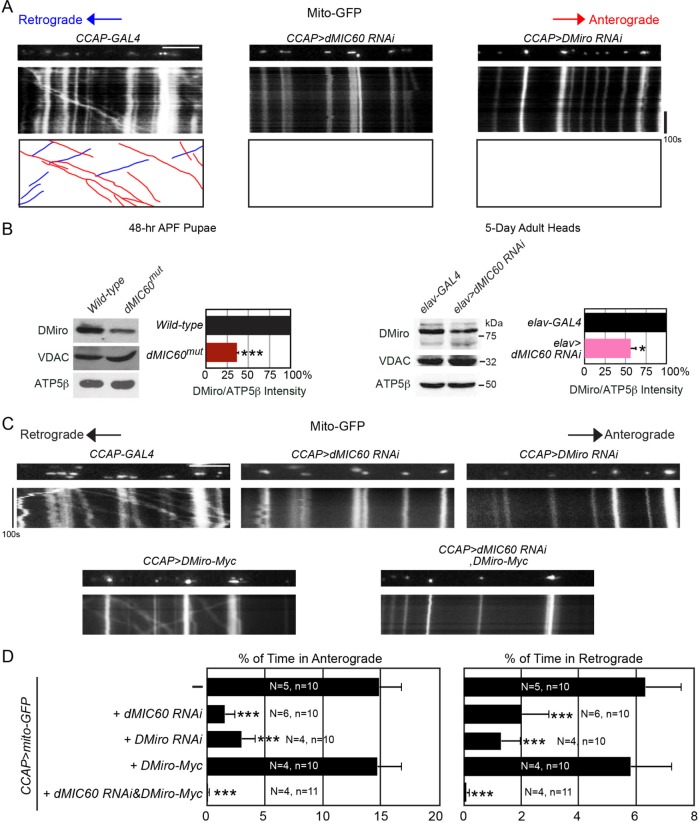
We next live-imaged mitochondrial movement by labeling mitochondria with mito-GFP driven by neuropeptidergic CCAP-GAL4 in third instar larvae, which allows tracing of individual mitochondria within a single axon in a semi-intact in vivo environment (Wang and Schwarz, 2009a). Notably, we found that axonal mitochondrial motility was significantly decreased in both anterograde and retrograde directions in axons of dMIC60 RNAi larvae (Figure 4, A and D). Interestingly, the same impairment occurred in Drosophila Miro (DMiro) RNAi larvae (Figure 4, A and D). Miro is an OMM protein that anchors mitochondria to the microtubule motors. Reduction of Miro protein causes dissociation of mitochondria from the motors and microtubules and thus mitochondrial motility ceases (Guo et al., 2005; Wang and Schwarz, 2009b; Wang et al., 2011). We then considered the possibility that DMiro protein level was decreased in dMIC60 mutant flies. Strikingly, endogenous DMiro protein was significantly reduced in both dMIC60mut and in dMIC60 RNAi flies (Figure 4B and Supplemental Figure S1B). In contrast, another OMM protein, VDAC, and the matrix protein ATP5β were not affected by lack of dMIC60 (Figure 4B), demonstrating that the decrease of DMiro protein is selective and not due to a general mitochondrial destruction. We also found that endogenous human Miro1 and MIC60 associated in the same complex in HEK293T cells (Supplemental Figure S1C). It has been shown that MIC60 can form a complex with OMM proteins such as Sam50 through scaffolding proteins in the intermembrane space (Xie et al., 2007; Darshi et al., 2011; Korner et al., 2012; van der Laan et al., 2012; Zerbes et al., 2012; Ding et al., 2015). Interestingly, it has also been shown that disruption of the MIC60 scaffolding complex or of MIC60 itself destabilizes and down-regulates Sam50 (Darshi et al., 2011; Ding et al., 2015). It is possible that loss of dMIC60 may dissociate the dMIC60/DMiro complex and destabilize and release DMiro from the OMM causing mitochondrial arrest, and DMiro is subsequently degraded in the cytosol (Figure 4, A and B). Consistent with this hypothesis, overexpression of DMiro in dMIC60 RNAi axons did not rescue their phenotype of mitochondrial arrest (Figure 4, C and D), although other scenarios are possible. Collectively, we have discovered a second mitochondrial function of dMIC60 to regulate mitochondrial motility and DMiro protein levels.
FIGURE 4:
dMIC60 regulates mitochondrial motility and DMiro. (A, C) Mitochondrial movement labeled by mito-GFP driven by CCAP-GAL4 in representative axons passing segment A3. The first frame of each live-imaging series is shown above a kymograph generated from the movie. The x-axis of each is mitochondrial position, and the y-axis corresponds to time (moving from top to bottom). Vertical white lines represent stationary mitochondria and diagonal lines are moving mitochondria. For A, the bottom kymograph extracts moving mitochondria, red in anterograde and blue in retrograde. Scale bars: 10 μm. (B) Whole-cell lysates of pupae or of adult heads were blotted as indicated. The band intensity of DMiro is normalized to that of ATP5β, a matrix loading control, and expressed as a percentage of the values of “Wild-type” or “elav-GAL4.” The raw intensities of VDAC or ATP5beta are not significantly different between the two genotypes, p > 0.05. n = 6 independent experiments. (D) From kymographs as in A and C, the percentage of time each mitochondrion in motion was determined and averaged. n = 100–232 mitochondria from 10 to 11 axons from for to six larvae. “N” is the number of larvae, and “n” is the number of axons used for each genotype. Comparisons with “CCAP>mito-GFP.”
dMIC60mut disrupts synaptic structure and function at the neuromuscular junctions
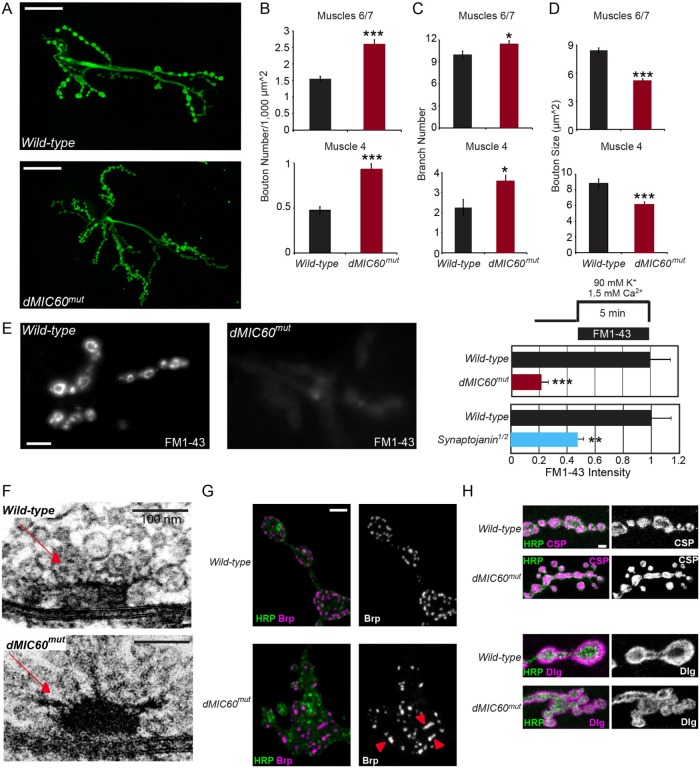
We next explored the functional consequences of disrupting the dual roles of dMIC60 in mitochondrial crista structure and motility at a cellular level. We found that at the neuromuscular junctions (NMJs) of third instar larvae the numbers of boutons and NMJ branches were significantly increased at both muscles 6/7 and 4 in dMIC60mut (Figure 5, A–C). The bouton size was notably smaller and boutons tended to cluster, causing a “cauliflower”-like appearance (Figure 5, A and D). Loading of synaptic vesicles at the NMJs was significantly compromised in dMIC60mut as demonstrated using the FM1-43 loading assay (Figure 5E) (Verstreken et al., 2008), which could be due to impaired synaptic vesicle endocytosis and/or exocytosis and indicates that synaptic transmission is dysfunctional (Verstreken et al., 2003; Dickman et al., 2005). We also examined the ultrastructure of NMJ boutons under TEM. We observed that in dMIC60mut third instar larvae the T-bar shape of some presynaptic active zones was abnormal. The typical “T” shape turned into a hemisphere shape, with threadlike structures pointing in different directions (Figure 5F). By immunostaining, Brp-labeled active zones appeared disorganized in dMIC60mut; Brp puncta were fused and clustered, while other pre- and postsynaptic markers including CSP and Dlg were normal (Figure 5, G and H). These impairments in presynaptic active zones may interfere with neurotransmitter release. Interestingly, dMIC60 RNAi did not affect bouton number, size, branch number, or FM1-43 loading at the NMJs (Supplemental Figure S1, D–J), suggesting that a partial reduction of dMIC60 is insufficient to cause obvious NMJ phenotypes. In summary, dMIC60 regulates synaptic structure and function in Drosophila.
FIGURE 5:
dMIC60mut impairs synaptic structure and function at the NMJs. (A) Confocal stack images show NMJ boutons visualized by anti-HRP which labels bouton membranes, at muscles 6/7 hemisegment A2 of third instar larvae. (B–D) Quantifications of the bouton number normalized to the muscle size (B), the NMJ branch number (C), and the bouton size (D), as shown in A. (B, C) n = 8–23 NMJs from 8 to 23 larvae. (D) n = 549 (Wild-type)-823 (dMIC60mut) boutons (muscle 6/7), or 147 (Wild-type)-221 (dMIC60mut) boutons (muscle 4), from 6 NMJs from 6 larvae. (E) FM1-43 labeling at muscle 4 hemisegment A3 of third instar larvae. The fluorescence intensity was measured and normalized to the mean of “Wild-type.” Synaptojanin1/2 that blocks synaptic transmission (Verstreken et al., 2003; Vanhauwaert et al., 2017) was used as a positive control. n = 10–15 NMJs from seven to eight larvae. (F) TEM images of thin sections, performed on the NMJs of late third instar larvae. Red arrows point to the T-bar shape of presynaptic active zones. (G, H) Confocal stack images show immunostaining at the NMJs at muscle 4 hemisegment A2 of third instar larvae. (G) Brp, red arrowheads show clustered Brp puncta; (H) CSP and Dlg. Scale bars: (A) 30 μm; (E) 10 μm; (F) 100 nm; (G, H) 10 μm.
In this study, we have revealed dual roles of dMIC60 in mitochondrial crista structure and motility in vivo. The fundamental role of dMIC60 in maintaining crista structure must be crucial for a rapid and sustained supply of mitochondrial ATP. The new role in regulating mitochondrial motility that we have discovered here could facilitate spatial mitochondrial redistribution. Efficient recycling and release of synaptic vesicles rely heavily on sufficient local ATP. The demands for local mitochondria are ever changing and heterogeneous, even among the microdomains of the same neuron (Wang and Schwarz, 2009a; Course and Wang, 2016). The ability of dMIC60 to redistribute mitochondria and maintain ATP production within synaptic compartments in response to swift changes in neuronal activities could be essential to synaptic structure and transmission (Stowers et al., 2002; Guo et al., 2005; Dickman et al., 2006). The phenotypes in dMIC60 mutants including impairments in NMJ bouton morphology, mitochondrial motility and ultrastructure, and behavior have also been observed in fly models of neurodegenerative diseases such as Parkinson’s disease and amyotrophic lateral sclerosis (Clark et al., 2006; Park et al., 2006; Liu et al., 2012; Kim et al., 2013; Godena et al., 2014). Our results highlight the importance of mitochondrial structure, transport, and distribution to neuronal homeostasis and survival.
Miro resides in a central hub to allow multiple cellular signals to instruct mitochondrial motility (Lovas and Wang, 2013; Course and Wang, 2016). These signals include intracellular Ca2+, glucose, hypoxia, and so on. Our finding adds a new player, MIC60, to these pathways. However, unlike the aforementioned signals that generally cause an effect outside the mitochondria, MIC60 localizes inside the mitochondria. It is likely that MIC60 can sense signals inside the mitochondria and relay them to Miro. For example, when mitochondria need more nutrients and substrates for ATP production, this mechanism may assist the Miro/milton complex to move mitochondria to places where glucose concentration is higher (Pekkurnaz et al., 2014). Our model provides a possibility that allows the motor/adaptor complex to follow instructions from the internal mitochondria to distribute mitochondria when and where needed. Future studies are warranted to unravel how the intrinsic signals coming from the inside of mitochondria influence Miro by MIC60.
MIC60 is one of the components of a large complex known as MICOS, which has a primary role in maintaining crista structure (John et al., 2005; Xie et al., 2007; Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Zerbes et al., 2012; Ding et al., 2015; Friedman et al., 2015). Although this complex is best characterized in yeast, many of its components have homologues in flies, and their mutants could be studied. It would be interesting to examine whether knockout of other components in the complex in flies causes cellular and behavioral phenotypes similar to dMIC60 mutants. This will gauge the importance of each component to the central role of the complex over evolution and explore their physiological functions. A partial reduction of dMIC60 by RNAi does not affect crista structure but impairs mitochondrial motility and adult behavior, demonstrating that dMIC60 has additional roles in vivo. The fly could serve as a powerful model system to better understand the functional consequences of the MICOS complex and dissect its neuronal and signaling roles.
MATERIALS AND METHODS
Fly stocks
The following fly stocks were used: MHC-GAL4, elav-GAL4, da-GAL4, Actin-GAL4, Tubulin-GAL4 (Bloomington Drosophila Stock Center, BDSC); CCAP-GAL4 (Wang et al., 2011); dMIC60LL02849 (Drosophila Genomics Resource Center, DGRC, Kyoto) (Schuldiner et al., 2008); Df(3R)Ex6118 (BL #7597); UAS-DMiro RNAi (VDRC #106683); UAS-dMIC60 RNAi (VDRC #47615); UAS-DMiro-Myc, DMirosd32 (Guo et al., 2005); and synaptojanin1/2 (Vanhauwaert et al., 2017). UAS-dMIC60-Myc were generated using the PhiC31 integrase-mediated transgenesis system, with an insertion at an estimated position of 25C6 at the attP40 site (BestGene) (Markstein et al., 2008).
Constructs
pUASTattB-dMIC60-Myc was generated by PCR amplifying the Drosophila MIC60 cDNA from pOT-dMIC60 (Flybase ID: GH04666), engineered with a C-terminal Myc tag and EcoRI/SalI restriction sites at either side, and cloning it (New England BioLabs) into a pUASTattB vector (Groth et al., 2004).
RT–PCR and qPCR
Total RNA was extracted from five 48-h after puparium formation (APF) pupae by homogenization in TRIzol (Thermo Fisher) and mixing with chloroform vigorously. Samples were centrifuged at 12,000 × g at 4°C for 15 min. The aqueous phase was then mixed with 100% isopropanol at 1:1 ratio to precipitate RNA. RNA pellets were washed with 70% ethanol and then resuspended in nuclease-free water. For RT–PCR, 100 ng of total RNA was reverse transcribed and amplified by an Access RT–PCR System (Promega), and run in a 1.5% agarose gel. For qPCR, 250 ng of total RNA was mixed with SYBR Green Real Time PCR Reagents (Life Technologies) and analyzed by a Step One Plus Real-Time PCR System (Applied Biosystems). Each data point was normalized to the expression level of the housekeeping gene RPL19. The following primers were used:
dMIC60–5′: GATAAGCTGCTGCGCTTGCAGCTCAAAAAG
dMIC60–3′: GCCACCTTGGCAATGGCATTGATCTCGTTC
RPL19–5′: CTAACACAAACTCGCGTCAGAACATTCGCA
RPL19–3′: CGATGAGCTCCTGCTTCTTGGTGGCAATAC
mRPL35–5′: CTTTTGTGACAAGCGCGCTGCGAAGCGTTT
mRPL35–3′: GGTCTTTCGTTTACCCTTCACCAGCGAGAA
Western blotting
Larvae, pupae, and adult flies were lysed as previously described (Wang et al., 2011; Tsai et al., 2014). Lysates were analyzed by SDS–PAGE and immunoblotted with mouse anti-ATP5β (ab14730; AbCam) at 1:5000, guinea pig anti-DMiro (GP5) at 1:20,000 (Tsai et al., 2014), or mouse anti-VDAC (4661S; Cell Signaling Technology) at 1:1000, and HRP-conjugated-goat anti-guinea pig or mouse Immunoglobulin G (IgG; Jackson ImmunoResearch Laboratories) at 1:3000. Coimmunoprecipitation experiments in HEK293T cells were performed using mouse anti-hMIC60 (ab110329; Abcam), mouse anti-hMiro1 (WH0055288M1; Sigma-Aldrich), or normal mouse IgG (sc-2025; Santa Cruz), as previously described (Wang et al., 2011). For Western blotting, the following antibodies were used: mouse anit-hMIC60 (ab110329; Abcam) at 1:1000, mouse anti-hMiro1 (WH0055288M1; Sigma-Aldrich) at 1:1000, and mouse anti-tubulin (ab7291; Abcam) at 1:3000.
Live-image acquisition and quantification
Third instar wandering larvae were dissected in Schneider’s medium (Sigma) with 5 mM ethylene glycol-bis(b-aminoethyl ether)-N,N,N’,N’-tetraacetic acid (EGTA) at room temperature (22°C) in a chamber on a glass slide and then washed and incubated for 20 min with fresh Schneider’s medium with 5 mM EGTA, 20 nM TMRM (Molecular Probes), 1 μM MitoTracker Green FM (Molecular Probes), and Cy5-conjugated-rabbit anti-HRP (Jackson ImmunoResearch Laboratories) at 1:100, and then the solution was replaced with Schneider’s medium for live imaging. Samples were imaged at room temperature with a 63×/N.A.1.30 water Plan-Apochromat objective on a Leica SPE laser scanning confocal microscope (JH Technologies). For mitochondrial motility, mito-GFP was excited by a mercury lamp (HBO100), and images were captured every 2 s using a Leica DFC365 FX charge-coupled device (CCD) camera for SPE II system with an I3 filter LP 515 nm (JH Technologies). Kymographs were generated and analyzed as previously described (Wang and Schwarz, 2009a, b; Wang et al., 2011). FM1-43 loading assay was as described in Verstreken et al. (2008). All images were processed with Adobe Photoshop CS4 or National Institutes of Health ImageJ using only linear adjustments of contrast and brightness.
Immunocytochemistry and confocal microscopy
Third instar wandering larvae were dissected in phosphate-buffered saline (PBS) or 1 × Ca2+ free saline (0.128 M NaCl, 2 mM KCl, 5 mM EGTA, 4 mM MgCl2, 5 mM HEPES, 0.0355 M sucrose), incubated with fixative solution (4% formaldehyde in PBS or Ca2+ free saline) for 20 min, and immunostained with mouse anti-CSP (Developmental Studies Hybridoma Bank) at 1:50, mouse anti-Dlg (Developmental Studies Hybridoma Bank) at 1:250 or mouse anti-Brp (nc82, Developmental Studies Hybridoma Bank) at 1:100, and Alexa 488-conjugated-goat anti-mouse IgG (Jackson ImmunoResearch Laboratories) at 1:500 or Alexa Fluor 647/Cy3-conjugated-rabbit anti-HRP (Jackson ImmunoResearch Laboratories) at 1:50–1:100. Samples were imaged at room temperature (22°C) with a 20 ×/N.A.0.60 (for measuring muscle sizes) or a 63 ×/N.A.1.30 oil Plan-Apochromat objective on a Leica SPE laser scanning confocal microscope (JH Technologies), with identical imaging parameters among different genotypes in a blind manner. Images were processed with Photoshop CS4 using only linear adjustments of contrast and color.
Detection of ATP levels
ATP levels were measured using a luciferase-based bioluminescence assay (ATP Determination Kit; Life Technologies). For each experiment, pupae were homogenized in 100 µl lysis buffer (6 M guinidine-HCl, 100 M Tris, pH 8.0, and 4 mM EDTA). Lysates were boiled for 5 min and cooled down on ice for 5 min. Then lysates were centrifuged at 20,000 × g for 15 min. Supernatant was next diluted to 1:1000 with reaction buffer (provided by the kit) and luciferase was added for 1 min at 25°C. The luminescence was immediately measured using a Glomax Multi Jr. Reader (Promega). Each reading was normalized to protein concentration measured by bicinchoninic acid (BCA) assay (Thermo Scientific).
Behavioral assays
Crawling ability was defined as the distance a larva crawled in 30 s on a 55-mm grape agar plate poured over a 0.2-cm grid paper. Climbing ability was defined as the distance a fly climbed in 5 s. Flying ability was defined as the ability of a fly to fly when it was freed. If the fly was able to perform the action, it was scored as a 1, and if not, then it was scored as a 0; the flying ability was given as a fraction of total flies that scored a 1.
Transmission electron microscopy (TEM)
Modified Trump’s fixative (0.1 M sodium cacodylate buffer, 1% glutaraldehyde, and 4% formaldehyde) was used to fix dissected larvae at room temperature (22°C) for 30 min and then at 4°C overnight. Specimens were washed three times in 0.1 M sodium cacodylate buffer for 10 min to remove fixative, postfixed with 2% osmium tetroxide in 0.1 M sodium cacodylate buffer for 1 h, and then rinsed three times in ddH2O for 10 min. Specimens were next stained en bloc with 1% aqueous uranyl acetate at 4°C overnight, dehydrated in a graded ethanol series, and subsequently set into Embed812 embedding medium. For immunogold labeling, third instar larvae were dissected in Schneider’s medium with 5 mM EGTA and fixed by prefix solution (0.1% glutaraldehyde, 0.1 M sodium cacodylate buffer, 4% formaldehyde, and 2 mM MgCl2) for 1 h at room temperature. The samples were incubated with mouse anti-DNA (1:50; AC-30-10; EMD Millipore) overnight at 4°C, then incubated with anti-mouse IgG 5-nm samples nanogold (1:50; Nanoprobes) for 1 h at room temperature, and washed with ddH2O. The signal was intensified by incubation with Goldenhance EM Reagents (Nanoprobes) in the dark for 7 min. The specimens were then rinsed with 0.1 M sodium cacodylate for 10 min three times. Samples were then postfixed by Trump’s fixative (0.1 M sodium cacodylate buffer, 1% glutaraldehyde, and 4% formaldehyde) for 1 h at 4°C. The specimens were stained en bloc in 1% aqueous uranyl acetate overnight, dehydrated in a graded ethanol series, and subsequently set into Spurr’s embedding medium. This sections (50 nm) were stained with uranyl acetate and lead citrate and imaged with a TEM1400 electron microscope (JEOL) and a 967 slow-scan, cooled CCD camera (Gatan). TEM images were processed with Photoshop CS4.
Statistical analysis
Throughout this paper, the distribution of data points is expressed as mean ± SEM, except as otherwise stated. The Mann–Whitney U test was used for statistical comparisons between two groups, one-way analysis of variance post-hoc Tukey test was performed for comparisons among multiple groups (adjustment applied), and the chi-square test was used for behavior tests due to categorical data. Statistical tests (two-sided) were performed using Statistical Package for the Social Sciences (SPSS).
Supplementary Material
Acknowledgments
We thank Roeland Vanhauwaert and Patrik Verstreken for flies and John Perrino and the Stanford Cell Science Imaging EM Facility (1S10RR026780-01, National Center for Research Resources) for assistance with the EM work. This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS) (X.W., R00 NS067066), the Department of Defense (X.W., PR150380), the Alfred P. Sloan Foundation (X.W.), the Klingenstein Foundation (X.W.), the Shurl and Kay Curci Foundation (X.W.), the Postdoctoral Research Abroad Program of the National Science Council, Taiwan (P.T.), and the Graduate Research Fellowship Program of the National Science Foundation (A.M.P).
Abbreviations used:
- ETC
electron transport chain
- IMM
inner mitochondrial membranes
- MICOS
mitochondrial contact site and cristae organizing system
- OMM
outer mitochondrial membranes
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0177) on September 13, 2017.
REFERENCES
- Cameron JM, Levandovskiy V, Mackay N, Ackerley C, Chitayat D, Raiman J, Halliday WH, Schulze A, Robinson BH. Complex V TMEM70 deficiency results in mitochondrial nucleoid disorganization. Mitochondrion. 2011;11:191–199. doi: 10.1016/j.mito.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Course MM, Wang X. Transporting mitochondria in neurons. F1000Research. 2016;5 doi: 10.12688/f1000research.7864.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286:2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devireddy S, Sung H, Liao PC, Garland-Kuntz E, Hollenbeck PJ. Analysis of mitochondrial traffic in Drosophila. Methods Enzymol. 2014;547:131–150. doi: 10.1016/B978-0-12-801415-8.00008-4. [DOI] [PubMed] [Google Scholar]
- Dickman DK, Horne JA, Meinertzhagen IA, Schwarz TL. A slowed classical pathway rather than kiss-and-run mediates endocytosis at synapses lacking synaptojanin and endophilin. Cell. 2005;123:521–533. doi: 10.1016/j.cell.2005.09.026. [DOI] [PubMed] [Google Scholar]
- Dickman DK, Lu Z, Meinertzhagen IA, Schwarz TL. Altered synaptic development and active zone spacing in endocytosis mutants. Curr Biol. 2006;16:591–598. doi: 10.1016/j.cub.2006.02.058. [DOI] [PubMed] [Google Scholar]
- Ding C, Wu Z, Huang L, Wang Y, Xue J, Chen S, Deng Z, Wang L, Song Z, Chen S. Mitofilin and CHCHD6 physically interact with Sam50 to sustain cristae structure. Sci Rep. 2015;5:16064. doi: 10.1038/srep16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson S, Ruusala A, Aspenstrom P. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006;344:500–510. doi: 10.1016/j.bbrc.2006.03.163. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Mourier A, Yamada J, McCaffery JM, Nunnari J. MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. eLife. 2015;4:07739. doi: 10.7554/eLife.07739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerencser AA, Chinopoulos C, Birket MJ, Jastroch M, Vitelli C, Nicholls DG, Brand MD. Quantitative measurement of mitochondrial membrane potential in cultured cells: calcium-induced de- and hyperpolarization of neuronal mitochondria. J Physiol. 2012;590:2845–2871. doi: 10.1113/jphysiol.2012.228387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godena VK, Brookes-Hocking N, Moller A, Shaw G, Oswald M, Sancho RM, Miller CC, Whitworth AJ, De Vos KJ. Increasing microtubule acetylation rescues axonal transport and locomotor deficits caused by LRRK2 Roc-COR domain mutations. Nat Commun. 2014;5:5245. doi: 10.1038/ncomms6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron. 2005;47:379–393. doi: 10.1016/j.neuron.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Shaltouki A, Gonzalez AE, Bettencourt da Cruz A, Burbulla LF, St Lawrence E, Schule B, Krainc D, Palmer TD, Wang X. Functional impairment in Miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell. 2016;19:709–724. doi: 10.1016/j.stem.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JM, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell. 2005;16:1543–1554. doi: 10.1091/mbc.E04-08-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NC, Tresse E, Kolaitis RM, Molliex A, Thomas RE, Alami NH, Wang B, Joshi A, Smith RB, Ritson GP, et al. VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron. 2013;78:65–80. doi: 10.1016/j.neuron.2013.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner C, Barrera M, Dukanovic J, Eydt K, Harner M, Rabl R, Vogel F, Rapaport D, Neupert W, Reichert AS. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol Biol Cell. 2012;23:2143–2155. doi: 10.1091/mbc.E11-10-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsopoulos OS, Laine D, Osellame L, Chudakov DM, Parton RG, Frazier AE, Ryan MT. Human Miltons associate with mitochondria and induce microtubule-dependent remodeling of mitochondrial networks. Biochim Biophys Acta. 2010;1803:564–574. doi: 10.1016/j.bbamcr.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Liu S, Sawada T, Lee S, Yu W, Silverio G, Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al. Parkinson’s disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genet. 2012;8:e1002537. doi: 10.1371/journal.pgen.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovas JR, Wang X. The meaning of mitochondrial movement to a neuron’s life. Biochim Biophys Acta. 2013;1833:184–194. doi: 10.1016/j.bbamcr.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella CA, Lederer WJ, Jafri MS. The connection between inner membrane topology and mitochondrial function. J Mol Cell Cardiol. 2013;62:51–57. doi: 10.1016/j.yjmcc.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JY, Lee TH, Kim JH, Yoo BH, Bahk YY, Koo HS, Han SS. Caenorhabditis elegans mitofilin homologs control the morphology of mitochondrial cristae and influence reproduction and physiology. J Cell Physiol. 2010;224:748–756. doi: 10.1002/jcp.22177. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Pekkurnaz G, Trinidad JC, Wang X, Kong D, Schwarz TL. Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell. 2014;158:54–68. doi: 10.1016/j.cell.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev-Neumeyer A, Korner C, Jagasia R, Keil T, Baumeister W, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner O, Berdnik D, Levy JM, Wu JS, Luginbuhl D, Gontang AC, Luo L. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Tsai PI, Course MM, Lovas JR, Hsieh CH, Babic M, Zinsmaier KE, Wang X. PINK1-mediated phosphorylation of Miro inhibits synaptic growth and protects dopaminergic neurons in Drosophila. Sci Rep. 2014;4:6962. doi: 10.1038/srep06962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Bohnert M, Wiedemann N, Pfanner N. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 2012;22:185–192. doi: 10.1016/j.tcb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Vanhauwaert R, Kuenen S, Masius R, Bademosi A, Manetsberger J, Schoovaerts N, Bounti L, Gontcharenko S, Swerts J, Vilain S, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017;36:1392–1411. doi: 10.15252/embj.201695773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Koh TW, Schulze KL, Zhai RG, Hiesinger PR, Zhou Y, Mehta SQ, Cao Y, Roos J, Bellen HJ. Synaptojanin is recruited by endophilin to promote synaptic vesicle uncoating. Neuron. 2003;40:733–748. doi: 10.1016/s0896-6273(03)00644-5. [DOI] [PubMed] [Google Scholar]
- Verstreken P, Ohyama T, Bellen HJ. FM 1–43 labeling of synaptic vesicle pools at the Drosophila neuromuscular junction. Methods Mol Biol. 2008;440:349–369. doi: 10.1007/978-1-59745-178-9_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. Imaging axonal transport of mitochondria. Methods Enzymol. 2009a;457:319–333. doi: 10.1016/S0076-6879(09)05018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009b;136:163–174. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ, Schwarz TL. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581:3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- Zerbes RM, van der Klei IJ, Veenhuis M, Pfanner N, van der Laan M, Bohnert M. Mitofilin complexes: conserved organizers of mitochondrial membrane architecture. Biol Chem. 2012;393:1247–1261. doi: 10.1515/hsz-2012-0239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.