Abstract
The only cytogenetic aberration defining a myelodysplastic syndrome subtype is the deletion of the long arm of chromosome 5, which, along with morphological features, leads to the diagnosis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5. These patients show a good prognosis and respond to treatment such as lenalidomide, but some cases progress to acute myeloid leukemia; however, the molecular mutation pattern is rarely characterized. Therefore, we investigated a large cohort of 123 myelodysplastic syndrome patients with isolated deletion of the long arm of chromosome 5, diagnosed following the World Health Organization classifications 2008 and 2016, by sequencing 27 genes. A great proportion of patients showed no or only one mutation. Only seven genes showed mutation frequencies >5% (SF3B1, DNMT3A, TP53, TET2, CSNK1A1, ASXL1, JAK2). However, the pattern of recurrently mutated genes was comparable to other myelodysplastic syndrome subtypes by comparison to a reference cohort, except that of TP53 which was significantly more often mutated in myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5. As expected, SF3B1 was frequently mutated and correlated with ring sider-oblasts, while JAK2 mutations correlated with elevated platelet counts. Surprisingly, SF3B1 mutations led to significantly worse prognosis within cases with isolated deletion of the long arm of chromosome 5, but showed a comparable outcome to other myelodysplastic syndrome subtypes with SF3B1 mutation. However, addressing genetic stability in follow-up cases might suggest different genetic mechanisms for progression to secondary acute myeloid leukemia compared to overall myelodysplastic syndrome patients.
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous group of clonal bone marrow neoplasms characterized by ineffective hematopoiesis, morphologic dysplasia and peripheral cytopenias. While the degree of dysplasia and blast percentage are disease classifying, the specific types of cytopenias have a minor impact on MDS classification. The diagnosis of MDS is driven by the number of dysplastic cell lineages as well as blast counts <1% in the peripheral blood (PB) or <5% in the bone marrow (BM), or <19% in PB or BM for subtypes with excess blasts and at least one cytopenia.1 The risk stratification for MDS patients is categorized according to the Revised International Prognostic Scoring System (IPSS-R), whereby, along with morphological features, cytogenetics also plays a crucial role. Some cytogenetic aberrations can also define an MDS even in the absence of morphologic dysplasia.
However, also according to the new World Health Organization (WHO) 2016 classification, the only cytogenetic aberration defining a MDS subtype is the deletion of the long arm of chromosome 5 (del (5q)), giving the diagnosis of MDS with isolated del(5q). Overall, less than 50% of MDS patients show an aberrant karyotype, of which del(5q) is the most common aberration presenting in 10–20% of MDS patients.2,3 In about 55% of patients with del(5q) this aberration appears as a sole abnormality, in 17% it presents with one additional aberration, while in another 28% del(5q) appears with two or more additional cytogenetic lesions, leading by definition to a complex karyotype.4 This is shifted towards sole del(5q) (82%) when addressing only low- and intermediate-1-risk MDS according to the IPSS.5 However, the WHO classification emphasizes that the main impact of cytogenetics is the prognostic rather than the classifying information. It was demonstrated that MDS with isolated del(5q) shows a good prognosis, while MDS with del(5q) within a complex karyotype shows a poor prognosis.2,4 In recent years the prognostic impact of additional aberrations was addressed, showing that the presence of only one aberration in addition to del(5) (excluding the abnormality −7 or del(7q)) had no adverse effect on prognosis.6
Therefore, in the new WHO classification 2016 a diagnosis of MDS with isolated del(5q) allows the presence of one additional aberration excluding aberrations affecting chromosome 7.1 In addition to the good prognosis of these patients, a sensitivity to specific treatments, such as lenalidomide, was demonstrated.7,8 Morphologically, MDS with isolated del(5q) is defined by blast counts <1% in the PB and <5% in the BM, severe macrocytic anemia and frequent thrombocytosis. Patients who have MDS with isolated del(5q) show a lower risk for progression to acute myeloid leukemia (AML) than those with other MDS. However, about 10% of these patients evolve to secondary (s)-AML.4,9,10 The underlying pathobiological mechanisms are still being debated, while recent studies indicate that TP53 mutation as well as karyotype evolution predict disease progression.11,12
Furthermore, the separation of MDS from reactive causes remains a challenge. In recent years large data sets became available showing that a limited number of genes were mutated in patients with MDS. Eighty to ninety percent of patients show at least one mutation in one of the >100 addressed genes, supporting the clonal hematopoiesis of the disease and with that the diagnosis.13,14 Moreover, it was demonstrated that the increasing number of gene mutations correlates with the disease outcome in MDS patients; the addition of this data improves the existing risk stratifications.13,14
Mutations in TP53 are generally associated with adverse outcome and an aggressive disease course in MDS. Furthermore, in MDS with isolated del(5q), a TP53 mutation seems to predict a poorer response to lenalidomide and a higher risk of transformation to AML.11 Thus, the respective mutation status should be addressed not only at the time of diagnosis, but also before treatment decisions are made, as recommended in the new WHO 2016 classification.1
Therefore, the aim of the study herein was to determine the frequency of mutations in a large cohort of 123 patients with MDS and isolated del(5q) with a 27 gene panel, and to combine clinical data and prognostic information.
Methods
Patients cohort
We investigated 123 patients (35 male, 88 female, ratio: 1:2.5) diagnosed as having MDS with isolated del(5q), strictly classified according to the WHO classification of 2008 and including the added guidelines of 2016 with respect to cytomorphology and cytogenetics (blast counts below 5% in the BM and below 1% in the PB, and isolated 5q deletion or one additional aberration that does not affect chromosome 7).1 All patients’ samples, taken between 2005 and 2013, were sent from different hematological centers to the MLL Munich leukemia laboratory for diagnostic purposes. The median age of the patients was 75 years (range: 35–93 years). One hundred and nineteen patients showed an isolated del(5q) while four cases appeared with only one additional cytogenetic aberration (two patients with del(13q), one with del(9q) and one with a trisomy 8). Follow-up data was available in 111/123 cases with a median follow up of 41 months. One hundred and twelve cases from the cohort presented herein were also included in the study by Heuser et al.15 The mutation patterns and prognostic impact of MDS with del(5q) were compared to a MDS cohort of 944 patients representing all MDS subtypes.13 Of the 123 patients diagnosed with MDS with isolated del(5q), 39 were already included in this recent study, resulting in a reference cohort of 905 patients after excluding these 39 cases.13 This reference cohort contained the following MDS subtypes: 41 refractory anemia (RA), 81 refractory anemia with ringed sideroblasts (RARS), 27 RARS with thrombocytosis (RARS-T), 194 refractory cytopenia with multilineage dysplasia (RCMD), 183 refractory cytopenia with multilineage dysplasia and ringed sideroblasts (RCMD-RS), 191 refractory anemia with excess blasts-1 (RAEB-1), and 188 refractory anemia with excess blasts-2 (RAEB-2), diagnosed according to the WHO 2008 classification guidelines.16 The median age was 73 years (range: 23–91 years), the male to female ratio was 1:7. Follow-up data in the reference cohort was available in 869 cases with a median follow up of 62 months.
All patients gave their consent for genetic analyses and the use of laboratory results for research purposes. The study adhered to the tenets of the Declaration of Helsinki and was approved by the institutional review board of the laboratory.
Cytomorphology
Cytomorphology of BM and/or PB samples was performed in all cases following May-Grünwald Giemsa staining, cytochemistry with myeloperoxidase (MPO), non-specific esterase (NSE) and iron staining (Fe).17
Cytogenetics
Chromosome preparations and banding analysis of BM and/or PB samples were performed for all 123 cases as previously described according to standard methods.18 For classification of abnormalities and karyotypes, the 2016 International System for Human Cytogenetic Nomenclature (ISCN) guidelines were used.19
Next-generation sequencing (NGS)
All patients were analyzed via a myeloid gene panel containing ASXL1, BCOR, BRAF, CSNK1A1, CBL, DNMT3A, ETV6, EZH2, FLT3-TKD, GATA1, GATA2, IDH1, IDH2, JAK2, KIT, NRAS, KRAS, MPL, NPM1, PHF6, RUNX1, SF3B1, SRSF2, TET2, TP53, U2AF1 and WT1. The library of 26 genes was generated with the ThunderStorm (RainDance Technologies, Billerica, MA, USA), and CSNK1A1 with the Access Array System (Fluidigm, San Francisco, CA, USA). Both libraries were sequenced and demultiplexed on a MiSeq instrument (Illumina, San Diego, CA, USA) as described previously.20 The FASTQ files were further processed using the Sequence Pilot software version 4.1.1 Build 510 (JSI Medical Systems, Ettenheim, Germany) for alignment and variant calling. Analysis parameters were set according to the manufacturer’s default recommendation. The validity of the somatic mutations was checked against the publicly accessible Catalogue Of Somatic Mutations In Cancer (COSMIC) v69 database, and functional interpretation was performed using SIFT 1.03, PolyPhen 2.0 and MutationTaster 1.0 algorithms.21 Additionally, TP53 variants were verified using the International Agency for Research on Cancer (IARC) repository.22 Single-nucleotide polymorphisms (SNP) were annotated according to the National Center for Biotechnology Information Single Nucleotide Polymorphism Database (NCBI dbSNP). The detection limit for small nuclear variants was 3% variant allele frequency (VAF). Variants (n=9) not yet described in any public database were excluded from statistical analyses.
Statistical analysis
Dichotomous variables were compared between different groups using the χ2-test and continuous variables by the Student’s t-test; results were considered significant at P<0.05. Adjustment for multiple testing was not done. Statistical analyses were performed using SPSS version 19.0 (IBM Corporation, Armonk, NY, USA); the reported P-values are two-sided. Survival curves were calculated for overall survival (OS) according to Kaplan-Meier and compared using the two-sided log-rank test. OS was considered as being the time from diagnosis to death or last follow up.
Results
Incidence of MDS with isolated del(5q) following the new WHO classification
Following the recently revised 2016 version of the WHO classification for myeloid neoplasms, one additional chromosomal aberration (other than aberration of chromosome 7), in addition to the deletion of the long arm of chromosome 5, allows for the diagnosis of MDS with isolated del(5q).1 Therefore, we analyzed a well characterized MDS cohort for incidences of del(5q) and other diagnostic criteria leading to a diagnosis of MDS with isolated del(5q).13 Of 944 MDS patients, 84 (9%) carried a del(5q) either as a sole abnormality or co-occurring with other lesions. Of 84 cases, 53 (63%) showed a sole del(5q), while only five cases (6%) appeared with one additional aberration and 26 (31%) showed a complex karyotype. Of the 53 patients with isolated del(5q), 44 fulfilled the diagnostic criteria of <1% blasts in the PB and <5% in the BM. Furthermore, 4/5 cases with one additional lesion were also diagnosed as MDS with isolated del(5q), while the fifth case harbored a −7 as an additional aberration. Therefore, 48 cases were diagnosed as MDS with isolated del(5q), whereof only 4/48 cases (8%) were grouped to these patients based on the new WHO classification and its additional criteria, while the main group was diagnosed using the criteria of the WHO 2008 classification, without allowing for additional lesions.1
Molecular genetic characterization of MDS with isolated del(5q)
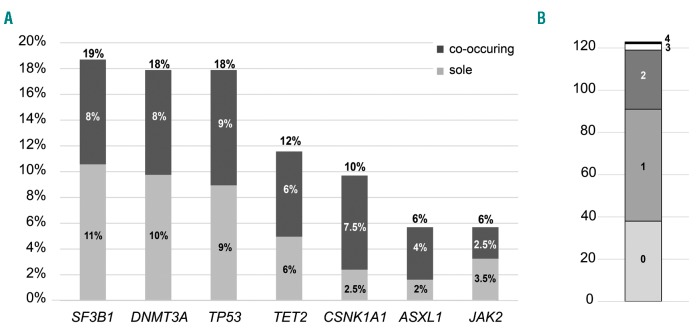
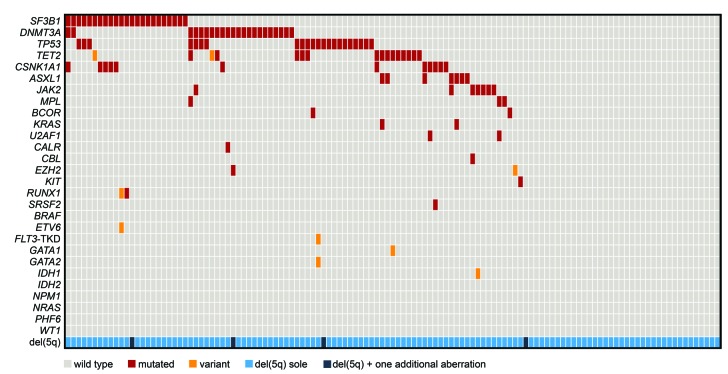
For a comprehensive molecular characterization we assembled a cohort of 123 patients diagnosed with MDS with isolated del(5q) and investigated 27 genes by mutation analyses. The most frequently mutated gene was SF3B1 (23/123; 19%), followed by DNMT3A (22/123; 18%), TP53 (22/123; 18%), TET2 (14/121; 12%), CSNK1A1 (12/123; 10%), ASXL1 and JAK2 (both 7/123; 6%) (Figure 1A). All other analyzed genes showed mutation frequencies below 5%, with BRAF, ETV6, FLT3-TKD, GATA1, GATA2, IDH1, IDH2, NPM1, NRAS, PHF6 and WT1 showing no mutations at all. Nearly all mutations were heterozygous with a median VAF for six of the most frequently mutated genes of 15–30% (range: 2–50% VAF). Solely CSNK1A1 showed VAF of 3–78%, mimicking a homozygous mutation status, caused by the location of CSNK1A1 on chromosome 5q in the commonly deleted region. Addressing the number of mutations per patient revealed a large number of patients with no mutation (38/123; 31%) or one mutation (53/123; 43%) in any of the 27 analyzed genes. Two genes were mutated in 23% (28/123) of patients, while three and four mutations were detected in only 2% (3/123) and 1% (1/123) of patients, respectively, (Figure 1B). Looking at the co-occurrence of gene mutations resulted in a single association of CSNK1A1 and SF3B1 mutations. Only 16% of CSNK1A1 wild-type (wt) cases showed a SF3B1 mutation (18/111), while 42% of CSNK1A1-mutated patients displayed a co-occurring SF3B1 mutation (5/12; P=0.047). Albeit the most frequent gene mutations rarely overlapped and occurred frequently as sole mutations, they were not completely mutually exclusive (Figure 1A and Figure 2).
Figure 1.
Mutation frequencies and distributions. (A) The frequencies of the seven most often mutated genes are given, as well as their appearance as a sole abnormality. (B) Illustration of the distribution of the number of mutations per patient.
Figure 2.
Molecular and cytogenetic characterization of patients with MDS with isolated del(5q). Illustration of all 123 samples, each column represents one patient. All 27 analyzed genes as well as the occurrence of del(5q) as a sole aberration or with one additional cytogenetic aberration are given for each patient. Light gray: wild-type; red: mutated; orange: variant; light blue: del(5q) sole; dark blue: del(5q) and one additional lesion.
Comparison of mutation frequencies in MDS with isolated del(5q) and other MDS subtypes
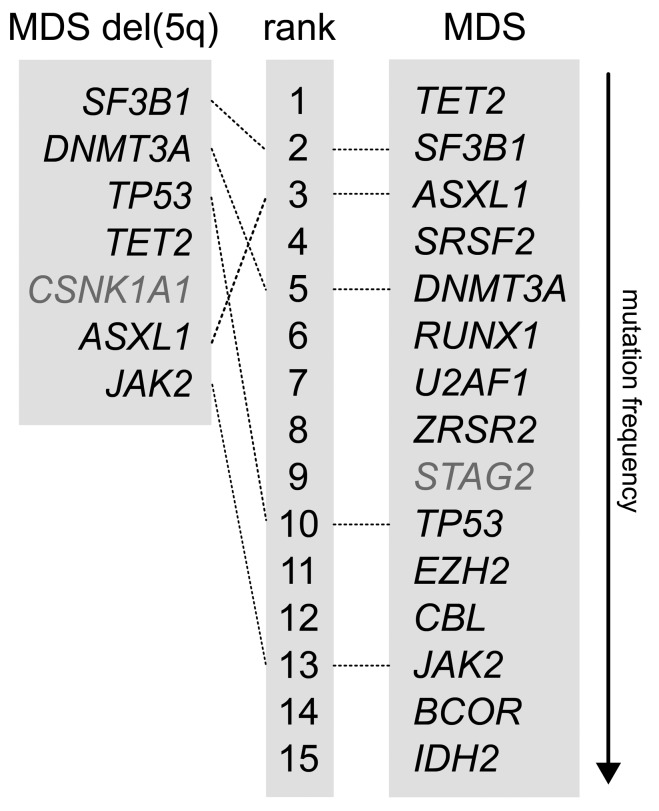
In order to analyze the differences in the mutation patterns of MDS with isolated del(5q) and all other MDS subtypes, we investigated the most frequently mutated genes in comparison to a reference cohort, represented by the previously published MDS cohort after excluding the cases diagnosed as MDS with isolated del(5q).13 Comparing the mutation frequencies of the seven most frequently mutated genes in MDS with isolated del(5q) with all other MDS subtypes revealed that the respective six mutated genes, SF3B1, DNMT3A, TP53, TET2, ASXL1 and JAK2, were also ranked within the 15 most frequently mutated genes in all MDS subtypes (Figure 3; CSNK1A1 was not addressed due to missing data in the MDS reference cohort). Of note, TP53 mutations were found significantly more often in MDS with isolated del(5q) in comparison to all other MDS subtypes, with a mutation frequency of 18% (22/123) compared to 6% in the MDS reference cohort (52/905; P<0.001).
Figure 3.
Comparison of mutation frequencies between MDS with isolated del(5q) and all MDS subtypes. Mutation frequencies for MDS were taken from the previously published reference cohort.13 The six most frequently mutated genes in MDS with isolated del(5q) are compared to the 14 most frequently mutated genes in the MDS reference cohort. CSNK1A1 and STAG2 (typed in gray) were not analyzed in the MDS reference cohort or MDS del(5q) cohort, respectively. MDS: myelodysplastic syndromes; MDS del(5q): MDS with isolated del(5q).
Clinical and morphological correlations
Upon dividing the patients into groups defined by a BM blast count of <2% and 2–5%, as described in the IPSS-R, we could not detect any correlation with the number of mutations per patient. However, patients who had no mutation (n=38) were younger compared to patients with at least one mutation (n=85; 70 vs. 76 years, P=0.018). There was no difference between these two patient groups as regards to sex, white blood cell count, hemoglobin level or platelet count. Taking single gene mutations into account revealed that TP53 mutations correlated with older age (78 vs. 73 years, P=0.038). All 22 patients with a TP53 mutation were ≥67 years of age, with only two patients under 70; 11 patients were between 70 and 79 years of age, eight patients were between 80 and 89 years of age, and one patient in the group was ≥90 years of age. Addressing the correlation between percentages of ring sideroblasts (RS) and SF3B1 mutations also showed that in MDS with isolated del(5q) these two parameters significantly correlated with a mean of 17% RS (range: 0–80%) in SF3B1-mutated patients and only 1% RS in SF3B1 wt patients (range: 0–12%, P=0.004). Furthermore, JAK2 mutations correlated with a platelet count of >450,000/μl (80% in JAK2 mutated vs. 12% in JAK2 wt, P=0.002).
Prognostic impact of gene mutations
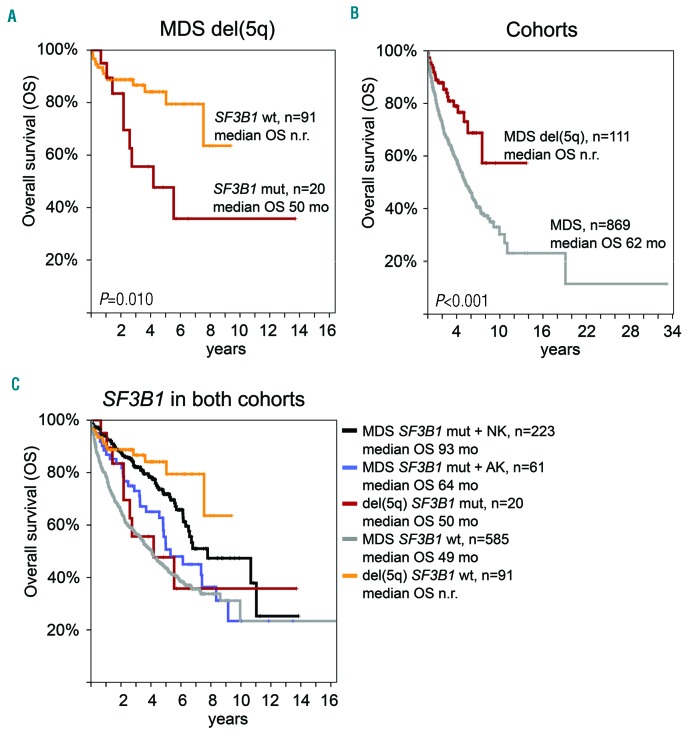
Looking at the prognostic relevance of gene mutations showed, surprisingly, that SF3B1-mutated patients (n=20 with follow-up data) had a significantly inferior outcome than that of SF3B1 wt patients (n=91; median OS: 50 months vs. not reached, P=0.010; Figure 4A). Furthermore, we analyzed the OS compared to the MDS reference cohort representing all other than isolated del(5q) MDS subtypes (n=869 with follow-up data). Overall the median OS was significantly favorable for patients with MDS with isolated del(5q) (62 months vs. not reached, P<0.001; Figure 4B). To better characterize the unexpected worse impact of SF3B1 mutations we analyzed both cohorts with respect to their SF3B1 mutation status as well as cytogenetics in terms of normal or aberrant karyotypes (Figure 4C). SF3B1 mutations within a normal karyotype showed favorable outcomes in MDS patients (median OS: 93 months), however, this effect was reduced by an additional aberrant karyotype (median OS: 64 months). Therefore, SF3B1-mutated patients possessing aberrant karyotypes showed comparable outcomes (median OS: 50 vs. 64 months), irrespective of diagnoses of MDS with isolated del(5q) or any other subtype of MDS, while SF3B1 wt patients with isolated del(5q) had a favorable outcome compared to all other SF3B1 wt MDS patients (median OS: not reached vs. 49 months). However, we did not find a prognostic impact of TP53 mutations in our cohort (Online Supplementary Figure S1; median OS was not reached in TP53 wt (n=92) and TP53 mutated (n=19) patients; P=0.094).
Figure 4.
Overall survival (OS) analyses. Case numbers, median OS and P-values are given. (A) Prognostic impact of SF3B1 mutations in MDS with isolated del(5q). (B) OS of MDS with isolated del(5q) in comparison to the MDS reference cohort. (C) OS of SF3B1 mutation with additional cytogenetic aberrations in MDS with isolated del(5q) in comparison to the MDS reference cohort. MDS: myelodysplastic syndromes; MDS del(5q): MDS with isolated del(5q); NK: normal karyotype; AK: aberrant karyotype; wt: wild-type; n.r.: not reached; mo: months; mut: mutated.
Molecular genetics during follow up and progression to AML
Follow-up samples were available in 13 patients with a median time between two investigations of 22 months (range: 5–47 months, cases with less than four months between two investigations were excluded). In a further 6/123 (5%) cases we could follow a progression to AML with a median transformation time to s-AML of 24 months (range: 10–53 months).
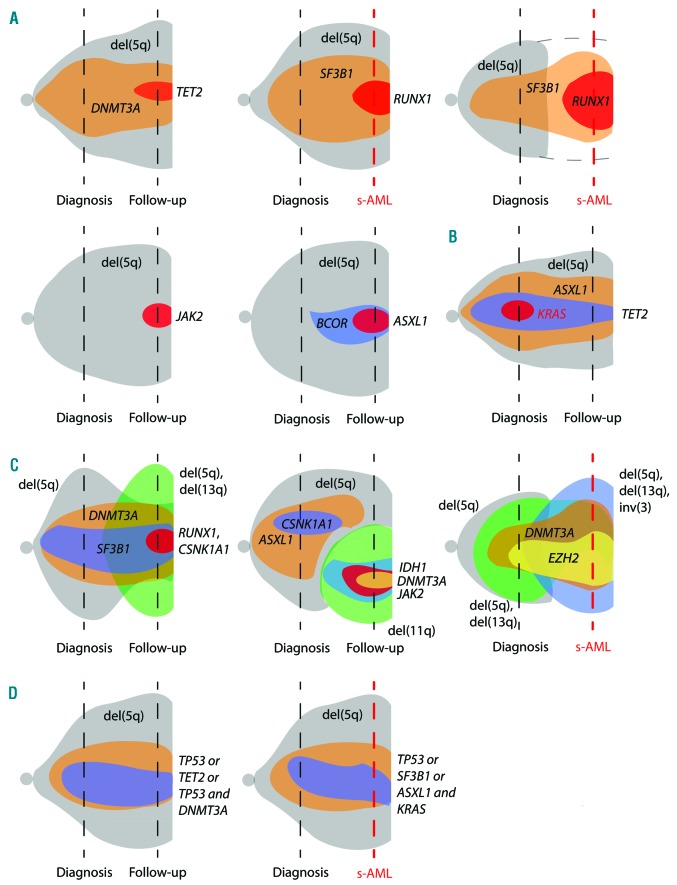
In these 19 cases with a follow-up sample mutation analyses were performed, in 18/19 cases cytogenetics were also available. Overall, in 5/13 (38%) patients showing no transformation to AML no clonal evolution was detectable, neither in karyotype nor in the mutation pattern, leaving 8/13 (62%) patients with clonal evolution. In 4/13 cases (31%) a change in cytogenetics occurred and in 7/13 patients (54%) additional gene mutations appeared; in 3/13 patients clonal evolution occurred in both instances. Studying patients progressing to AML after MDS revealed a similar pattern regarding stability in genetics with 3/5 cases (60%) showing no genetic evolution, 1/5 (20%) a clonal evolution in cytogenetics and 2/6 (33%) exhibiting a change in the mutation pattern. Of note, none of the patients gained a mutation in TP53 or FLT3 during progression to AML. Addressing the gains of genetic lesions in the disease course showed no specific pattern. Cytogenetic gains included –Y, del(11q), del(13q) and inversion 3 (inv(3)), and gained mutations affected ASXL1, BCOR, CSNK1A1, DNMT3A, IDH1, JAK2, RUNX1 and TET2. However, recurrent gains were limited to JAK2 (n=2) and RUNX1 (n=3), with two cases showing a gain of RUNX1 mutation progressing to AML and one progressing to MDS RAEB-1. Furthermore, the VAF of all gained mutations were <10% (median 4%, range: 3–7%), except for those of RUNX1 during transformation to AML, which showed VAF of 14% and 25%, respectively. An illustration of the different types of clonal evolution is given in Figure 5.
Figure 5.
The dynamic of variant allele frequency (VAF) and acquisition and loss of mutated genes as well as cytogenetic lesions at diagnosis and follow up. VAF and clone size are indicated by the height of the shapes. Cases evolving to s-AML are indicated by the red dashed time line, while follow up without progression to s-AML is indicated by a black dashed time line. Disease course showing (A) gain of gene mutations, (B) loss of a gene mutation, (C) cytogenetic evolution, and (D) no significant change in del(5q) clone size as well as gene mutation (VAF). In (D) both illustrations represent three patients. For the third patient in the first row no cytogenetics was available at s-AML stage. s-AML: secondary acute myeloid leukemia; del(5q): deltion of the long arm of chromosome 5.
Discussion
In the study herein, we investigated the molecular mutation pattern of a large cohort of 123 MDS patients with isolated del(5q) classified according to the WHO classification of 2008 and including the added guidelines of 2016. A large proportion of patients showed no or only one mutation in 27 analyzed genes, with only seven genes showing mutation frequencies >5% (SF3B1, DNMT3A, TP53, TET2, CSNK1A1, ASXL1, JAK2). Albeit in other studies more genes were investigated for mutation analyses, in overall MDS the median number of mutations is higher with three mutations per patient13 or nearly half of the patients showing two or three mutations14 in contrast to MDS with isolated del(5q). This indicates the specific and narrow spectrum of genetic lesions in this specific MDS subtype. However, the most frequently mutated genes were comparable to all other MDS subtypes investigated in a large MDS reference cohort.13 The 15 most frequently mutated genes have also been identified by another large MDS study and by meta-analysis showing consistent data for the MDS mutation landscape.14,23 Furthermore, SF3B1 was frequently mutated and correlated with the presence of RS, which is in line with previously published data on other MDS entities.24–26
In contrast, TP53 was mutated significantly more often in MDS with isolated del(5q). In a number of studies mutations of TP53 were shown to occur in high-risk or therapy-related MDS, MDS-derived leukemia or within a complex karyotype.27–29 Incidences of TP53 mutations in MDS with isolated del(5q) were found to be 5%, much lower than in other MDS subtypes.12 However, in another previous study the incidence of TP53 mutations, investigated by back tracking TP53-mutated samples in low-risk MDS with del(5q), was 18%, suggesting a previous underestimation of TP53 subclonal mutations.11 It was shown that the TP53 mutation was already present in the early stages of the disease and increased during the course of the disease.11 This is in line with the present data showing a TP53 mutation incidence of 18%, with 7/22 cases having VAF of <10%.
Surprisingly, SF3B1 mutations led to significantly worse OS, but ultimately showed a comparable outcome, still favorable, to all other MDS subtypes with a SF3B1 mutation and an aberrant karyotype. Since a high frequency of MDS patients show a normal karyotype, the need to analyze the impact of karyotype information on SF3B1 mutations in overall MDS in our reference cohort seemed the next logical step. This demonstrated that an accompanying altered karyotype also reduces the favorable impact of SF3B1 mutations. Therefore, the highly favorable impact of SF3B1 mutation was limited to sole SF3B1 mutations without any additional lesions, and might be an explanation as to why the favorable impact is reduced and the overall good prognosis is not driven by SF3B1 mutations alone in MDS with isolated del(5q). However, this finding is in contrast to previous studies which demonstrate that the prognostic effect of SF3B1 mutations is independent of variables that could coexist, such as age, sex and cytogenetics.24,26 In other studies, however, SF3B1 mutations did not keep independent prognostic significance in multivariate analyses or showed no prognostic impact whatsoever, indicating that perhaps MDS represents a heterogeneous disease and as such should be analyzed in respective subgroups.30–33
In MDS it was evidenced that transformation to s-AML is frequently accompanied by mutations in the FLT3 and RAS genes and karyotype evolution.34–38 In addition, RUNX1, GATA2 and CEBPA were identified by progression to s-AML, abrogating normal differentiation.39 Furthermore, some genes have been identified as being affected by mutations occurring as late events in MDS patients, thus giving rise to a potential progression to AML, such as mutations in ASXL1, RUNX1, SRSF2, IDH2 or NRAS.34,35,38 However, most of these genes were not identified to be affected in MDS with isolated del(5q), arguing for an alternative progression mechanism to that found in all other MDS patients. Nevertheless, the recurrent gain of RUNX1 mutations in the disease course might indicate transformation to AML. In a comprehensive study of the mutation patterns in AML, it became obvious that some genes were specific for s-AML, covering the spliceosomal genes as well as chromatin modifiers.38,39 The high incidence of SF3B1 mutations in MDS with isolated del(5q) would therefore reflect the MDS origin of the disease rather than a high SF3B1 mutation frequency indicating disease progression. In previous studies, mutations in TP53 and gain of cytogenetic aberrations were speculated to indicate disease progression and transformation to AML.11,12 However, the clonal evolution was comparable in MDS with isolated del(5q) to that described for overall MDS, with a karyotype evolution in 34% and mutation pattern in 53% of MDS cases transforming to AML.34 Moreover, the clonal evolution appeared in the same manner in cases without progression to AML. Additionally, TP53 mutations were never gained during the disease course, but the mutation burden increased (from 11% to 75%) in the one case which transformed to AML, while in follow-up cases without progression (n=2) the burden did not change. Therefore, one might suppose that in patients with clonal evolution clinical progress will be encountered, while stable patients will also remain clinically stable. It was shown that clinical stability accompanies mutational stability, while developing new mutations resulted in AML progression in patients with MDS with isolated del(5q).40 The previously published worse impact prognosis of TP53 mutations was not observed in the present cohort. Our patients were unselected and the median follow up was 41 months. Thus, our cohort may include a larger proportion of patients earlier in their clinical course compared to cohorts enrolled in treatment studies. Therefore, the negative impact of TP53 mutations may become obvious in later stages of the disease course or even subsequent to the need for starting treatment, a phenomenon that is not represented by the collection of patients presented herein. Of note, 60% (61/101) of our patients were under observation alone, or only received red blood cells or erythropoietin. However, the presence, in advance, of TP53 mutations in the early stages of the disease would therefore be in line with our data, supported by the observed increase of clone size rather than the gain of TP53 mutation during the disease course.11 However, this needs to be further proven in larger cohorts as we had a small number of cases for follow up.
In summary, MDS with isolated del(5q) shows a very limited mutation spectrum of the recurrently mutated genes in comparison to all other MDS subtypes. However, the seven frequently mutated genes resemble the MDS mutation landscape, with the exception of TP53 mutations, which are more often found in MDS with isolated del(5q). Therefore, progression to s-AML is driven by the worse prognostic impact of TP53 mutation in combination with del(5q) and potentially the gain of RUNX1 mutations, rather than the otherwise identified accompanying mutations during MDS progression. The diagnostic and prognostic approach of MDS with isolated del(5q) should therefore include the mutation status of the seven most frequently mutated genes, including SF3B1, and TP53 and RUNX1 as potential progression markers, investigated with deep sequencing to catch all appearing mutations including those of a subclonal nature with low VAF.
Supplementary Material
Acknowledgments
We thank all clinicians for sending samples to our laboratory for diagnostic purposes and for providing clinical information and follow-up data. In addition, we would like to thank all our coworkers at the MLL Munich Leukemia Laboratory for jointly approaching many aspects in the field of leukemia diagnostics and research. We also thank Karolína Perglerová for analyzing FLT3-ITD in the follow-up samples at MLL2 s.r.o., Prague, Czech Republic.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/9/1502
References
- 1.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 2.Haase D, Germing U, Schanz J, et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood. 2007;110(13):4385–4395. [DOI] [PubMed] [Google Scholar]
- 3.Bernasconi P, Klersy C, Boni M, et al. World Health Organization classification in combination with cytogenetic markers improves the prognostic stratification of patients with de novo primary myelodysplastic syndromes. Br J Haematol. 2007;137(3):193–205. [DOI] [PubMed] [Google Scholar]
- 4.Mallo M, Cervera J, Schanz J, et al. Impact of adjunct cytogenetic abnormalities for prognostic stratification in patients with myelodysplastic syndrome and deletion 5q. Leukemia. 2011;25(1):110–120. [DOI] [PubMed] [Google Scholar]
- 5.Germing U, Lauseker M, Hildebrandt B, et al. Survival, prognostic factors and rates of leukemic transformation in 381 untreated patients with MDS and del(5q): A multi-center study. Leukemia. 2012;26(6):1286–1292. [DOI] [PubMed] [Google Scholar]
- 6.Schanz J, Tuchler H, Sole F, et al. New Comprehensive cytogenetic scoring system for primary myelodysplastic syndromes (MDS) and oligoblastic acute myeloid leukemia after MDS derived from an international database merge. J Clin Oncol. 2012;30(8):820–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallo M, del Rey M, Ibanez M, et al. Response to lenalidomide in myelodysplastic syndromes with del(5q): influence of cytogenetics and mutations. Br J Haematol. 2013;162(1):74–86. [DOI] [PubMed] [Google Scholar]
- 8.List AF, Dewald GW, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456–1465. [DOI] [PubMed] [Google Scholar]
- 9.Giagounidis AA, Germing U, Haase S, et al. Clinical, morphological, cytogenetic, and prognostic features of patients with myelodysplastic syndromes and del(5q) including band q31. Leukemia. 2004;18(1):113–119. [DOI] [PubMed] [Google Scholar]
- 10.Side LE, Curtiss NP, Teel K, et al. RAS, FLT3, and TP53 mutations in therapy-related myeloid malignancies with abnormalities of chromosomes 5 and 7. Genes Chromosomes Cancer. 2004;39(3):217–223. [DOI] [PubMed] [Google Scholar]
- 11.Jadersten M, Saft L, Smith A, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29(15):1971–1979. [DOI] [PubMed] [Google Scholar]
- 12.Fernandez-Mercado M, Burns A, Pellagatti A, et al. Targeted re-sequencing analysis of 25 genes commonly mutated in myeloid disorders in del(5q) myelodysplastic syndromes. Haematologica. 2013;98(12):1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heuser M, Meggendorfer M, Cruz MM, et al. Frequency and prognostic impact of casein kinase 1A1 mutations in MDS patients with deletion of chromosome 5q. Leukemia. 2015;29(9):1942–1945. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues, 4th ed. Lyon: International Agency for Research on Cancer (IARC), 2008. [Google Scholar]
- 17.Löffler H, Rastetter J, Haferlach T. Atlas of clinical hematology, 6th ed. Heidelberg: Springer, 2010. [Google Scholar]
- 18.Schoch C, Schnittger S, Bursch S, et al. Comparison of chromosome banding analysis, interphase- and hypermetaphase-FISH, qualitative and quantitative PCR for diagnosis and for follow-up in chronic myeloid leukemia: a study on 350 cases. Leukemia. 2002;16(1):53–59. [DOI] [PubMed] [Google Scholar]
- 19.McGowan-Jordan J, Simons A, Schmid M. ISCN 2016: An International System for Human Cytogenomic Nomenclature. Basel, New York: Karger, 2016. [Google Scholar]
- 20.Delic S, Rose D, Kern W, et al. Application of an NGS-based 28-gene panel in myeloproliferative neoplasms reveals distinct mutation patterns in essential thrombocythaemia, primary myelofibrosis and polycythaemia vera. Br J Haematol. 2016;175(3):419–426. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–576. [DOI] [PubMed] [Google Scholar]
- 22.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28(6):622–629. [DOI] [PubMed] [Google Scholar]
- 23.Rose D, Kohlmann A, Nagata Y, et al. A robust molecular pattern for myelodysplastic syndromes in two independent cohorts investigated by next-generation sequencing can be revealed by comparative bioinformatic analyses. Br J Haematol. 2014;167(2): 278–281. [DOI] [PubMed] [Google Scholar]
- 24.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. [DOI] [PubMed] [Google Scholar]
- 26.Malcovati L, Papaemmanuil E, Bowen DT, et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118(24):6239–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haferlach C, Dicker F, Herholz H, et al. Mutations of the TP53 gene in acute myeloid leukemia are strongly associated with a complex aberrant karyotype. Leukemia. 2008:1539–1541. [DOI] [PubMed] [Google Scholar]
- 28.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patnaik MM, Lasho TL, Hodnefield JM, et al. SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119(2):569–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Damm F, Thol F, Kosmider O, et al. SF3B1 mutations in myelodysplastic syndromes: clinical associations and prognostic implications. Leukemia. 2012;26(5):1137–1140. [DOI] [PubMed] [Google Scholar]
- 32.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. [DOI] [PubMed] [Google Scholar]
- 33.Cazzola M, Rossi M, Malcovati L. Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood. 2013;121(2):260–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meggendorfer M, de AA, Nadarajah N, et al. Karyotype evolution and acquisition of FLT3 or RAS pathway alterations drive progression of myelodysplastic syndrome to acute myeloid leukemia. Haematologica. 2015;100(12):e487–e490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi K, Jabbour E, Wang X, et al. Dynamic acquisition of FLT3 or RAS alterations drive a subset of patients with lower risk MDS to secondary AML. Leukemia. 2013;27(10):2081–2083. [DOI] [PubMed] [Google Scholar]
- 37.Murphy DM, Bejar R, Stevenson K, et al. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013;27(10):2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17(1):5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsley RC, Mar BG, Mazzola E, et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125(9):1367–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woll PS, Kjallquist U, Chowdhury O, et al. Myelodysplastic syndromes are propagated by rare and distinct human cancer stem cells in vivo. Cancer Cell. 2014;25(6):794–808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.