Summary
Glaucoma-associated myocilin is a member of the olfactomedins, a protein family involved in neuronal development and human diseases. Molecular studies of the myocilin N-terminal coiled-coils demonstrate a unique tripartite architecture: a Y-shaped parallel dimer-of-dimers with distinct tetramer and dimer regions. The structure of the C-terminal 7-heptad repeat dimer elucidates an unexpected repeat pattern involving inter-strand stabilization by oppositely charged residues. Molecular dynamics simulations reveal an alternate accessible conformation in which the terminal inter-strand disulfide limits the extent of unfolding and results in a kinked configuration. By inference, full-length myocilin is also branched, with two pairs of C-terminal olfactomedin domains. Selected variants within the N-terminal region alter the apparent quaternary structure of myocilin but do so without compromising stability or causing aggregation. In addition to increasing our structural knowledge of naturally-occurring extracellular coiled-coils and biomedically-important olfactomedins, this work broadens the scope of protein misfolding in the pathogenesis of myocilin-associated glaucoma.
eTOC blurb
Hill et al. describe the structure of the coiled-coil region of myocilin, the extracellular olfactomedin family member closely associated with the ocular disorder glaucoma. Myocilin’s coiled-coil adopts a unique Y-shaped parallel dimer-of-dimers employing an unusual heptad repeat pattern. Selected disease variants only alter quaternary structure.
Introduction
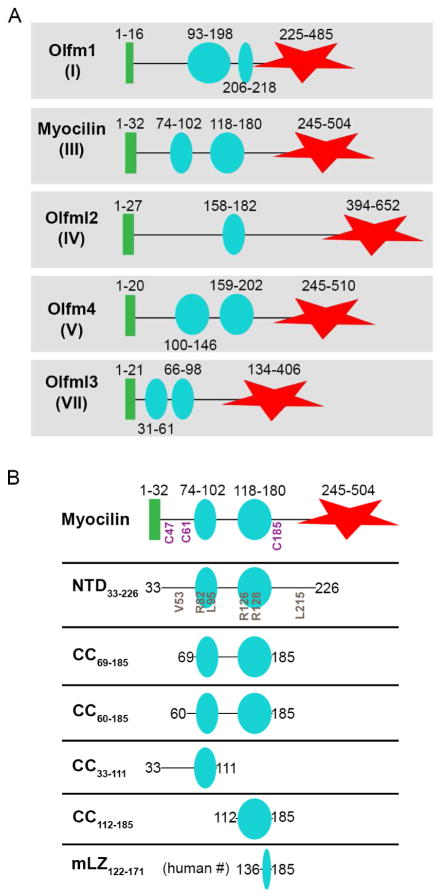
Olfactomedins (PFAM: PF02191), a family of extracellular, modular, multidomain proteins each with a 5-bladed β-propeller (Donegan et al., 2015) olfactomedin (OLF) domain, are broadly involved in eukaryotic development (Tomarev and Nakaya, 2009; Zeng et al., 2005). The majority of olfactomedin subfamilies also contain a high probability signal sequence for cellular secretion followed by homo-oligomerizing coiled-coil (CC) domains of varying lengths (Figure 1A). Despite their prevalence in multicellular organisms and implication in a variety and increasing number of human diseases (Anholt, 2014), the molecular architecture and specific functions of olfactomedins remain largely unknown.
Figure 1. Schematic of olfactomedin family domain organization and constructs used in this study.
(A) Distinctive domain organization among CC-containing olfactomedin subfamilies. Not shown: subfamily II (membrane-anchored); subfamily VI (collagen domain). (B) Schematic of myocilin constructs used in this study. Positions of key disease-associated mutations and cysteines are indicated. Green bar, predicted signal sequence for cellular secretion; cyan circles, predicted CC regions; red star, olfactomedin domain. See also Figure S1.
The structures of CCs, one of the most common structural domains in nature (Wolf et al., 1997) and of considerable interest for protein design and biomaterials (Lupas and Bassler, 2017; Woolfson, 2005), can be challenging to predict in silico, particularly beyond dimers that are seen in extracellular CCs (Kammerer, 1997). Common among CCs is a regular heptad repeat pattern (abcdefg)n period where hydrophobic residues are usually found in positions ‘a’ and ‘d’ and electrostatic residues at positions ‘e’ and ‘g’ (Mason and Arndt, 2004). CCs encoded within olfactomedins diverge somewhat from this general repeat pattern. Thus, bioinformatics programs detect CCs in olfactomedins, but vary in predictions for register, stoichiometry, and orientation. Olfactomedin-1 (subfamily 1) is the only CC-containing olfactomedin for which a supramolecular organization has been structurally characterized: an apparent V-shaped molecule seen by transmission electron microscopic imaging and small angle X-ray scattering (SAXS) envelope of the full-length protein (Pronker et al., 2015). However, because olfactomedin family CC domains share very low sequence similarity (< 15% overall, ~22% pairwise identity, Figure S1), results from olfactomedin-1 are not predictive of other olfactomedin family members.
Here we present the unique architecture of full-length myocilin (olfactomedin subfamily 3), one of the best-studied olfactomedins, by characterizing molecular details of the myocilin N-terminal region. Myocilin is expressed at high levels in the trabecular meshwork (TM), an extracellular matrix in the anterior segment of the eye that forms part of the anatomical ultrafiltration system for aqueous humor (Abu-Hassan et al., 2014), and is a critical tissue for maintaining intraocular pressure (IOP) (Acott et al., 2014). The myocilin CC-containing region has been reported to interact with matrix components (Filla et al., 2002; Ueda et al., 2002; Wentz-Hunter et al., 2004), associate with membranes (Stamer et al., 2006), and confer its cell adhesion properties (Goldwich et al., 2009; Wentz-Hunter et al., 2004), but there is still much to learn about its structure and biological function. Myocilin is also associated with glaucoma (Polansky et al., 1997; Rozsa et al., 1998; Stone et al., 1997), a leading cause of blindness worldwide typically associated with elevated IOP (Quigley and Broman, 2006). Mutations in myocilin are causative for Mendelian-inherited, familial open angle glaucoma (~4% of ~70 million patients), the most common subtype (Hewitt et al., 2006). OLF-resident myocilin variants are destabilized (Burns et al., 2010; Burns et al., 2011) and prone to amyloid-like aggregation (Hill et al., 2014; Orwig et al., 2012), which is believed to cause a toxic gain of function, leading to early-onset glaucoma (Stothert et al., 2016).
We present biophysical and biochemical experiments demonstrating that the myocilin CC domain is a distinct Y-shaped biopolymer composed of two parallel dimer-of-dimers that are stabilized on the three ends by disulfide bonds. The 1.9 Å resolution X-ray structure of the C-terminal 7 heptad repeats of the CC further reveals an unexpected repeat pattern involving inter-strand stabilization between oppositely charged residues at heptad positions ‘a’ and ‘g’. Molecular dynamics (MD) simulations demonstrate that the C-terminal capping disulfide bond limits unfolding of the CC and an alternative kinked structure is predicted to form. Selected glaucoma-associated mutations within the N-terminal CC region adopt non-native structures but are not destabilized or aggregation prone. Our study increases structural knowledge of extracellular CC, biomedically important olfactomedins, and broadens our appreciation of the extent of protein misfolding in the pathogenesis of myocilin-associated glaucoma.
Results
The myocilin CC imparts a unique Y-shaped supramolecular architecture
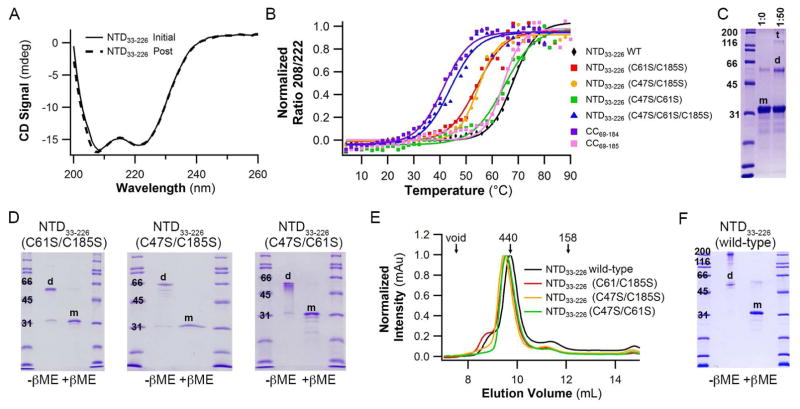
After unsuccessful attempts to express soluble full-length myocilin in E. coli or purify sufficient quantities from spent media of primary human TM cells (not shown), we generated well-behaved constructs of myocilin (Figure 1B, described in detail in STAR Methods) beyond the 30 kDa OLF domain whose structure we solved previously (Donegan et al., 2015). Biophysical characterization of N-terminal domain (NTD) construct NTD33-226, which includes three Cys residues and linker regions before and after the predicted CC domain (Figure 1B), reveals the typical circular dichroism (CD) signatures for α-helices. Thermal unfolding is reversible (Figure 2A) and the melting temperature ((Tm) ~ 67 °C, Figure 2B and Table 1) is considerably higher than the ~52 °C measured for the OLF domain (Orwig and Lieberman, 2011).
Figure 2. Biophysical characterization of NTD33-226.
(A) Initial NTD33-226 CD spectrum reveals α-helical signature. Overlay with post-melt NTD33-226 spectrum confirms thermal unfolding is reversible. (B) CD melt curves for selected CC constructs. See also Table 1. (C) Chemical crosslinking of NTD33-226 traps both dimer and tetramer species. (D) SDS-PAGE analysis of NTD33-226 Cys-to-Ser variants under non-reducing conditions show predominantly dimer (d) species. (E) SEC elution profiles demonstrate unchanged quaternary structure for wild type and Cys-to-Ser NTD33-226 variants. (F) SDS-PAGE analysis of wild-type NTD33-226 under non-reducing conditions show dimer and high molecular weight species consistent with octamer. Calculated molecular weight for the NTD33-226 monomer (m) = 27 kDa; d=dimer, t=tetramer. See also Figure S3.
Table 1.
Tm values for constructs in this study.
| Construct | Tm (°C) | Rationale |
|---|---|---|
| NTD33-226 | 66.7 ± 1.8 | All except OLF domain |
| NTD33-226 (C61S/C185S) | 56.8 ± 1.0 | Cys47 only |
| NTD33-226 (C47S/C185S) | 56.3 ± 0.9 | Cys61 only |
| NTD33-226 (C47S/C61S) | 64.4 ± 1.1 | Cys185 only |
| NTD33-226 (C47S/C61S/C185S) | 44.2 ± 2.5 | Cys-free construct |
| CC69-184 | 44.2 ± 0.2 | |
| CC69-185 | 64.5 ± 0.6 | +Cys185 |
| NTD33-226 (V53A) | 67.0 ± 0.4 | GCM*, within linker |
| NTD33-226 (R82C) | 68.0 ± 0.4 | GCM, within tetramer |
| NTD33-226 (L95P) | 66.1 ± 0.8 | Neutral, within tetramer |
| NTD33-226 R126W) | 66.2 ± 0.4 | GCM, within dimer |
| NTD33-226 (R128W) | 68.1 ± 0.7 | Neutral, within dimer |
| NTD33-226 (L215Q) | 67.3 ± 0.5 | GCM, within linker |
GCM (glaucoma causing mutation) or Neutral as annotated in the database myocilin.com
Chemical crosslinking of NTD33-226 indicates an oligomeric state of tetramer and/or dimer (Figure 2C). To investigate individual contributions of disulfide bonds to the observed oligomeric state, we visualized single Cys-containing variants of NTD33-226 by gel electrophoresis under non-reducing conditions. In all cases, the predominant species present upon disulfide bond formation of a single Cys is a dimer (Figure 2D). The Cys residues thermally stabilize NTD33-226 nearly 20 °C, with Cys185 contributing the biggest thermal increase (Figure 2B and Table 1). These mutations do not alter the elution profiles from size exclusion chromatography (SEC) and thus the apparent quaternary structure in solution (Figure 2E). Surprisingly, non-reducing SDS-PAGE analysis of wild-type (WT) NTD33-226, which contains all three Cys residues, reveals a dimer plus a higher ordered species consistent with an octamer (Figure 2F), which has been seen by others (Dismuke et al., 2012; Fautsch and Johnson, 2001; Fautsch et al., 2004; Russell et al., 2001).
To clarify the oligomeric state and shape of the structural CC of myocilin, we next prepared a CC construct bracketed by the two Cys residues closest to the predicted CC (CC60-185, 14.6 kDa calculated mass of monomer). The SEC elution profile (Figure S2A) contains two species, one in the void volume and another with a molecular mass of ~70 kDa, a size consistent with a 4- or 5-mer. The molecular weight determined by SEC-SAXS analysis of the ~70 kDa species (Figures S2B-S2D and Table 2) corroborates this stoichiometry, and the molecular envelope reveals a branched, tripartite structure that resembles the letter ‘Y’ (Figure 3A and Table 2). There is a short stem (3–4 nm wide × 4 nm long) and two arms (each 3–4 nm wide and 9–11 nm long) ~130° apart. The molecule appears nearly symmetric, but apparent symmetry is broken due to apparent flexibility of the arms (Figure 3A).
Table 2.
SEC-SAXS statistics.
| Construct | Rg (Å) | Monomer MW | SAXS MoW (Oligomeric Species) | Chi2 | NSD |
|---|---|---|---|---|---|
| CC60-185 | 60.2 | 14.6 kDa | 72 kDa (4.9-mer) | 0.9692 | 0.900 |
| CC69-185 | 56.5 | 13.7 kDa | 49 kDa (3.6-mer) | 1.104 | 0.695 |
| 1.181 (P2) | 1.532 (P2) | ||||
| CC33-111 | 29.4 | 8.8 kDa | 25 kDa (2.8-mer) | 1.006 | 0.606 |
| CC112-185 | 32.9 | 8.9 kDa | 21 kDa (2.4-mer) | 1.036 | 0.526 |
| 1.118 (P2) | 1.801 (P2) | ||||
| mLZ93-171 | 32.2 | 9.1 kDa | 20 kDa (2.2-mer) | 1.072 | 0.586 |
| 1.078 (P2) | 1.268 (P2) |
Chi2 and normalized spatial discrepancy (NSD) values are from ab initio modeling. P2 indicates values obtained from ab initio modeling when constrained to P2 symmetry. Rg, radius of gyration; MoW= molecular weight calculated from SAXS.
Figure 3. Ab initio models from SEC-SAXS reveal tripartite structure for myocilin CC.
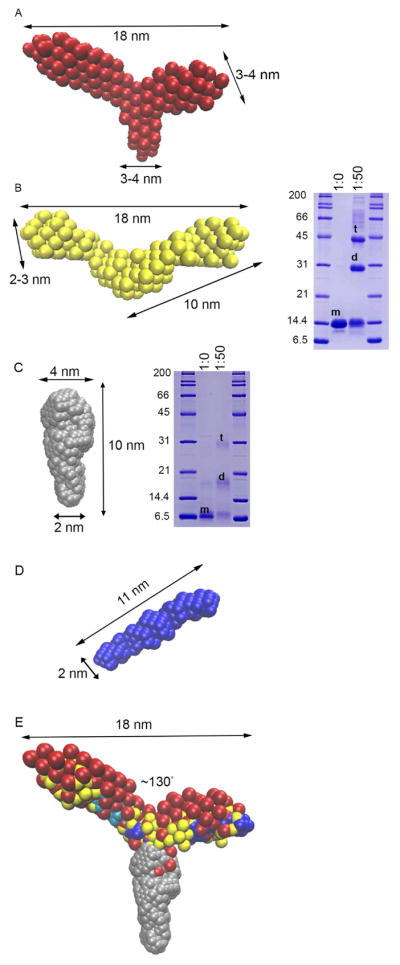
(A) CC60-185 is Y-shaped with arms spanning 18 nm and 130° apart from a stem ~3–4 nm in width. (B) Left: CC69-185 is a P2-symmetric V-shaped molecule with similar dimensions to arms of CC60-185. Right: Chemical crosslinking of CC69-185 reveals both dimer and tetramer species. m=monomer, d=dimer, t=tetramer. (C) Left: CC33-111 is an oblong molecule with varying widths of 2–4 nm. Right: Chemical crosslinking of CC33-111 indicates dimer and tetramer species with labels as in (B). (D) CC112-185 is a P2-symmetric, ~11 nm long rod. (E) Overlay of SAXS molecular envelopes from A–D. Molecular envelopes are overlaid on the same scale. Two orientations for CC112-185 are shown in dark and light blue. See also Figures S2, S4, and Table 2.
Identification of parallel tetramer and dimer CC subdomains within Y-shaped molecule
To probe the molecular principles governing the adoption of the Y-shaped structure, we next tested the hypothesis that the shape apparent from the Y-shaped molecule of CC60-185 is composed of distinct tetramer and dimer regions corresponding to the two CC sub-regions predicted by Marcoil (Delorenzi and Speed, 2002) and Logicoil (Vincent et al., 2013), residues ~74-102 and ~118-180 (Figure 1). The former stretch escapes detection by some CC prediction programs but even when predicted, assignment of its oligomeric state is of low confidence (not shown). The latter stretch is readily predicted as it has regularly spaced Leu residues characteristic of the leucine zipper CC subtype.
The Cys-free CC69-184 exhibits moderate stability (Figure 2B and Table 1), high helical content (Figure S3A), and elutes from SEC at a volume consistent with a molecular mass of a tetramer (Figure S3B). The addition of Cys185 to yield CC69-185 increases expression yields and increases thermal stability by 20 °C (Figure 2B and Table 1) but secondary and quaternary structures do not change (Figures S3B and S3C). Non-reducing gel analysis of CC69-185 to deduce disulfide bond contribution to the oligomeric state reveals only dimers, not higher ordered species (Figure S3D). Chemical crosslinking of CC69-185 shows masses consistent with dimer and tetramer (Figure 3B). The calculated molecular weight from SAXS is consistent with a tetramer (Table 2), corroborating preparative SEC and crosslinking results. The molecular envelope of CC69-185 is a P2 symmetric, V-shaped molecule with dimensions 18 nm × 10 nm (Figures S2, S4A, and Table 2). The envelope overlays well with the larger CC60-185 construct (Figure S4B), but appears to be missing the third ‘stem’. We propose that the stem is not resolved in this construct because, without the disulfide bond from Cys61, the protein is susceptible to proteolysis (Daniel et al., 1982; Reddy et al., 1988).
To clarify the orientation of CC69-185, relative to the Y-shaped CC60-185, we characterized two additional constructs, CC33-111 and CC112-185, biochemically and by SEC-SAXS (Figures S2, S3E-L, S4, and Table 2). Both proteins are, as expected, helical (Figures S3E and S3I). SEC-SAXS envelopes reveal distinct rods for each construct (Figures 3C and 3D). A tetramer is seen in analysis of the SEC profile of CC33-111 (a single ~44 kDa species composed of 8.8 kDa monomer) (Figure S2), chemical crosslinking (Figure 3C), and non-reducing electrophoresis (Figure S3H), a surprise based on results from single Cys variants (Figure 2D). The corresponding rod is not perfectly symmetric, with half of the rod length ~4 nm wide and half ~2 nm wide (Figure 3C), perhaps reflecting structural differences in the predicted unstructured N-terminal region and the CC region starting near Gln75 (Figure 1B). By comparison, the preparative SEC profile (Figure S3J) and molecular weight calculation of CC112-185 from SAXS (Table 2) indicate a dimer, which is disulfide bonded (Figures S3K and S3L). CC112-185 is a rod that best matches the length of the arms of the V-shaped CC69-185 (Figures 3D, S4C, and S4D). A bulge is observed approximately in the middle of the CC112-185 rod, a feature seen in the pairwise distribution (P(r)) plot of the SEC-SAXS data (Figures S2D and S4E, and see below). Taken together, these data indicate that CC60-185 is composed of two sequentially-identical C-terminally disulfide-bonded dimers, which converge at the N-terminal region to form a tetrameric stem (Figure 3E).
Crystal structure of the 7 C-terminal heptad repeats reveals atypical stabilization for a leucine zipper
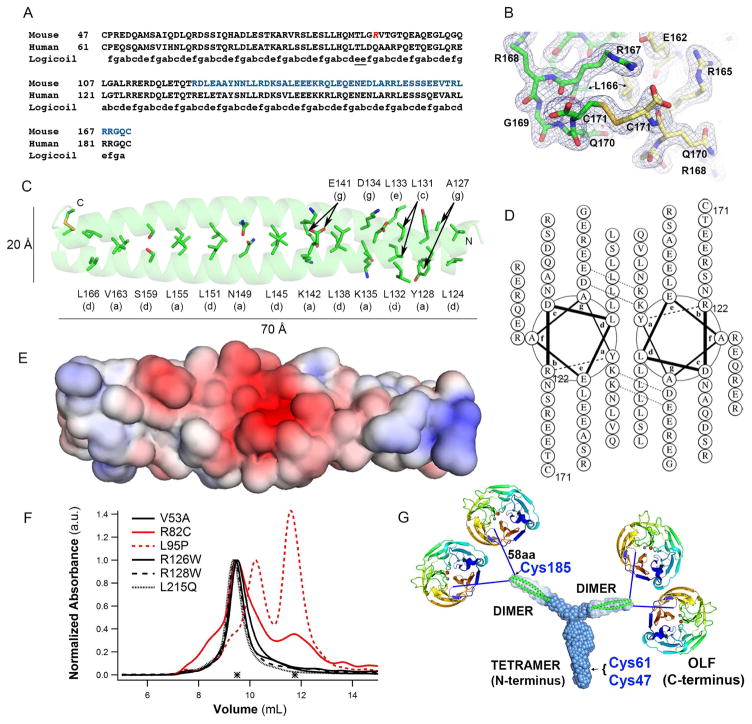
Next, we solved a 1.9 Å resolution crystal structure of the C-terminal 7 heptad repeats of the mouse myocilin leucine zipper (LZ) region, mLZ122-171 (residues 136-185 in human myocilin, 80% sequence identity, Figure 4A and Table 3). This crystal grew after in situ proteolysis of mLZ93-171, a construct nearly identical to CC112-185 in sequence and structure by SEC-SAXS (Figures 3C and S4E-F), which failed to crystallize under conditions tested. The susceptibility of mLZ93-171 to further proteolysis is not immediately clear, but the presence of a so-called skip residue at Thr89/Thr103 (Figure 4A) suggests the possibility of extended unraveling as seen in myosin (Taylor et al., 2015). The structure of mLZ122-171, which was solved by combining ab initio modeling with molecular replacement, reveals the C-terminal disulfide bond (Figure 4B and S5A) anticipated from biochemical experiments (Figures 2D, S3D, S3L, and Table 1) capping a parallel dimer with a left-handed supercoiled twist (Figure 4C). The two CCs in the asymmetric unit are nearly identical to each other, as are the two helices within each CC, with root mean squared deviations (r.m.s.d.) ~ 1.5 Å (not shown).
Figure 4. Crystal structure of mLZ122-171, overall myocilin model, and effects of selected glaucoma-relevant variants.
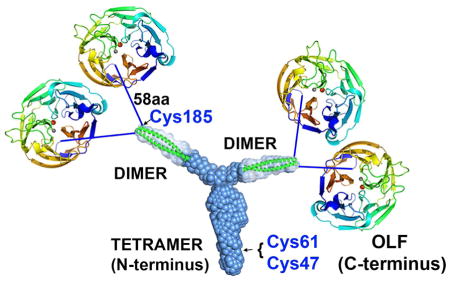
(A) Partial sequence alignment of human and mouse myocilin with predicted register of heptad repeats. Red Arg, start of the construct used for crystallization (mLZ93-171). Blue stretch, crystallized fragment mLZ122-171. (B) Two mLZ122-171 helices are covalently tethered by a C-terminal disulfide bond. Final 2Fo-Fc (blue) electron density map is contoured at 1.0 σ and Fo-Fc (red/green) is contoured at ±3.5 σ. Chains A and B are labeled green and yellow. Stereo image presented in Figure S5A. (C) Ribbon diagram of mLZ122-171 parallel dimer with labeled heptad positions (a) and (d) and disulfide represented as sticks. (D) mLZ122-171 in helical wheel representation. (E) Electrostatic surface (negative (red, -5 kT/e-) to positive (blue, + 5 kT/e-)) of mLZ122-171 dimer. (F) SEC profile comparison of six NTD33-226 glaucoma-relevant variants reveals altered elution and thus quaternary structure for R82C and L95P (red traces). (G) Model of full-length myocilin: Y-shaped dimer-of-dimers composed of an N-terminal parallel tetrameric domain leading to two parallel CC dimers, followed by 58 amino acid long linkers, and C-terminal OLF domains. Cys185 is located just prior to the start of the 58aa linker; Cys47 and Cys 61 are within the N-terminal tetramer but locations within the stem are not precisely identified. See also Figure S1, S5.
Table 3.
Crystallographic statistics.
| Wavelength | 1 Å |
| Resolution range | 41.9-1.92(1.99-1.92) |
| Space group | C 1 2 1 |
| Unit cell | 85.13 48.75 55.80 90 105.97 90 |
| Total reflections | 16652 |
| Unique reflections | 16650(1447) |
| Multiplicity | 7.0(5.1) |
| Completeness (%) | 96.1(79.9) |
| Mean I/sigma(I) | 21.7 |
| Wilson B-factor | 28.2 |
| R-meas | 0.077(0.339) |
| R-pim | 0.032(0.141) |
| Reflections used in refinement | 16259(1337) |
| Reflections used for R-free | 1616(133) |
| R-work | 0.187(0.236) |
| R-free | 0.240(0.313) |
| Number of non-hydrogen atoms | 1824 |
| macromolecules | 1646 |
| solvent | 178 |
| Protein residues | 200 |
| RMS(bonds) | 0.006 |
| RMS(angles) | 0.75 |
| Ramachandran favored (%) | 97.4 |
| Ramachandran allowed (%) | 2.1 |
| Ramachandran outliers (%) | 0.5 |
| Rotamer outliers (%) | 0.5 |
| Clashscore | 6.8 |
| Average B-factor | 42.0 |
| macromolecules | 41.9 |
| solvent | 43.7 |
| Number of TLS groups | 8 |
Statistics for the highest-resolution shell are shown in parentheses.
Analysis of the mLZ122-171 structure reveals key deviations from expected general patterns found in the stereotypical “knobs-in-holes” CC (Parry et al., 2008) (Figures 4C and 4D). Overall, mLZ122-171 follows the pattern of a hydrophobic residue, Leu, in position ‘d’ except for the penultimate heptad in which it is a polar Ser159. By contrast, fewer than half of the ‘a’ register residues are hydrophobic. The four remaining residues at ‘a’ positions (Figure 4D), which are highly conserved among myocilin from human, mouse, rat, fish and cow (Figure S1F), are positively charged or polar. Two Lys residues are in positions poised to stabilize negatively charged aspartate and glutamates found one residue N-terminal in the adjacent helix at the ‘g’ position, though we note these distances are somewhat long to be considered robust electrostatic interactions (~3–5 Å). Consensus from prior studies of CCs has suggested that ‘a’ positions can tolerate polar residues because they are less buried. Polar residues like Asn are specifically associated with parallel dimers (Mason and Arndt, 2004); in our structure, Asn149 is in position ‘a’. Charged residues in ‘a’, particularly the frequency of their occurrence, is unexpected. One protein design study identified this atypical pairing as important for the specificity of intramolecular interactions among closely related CCs (Grigoryan et al., 2009), although the need for such specificity is not clear yet for myocilin. In support of the importance of the a-g pairing for specificity, and not the traditional ‘e’ and ‘g’ positions, there are three negatively charged residues in position ‘e’ in mLZ122-171 and four in ‘g’, with just one positively charged residue in ‘e’ and ‘g’ (Figure 4D). Two of the acidic residues in ‘g’, which do not participate in salt bridges with ‘core’ Lys residues in ‘a’ positions, are proximal to Asn149 and impart the overall negative electrostatic surface charge of mLZ122-171 (Figure 4E). Remarkably, despite these non-canonical repeat features, the mLZ122-171 structure overlays well with the sequentially-distant structural homologs, the prototypical leucine zipper GCN4 and geminin (24% and 19% sequence homology across 40 residues, and r.m.s.d.= 1.05 Å and 1.54 Å, respectively, not shown).
Molecular dynamics (MD) simulations suggest the potential for mLZ122-171 to adopt an alternative structure
After 50 ns of equilibration at 310 K, the helical structures of mLZ122-171 and the disulfide-deletion variant mLZ122-170 remain essentially unchanged (Figure S5B). However, after 100 ns at 400 K, a bend emerges in the structure of mLZ122-171 near the C-terminus (Ser159-Gln170), concomitant with a shift in paired interactions between oppositely charged residues on either helix (Figures S5C and S5D). This region of the leucine zipper includes paired Ser159 residues located in the heptad ‘d’ position as well as other polar/charged residues. Without the disulfide formed by Cys171 (Cys185 in human myocilin) and consistent with stability measurements (Table 1), the mLZ122-170 dimer is considerably more susceptible to unfolding. For example, in the 400 K simulation, the distinct kinked structure was not formed. In steered molecular dynamics simulations at 310 K in which the centers-of-mass of each coil were pulled apart at a constant velocity of 1 Å/ns for 10 ns, the C-terminal ends separated completely (Figure S5E). In line with its more hydrophobic character, the N-terminal region of mLZ122-171 and mLZ122-170 dimers remained unaltered in all simulations. In sum, the disulfide bond at the C-terminus of the myocilin leucine zipper appears to limit the extent to which each protomer can unfold and the prevalence of complementary charged residues allows the coil to adopt an alternative structure. Such resistance to unfolding likely helps myocilin cope with the regular mechanical distortions of the TM ocular tissue (Acott et al., 2014).
Structural defects imparted by single nucleotide polymorphism and glaucoma-associated myocilin variants
In contrast to mutations found within the myocilin OLF domain, many of which are associated with early-onset and familial cases of glaucoma (Hewitt et al., 2006), the vast majority of mutations in the N-terminal module have been identified via case-control genome sequencing studies. As such, their annotation as pathogenic is predicted statistically. The extent to which the toxic gain-of-function hypothesis for myocilin-associated glaucoma (misfolding/non-secretion/aggregation) holds for mutations found beyond OLF is unclear because such variants have not been systematically studied. Just one study evaluated cellular trafficking and adhesion properties of two annotated N-terminal disease variants (R82C and R126W, a rare late-onset familial mutation (Faucher et al., 2002)) and one neutral (L95P) mutant (Gobeil et al., 2006). Compared to most OLF-resident mutations, which were retained intracellularly (Gobeil et al., 2006; Liu and Vollrath, 2004; Vollrath and Liu, 2006; Zhou and Vollrath, 1999), all three N-terminal variants tested were secreted (Gobeil et al., 2006). For R82C and L95P variants, however, partial intracellular sequestration was observed. R82C- and L95P- mutant myocilins also lost adhesion properties, but R126W did not (Gobeil et al., 2006).
On the basis of the examined WT CC constructs, which exhibit reversible thermal unfolding (Figures 2A, S3A, and S3C), it is apparent that the CC domain is not intrinsically prone to aggregation like OLF, but disease-associated N-terminal mutants might still misfold, for example, by exhibiting structural anomalies without aggregation. To test our hypothesis, we biophysically characterized six NTD33-226 variants (Figures 4F and S3M-T, and Table 1). Linker-resident, conservative substitutions V53A and L215Q are annotated as glaucomatous (Hewitt et al., 2007). Biophysically, however, they are indistinguishable from WT in stability, thermal unfolding reversibility, and SEC elution profile, indicating no major structural changes have occurred. In the dimer region of the CC, the two changes R126W and R128W, annotated as glaucomatous and neutral, respectively, are expected to be poorly tolerated. This is especially true for R128W, in which the Arg appears to fall in the electrostatically positive ‘a’ position for pairing with a negatively charged Glu ‘g’ in an adjacent helix. Surprisingly, both variants behaved as WT. In the tetramer region, which has thus far eluded crystallization, similarly nonconservative substitutions R82C and L95P previously mentioned, were also not expected to be well tolerated. Both variants exhibited WT like stability and reversible thermal unfolding, with no evidence for aggregation (Figure S3O–T). The elution profiles from SEC differ from WT, however, in that they are both mixtures of WT and smaller, non-native species (Figure 4F), suggesting disruption of the tetramer.
Discussion
With the molecular structures of the myocilin CC and OLF domains (Donegan et al., 2015) in hand, we infer that full-length myocilin consists of an N-terminal tetrameric stalk that branches at an obtuse angle into two parallel dimer-of-dimers affixed to paired C-terminal OLF domains. The transition from tetramer to the two dimers begins between Ser69 and Glu112, and most likely includes the lower-CC probability region containing the skip residue, Thr103 (Figure 4A). Two pairs of C-terminal OLF domains are connected to the CC region by a 58-amino acid linker with unknown structure and functional significance (Figure 4G).
To the best of our knowledge, the Y-shape topology is unprecedented among both naturally occurring and designed CCs. Modular proteins containing repeat structures are common within the extracellular matrix proteome (Hynes and Naba, 2012), and the addition of the myocilin CC diversifies the modular arrangement. The myocilin CC further expands our appreciation for the versatility of deceptively-straightforward CCs and should motivate the design of bio-inspired materials with novel supramolecular arrangements, a highly active research area of protein science (Lupas and Bassler, 2017).
Previous studies of myocilin have detected a tetramer, by crosslinking with glutaraldehyde (Nguyen et al., 1998) and non-reducing Western blot analysis (Gobeil et al., 2004), as well as a dimer (Dismuke et al., 2012; Fautsch and Johnson, 2001; Gobeil et al., 2004; Nguyen et al., 1998; Russell et al., 2001; Stamer et al., 2006) and higher ordered states (Dismuke et al., 2012; Fautsch et al., 2004; Russell et al., 2001). Based on our study, tetramerization occurs as a result of the amino acid sequence in the N-terminal stalk, and is independent of the disulfide bonds, which serve to stabilize individual dimers. Alternative oligomeric states of myocilin might form in vivo, perhaps mediated by a binding partner not present in our purified samples, or by mechanical changes in the TM, or by fluctuations in the redox or other chemical properties of aqueous humor fluid (Richer and Rose, 1998). For example, changes in the redox state of aqueous humor might lead to a higher ordered state via inter-molecular disulfide bond formation, such as seen in non-reducing SDS-PAGE analysis of CC33-111. Whether inter-molecular disulfide swapping of Cys47 and Cys61 at the unstructured N-terminus, which we envision also leading to the octamer seen in non-reducing SDS-PAGE analysis of NTD33-226 and previously (Fautsch et al., 2004), occurs as a feature or is a misfolded state of myocilin, remains to be clarified.
The tetrameric arrangement subsequently imparts multivalency to the otherwise monomeric C-terminal OLF domain (Burns et al., 2010), simultaneously increasing avidity and spacing OLF pairs apart from one another. The long linker extending from its N-terminus may assist OLF in binding to partners. Like for gliomedin, another OLF domain-containing protein whose oligomerization state is mediated by trimerizing collagen domains (Maertens et al., 2007), avidity likely underlies OLF-mediated binding partners. The regular gap between dimers within one molecule implies a particular distribution in the TM tissue, and may be further modulated by the intervening linker.
The N-terminal coiled regions may have a purpose beyond a molecular spacer for the OLF domain. We observed that our recombinant myocilin CC constructs, despite exhibiting robust stability and reversible folding upon denaturation, are adherent to membrane filters and other labware if glycerol is omitted from the buffer. Such natural stickiness, which was noted previously (Russell et al., 2001), is not completed abrogated by the presence of glycerol, and thus may be an asset of myocilin that contributes mechanical stiffness to the TM extracellular tissue (Wang et al., 2017). This feature is also compatible with the previously suggested cell adhesion function for myocilin (Goldwich et al., 2009; Wentz-Hunter et al., 2004), and may tie into the role of myocilin in elevating IOP (Fautsch et al., 2006; Fautsch and Johnson, 2001; Resch and Fautsch, 2009). However, this property also may be impeding the identification of bona fide functional binding partners. Previously reported binding partners localized to the myocilin CC (Resch and Fautsch, 2009) could be experimental artifacts stemming from adhesiveness and should be reevaluated. Previous reports also indicate an association with lipid membranes (Stamer et al., 2006). Our data support the prior proposal that such association would be mediated by another membrane-bound protein (Stamer et al., 2006) because the negative electrostatic surface potential of mLZ122-171 would prevent its direct insertion into the membrane.
The high level of sequence conservation across myocilin-like subfamily 3 members (Figure S1G) indicates that the Y-shape architecture is shared across eukaryotes. The topology of myocilin is distinct from the dimer-of-dimers V of olfactomedin-1 (subfamily 1), likely a result of the different coil prediction pattern (Figures 1A and S1B) and low overall sequence similarity. The angle of the V of olfactomedin-1 appears acute, not obtuse like measured for CC69-185. In addition, the C-terminal region of the olfactomedin-1 CC, which harbors a disulfide-forming Cys found within the CC (Pronker et al., 2015), is not aligned with the CC-capping residue Cys 185 in myocilin (Figures 4B and S1B). In olfactomedin-1, the end of the disulfide-harboring CC is immediately adjacent to the start of the OLF domain (Pronker et al., 2015) whereas Cys185 in myocilin is remote from the start of the structural OLF near Cys245. Whether the remaining three CC-containing olfactomedin subfamilies adopt a dimer-of-dimers and to what extent they are similar to myocilin or olfactomedin-1, remains to be seen. Other open questions across the olfactomedin family surround the functional consequences of their unusual topologies. A naive interpretation would suggest the CCs are tailored “molecular rulers,” to space pairs of OLF domains, but mechanotransduction, or molecular scaffold for complex protein-protein interactions, are intriguing alternative possibilities.
In sum, our study of the myocilin CC regions expands the scope of naturally occurring protein shapes to include a tripartite dimer-of-dimers. The arrangement offers new molecular clues to biological and/or biomechanical function that should prompt innovative experiments, and provides insight into glaucoma-associated mutations found outside the myocilin OLF domain. The tetramer is sensitive to mutation, as changes in quaternary structure were observed for R82C and L95P, with a consequence of non-native cellular secretion and poor cellular adhesion observed previously by others (Gobeil et al., 2006). Amyloid-like aggregation seen for OLF-resident mutations (Hill et al., 2014; Orwig et al., 2012) did not occur. Further studies will be required to assess whether both variants are glaucoma-causing, and to what extent gain-of-function (misfolding, intracellular sequestration) and/or loss-of-function (adhesion, mechanical properties, other) mechanisms are at play. By contrast, the dimer region appears relatively resilient to mutation and unfolding, and may intentionally access multiple distinct conformations. Linker-resident mutations, which are not expected to affect quaternary structure nor located at positions with high likelihood for posttranslational modification, may not be causative for glaucoma. Knowledge of the biophysical misfolding properties associated with variants within the N-terminal portion of myocilin will inform likelihood of pathogenicity for new mutations identified. Such work is synergistic with efforts to identify at-risk patients to manage glaucoma prior to onset of vision loss (Souzeau et al., 2017) and to develop personalized medicine for glaucoma.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Raquel L. Lieberman (Raquel.lieberman@chemistry.gatech.edu).
Experimental Model and Subject Details
Proteins were expressed in E. coli BL21(DE3)plysS cells using the same protocol: A single transformed colony was inoculated into a selective 150–250 mL Luria-Bertani culture (LB, Fisher, supplemented with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol) and agitated at 225 r.p.m., 37 °C, overnight (16–24 hours). The starter culture (20 mL) was used to inoculate 1L of Superior Broth media (U.S. Biological), which was then agitated at 225 r.p.m., 37 °C until an optical density at 600 nm of 1.0–2.0 was reached. Protein expression was induced with 0.5 mM ispropyl β-D-thiogalactopyranoside (IPTG) at 18 °C and induced cells were grown overnight (16–20 hours) by agitation at 225 r.p.m., 18 °C. Cells were harvested by centrifugation and flash frozen in liquid nitrogen for storage at -80 °C.
Method Details
Molecular Cloning
Constructs of N-terminal portions of human myocilin were obtained by subcloning from an E. coli codon-optimized construct of full-length human myocilin (DNA 2.0). Primers for NTD33-226, CC60-185, CC69-184, CC33-111, and CC112-184, were designed to incorporate a C-terminal TAA stop codon as listed in Figure S6E. Amplified DNA was purified via a Promega PCR purification procedure, treated with T4 DNA polymerase, and annealed into the pET-30 Xa/LIC vector following the manufacturer’s directions. Cys-to-Ser variants of NTD33-226 (C61S/C185S, C47S/C185S, C47S/C61S, C47S/C61S/C185S), CC69-185, CC112-185, and NTD33-226 disease-causing variants were generated by site directed mutagenesis (QuikChange Lightning mutagenesis kit, primers listed in Figure S6E). Plasmids were propagated by E. coli NovaBlue GigaSingles and the correct in-frame DNA sequence was confirmed (MWG Operon). The plasmid for mouse LZ55-171, designed to be identical to the pET-30 Xa/LIC vector with a Factor Xa protease cleavage site was purchased from Genscript. After proteins were expressed in E. coli BL21(DE3)plysS cells (detailed in Experimental Model section), they were purified as detailed below.
Protein Purification
NTD33-226 (WT))
Cell pellets (20 g) were resuspended using a serological pipette into 40 mL Ni2+-affinity purification wash buffer (50 mM Hepes pH 7.5, 200 mM NaCl, 40 mM imidazole, 10% glycerol) supplemented with 2 EDTA-free Roche Protease Inhibitor Cocktail tablets. Cells were lysed by 2–3 passages through a French press (12,000 psi). To increase protein yield, 1% Trion X-100 was added and lysed cells were rocked gently in a cold room for 1 hour. Cellular debris was removed by ultracentrifugation at 130,000 × g, 4 °C for 1 hour and the supernatant loaded onto a 1 mL Ni2+-affinity purification column (GE Healthcare) using an AKTA purification system (GE Healthcare). After contaminant proteins were removed by 120 column volumes of wash buffer, NTD33-226 was eluted with a gradient from 40–500mM imidazole using a mixture of wash buffer and elution buffer composed of 50mM Hepes pH 7.5, 200mM NaCl, 500 mM imidazole, 10% glycerol. Fractions containing NTD33-226 were concentrated using Amicon 100K MWCO filtration devices (EMD Millipore) and loaded onto a Superdex 200 10/300 GL (GE Healthcare) size exclusion column equilibrated with gel filtration buffer (50 mM Hepes pH 7.5, 200 mM NaCl, 10% glycerol). The fractions containing NTD33-226 were identified by SDS-PAGE analysis (15% polyacrylamide) with Coomassie staining and found to have a contaminating band of DnaK, a result confirmed by trypsin digest mass spectrometry (Figure S6A, Systems Mass Spectrometry Core Facility, Georgia Tech). Taking advantage of the finding that NTD33-226 can unfold reversibly (Figure 2A), an on-column unfolding refolding protocol (see below) was used to remove the contaminating DnaK for selected preparations (e.g. Figure 2 and NTD33-226 Cys-to-Ser variants below). Removal of DnaK did not alter the elution profile of NTD33-226 from Superdex 200 (Figures S6B and S6C). Protein concentration was estimated by absorbance at 280 nm using a molecular extinction coefficient (8,605 M−1cm−1) and a molecular weight (27,014.8 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005)
NTD33-226 Cys-to-Ser Variants (C47S/C61S, C61S/C185S, C47S/C185S, C47S/C61S/C185S)
Cell pellets (20 g) were lysed using the Triton X-100 step and purified by Ni2+-affinity column chromatography as outlined above. Fractions containing NTD33-226 were concentrated to 1 mL using Amicon 100K MWCO filtration devices followed by dilution into 50mL urea unfolding buffer (50 mM Hepes pH 7.5, 500 mM NaCl, 8 M Urea). Unfolded NTD33-226 was reloaded onto the Ni2+-affinity column, washed with 20 column volumes of urea unfolding buffer to remove contaminating DnaK, and then refolded on the column by a gradient into refolding buffer (50mM Hepes pH 7.5, 200mM NaCl, 10% glycerol). Folded NTD33-226 was eluted by a gradient from 0–500 mM imidazole also containing 50mM Hepes pH 7.5, 200mM NaCl, 10% glycerol, fractions containing NTD33-226 were concentrated using Amicon 30K MWCO filtration devices (EMD Millipore), and samples were then purified by Superdex 200 10/300 GL as described above. Purified proteins were assessed and quantified as for WT NTD33-226.
NTD33-226 Variants (V53A, R82C, L95P, R126W, R128W, L215Q)
Cell pellets (10 g) were resuspended using a serological pipette into 20 mL Ni2+-affinity purification wash buffer supplemented with 1 EDTA-free Roche Protease Inhibitor Cocktail tablet. Cells were lysed, debris removed, and protein purified as described for NTD33-226. Fractions containing NTD33-226 were assessed by SDS-PAGE analysis, concentrated with Amicon 100K MWCO filtration devices, and re-loaded onto a Superdex 200 10/300 GL column equilibrated with gel filtration buffer to obtain the final presented chromatogram.
CC60-185
Cell pellets (20 g) were resuspended, lysed, and purified by Ni2+-affinity and SEC as for NTD33-226, except that the initial cell resuspension buffer was further supplemented with 1 mL of 10 mg/mL DNase, 200 μL of 1 M CaCl2, 200 μL of 1 M MgCl2 and the Triton X-100 step was not used. To remove tags, purified CC60-185 was incubated with Factor Xa (Roche, ~50:1 mass ratio) plus ~5–10 mM CaCl2 for 96 hours at 4 °C. Digested CC60-185 was diluted ~20 fold into heparin wash buffer (20 mM Hepes, pH 7.5, 10% glycerol), loaded onto a 5 mL HiTrap Heparin HP column (GE Healthcare), and eluted by a gradient from 0–1 M NaCl with buffers also containing 20 mM Hepes pH 7.5, 10% glycerol, which also removed Factor Xa and DnaK contaminants. Heparin elution fractions were assessed by SDS-PAGE analysis as above, and those containing CC60-185 were concentrated with Amicon 10K MWCO filtration devices (EMD Millipore) and loaded onto a HiLoad 16/600 Superdex 75 prep grade column (GE Healthcare) equilibrated with gel filtration buffer. Fractions corresponding to a molecular weight of ~60 kDa (based on a calibration curve with standards (GE Healthcare)), containing pure CC60-185 as assessed by SDS-PAGE analysis with Coomassie staining, were concentrated in Amicon 10K MWCO filtration devices. Protein concentration was measured by absorbance at 280 nm using a molecular extinction coefficient (1,490 M−1cm−1) and a molecular weight (14,643 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005).
CC69-185
Cell pellets (20 g) were resuspended and lysed as for CC60-185 except that the 1% Triton X-100 procedure was performed as for WT NTD33-226. Purification by Ni2+ affinity and SEC proceeded as described for WT NTD33-226. After assessment by SDS-PAGE, fractions containing CC69-185 were digested and purified by heparin affinity and Superdex 75 following procedures for CC60-185. Fractions corresponding to a molecular weight of ~50 kDa (based on a calibration curve with standards (GE Healthcare)) containing pure CC69-185 as assessed by SDS-PAGE analysis with Coomassie staining were concentrated in an Amicon 10K MWCO filtration devices. Protein concentration was measured by absorbance at 280 nm using a molecular extinction coefficient (1,490 M−1cm−1) and a molecular weight (13,681 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005).
CC33-111
Cell pellets (20 g) were resuspended, lysed, and purified as described for CC60-185 except Amicon 10K MWCO filtration devices were used to concentrate the protein. Following assessment by SDS-PAGE, fractions containing pure CC33-111 were incubated with Factor Xa (Roche, ~50:1 mass ratio) plus ~5-10 mM CaCl2 for 96 hours at 4 °C. Digested CC33-111 was concentrated with Amicon 10K MWCO filtration devices and loaded onto a HiLoad 16/600 Superdex 75 prep grade column (GE Healthcare) equilibrated with gel filtration buffer. Fractions corresponding to a molecular weight of 40 kDa (based on a calibration curve with standards (GE Healthcare)) containing pure CC33-111 as assessed by SDS-PAGE analysis with Coomassie staining were concentrated using Amicon 10K MWCO filtration devices. Protein concentration was measured by absorbance at 280 nm using a molecular extinction coefficient (1,490 M−1cm−1) and a molecular weight (8,801 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005).
CC112-185
Cell pellets (20 g) were resuspended, lysed, and purified as described for CC33-111. Purified CC112-185 was digested and further purified using the procedure described for CC60-185. Fractions corresponding to a molecular weight of 23 kDa (based on a calibration curve with standards (GE Healthcare)) containing pure CC112-185 as assessed by SDS-PAGE analysis were concentrated in Amicon 10K MWCO filtration devices. Protein concentration was measured by absorbance at 280 nm using a molecular extinction coefficient (1,490 M−1cm−1) and a molecular weight (8,858 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005).
Mouse LZ55-171 and LZ93-171
Cell pellets (20 g) were resuspended, lysed, and purified as described for CC60-85 except 100K MWCO filtration devices were used to concentrate the protein. After assessment by SDS-PAGE, fractions containing pure mouse LZ55-171 were incubated with Factor Xa (Roche, ~50:1 mass ratio) plus ~5–10 mM CaCl2 for 72 hours at 4 °C. Digested mouse LZ55-171 was diluted ~30 fold into heparin wash buffer (20 mM Hepes, pH 7.5, 10% glycerol), loaded onto a 5 mL HiTrap Heparin HP column (GE Healthcare). Unlike the human constructs, mouse LZ55-171 did not bind the heparin column, so flow through fractions were concentrated with Amicon 10K MWCO filtration devices and loaded onto a HiLoad 16/600 Superdex 75 prep grade column (GE Healthcare) equilibrated with gel filtration buffer. The majority of protein was found in fractions corresponding to a molecular weight of ~30 kDa based on a calibration curve; however, after assessment by SDS-PAGE the dominant band on the gel was below the predicted 13,500 g/mol molecular weight for cleaved mouse LZ55-171. Mass spectrometry (Figure S6D) confirmed mouse LZ55-171 was cleaved by Factor Xa after a second Gly-Arg amino acid within the amino acid sequence, yielding mouse LZ93-171 used in subsequent experiments. Fractions containing mouse LZ93-171 were concentrated in Amicon 10K MWCO filtration devices. Protein concentration was measured by absorbance at 280 nm using a molecular extinction coefficient (1,490 M−1cm−1) and a molecular weight (9,144 g/mol) calculated by ExPASy ProtParam (Gasteiger et al., 2005) for mouse LZ93-117.
Circular Dichroism
Far-UV CD spectra and thermal melts were acquired on a Jasco J-815 spectropolarimeter equipped with a Jasco PTC-4245/15 temperature control system. Protein samples at a concentration range of 5–30 μM were measured in 50 mM Tris pH 7.5, 200 mM NaCl buffer at 4 °C. Scans were measured from 200 nm to 300 nm at a rate of 200 nm/min and a data pitch of 1 nm using a 0.1 cm cuvette. Each measurement was blank-subtracted with buffer and is an average of 10 scans. Far-UV thermal melts were performed from 4 to 80 °C at a rate of 1 °C min−1 increase in temperature and a data pitch of 2 °C. Ten scans from 300 to 200 nm at a 200-nm min−1 scan rate were averaged for each temperature. The Tm was determined by plotting the ratio of values recorded at 208 nm and 222 nm versus temperature and analyzed by Boltzmann Sigmoid analysis using Igor Pro. Recorded values are an average and standard deviation of two independent measurements, except for NTD33-226 glaucoma-associated variants standard deviation associated with the coefficient from curve fitting.
Chemical Crosslinking
Crosslinking reactions of NTD33-226 (30 μM), CC69-185 (75 μM), and CC33-111 (35 μM) were carried out at room temperature in 10 mM Na2HPO4/KH2PO4, pH 7.2, 200 mM NaCl with or without 50-fold molar excess of bis(sulfosuccinimidyl)suberate (BS3) crosslinker for 4 hours, 45 minutes, and 2 hours, respectively. The reactions were then quenched with an equal volume of 2x Laemmli sample loading dye and visualized by standard SDS-PAGE.
SEC-SAXS
SEC–SAXS experiments were performed at BioCAT (beamline 18ID, Advanced Photon Source at Argonne National Labs). The camera included a focused 12 KeV (1.03 Å) X-ray beam, a 1.5 mm quartz capillary sample cell, a sample to detector distance of ~3.5 m, and a Pilatus 3 1M detector (Dectris). The Q-range sampled was ~0.0042–0.4 Å−1. A size exclusion chromatography setup in line with the SAXS camera was used to separate the sample from potential aggregates, breakdown products and other contaminants using a Superdex 75 10/300 GL (GE Healthcare Lifesciences) column equilibrated with gel filtration buffer (50 mM Hepes pH 7.5, 200 mM NaCl, 10% glycerol). The elution trajectory after the UV monitor was redirected to the SAXS sample flow-cell. Exposures (1 s) were collected every 2 s during the chromatography run. Data reduction to generate the I(q) vs. q curves for all constructs except CC60-185 was performed with the BioCAT beam line specific pipeline, which uses the ATSAS program suite (Petoukhov et al., 2012). For CC60-185 multiple I(q) vs. q curves were manually generated and inspected to optimize frame choice, eliminating frames for which signal was being derived from aggregated protein. Exposures flanking the elution peak were averaged and used as the buffer curve for each run, and the exposures corresponding to the UV peak on the chromatogram were treated as protein plus buffer curves. Data were corrected for background scattering by subtracting the buffer curve from protein plus buffer curves. The radius of gyration (Rg) was calculated using Guinier approximation and a pairwise (P(r)) distribution plot, were both done using PRIMUS (Konarev et al., 2003). Dmax was also calculated using the P(r) curve and the output was used to calculate molecular envelopes using DAMMIN (Svergun, 1999) The envelopes were then averaged using DAMAVER and DAMFILT (Volkov and Svergun, 2003). For CC69-185, CC112-185, and mLZ93-171 results of imposing two-fold symmetry (P2) was compared to results with no imposed symmetry (P1). For all constructs and symmetry constraints presented, at least 9 out of the 10 envelopes were accepted by DAMAVER on the basis of the Normalized Spatial Discrepancy (NSD). SUPCOMB (Kozin and Svergun, 2001) was used to overlay envelopes of differing symmetry, compare CC60-185 with CC69-185, and compare CC112-185 to mLZ93-171 (Figure S4). Overlay of CC112-185 and CC33-111 (Figure 3E and S4) was performed manually within the PyMOL Molecular Graphic System v. 1.7.6.0. Final ab initio models were rendered using Visual Molecular Dynamics (Humphrey et al., 1996). Molecular masses were calculated using SAXS MoW2 (Fischer et al., 2010).
Crystallization, data collection, structure determination
Crystals of mouse LZ93-171 (later determined to be mouse LZ122-171) at 20 mg/mL in 10 mM Hepes pH 7.5 No NaCl grew after 90 days in 0.1 M ammonium acetate, 0.1 M BisTris pH 5.5, and 17% PEG 10,000 from a Hampton Index, HR2-134, sparse matrix sitting drop tray. Crystals were cryoprotected in 0.1 M ammonium acetate, 0.1 M BisTris pH 5.5, 25% PEG 10,000, and 25% glycerol. Data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline and processed using HKL-2000 (Otwinowski and Minor, 1997). An initial molecular replacement solution was found using AMPLE (Bibby et al., 2012), which consisted of three ~3 kDa helical poly-alanine chains with reasonable electron density. Using one of these ~3 kDa chains another round of molecular replacement was performed with Phaser (McCoy et al., 2007) but with a search criteria of eight molecules in the asymmetric unit as suggested by analysis of Matthews Coefficient (Kantardjieff and Rupp, 2003). Visualization of neighboring unit cells in Coot (Emsley et al., 2010) suggested a probable connection of pairs of chains, leading to the final solution of four ~6 kDa poly-alanine chains in the asymmetric unit. The poly-alanine chains were iteratively built and refined using Coot (Emsley et al., 2010) and Phenix.refine (Afonine et al., 2012) by slowly replacing alanines with respective amino acids to fit the electron density. The final model consists of mouse residues 122-171. The structure has been deposited to the PDB with accession number 5VR2.
Molecular Dynamics (MD) Simulations
Two structures were prepared for MD simulations: the dimer of mLZ122-171 both with (WT) and mLZ122-171 without (ΔC171) the C-terminal cysteines. Each structure was solvated in a water box of dimensions 105 × 105 × 105 Å3 using VMD (Humphrey et al., 1996). The system was neutralized with 0.15 M NaCl, resulting in a total of ~110,000 atoms for each system. MD simulations were carried out using both the NAMD 2.12 (Phillips et al., 2005) and Amber16 (Case et al., 2016) programs with the CHARMM36 all-atom force field (Best et al., 2012a; Best et al., 2012b). The temperature was fixed using Langevin dynamics; the pressure was kept constant at 1 atm using the Langevin piston method (Feller et al., 1995). The equations of motion were integrated using the RESPA multiple time-step algorithm with a time step of 2 fs used for all bonded interactions, 2 fs for short-range non-bonded interactions, and 4 fs for long-range electrostatic interactions. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method (Darden et al., 1993). Bonds involving hydrogen atoms were constrained to their equilibrium length employing the Rattle algorithm (Andersen, 1983).
Bioinformatics
Multiple sequence alignments were generated in CLUSTAL Omega (Sievers and Higgins, 2014). Myocilin sequences from multiple organisms were identified using the human myocilin OLF domain sequence as a query within BLAST (Altschul et al., 1990). The list was manually culled to remove partial sequences and non-myocilin proteins, which were identified either by annotation or by non-myocilin sequence stretches recognized by us through other related structures of OLF domain proteins (Hill et al., 2015), leaving 75 N-terminal sequences intact. Then the OLF domain sequences were deleted from the file, beginning with the first Cys (Donegan et al., 2015). After multiple sequence alignments, the consensus sequence was visualized using Skylign (Wheeler et al., 2014). CC prediction servers Marcoil (Delorenzi and Speed, 2002) and Logicoil (Vincent et al., 2013) were utilized to generate models for the CC domain of myocilin; a consensus cutoff of 80% probability was used to label the limits of the domains presented in Figure 1. Logicoil was used to obtain the coil register for Figure 4A and DrawCoil 1.0 (Grigoryan and Keating, 2008) was used to generate the image presented in Figure 4D. A structure similarity search was conducted with PDBeFOLD (Krissinel and Henrick, 2005) and sequence similarity searches limited to CCs were conducted using CC+ Database (Testa et al., 2009). Structures were aligned using the secondary structure matching algorithm (Krissinel and Henrick, 2004) to obtain r.m.s.d. values in Coot (Emsley et al., 2010). Electrostatic surfaces were generated using default options within the PDB2PQR web server (Dolinsky et al., 2007) and the APBS plugin (Baker et al., 2001) within PyMOL.
Data and Software Availability
Coordinates have been deposited to the Protein Data Bank under the accession code 5VR2.
Supplementary Material
Highlights.
Glaucoma-causing extracellular myocilin is associated with amyloid formation
Coiled-coil domain adopts a tripartite parallel dimer-of-dimers structure
Leucine zipper exhibits non-canonical heptad repeat pattern with disulfide cap
Glaucoma-associated variants in tetramer region alter structure but not stability
Acknowledgments
This work was funded by NIH R01EY021205 to R.L.L. and NSF MCB-1452464 to J.C.G. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Work performed at Bio-CAT was supported by NIH NIGMS 9P41 GM103622. Use of the Pilatus 3 1M detector was provided by NIGMS 1S10OD018090-01. We thank Srinivas Chakravarthy for SEC-SAXS data collection.
Footnotes
Author Contributions
Conceptualization, Methodology, S.E.H., R.L.L., J.C.G.; Investigation, S.E.H., E.N., R.K.D., A.C.P.O., and A.H.; Writing – Original Draft, S.E.H., R.L.L., J.C.G., A.H.; Writing – Review & Editing, S.E.H., R.L.L., A.C.P.O., Funding Acquisition and Supervision, R.L.L. and J.C.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Hassan DW, Acott TS, Kelley MJ. The trabecular meshwork: a basic review of form and function. J Ocul Biol. 2014;2:9–18. doi: 10.13188/2334-2838.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acott TS, Kelley MJ, Keller KE, Vranka JA, Abu-Hassan DW, Li X, Aga M, Bradley JM. Intraocular pressure homeostasis: maintaining balance in a high-pressure environment. J Ocul Pharmacol Ther. 2014;30:94–101. doi: 10.1089/jop.2013.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. 2012;D68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen HC. Rattle - a velocity version of the shake algorithm for molecular-dynamics calculations. J Comput Phys. 1983;52:24–34. [Google Scholar]
- Anholt RR. Olfactomedin proteins: central players in development and disease. Front Cell Dev Biol. 2014;2 doi: 10.3389/fcell.2014.00006. article 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Mittal J, Feig M, MacKerell AD., Jr Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of alpha-helix and beta-hairpin formation. Biophys J. 2012a;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput. 2012b;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby J, Keegan RM, Mayans O, Winn MD, Rigden DJ. AMPLE: a cluster-and-truncate approach to solve the crystal structures of small proteins using rapidly computed ab initio models. Acta Crystallogr. 2012;D68:1622–1631. doi: 10.1107/S0907444912039194. [DOI] [PubMed] [Google Scholar]
- Burns JN, Orwig SD, Harris JL, Watkins JD, Vollrath D, Lieberman RL. Rescue of glaucoma-causing mutant myocilin thermal stability by chemical chaperones. ACS Chem Biol. 2010;5:477–487. doi: 10.1021/cb900282e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JN, Turnage KC, Walker CA, Lieberman RL. The stability of myocilin olfactomedin domain variants provides new insight into glaucoma as a protein misfolding disorder. Biochemistry. 2011;50:5824–5833. doi: 10.1021/bi200231x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Betz RM, Botello-Smith W, Cerutti DS, Cheatham TEI, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, et al. Amber. 2016:16. [Google Scholar]
- Daniel RM, Cowan DA, Morgan HW, Curran MP. A correlation between protein thermostability and resistance to proteolysis. Biochem J. 1982;207:641–644. doi: 10.1042/bj2070641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T, York D, Pedersen L. Particle mesh Ewald - an N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- Delorenzi M, Speed T. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics. 2002;18:617–625. doi: 10.1093/bioinformatics/18.4.617. [DOI] [PubMed] [Google Scholar]
- Dismuke WM, McKay BS, Stamer WD. Myocilin, a component of a membrane-associated protein complex driven by a homologous Q-SNARE domain. Biochemistry. 2012;51:3606–3613. doi: 10.1021/bi300073r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donegan RK, Hill SE, Freeman DM, Orwig SD, Turnage KC, Lieberman RL. Structural basis for misfolding in glaucoma-associated myocilin. Hum Mol Genet. 2015;24:2111–2124. doi: 10.1093/hmg/ddu730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. 2010;D66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher M, Anctil JL, Rodrigue MA, Duchesne A, Bergeron D, Blondeau P, Cote G, Dubois S, Bergeron J, Arseneault R, et al. Founder TIGR/myocilin mutations for glaucoma in the Quebec population. Hum Mol Genet. 2002;11:2077–2090. doi: 10.1093/hmg/11.18.2077. [DOI] [PubMed] [Google Scholar]
- Fautsch MP, Bahler CK, Vrabel AM, Howell KG, Loewen N, Teo WL, Poeschla EM, Johnson DH. Perfusion of his-tagged eukaryotic myocilin increases outflow resistance in human anterior segments in the presence of aqueous humor. Invest Ophthalmol Vis Sci. 2006;47:213–221. doi: 10.1167/iovs.05-0334. [DOI] [PubMed] [Google Scholar]
- Fautsch MP, Johnson DH. Characterization of myocilin-myocilin interactions. Invest Ophthalmol Vis Sci. 2001;42:2324–2331. [PubMed] [Google Scholar]
- Fautsch MP, Vrabel AM, Peterson SL, Johnson DH. In vitro and in vivo characterization of disulfide bond use in myocilin complex formation. Mol Vis. 2004;10:417–425. [PubMed] [Google Scholar]
- Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant-pressure molecular-dynamics simulation - the Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- Filla MS, Liu X, Nguyen TD, Polansky JR, Brandt CR, Kaufman PL, Peters DM. In vitro localization of TIGR/MYOC in trabecular meshwork extracellular matrix and binding to fibronectin. Invest Ophthalmol Vis Sci. 2002;43:151–161. [PubMed] [Google Scholar]
- Fischer H, Neto MD, Napolitano HB, Polikarpov I, Craievich AF. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J Appl Crystallogr. 2010;43:101–109. [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gobeil S, Letartre L, Raymond V. Functional analysis of the glaucoma-causing TIGR/myocilin protein: integrity of amino-terminal coiled-coil regions and olfactomedin homology domain is essential for extracellular adhesion and secretion. Exp Eye Res. 2006;82:1017–1029. doi: 10.1016/j.exer.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Gobeil S, Rodrigue MA, Moisan S, Nguyen TD, Polansky JR, Morissette J, Raymond V. Intracellular sequestration of hetero-oligomers formed by wild-type and glaucoma-causing myocilin mutants. Invest Ophthalmol Vis Sci. 2004;45:3560–3567. doi: 10.1167/iovs.04-0300. [DOI] [PubMed] [Google Scholar]
- Goldwich A, Scholz M, Tamm ER. Myocilin promotes substrate adhesion, spreading and formation of focal contacts in podocytes and mesangial cells. Histochem Cell Biol. 2009;131:167–180. doi: 10.1007/s00418-008-0518-4. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Keating AE. Structural specificity in coiled-coil interactions. Curr Opin Struct Biol. 2008;18:477–483. doi: 10.1016/j.sbi.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt AW, Craig JE, Mackey DA. Complex genetics of complex traits: the case of primary open-angle glaucoma. Clin Experiment Ophthalmol. 2006;34:472–484. doi: 10.1111/j.1442-9071.2006.01268.x. [DOI] [PubMed] [Google Scholar]
- Hewitt AW, Mackey DA, Craig JE. Myocilin allele-specific glaucoma phenotype database. Hum Mutat. 2007;29:207–211. doi: 10.1002/humu.20634. [DOI] [PubMed] [Google Scholar]
- Hill SE, Donegan RK, Lieberman RL. The glaucoma-associated olfactomedin domain of myocilin forms polymorphic fibrils that are constrained by partial unfolding and peptide sequence. J Mol Biol. 2014;426:921–935. doi: 10.1016/j.jmb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SE, Donegan RK, Nguyen E, Desai TM, Lieberman RL. Molecular details of olfactomedin domains provide pathway to structure-function studies. PLoS One. 2015;10:e0130888. doi: 10.1371/journal.pone.0130888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb Perspect Biol. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer RA. Alpha-helical coiled-coil oligomerization domains in extracellular proteins. Matrix Biol. 1997;15:555–568. doi: 10.1016/s0945-053x(97)90031-7. [DOI] [PubMed] [Google Scholar]
- Kantardjieff KA, Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Multiple alignment of protein structures in three dimensions. Lect Notes Comput Sc. 2005;3695:67–78. [Google Scholar]
- Liu Y, Vollrath D. Reversal of mutant myocilin non-secretion and cell killing: implications for glaucoma. Hum Mol Genet. 2004;13:1193–1204. doi: 10.1093/hmg/ddh128. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Bassler J. Coiled coils - a model system for the 21st Century. Trends Biochem Sci. 2017;42:130–140. doi: 10.1016/j.tibs.2016.10.007. [DOI] [PubMed] [Google Scholar]
- Maertens B, Hopkins D, Franzke CW, Keene DR, Bruckner-Tuderman L, Greenspan DS, Koch M. Cleavage and oligomerization of gliomedin, a transmembrane collagen required for node of ranvier formation. J Biol Chem. 2007;282:10647–10659. doi: 10.1074/jbc.M611339200. [DOI] [PubMed] [Google Scholar]
- Mason JM, Arndt KM. Coiled coil domains: stability, specificity, and biological implications. Chembiochem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TD, Chen P, Huang WD, Chen H, Johnson D, Polansky JR. Gene structure and properties of TIGR, an olfactomedin-related glycoprotein cloned from glucocorticoid-induced trabecular meshwork cells. J Biol Chem. 1998;273:6341–6350. doi: 10.1074/jbc.273.11.6341. [DOI] [PubMed] [Google Scholar]
- Orwig SD, Lieberman RL. Biophysical characterization of the olfactomedin domain of myocilin, an extracellular matrix protein implicated in inherited forms of glaucoma. PLoS One. 2011;6:e16347. doi: 10.1371/journal.pone.0016347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwig SD, Perry CW, Kim LY, Turnage KC, Zhang R, Vollrath D, Schmidt-Krey I, Lieberman RL. Amyloid fibril formation by the glaucoma-associated olfactomedin domain of myocilin. J Mol Biol. 2012;421:242–255. doi: 10.1016/j.jmb.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–269. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HDT, Konarev PV, Svergun DI. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polansky JR, Fauss DJ, Chen P, Chen H, Lutjen-Drecoll E, Johnson D, Kurtz RM, Ma ZD, Bloom E, Nguyen TD. Cellular pharmacology and molecular biology of the trabecular meshwork inducible glucocorticoid response gene product. Ophthalmologica. 1997;211:126–139. doi: 10.1159/000310780. [DOI] [PubMed] [Google Scholar]
- Pronker MF, Bos TG, Sharp TH, Thies-Weesie DM, Janssen BJ. Olfactomedin-1 has a V-shaped disulfide-linked tetrameric structure. J Biol Chem. 2015;290:15092–15101. doi: 10.1074/jbc.M115.653485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy IM, Kella NKD, Kinsella JE. Structural and conformational basis of the resistance of beta-lactoglobulin to peptic and chymotryptic Digestion. J Agr Food Chem. 1988;36:737–741. [Google Scholar]
- Resch ZT, Fautsch MP. Glaucoma-associated myocilin: a better understanding but much more to learn. Exp Eye Res. 2009;88:704–712. doi: 10.1016/j.exer.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer SP, Rose RC. Water soluble antioxidants in mammalian aqueous humor: interaction with UV B and hydrogen peroxide. Vision Res. 1998;38:2881–2888. doi: 10.1016/s0042-6989(98)00069-8. [DOI] [PubMed] [Google Scholar]
- Rozsa FW, Shimizu S, Lichter PR, Johnson AT, Othman MI, Scott K, Downs CA, Nguyen TD, Polansky J, Richards JE. GLC1A mutations point to regions of potential functional importance on the TIGR/MYOC protein. Mol Vis. 1998;4 article 20. [PubMed] [Google Scholar]
- Russell P, Tamm ER, Grehn FJ, Picht G, Johnson M. The presence and properties of myocilin in the aqueous humor. Invest Ophthalmol Vis Sci. 2001;42:983–986. [PubMed] [Google Scholar]
- Sievers F, Higgins DG. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Souzeau E, Tram KH, Witney M, Ruddle JB, Graham SL, Healey PR, Goldberg I, Mackey DA, Hewitt AW, Burdon KP, et al. Myocilin predictive genetic testing for primary open-angle glaucoma leads to early identification of at-tisk individuals. Ophthalmology. 2017;124:303–309. doi: 10.1016/j.ophtha.2016.11.011. [DOI] [PubMed] [Google Scholar]
- Stamer WD, Perkumas KM, Hoffman EA, Roberts BC, Epstein DL, McKay BS. Coiled-coil targeting of myocilin to intracellular membranes. Exp Eye Res. 2006;83:1386–1395. doi: 10.1016/j.exer.2006.07.018. [DOI] [PubMed] [Google Scholar]
- Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- Stothert AR, Fontaine SN, Sabbagh JJ, Dickey CA. Targeting the ER-autophagy system in the trabecular meshwork to treat glaucoma. Exp Eye Res. 2016;144:38–45. doi: 10.1016/j.exer.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KC, Buvoli M, Korkmaz EN, Buvoli A, Zheng Y, Heinze NT, Cui Q, Leinwand LA, Rayment I. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc Natl Acad Sci U S A. 2015;112:E3806–3815. doi: 10.1073/pnas.1505813112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa OD, Moutevelis E, Woolfson DN. CC+: a relational database of coiled-coil structures. Nucleic Acids Res. 2009;37:D315–322. doi: 10.1093/nar/gkn675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Wentz-Hunter K, Yue BY. Distribution of myocilin and extracellular matrix components in the juxtacanalicular tissue of human eyes. Invest Ophthalmol Vis Sci. 2002;43:1068–1076. [PubMed] [Google Scholar]
- Vincent TL, Green PJ, Woolfson DN. LOGICOIL--multi-state prediction of coiled-coil oligomeric state. Bioinformatics. 2013;29:69–76. doi: 10.1093/bioinformatics/bts648. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollrath D, Liu Y. Temperature sensitive secretion of mutant myocilins. Exp Eye Res. 2006;82:1030–1036. doi: 10.1016/j.exer.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Wang K, Read AT, Sulchek T, Ethier CR. Trabecular meshwork stiffness in glaucoma. Exp Eye Res. 2017;158:3–12. doi: 10.1016/j.exer.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentz-Hunter K, Kubota R, Shen X, Yue BY. Extracellular myocilin affects activity of human trabecular meshwork cells. J Cell Physiol. 2004;200:45–52. doi: 10.1002/jcp.10478. [DOI] [PubMed] [Google Scholar]
- Wheeler TJ, Clements J, Finn RD. Skylign: a tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinformatics. 2014;15:7. doi: 10.1186/1471-2105-15-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf E, Kim PS, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfson DN. The design of coiled-coil structures and assemblies. Adv Protein Chem. 2005;70:79–112. doi: 10.1016/S0065-3233(05)70004-8. [DOI] [PubMed] [Google Scholar]
- Zeng LC, Han ZG, Ma WJ. Elucidation of subfamily segregation and intramolecular coevolution of the olfactomedin-like proteins by comprehensive phylogenetic analysis and gene expression pattern assessment. FEBS Lett. 2005;579:5443–5453. doi: 10.1016/j.febslet.2005.08.064. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Vollrath D. A cellular assay distinguishes normal and mutant TIGR/myocilin protein. Hum Mol Genet. 1999;8:2221–2228. doi: 10.1093/hmg/8.12.2221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.