Abstract
Reactive oxygen species (ROS) are increasingly recognized as critical determinants of cellular signaling and a strict balance of ROS levels must be maintained to ensure proper cellular function and survival. Notably, ROS is increased in cancer cells. The superoxide dismutase family plays an essential physiological role in mitigating deleterious effects of ROS. Due to the compartmentalization of ROS signaling, EcSOD, the only superoxide dismutase in the extracellular space, has unique characteristics and functions in cellular signal transduction. In comparison to the other two intracellular SODs, EcSOD is a relatively new comer in terms of its tumor suppressive role in cancer and the mechanisms involved are less well understood. Nevertheless, the degree of differential expression of this extracellular antioxidant in cancer versus normal cells/tissues is more pronounced and prevalent than the other SODs. A significant association of low EcSOD expression with reduced cancer patient survival further suggests that loss of extracellular redox regulation promotes a conducive microenvironment that favors cancer progression. The vast array of mechanisms reported in mediating deregulation of EcSOD expression, function, and cellular distribution also supports that loss of this extracellular antioxidant provides a selective advantage to cancer cells. Moreover, overexpression of EcSOD inhibits tumor growth and metastasis, indicating a role as a tumor suppressor. This review focuses on the current understanding of the mechanisms of deregulation and tumor suppressive function of EcSOD in cancer.
Keywords: EcSOD, SOD3, cancer, reactive oxygen species, heparin binding domain, tumor suppressor, metastasis, recurrence, relapse free survival, epigenetic, loss of heterozygosity, single nucleotide polymorphism, microRNA-21, oxidative tumor microenvironment
Graphical Abstract
1. Introduction
Reactive oxygen species (ROS) and reactive nitrogen species, including superoxide (O2•− ), hydrogen peroxide (H2O2), peroxynitrite (ONOO−) and nitric oxide (NO•), are integrally engrained into signal propagation within eukaryotic physiology and have essential roles in metabolism, innate immunity, differentiation and cell survival. As such, ROS signaling has been linked to aging, cardiovascular pathologies, inflammation, neurodegeneration and cancer [1–5]. Insights into ROS produced by regulated processes and the defenses evolved to protect against these disruptions of homeostatic redox states have fueled intense interest in the search for drug targets and clinical antioxidants.
At physiological levels ROS are important modulators of metabolism, signal transduction and stress response by acting as secondary messengers to redox sensitive substrates, altering protein structure and function [6–8]. ROS are spatially restricted due to limited diffusion distance granted by short half-lives [6]. Due primarily to its reactivity, the estimated half-life of O2•− ion is 10−9 seconds, and tends to interact with directly proximal substrates. In contrast, H2O2 catalyzed enzymatically or derived spontaneously from O2•−, is orders of magnitude more stable. The cytosolic lifespan of H2O2 has been determined to be one millisecond, enabling diffusion from endogeneous sources approximately 1 μm from its origin [8 9]. The lifespan is likely be dependent on the environment. In a bolus addition experiment, while the intracellular concentration of H2O2 requires only milliseconds to reach a pseudo-steady state, the extracellular concentration takes significantly longer to be consumed by cells (~ 12 minutes) [9].
Furthermore, H2O2 permeates passively between membranous compartments through aquaporin channels, and its lifespan is dependent on the available pool of reactive substrates [10–12]. These diffusibility limitations impart compartmentalization of ROS signaling, which is influenced by the redox state native within individual organelles. For example, disulfide bond formation in the ER is particularly sensitive to ROS stress [13]. Disruptions in redox homeostasis, tending towards a more oxidizing state, termed oxidative stress, constitute an imbalance between the production of ROS and the available systemic mechanisms of detoxification, whereby oxidants adversely modify DNA, lipids and proteins, generating toxic adducts deleterious to organismal health and contributing to genomic instability. Therefore, levels of these ROS need to be tightly regulated in a temporal and spatial manner, via a sophisticated cellular antioxidant network comprised of both enzymatic and non-enzymatic molecules.
2. Distribution and Function of the Three Sods in Mammalian Cells
One of the essential enzymatic components of the antioxidant defense system are the metal ion-dependent superoxide dismutases (SODs). There are three members of the SOD family present in mammalian physiology, with tightly regulated localization patterns. Of these, there are two copper/zinc containing members, CuZnSOD (SOD1) within the cytosol, mitochondrial inter membrane space, and nucleus, and EcSOD (SOD3) is the predominant antioxidant enzyme secreted into the extracellular space [14 15]. The manganese-containing MnSOD (SOD2), localizes to the mitochondrial matrix, is the most divergent and displays minimal similarity to the other SODs, which share 60% homology around the catalytic and metal-binding domains [16]. All SOD family members require metal cofactors for catalyzing one-electron oxidation followed by one-electron reduction of two O2•− anions to affect disproportionation. Due to their distinc t localizations, and membrane impermeability of O2•−, each member of the SOD family is expected to have specific compartmentalized roles, such as regulation of redox sensitive transcription factors [17], mitochondrial oxygen level sensing [18], or protection of surrounding tissue from oxidative inflammation during infection [19]. This implies SOD functions are non-redundant, despite having similar rate constants. Tumor suppressive effects of Cu/ZnSOD and MnSOD have been well described [20–22]. In this review, we will focus on EcSOD, its features and potential role in oncogenesis.
3. EcSOD Tissue-Specific Expression and Localization
While the other SODs are ubiquitously expressed, EcSOD is more restricted in a cell type and tissue dependent manner. EcSOD demonstrates high levels of protein expression within the cardiovascular endothelium, lungs, and placenta, displays moderately within kidney, pancreas and uterus, cartilage, skeletal muscle, adipose tissue, brain, and eye [15]. Abundantly secreted into the extracellular compartment, presence of EcSOD is detectable in milk, plasma, synovium, and lymph [14 23]. Once secreted, EcSOD is bound to cell surface proteoglycans through its positively charged heparin-binding domain (HBD). A portion of secreted EcSOD is subjected to intracellular proteolytic cleavage removing the HBD, which precludes tethering to the cell surface, and facilitates distribution in the extracellular milieu and circulation [24 25]. The secreted full length enzyme has been observed to be taken up by cells via endocytosis, facilitated by surface binding to proteoglycans and internalized through clathrin-coated pits [26]. In addition, EcSOD has been detected in nuclei associated with chromatin [27 28], and trafficking through the endo-lysosome system has been suggested [26–28]. Thus, although EcSOD, as the name implies, mainly resides in the extracellular space this enzyme has other intracellular localizations.
4. Molecular Characteristics: EcSOD
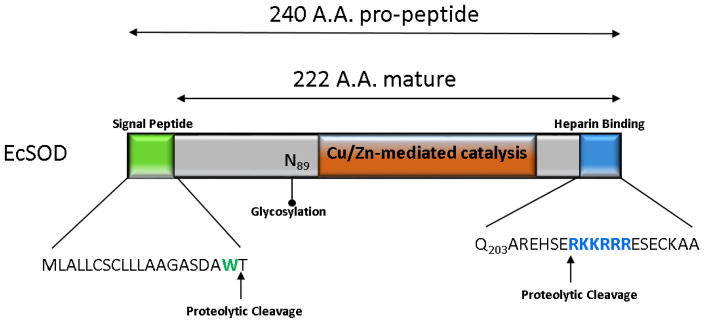
The monomeric subunit of human EcSOD is synthesized as a 32 kDa monomeric protein that exhibits higher order dimers, tetramers and octamers cross-linked through disulfide bridges between cysteine residues in the C-terminal region [29–32]. A 240 amino acid propeptide contains a signal peptide on the N-terminal required for secretion and is cleaved to generate a 222 amino acid protein making up the mature form [33] (Figure 1). Mature EcSOD can be separated into 3 regions, the amino-terminal features an asparagine at position 89 revealed by mass spectrometry to be a singular glycosylated residue greatly enhancing protein solubility and has been demonstrated as required for secretion but not activity [34–36]. The second domain bears 60% homology to CuZnSOD and contains the conserved active site and ion binding folds for the singular copper and zinc ions required for catalysis [37 38]. EcSOD is remarkably stable and resistant to extreme temperature, pH, urea and guanidinium chloride concentrations [37].
Figure 1.
Schematic illustration of human EcSOD protein structure. The 240 amino acid pro-peptide features an N-terminal signal peptide (green) cleaved to generate the 222 amino acid matured protein. Asparagine 89 is depicted as the singular glycosylated residue. The C-terminal heparin binding domain is depicted (blue).
4.1 Heparin Binding Domain
The third domain is unique to EcSOD and tethers it to the surface glycocalyx with high affinity, via a cleavable C-terminal heparin-binding domain (HBD). The HBD comprises a cluster of positively-charged arginine and lysine residues (SERKKRRR) which associates electrostatically with proteoglycans, notably heparin but also collagen type I and fibulin-5, on endothelial surfaces and tissue matrix, where it protects against oxidative fragmentation [33 39–41]. Intracellular proteolytic cleavage by a furin-like protease followed by carboxypeptidase activity within the HBD is expected to account for release of EcSOD into plasma and fluids [24 42 43]. Secreted distribution of hetero-oligomers possess a range of binding affinities and enzymatic activity. Tetramers separate into three fractions entirely with or without the HBD, or as a mixture of intact and truncated monomers [24 25 35 44]. Secretion of both full length and truncated forms of EcSOD with varying degrees of heparin affinity allows for differential tissue distribution in regulating specific protection from ROS [44]. Endothelial cells prominently feature EcSOD bound to the surface, do not synthesize the protein on their own, and acquire secreted EcSOD from vascular smooth muscle cells [45]. The positively charged HBD resembles a nuclear localization signal. Heparin binding not only enables O2•− scavenging on cell surfaces and where EcSOD is tethered, but also mediates endocytosis [28]. Internalized EcSOD has been observed to be associated with lysosomes, suggesting a possible degradation pathway. This HBD-mediated endocytosis has also been linked to localization of EcSOD to the nucleus [27 28]. The EcSOD crystal structure has been solved at a resolution of 1.7 angstroms. The overall tetramer is held together by disulfide bonds positioned towards the N-terminal, and uniquely features two grooves (‘major’ and ‘minor’) at polar ends, a result of dimeric organization [46]. Molecular modeling indicates the major groove accommodates binding of the heparin molecule some distance from the active site, consistent with previous reports that enzymatic activity is not inhibited while retained at the cell surface [39]. While missing from the overall structure, the C-terminal HBD is expected to be located along the top of this major groove to facilitate heparin binding, a conformational change ‘locking’ heparin into place protects EcSOD from proteolysis [47]. The minor groove on the reverse face is proposed to interact with collagen and involves electrostatic interaction with the C-terminus.
A naturally occurring polymorphism at arginine 213 (R213G) located in the HBD impairs heparin binding, increasing the concentration of EcSOD released into plasma. This results in no overt clinical phenotype, and does not appear to affect its intracellular distribution [48 49]. Interestingly, this renders EcSOD resistant to trypsin-like proteases [50]. The R213G polymorphism also confers resistance to furin protease activity, promoting secretion of an active, full-length molecule unable to bind heparin [43]. Full length EcSOD has a significantly longer tissue half-life than the truncated and released form (85 versus 7 hours respectively), while the HBD-null form shows less clearance by the liver due to its reduced ability to be endocytosed [51].
In diabetic patients, elevated blood glucose promotes non-enzymatic glycation products of EcSOD disrupting heparin binding but not catalytic activity [52 53]. While the R213G polymorphism has not been directly linked to diabetic susceptibility, diabetic patients on hemodialysis exhibit increased risk of ischemic complications of the cardiovascular system due to diminished EcSOD protection when absent from the surface endothelium [54 55]. Presence of R213G correlates with decline in lung function and susceptibility to chronic obstructive pulmonary disease [56]. Controversially, other studies have suggested a protective role, the polymorphism being more common among smokers resistant to COPD development [57].
Development of therapeutic intervention strategies for ameliorating the negative effects of oxidative stress have produced both pharmacological and gene therapy based approaches for preventing tissue injury in disease models [58 59]. SODs have shown promise as treatment in the laboratory, yet clinical efforts are stymied by source purification, clearance rate, and distribution [60–62]. To circumvent these challenges, a chimeric fusion of MnSOD with the EcSOD HBD, designed to promote its cellular internalization, successfully prevented vascular edema in two models of inflammation [62]. The highly basic residues of the HBD domain of EcSOD that form a predominantly helical structure is similar to the features described for cell penetrating peptides (CPPs), such as the HIV transactivator protein (TAT) and other experimental CPPs [63 64]. This unique domain of EcSOD has been demonstrated to possess efficacy as a CPP, where synthetic peptide corresponding to this region translocates into the cytoplasm and nucleus when added exogenously [64]. When this peptide is linked with apoptin (chicken anemia virus-derived protein), the recombinant apoptin-HBD fusion protein exhibited a significant and specific anti-tumor effect versus the apoptin protein alone, in the Lewis Lung carcinoma model in mice [64]. Although the uptake mechanism and intracellular fate of this highly basic EcSOD HBD peptide is not clear at this point, EcSOD HBD exhibits potential clinical application as a delivery tool to translocate cargo molecules into cells.
4.2 Catalytic Domain and Reaction Mechanism
EcSOD scavenges O2•− through the catalyzed dismutation of two molecules of O2•− to bimolecular oxygen and H2O2, which is subsequently reduced to water by catalase, peroxiredoxins and other enzymes [65]. EcSOD requires a redox-active Cu1+/2+ ion at its active site. Three histidine residues anchor the catalytic copper in place. One of these, the “bridging histidine”, also ligates the zinc ion, which itself requires an aspartic acid and additional electrostatic contributions from three separate histidines. The zinc ion is not required for catalytic activity, but facilitates protonation between the bridging histidine and confers thermal stability [66]. Electrostatic guidance of O2•− into the positively-charged catalytic active site requires Lys134 and Glu131 for approach from longer-range, and Arg141 for a more local orienting effect [67].
Superoxide disproportionation occurs by a ping-pong mechanism which proceeds in two steps. Step one begins as the O2•− substrate binds to copper2+. Superoxide anion donates an electron to become molecular oxygen, in the process reducing copper2+ to copper1+. The bond between copper and its anchoring histidine is broken, and histidine becomes protonated. In step two, this proton is donated along with the electron from copper1+ to a second O2•− anion, forming H2O2, and copper2+ reforms its histidine bond [68]. A transfer of one electron from O2•− to copper reduces the oxidized copper ion. The reduced copper is oxidized, donating an electron to a second O2•− anion at rates close to diffusion limits [69]. The overall stoichiometry results in formation of molecular oxygen and H2O2 from two O2•− molecules. The remarkable structure of SODs have pioneered mechanistic studies of ‘electrostatic guidance’, described in intricate detail within an excellent review [70].
Cu2+ZnSOD + O2•− → Cu+ZnSOD + O2
Cu+ZnSOD + O2•− + 2H+ → Cu2+ZnSOD + H2O2
Despite abundant documentation on SODs as anti-oxidants, a theme suggesting a pro-oxidant role is also coming into focus, with generation of H2O2 suggested to account for deleterious effects seen in some SOD overexpression systems [71]. In consideration, dismutation, spontaneous or catalyzed, yields identical amounts of H2O2, though with significantly different kinetics. In single-celled E.coli, endogenous SOD expression levels conferred 95% protection from O2•− induced damage to sensitive targets, this suggests the effects of SOD overexpression on H2O2 production should be negligible as O2•− concentrations would already be limited [71 72]. This however does not account for the rapidity of H2O2 formation or subcellularly localized bursts of increased H2O2 concentration. Such phenomena arguably would result from the presence of NADPH oxidase (NOX) enzymes not found in prokaryotic organisms, having made their first appearance later in evolution and function as compartmentalized producers of O2•− [73]. Whether SODs have pro-oxidant effects is likely to remain a conflicting issue. It is critical to know the answer to this antioxidant conundrum as there is accumulating evidence to support divergent effects on cell proliferation and death signaling for the two major intracellular ROS, O2•− and H2O2, as discussed in a later section. In addition to scavenging O2•−, SODs exhibit less efficient, non-specific peroxidase activity [74–77]. Here, the Cu/Zn-containing enzymes require CO2, ultimately generating a diffusible carbonate radical. This in turn disables the enzyme and thus the peroxidase action of both CuZnSOD and EcSOD functions as a ‘suicide reaction’ [74–77]. Determination of the precise mechanism has been notably contentious, and it remains to be seen whether SOD peroxidase activity is significant within living organisms, or can occur at physiological concentrations of H2O2 [71 78].
5. EcSOD as a primary defense against other ROS and RNS
The primary function of EcSOD is a superoxide scavenger, as the name implies. However, its redox modulation effect is not limited to controlling the levels of this radical. Superoxide is a precursor of many ROS and RNS as described [79]. By suppressing the accumulation of superoxide, EcSOD also prevents spontaneous dismutation of superoxide into H2O2. Generation of •OH via Fenton reaction and Harber Weiss reaction, can also be inhibited by EcSOD. Furthermore, by preventing the superoxide-mediated oxidation of NO•, EcSOD also controls the formation of ONOO−.
NO•, generated by nitric oxide synthases (NOSs) plays an important role as neurotransmitter, in regulating vessel relaxation in endothelial cells, and in mediating neutrophil and macrophage functions. Since superoxide reacts at a diffusion-limit rate with NO−, oxidation of this NO• into ONOO− results in alterations of cellular function as described [80]. The actions of NO• are mainly mediated through cGMP-dependent manner. In circumstances where cGMP is not available, the actions of NO• are carried out mainly in three ways: (i) interaction with proteins containing transition metal, (ii) interaction with proteins without the attached NO• group, and (iii) modulation of cell signaling by forming S-nitrosothiol- (SNO) modification on proteins. NO• has been shown to either facilitate cancer-promoting effects or act as an anti-cancer agent. Pro-tumor effects of NO• were linked to the expression of NO•-producing enzymes in tumor progression [81]. NO• also portrays anti-tumor effects by utilizing the immune defense mechanisms in animal models of various human cancers [82]. Recent evidence indicates that most of the cytotoxicity attributed to NO• is rather due to ONOO−, produced from the diffusion-controlled reaction between NO• and superoxide anion. Peroxynitrite interacts with lipids, DNA, and proteins via direct oxidative reactions or via indirect, radical-mediated mechanisms. These reactions mediate cellular responses ranging from modulations of cell signaling to overwhelming oxidative injury, eventually triggering necrotic or apoptotic cell death. In vivo, ONOO− generation represents a crucial pathogenic mechanism in conditions such as stroke, myocardial infarction, chronic heart failure, diabetes, circulatory shock, chronic inflammatory diseases, cancer, and neurodegenerative disorders. Hence, novel pharmacological strategies aimed at removing ONOO− might represent powerful therapeutic tools in the future. EcSOD, in preserving bioavailability of NO•, is expected to have an indirect consequential effect in cancer through alterations of these NO /ONOO− signaling. The seemingly paradoxical role of NO• in cancer has been quite extensively covered in other reviews [83] and is not discussed in detail here.
6. Role of superoxide versus H2O2 signaling in oncogenesis
Deregulation of redox homeostasis has long been implicated in a variety of diseases and the role of ROS in oncogenesis remains an area of major interest. Nevertheless, ROS should not be considered as one single biochemical entity that has a single global effect in cancer. Rather, ROS function as cellular secondary messengers, with each reactive species orchestrating unique signaling events, in a temporal and spatial manner. Although H2O2 has been the main focus as the ROS-mediated signaling molecule, partly due to the conception that it is more stable and longer lived than superoxide, the insight starts to emerge that O2•− may also be an important mediator of cellular effects. This is further supported by the fact that most cells possess enzymatic systems that are capable of producing O2•−, whereas to date no cellular system is known that exclusively generates H2O2 [84].
Superoxide is believed to function as a signaling molecule in a distinct manner from those mediated by H2O2, •OH, and ONOO−, although the mechanism is not fully understood. The name, superoxide is misleading in a sense that it is not a super-oxidant but a relatively moderate reductant. However, superoxide, being both a radical and an anion, can react with organic molecules by nucleophilic mechanism. Owing to this nucleophilic property, superoxide is able to rapidly deprotonize alcohols, phenols, and thiols, and hydrolyze esters as proposed [85]. By deprotonation of protein serine or threonine residues, superoxide is able to mediate phosphorylation of numerous proteins by protein kinases, thereby accelerating the rates of nucleophilic reaction between kinases and phosphorylating proteins. For examples, superoxide has been shown to mediate the activation of many protein kinases including PKC, PKD (protein kinase D), PKB (Akt) (protein kinaseB), and mitogen-activated (MAPK) kinases, p42/44, p38, and ERK [86–89]. Another important stimulus of enzymatic phosphorylation by superoxide signaling is via phosphatidylinositol 3-kinase activation, which subsequently activates PKB and MAPK [87 90].
Furthermore, O2•− can promote protein phosphorylation by inhibiting dephosphorylation catalyzed by protein phosphatases. Superoxide affects both serine/threonine protein phosphatases (PPs) and protein tyrosine phosphatases (PTPs), by oxidizing the metal ion center of the former class of phosphatases and via nucleophilic attack of the cysteine residue in the later class [91 92]. While H2O2 has also been demonstrated to inactivate PTPs, the rate of superoxide signaling is about 10–100 times higher than that of hydrogen peroxide signaling [93 94]. In addition to being kinetically more efficient, O2•− is chemically more specific than H2O2•− in this process as the catalytic site of PTP-1B is surrounded by positively charged residues [95]. Moreover, O2•−-inactivated PTP-1B is more reversible than that of H2O2•− since significantly more methionine residues are oxidized by H2O2. This provides an efficient fine-tuning ability of O2•− in regulating PTP-1B in signal transduction. This emphasizes an importance of O2•− signaling in many oncogenic signaling processes and the potential application of the specific superoxide inhibitors for their regulation.
It has long been recognized that low levels of O2•− and H2O2 are involved in proliferative signaling, partly via alterations in the activities of protein kinases and oxidative inactivation of phosphatases, as discussed earlier. Although these two major ROS are considered oncogenic ROS, there is strong evidence to support that these two ROS diverge in their roles in cellular survival/death pathways. Indeed, it is the ratio of intracellular superoxide to H2O2 that could dictate the fate of cells as discussed in detail by Pervaiz and Clement [96]. A prominent increase in superoxide in the absence of cytotoxic levels of H2O2 supports cell survival and promotes oncogenesis by inhibiting activation of the pro-apoptotic pathway [97 98]. In contrast, a rise in H2O2 levels with an accompanying decrease in superoxide facilitates apoptotic execution by activating caspase proteases [99 100]. Superoxide, due to its specific anti-apoptotic effect in creating an environment conducive for cellular survival and proliferation in favor of oncogenesis, has been termed “Onco-ROS” [96]. However, competition between superoxide and hydrogen peroxide in cells might be more complicated. The role of superoxide and its ROS derivatives in cellular outcome, i. e. cancer is clearly more diverse and complex than anticipated.
7. Gene Regulation and Transcription
Human SOD3 is located on chromosome 4p with approximately 5.9 kilobase pairs, and contains two exons and one intron [101]. The entirety of the 722 bp coding region is within exon 2. The promoter contains two CAAT-box elements but is without a TATA promoter sequence [101]. Features in common with the other two SOD family members include antioxidant response elements (ARE), AP-1 and AP-2 binding sites, and NF-κB motifs. A more recent study of EcSOD transcriptional regulation that looked at tissue-specific expression in the mouse revealed a repressor role for myeloid zinc finger 1 and Krüppel-like transcription factors, whereas Ets family members, Elf-1 and GA-binding protein α and β, were transcriptional activators [102]. The farnesoid x receptor was found to bind an inverted repeat, IR-1 element promoting transcription [103].
Various growth factors, cytokines and ions likely play a role in transcriptional control of EcSOD mRNA expression. EcSOD was reportedly induced in response to interferon-γ and IL-4, but downregulated by TNF-α [104]. Rat brain astrocytes were protected from H2O2 by purinergic receptor agonists, which increased expression of both MnSOD and EcSOD, an effect likely dependent on intracellular calcium ion, cyclic AMP and PKA activity [105]. Leukemia inhibitor factor increases EcSOD expression and activity in brain tissue, protecting neurons from ischemic damage [106]. Exendin-4, a glucagon-like peptide-1 receptor agonist, induced the expression of EcSOD through epigenetic regulation at its proximal promoter by influencing the acetylation of histone H3 [107]. Lastly, the presence of metal ions bear influence on modulating EcSOD expression. Dietary copper/zinc levels [108 109] influence EcSOD levels, and the intrinsic transcriptional activity of copper chaperone Atox1, can be stimulated to translocate to the nucleus and bind a response element in the EcSOD promoter [110]. Unexpectedly, the application of various oxidizing conditions reduced EcSOD expression levels, although these results may be confounded by general toxicity [111].
8. Sod3 KO Models
Whole body EcSOD null mice are born at Mendelian ratios, are fertile, appear to develop normally, and adult mice are healthy to 14 months of age [112]. Other antioxidant genes (catalase, glutathione peroxidase, glutathione reductase, glucose-6-phosphate dehydrogenase), were also reported as unchanged. Common hematological markers reported as normal. Significantly, Sod3−/− mice were more susceptible to oxidative stress resulting from hyperoxic exposure, with considerably diminished survival rates to wildtype counterparts. When exposed to hyperoxic conditions whole body KO mice developed severe and sudden, inflammatory pulmonary edema, with enhanced neutrophil recruitment, signs of intra-alveolar hemorrhage, vascular congestion and thickening of alveolar septa. The expression levels of the other two SOD family members were examined, but no compensatory changes were observed. Moreover, a double knockout model for both SOD1 and SOD3 reported mild phenotypes, suggesting limited or no functional redundancy exists between these two family members [113].
Whole body KO mice do not have increased tumor incidence. Possibly, these conditions are not ideal to study the role of EcSOD in tumorigenesis. In dramatic contrast to the whole body knockout, an inducible deletion of EcSOD in adult mice exhibited an 85% mortality rate in ambient air conditions, 3–7 days after tamoxifen injection, and demonstrated severe respiratory distress, highlighting the critical role of EcSOD at the oxygen interface in the lungs [114]. This suggests that animals with an embryonic deletion of EcSOD have compensated/adapted, and will not demonstrate accurate phenotypic identities as reflected upon deletion in adult mice. While the whole body KO model suggests loss of EcSOD expression alone is not an initiating factor in inducing tumorigenesis, reduced function of this antioxidant will likely contribute to oncogenesis as demonstrated by numerous studies further discussed in the following.
9. Expression levels and effects of EcSOD in cancers
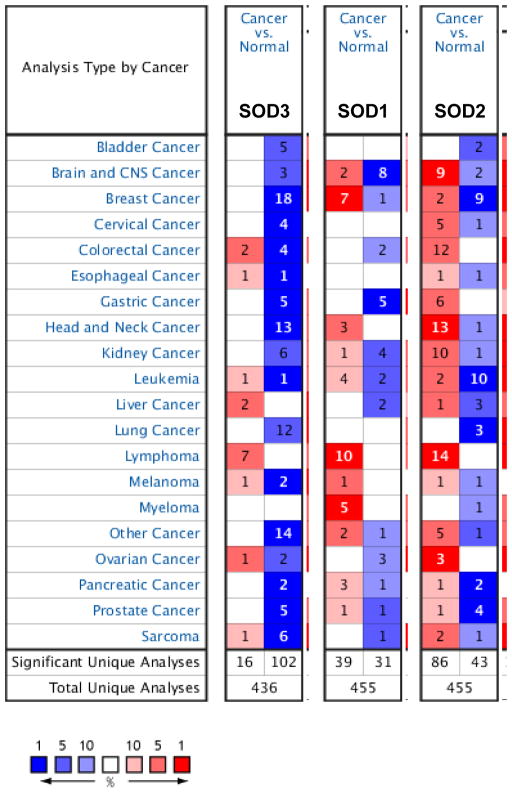
Oncomine analysis of neoplastic versus normal tissues showed that EcSOD (or SOD3) expression levels were significantly down-regulated across a majority of cancers including breast cancer, head and neck cancer, lung cancer, and sarcoma (Figure 2), suggesting that loss of EcSOD could contributes to oncogenesis. Amongst the Oncomine datasets, the breast cancer category shows the highest number of analyses that met the threshold, where 18 out of 53 analyses in 5 out of 14 breast cancer datasets met the thresholds for P-value < 0.01 and changes in EcSOD expression is scored in the top 10% of gene rank for most significantly under-expressed genes. Specific studies examining the expression level and function of EcSOD in various cancers are further discussed in the following.
Figure 2.
Oncomine gene summary view comparing the number of datasets that had significant changes in mRNA expression for EcSOD (SOD3), CuZnSOD (SOD2), and MnSOD (SOD2) in cancer versus normal tissues. Thresholds were set for P<0.01 and top 10% score in gene rank for most significantly changed genes. Blue boxes indicate downregulation and red represents upregulation. Number in each box shows the number of analyses that met the thresholds.
9.1 Breast cancer
In breast cancer cells, overexpression of EcSOD inhibited in vitro proliferation, clonogenic survival, and invasion of a triple negative breast cancer cell line partly via suppressing heparanase-mediated fragmentation of cell surface proteoglycans and by reducing VEGF bioavailability [115]. Overexpression of EcSOD also significantly inhibited tumor metastasis in both an experimental lung and a spontaneous metastasis mouse model [116], further suggesting a role for this extracellular enzyme as in suppressing tumor progression. Concurrently, in a normal mammary epithelial cell line, siRNA-mediated knockdown of EcSOD promoted clonogenic capacity, tumorsphere formation, and wound healing in MCF10A cells [117]. In addition to the Oncomine analysis showing a prominent down-regulation of EcSOD expression in breast carcinomas, an inverse correlation between the mRNA expression levels of EcSOD and breast cancer stage has been reported [116 118]. Immunohistochemistry data also show a significant decrease in EcSOD protein expression in both ductal carcinoma in situ and invasive breast carcinoma compared to normal tissue [116], further suggesting that loss of EcSOD provides a selective advantage in cancer cells. In contrast to the tumor suppressive role, EcSOD may be an important mediator in VEGF-C-promoted oncogenesis. Expression levels of EcSOD were found to be down-regulated when VEGF-C is knocked down in claudin-low breast cancers, and EcSOD was partly required for VEGF-C-mediated cell survival in response to oxidative stress and for VEGF-C-mediated metastasis [119]. This apparent discrepancy in the role of EcSOD in breast cancer hints at the complexity of redox-mediated cellular processes. It should be emphasized that these experiments were performed in a single murine mammary carcinoma cell line that constitutively expressed EcSOD. Since EcSOD expression is downregulated or absent in the majority of human breast cancer cases, the relevance of these studies with respect to the role of EcSOD in human breast cancer progression remain unclear.
Interesting, long-term estradiol stimulation resulted in significantly downregulated EcSOD in normal mammary epithelial cells, and in tumors derived from the ACI rat model of breast cancer [117 120 121]. The exact mechanism involved in the estrogen-mediated loss of EcSOD is not clear. The induction of neoplastic transformation by estrogen can be mediated through non-receptor alpha regulated mechanisms, via a direct genotoxic effect. Russo et al [122] found that loss of chromosome 4 is associated with a tumorigenic phenotype by using an in vitro transformation model of normal mammary epithelial cells treated with 17-beta estradiol to elucidate a sequence of chromosomal changes correlating with specific stages of neoplastic progression. The authors implicated Slit2 as a candidate tumor suppressor located at 4p15.2 that was silenced in the estrogen-induced tumors. However, considering the fact that the EcSOD gene or SOD3 is also located in the chromosome 4p15.2 region, it is tempting to speculate that re-expression of EcSOD in their C5 cells (tumorigenic cell line derived from estrogen treated MCF10A cells) would inhibit tumorigenicity. Chromosome 4p15.1-15.3 is one of the most commonly deleted region (57%) reported in breast cancer [123], which would deplete expression of EcSOD in these cancers.
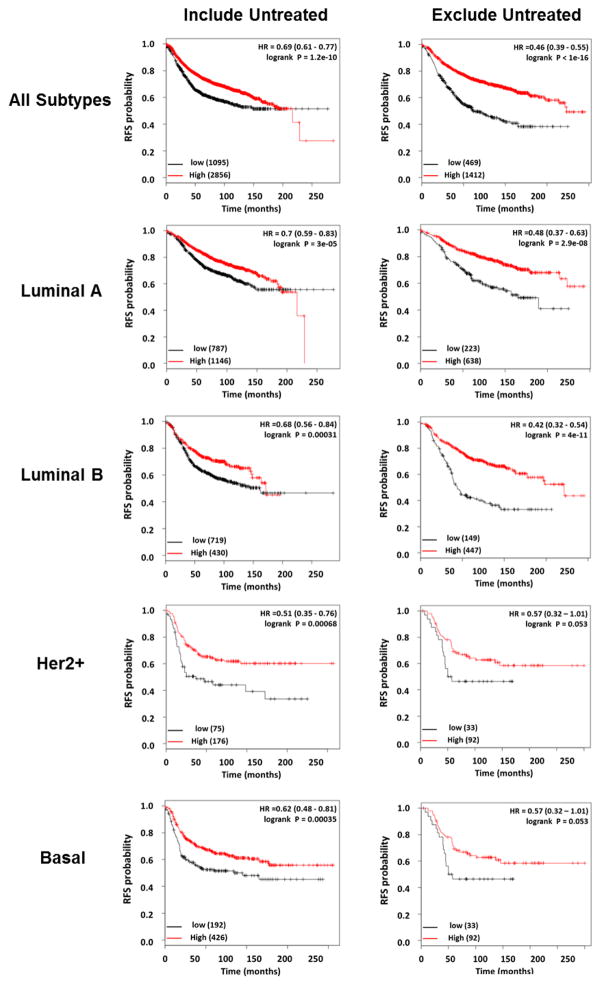
Loss of EcSOD expression, in addition to having a tumor promoting effect, also contributes to tumor recurrence and poor patient outcome. Local relapse remains a significant issue for breast cancer patients who have undergone breast conserving surgery [124]. In a murine 4T1 cytoreductive surgery model study aimed at identifying mechanisms driving local recurrence, EcSOD was found to be one of the top 40 genes underexpressed in the recurrent tumors versus primary tumors (fold change = 4.8) [125]. The recurrent tumors grew at a significantly accelerated rate compared to controls, suggesting that down-regulation of EcSOD is one of the contributing factors leading to tumor aggressiveness. More significantly, low expression levels of EcSOD are associated with reduced relapse-free survival in multiple subtypes of breast cancer. An integrative microarray data analysis using the Kaplan Meier Plotter [126] shows that low EcSOD expression is associated with significantly reduced relapse free survival in all breast cancers, as well as in Luminal A (ER+ and/or PR+, Her2−), Luminal B (ER+ and/or PR+, Her2+), Her2+, or basal-like (ER−, PR−, Her2−, CK 5/6+, and/or EGFR+) breast cancers (Figure 3). More profound changes in the hazard ratio (HR) for the association are seen when restriction was set to exclude untreated patients (systemic therapies) in the analysis (right panel). Taken together, all of the evidence discussed here underscores the importance of EcSOD as a potential tumor suppressor gene, inhibiting the progression of malignant phenotype in human breast cancer.
Figure 3.
Analysis of breast cancer data by Kaplan-Meier Plotter (http://kmplot.com) shows that low EcSOD expression is significantly associated with poor outcome (relapse free survival, RFS) in all types of breast cancer examined. Left panel shows analyses performed on all patients regardless of treatments while systemically untreated patients were excluded in analyses shown on the right. In red, patients with expression above the median and in black, patients with expressions below the median. The numbers of samples in each group are indicated in parentheses, and the hazard ratios (HR) and log rank p values are shown. Gene expression data and survival information are downloaded from GEO (Affymetrix HGU133A and HGU133+2 microarrays), EGA and TCGA.
9.2 Lung cancer
Despite the protective role of EcSOD in normal lung function, relatively little information describes its role in lung cancer. EcSOD expression is significantly down-regulated in primary human lung cancer compared to normal lung tissue, with further reduction occurring between stages I and IV [127–129]. Overexpression of this extracellular enzyme in lung cancer cells reduced clonogenic survival and invasion via inhibition of NF-κB activation [127 130], suggesting a tumor suppressive role of EcSOD in lung cancer. EcSOD may also play a role in gemcitabine resistance, as it is was identified amongst the gene set that was found to be up-regulated in resistant non-small cell lung cancer cell lines [131]. However, it is not known if EcSOD directly confers resistance of cells against gemcitabine or up-regulation of this gene is merely an indirect response to the cytotoxic effects of gemcitabine. In summation, EcSOD likely functions as a tumor suppressor and further investigation of its direct involvement in mediating sensitivity to gemcitabine would help to shed light on its role in drug resistance in lung cancer.
9.3 Prostate cancer
Confirming Oncomine analysis, IHC studies of prostate tissue revealed a significant reduction of EcSOD expression in cancer tissue compared to the normal counterparts [132]. Additionally, EcSOD levels and activity significantly decreased between high and intermediate grade prostate carcinomas, as well as in prostate cancer cell lines compared to normal prostate epithelial cells [133]. Migration and cell growth was also inhibited in a dose-dependent manner by overexpression of EcSOD or addition of recombinant human EcSOD [132]. The authors further showed that the inhibitory effects of EcSOD are due to reduced MMP2/MMP9 expression and activity, as well as increased H2O2 production. Other groups have since further correlated EcSOD expression with reduction of MMP2 activity and invasion in prostate cancer [133 134]. These studies highlight the role of this secreted antioxidant in regulating key extracellular enzymatic activities that promote invasion and metastasis.
9.4 Pancreatic cancer
EcSOD mRNA expression and IHC analysis reveal significantly decreased levels of EcSOD in tumor tissue compared to normal pancreatic ductal epithelium in paired and unpaired samples [135]. EcSOD loss is associated with a reduction in mean survival from 11.0 to 6.5 months in patients with pancreatic adenocarcinoma [135]. EcSOD over-expression inhibits in vitro cell proliferation, invasion, and clonogenic capacity of pancreatic cancer cells in a dose-dependent manner [135–137]. Furthermore, both transient and stable over-expression of EcSOD inhibited primary tumor growth and increased survival in tumor xenograft models [135–137]. EcSOD was also found to decrease levels of VEGF and HIF-1α protein levels in pancreatic cancer cell lines [136 137]. Since HIF-1α and VEGF promote angiogenesis, EcSOD is likely inhibitory, restricting blood flow to the tumor. However, EcSOD may also promote survival of quiescent pancreatic cancer cells as revealed by Deng et al [138] where they showed that Mirk/Dyrk1B kinase maintains the viability of quiescent pancreatic cancer cells by up-regulation of both CuZnSOD and EcSOD thereby lowering ROS levels in quiescent SU86.86 and Panc1 cells.
9.5 Thyroid cancer
Expression levels of EcSOD have been shown to be slightly increased in a benign thyroid tumor goiter model but gradually downregulated in cell lines that model advanced papillary and anaplastic thyroid cancers correlating with the level of Ras oncogene activation [139 140]. Although growth promoting effects of EcSOD at lower levels have been shown in a thyroid cancer cell line, PCCL3 [141 142], inhibitory effects of this antioxidant in cells harboring p53 mutations resulted in reductions in cell growth, invasion, and soft agar colony formation [143 144]. Thus, EcSOD may have biphasic effects on tumor progression switching from a tumor promoter during tumorigenesis to a tumor suppresser shortly after transformation. The authors further implied that disparate effects seen with EcSOD overexpression are due to the dose-dependent responses. Interestingly, high levels of EcSOD, although inhibited cellular proliferation were found to promote phosphorylation of various receptor tyrosine and non-receptor tyrosine kinases such as EGFR, ERBB2, and FLT-3 [140 143]. Propagation of cellular signaling was halted due to inhibition of small GTPases, (Ras, Rac, Rho, and CDC42), thereby decreasing activation of downstream effectors MEK and Erk [140 143]. Thus, EcSOD overexpression can promote cell cycle arrest and apoptosis via activation of p53 and reduce cell growth via promoting inactivation of GTPases. Intriguingly, secretion of EcSOD in the tumor stroma by mesenchymal stem cells (MSCs) increased the growth of thyroid cancer cells in co-culture models despite having an inhibitory effect on their migration (106). Further studies are needed to assess the extent and role of stromal-derived EcSOD versus cancer cell-expressed EcSOD in thyroid cancer.
9.6 Melanoma
Overexpression of EcSOD showed a prominent inhibitory effect in melanomas. Inhibition of cell proliferation by IFNγ was mediated through increased expression of EcSOD [145]. Moreover, muscle cell-mediated EcSOD secretion inhibited the growth of B16 melanoma in mice via a decrease in VEGF expression [146]. Similarly, EcSOD overexpressing transgenic mice demonstrated reduced growth of metastatic cancer cells despite no effect on infiltration to the lungs after tail vein injection [145]. In a DMPA/TPA-induced skin carcinogenic model, skin-specific EcSOD transgenic mice showed half the number of tumors compared with the nontransgenic mice by reducing DNA damage associated with the carcinogens [147]. A metabolomics and transcriptomics analysis however, revealed EcSOD as one of the up-regulated genes in tumors recovering from chemotherapy treatment [148]. This is likely due to the severe oxidative stress in both localized chemotherapy-treated and bystander tumors as suggested by the authors. Overall, EcSOD has a clear anti-proliferative and anti-tumor role in melanoma.
9.7 Additional Cancers
In addition to the cancer models described above, EcSOD expression is also decreased in colorectal cancer compared to paired normal controls [149]. In a liver cancer study, expression of EcSOD was increased by Farnesoid X receptor activity, which by inhibiting JNK activation, inhibited liver carcinogenesis, providing indirect evidence of the tumor suppressive role of EcSOD [103]. In renal cell carcinoma tissues, higher EcSOD expression correlated significantly with higher levels of apoptosis, as indicated by TUNEL staining [150]. Although down-regulation of EcSOD is reported in a majority of cancers, serum levels of EcSOD were observed to be increased in patients with gastric adenocarcinoma and prolactinoma, a benign pituitary gland tumor, compared to healthy controls [151 152]. However, the cause or role of increased serum EcSOD in either case remains unclear.
Other studies further indicate a potential role of EcSOD in therapy response and tumorigenesis. EcSOD expression can promote inhibition of radiation induced lung damage, such as myofibroblast expansion, oxidative stress, and fibrosis, via injection of mesenchymal stromal cells [153]. Additionally, EcSOD may play a role in obesity induced tumorigenesis, as EcSOD gene transfer in mice inhibited high fat diet-induced obesity and fatty liver [154]. It also reduced pro-inflammatory crowns, which are formed by the recruitment of macrophages to hypertrophic and necrotic adipose cells [154]. Both obesity and crown-like structures are associated with increased risk of breast cancer [155 156]. These studies highlight the potential role of EcSOD in protecting healthy tissue from chronic inflammation and reducing the risk of cancer.
10. Plasma EcSOD in cancer
Since EcSOD is a secreted protein, whether there is any correlations between the plasma levels of this antioxidant with cancer progression are of a high interest. Numerous studies have assessed the activity of serum/plasma SOD in cancer patients versus the normal patients but the results are controversial and largely inconclusive. For example, in breast cancer studies, plasma SOD activity has been found to be lower in patients with malignancy in comparison with the control group [157–159], while a reverse observation was reported showing higher SOD activities in patients with breast cancer versus the control patients [160–162]. Similar opposing results were also reported for prostate cancer [163 164]. Importantly, these studies relied on commercially available SOD activity kit assays which do not discern the 3 distinct forms of SODs. Specific activity of EcSOD can be further differentiated from the total SOD activities by including a Concanavalin A based purification step. However, since Concanavalin A only binds to the full length ECSOD, this will exclude the detection of the truncated form of EcSOD, and therefore may not be a reliable method to account for the total EcSOD activity levels. Nevertheless, since circulating levels of EcSOD could be contributed by other tissues and organs, assessing plasma levels of this antioxidant is likely not an accurate measurement of the specific expression levels of EcSOD in localized tumor tissues. Furthermore, EcSOD exists in both tissue-bound and freely circulating form due to its unique HBD. A significant portion of the full length EcSOD, despite having a strong affinity for negatively charged ECM molecules, is also released into the circulation. Various factors such as heparin, a commonly used blood-thinning agent, also influence this dynamic redistribution of EcSOD. Therefore, a snap-shot measurement of this extracellular antioxidant in the blood is likely not a true reflection of the total levels of EcSOD secreted by cancer cells. Perhaps determining the released form of EcSOD locally i.e. in nipple aspiration fluid (NAF) may provide a better assessment in the breast cancer model. The levels of SOD1 protein expression in NAF has been reported to be similar between normal patients and breast cancer patients [165]. Although one would speculate an alteration in the extracellular levels of EcSOD due to its secretory nature, levels of EcSOD in NAF of breast cancer patients remain to be evaluated.
11. Deregulation of EcSOD in cancers
The degree of difference in EcSOD expression in cancer versus normal cells/tissues is more pronounced and prevalent than for other SODs as shown in Oncomine analysis (Figure 2). Down-regulation of EcSOD expression in cancer has been associated with epigenetic silencing, up-regulation of oncomir microRNA-21, Ras oncogene-mediated gene silencing, chronic estrogen-induced gene suppression, single nucleotide polymorphisms, DNA copy number variation, and loss of heterozygosity. All of these observations imply that deregulation of EcSOD expression, distribution, or function has a clinical significance. In view of predominantly down-regulated EcSOD expression in a majority of cancers and a tumor suppressive role as supported by a large number of in vitro and in vivo models, understanding the mechanisms involved in deregulation of its expression could provide a tool towards therapeutic interventions. The mechanisms known to regulate EcSOD expression in cancers is discussed in the following sections.
11.1 Epigenetic
One of the most well studied mechanisms regulating EcSOD silencing is DNA methylation. EcSOD lacks a standard CpG island but contains a cluster of 18 CpG sites surrounding the transcriptional start site (−550 bp upstream to 100 bp downstream) with known transcription factor binding sites, such as Sp1/Sp3. SOD3 CpG sites have been reported to be hypermethylated in tumor tissue from gallbladder, liver, prostate, lung, and breast cancer samples [116 127 166–168]. Furthermore, SOD3 promoter hypermethylation correlated with decreased mRNA expression indicating epigenetic silencing via promoter DNA methylation [166]. SOD3 is also hypermethylated and downregulated in other diseases, such as coronary artery disease [169]. Highlighting the functional role of epigenetic silencing of EcSOD, treatment with 5-aza-2′-deoxycytidine (5-aza-dC), an inhibitor of DNA methylation increased EcSOD expression in both normal and cancer cells [116 127 133 170–172]. This methyltransferase inhibitor increased DNA accessibility via nucleosome remodeling thereby increasing RNA polymerase II and Sp3 binding to the SOD3 promoter [172]. In Ras-driven thyroid cancers, loss of EcSOD expression has also been shown to be affected with 5-aza-dC treatment, where mutant H-RasV12-mediated suppression of EcSOD was reverted [140].
Interestingly, methylation status of the EcSOD promoter can be influenced by the presence of extracellular matrix (ECM) or Matrigel in culture. While significant expression of EcSOD is detected in mammary epithelial cells in normal tissues, once the human mammary epithelial cells (HMECs) were isolated and plated as monolayer cells, there was a progressive loss of EcSOD mRNA expression as the cells were passaged without the ECM stimuli [116]. This was due to hypermethylated promoter of EcSOD when cells lose their polarity and acinari architecture. On the other hand, restoring the three dimensional (3D) acinar morphogenesis by culturing the HMEC cells in Matrigel induces re-expression of EcSOD and its promoter region became largely unmethylated [116]. Similarly, the expression pattern of another mammary-tissue specific gene, milk casein has also been shown to be regulated in this manner, where cell culture content and context can dictate gene expression via epigenetic mechanisms [173 174]. Interestingly, the change to 3D culture did not restore EcSOD expression in the breast cancer cells, presumably due to inability to form normal acini but instead displayed an disorganized stellate growth in 3D culture [116]. These studies reveal a novel process by which ECM integrates structure and function in mammary epithelial cells through alterations of chromatin structure and epigenetic codes.
Additionally, SOD3 methylation patterns may be impaired via a reduced ability to remove DNA methylation. DNA methylation removal is initiated by the ten-eleven translocation (Tet) family members, which are dioxygenases of 5-methylcytosine (5-mC). EcSOD expression is positively correlated with Tet1 expression in several tumor cell types, where Tet1 expression increases EcSOD expression via de-methylation of its CpG sites [175]. Interestingly, Tet1 is often downregulated in several types of solid cancer, such as breast, lung, colorectal, and gastric cancer [176], suggesting a potential mechanism of EcSOD silencing in cancer via downregulation of Tet1 [175].
Alterations to the glutamate carboxypeptidase II (GCPII) also modify methylation patterns resulting in changes in EcSOD expression. GCPII promotes folate uptake, which promotes DNA methylation by regenerating the methyl donor S-adenosyl methionine. In prostate cancer, EcSOD expression correlates with mutations in GCPII [167]. The GCPII D191V mutation promotes SOD3 hypermethylation and is associated with an increased risk of breast cancer [167 177]. Additionally, the GCPII H475Y variant is associated with decreased SOD3 methylation and decreased risk of breast and prostate cancer [167 178]. These studies imply an interplay between EcSOD and GCPII variants associated with cancer risk via changes in the folate cycle and DNA methylation status of EcSOD.
Histone modifications and histone variants can also play a pivotal role in epigenetic regulation. Histone acetylation typically promotes gene expression by relaxing the condensed nucleosome complex allowing for easier access to genes. Inhibitors against histone acetyltransferases (HATs), such as GCN5, p300, and PCAF, prevented EcSOD increases in a stimulated monocytic leukemia cell line, THP-1 [170]. Additionally, several studies have demonstrated increased EcSOD expression with histone deacetylase (HDAC) inhibitors [116 170 171 179 180]. Specifically, inhibition or knockdown of HDAC3 led to increased EcSOD expression in pulmonary artery smooth muscle cells [179]. HDAC3 is up-regulated in many solid cancers, such as lung, breast, pancreatic, liver, and colon, indicating another potential mechanism of EcSOD silencing in cancer [181–185]. Although, HDAC inhibition may also indirectly increase EcSOD expression via loss of thyroid stimulating hormone autocrine signaling [140 142], these studies indicate that decreased histone acetylation can contribute to silencing of EcSOD in cancer.
Not only are histone modifications able to modify EcSOD expression, but changes in histone variants have recently been shown to play a role in expression of this antioxidant. Histone variants mediate a variety of functions, such as modifying expression, controlling chromatin condensation, sensing DNA damage, and controlling the cellular response toward DNA damage repair or apoptosis [186]. MacroH2A isoforms are unique H2A histone variants due to the presence of a 30-kDa non-histone domain (macro domain) at their C-termini. MacroH2A variants are generally considered transcriptionally repressive in nature due to their association with forms of condensed chromatin such as the inactive X chromosome (Xi) and inactive genes [187 188]. MacroH2A1, a variant in the H2A family, has dramatic effects on cancer progression dependent on its splice variant expression. The splice variant, macroH2A1.1, inhibits proliferation, invasion, migration, and is associated with better prognosis, while macroH2A1.2 is associated with cancer progression [186]. Interestingly, altered RNA splicing via increases in RNA helicase, Ddx5 and Ddx17 promoted macroH2A1.2 which resulted in suppression of EcSOD expression in a mouse mammary tumor cell line, 4T1 [189]. These macroH2A1 splice variants had opposing effects on SOD3 expression. MacroH2A1.1 increased SOD3 expression in 4T1 cells, while macroH2A1.2 decreased its expression [189]. Ddx5 and/or Ddx17 is overexpressed in a variety of solid cancers, such as breast, colon, prostate, non-small cell lung, head and neck, glioma, and leukemia [190–196]. Therefore, increases in Ddx5 and Ddx17 leading to higher macroH2A1.2 levels are an additional mechanism of EcSOD down-regulation in cancer.
11.2 Single Nucleotide Polymorphisms
Alterations of EcSOD expression, tissue distribution, and/or function can also occur via single nucleotide polymorphisms (SNPs). Here, we will highlight the research into EcSOD SNPs and their effect on cancer risk and progression. These SNPs include rs1799895, rs2536512, rs2284659, and rs699473 as shown in Table 1.
Table 1.
Summary of EcSOD SNPs with associated cancer risk.
| SNP | Location | Mutation | Effect on Cancer |
|---|---|---|---|
| rs1799895 (Ex3-631C>G) | Coding region | C→G; R213G | Decreased risk of lung cancer [198] Increased number of lesions in metastatic gastric cancer [200] |
| rs2536512 | Coding region | A→G; A58T | Increased risk of hepatocellular carcinoma [204] Increased risk of glioma [205] Associated with estrogen/progesterone receptor expression in breast cancer [118] |
| rs2284659 | Promoter | G→T | In a 4 SNP set that results in increased risk of breast cancer [209] |
| rs699473 (IVS1+186C>T) | Promoter; AhR-XRE binding site, CpG Island | C→T | Enriched in high-grade prostate cancer [211] Decreased progression free survival in breast cancer [118] |
The most extensively studied SNP is rs1799895, which occurs in the protein coding region and results in an arginine to glycine mutation at position 213 (R213G). This missense mutation occurs within the heparin binding domain of EcSOD drastically reducing the binding of EcSOD to heparin, as a result increasing circulating EcSOD [48]. The R213G mutation also inhibits the ability of EcSOD to bind lipoprotein receptor-related protein (LRP), which promotes its uptake by LRP-mediated endocytosis and eventual clearance by the liver [51]. As a result, patients with R213G have a ~10-fold increase in plasma EcSOD activity levels [48]. It is also associated with decreased development of chronic obstructive pulmonary disease (COPD) in smokers [197]. Meta-analysis of many studies indicate smokers with COPD are associated with an increased risk of lung cancer [198]. These studies indicate it may provide protection to smokers from lung cancer development. Indeed, the R213G SNP in lung cancer is associated with a protective phenotype as it was enriched in healthy control smokers compared to lung cancer patients [199]. It is also associated with the number of lesions in metastatic gastric cancer [200]. Although, in a large study assessing the role of R213G SNP within the Danish population, the mutation was found to have no effect on the overall risk of cancer [201]. Despite the dramatic change in localization of SOD3 caused by the R213G mutation, these studies suggest that it likely has no effect on cancer risk, except for a potential protective role in lung cancer.
The SNP, rs2536512, similarly occurs within the coding region of SOD3 and results in a missense mutation of alanine to threonine at residue 58 (A58T). This mutation occurs within the oligomerization domain of EcSOD and is hypothesized to play a role in protein tetramerization. There are conflicting results about its effect on plasma EcSOD activity [202 203]. However, to date there is little known about the effect of the A58T mutation on EcSOD activity, localization, or oligomerization. In cancer, it is associated with increased risk of hepatocellular carcinoma and glioma [204 205]. Similarly, it is correlated with estrogen and progesterone receptor expression in breast cancer patients [118]. The A58T mutation also increases risk of other diseases, such a cerebral infarction in women, type 2 diabetes mellitus, and diminished lung function in children [206–208]. The functional effect of this mutation remains unclear and requires further study to fully understand its role in cancer.
The SNP rs2284659 occurs within the SOD3 promoter. Due to its location in the promoter, it may modify expression of EcSOD. However, the effect of this SNP on EcSOD expression is unclear as it does not occur within any known transcription factor binding domains. In cancer, the SNP rs2284659 is associated with an increase in breast cancer incidence [209]. It is also associated with higher plasma levels of EcSOD in diabetic patients [210]. However, the mechanism behind the increase in patients with the SNP rs2284659 remains unclear.
Another EcSOD SNP shown to affect cancer risk is rs699473. This SNP occurs within the aryl hydrocarbon receptor-xenobiotic response element (AhR-XRE) binding motif of the EcSOD promoter. The mutation causes reduced binding capacity of nuclear proteins and alteration of a CpG site potentially perturbing DNA methylation at this location [127 206]. The resulting effect on EcSOD expression remains unclear. In cancer, the SNP rs699473 is associated with increased risk of high-grade prostate cancer [211]. Interestingly, this SNP was also found to protect patients from increased risk of high grade prostate cancer after selenium supplementation in the Selenium and Vitamin E Cancer Prevention Trial (SELECT) [212]. In breast cancer, SNP rs699473 is associated with a significant reduction in the progression-free survival in breast cancer compared to patients with wild type EcSOD [118]. This SNP has also been linked to an increased risk of adult brain cancer, specifically meningioma and possibly glioma [213].
11.3 Loss of Heterozygosity
SNPs are not the only genetic abnormality regulating EcSOD in cancer. Loss of heterozygosity (LOH), also known as allelic deletion, is the process by which a cell deletes one of the two copies of a gene. Allelic deletion in cancer cells results in hemizygotes containing only one parental copy of that gene. As EcSOD has several SNPs, it is likely that parentally inherited SNPs of EcSOD exist in a heterozygous state. Therefore, LOH may help expose functional roles of SNPs in EcSOD previously masked by the deleted allele. In addition to unmasking SNP phenotypes, allelic deletion often results in reduced gene expression. Several reports indicate the SOD3 gene, located on chromosome 4p15.3-4p15.1, is a hotspot for LOH in cancer. The deletion of chromosome 4p15.1-15.3 has been observed in many types of solid cancers, such as cervical, breast, head and neck, liver, colorectal, lung, and bladder [123 127 214–222]. These loses range from 30% in bladder cancer up to 60% in lung cancer [219 222]. LOH of this region increases dramatically between cervical intraepithelial neoplasia and grade I cervical cancer suggesting that allelic deletion occurs early in tumorigenesis [214]. LOH plays a role in EcSOD suppression in the lung cancer cell line, A549 indicating both LOH and DNA methylation mediate EcSOD silencing in these cells [127]. Moreover, 17β-estradiol induced transformation of the normal breast epithelial cell line, MCF10F, resulted in loss of chromosome 4p [223]. Thus, indicating a potential role of estrogen in EcSOD deletion in breast cancer.
11.4 miRNA
Expression of EcSOD can also be regulated by microRNA, which are a class of small (~22 nucleotides) non-coding RNAs that negatively regulate gene expression post-transcriptionally [224]. Currently, miR-21 is the only known microRNA to directly target EcSOD, as it binds a 3′ UTR site in EcSOD mRNA [225]. MiR-21 is considered an oncomir and is upregulated in both leukemias and solid cancers of the lung, breast, prostate, pancreas, stomach, colon, ovaries, cervix, and thyroid [226–230]. MiR-21 has diagnostic and prognostic value, as its expression typically increases with tumor grade [230]. Other targets of miR-21 are tropomyosin 1 (TPM-1), programmed cell death protein 4 (PCDCD4), reversion-inducing cysteine-rich protein with kazal (RECK), maspin, NFIB, Sporouty2, and PTEN [230]. Overexpression of miR-21 in non-transformed immortalized human bronchial epithelial cells (NL20) reduced EcSOD levels and increased transformation as shown via colony formation in soft agar [225]. Transformation was blunted by re-expression of EcSOD, supporting a tumor suppressive role for EcSOD [225]. Additionally, miR-21 levels were significantly increased after >10 fold increases in Ras activity via expression of mutant H-RasV12 leading to decreased EcSOD expression [140]. Therefore, cancer cells can also silence EcSOD expression via upregulation of the oncomir miR-21.
12. Conclusion
In conclusion, the preponderance of studies have demonstrated tumor suppressive effects of EcSOD in cancers with a few exceptions. A recent review makes the case for EcSOD as a growth promoting enzyme during the early stages of tumorigenesis based on EcSOD overexpression promoting mouse embryonic fibroblasts transformation [231]. However, EcSOD inhibited tumorigenesis in a chemically induced melanoma model using EcSOD overexpressing transgenic mice [147]. Therefore, further studies are required to fully elucidate the role of EcSOD during tumorigenesis. The role of EcSOD in established tumor progression is more clearly understood as vast majority of evidence indicates EcSOD is downregulated in established tumors and low expression of this antioxidant is significantly associated with poor outcome in breast cancer and pancreatic cancer patients. Inhibition of cancer cell growth and metastasis in EcSOD overexpression studies also supported its tumor suppressor function. The vast array of mechanisms involved in deregulating EcSOD further suggest its loss is not an artifact or merely casually associated with cancer progression. Future studies into the mechanism of action are required to elucidate remaining apparent discrepancies in the role of EcSOD during tumorigenesis and in therapeutic responses of different cancers.
The degree of difference between cancer cells and the normal counterparts in their metabolic and oxidative status provides a therapeutic rational to exploit these characteristics. Currently, the redox approach to cancer therapy has focused on two main paths. One approach aims at promoting ROS level beyond the threshold in cancer cells, while the other takes the opposite direction of scavenging oncogenic superoxide radical. Since SOD is the only enzyme that scavenges superoxide, an essential primary step in eliminating the toxic free radical and its derivatives, SOD is an attractive target for pharmacological intervention. Application of exogenous SODs would exhibit clinical limitations and a variety of innovative approaches should be explored. Among these are SOD mimetics, which are reviewed here [232]. Based on recent cancer clinical trials showing mostly underwhelming performance of antioxidant supplements, it is premature to use SOD mimetics for cancer therapy. Preclinical models however, showed that the use of these mimetics provides a greater potential when used in combination with chemotherapies or with radiotherapy, than its use as a single anti-cancer agent [232]. Another strategy to restore EcSOD activity in cancer cells is by an epigenetic approach. Inhibitors of histone deacetylases have been synthesized and tested in numerous clinical trials, although the lack of their selectivity often yields favorable results only in a small set of patients. Of interesting note, the unique HBD of EcSOD could be further exploited as a cell penetrating drug-delivery tool.
As an extracellular protein, EcSOD also plays an important role in ECM remodeling. The ECM not only provides the structural foundation for tissue function and mechanical integrity, it regulates the availability of growth factors and cytokines to direct cell growth, survival, migration, differentiation, and immune function. Deregulated ECM dynamics in the molecular etiology of cancer development and progression has been well established [233]. Negatively charged ECM components, such as heparan sulfate, collagen type I, and fibulin-5, which are sensitive to oxidative fragmentation, are known to form ionic interactions with the basic residues in the HBD of EcSOD [33 39–41]. Numerous reports have demonstrated the protective function of EcSOD against ROS/RNS-induced fragmentation of heparan sulfate, elastin, collagen, and hyaluronan in suppressing oxidative-mediated lung injuries [56 234 235], pointing to a potential role of EcSOD in regulating the ECM dynamics in cancer. In support of this, a breast cancer study showed that overexpression of EcSOD prevented oxidative-mediated heparan sulfate cleavage from cell surfaces [115]. Although the mechanism remains to be elucidated, a down-regulation of heparanase expression is associated with EcSOD in these breast cancer cells. Heparanase is elevated in a wide variety of human cancer cells, including breast cancer. By degrading the constituents of basement membranes and ECM, heparanase participates in promoting invasion of cancer cells into the underlying stroma and metastasis to distal sites via vascular and lymphatic routes [236]. In addition, EcSOD has been shown to inhibit metalloproteinases (MMPs), which is another major determinant of the ECM dynamics. In a prostate cancer study, by scavenging O2•− and preventing the oxidation of NO• into ONOO−, EcSOD inhibited MT1-MMP activity and subsequently reduced MMP2 activation [132–134]. Considering the fact that the extracellular microenvironment of most cells is exposed to higher degrees of oxidative insult than the intracellular environment, due to limited amounts of secreted antioxidant enzymes, a down-regulation of EcSOD expression or activity would promote cancer-associated ECM remodeling.
Due to its secreted nature and the ability to be transported to recipient cells through its binding with heparin sulfate proteoglycans on the cell surface, its expression in the surrounding tumor stroma needs to be addressed. Much focus has been placed in determining the role of EcSOD in neoplastic epithelial cells but little is known about this antioxidant enzyme in the tumor stroma. A recent report showed that mesenchymal stem cells could be an important source of EcSOD secretion in thyroid tumors. In breast cancer, down-regulation of EcSOD protein expression was observed in the tumor stroma when compared to the normal stroma (unpublished data). The implication of this reduction is not clear at this point although understanding the contributing role of stroma EcSOD would provide a window into the cross talks between cancer cells and their stromal cell partners. In view of its ability to regulate receptor tyrosine kinase signaling, which suggests that EcSOD can regulate heterotypic cell-cell interactions, the role of EcSOD in regulating cancer-stroma interactions is an additional area of interest. This is particularly important considering the oxidative nature of tumor microenvironment in most solid tumors. Since EcSOD is an extracellular protein, it is possible that the effect in cancer is mediated through the tumor microenvironment, explaining why downregulation of EcSOD promotes cancer metastasis, which is in contrast to some studies reported for CuZnSOD and MnSOD.
Furthermore, the oxidative tumor microenvironment can play a role in the tumor immune response, as EcSOD can regulate both innate and adaptive immune cell infiltration and function. Macrophage infiltration, which is correlated with poor prognosis in cancer [237], is reduced with EcSOD overexpression in acute inflammation mouse models [238]. In addition to its role in the innate immune system, EcSOD selectively inhibited Th2 and Th17 differentiation with no effect on Th1 [239], which are implicated in tumor progression [240]. Additionally, macrophage-derived superoxide induces Treg formation [241], indicating EcSOD may serve a potential inhibitory role for Tregs in cancer. With the rapid expansion of the tumor immunology field due to limited yet promising efficacy of targeted immunotherapies, further studies into the role of EcSOD on the tumor immune response, such as its effects on the PD1/PDL1 checkpoint pathway, are required due to its unique ability to regulate the oxidative tumor microenvironment and its potential for combinatorial treatment with immunotherapies.
Highlights.
Expression levels of EcSOD are significantly downregulated across a vast majority of cancers.
Overexpression of EcSOD inhibits cancer cell growth, invasion, and metastasis in numerous in vitro and in vivo studies, suggesting a tumor suppressive role.
Low EcSOD expression confers poor prognosis in breast cancer and pancreatic cancer patients.
Deregulation of EcSOD expression, distribution, or function in cancer has been associated with a vast array of genetic and epigenetic mechanisms.
Acknowledgments
This work was financially supported by grants from the NIH R01 CA115438 (FE Domann), NIH R0I-CA182086A (Teoh-Fitzgerald), Redox Biology Pilot Project Fund (NTSBRDF, Uni. of Nebraska, Lincoln) (Teoh-Fitzgerald), and Nebraska Department of Health and Human Services Award LB506 (Teoh-Fitzgerald). Brandon Griess was supported by the Eppley Institute in Cancer Biology Training Grant (NCI T32CA009476).
Footnotes
The authors declare no potential conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones DP. Redox theory of aging. Redox biology. 2015;5:71–9. doi: 10.1016/j.redox.2015.03.004. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgoyne JR, Mongue-Din H, Eaton P, Shah AM. Redox signaling in cardiac physiology and pathology. Circulation research. 2012;111(8):1091–106. doi: 10.1161/circresaha.111.255216. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 3.Lei Y, Wang K, Deng L, Chen Y, Nice EC, Huang C. Redox regulation of inflammation: old elements, a new story. Medicinal research reviews. 2015;35(2):306–40. doi: 10.1002/med.21330. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 4.Kovacic P, Somanathan R. Redox processes in neurodegenerative disease involving reactive oxygen species. Current neuropharmacology. 2012;10(4):289–302. doi: 10.2174/157015912804143487. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chio IIC, Tuveson DA. ROS in Cancer: The Burning Question. Trends in molecular medicine. 2017;23(5):411–29. doi: 10.1016/j.molmed.2017.03.004. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nature reviews Molecular cell biology. 2007;8(10):813–24. doi: 10.1038/nrm2256. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 7.Wani R, Nagata A, Murray BW. Protein redox chemistry: post-translational cysteine modifications that regulate signal transduction and drug pharmacology. Frontiers in pharmacology. 2014;5:224. doi: 10.3389/fphar.2014.00224. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickinson BC, Chang CJ. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nature chemical biology. 2011;7(8):504–11. doi: 10.1038/nchembio.607. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim JB, Langford TF, Huang BK, Deen WM, Sikes HD. A reaction-diffusion model of cytosolic hydrogen peroxide. Free Radic Biol Med. 2016;90:85–90. doi: 10.1016/j.freeradbiomed.2015.11.005. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 10.Adimora NJ, Jones DP, Kemp ML. A model of redox kinetics implicates the thiol proteome in cellular hydrogen peroxide responses. Antioxid Redox Signal. 2010;13(6):731–43. doi: 10.1089/ars.2009.2968. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bienert GP, Chaumont F. Aquaporin-facilitated transmembrane diffusion of hydrogen peroxide. Biochimica et biophysica acta. 2014;1840(5):1596–604. doi: 10.1016/j.bbagen.2013.09.017. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 12.Appenzeller-Herzog C, Banhegyi G, Bogeski I, et al. Transit of H2O2 across the endoplasmic reticulum membrane is not sluggish. Free Radic Biol Med. 2016;94:157–60. doi: 10.1016/j.freeradbiomed.2016.02.030. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 13.Zeeshan HM, Lee GH, Kim HR, Chae HJ. Endoplasmic Reticulum Stress and Associated ROS. International journal of molecular sciences. 2016;17(3):327. doi: 10.3390/ijms17030327. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marklund SL, Holme E, Hellner L. Superoxide dismutase in extracellular fluids. Clinica chimica acta; international journal of clinical chemistry. 1982;126(1):41–51. doi: 10.1016/0009-8981(82)90360-6. [DOI] [PubMed] [Google Scholar]
- 15.Marklund SL. Extracellular superoxide dismutase in human tissues and human cell lines. J Clin Invest. 1984;74(4):1398–403. doi: 10.1172/jci111550. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33(3):337–49. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 17.Brigelius-Flohe R, Flohe L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal. 2011;15(8):2335–81. doi: 10.1089/ars.2010.3534. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins Y, Chouchani ET, James AM, Menger KE, Cocheme HM, Murphy MP. Mitochondrial redox signalling at a glance. Journal of cell science. 2012;125(Pt 4):801–6. doi: 10.1242/jcs.098475. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 19.Mittal M, Siddiqui MR, Tran K, Reddy SP, Malik AB. Reactive oxygen species in inflammation and tissue injury. Antioxid Redox Signal. 2014;20(7):1126–67. doi: 10.1089/ars.2012.5149. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim A. Modulation of MnSOD in Cancer:Epidemiological and Experimental Evidence. Toxicological research. 2010;26(2):83–93. doi: 10.5487/tr.2010.26.2.083. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhar SK, St Clair DK. Manganese superoxide dismutase regulation and cancer. Free Radic Biol Med. 2012;52(11–12):2209–22. doi: 10.1016/j.freeradbiomed.2012.03.009. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 22.Papa L, Manfredi G, Germain D. SOD1, an unexpected novel target for cancer therapy. Genes & cancer. 2014;5(1–2):15–21. doi: 10.18632/genesandcancer.4. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicks CL. Occurrence and consequence of superoxide dismutase in milk products: a review. Journal of dairy science. 1980;63(7):1199–204. doi: 10.3168/jds.S0022-0302(80)83065-7. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 24.Sandstrom J, Karlsson K, Edlund T, Marklund SL. Heparin-affinity patterns and composition of extracellular superoxide dismutase in human plasma and tissues. The Biochemical journal. 1993;294( Pt 3):853–7. doi: 10.1042/bj2940853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen DA, Petersen SV, Oury TD, et al. The intracellular proteolytic processing of extracellular superoxide dismutase (EC-SOD) is a two-step event. J Biol Chem. 2004;279(21):22152–7. doi: 10.1074/jbc.M401180200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 26.Chu Y, Piper R, Richardson S, Watanabe Y, Patel P, Heistad DD. Endocytosis of extracellular superoxide dismutase into endothelial cells: role of the heparin-binding domain. Arteriosclerosis, thrombosis, and vascular biology. 2006;26(9):1985–90. doi: 10.1161/01.ATV.0000234921.88489.5c. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 27.Combelles CM, Holick EA, Paolella LJ, Walker DC, Wu Q. Profiling of superoxide dismutase isoenzymes in compartments of the developing bovine antral follicles. Reproduction. 2010;139(5):871–81. doi: 10.1530/rep-09-0390. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ookawara T, Kizaki T, Takayama E, et al. Nuclear translocation of extracellular superoxide dismutase. Biochem Biophys Res Commun. 2002;296(1):54–61. doi: 10.1016/s0006-291x(02)00804-5. [DOI] [PubMed] [Google Scholar]
- 29.Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med. 2003;35(3):236–56. doi: 10.1016/s0891-5849(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 30.Oury TD, Crapo JD, Valnickova Z, Enghild JJ. Human extracellular superoxide dismutase is a tetramer composed of two disulphide-linked dimers: a simplified, high-yield purification of extracellular superoxide dismutase. The Biochemical journal. 1996;317( Pt 1):51–7. doi: 10.1042/bj3170051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fattman CL, Enghild JJ, Crapo JD, Schaefer LM, Valnickova Z, Oury TD. Purification and characterization of extracellular superoxide dismutase in mouse lung. Biochem Biophys Res Commun. 2000;275(2):542–8. doi: 10.1006/bbrc.2000.3327. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 32.Due AV, Petersen SV, Valnickova Z, et al. Extracellular superoxide dismutase exists as an octamer. FEBS Lett. 2006;580(5):1485–9. doi: 10.1016/j.febslet.2006.01.081. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hjalmarsson K, Marklund SL, Engstrom A, Edlund T. Isolation and sequence of complementary DNA encoding human extracellular superoxide dismutase. Proc Natl Acad Sci U S A. 1987;84(18):6340–4. doi: 10.1073/pnas.84.18.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stromqvist M, Holgersson J, Samuelsson B. Glycosylation of extracellular superoxide dismutase studied by high-performance liquid chromatography and mass spectrometry. Journal of chromatography. 1991;548(1–2):293–301. doi: 10.1016/s0021-9673(01)88611-8. [DOI] [PubMed] [Google Scholar]
- 35.Edlund A, Edlund T, Hjalmarsson K, et al. A non-glycosylated extracellular superoxide dismutase variant. The Biochemical journal. 1992;288( Pt 2):451–6. doi: 10.1042/bj2880451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ota F, Kizuka Y, Kitazume S, Adachi T, Taniguchi N. N-Glycosylation is essential for the secretion of extracellular superoxide dismutase. FEBS Lett. 2016;590(19):3357–67. doi: 10.1002/1873-3468.12378. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 37.Tibell L, Aasa R, Marklund SL. Spectral and physical properties of human extracellular superoxide dismutase: a comparison with CuZn superoxide dismutase. Archives of biochemistry and biophysics. 1993;304(2):429–33. doi: 10.1006/abbi.1993.1371. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 38.Marklund SL. Human copper-containing superoxide dismutase of high molecular weight. Proc Natl Acad Sci U S A. 1982;79(24):7634–8. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adachi T, Marklund SL. Interactions between human extracellular superoxide dismutase C and sulfated polysaccharides. J Biol Chem. 1989;264(15):8537–41. [PubMed] [Google Scholar]
- 40.Petersen SV, Oury TD, Ostergaard L, et al. Extracellular superoxide dismutase (EC-SOD) binds to type i collagen and protects against oxidative fragmentation. J Biol Chem. 2004;279(14):13705–10. doi: 10.1074/jbc.M310217200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen AD, Itoh S, Jeney V, et al. Fibulin-5 is a novel binding protein for extracellular superoxide dismutase. Circulation research. 2004;95(11):1067–74. doi: 10.1161/01.res.0000149568.85071.fb. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 42.Adachi T, Yamada H, Futenma A, Kato K, Hirano K. Heparin-induced release of extracellular-superoxide dismutase form (V) to plasma. Journal of biochemistry. 1995;117(3):586–90. doi: 10.1093/oxfordjournals.jbchem.a124748. [DOI] [PubMed] [Google Scholar]
- 43.Bowler RP, Nicks M, Olsen DA, et al. Furin proteolytically processes the heparin-binding region of extracellular superoxide dismutase. J Biol Chem. 2002;277(19):16505–11. doi: 10.1074/jbc.M105409200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 44.Karlsson K, Sandstrom J, Edlund A, Marklund SL. Turnover of extracellular-superoxide dismutase in tissues. Lab Invest. 1994;70(5):705–10. [PubMed] [Google Scholar]
- 45.Oury TD, Day BJ, Crapo JD. Extracellular superoxide dismutase: a regulator of nitric oxide bioavailability. Lab Invest. 1996;75(5):617–36. [PubMed] [Google Scholar]
- 46.Antonyuk SV, Strange RW, Marklund SL, Hasnain SS. The structure of human extracellular copper-zinc superoxide dismutase at 1. 7 A resolution: insights into heparin and collagen binding. Journal of molecular biology. 2009;388(2):310–26. doi: 10.1016/j.jmb.2009.03.026. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 47.Karlsson K, Edlund A, Sandstrom J, Marklund SL. Proteolytic modification of the heparin-binding affinity of extracellular superoxide dismutase. The Biochemical journal. 1993;290( Pt 2):623–6. doi: 10.1042/bj2900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandstrom J, Nilsson P, Karlsson K, Marklund SL. 10-fold increase in human plasma extracellular superoxide dismutase content caused by a mutation in heparin-binding domain. J Biol Chem. 1994;269(29):19163–6. [PubMed] [Google Scholar]
- 49.Gottfredsen RH, Goldstrohm DA, Hartney JM, Larsen UG, Bowler RP, Petersen SV. The cellular distribution of extracellular superoxide dismutase in macrophages is altered by cellular activation but unaffected by the naturally occurring R213G substitution. Free Radic Biol Med. 2014;69:348–56. doi: 10.1016/j.freeradbiomed.2014.01.038. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi T, Morihara N, Yamazaki N, et al. An arginine-213 to glycine mutation in human extracellular-superoxide dismutase reduces susceptibility to trypsin-like proteinases. Journal of biochemistry. 1996;120(1):184–8. doi: 10.1093/oxfordjournals.jbchem.a021383. [DOI] [PubMed] [Google Scholar]
- 51.Petersen SV, Thøgersen IB, Valnickova Z, et al. The concentration of extracellular superoxide dismutase in plasma is maintained by LRP-mediated endocytosis. Free Radic Biol Med. 2010;49(5):894–9. doi: 10.1016/j.freeradbiomed.2010.06.019. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 52.Adachi T, Ohta H, Hirano K, Hayashi K, Marklund SL. Non-enzymic glycation of human extracellular superoxide dismutase. The Biochemical journal. 1991;279( Pt 1):263–7. doi: 10.1042/bj2790263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adachi T, Ohta H, Hayashi K, Hirano K, Marklund SL. The site of nonenzymic glycation of human extracellular-superoxide dismutase in vitro. Free Radic Biol Med. 1992;13(3):205–10. doi: 10.1016/0891-5849(92)90016-a. [DOI] [PubMed] [Google Scholar]
- 54.Tamai M, Furuta H, Kawashima H, et al. Extracellular superoxide dismutase gene polymorphism is associated with insulin resistance and the susceptibility to type 2 diabetes. Diabetes research and clinical practice. 2006;71(2):140–5. doi: 10.1016/j.diabres.2005.05.006. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 55.Yamada H, Yamada Y, Adachi T, et al. Protective role of extracellular superoxide dismutase in hemodialysis patients. Nephron. 2000;84(3):218–23. doi: 10.1159/000045580. doi:45580. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 56.Yao H, Arunachalam G, Hwang JW, et al. Extracellular superoxide dismutase protects against pulmonary emphysema by attenuating oxidative fragmentation of ECM. Proc Natl Acad Sci U S A. 2010;107(35):15571–6. doi: 10.1073/pnas.1007625107. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sorheim IC, DeMeo DL, Washko G, et al. Polymorphisms in the superoxide dismutase-3 gene are associated with emphysema in COPD. Copd. 2010;7(4):262–8. doi: 10.3109/15412555.2010.496821. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahmad KA, Yuan Yuan D, Nawaz W, et al. Antioxidant therapy for Management of Oxidative stress Induced Hypertension. Free radical research. 2017:1–18. doi: 10.1080/10715762.2017.1322205. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 59.Schiffrin EL. Antioxidants in hypertension and cardiovascular disease. Molecular interventions. 2010;10(6):354–62. doi: 10.1124/mi.10.6.4. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 60.Laukkanen MO, Leppanen P, Turunen P, Tuomisto T, Naarala J, Yla-Herttuala S. EC-SOD gene therapy reduces paracetamol-induced liver damage in mice. The journal of gene medicine. 2001;3(4):321–5. doi: 10.1002/jgm.194. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 61.Jiang W, Tang L, Zeng J, Chen B. Adeno-associated virus mediated SOD gene therapy protects the retinal ganglion cells from chronic intraocular pressure elevation induced injury via attenuating oxidative stress and improving mitochondrial dysfunction in a rat model. American journal of translational research. 2016;8(2):799–810. [PMC free article] [PubMed] [Google Scholar]
- 62.Gao B, Flores SC, Leff JA, Bose SK, McCord JM. Synthesis and anti-inflammatory activity of a chimeric recombinant superoxide dismutase: SOD2/3. Am J Physiol Lung Cell Mol Physiol. 2003;284(6):L917–25. doi: 10.1152/ajplung.00374.2002. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 63.Ho A, Schwarze SR, Mermelstein SJ, Waksman G, Dowdy SF. Synthetic protein transduction domains: enhanced transduction potential in vitro and in vivo. Cancer Res. 2001;61(2):474–7. [PubMed] [Google Scholar]
- 64.Zhao J, Gao P, Xiao W, et al. A novel human derived cell-penetrating peptide in drug delivery. Molecular biology reports. 2011;38(4):2649–56. doi: 10.1007/s11033-010-0406-6. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 65.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox biology. 2017;11:613–19. doi: 10.1016/j.redox.2016.12.035. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furukawa Y, Torres AS, O’Halloran TV. Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. Embo j. 2004;23(14):2872–81. doi: 10.1038/sj.emboj.7600276. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Getzoff ED, Tainer JA, Weiner PK, Kollman PA, Richardson JS, Richardson DC. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature. 1983;306(5940):287–90. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 68.Tainer JA, Getzoff ED, Richardson JS, Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983;306(5940):284–7. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 69.Sheng Y, Abreu IA, Cabelli DE, et al. Superoxide dismutases and superoxide reductases. Chemical reviews. 2014;114(7):3854–918. doi: 10.1021/cr4005296. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochimica et biophysica acta. 2010;1804(2):245–62. doi: 10.1016/j.bbapap.2009.11.004. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fridovich I. Superoxide dismutases: anti- versus pro- oxidants? Anti-cancer agents in medicinal chemistry. 2011;11(2):175–7. doi: 10.2174/187152011795255966. [DOI] [PubMed] [Google Scholar]
- 72.Liochev SI, Fridovich I. Lucigenin luminescence as a measure of intracellular superoxide dismutase activity in Escherichia coli. Proc Natl Acad Sci U S A. 1997;94(7):2891–6. doi: 10.1073/pnas.94.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawahara T, Quinn MT, Lambeth JD. Molecular evolution of the reactive oxygen-generating NADPH oxidase (Nox/Duox) family of enzymes. BMC evolutionary biology. 2007;7:109. doi: 10.1186/1471-2148-7-109. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hink HU, Santanam N, Dikalov S, et al. Peroxidase properties of extracellular superoxide dismutase: role of uric acid in modulating in vivo activity. Arteriosclerosis, thrombosis, and vascular biology. 2002;22(9):1402–8. doi: 10.1161/01.atv.0000027524.86752.02. [DOI] [PubMed] [Google Scholar]
- 75.Yim MB, Chock PB, Stadtman ER. Copper, zinc superoxide dismutase catalyzes hydroxyl radical production from hydrogen peroxide. Proc Natl Acad Sci U S A. 1990;87(13):5006–10. doi: 10.1073/pnas.87.13.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liochev SI, Fridovich I. Mechanism of the peroxidase activity of Cu, Zn superoxide dismutase. Free Radic Biol Med. 2010;48(12):1565–9. doi: 10.1016/j.freeradbiomed.2010.02.036. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 77.Ansenberger-Fricano K, Ganini D, Mao M, et al. The peroxidase activity of mitochondrial superoxide dismutase. Free Radic Biol Med. 2013;54:116–24. doi: 10.1016/j.freeradbiomed.2012.08.573. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mason RP. Two hypotheses for the peroxidase activity of Mn-superoxide dismutase. Free Radic Biol Med. 2013;65:1533. doi: 10.1016/j.freeradbiomed.2013.02.013. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 79.Halliwell B, Gutteridge JM. Role of free radicals and catalytic metal ions in human disease: an overview. Methods in enzymology. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 80.Adams L, Franco MC, Estevez AG. Reactive nitrogen species in cellular signaling. Exp Biol Med (Maywood) 2015;240(6):711–7. doi: 10.1177/1535370215581314. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fukumura D, Kashiwagi S, Jain RK. The role of nitric oxide in tumour progression. Nature reviews Cancer. 2006;6(7):521–34. doi: 10.1038/nrc1910. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 82.Burke AJ, Sullivan FJ, Giles FJ, Glynn SA. The yin and yang of nitric oxide in cancer progression. Carcinogenesis. 2013;34(3):503–12. doi: 10.1093/carcin/bgt034. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 83.Vahora H, Khan MA, Alalami U, Hussain A. The Potential Role of Nitric Oxide in Halting Cancer Progression Through Chemoprevention. J Cancer Prev. 2016;21(1):1–12. doi: 10.15430/JCP.2016.21.1.1. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buetler TM, Krauskopf A, Ruegg UT. Role of superoxide as a signaling molecule. News Physiol Sci. 2004;19:120–3. doi: 10.1152/nips.01514.2003. [DOI] [PubMed] [Google Scholar]
- 85.Afanas’ev IB. On mechanism of superoxide signaling under physiological and pathophysiological conditions. Med Hypotheses. 2005;64(1):127–9. doi: 10.1016/j.mehy.2004.05.009. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Schattenberg JM, Rigoli RM, Storz P, Czaja MJ. Hepatocyte resistance to oxidative stress is dependent on protein kinase C-mediated down-regulation of c-Jun/AP-1. J Biol Chem. 2004;279(30):31089–97. doi: 10.1074/jbc.M404170200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 87.Dong-Yun S, Yu-Ru D, Shan-Lin L, Ya-Dong Z, Lian W. Redox stress regulates cell proliferation and apoptosis of human hepatoma through Akt protein phosphorylation. FEBS Lett. 2003;542(1–3):60–4. doi: 10.1016/s0014-5793(03)00338-7. [DOI] [PubMed] [Google Scholar]
- 88.Lee SL, Wang WW, Finlay GA, Fanburg BL. Serotonin stimulates mitogen-activated protein kinase activity through the formation of superoxide anion. Am J Physiol. 1999;277(2 Pt 1):L282–91. doi: 10.1152/ajplung.1999.277.2.L282. [DOI] [PubMed] [Google Scholar]
- 89.Wang W, Wang S, Nishanian EV, Del Pilar Cintron A, Wesley RA, Danner RL. Signaling by eNOS through a superoxide-dependent p42/44 mitogen-activated protein kinase pathway. Am J Physiol Cell Physiol. 2001;281(2):C544–54. doi: 10.1152/ajpcell.2001.281.2.C544. [DOI] [PubMed] [Google Scholar]
- 90.Jin Q, Jhun BS, Lee SH, et al. Differential regulation of phosphatidylinositol 3-kinase/Akt, mitogen-activated protein kinase, and AMP-activated protein kinase pathways during menadione-induced oxidative stress in the kidney of young and old rats. Biochem Biophys Res Commun. 2004;315(3):555–61. doi: 10.1016/j.bbrc.2004.01.093. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 91.Meng TC, Buckley DA, Galic S, Tiganis T, Tonks NK. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. J Biol Chem. 2004;279(36):37716–25. doi: 10.1074/jbc.M404606200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 92.Namgaladze D, Hofer HW, Ullrich V. Redox control of calcineurin by targeting the binuclear Fe(2+)-Zn(2+) center at the enzyme active site. J Biol Chem. 2002;277(8):5962–9. doi: 10.1074/jbc.M111268200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 93.Afanas’ev IB. Competition between superoxide and hydrogen peroxide signaling in heterolytic enzymatic processes. Med Hypotheses. 2006;66(6):1125–8. doi: 10.1016/j.mehy.2005.11.046. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 94.Wang Q, Dube D, Friesen RW, et al. Catalytic inactivation of protein tyrosine phosphatase CD45 and protein tyrosine phosphatase 1B by polyaromatic quinones. Biochemistry. 2004;43(14):4294–303. doi: 10.1021/bi035986e. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 95.Barrett WC, DeGnore JP, Keng YF, Zhang ZY, Yim MB, Chock PB. Roles of superoxide radical anion in signal transduction mediated by reversible regulation of protein-tyrosine phosphatase 1B. J Biol Chem. 1999;274(49):34543–6. doi: 10.1074/jbc.274.49.34543. [DOI] [PubMed] [Google Scholar]
- 96.Pervaiz S, Clement MV. Superoxide anion: oncogenic reactive oxygen species? Int J Biochem Cell Biol. 2007;39(7–8):1297–304. doi: 10.1016/j.biocel.2007.04.007. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 97.Clement MV, Hirpara JL, Pervaiz S. Decrease in intracellular superoxide sensitizes Bcl-2-overexpressing tumor cells to receptor and drug-induced apoptosis independent of the mitochondria. Cell death and differentiation. 2003;10(11):1273–85. doi: 10.1038/sj.cdd.4401302. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 98.Clement MV, Pervaiz S. Intracellular superoxide and hydrogen peroxide concentrations: a critical balance that determines survival or death. Redox report: communications in free radical research. 2001;6(4):211–4. doi: 10.1179/135100001101536346. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 99.Yamakawa H, Ito Y, Naganawa T, et al. Activation of caspase-9 and -3 during H2O2-induced apoptosis of PC12 cells independent of ceramide formation. Neurol Res. 2000;22(6):556–64. doi: 10.1080/01616412.2000.11740718. [DOI] [PubMed] [Google Scholar]
- 100.Hampton MB, Orrenius S. Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis. FEBS Lett. 1997;414(3):552–6. doi: 10.1016/s0014-5793(97)01068-5. [DOI] [PubMed] [Google Scholar]
- 101.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics. 1994;22(1):162–71. doi: 10.1006/geno.1994.1357. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 102.Zelko IN, Folz RJ. Myeloid zinc finger (MZF)-like, Kruppel-like and Ets families of transcription factors determine the cell-specific expression of mouse extracellular superoxide dismutase. The Biochemical journal. 2003;369(Pt 2):375–86. doi: 10.1042/bj20021431. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y-D, Chen W-D, Li C, et al. Farnesoid X receptor antagonizes JNK signaling pathway in liver carcinogenesis by activating SOD3. Molecular endocrinology (Baltimore, Md) 2015;29(2):322–31. doi: 10.1210/me.2014-1225. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stralin P, Marklund SL. Multiple cytokines regulate the expression of extracellular superoxide dismutase in human vascular smooth muscle cells. Atherosclerosis. 2000;151(2):433–41. doi: 10.1016/s0021-9150(99)00427-x. [DOI] [PubMed] [Google Scholar]
- 105.Forster D, Reiser G. Nucleotides protect rat brain astrocytes against hydrogen peroxide toxicity and induce antioxidant defense via P2Y receptors. Neurochemistry international. 2016;94:57–66. doi: 10.1016/j.neuint.2016.02.006. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 106.Davis SM, Collier LA, Leonardo CC, Seifert HA, Ajmo CT, Jr, Pennypacker KR. Leukemia Inhibitory Factor Protects Neurons from Ischemic Damage via Upregulation of Superoxide Dismutase 3. Molecular neurobiology. 2017;54(1):608–22. doi: 10.1007/s12035-015-9587-2. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yasuda H, Ohashi A, Nishida S, et al. Exendin-4 induces extracellular-superoxide dismutase through histone H3 acetylation in human retinal endothelial cells. Journal of clinical biochemistry and nutrition. 2016;59(3):174–81. doi: 10.3164/jcbn.16-26. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Davis CD, Milne DB, Nielsen FH. Changes in dietary zinc and copper affect zinc-status indicators of postmenopausal women, notably, extracellular superoxide dismutase and amyloid precursor proteins. The American journal of clinical nutrition. 2000;71(3):781–8. doi: 10.1093/ajcn/71.3.781. [DOI] [PubMed] [Google Scholar]
- 109.Olin KL, Golub MS, Gershwin ME, Hendrickx AG, Lonnerdal B, Keen CL. Extracellular superoxide dismutase activity is affected by dietary zinc intake in nonhuman primate and rodent models. The American journal of clinical nutrition. 1995;61(6):1263–7. doi: 10.1093/ajcn/61.6.1263. [DOI] [PubMed] [Google Scholar]
- 110.Itoh S, Ozumi K, Kim HW, et al. Novel mechanism for regulation of extracellular SOD transcription and activity by copper: role of antioxidant-1. Free Radic Biol Med. 2009;46(1):95–104. doi: 10.1016/j.freeradbiomed.2008.09.039. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stralin P, Marklund SL. Effects of oxidative stress on expression of extracellular superoxide dismutase, CuZn-superoxide dismutase and Mn-superoxide dismutase in human dermal fibroblasts. The Biochemical journal. 1994;298( Pt 2):347–52. doi: 10.1042/bj2980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carlsson LM, Jonsson J, Edlund T, Marklund SL. Mice lacking extracellular superoxide dismutase are more sensitive to hyperoxia. Proc Natl Acad Sci U S A. 1995;92(14):6264–8. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sentman ML, Granstrom M, Jakobson H, Reaume A, Basu S, Marklund SL. Phenotypes of mice lacking extracellular superoxide dismutase and copper- and zinc-containing superoxide dismutase. J Biol Chem. 2006;281(11):6904–9. doi: 10.1074/jbc.M510764200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 114.Gongora MC, Lob HE, Landmesser U, et al. Loss of extracellular superoxide dismutase leads to acute lung damage in the presence of ambient air: a potential mechanism underlying adult respiratory distress syndrome. Am J Pathol. 2008;173(4):915–26. doi: 10.2353/ajpath.2008.080119. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Teoh ML, Fitzgerald MP, Oberley LW, Domann FE. Overexpression of extracellular superoxide dismutase attenuates heparanase expression and inhibits breast carcinoma cell growth and invasion. Cancer Res. 2009;69(15):6355–63. doi: 10.1158/0008-5472.CAN-09-1195. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Teoh-Fitzgerald ML, Fitzgerald MP, Zhong W, Askeland RW, Domann FE. Epigenetic reprogramming governs EcSOD expression during human mammary epithelial cell differentiation, tumorigenesis and metastasis. Oncogene. 2014;33(3):358–68. doi: 10.1038/onc.2012.582. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh B, Bhat HK. Superoxide dismutase 3 is induced by antioxidants, inhibits oxidative DNA damage and is associated with inhibition of estrogen-induced breast cancer. Carcinogenesis. 2012;33(12):2601–10. doi: 10.1093/carcin/bgs300. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hubackova M, Vaclavikova R, Ehrlichova M, et al. Association of superoxide dismutases and NAD(P)H quinone oxidoreductases with prognosis of patients with breast carcinomas. International journal of cancer Journal international du cancer. 2012;130(2):338–48. doi: 10.1002/ijc.26006. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 119.Wang C-A, Harrell JC, Iwanaga R, Jedlicka P, Ford HL. Vascular endothelial growth factor C promotes breast cancer progression via a novel antioxidant mechanism that involves regulation of superoxide dismutase 3. Breast Cancer Res. 2014;16(5):462. doi: 10.1186/s13058-014-0462-2. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chatterjee A, Ronghe A, Singh B, Bhat NK, Chen J, Bhat HK. Natural antioxidants exhibit chemopreventive characteristics through the regulation of CNC b-Zip transcription factors in estrogen-induced breast carcinogenesis. Journal of biochemical and molecular toxicology. 2014;28(12):529–38. doi: 10.1002/jbt.21594. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh B, Shoulson R, Chatterjee A, et al. Resveratrol inhibits estrogen-induced breast carcinogenesis through induction of NRF2-mediated protective pathways. Carcinogenesis. 2014;35(8):1872–80. doi: 10.1093/carcin/bgu120. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Russo J, Russo IH. The role of estrogen in the initiation of breast cancer. J Steroid Biochem Mol Biol. 2006;102(1–5):89–96. doi: 10.1016/j.jsbmb.2006.09.004. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shivapurkar N, Maitra A, Milchgrub S, Gazdar AF. Deletions of chromosome 4 occur early during the pathogenesis of colorectal carcinoma. Human pathology. 2001;32(2):169–77. doi: 10.1053/hupa.2001.21560. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 124.Vicini FA, Kestin L, Huang R, Martinez A. Does local recurrence affect the rate of distant metastases and survival in patients with early-stage breast carcinoma treated with breast-conserving therapy? Cancer. 2003;97(4):910–9. doi: 10.1002/cncr.11143. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 125.Smith MJ, Culhane AC, Killeen S, et al. Mechanisms driving local breast cancer recurrence in a model of breast-conserving surgery. Annals of surgical oncology. 2008;15(10):2954–64. doi: 10.1245/s10434-008-0037-5. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 126.Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–31. doi: 10.1007/s10549-009-0674-9. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 127.Teoh-Fitzgerald MLT, Fitzgerald MP, Jensen TJ, Futscher BW, Domann FE. Genetic and epigenetic inactivation of extracellular superoxide dismutase promotes an invasive phenotype in human lung cancer by disrupting ECM homeostasis. Mol Cancer Res. 2012;10(1):40–51. doi: 10.1158/1541-7786.mcr-11-0501. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yoo DG, Song YJ, Cho EJ, et al. Alteration of APE1/ref-1 expression in non-small cell lung cancer: the implications of impaired extracellular superoxide dismutase and catalase antioxidant systems. Lung cancer (Amsterdam, Netherlands) 2008;60(2):277–84. doi: 10.1016/j.lungcan.2007.10.015. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 129.Svensk AM, Soini Y, Paakko P, Hiravikoski P, Kinnula VL. Differential expression of superoxide dismutases in lung cancer. American journal of clinical pathology. 2004;122(3):395–404. doi: 10.1309/a45q-hb0q-rrx6-ct9a. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 130.Hoesel B, Schmid JA. The complexity of NF-kappaB signaling in inflammation and cancer. Molecular cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang H-H, Zhang Z-Y, Che C-L, Mei Y-F, Shi Y-Z. Array analysis for potential biomarker of gemcitabine identification in non-small cell lung cancer cell lines. International journal of clinical and experimental pathology. 2013;6(9):1734–46. [PMC free article] [PubMed] [Google Scholar]
- 132.Kim J, Mizokami A, Shin M, et al. SOD3 acts as a tumor suppressor in PC-3 prostate cancer cells via hydrogen peroxide accumulation. Anticancer Res. 2014;34(6):2821–31. [PubMed] [Google Scholar]
- 133.Chaiswing L, Zhong W, Oberley TD. Increasing discordant antioxidant protein levels and enzymatic activities contribute to increasing redox imbalance observed during human prostate cancer progression. Free Radic Biol Med. 2014;67:342–52. doi: 10.1016/j.freeradbiomed.2013.11.006. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chaiswing L, Zhong W, Cullen JJ, Oberley LW, Oberley TD. Extracellular redox state regulates features associated with prostate cancer cell invasion. Cancer Res. 2008;68(14):5820–6. doi: 10.1158/0008-5472.can-08-0162. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 135.O’Leary BR, Fath MA, Bellizzi AM, et al. Loss of SOD3 (EcSOD) Expression Promotes an Aggressive Phenotype in Human Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2015;21(7):1741–51. doi: 10.1158/1078-0432.ccr-14-1959. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Teoh ML, Sun W, Smith BJ, Oberley LW, Cullen JJ. Modulation of reactive oxygen species in pancreatic cancer. Clin Cancer Res. 2007;13(24):7441–50. doi: 10.1158/1078-0432.CCR-07-0851. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 137.Sibenaller ZA, Welsh JL, Du C, et al. Extracellular superoxide dismutase suppresses hypoxia-inducible factor-1alpha in pancreatic cancer. Free Radic Biol Med. 2014;69:357–66. doi: 10.1016/j.freeradbiomed.2014.02.002. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng X, Ewton DZ, Friedman E. Mirk/Dyrk1B maintains the viability of quiescent pancreatic cancer cells by reducing levels of reactive oxygen species. Cancer Res. 2009;69(8):3317–24. doi: 10.1158/0008-5472.can-08-2903. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Laatikainen LE, Castellone MD, Hebrant A, et al. Extracellular superoxide dismutase is a thyroid differentiation marker down-regulated in cancer. Endocrine-related cancer. 2010;17(3):785–96. doi: 10.1677/erc-10-0021. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 140.Cammarota F, de Vita G, Salvatore M, Laukkanen MO. Ras oncogene-mediated progressive silencing of extracellular superoxide dismutase in tumorigenesis. Biomed Res Int. 2015;2015:780409. doi: 10.1155/2015/780409. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cammarota F, Fiscardi F, Esposito T, de Vita G, Salvatore M, Laukkanen MO. Clinical relevance of thyroid cell models in redox research. Cancer cell international. 2015;15:113. doi: 10.1186/s12935-015-0264-3. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Laatikainen LE, Castellone MD, Hebrant A, et al. Extracellular superoxide dismutase is a thyroid differentiation marker down-regulated in cancer. Endocrine-related cancer. 2010;17(3):785–96. doi: 10.1677/erc-10-0021. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 143.Laukkanen MO, Cammarota F, Esposito T, Salvatore M, Castellone MD. Extracellular superoxide dismutase regulates the expression of small gtpase regulatory proteins GEFs, GAPs, and GDI. PLoS One. 2015;10(3):e0121441. doi: 10.1371/journal.pone.0121441. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Castellone MD, Langella A, Cantara S, et al. Extracellular superoxide dismutase induces mouse embryonic fibroblast proliferative burst, growth arrest, immortalization, and consequent in vivo tumorigenesis. Antioxid Redox Signal. 2014;21(10):1460–74. doi: 10.1089/ars.2013.5475. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 145.Jeon YJ, Yoo H, Kim BH, et al. IFNgamma-mediated inhibition of cell proliferation through increased PKCdelta-induced overexpression of EC-SOD. BMB reports. 2012;45(11):659–64. doi: 10.5483/BMBRep.2012.45.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wheeler MD, Smutney OM, Samulski RJ. Secretion of extracellular superoxide dismutase from muscle transduced with recombinant adenovirus inhibits the growth of B16 melanomas in mice. Mol Cancer Res. 2003;1(12):871–81. [PubMed] [Google Scholar]
- 147.Kim SH, Kim MO, Gao P, et al. Overexpression of extracellular superoxide dismutase (EC-SOD) in mouse skin plays a protective role in DMBA/TPA-induced tumor formation. Oncology research. 2005;15(7–8):333–41. doi: 10.3727/096504005776449725. [DOI] [PubMed] [Google Scholar]
- 148.Morvan D, Demidem A. Metabolomics and transcriptomics demonstrate severe oxidative stress in both localized chemotherapy-treated and bystander tumors. Biochimica et biophysica acta. 2014;1840(3):1092–104. doi: 10.1016/j.bbagen.2013.11.022. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 149.Liu X, Xu Y, Meng Q, et al. Proteomic analysis of minute amount of colonic biopsies by enteroscopy sampling. Biochem Biophys Res Commun. 2016;476(4):286–92. doi: 10.1016/j.bbrc.2016.05.114. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 150.Soini Y, Kallio JP, Hirvikoski P, et al. Antioxidant enzymes in renal cell carcinoma. Histology and histopathology. 2006;21(2):157–65. doi: 10.14670/hh-21.157. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 151.Yang J, Zhang C, Zhang W, Shi R, Zhang Z. Extracellular superoxide dismutase, a potential extracellular biomarker candidate for prolactinoma. West Indian Med J. 2012;61(7):665–9. [PubMed] [Google Scholar]
- 152.Subbannayya Y, Mir SA, Renuse S, et al. Identification of differentially expressed serum proteins in gastric adenocarcinoma. Journal of proteomics. 2015;127(Pt A):80–8. doi: 10.1016/j.jprot.2015.04.021. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wei L, Zhang J, Yang ZL, You H. Extracellular superoxide dismutase increased the therapeutic potential of human mesenchymal stromal cells in radiation pulmonary fibrosis. Cytotherapy. 2017;19(5):586–602. doi: 10.1016/j.jcyt.2017.02.359. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 154.Cui R, Gao M, Qu S, Liu D. Overexpression of superoxide dismutase 3 gene blocks high-fat diet-induced obesity, fatty liver and insulin resistance. Gene Ther. 2014;21(9):840–8. doi: 10.1038/gt.2014.64. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chan DS, Vieira AR, Aune D, et al. Body mass index and survival in women with breast cancer-systematic literature review and meta-analysis of 82 follow-up studies. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2014;25(10):1901–14. doi: 10.1093/annonc/mdu042. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Iyengar NM, Zhou XK, Gucalp A, et al. Systemic Correlates of White Adipose Tissue Inflammation in Early-Stage Breast Cancer. Clin Cancer Res. 2016;22(9):2283–9. doi: 10.1158/1078-0432.ccr-15-2239. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Negahdar M, Djalali M, Abthahi H, et al. Blood superoxide dismutase and catalase activities in women affected with breast cancer. Iran J Public Health. 2005;34(3):39–43. [Google Scholar]
- 158.Sinha RJ, Singh R, Mehrotra S, Singh RK. Implications of free radicals and antioxidant levels in carcinoma of the breast: a never-ending battle for survival. Indian journal of cancer. 2009;46(2):146–50. doi: 10.4103/0019-509x.49153. [DOI] [PubMed] [Google Scholar]
- 159.Eliyasinia F, Chegini V, Olfat-Bakhsh A, Pasalar P. Superoxide dismutase activities in plasma of patients with breast cancer. Archives Breast Cancer. 2014;1(2):29–32. [Google Scholar]
- 160.Khanzode SS, Muddeshwar MG, Khanzode SD, Dakhale GN. Antioxidant enzymes and lipid peroxidation in different stages of breast cancer. Free radical research. 2004;38(1):81–5. doi: 10.1080/01411590310001637066. [DOI] [PubMed] [Google Scholar]
- 161.Ray G, Batra S, Shukla NK, et al. Lipid peroxidation, free radical production and antioxidant status in breast cancer. Breast Cancer Res Treat. 2000;59(2):163–70. doi: 10.1023/a:1006357330486. [DOI] [PubMed] [Google Scholar]
- 162.Yeh C-C, Hou M-F, Tsai S-M, et al. Superoxide anion radical, lipid peroxides and antioxidant status in the blood of patients with breast cancer. Clinica Chimica Acta. 2005;361(1–2):104–11. doi: 10.1016/j.cccn.2005.05.002. doi: https://doi.org/10.1016/j.cccn.2005.05.002. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 163.Battisti V, Maders LD, Bagatini MD, et al. Oxidative stress and antioxidant status in prostate cancer patients: relation to Gleason score, treatment and bone metastasis. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2011;65(7):516–24. doi: 10.1016/j.biopha.2011.06.003. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 164.Sandhya B, Moanoharan S, Sirisha Lavanya G, Ch RM. Lipid peroxidation and antioxidant status in prostate cancer patients. Indian J Science Technology. 2010;3(1):83–86. [Google Scholar]
- 165.Shidfar A, Fatokun T, Ivancic D, Chatterton RT, Khan SA, Wang J. Protein Biomarkers for Breast Cancer Risk Are Specifically Correlated with Local Steroid Hormones in Nipple Aspirate Fluid. Hormones & cancer. 2016;7(4):252–9. doi: 10.1007/s12672-016-0264-3. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sharma P, Bhunia S, Poojary SS, et al. Global methylation profiling to identify epigenetic signature of gallbladder cancer and gallstone disease. Tumour Biol. 2016;37(11):14687–99. doi: 10.1007/s13277-016-5355-9. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 167.Divyya S, Naushad SM, Murthy PVLN, Reddy CR, Kutala VK. GCPII modulates oxidative stress and prostate cancer susceptibility through changes in methylation of RASSF1, BNIP3, GSTP1 and Ec-SOD. Molecular biology reports. 2013;40(10):5541–50. doi: 10.1007/s11033-013-2655-7. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 168.Chiba T, Yokosuka O, Fukai K, et al. Cell growth inhibition and gene expression induced by the histone deacetylase inhibitor, trichostatin A, on human hepatoma cells. Oncology. 2004;66(6):481–91. doi: 10.1159/000079503. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 169.Lakshmi SV, Naushad SM, Reddy CA, et al. Oxidative stress in coronary artery disease: epigenetic perspective. Molecular and cellular biochemistry. 2013;374(1–2):203–11. doi: 10.1007/s11010-012-1520-7. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 170.Kamiya T, Machiura M, Makino J, Hara H, Hozumi I, Adachi T. Epigenetic regulation of extracellular-superoxide dismutase in human monocytes. Free Radic Biol Med. 2013;61:197–205. doi: 10.1016/j.freeradbiomed.2013.04.013. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 171.Zelko IN, Stepp MW, Vorst AL, Folz RJ. Histone acetylation regulates the cell-specific and interferon-gamma-inducible expression of extracellular superoxide dismutase in human pulmonary arteries. American journal of respiratory cell and molecular biology. 2011;45(5):953–61. doi: 10.1165/rcmb.2011-0012OC. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zelko IN, Mueller MR, Folz RJ. CpG methylation attenuates Sp1 and Sp3 binding to the human extracellular superoxide dismutase promoter and regulates its cell-specific expression. Free Radic Biol Med. 2010;48(7):895–904. doi: 10.1016/j.freeradbiomed.2010.01.007. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105(2):223–35. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Le Beyec J, Xu R, Lee SY, et al. Cell shape regulates global histone acetylation in human mammary epithelial cells. Experimental cell research. 2007;313(14):3066–75. doi: 10.1016/j.yexcr.2007.04.022. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kamiya T, Nakahara R, Mori N, Hara H, Adachi T. Ten-eleven translocation 1 functions as a mediator of SOD3 expression in human lung cancer A549 cells. Free radical research. 2017:1–33. doi: 10.1080/10715762.2017.1313415. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 176.Li L, Li C, Mao H, et al. Epigenetic inactivation of the CpG demethylase TET1 as a DNA methylation feedback loop in human cancers. Scientific reports. 2016;6:26591. doi: 10.1038/srep26591. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Divyya S, Naushad SM, Addlagatta A, et al. Association of glutamate carboxypeptidase II (GCPII) haplotypes with breast and prostate cancer risk. Gene. 2013;516(1):76–81. doi: 10.1016/j.gene.2012.11.047. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 178.Divyya S, Naushad SM, Addlagatta A, et al. Paradoxical role of C1561T glutamate carboxypeptidase II (GCPII) genetic polymorphism in altering disease susceptibility. Gene. 2012;497(2):273–9. doi: 10.1016/j.gene.2012.01.055. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 179.Nozik-Grayck E, Woods C, Stearman RS, et al. Histone deacetylation contributes to low extracellular superoxide dismutase expression in human idiopathic pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2016;311(1):L124–34. doi: 10.1152/ajplung.00263.2015. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang Q, Dahl MJ, Albertine KH, Ramchandran R, Sun M, Raj JU. Role of histone deacetylases in regulation of phenotype of ovine newborn pulmonary arterial smooth muscle cells. Cell Prolif. 2013;46(6):654–64. doi: 10.1111/cpr.12076. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Muller BM, Jana L, Kasajima A, et al. Differential expression of histone deacetylases HDAC1, 2 and 3 in human breast cancer--overexpression of HDAC2 and HDAC3 is associated with clinicopathological indicators of disease progression. BMC cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wilson AJ, Byun DS, Popova N, et al. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281(19):13548–58. doi: 10.1074/jbc.M510023200. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 183.Liu C, Liu L, Shan J, et al. Histone deacetylase 3 participates in self-renewal of liver cancer stem cells through histone modification. Cancer Lett. 2013;339(1):60–9. doi: 10.1016/j.canlet.2013.07.022. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 184.Jiao F, Hu H, Yuan C, et al. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int J Oncol. 2014;45(4):1523–30. doi: 10.3892/ijo.2014.2568. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 185.Weichert W, Roske A, Gekeler V, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98(3):604–10. doi: 10.1038/sj.bjc.6604199. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Monteiro FL, Baptista T, Amado F, Vitorino R, Jeronimo C, Helguero LA. Expression and functionality of histone H2A variants in cancer. Oncotarget. 2014;5(11):3428–43. doi: 10.18632/oncotarget.2007. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hernandez-Munoz I, Lund AH, van der Stoop P, et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci U S A. 2005;102(21):7635–40. doi: 10.1073/pnas.0408918102. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buschbeck M, Uribesalgo I, Wibowo I, et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nature structural & molecular biology. 2009;16(10):1074–9. doi: 10.1038/nsmb.1665. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 189.Dardenne E, Pierredon S, Driouch K, et al. Splicing switch of an epigenetic regulator by RNA helicases promotes tumor-cell invasiveness. Nature structural & molecular biology. 2012;19(11):1139–46. doi: 10.1038/nsmb.2390. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 190.Wang Z, Luo Z, Zhou L, Li X, Jiang T, Fu E. DDX5 promotes proliferation and tumorigenesis of non-small-cell lung cancer cells by activating beta-catenin signaling pathway. Cancer science. 2015;106(10):1303–12. doi: 10.1111/cas.12755. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Mazurek A, Luo W, Krasnitz A, Hicks J, Powers RS, Stillman B. DDX5 regulates DNA replication and is required for cell proliferation in a subset of breast cancer cells. Cancer discovery. 2012;2(9):812–25. doi: 10.1158/2159-8290.cd-12-0116. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Alqahtani H, Gopal K, Gupta N, et al. DDX17 (P72), a Sox2 binding partner, promotes stem-like features conferred by Sox2 in a small cell population in estrogen receptor-positive breast cancer. Cellular signalling. 2016;28(2):42–50. doi: 10.1016/j.cellsig.2015.11.004. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 193.Wortham NC, Ahamed E, Nicol SM, et al. The DEAD-box protein p72 regulates ERalpha-/oestrogen-dependent transcription and cell growth, and is associated with improved survival in ERalpha-positive breast cancer. Oncogene. 2009;28(46):4053–64. doi: 10.1038/onc.2009.261. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shin S, Rossow KL, Grande JP, Janknecht R. Involvement of RNA helicases p68 and p72 in colon cancer. Cancer Res. 2007;67(16):7572–8. doi: 10.1158/0008-5472.can-06-4652. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 195.Janknecht R. Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17) American journal of translational research. 2010;2(3):223–34. [PMC free article] [PubMed] [Google Scholar]
- 196.Clark EL, Coulson A, Dalgliesh C, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res. 2008;68(19):7938–46. doi: 10.1158/0008-5472.can-08-0932. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 2006;173(8):858–64. doi: 10.1164/rccm.200509-1387OC. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 198.Brenner DR, McLaughlin JR, Hung RJ. Previous lung diseases and lung cancer risk: a systematic review and meta-analysis. PLoS One. 2011;6(3):e17479. doi: 10.1371/journal.pone.0017479. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Young RP, Hopkins RJ, Hay BA, et al. Lung cancer susceptibility model based on age, family history and genetic variants. PLoS One. 2009;4(4):e5302. doi: 10.1371/journal.pone.0005302. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Geng R, Chen Z, Zhao X, et al. Oxidative stress-related genetic polymorphisms are associated with the prognosis of metastatic gastric cancer patients treated with epirubicin, oxaliplatin and 5-fluorouracil combination chemotherapy. PLoS One. 2014;9(12):e116027. doi: 10.1371/journal.pone.0116027. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kobylecki CJ, Afzal S, Nordestgaard BG. Does SOD3 R213G Homozygosity Influence Morbidity, Mortality, and Lung Function in the General Population? Antioxid Redox Signal. 2016;24(15):884–91. doi: 10.1089/ars.2016.6629. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 202.Dong X, Li D, Liu H, Zhao Y. SOD3 and eNOS genotypes are associated with SOD activity and NOx. Experimental and therapeutic medicine. 2014;8(1):328–34. doi: 10.3892/etm.2014.1720. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Iida R, Tsubota E, Takeshita H, Yasuda T. Multiplex single base extension method for simultaneous genotyping of non-synonymous SNP in the three human SOD genes. Electrophoresis. 2008;29(23):4788–94. doi: 10.1002/elps.200800332. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 204.Su S, He K, Li J, et al. Genetic polymorphisms in antioxidant enzyme genes and susceptibility to hepatocellular carcinoma in Chinese population: a case-control study. Tumour Biol. 2015;36(6):4627–32. doi: 10.1007/s13277-015-3110-2. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 205.Zhao P, Zhao L, Zou P, et al. Genetic oxidative stress variants and glioma risk in a Chinese population: a hospital-based case-control study. BMC Cancer. 2012;12:617. doi: 10.1186/1471-2407-12-617. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Ganguly K, Depner M, Fattman C, et al. Superoxide dismutase 3, extracellular (SOD3) variants and lung function. Physiological genomics. 2009;37(3):260–7. doi: 10.1152/physiolgenomics.90363.2008. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Naganuma T, Nakayama T, Sato N, et al. Association of extracellular superoxide dismutase gene with cerebral infarction in women: a haplotype-based case-control study. Hereditas. 2008;145(6):283–92. doi: 10.1111/j.1601-5223.2008.02086.x. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 208.Yang YM, Xie XR, Jin AL. Genetic polymorphisms in extracellular superoxide dismutase Leu53Leu, Arg213Gly, and Ala40Thr and susceptibility to type 2 diabetes mellitus. Genetics and molecular research: GMR. 2016;15(4) doi: 10.4238/gmr15048418. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 209.Rodrigues P, de Marco G, Furriol J, et al. Oxidative stress in susceptibility to breast cancer: study in Spanish population. BMC Cancer. 2014;14:861. doi: 10.1186/1471-2407-14-861. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Mohammedi K, Bellili-Munoz N, Marklund SL, et al. Plasma extracellular superoxide dismutase concentration, allelic variations in the SOD3 gene and risk of myocardial infarction and all-cause mortality in people with type 1 and type 2 diabetes. Cardiovascular diabetology. 2015;14:845. doi: 10.1186/s12933-014-0163-2. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bauer SR, Richman EL, Sosa E, et al. Antioxidant and vitamin E transport genes and risk of high-grade prostate cancer and prostate cancer recurrence. Prostate. 2013;73(16):1786–95. doi: 10.1002/pros.22717. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chan JM, Darke AK, Penney KL, et al. Selenium- or Vitamin E-Related Gene Variants, Interaction with Supplementation, and Risk of High-Grade Prostate Cancer in SELECT. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1050–8. doi: 10.1158/1055-9965.epi-16-0104. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rajaraman P, Hutchinson A, Rothman N, et al. Oxidative response gene polymorphisms and risk of adult brain tumors. Neuro Oncol. 2008;10(5):709–15. doi: 10.1215/15228517-2008-037. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Singh RK, Indra D, Mitra S, et al. Deletions in chromosome 4 differentially associated with the development of cervical cancer: evidence of slit2 as a candidate tumor suppressor gene. Human genetics. 2007;122(1):71–81. doi: 10.1007/s00439-007-0375-6. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 215.Beder LB, Gunduz M, Ouchida M, et al. Genome-wide analyses on loss of heterozygosity in head and neck squamous cell carcinomas. Lab Invest. 2003;83(1):99–105. doi: 10.1097/01.lab.0000047489.26246.e1. [DOI] [PubMed] [Google Scholar]
- 216.Dallol A, Da Silva NF, Viacava P, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62(20):5874–80. [PubMed] [Google Scholar]
- 217.Kim GJ, Cho SJ, Won NH, et al. Genomic imbalances in Korean hepatocellular carcinoma. Cancer genetics and cytogenetics. 2003;142(2):129–33. doi: 10.1016/s0165-4608(02)00834-8. [DOI] [PubMed] [Google Scholar]
- 218.Pershouse MA, El-Naggar AK, Hurr K, Lin H, Yung WK, Steck PA. Deletion mapping of chromosome 4 in head and neck squamous cell carcinoma. Oncogene. 1997;14(3):369–73. doi: 10.1038/sj.onc.1200836. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 219.Polascik TJ, Cairns P, Chang WY, Schoenberg MP, Sidransky D. Distinct regions of allelic loss on chromosome 4 in human primary bladder carcinoma. Cancer Res. 1995;55(22):5396–9. [PubMed] [Google Scholar]
- 220.Sherwood JB, Shivapurkar N, Lin WM, et al. Chromosome 4 deletions are frequent in invasive cervical cancer and differ between histologic variants. Gynecologic oncology. 2000;79(1):90–6. doi: 10.1006/gyno.2000.5922. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 221.Shivapurkar N, Sood S, Wistuba II, et al. Multiple regions of chromosome 4 demonstrating allelic losses in breast carcinomas. Cancer Res. 1999;59(15):3576–80. [PubMed] [Google Scholar]
- 222.Shivapurkar N, Virmani AK, Wistuba II, et al. Deletions of chromosome 4 at multiple sites are frequent in malignant mesothelioma and small cell lung carcinoma. Clin Cancer Res. 1999;5(1):17–23. [PubMed] [Google Scholar]
- 223.Russo J, Fernandez SV, Russo PA, et al. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2006;20(10):1622–34. doi: 10.1096/fj.05-5399com. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 224.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nature reviews Genetics. 2015;16(7):421–33. doi: 10.1038/nrg3965. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 225.Zhang X, Ng W-L, Wang P, et al. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012;72(18):4707–13. doi: 10.1158/0008-5472.can-12-0639. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 226.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–61. doi: 10.1073/pnas.0510565103. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Nam EJ, Yoon H, Kim SW, et al. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–5. doi: 10.1158/1078-0432.ccr-07-1731. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 228.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–43. doi: 10.1158/0008-5472.can-06-0561. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 229.Tetzlaff MT, Liu A, Xu X, et al. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocrine pathology. 2007;18(3):163–73. doi: 10.1007/s12022-007-0023-7. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 230.Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. Journal of cellular and molecular medicine. 2009;13(1):39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Laukkanen MO. Extracellular Superoxide Dismutase: Growth Promoter or Tumor Suppressor? Oxid Med Cell Longev. 2016;2016:3612589. doi: 10.1155/2016/3612589. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Batinic-Haberle I, Tovmasyan A, Spasojevic I. An educational overview of the chemistry, biochemistry and therapeutic aspects of Mn porphyrins – From superoxide dismutation to H(2)O(2)-driven pathways. Redox biology. 2015;5:43–65. doi: 10.1016/j.redox.2015.01.017. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO reports. 2014;15(12):1243–53. doi: 10.15252/embr.201439246. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Gao F, Koenitzer JR, Tobolewski JM, et al. Extracellular superoxide dismutase inhibits inflammation by preventing oxidative fragmentation of hyaluronan. J Biol Chem. 2008;283(10):6058–66. doi: 10.1074/jbc.M709273200. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kliment CR, Tobolewski JM, Manni ML, Tan RJ, Enghild J, Oury TD. Extracellular superoxide dismutase protects against matrix degradation of heparan sulfate in the lung. Antioxid Redox Signal. 2008;10(2):261–8. doi: 10.1089/ars.2007.1906. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Vlodavsky I, Elkin M, Abboud-Jarrous G, et al. Heparanase: one molecule with multiple functions in cancer progression. Connective tissue research. 2008;49(3):207–10. doi: 10.1080/03008200802143281. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 237.Goswami KK, Ghosh T, Ghosh S, Sarkar M, Bose A, Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cellular immunology. 2017 doi: 10.1016/j.cellimm.2017.04.005. [published Online First: Epub Date]|. [DOI] [PubMed] [Google Scholar]
- 238.Laurila JP, Laatikainen LE, Castellone MD, Laukkanen MO. SOD3 reduces inflammatory cell migration by regulating adhesion molecule and cytokine expression. PLoS One. 2009;4(6):e5786. doi: 10.1371/journal.pone.0005786. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kwon MJ, Jeon YJ, Lee KY, Kim TY. Superoxide dismutase 3 controls adaptive immune responses and contributes to the inhibition of ovalbumin-induced allergic airway inflammation in mice. Antioxid Redox Signal. 2012;17(10):1376–92. doi: 10.1089/ars.2012.4572. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99. doi: 10.1016/j.cell.2010.01.025. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kraaij MD, Savage ND, van der Kooij SW, et al. Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(41):17686–91. doi: 10.1073/pnas.1012016107. [published Online First: Epub Date]|. [DOI] [PMC free article] [PubMed] [Google Scholar]