Abstract
Glioblastomas are among the most lethal cancers; however, recent advances in survival have increased the need for better prognostic markers. microRNAs (miRNAs) hold great prognostic potential being deregulated in glioblastomas and highly stable in stored tissue specimens. Moreover, miRNAs control multiple genes representing an additional level of gene regulation possibly more prognostically powerful than a single gene. The aim of the study was to identify a novel miRNA signature with the ability to separate patients into prognostic subgroups. Samples from 40 glioblastoma patients were included retrospectively; patients were comparable on all clinical aspects except overall survival enabling patients to be categorized as short-term or long-term survivors based on median survival. A miRNome screening was employed, and a prognostic profile was developed using leave-one-out cross-validation. We found that expression patterns of miRNAs; particularly the four miRNAs: hsa-miR-107_st, hsa-miR-548x_st, hsa-miR-3125_st and hsa-miR-331-3p_st could determine short- and long-term survival with a predicted accuracy of 78%. Heatmap dendrograms dichotomized glioblastomas into prognostic subgroups with a significant association to survival in univariate (HR 8.50; 95% CI 3.06–23.62; p<0.001) and multivariate analysis (HR 9.84; 95% CI 2.93–33.06; p<0.001). Similar tendency was seen in The Cancer Genome Atlas (TCGA) using a 2-miRNA signature of miR-107 and miR-331 (miR sum score), which were the only miRNAs available in TCGA. In TCGA, patients with O6-methylguanine-DNA-methyltransferase (MGMT) unmethylated tumors and low miR sum score had the shortest survival. Adjusting for age and MGMT status, low miR sum score was associated with a poorer prognosis (HR 0.66; 95% CI 0.45–0.97; p = 0.033). A Kyoto Encyclopedia of Genes and Genomes analysis predicted the identified miRNAs to regulate genes involved in cell cycle regulation and survival. In conclusion, the biology of miRNAs is complex, but the identified 4-miRNA expression pattern could comprise promising biomarkers in glioblastoma stratifying patients into short- and long-term survivors.
Introduction
Glioblastomas are the most common primary malignant brain tumors in adults. Patients diagnosed with glioblastoma have a poor prognosis, but improvements in overall survival have been made over the last decade [1, 2] increasing the necessity for better prognostic markers. Histology combined with new molecular techniques is now the gold standard in glioma diagnostics [3]; as several molecular alterations have proved to be important as diagnostic and prognostic tools e.g. mutations in the isocitrate dehydrogenase 1/2 (IDH1/2) genes and the promoter of telomerase reverse transcriptase (TERT) as well as methylations of the O6-methylguanine-DNA-methyltransferase (MGMT) promoter [3, 4]. However, glioblastoma patients with tumors of similar histological appearance and molecular pattern still show great differences in overall survival. Better separation of patients could help select candidates for more aggressive treatment and active rehabilitation.
A group of non-coding RNAs called microRNAs (miRNAs) can base-pair to target messenger RNA (mRNA) causing translational repression or mRNA degradation based on the level of complementarity between strands. miRNAs originate from endogenous miRNA gene transcripts (pri-miRNAs) or from introns of protein-coding genes [5]. In mammalian cells, miRNAs mainly inhibit mRNA translation under imperfect binding to miRNA-recognition elements (MRE) within the 3’-untranslated region (UTR) of target mRNAs [6, 7].
miRNAs are excellent biomarker candidates as they are more robust than mRNA [8–11], are deregulated in glioblastomas [12], and may control numerous targets [13]. Furthermore, global expression profiling of miRNAs generates more simple data sets than mRNA (2000 miRNAs vs. >40000 mRNAs). Several miRNAs and miRNA signatures have been interrogated to evaluate their diagnostic, prognostic, predictive and/or therapeutic potential in glioblastomas as recently reviewed by Areeb et al. [14]. Dependent on their prognostic impact, some miRNAs have been characterized as pro-oncogenic and others as tumor-suppressive. High levels of miR-21 [15–17], miR-182 [18], and miR-196a/miR-196b [19] as well as low levels of miR-181b [16], miR-195 [20], and miR-196b [20] have been associated with poor prognosis in glioma. miR-196a/b has been found to hold both positive and negative prognostic impact [19, 20]. miRNA signatures, comprised of a combination of miRNAs, have been suggested for prediction of patient prognosis, but signatures found by different studies share no or only few common miRNAs [21–25] warranting further investigation to enable clinical usefulness.
To find new biomarkers allowing separation of prognostic subgroups in glioblastoma, we profiled 2016 miRNAs in 40 patients using formaldehyde-fixed and paraffin-embedded (FFPE) material. Biomarkers developed using FFPE samples have several advantages being available in large amounts and readily accessible for retrospective studies. Since FFPE is the most common way to process tissue in routine pathology, these biomarkers can subsequently be applied to most patient samples. We utilized the leave-one-out cross-validation (LOOCV) method in training and validation sets as well as the entire cohort to produce the best prognostic profile and for most efficient use of data [26].
Materials and methods
Patients
Investigation was carried out using FFPE sections from 40 glioblastoma patients who underwent initial surgical resection between December 1992 and April 2005 at the Department of Neurosurgery, Odense University Hospital, Denmark. No treatment was received prior to surgery. Eligible patients had a documented survival of at least 5 months from initial diagnosis to reduce the impact of post-surgical complications. Patients were categorized as short-term (STS) or long-term survivors (LTS) based on the median survival (13 months), and difference in survival between the two groups was significant using Student’s t-test (P < 0.0001). The pathological specimens had ≥60% vital tumor tissue and a minimum tumor tissue area of 20 mm2. Two neuropathologists diagnosed all samples according to the World Health Organization 2007 guidelines [27].
The use of human tissue was approved by the official Danish ethical review board named the Regional Scientific Ethical Committee of the Region of Southern Demark (Project-ID: S2DO9Oo8O) and the official Danish data registration authority named the Data Protection Authority (file number: 2009-41-3070) and was performed in accordance with the Declaration of Helsinki. As this study was retrospective using archival brain tumor tissue, no written or verbal consent should be obtained, and none of the patients had prohibited the use of their tissue according to the Danish Tissue Application Register.
Immunohistochemistry
mIDH1 status was determined using the BenchMark Ultra instrument (Ventana Medical Systems, Inc) with anti-IDH1 R132H H09 antibody (1:100, Dianova) as previously described [28].
Tissue preparation
Fresh tissue biopsies and cell cultures were fixed in 4% neutral buffered formaldehyde and subsequently paraffin embedded. Four 20 μm sections were cut from each specimen, placed in RNase-free cryotubes, and stored at -20°C.
RNA extraction and purification
Total RNA was extracted from FFPE sections using the RecoverAll™Total Nucleic Acid Isolation Kit (Ambion, AM1975) according to manufacturer’s protocol.
Microarray
RNA was biotin-labeled using the FlashTag™ Biotin HSR RNA Labeling Kit (Affymetrix). An input of 400 nanograms total RNA was used for each reaction. Hybridization, washing and staining were performed using the Affymetrix GeneChip Hybridization, Wash and Stain Kit. All samples were hybridized to the Affymetrix GeneChip miRNA 2.0 Array, which returns expression data of 1105 human mature miRNAs and 911 precursor miRNAs (miRBase v. 15). We have used the Affymetrix platform in several published studies [29–33]. Expression data was normalized using the robust multi-array average (justRMA) method where the raw intensity values are background-corrected, log2-transformed and then quartile-normalized [34]. A linear model was fit to the normalized data to obtain an expression measure for each probe set on each array.
Experimental and statistical setup
Useable miRNA expression data were obtained from 39 patients. One sample (sample 10) was omitted due to low intensity on chip and categorization as an outlier in the principal component analysis (Fig 1A). Another sample (sample 21) had mutated IDH1 (mIDH1) and was omitted from further analysis due to potential confounding of the results [35, 36]. Patients were allocated in two sets: a training set and an independent validation set. The training set consisted of 19 patients of which ten were STS (overall survival 5–9 months) and nine LTS (overall survival > 17 months). The lower cutoff of 9 months and upper cutoff of 17 months was determined based on the median survival of glioblastoma patients being between 12 and 14 months [37], thus both cutoffs are approx. 3 months below and above the reported median survival, respectively. The validation set comprised 19 patients with continuous survival (overall survival 5–21 months). The purpose of the training set was to identify which miRNAs were associated with survival followed by testing the prognostic value of the miRNAs on the validation set. The prognostic miRNA profile was generated from the training set using LOOCV utilizing Student’s t-test for selecting probes in each loop. The t-test compared the expression of genes in LTS and STS patients and ranked genes according to their t-statistic. The number of genes to use for prediction was determined in a nested LOOCV using the inner loop to determine the optimal number of genes and the outer loop to test the performance of the optimally selected genes by a support vector machine (r-project.org package e1071). The nesting was necessary to avoid overfitting the model to the data. The resulting prognostic profile was applied as a support vector machine to the validation set of to make the validation as generalizable as possible. It categorized patients as STS or LTS. Results were subsequently checked against the clinical data. Overall survival based on the prediction was compared using log-rank testing, and prognosticator accuracy was calculated using Fisher’s exact test. An association with overall survival was tested using an unsupervised approach where the prognosticator sorted the validation set without prior grouping into STS and LTS. A heatmap of top ten deregulated miRNAs was generated using Euclidean distance measure and hierarchical clustering. Overall survival between patterns was compared using log-rank testing and Cox proportional hazard regression analysis. A volcano plot was generated to visualize important changes between STS and LTS datasets. Subsequently, LOOCV was carried out using all eligible samples, and the accuracy of the predictive model was calculated. STS and LTS groups were balanced with regards to age, treatment, extent of resection, and age of FFPE material (Table 1). Difference between means was computed using one-way analysis of variance (ANOVA). The raw microarray data files and data underlying the survival analysis have been deposited at Gene Expression Omnibus (GEO) under accession number GSE104554.
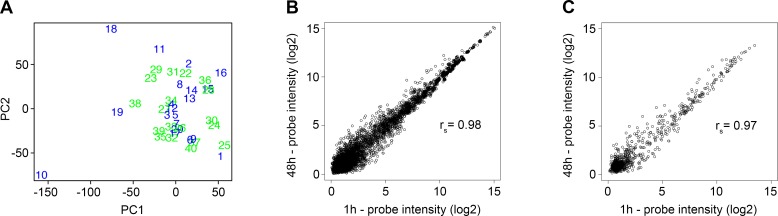
Fig 1. Principal component analysis and effect of fixation time.
(A) Principal component analysis (PCA) plot showing differences between patients. The data supports our initial observation that sample 10 was a technical outlier. (B, C) Fixation time did not significantly affect the miRNA array data as miRNA data, obtained from a glioblastoma short-term fixated 1 hour and 48 hours, showed a strong correlation for (B) all probes (rs = 0.98) and (C) human probes (rs = 0.97).
Table 1. Glioblastoma patient characteristics.
| Parameter | Training set | Validation set | |
|---|---|---|---|
| STS (n = 10) |
LTS (n = 9) |
Continuous (n = 19) | |
| Patient age | |||
| Mean (range) | 59.6 (49.0–68.0) | 56.8 (37.0–72.0) | 55.8 (42.0–69.0) |
| Overall survival (months) | |||
| Mean (range) | 7 (5–9) | 21 (17–32) | 13 (5–21) |
| Radiation | |||
| Yes | 8 | 8 | 19 |
| No | 1 | 1 | 0 |
| Unknown | 1 | 0 | 0 |
| Temozolomide | |||
| Concomitant | 0 | 0 | 0 |
| Adjuvant (at tumor relapse) | 0 | 4 | 1 |
| Unknown | 0 | 0 | 0 |
| Resection | |||
| Partial | 4 | 5 | 11 |
| Radical | 4 | 3 | 5 |
| Unknown | 2 | 1 | 3 |
| Tumor tissue (%) | |||
| Mean (range) | 77.0 (60.0–100.0) | 82.8 (70.0–100.0) | 78.4 (60.0–100.0) |
| Specimen age | |||
| Mean (range) | 13.5 (8.4–19.7) | 12.4 (7.5–17.6) | 13.1 (7.1–20.5) |
Abbreviations: LTS: long-term survivors; STS short-time survivors
Cell cultures and fixation experiment
A glioblastoma short-term culture, T78, established in our laboratory [38], was cultured in serum-free medium and grown as spheroids as previously described [39]. To test the influence of fixation time, we did a correlation test between miRNA profiles of the glioblastoma short-term culture that underwent fixation for 1 hour, 12, 24, or 48 hours. An acceptable Spearman correlation existed between miRNA arrays at 1 hour and 48 hours (rs = 0.98 all probes, rs = 0.97 human probes) validating that fixation time did not affect the results (Fig 1B and 1C).
The Cancer Genome Atlas (TCGA)
miRNA signatures were derived for 533 patients in The Cancer Genome Atlas (TCGA [40], available at https://tcga-data.nci.nih.gov/tcga/) based on expression of miR-107 and miR-331. Clinical data was retrieved from the study by Brennan et al [41]. To make the TCGA dataset comparable to our dataset, only patients with primary glioblastoma, wildtype IDH1 (wtIDH1), and an overall survival between 5–33 months were included in the analysis (n = 247). Patient characteristics of the complete set used in this study (n = 38) and the TCGA dataset (n = 247) is presented in S1 Table. Of the 247 patients, MGMT status was available for only 180 patients. Using the median as cutoff value, patients were divided into those with a high or low score based on the expression levels of miRNA-107, miRNA-331, and the summed expression levels of miRNA-107 and miRNA-331s (miR sum score). Differences in survival were analyzed with log-rank testing. Multivariate analysis was performed including the following variables: age at time of surgery, MGMT status, and miR sum score. Difference in miR sum score between patients with methylated and unmethylated tumors was compared using Student’s unpaired t-test. Data underlying the statistical analysis is available in S1 File.
Target prediction and pathway analysis
Targets of the four hsa-miRNAs used in the 4-miRNA signature were predicted with DIANA-miRPath v.3 provided by the DIANA-microT-CDS algorithm and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway [42–44].
Statistics
Statistical analyses were performed in Prism 5.0 (GraphPad Software, Inc.), STATA (StataCorp LP), and R (affy package from Bioconductor [45]). P-values <0.05 were considered significant.
Results
Developing the prognostic profile
We profiled miRNA expression levels in 40 glioblastoma patients of which 38 qualified for subsequent analyses (Tables 1 and 2). The prognostic profile was developed on the 19 eligible samples in the training set using LOOCV, where a single patient is used as validation and the remaining patients as the training set. This was repeated until every patient had been used once for validation generating a list of optimal predictor miRNAs for each sample. Lastly, selecting the miRNAs present in all lists resulted in an aggregated and final list of miRNAs. The LOOCV yielded a prognostic gene list consisting of ten miRNAs (hsa-miR-107_st, hsa-miR-3125_st, hsa-miR-331-3p_st, hp_hsa-mir-4315-2_s_st, hsa-miR-548x_st, hsa-miR-3126-5p_st, hp_hsa-mir-885_st, hsa-miR-4270_st, hsa-miR-103_st, hsa-miR-887_st) with a predicted accuracy of 68%. However, when tested against the validation set, this prognosticator predicted only one LTS correctly. The rest was predicted as STS (log-rank: P = 0.75; Fischer’s exact test: P = 0.47) corresponding to an accuracy of 58% (11 correct, 8 wrong). Due to the relatively small training and validation sets and very heterogeneous tumors, the accuracy of the predictive model was investigated using all 38 eligible samples. Applying the LOOCV approach to all 38 samples, the model predicted an accuracy of 78% with hsa-miR-107_st, hsa-miR-548x_st, hsa-miR-3125_st and hsa-miR-331-3p_st as optimal predictors. The wrongly categorized patients were STS categorized as LTS corresponding to false negatives (type 2 errors). The four miRNAs were all downregulated in STS (Table 2), and low levels of each of the four miRNAs were significantly associated with poorer prognosis both using the median as a predefined cutoff (S1 Fig) and the optimal cut-point (S2 Fig).
Table 2. Glioblastoma patient characteristics.
| ProbeID | Fold change | P-value (unadjusted) |
|---|---|---|
| hsa-miR-107_st | -0.3050 | 0.000024 |
| hsa-miR-3125_st | -0.3397 | 0.000092 |
| hsa-miR-331-3p_st | -1.0500 | 0.000101 |
| hp_hsa-mir-4315-2_s_st | -0.2556 | 0.000172 |
| hsa-miR-548x_st | -1.2050 | 0.000204 |
| hsa-miR-3126-5p_st | -0.6107 | 0.000227 |
| hp_hsa-mir-885_st | 0.3883 | 0.000292 |
| hsa-miR-4270_st | 1.1590 | 0.000676 |
| hsa-miR-103_st | -0.3822 | 0.000705 |
| hsa-miR-887_st | -0.7126 | 0.000873 |
The ten most significantly deregulated miRNAs in the 38 glioblastomas comparing STS to LTS depicted as fold change relative to LTS. Using the leave-one-out cross validation approach, the model predicted an accuracy of 78% with hsa-miR-107_st, hsa-miR-548x_st, hsa-miR-3125_st and hsa-miR-331-3p_st as optimal predictors (indicated with bold).
STS and LTS have different miRNA profiles
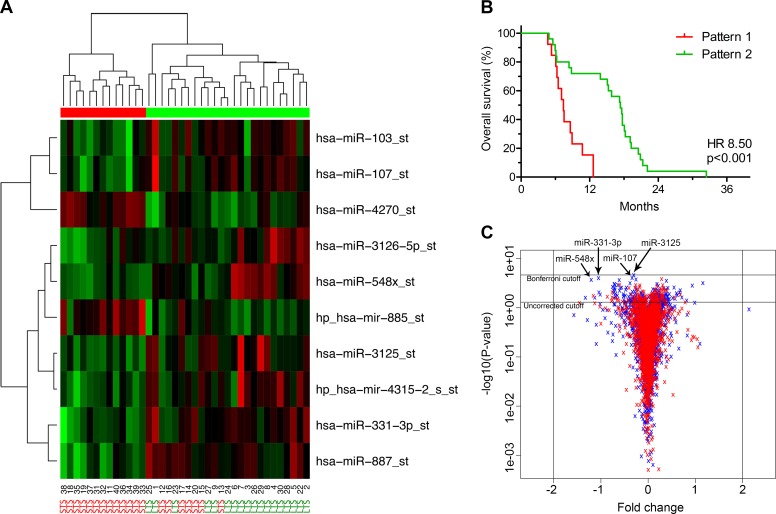
Plotting the ten most deregulated miRNAs in a heatmap with dendrograms suggested that two overall patterns existed within the glioblastomas (Table 2 and Fig 2A). Pattern one was characterized by 13 patients with an overall survival shorter than 13 months, i.e. STS, whereas pattern two mostly characterized patients with an overall survival longer than 13 months, i.e. LTS, and included 18 LTS and 7 STS. The Kaplan Meier plot and log-rank statistics showed a significant separation in overall survival between the two patterns (Hazard ratio (HR) 8.50; 95% confidence interval (CI) 3.06–23.62; P < 0.001) (Fig 2B), also independent of age, second line treatment, and degree of resection (HR 9.84; 95% CI 2.93–33.06; P < 0.001) (Table 3). The two patterns showed greater prognostic impact compared to the single miRNAs in the 4-miRNA signature (S1 and S2 Figs). Yet, no miRNAs were above the two-fold threshold while being statistically significant in the t-test before or after Bonferroni adjustment for multiple testing (Fig 2C).
Fig 2. Short-(STS) and long-term (LTS) glioblastoma survivors have different microRNA (miRNA) profiles.
(A) Heatmap of the ten most deregulated miRNAs in STS and LTS. STS and LTS are grouped into two overall patterns as shown by the dendrograms. Pattern one (red bar) was characterized by STS whereas pattern two (green bar) mostly characterized LTS (18 LTS and 7 STS). In the heatmap, red represents upregulated miRNAs and green represents downregulated miRNAs. (B) Kaplan Meier plot showing a significant separation in overall survival between the two patterns. (C) Volcano plot illustrating that no miRNAs were significantly deregulated above the two-fold threshold. Blue represent normal fold changes and p-values while red represent permutated values. The four miRNAs included in the signature are indicated with arrows.
Table 3. The two miRNA patterns and multivariate analysis.
| Variable | Hazard Ratio (95% CI) | P-value | |
|---|---|---|---|
| Pattern | 1/2 | 9.84 (2.93–33.06) | <0.001 |
| Age | Continuous | 0.99 (0.95–1.03) | 0.59 |
| Radiation therapy | No/Yes | 1.51 (0.18–12.76) | 0.70 |
| Chemotherapy | No/Yes | 1.74 (0.62–4.85) | 0.29 |
| Resection | Partial/Complete | 0.77 (0.33–1.79) | 0.54 |
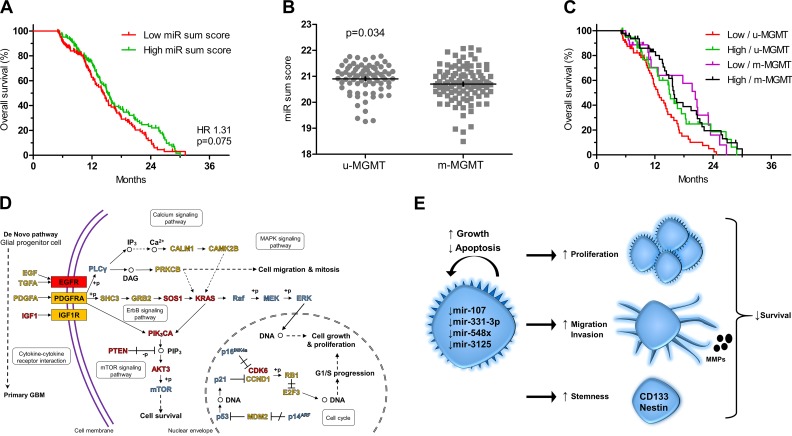
To further investigate the role of the miRNA profile, we evaluated the TCGA database with data available for miR-107 and miR-331. The included patients were all diagnosed with primary glioblastoma, had wtIDH1 and an overall survival between 5–33 months. No significant difference in overall survival was found for miR-107 or miR-331 alone (S3 Fig). However, combining the two miRNAs into a summed score, patients with low miR sum score tended to have a poorer survival than patients with high sum score (HR 1.31; 95% CI 0.97–1.77; P = 0.075) (Fig 3A). When only including patients with known MGMT status in the analysis, low miR sum score was significantly associated with poorer outcome (HR 1.59; 95% CI 1.09–2.30; P = 0.014), and looking at the relation between the sum score and MGMT methylation status, unmethylated tumors had significantly higher miR sum score compared to methylated tumors (P = 0.034) (Fig 3B). Patients with low sum score and unmethylated tumors had the shortest overall survival compared to patients with high sum score and unmethylated tumors (P = 0.025), patients with low sum score and methylated tumors (P = 0.014) as well as patients with high sum score and methylated tumors (P = 0.001) (Fig 3C). Adjusting for age and MGMT status, low miR sum score was independently associated with poorer prognosis (HR 1.52; 95% CI 1.04–2.22; P = 0.029) (Table 4).
Fig 3. In silico gene expression analysis and KEGG pathway analysis.
(A) Low summed scores for miR-107 and miR-331-3p (miR sum score) tended to associate with poorer prognosis in the TCGA data set (n = 247) when dichotomized at the median. (B) Glioblastomas with unmethylated MGMT promoter (u-MGMT) had higher miR sum score than glioblastomas with methylated MGMT promoter (m-MGMT). (C) Stratified into groups based on MGMT methylation status and miR sum score, patients with low miR sum score and u-MGMT had the shortest survival. (D) KEGG pathway analysis based on predicted targets of hsa-miR-107, hsa-331-3p, hsa-548x and hsa-3125 in the Glioma Pathway performed using the DIANA-mirPath tool. Genes regulated by at least two miRNAs are indicated with red, genes regulated by one miRNA with yellow, and genes not regulated by any of the miRNAs with blue. (E) Possible mechanisms by which downregulation of the 4-miRNA signature contribute to shorter survival in patients with glioblastoma.
Table 4. TCGA and multivariate analysis.
| Variable | Hazard Ratio (95% CI) | P-value | |
|---|---|---|---|
| Age | Continuous | 1.02 (1.00–1.03) | 0.007 |
| MGMT status | u-MGMTm-MGMT | 0.68 (0.46–1.00) | 0.045 |
| miR sum score | High/Low | 1.52 (1.04–2.22) | 0.029 |
KEGG pathway analysis
Target genes of the 4-signature miRNAs were predicted using DIANA-miRPath [42–44]. In the Glioma Pathway, genes involved in ErbB, mTOR, and MAPK signaling pathways as well as genes important for cell cycle were found to be most likely regulated by the miRNAs e.g. Kirsten rat sarcoma viral oncogene homolog (KRAS) (miR-3125, miR-548x-3p, miR-548x-5p), phosphatase and tensin homolog (PTEN) (miR-548x-3p, miR-548x-5p), V-akt murine thymoma viral oncogene homolog 3 (AKT3) (miR-107, miR-548x-3p), and cyclin-dependent kinase 6 (CDK6) (miR-107, miR-548x-3p) (Fig 3D). Generally, the miRNAs had predicted targets in many pathways important to cancer progression e.g. angiogenesis, invasion, proliferation, and survival (Table 5).
Table 5. KEGG pathway enriched for mRNAs predicted to be targeted by miRNAs in the 4-miRNA signature.
| KEGG pathway | P-value | # genes | # miRNAs | miRNAs |
|---|---|---|---|---|
| Adherens junction | 6.28e-11 | 45 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| Pathways in cancer | 8.82e-11 | 159 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| Proteoglycans in cancer | 1.13e-08 | 68 | 5 | mir-107, -331-3p, -3125, -548x-3p, -548x-5p |
| Signaling pathways regulating pluripotency of stem cells | 2.01e-08 | 68 | 5 | mir-107, -331-3p, -3125, -548x-3p, -548x-5p |
| ErbB signaling pathway | 2.85e-07 | 45 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| MAPK signaling pathway | 0.009 | 92 | 5 | mir-107, -331-3p, -3125, -548x-3p, -548x-5p |
| PI3K-Akt signaling pathway | 0.016 | 113 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| Glioma | 4.95e-07 | 33 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| Ras signaling pathway | 7.08e-05 | 86 | 5 | mir-107, -331-3p, -3125, -548x-3p, -548x-5p |
| p53 signaling pathway | 0.023 | 28 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
| Cell cycle | 0.016 | 48 | 4 | mir-107, -3125, -548x-3p, -548x-5p |
Discussion
We investigated whether a good prognostic miRNA profile could be generated from the most deregulated miRNAs in the training set consisting of 19 eligible glioblastomas resulting in an optimal profile of ten miRNAs. Applying the LOOCV procedure to the entire dataset, the predicted accuracy of the prognostic profile was 78% using the four miRNAs hsa-miR-107_st, hsa-miR-548x_st, hsa-miR-3125_st and hsa-miR-331-3p_st. These miRNAs were all slightly downregulated in STS suggesting a protective role in glioblastomas. Examining the TCGA data based on expression of miR-107 and miR-331, low miR sum score tended to associate with poor prognosis in the univariate analysis suggesting that miR-548 and miR-3125 may be important prognosticators in the miRNA profile. However, when including only patients with known MGMT methylation status, low miR sum score was an independent negative prognostic factor. Unfortunately, only two of the four miRs included in our signature were available in the TCGA dataset, thereby preventing complete validation of our 4-miRNA signature. Further, the TCGA 2-miR sum score may be a less strong prognosticator compared to the 4-miRNA signature. Also, the miR sum score appeared to have higher prognostic value in patients with unmethylated MGMT promoters.
Currently, glioblastoma STS can to some extent be identified using molecular markers e.g. IDH, MGMT, and TERT [4]. Similarly, dividing patients into risk classes using the Radiation Therapy Oncology Group-Recursive Partitioning Analysis (RTOG-RPA) has proven applicable for differentiating between LTS and STS [46, 47]. The 4-miRNA signature presented in this study may have a potential in daily pathology to further stratify patients into high or low risk groups. Hsa-miR-107 belongs to the miR-103 family both sharing homologous precursor. In the present study, hsa-miR-107 and hsa-miR-103 were among the top ten most significantly deregulated miRNAs with similar fold changes. Hsa-miR-107 has been shown to be a glioma suppressor in functional studies, and its downregulation has been reported in gliomas, glioma cell lines, and glioma stem-like cells [48–50]. Overexpression of hsa-miR-107 in glioma cells suppressed their proliferation and their migratory, invasive, and angiogenetic capabilities by targeting p53 [50], Notch-2 [48, 49], CDK6 [50], matrix metalloproteinase (MMP)-12 [48], and vascular endothelial growth factor (VEGF) [51]. Additionally, the glioma stem cell-like markers CD133 and nestin were downregulated when hsa-miR-107 was overexpressed [48]. A possible mechanism for the downregulation of hsa-miR-107 could be its localization on the long arm of chromosome 10 (10q), which is lost in up to 80% of glioblastomas alongside other tumor suppressors e.g. PTEN [35, 52]. Alternatively, downregulation could be explained by epigenetic silencing of the hsa-miR-107 promoter as seen in pancreatic cancer [53]. Recently, expression levels of hsa-miR-107 were reported to be diminished in gliomas compared to normal brain and in high-grade gliomas compared to low-grade gliomas. Further, low expression was associated with shorter overall and progression-free survival looking at all glioma grades combined [54], overall indicating that low hsa-miR-107 is involved in tumor aggressiveness. miR-331-3p was reported to be downregulated in glioblastoma cell lines compared to normal brain, and overexpression of miR-331-3p inhibited proliferation, clonogenic growth, and migration in vitro by reducing mRNA levels of neuropilin-2 [55]. Further, miR-331-3p was shown to regulate EGFR in glioblastoma cells resulting in reduced AKT activity [56]. The two remaining miRNAs in the 4-miRNA signature are largely unknown in glioma biology. Altogether, studies and target prediction in DIANA-mirPath suggest various tumor suppressive roles in pathways relating to cell survival, migration, and mitosis [43]. Theoretically, downregulation of the miRNAs in glioblastomas may lead to more aggressive tumors thereby resulting in poorer prognosis (Fig 3E).
Our findings that miRNA deregulation may predict prognosis in glioblastomas are consistent with three recent studies [21–23]. However, these studies used other miRNAs to generate their prognostic profiles. We measured miRNA expression levels using the Affymetrix platform. Zhang et al. used the Illumina miRNA Expression BeadChip [23], Niyazi et al. the Geniom Biochip [21], and Srinivasan et al. TCGA data that contain profiling data originating from the Agilent Human 8x15K miRNA platform [22]. Only Zhang et al. performed cross-platform validation by comparing data obtained from TaqMan assays and the Agilent Human 8x15K miRNA platform (TCGA data). Even though we used an equal amount of training samples as Zhang et al. (40 and 41, respectively), we did not identify the same miRNAs. This may be due to different ethnic groups. Although Zhang et al. were successful in validating their 5-miRNA profile in a different ethnic group (the TCGA); these records only contained expression data for three out of five miRNAs. The dissimilar miRNA profiles could also be a result of concluding on heterogeneous tumors with heterogeneous treatment histories and inclusion of patients with overall survival of weeks.
In the present study, profiling was done on a comparable patient cohort exploiting FFPE material, the most common way to store tissue samples. The patient sets used were balanced with regards to age, treatment, extent of resection, and age of FFPE material. Based on histological verification, all specimens had high tumor percentages. The objective was to study the impact of miRNAs on tumor biology and aggressiveness rather than the response to treatment including temozolomide. Whereas most patients receive radiotherapy, elderly patients with wtIDH and MGMT-unmethylated tumors are suggested not to receive temozolomide [57, 58]. We therefore decided to use tissue from before temozolomide was introduced to the standard of care to make the interpretation of results less complex. However, our analyses from the TCGA database also suggested that the miRNAs investigated in this study primarily has a prognostic influence in patients whose tumors have unmethylated MGMT promoters, and these patients do not have a significant survival benefit from TMZ treatment [59]. Further, all patients had a documented survival of at least 5 months from initial diagnosis to avoid confounding from death due to post-surgical complications. We used the LOOCV which is more powerful for distinguishing predefined classes than the clustering approach adopted by Niyazi et al. [21]. Further, the clustering analysis does not provide valid statistical identification of differentially expressed miRNAs. A drawback in our study is the limited number of patients. Further, the fold changes of the identified miRNAs were small and may not be sufficiently deregulated to be applied as solid biomarkers. To examine the prognostic strength of this 4-miRNA signature, the results should be validated on a larger patient cohort including patients who have received multimodal treatment with surgery followed by radio-chemotherapy.
In summary, we have identified a novel miRNA signature based on an independent cohort in which all the patients are clinically treated in an identical manner. Our data suggest that future identification of glioblastoma STS and LTS may consist of evaluating expression patterns of miRNAs; particularly the expression of the four miRNAs hsa-miR-107_st, hsa-miR-548x_st, hsa-miR-3125_st and hsa-miR-331-3p_st. Heatmap dendrograms dichotomized glioblastomas into prognostic subgroups that were significantly different in uni- and multivariate analyses. Using the TCGA dataset we could validate the prognostic impact of miR-107 and miR-331 with low levels being independently associated with shorter survival. The miRNAs identified in the current study were all linked to larger signaling pathways that work controlling key cellular phenotypes.
Although various reports support a great future for miRNAs as biomarkers, major discrepancies exist across studies, and improved evaluation in the future will require standardization of methods and normalization.
Supporting information
(A) Low expression of hsa-miR-107 was significantly associated with shorter overall survival. (B) The similar association was found for hsa-miR-331-3p, (C) hsa-miR-548x, and (D) hsa-miR-3125.
(TIF)
(A) Low expression of hsa-miR-107 was significantly associated with shorter overall survival. (B) The similar association was found for hsa-miR-331-3p, (C) hsa-miR-548x, and (D) hsa-miR-3125.
(TIF)
(A) Expression levels of miR-107 did not impact survival. (B) miR-331 levels did not correlate significantly with overall survival.
(TIF)
(XLSX)
(PDF)
Acknowledgments
We thankfully acknowledge Charlotte Aaberg-Jessen for providing cell culture material to this study as well as Medical Prognosis Institute A/S for providing arrays and reagents.
A part of this study is based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Data Availability
All relevant data are available from Gene Expression Omnibus at the following accession number: GSE104554.
Funding Statement
The work was supported by the Region of Southern Denmark and The Danish Council for Independent Research. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncology. 2009;10(5):459–66. doi: 10.1016/S1470-2045(09)70025-7 . [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–96. doi: 10.1056/NEJMoa043330 . [DOI] [PubMed] [Google Scholar]
- 3.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Ellison DW, Figarella-Branger D, et al. WHO classification of tumours of the central nervous system, Revised, 4th edition International Agency for Research on Cancer (IARC), Lyon: 2016. [Google Scholar]
- 4.Delgado-Lopez PD, Corrales-Garcia EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2016. Epub 2016/03/11. doi: 10.1007/s12094-016-1497-x . [DOI] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Molecular cell biology. 2009;10(2):126–39. doi: 10.1038/nrm2632 . [DOI] [PubMed] [Google Scholar]
- 6.Kiriakidou M, Nelson P, Lamprinaki S, Sharma A, Mourelatos Z. Detection of microRNAs and assays to monitor microRNA activities in vivo and in vitro. Methods in molecular biology (Clifton, NJ). 2005;309:295–310. doi: 10.1385/1-59259-935-4:295 . [DOI] [PubMed] [Google Scholar]
- 7.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & development. 2001;15(2):188–200. ; PubMed Central PMCID: PMC312613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doleshal M, Magotra AA, Choudhury B, Cannon BD, Labourier E, Szafranska AE. Evaluation and validation of total RNA extraction methods for microRNA expression analyses in formalin-fixed, paraffin-embedded tissues. The Journal of molecular diagnostics: JMD. 2008;10(3):203–11. doi: 10.2353/jmoldx.2008.070153 ; PubMed Central PMCID: PMC2329784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. Rna. 2007;13(10):1668–74. doi: 10.1261/rna.642907 ; PubMed Central PMCID: PMC1986820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC biotechnology. 2007;7:36 doi: 10.1186/1472-6750-7-36 ; PubMed Central PMCID: PMC1914054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall JS, Taylor J, Valentine HR, Irlam JJ, Eustace A, Hoskin PJ, et al. Enhanced stability of microRNA expression facilitates classification of FFPE tumour samples exhibiting near total mRNA degradation. British journal of cancer. 2012;107(4):684–94. doi: 10.1038/bjc.2012.294 ; PubMed Central PMCID: PMC3419950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermansen SK, Kristensen BW. MicroRNA biomarkers in glioblastoma. Journal of neuro-oncology. 2013;114(1):13–23. Epub 2013/05/24. doi: 10.1007/s11060-013-1155-x . [DOI] [PubMed] [Google Scholar]
- 13.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nature reviews Genetics. 2012;13(4):271–82. doi: 10.1038/nrg3162 . [DOI] [PubMed] [Google Scholar]
- 14.Areeb Z, Stylli SS, Koldej R, Ritchie DS, Siegal T, Morokoff AP, et al. MicroRNA as potential biomarkers in Glioblastoma. Journal of neuro-oncology. 2015;125(2):237–48. Epub 2015/09/24. doi: 10.1007/s11060-015-1912-0 . [DOI] [PubMed] [Google Scholar]
- 15.Hermansen SK, Dahlrot RH, Nielsen BS, Hansen S, Kristensen BW. MiR-21 expression in the tumor cell compartment holds unfavorable prognostic value in gliomas. Journal of neuro-oncology. 2013;111(1):71–81. Epub 2012/10/30. doi: 10.1007/s11060-012-0992-3 . [DOI] [PubMed] [Google Scholar]
- 16.Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. European journal of cancer (Oxford, England: 1990). 2010;46(9):1640–9. doi: 10.1016/j.ejca.2010.02.003 . [DOI] [PubMed] [Google Scholar]
- 17.Barbano R, Palumbo O, Pasculli B, Galasso M, Volinia S, D'Angelo V, et al. A miRNA signature for defining aggressive phenotype and prognosis in gliomas. PloS one. 2014;9(10):e108950 Epub 2014/10/04. doi: 10.1371/journal.pone.0108950 ; PubMed Central PMCID: PMCPmc4184816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L, Mao P, Song L, Wu J, Huang J, Lin C, et al. miR-182 as a prognostic marker for glioma progression and patient survival. The American journal of pathology. 2010;177(1):29–38. doi: 10.2353/ajpath.2010.090812 ; PubMed Central PMCID: PMC2893648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16(16):4289–97. Epub 2010/07/06. doi: 10.1158/1078-0432.ccr-10-0207 . [DOI] [PubMed] [Google Scholar]
- 20.Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, Lzicarova E, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O-6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer science. 2011;102(12):2186–90. doi: 10.1111/j.1349-7006.2011.02092.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niyazi M, Zehentmayr F, Niemoller OM, Eigenbrod S, Kretzschmar H, Schulze-Osthoff K, et al. MiRNA expression patterns predict survival in glioblastoma. Radiat Oncol. 2011;6:153 doi: 10.1186/1748-717X-6-153 ; PubMed Central PMCID: PMC3235977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivasan S, Patric IR, Somasundaram K. A ten-microRNA expression signature predicts survival in glioblastoma. PloS one. 2011;6(3):e17438 doi: 10.1371/journal.pone.0017438 ; PubMed Central PMCID: PMC3069027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Zhang J, Yan W, You G, Bao Z, Li S, et al. Whole-genome microRNA expression profiling identifies a 5-microRNA signature as a prognostic biomarker in Chinese patients with primary glioblastoma multiforme. Cancer. 2013;119(4):814–24. doi: 10.1002/cncr.27826 . [DOI] [PubMed] [Google Scholar]
- 24.Cheng W, Ren X, Cai J, Zhang C, Li M, Wang K, et al. A five-miRNA signature with prognostic and predictive value for MGMT promoter-methylated glioblastoma patients. Oncotarget. 2015;6(30):29285–95. Epub 2015/09/01. doi: 10.18632/oncotarget.4978 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes J, Thygesen H, Tumilson C, Droop A, Boissinot M, Hughes TA, et al. Prediction of clinical outcome in glioblastoma using a biologically relevant nine-microRNA signature. Molecular oncology. 2015;9(3):704–14. Epub 2014/12/17. doi: 10.1016/j.molonc.2014.11.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. Journal of the National Cancer Institute. 2003;95(1):14–8. . [DOI] [PubMed] [Google Scholar]
- 27.Louis DN, Ohgaki H., Wiestler O.D., Cavenee W.K. WHO Classification of Tumours of the Central Nervous System, Fourth Edition Lyon: International Agency for Research on Cancer (IARC); 2007. [Google Scholar]
- 28.Dahlrot RH, Kristensen BW, Hjelmborg J, Herrstedt J, Hansen S. A population-based study of high-grade gliomas and mutated isocitrate dehydrogenase 1. International journal of clinical and experimental pathology. 2013;6(1):31–40. Epub 2012/12/14. ; PubMed Central PMCID: PMCPMC3515987. [PMC free article] [PubMed] [Google Scholar]
- 29.Patnaik SK, Dahlgaard J, Mazin W, Kannisto E, Jensen T, Knudsen S, et al. Expression of microRNAs in the NCI-60 cancer cell-lines. PloS one. 2012;7(11):e49918 Epub 2012/12/05. doi: 10.1371/journal.pone.0049918 ; PubMed Central PMCID: PMCPmc3509128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carlsen AL, Joergensen MT, Knudsen S, de Muckadell OB, Heegaard NH. Cell-free plasma microRNA in pancreatic ductal adenocarcinoma and disease controls. Pancreas. 2013;42(7):1107–13. Epub 2013/09/21. doi: 10.1097/MPA.0b013e318296bb34 . [DOI] [PubMed] [Google Scholar]
- 31.Knudsen S, Hother C, Gronbaek K, Jensen T, Hansen A, Mazin W, et al. Development and blind clinical validation of a microRNA based predictor of response to treatment with R-CHO(E)P in DLBCL. PloS one. 2015;10(2):e0115538 Epub 2015/02/19. doi: 10.1371/journal.pone.0115538 ; PubMed Central PMCID: PMCPmc4333339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winther M, Knudsen S, Dahlgaard J, Jensen T, Hansen A, Jensen PB, et al. Clinical Impact of a Novel MicroRNA Chemo-Sensitivity Predictor in Gastrooesophageal Cancer. PloS one. 2016;11(2):e0148070 Epub 2016/02/18. doi: 10.1371/journal.pone.0148070 ; PubMed Central PMCID: PMCPmc4757421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prahm KP, Hogdall C, Karlsen MA, Christensen IJ, Novotny GW, Knudsen S, et al. Clinical validation of chemotherapy predictors developed on global microRNA expression in the NCI60 cell line panel tested in ovarian cancer. PloS one. 2017;12(3):e0174300 Epub 2017/03/24. doi: 10.1371/journal.pone.0174300 ; PubMed Central PMCID: PMCPmc5363866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31(4):e15 ; PubMed Central PMCID: PMC150247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer science. 2009;100(12):2235–41. doi: 10.1111/j.1349-7006.2009.01308.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kloosterhof NK, Bralten LB, Dubbink HJ, French PJ, van den Bent MJ. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? The lancet oncology. 2011;12(1):83–91. doi: 10.1016/S1470-2045(10)70053-X . [DOI] [PubMed] [Google Scholar]
- 37.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine. 2005;352(10):987–96. Epub 2005/03/11. doi: 10.1056/NEJMoa043330 . [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer cell. 2006;9(5):391–403. Epub 2006/05/16. doi: 10.1016/j.ccr.2006.03.030 . [DOI] [PubMed] [Google Scholar]
- 39.Jensen SS, Aaberg-Jessen C, Andersen C, Schroder HD, Kristensen BW. Glioma spheroids obtained via ultrasonic aspiration are viable and express stem cell markers: a new tissue resource for glioma research. Neurosurgery. 2013;73(5):868–86; discussion 86. Epub 2013/07/28. doi: 10.1227/NEU.0000000000000118 . [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. Epub 2008/09/06. doi: 10.1038/nature07385 ; PubMed Central PMCID: PMCPMC2671642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77. Epub 2013/10/15. doi: 10.1016/j.cell.2013.09.034 ; PubMed Central PMCID: PMCPmc3910500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, et al. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic acids research. 2012;40(Web Server issue):W498–504. Epub 2012/06/01. doi: 10.1093/nar/gks494 ; PubMed Central PMCID: PMCPmc3394305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: Integrating human and mouse microRNAs in pathways. Bioinformatics. 2009;25(15):1991–3. doi: 10.1093/bioinformatics/btp299 . [DOI] [PubMed] [Google Scholar]
- 44.Fazi B, Felsani A, Grassi L, Moles A, D'Andrea D, Toschi N, et al. The transcriptome and miRNome profiling of glioblastoma tissues and peritumoral regions highlights molecular pathways shared by tumors and surrounding areas and reveals differences between short-term and long-term survivors. Oncotarget. 2015;6(26):22526–52. Epub 2015/07/19. doi: 10.18632/oncotarget.4151 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. Epub 2004/02/13. doi: 10.1093/bioinformatics/btg405 . [DOI] [PubMed] [Google Scholar]
- 46.Simon JM, Noel G, Chiras J, Hoang-Xuan K, Delattre JY, Baillet F, et al. Radiotherapy and chemotherapy with or without carbogen and nicotinamide in inoperable biopsy-proven glioblastoma multiforme. Radiotherapy and oncology: journal of the European Society for Therapeutic Radiology and Oncology. 2003;67(1):45–51. Epub 2003/05/22. . [DOI] [PubMed] [Google Scholar]
- 47.Paravati AJ, Heron DE, Landsittel D, Flickinger JC, Mintz A, Chen YF, et al. Radiotherapy and temozolomide for newly diagnosed glioblastoma and anaplastic astrocytoma: validation of Radiation Therapy Oncology Group-Recursive Partitioning Analysis in the IMRT and temozolomide era. Journal of neuro-oncology. 2011;104(1):339–49. Epub 2010/12/25. doi: 10.1007/s11060-010-0499-8 ; PubMed Central PMCID: PMCPmc3151374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, Chen XR, Chen FF, Liu Y, Li P, Zhang R, et al. MicroRNA-107 inhibits U87 glioma stem cells growth and invasion. Cellular and molecular neurobiology. 2013;33(5):651–7. doi: 10.1007/s10571-013-9927-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Chen XR, Zhang R, Li P, Liu Y, Yan K, et al. MicroRNA-107 inhibits glioma cell migration and invasion by modulating Notch2 expression. Journal of neuro-oncology. 2013;112(1):59–66. doi: 10.1007/s11060-012-1037-7 . [DOI] [PubMed] [Google Scholar]
- 50.Chen L, Zhang R, Li P, Liu Y, Qin K, Fa ZQ, et al. P53-induced microRNA-107 inhibits proliferation of glioma cells and down-regulates the expression of CDK6 and Notch-2. Neuroscience letters. 2013;534:327–32. doi: 10.1016/j.neulet.2012.11.047 . [DOI] [PubMed] [Google Scholar]
- 51.Chen L, Li ZY, Xu SY, Zhang XJ, Zhang Y, Luo K, et al. Upregulation of miR-107 Inhibits Glioma Angiogenesis and VEGF Expression. Cellular and molecular neurobiology. 2015. Epub 2015/06/19. doi: 10.1007/s10571-015-0225-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolter M, Werner T, Malzkorn B, Reifenberger G. Role of microRNAs Located on Chromosome Arm 10q in Malignant Gliomas. Brain pathology (Zurich, Switzerland). 2015. Epub 2015/08/01. doi: 10.1111/bpa.12294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology: official journal of the International Association of Pancreatology. 2009;9(3):293–301. doi: 10.1159/000186051. ; PubMed Central PMCID: PMC2835374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Y, Wei Y, Wang J, Ao Q, Gong K, Zuo H. Decreased expression of microRNA-107 predicts poorer prognosis in glioma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(6):4461–6. Epub 2015/01/19. doi: 10.1007/s13277-015-3086-y . [DOI] [PubMed] [Google Scholar]
- 55.Epis MR, Giles KM, Candy PA, Webster RJ, Leedman PJ. miR-331-3p regulates expression of neuropilin-2 in glioblastoma. Journal of neuro-oncology. 2014;116(1):67–75. Epub 2013/10/22. doi: 10.1007/s11060-013-1271-7 ; PubMed Central PMCID: PMCPmc3889298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giles KM, Barker A, Zhang PM, Epis MR, Leedman PJ. MicroRNA regulation of growth factor receptor signaling in human cancer cells. Methods in molecular biology (Clifton, NJ). 2011;676:147–63. Epub 2010/10/12. doi: 10.1007/978-1-60761-863-8_11 . [DOI] [PubMed] [Google Scholar]
- 57.Hegi ME, Stupp R. Withholding temozolomide in glioblastoma patients with unmethylated MGMT promoter—still a dilemma? Neuro-oncology. 2015;17(11):1425–7. Epub 2015/09/17. doi: 10.1093/neuonc/nov198 ; PubMed Central PMCID: PMCPmc4648310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weller M, van den Bent M, Tonn JC, Stupp R, Preusser M, Cohen-Jonathan-Moyal E, et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. The lancet oncology. 2017;18(6):e315–e29. Epub 2017/05/10. doi: 10.1016/S1470-2045(17)30194-8 . [DOI] [PubMed] [Google Scholar]
- 59.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10(5):459–66. Epub 2009/03/10. doi: 10.1016/S1470-2045(09)70025-7 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Low expression of hsa-miR-107 was significantly associated with shorter overall survival. (B) The similar association was found for hsa-miR-331-3p, (C) hsa-miR-548x, and (D) hsa-miR-3125.
(TIF)
(A) Low expression of hsa-miR-107 was significantly associated with shorter overall survival. (B) The similar association was found for hsa-miR-331-3p, (C) hsa-miR-548x, and (D) hsa-miR-3125.
(TIF)
(A) Expression levels of miR-107 did not impact survival. (B) miR-331 levels did not correlate significantly with overall survival.
(TIF)
(XLSX)
(PDF)
Data Availability Statement
All relevant data are available from Gene Expression Omnibus at the following accession number: GSE104554.