Abstract
Background
We systematically reviewed the genetic variants associated with stroke in genome-wide association studies (GWAS) and examined the emerging priorities and opportunities for rapidly advancing stroke research in the era of Trans-Omics science.
Methods
Using the PRISMA guideline, we searched PubMed and NHGRI- EBI GWAS catalog for stroke studies from 2007 till May 2017.
Results
We included 31 studies. The major challenge is that the few validated variants could not account for the full genetic risk of stroke and have not been translated for clinical use. None of the studies included continental Africans. Genomic study of stroke among Africans presents a unique opportunity for the discovery, validation, functional annotation, trans-omics study and translation of genomic determinants of stroke with implications for global populations. This is because all humans originated from Africa, a continent with a unique genomic architecture and a distinctive epidemiology of stroke; as well as substantially higher heritability and resolution of fine mapping of stroke genes.
Conclusion
Understanding the genomic determinants of stroke and the corresponding molecular mechanisms will revolutionize the development of a new set of precise biomarkers for stroke prediction, diagnosis and prognostic estimates as well as personalized interventions for reducing the global burden of stroke.
Keywords: Stroke, Stroke Epidemiology, Genetic Research, Trans-omics, African Ancestry Population, GWAS, SNP, Genomics
1.0 INTRODUCTION
Stroke is a leading cause of long-term disability, depression and dementia globally.1, 2 For over 15 years, stroke remains the second most common cause of death worldwide with a higher burden among people of African ancestry compared to other populations.2–5 The lifetime risk of stroke has been estimated as one in five for middle-aged women and one in six for middle-aged men.6 Developing precise interventions to prevent and mitigate the devastating consequences of stroke requires a clear understanding of the environmental and genomic risk factors as well as the unique molecular pathways influencing its occurrence, type and outcome.2
We systematically reviewed the literature on genetic variants associated with stroke in genome-wide association studies (GWAS) and examined the emerging priorities and opportunities for rapidly advancing stroke research in the era of Trans-Omics science by including African ancestry populations, especially those from the continent of Africa.
2.0 METHODS
2.1 Search Strategy
Using the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline, we searched the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalog up till May 7, 2017. In addition, we searched PubMed for relevant stroke GWAS studies from 2007 till May 7, 2017. Search terms included “GWAS” “stroke”, “ischemic stroke”, “hemorrhagic stroke”, “lacunar stroke”/”small vessel stroke”, and “large artery atherosclerosis”, and “cardioembolic stroke”/”cardiogenic stroke”.
2.2 Eligibility Criteria
We included individual GWAS studies and consortia studies with meta-analysis of GWAS focusing on stroke or stroke subtypes. We excluded Matsushita et al (2010)7 and Zhang et al (2012)8 which were large candidate gene studies, Zhang et al (2014)9 which was a whole exome sequencing (WES) study, Cole et al (2012)10 and Zhou et al ( 2014) 11 which were small WES studies examining 10 and 9 subjects respectively; Opherk et al (2014) 12 which described white mater hyperintensity (WMH) volume in cerebral autosomal-dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL); Keene et al (2014)13 which described SNPs significantly associated with Vitamins B6 and B12 in stroke patients (not as a risk of stroke); and Traylor et al (2017)14 which was not a GWAS study, but an heritability study which also applied genetic risk score.
3.0 RESULTS
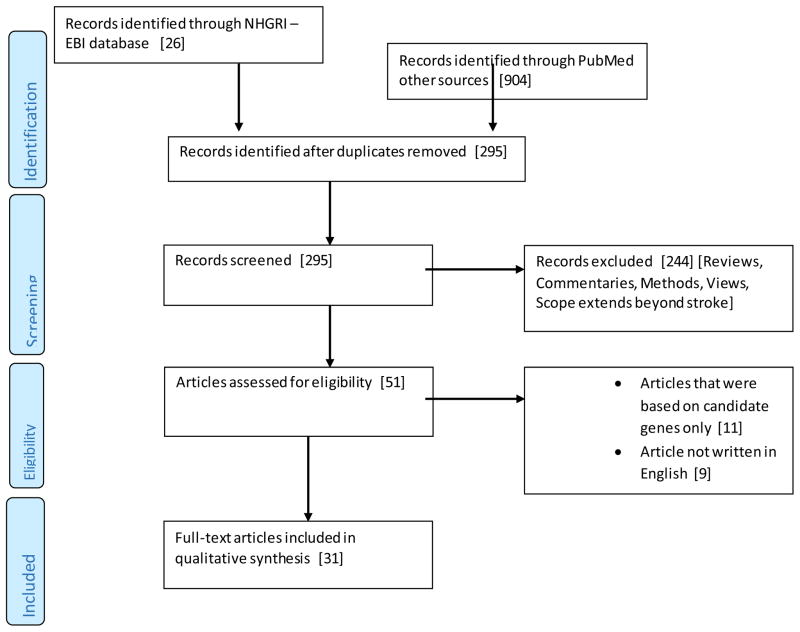
We included 31 studies (Figure 1) presented in Tables 1 and 2. Five studies were individual GWAS while 26 were meta-analyses. We provided information on year of publication, phenotype, sample size, sample ancestry, genetic loci, single nucleotide polymorphisms (SNPs), associated traits, odds ratio and p-values as well as replication status.
Figure 1.
PRISMA Flow Chart of the Search Process for this Review Article
Table 1.
Stroke GWAS Studies: Individual Funded Studies
| First Author | Year | Phenotype | Sample Size (cases/controls) | Sample Ancestry | Genes | Locus | SNPs | Associated trait | OR | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Matarin et al88 | 2007 | IS | 249/268 | European | IMPA2 | −18p11.2 | rs7506045 | IS | 5.39 | 7.07 × 10 (−7) |
|
| ||||||||||
| Gretasrdottir et al 89 | 2008 | IS | 1661/10815 replication | European | PITX2* | 4q25 | rs2200733 | CE (discov) | 1.54 | 8.05 × 10(−9) |
| rs2200733 | IS (All) | 1.26 | 2.18 × 10(−10) | |||||||
| 2,224/2,583;# | rs2200733 | CE (All) | 1.52 | 5.8 × 10(−12) | ||||||
| 2,327/16,760 | rs10033464 | CE (All) | 1.27 | 6.1 × 10(−4) | ||||||
|
| ||||||||||
| Yamada et al90 | 2009 | IS | 131/135 [Panel A] | Japanese | CELSR1 | 22q13 | rs6007897 | IS | 1.85 | 6.0 × 10 (−4) |
| rs4044210 | IS | 1.78 | 1.0 × 10 (−3) | |||||||
| 790/3435 [Panel B]# | LLGL2 | rs1671021 | IS | 0.74 | 4.0 × 10 (−4) | |||||
| 71/1779 [Panel C] | RUVBL2 | rs1062708 | IS | 0.97 | 6.9 × 10 (−1) | |||||
|
| ||||||||||
| Schürks et al91 | 2011 | IS | 164/4958 | European | MEPE | 4q22.1 | rs7698623 | IS | 6.37 | 2.7×10 (−7) |
| IRX4 | 5p15.33 | rs4975709 | IS | 5.06 | 7.7×10(−7 | |||||
|
| ||||||||||
| Hata, Kibo, and Kiyohara92 | 2011 | IS | 1112/1112 | Japanese | PRKCH | 14q22 | rs2230500 | IS | N/A | 9.84 × 10(−7) |
| AGTRL1 | 11q12 | rs9943582 | IS | 2.00 | 6.0 × 10(−3) | |||||
| ARHGEF 10 | rs4376531 | IS | 1.70 | 3.3 × 10(−2) | ||||||
IS-ischemic stroke CE-Cardioembolic stroke;
(pooled)
replicated genes
Table 2.
Stroke GWAS Studies – Consortium-based Meta Analytic Studies
| First Author |
Year | Phenotype | Sample Size (cases/contr ols) |
Sample Ancestry |
Genes | Locus | SNPs | Associated trait |
OR | P-value |
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Bilguvar et al93 | 2008 | IA | 2100/8000 | European; Japanese | PLCL1 | 2q33 | rs1429412 | IA | 1.22 | 5.8 × 10(−7) |
| rs700651 | IA | 1.24 | 4.4 × 10(−8) | |||||||
| SOX17* | 8q12 | rs10958409 | IA | 1.36 | 1.4 × 10(−10) | |||||
| CDKN2A/CDKN2B*, (ANRIL) | 9p21 | rs 9298506 | IA | 1.35 | 1.8 × 10(−9) | |||||
| rs1333040 | IA | 1.29 | 1.4 × 10(−10) | |||||||
|
| ||||||||||
| Ikram et al94 | 2009 | IS | 1164/18,058 | European, AA | NINJ2* | 12p13 | rs11833579 | IS | 1.41 | 2.3 × 10(−10) |
|
| ||||||||||
| Gudbjartss on et al95 | 2009 | IS | 6235/39898 | European | ZFHX3* | 16q22 | rs7193343 | IS (All) | 1.11 | 5.4 × 10 (−4) |
| rs7193343 | IS (CE) | 1.22 | 2.1 × 10 (−4) | |||||||
|
| ||||||||||
| Holliday et al21 | 2012 | IS | 1,162/1,244 | European | - | 6p21.1 | rs556621 | IS, LAA | 1.62 | 3.9×10(−8) |
|
| ||||||||||
| Bellenguez et al96 | 2012 | IS, LAA | 3548/5972 5,859/6,281 (replication) | European | HDAC9* | 7p21 | rs11984041 | IS, LAA | 1.42 | 1.87 × 10(−11) |
|
| ||||||||||
| Traylor et al97 | 2012 | IS | 12 389/62 00 | European | PITX2*, | rs6843082 | IS, CE | 1.36 | 2·’8 × 10(−16) | |
| ZFHX3* | rs879324 | IS,CE | 1.25 | 2·’28×10(−8) | ||||||
| 4 13 | HDAC9* | rs2107595 | IS,LVD | 1.39 | 2·’03×10(−16) | |||||
| 347/29083 (replication) | 9p21 | rs2383207 | IS,LVD | 1·’15 | 3·’32×10(−5) | |||||
| NINJ2* | rs11833579 | IS | 1.06 | 6·’1 × 10 (−4) | ||||||
|
| ||||||||||
| Foroud et al98 | 2012 | IA | 388/397 [1] | European | CDKN2BAS* | 9p21 | rs6475606 | IA | 1.35 | 3.6×10(−8) |
| 1095/1286 [2] | SOX17* | 8q12 | rs1072737 | IA | 1.25 | 8.7×10(−5) | ||||
|
| ||||||||||
| Arning et al99 | 2012 | Pediatric stroke, IS | 270 family-based trios | European | ADAMTS12 | 5 | rs1364044 | Pediatric stroke | 1.91 | 2.91 × 10(−6) |
| ADAMTS2 | 5 | rs469568 | 1.77 | 8.00 × 10(−6) | ||||||
| TRIM29 | 11 | rs2084898 | 2.37 | 4.30 × 10 (−6) | ||||||
| COPS8 | 2 | rs4663691 | 1.9 | 9.79 × 10 (−6) | ||||||
|
| ||||||||||
| Williams et al100 | 2013 | IS | 2,100 twins [1] 4,200 IS [2] 8,900/55,000 [3] |
European | ABO* | 9 | rs505922 | IS,LAA, CE | 0.94 | 2.3 × 10(−2) |
|
| ||||||||||
| Woo et al 25 | 2014 | ICH | 1,545/1,481 | European | TRHDE [PMF1, SLC25A44 ] | 12q21.1 | rs11179580 | ICH, Lobar | 1.56 | 7.0 × 10(−8) |
| 1,681/2,261 (replication) | AA | 1q22 | rs2984613 | ICH, | 1.44 | 1.6 × 10(−8) | ||||
| HA | 1q22 | rs2984613 | Non-Lobar | 1.33 | 2.2×10(−10)a | |||||
|
| ||||||||||
| Traylor et al101 | 2014 | IS,LAA | 6,778/12,095 1,881LAA/50,817 (validation) | European | MMP12 | 11 | rs 660599 | IS,LAA | 1.18 | 2.6 × 10(−8) |
|
| ||||||||||
| Williams et al102 | 2014 | Incident IS | 2710 (FHS) 2100 (VISP) |
European | ALDH1L1 | 3 | rs2364368 | Incident IS | 1.26 | 1.5 × 10(−2) |
|
| ||||||||||
| Foroud et al103 | 2014 | IA | 2617/2548 717/3004 799/2317 |
European | Chr7 (near HDAC9) | 7 | rs10230207 | IA | 1.21 | 9.91×10(−10) |
|
| ||||||||||
| Cotlarcius et al104 | 2014 | IS | 12,389/62,004 | European | SLC17A3 | 6 | rs9379800 | IS, overall, | 1.09 | 1.65 ×10 (−4) |
| SLC17A3 | 6 | rs17271121 | IS, overall | 1.09 | 2.8 × 10 (−4) | |||||
| SLC17A3 | 6 | rs12664474 | IS, overall | 1.08 | 2.86 × 10 (−4) | |||||
| FUT1 | 19 | rs2287921 | IS, overall | 1.06 | 2.09 × 10 (−4) | |||||
| MUT | 6 | rs566295 | IS, SVD | 1.13 | 2.2 × 10 (−4) | |||||
| MTHFR | 1 | rs1801131 | IS, LVD | 1.15 | 1.92 × 10 (−4) | |||||
|
| ||||||||||
| Carty et al24@ | 2015 | ICH, IS | 14 746 AA (1365 IS and 1592 total stroke cases) | AA | ALDH1A2, | 15q21.3 | rs4471613 | Stroke, total | 2.26 | 3.94 × 10 (−8) |
| AQP9, | ||||||||||
| LIPC | rs9899375 | 3.30 | 5.23 × 10 (−9) | |||||||
| CHD3 | 17p13.1 | rs9899375 | IS | 2.57 | 1.20 × 10 (−8) | |||||
| CHD3 | rs704341 | Stroke, total | 1.73 | 5.53 × 10 (−7) | ||||||
| PTPRG | 17p13.1 | rs781542 | 1.35 | 9.64 × 10 (−7) | ||||||
| SPINK2 | rs6880837 | Stroke, total | 1.33 | 3.88 × 10 (−7) | ||||||
| TGFBI | 3p14.2 | rs9345396 | 5.78 | 1.03 × 10 (−7) | ||||||
| TSG1 | rs17145593 | Stroke, total | 1.60 | 2.62 × 10 (−7) | ||||||
| GATA3 | 4q12 | 1.62 | 1.36 × 10 (−7) | |||||||
| GATA3 | 5q31.2 | rs768606 | Stroke, total | 4.29 | 8.94 × 10 (−8) | |||||
| WDFY4 | rs17771318 | 1.45 | ||||||||
| MICAL2 | 6q16.1 | Stroke, total | 1.39 | |||||||
| AGBL1 | rs12291066 | 0.42 | ||||||||
| C21orf81 | 10p14 | Stroke, total | 2.69 | |||||||
| CLDN17 | 10p14 | rs12438353 | ||||||||
| 10q11.22 | rs2822388 | Stroke, total | ||||||||
| 11p15.3 | rs7283054 | Stroke, total | ||||||||
| 15q25.3 | Stroke, total | 8.23 × 10 (−7) | ||||||||
| 21q11.2 | IS | |||||||||
| Stroke, total | 2.28 × 10 (−7) | |||||||||
| 21q21.3 | Stroke, total | 4.96 × 10 (−7) | ||||||||
| 9.75 × 10 (−7) | ||||||||||
|
| ||||||||||
| Holliday et al22 | 2015 | IS | 12,389/62,004 | European | OPRM1 | 6q25.2 | rs17084671 | LAA,SVD | 0.84 | 1.3 × 10 (−7) |
| 6q25.2 | rs6938958 | LAA,SVD | 1.19 | 1.8 × 10 (−7) | ||||||
| 6q25.2 | rs7763080 | LAA,SVD | 0.84 | 1.7 × 10 (−7) | ||||||
|
| ||||||||||
| Pulit et al86 | 2015 | Stroke | 14,300 cases | European | ABCC1 | 16 | rs74475935 | UND | 4·’63 | 5.0 × 10 (−11) |
| IS, ICH | 1,609 cases | AA | ALDH2 | 12 | rs10744777 | IS, SVD | 1·’17 | 2.92 × 10 (−9) | ||
| HDAC9* | 7 | rs11984041 | IS, LAA | 1·’24 | 4·’52 × 10(−9) | |||||
| PITX2* | 4 | rs2634074 | IS, CE | 1·’37 | 2·’79 ×10 (−32) | |||||
| TSPAN2 | 1p13.2 | rs12122341 | IS, LAA | 1·’19 | 1.3 × 10 (−9) | |||||
| ZFHX3 | 16 | rs7193343 | IS, CE | 1·’17 | 2.0 × 10 (−10) | |||||
|
| ||||||||||
| Rannikmae et al105 | 2015 | SVD | 1,545/1,485 - ICH (deep, lobar); (12,389/62,004 [IS] | European | COL4A2* | 13 | rs9521732 | ICH, deep | 1.28 | 7.0 × 10 (−5) |
| 2,733 – IS/WMH | 13 | rs9521733 | ICH, deep | 1.29 | 3.0 × 10 (−5) | |||||
| 9,361-Popn/WMH | 13 | rs9515199 | ICH, deep | 1.28 | 6.0 × 10 (−5) | |||||
|
| ||||||||||
| Anderson et al106 | 2016 | ICH | 1149/1238 | European | CETP | 16 | rs173539 | ICH | 1.25 | 6.0 × 10 (−4) |
| 1625/1845 (replication) | 16 | rs247617 | ICH | 1.24 | 8.74 × 10 (−4) | |||||
| 16 | rs17231506 | ICH | 1.23 | 9.13 × 10 (−4) | ||||||
| 16 | rs711752 | ICH | 1.15 | 2.08 × 10 (−2) | ||||||
| 16 | rs708272 | ICH | 1.15 | 2.23 × 10 (−2) | ||||||
|
| ||||||||||
| Chauhan et al107 | 2016 | Stroke, SVD | 4348/80613 | Multiple | FOXF2 | 6p25 | rs12204590 | Stroke, SVD | 1·’08 | 1·;48 × 10 (−8) |
|
| ||||||||||
| Cheng et al 108 | 2016 | IS, young onset (<60yrs) | 4505/21968 1003/7745 (replication) | European, South Asian AA | HABP2 | 10q25 | rs11196288 | IS, young onset | 1.41 | 9.5× 10 (−9) |
|
| ||||||||||
| Traylor et al 109 | 2016 | Stroke and WMH | 3,670 cases | European | TRIM65 | 17 | rs7214628 | Stroke/W MH | 1.08 | 2.4 × 10 (−15) |
| EFEMP1 | 2 | rs78857879 | Stroke/WMH | 1.11 | 5.0 × 10 (−8) | |||||
| LOC10050584 | 5 | rs17148926 | Stroke/WMH | 1.11 | 9.9 × 10 (−8) | |||||
| EVL | 14 | rs941898 | Stroke/WMH | 1.10 | 4.0 × 10 (−8) | |||||
| C1QL1 | 17 | rs962888 | Stroke/WMH | 1.09 | 1.1 × 10 (−8) | |||||
| COL4A2* | 13 | rs9515201 | Stroke/WMH | 1.09 | 6.9 × 10 (−9) | |||||
| NBEAL1 | 2 | rs72934505 | Stroke/WMH | 1.10 | 2.2 × 10 (−8) | |||||
|
| ||||||||||
| Malik et al 110 | 2017 | LAA | 3127/9778 | European | SERPINA 1 | 14 | rs6647 | LAA | 1.22 | 6.0 × 10 (−9) |
| South Asian | HDAC9* | 7 | rs2023938 | LAA | 1.28 | 7.8 × 10 (−7) | ||||
|
| ||||||||||
| Traylor et al111 | 2017 | SVD | 4203/50728 | European | ZCCHC14 | 16q24.2 | rs12445022 | SVD | 1.16 | 3.2 × 10 (−9) |
IA-intracranial aneurysm; IS-ischemic stroke; ICH-intracerebral hemorrhage; LAA-Large Artery Atherosclerosis; LVD-Large Vessel Disease; SVD-Small Vessel Disease; CE-Cardioembolic stroke; AA-African American; HA-Hispanic American UND-undetermined; Tables 1 and 2 are not limited to associations which attain genome wide significance alone (p < 5.0 × 10 (−8).
indicates genetic associations which have been replicated; @ Carty et al: Table reported findings from primary discovery analysis.
Based on this systematic review, only 24 genetic loci have been found to be associated with stroke. Many of them have neither been validated nor fine-mapped. Up to 61% have small to moderate effect sizes with odds ratios < 1.5. About 87% of the study subjects were European Caucasian, while only 10% were African Americans; however none was from the African continent.”
4.0 DISCUSSION
4.1 Genomic variants associated with stroke
Stroke is a heterogenous trait which includes brain infarction (ischemic stroke with blockage of blood flow); spontaneous intracerebral hemorrhage (bleeding in the brain) or subarachnoid haemorrhage.15 It is defined based on pathological, imaging, or other objective evidence of vascular brain injury in a defined vascular distribution; or clinical evidence of focal vascular brain injury based on symptoms persisting beyond 24 hours or until death, with other etiologies excluded.15 The pathway to the occurrence of stroke involves the cardiovascular cascade including cardiovascular risk factors (e.g. hypertension, diabetes, dyslipidemia, obesity16), and intermediate phenotypes such as atherosclerosis and microaneurysms.2, 17 This makes unravelling the precise genomic determinants of its occurrence, type, severity and outcome and the delineation of its underlying mechanisms challenging.
Nevertheless, several genetic loci [PITX2, HDAC9, ZFHX3, 12q24.2 near ALDH2, 1p13.2 near TSPAN2, CDKN2A/CDKN2B, and others in Tables 1 and 2] have been associated with ischemic stroke or its sub-types from genome-wide association studies (GWAS).5, 18–23 Although some of these loci have been associated with specific subtypes, genetic overlaps have also been reported among them.20, 22 For example, large artery atherosclerosis and small vessel disease, which were previously treated as genetically distinct, may share a substantial genetic component. Thus, combined analyses of both subtypes may increase power to identify small-effect alleles influencing shared pathophysiological processes.20, 22 Furthermore, the novel locus 15q21.3 [AQP9-LIPC gene region] was associated with both ischemic and hemorrhagic stroke in African Americans.24
Genetic variation plays a substantial role in intracerebral hemorrhage (ICH) risk, hematoma volume and outcome,.25 Promising candidates for risk alleles in ICH identified in populations (which had no continental Africans), include variants of the genes Apolipoprotein E (APOE), ACE, PMF1/SLC25A44, COL4A2, and MTHFR 18, 23, 26. Other genetic variants related to hemostasis, lipid metabolism, inflammation, and the central nervous system microenvironment as well as the locus 1q22 25 have also been linked to ICH in single candidate gene studies.26
However, previously reported risk variants, many of which are yet to be validated or fine-mapped, account for only a portion of inherited genetic influence on ischemic stroke or ICH pathophysiology, pointing to additional loci yet to be identified. 27,28
4.2 Evidence Gaps
Although evidence for genetic contributions to stroke exist, the scientific community lacks the evidence required to translate the current knowledge to clinical and community settings for stroke prevention and interventions to improve the health of individuals and populations at risk. Translating current findings into causal variants and target genes, is a major challenge along this pathway. Additional validation and fine-mapping to improve functional annotation for variants, especially those in non-coding regions of the genome, is a necessary step to address this challenge.29 This would require a global effort to expand stroke research to include more diverse populations, particularly African ancestry population, to elucidate the genetics of stroke.
Furthermore, expanding the genetic study of stroke in African populations, a population that has significantly higher susceptibility to stroke, will help elucidate the global heritability of stroke; as well as assist in developing new, and broadening existing, potential therapeutic options. This is because novel variants (SNPs/copy number variants CNVs/insertion-deletions InDels) that contribute to the higher burden, earlier age of onset and poorer outcomes of stroke in people of African ancestry are yet to be characterized. 2, 5, 18
4.3 Advantages of conducting stroke genomics research in African populations
Significant advantages exist to conduct stroke research in African ancestry populations. Below we outline the reasons for conducting said research:
4.3.1. Universal implications
All modern humans originated in Africa before migrating to populate the rest of the world in the last 100,000 years, hence the expression ‘we are all Africans beneath our skin’. 30–35 Therefore combining phenotype and genomic information from continental Africans (old genes in same environment) and the African Diaspora (interactions of old genes with modern environments) could lend novel insights into human evolution and adaptation, health disparities research, disease etiology, pathophysiology, and ancestry-based disease gene mapping. This could have significant universal implications for global populations.30–35
4.3.2. Unique genomic architecture
Genetic adaptations that took place across Africa, particularly against fatal pathogens and ecological forces, have resulted in elevated frequencies of alleles conferring survival advantages detectable in present-day continental Africans and those in the Diaspora.5, 32, 33 Unfortunately, some of these alleles are maladaptive contributing to the disproportionately high burden of some chronic diseases in these groups.5, 32, 33 The power to detect pathogenic genomic variants associated with such alleles is therefore substantially higher among people of African ancestry.
One important example is the 2 missense haplotype variants G1 and G2 in the apolipoprotein L1 (APOL1) gene, which have been found to be associated with end-stage renal disease in people of African ancestry.5, 18, 32, 33, 36 These haplotypes are common in African ancestry populations but absent in European Caucasians and Asians, presumably due to evolutionary positive selection because these APOL1 variants confer resistance to lethal Trypanosoma brucei infections, which cause African sleeping sickness.5, 18, 32, 33 Only by studying people of African ancestry, were the associations of the G1 and G2 APOL1 variants identified, and these variants turned out to account for the large amount of the ethnic disparity in end-stage renal disease that had long been recognized between African and European ancestry populations.5, 18, 32, 33
Novel insights into disease etiology have also been gained by comparing diasporan populations to their ancestral populations in sub-Saharan Africa and by characterizing local admixture at disease risk loci.33 A study found that the association between the LPL SNP rs328 and lipid levels was stronger and the levels of HDL-cholesterol were higher among African Americans with predominantly European ancestry than among those with predominantly African ancestry at this locus. 33 Lipid levels and their association with the LPL variant in African Americans with two African ancestry alleles at this locus were similar to those of West Africans despite widely different lifestyles and diets. 33 This mutation possibly occurred centuries ago and was then transported to America through the trans-Atlantic slave trade. 33
4.3.3. Greater understanding of stroke epidemiology
Substantial disparities in stroke epidemiology exist between people of African ancestry compared to other populations. The burden of stroke in Africa is higher and increasing, with stroke being among the leading causes of morbidity and mortality throughout continental Africa while the burden is less and appears to be declining in many high-income countries.2 Globally, Africa has the highest burden of stroke with age standardized incidence of up to 316/100,000 37; age-adjusted prevalence of as high as1,460/100,000; 38, 39 one month fatality of up to 43% 40, (compared to 4% North America)2, 16, 41, and 3 year fatality of 84% 42. Furthermore, stroke afflicts the younger population (up to 15 years younger2, 43), and presents with a higher proportion of the hemorrhagic type (34% in Africans, vs 9 % in high income countries). 41 Similarly, African Americans experience a higher burden, more hemorrhagic type, younger age of onset and worse outcome compared to European Americans. 2, 5
Some of the racial disparity and higher burden of stroke in people of African ancestry can be attributed to lifestyle and cardio-metabolic factors (eg diabetes mellitus, and hypertension 2, 44) as well as socioeconomic factors such as inadequate access to health care. 5 However, even after adjustment for these factors, substantial risk remains, suggesting a strong genetic influence.5, 32, 33, 45, 46
Studying the genetics of stroke in African ancestry populations may elucidate genetic mechanisms that could not be otherwise revealed in studies of non-African populations (e.g. European ancestry populations). However, most large stroke genomic consortia are based on subjects of predominantly European ancestry. As of May 7, 2017, the National Human Genome Research Institute-European Bioinformatics Institute GWAS catalog included 12 GWAS of stroke; in aggregate, these included 40,801 European Caucasian subjects but only 4,566 African American subjects. No subjects in these studies were from Africa despite the higher heritability of stroke in Africans.5, 14
4.3.4. Substantially higher heritability and greater potential for discovery of novel genes
It has been demonstrated that there is higher stroke genetic heritability among UK African ancestry sample (h2 = 0.35 [SE = 0.19], p = 0.043) compared to European populations (h2=0.16). 14 Thus the sample size requirement for detecting significant SNPs/CNVs/InDels associations may not be as large as what is often required among European populations. Therefore the potential and statistical power for discovery and functional investigation of genomic variants associated with stroke appears to be best among people of African ancestry.
4.3.5. Substantially better resolution for fine mapping
African ancestry populations have been shown to substantially increase the resolution of fine-mapping 5, 47 due to its low linkage disequilibrium and high genetic heterogeneity.18, 19, 35 This is the reason African ancestry data is critically needed (in independent and trans-ethnic analyses) to fine-map loci previously identified in GWAS studies of European populations so as to pinpoint specific causal variant(s) and gene(s) for various complex diseases including stroke. 5, 47
Genomic data for unravelling causative variants can take the form of dense-GWAS29 or whole exome sequencing (WES) or whole genome sequencing (WGS). Dense-GWAS using novel chips such as the 2.5 million SNPs array H3Africa chip48, 49 with 850K custom African content combined with imputation based on 5,500 WGS reference genomes can facilitate functional association analysis. 50 (The reference genomes comprise the Genome Diversity in Africa Project (GDAP)51, Uganda 2000 Genomes52, African Genome Variation Project53, 1000 Genomes54, 55, TrypanoGEN, 48, 49 Baylor College48, 49, and South African Human Genome Programme SAHGP48, 49, 56) The advantage of GWAS is that it is cost-efficient and can cover common variations of small to moderate effects (minor allelic frequency MAF>1 %) in large populations. Performing GWAS in people of African ancestry will substantially increase the diversity within the GWAS catalogue.50
Whole genome sequencing (WGS) has several advantages over GWAS. WGS covers the entire genome and can detect rare variants (with MAF<1 %), as well as other types of functionally important variations (copy number variations, insertions/deletions). 57, 58 Importantly, WGS gives single base resolution of the entire genome which is ~100–300-fold finer resolution than that provided by GWAS chips. Also, WGS is not biased by, or restricted to, the SNPs that have been placed on commercially available SNP chips (assumes commonly used chip-based methods).57, 58 It provides the best possible resource for linkage disequilibrium (LD) mapping due to the maximal marker density and lack of ascertainment bias.
Nevertheless WGS is at least 10 times more expensive than GWAS, and produces massive amounts of data for analysis (100 to 300 fold more than GWAS data), some of which are of uncertain significance.59 With future costs to generate WGS data reducing, WGS in large populations of continental Africans with stroke for discovery39; followed by validation in African Americans, is the most plausible approach to discover and pinpoint causal variants for stroke with implications for the entire human race. This is due to the African origin of all humans and the substantially higher heritability of stroke among African ancestry populations. Meanwhile, dense GWAS with novel chips and imputation using WGS data can also produce useful results, while samples can be stored for future WGS as prices reduce.
4.4 Understanding Complex traits in the era of Trans-Omics science
Although GWAS approach is a very powerful tool to tag genetic loci related to stroke etiology, it may not pinpoint the exact causal genetic variation or delineate the mechanistic path from DNA variation to regulation of gene transcription, translation and post-translation modification, all the way to the pathogenesis of stroke. It is necessary to combine the emerging tools of genomics (including genome wide genotyping with novel high throughput chips, and next generation whole exome sequencing and whole genome sequencing) with functional omics including transcriptomics, epigenomics, proteomics, and metabolomics in various integrative manners. Functional omics can further GWAS findings in two ways. First, functional omics can offer insight into which and how the static DNA variation leads to dynamic functional changes at RNA and protein level. One common application of Trans-Omics approach is to annotate the expression and methylation quantitative trait loci (eQTL and mQTL) for fine-mapping GWAS causal signals. For example, schizophrenia risk-associated SNPs in the Disrupted In Schizophrenia 1 (DISC1) gene has been shown to regulate RNA splicing and thus lead to expression changes of certain isoforms of DISC1 transcripts60. Prostate cancer risk allele rs11986220 found by GWAS locates in an enhancer region and can result in stronger FoxA1 binding and stronger androgen responsiveness.61 Second, functional omics reflect the results of both endogenous DNA genetic coding and exogenous dynamic environmental factors. Thus, it provides a unique opportunity to quantify complex gene-gene and gene-environment interactions for complex diseases, to which both genetics and environmental factors contribute substantially. Only through Trans-Omics approach can we draw a complete picture of the biological mechanisms of stroke and its various subtypes as well as its outcome. For example a recent publication conducted a GWAS of blood pressure in continental Africans and African Americans to identify genes conferring susceptibility to increased blood pressure. This research identified three novel genomic regions associated with blood pressure and utilized transomic techniques to unravel the associated pathways, which have not been previously reported in studies of other race/ethnicity. 62 Likewise, stroke research utilizing Africans and African ancestry populations can produce significant dividends because of the association between high blood pressure and stroke in African ancestry populations. 62
Trans-Omics approach has been utilized successfully in immunology,63, 64 oncology 63, 65 cardiovascular diseases66, and some neurological diseases such as Alzheimer’s diseases67, schizophrenia68 and autism.69 Although still rudimentary, this approach is also being employed in stroke research too, mostly using transcriptome and more recently epigenomics. For instance, transcriptome study observed that genes regulated in large-vessel atherosclerotic stroke are expressed in platelets and monocytes and modulate hemostasis while genes regulated in cardioembolic stroke are expressed in neutrophils and modulate immune responses to infectious-like stimuli.70 In the development of intracranial aneurysms, genomic DNA methylation has been found to play an important role in the genetic expression regulation involved in immune and inflammatory reactions, cell function, cell maintenance, and cell signal transduction.71 In ischemic stroke patients, CDKN2B methylation in peripheral blood leukocytes was found to be associated with carotid artery calcification. 72. Epigenomic study discovered that DNA methylation level for TRAF3 gene is associated with vascular recurrence in ischemic stroke patients treated with clopidogrel. 73
4.5 Methodological Standards, Current Research Priorities and Opportunities in Stroke Genomics and Trans-Omics
Important methodological considerations for stroke genomic research include adequate sample size, the selection of appropriate controls, careful clinical phenotyping using standardized classification systems, and determining associations with stroke subtypes as well as stroke as a whole. It is essential that positive associations are replicated in independent (but similar) populations,24 and appropriate methodology is used in such studies.74 Top priorities identified for global stroke genomic research include greater sample size and research within subtypes of stroke to aid discoveries, translation of existing findings, and utilization of novel techniques.75 African ancestry populations are critically needed for discovery of novel loci associated with stroke and fine-mapping of known loci discovered in American and other populations to pinpoint the precise causal variant(s) and gene(s) and translate to clinical applications.5, 47 The largest population of (continental) Africans with stroke in a single funded study are included in the Stroke Investigative Research and Education Network (SIREN) study76, part of the H3Africa Project, 31, 32, 49 which has >3000 case-control pairs, (with a targeted recruitment of 5000 case-control pairs) accurately phenotyped with the pictographic questionnaire for verification of stroke-free status (pQVSFS)77, 78, and ACCESS tool (for standardized phenotyping of ischemic and hemorrhagic strokes) 79, 80. Cases and controls are comprehensively evaluated for stroke risk factors to ensure appropriate co-variate adjustment. Furthermore, the controls, who are recruited from the same catchment population that produced the cases are matched for age, sex and ethnicity to minimize confounding due to population structure.2, 5, 44, 76–79. Preliminary data (n=917) from candidate gene studies (24 SNPs) in SIREN show that the IL-6 rs1800796 was significantly associated with ischemic stroke in West African men (OR = 2.006, 95% CI = [1.065, 3.777]. In addition, rs2383207 in CDKN2A/CDKN2B was also associated with ischemic stroke in men (OR = 2.550, 95% CI = [1.027, 6.331].81–83 However, these findings need to be explored further in the entire SIREN dataset.
4.6 Next Steps
To advance stroke genomics with existing opportunities and resources, the following steps are most likely to yield the best outcome.
4.6.1 Discovery of novel variants in the continental African dataset
Increasing the sample size of the SIREN study with additional 2000 case-control pairs will make it the largest ever stroke genomic dataset from a single study (10,000 subjects, 5000 case-control pairs). This will give over 81% power to detect Genetic Relative Risk of at least 1.46 with minimum allele frequency of 0.05. Thus, the statistical power, the genomic diversity and unique allelic frequencies of certain variants in Africans5 as well as substantially higher heritability14 will maximize the opportunity for discovery of novel variants (SNPs/CNVs/InDels) for total stroke, and its predominant subtypes, severity and outcome of relevance to African Americans ( 71% of whom migrated from West Africa several centuries ago) and indeed the entire human species. 32, 33, 35 Moreover, certain variants may be unique to populations of African Ancestry thus accounting for some of the observed racial disparities in stroke. The power for discovery will be further enhanced with pathway-based analyses.
4.6.2 Validation and Fine-mapping
Because African subjects are best suited for validation and fine-mapping, 5, 47 the phenotype and genomic dataset of 5000 case-control pairs (WGS or dense GWAS data with imputation ) should also be utilized for validation; and fine-mapping of variants previously associated with stroke in GWAS studies of Europeans and other populations. 29, 47, 84, 85 Secondly, variants discovered in continental Africans can be validated among African Americans within the Consortium of Minority Population Genome-Wide Association Studies of Stroke (COMPASS)24, International Stroke Genetics Consortium (ISGC) and other consortia.
4.6.3 Trans-ancestry big data meta-analyses including large datasets from people of African ancestry
The data generated in 4.6.1 and 4.6.2 above can be utilized in trans-ancestry meta-analysis and fine-mapping within harmonized consortia including NINDS-funded Stroke Genetics Network (SiGN)86, METASTROKE, 75 ISGC 75 and SIREN-Reasons for Geographic and Racial Differences in Stroke (REGARDS) collaboration44 to discover, validate and fine-map additional novel variants associated with stroke across European, African and other ancestry populations. This could provide more statistical power for discovery, fine-mapping and validation of loci with novel causal variant(s) and gene(s) for stroke and stroke types especially when they include large-scale African ancestry data.29, 87 It is crucial to promote opportunities for generation of high quality data from the African continent and dialog to facilitate mutual data sharing among international consortia of stroke genomics studies and investigators from the African continent, especially those currently involved in the SIREN study, through conferences and funded research collaborations.
4.6.4 Functional Genomics
Identified variants and target genomic loci can be investigated further with functional genomics to unravel the molecular mechanisms mediating their action (expression) using a combination of transcriptomics, epigenomics and metabolomics studies. Epigenomics, transcriptomics, and metabolomics can be selected as priority compared to other Omics mainly due to their relative maturity and feasibility of implementing in large international consortium settings.
5.0 Conclusions
To revolutionize the understanding of genomic contributions to stroke occurrence, severity, type and outcome, the best potential is provided by expanding available high quality, comprehensive, accurate phenotype and genomic data in continental Africans for independent trans-omics analyses, and then combining this with big data from African Americans and other populations in trans-ethnic trans-Omics meta-analyses. Delineation of molecular mechanisms of validated genomic determinants of stroke can facilitate the development of a new array of precise biomarkers for stroke prediction, diagnosis and prognostic estimates as well as discovery of new therapeutic targets for personalized interventions 19. These developments could revolutionize stroke prevention, treatment and recovery globally.
Acknowledgments
This work is supported by U54 HG007479 from the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pendlebury ST, Rothwell PM. Prevalence, incidence, and factors associated with pre-stroke and post-stroke dementia: a systematic review and meta-analysis. Lancet Neurol. 2009 Nov;8(11):1006–18. doi: 10.1016/S1474-4422(09)70236-4. [DOI] [PubMed] [Google Scholar]
- 2.Owolabi MO, Akarolo-Anthony S, Akinyemi R, Arnett D, Gebregziabher M, Jenkins C, Tiwari H, Arulogun O, Akpalu A, Sarfo FS, Obiako R, Owolabi L, Sagoe K, Melikam S, Adeoye AM, Lackland D, Ovbiagele B. The burden of stroke in Africa: a glance at the present and a glimpse into the future. Cardiovasc J Afr. 2015 Mar;26(2 Suppl 1):S27–S38. doi: 10.5830/CVJA-2015-038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de FS, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Judd SE, Kissela BM, Lackland DT, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Matchar DB, McGuire DK, Mohler ER, III, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Willey JZ, Woo D, Yeh RW, Turner MB. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation. 2015 Jan 27;131(4):e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 4.WHO. The top ten causes of death. 2017 http://www.whoint/mediacentre/factsheets/fs310/en/ Available at: URL: http://www.who.int/mediacentre/factsheets/fs310/en/
- 5.Xu H, Mitchell BD, Peprah E, Kittner SJ, Cole JW. The Importance of Conducting Stroke Genomics Research in African Ancestry Populations. Glob Heart. 2017 Mar 20; doi: 10.1016/j.gheart.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Seshadri S, Wolf PA. Lifetime risk of stroke and dementia: current concepts, and estimates from the Framingham Study. Lancet Neurol. 2007 Dec;6(12):1106–14. doi: 10.1016/S1474-4422(07)70291-0. [DOI] [PubMed] [Google Scholar]
- 7.Matsushita T, Ashikawa K, Yonemoto K, Hirakawa Y, Hata J, Amitani H, Doi Y, Ninomiya T, Kitazono T, Ibayashi S, Iida M, Nakamura Y, Kiyohara Y, Kubo M. Functional SNP of ARHGEF10 confers risk of atherothrombotic stroke. Hum Mol Genet. 2010 Mar 15;19(6):1137–46. doi: 10.1093/hmg/ddp582. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Tuomilehto J, Jousilahti P, Wang Y, Antikainen R, Hu G. Total and high-density lipoprotein cholesterol and stroke risk. Stroke. 2012 Jul;43(7):1768–74. doi: 10.1161/STROKEAHA.111.646778. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Tong Y, Zhang Y, Ding H, Zhang H, Geng Y, Zhang R, Ke Y, Han J, Yan Z, Zhou L, Wu T, Hu FB, Wang D, Cheng J. Two novel susceptibility SNPs for ischemic stroke using exome sequencing in Chinese Han population. Mol Neurobiol. 2014 Apr;49(2):852–62. doi: 10.1007/s12035-013-8561-0. [DOI] [PubMed] [Google Scholar]
- 10.Cole JW, Stine OC, Liu X, Pratap A, Cheng Y, Tallon LJ, Sadzewicz LK, Dueker N, Wozniak MA, Stern BJ, Meschia JF, Mitchell BD, Kittner SJ, O’Connell JR. Rare variants in ischemic stroke: an exome pilot study. PLoS One. 2012;7(4):e35591. doi: 10.1371/journal.pone.0035591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q, Yang D, Ombrello AK, Zavialov AV, Toro C, Zavialov AV, Stone DL, Chae JJ, Rosenzweig SD, Bishop K, Barron KS, Kuehn HS, Hoffmann P, Negro A, Tsai WL, Cowen EW, Pei W, Milner JD, Silvin C, Heller T, Chin DT, Patronas NJ, Barber JS, Lee CC, Wood GM, Ling A, Kelly SJ, Kleiner DE, Mullikin JC, Ganson NJ, Kong HH, Hambleton S, Candotti F, Quezado MM, Calvo KR, Alao H, Barham BK, Jones A, Meschia JF, Worrall BB, Kasner SE, Rich SS, Goldbach-Mansky R, Abinun M, Chalom E, Gotte AC, Punaro M, Pascual V, Verbsky JW, Torgerson TR, Singer NG, Gershon TR, Ozen S, Karadag O, Fleisher TA, Remmers EF, Burgess SM, Moir SL, Gadina M, Sood R, Hershfield MS, Boehm M, Kastner DL, Aksentijevich I. Early-onset stroke and vasculopathy associated with mutations in ADA2. N Engl J Med. 2014 Mar 6;370(10):911–20. doi: 10.1056/NEJMoa1307361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Opherk C, Gonik M, Duering M, Malik R, Jouvent E, Herve D, Adib-Samii P, Bevan S, Pianese L, Silvestri S, Dotti MT, De SN, Liem M, Boon EM, Pescini F, Pachai C, Bracoud L, Muller-Myhsok B, Meitinger T, Rost N, Pantoni L, Lesnik OS, Federico A, Ragno M, Markus HS, Tournier-Lasserve E, Rosand J, Chabriat H, Dichgans M. Genome-wide genotyping demonstrates a polygenic risk score associated with white matter hyperintensity volume in CADASIL. Stroke. 2014 Apr;45(4):968–72. doi: 10.1161/STROKEAHA.113.004461. [DOI] [PubMed] [Google Scholar]
- 13.Keene KL, Chen WM, Chen F, Williams SR, Elkhatib SD, Hsu FC, Mychaleckyj JC, Doheny KF, Pugh EW, Ling H, Laurie CC, Gogarten SM, Madden EB, Worrall BB, Sale MM. Genetic Associations with Plasma B12, B6, and Folate Levels in an Ischemic Stroke Population from the Vitamin Intervention for Stroke Prevention (VISP) Trial. Front Public Health. 2014;2:112. doi: 10.3389/fpubh.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traylor M, Rutten-Jacobs L, Curtis C, Patel H, Breen G, Newhouse S, Lewis CM, Markus HS. Genetics of stroke in a UK African ancestry case-control study: South London Ethnicity and Stroke Study. Neurol Genet. 2017 Apr;3(2):e142. doi: 10.1212/NXG.0000000000000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A, Elkind MS, George MG, Hamdan AD, Higashida RT, Hoh BL, Janis LS, Kase CS, Kleindorfer DO, Lee JM, Moseley ME, Peterson ED, Turan TN, Valderrama AL, Vinters HV. An updated definition of stroke for the 21st century: a statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2013 Jul;44(7):2064–89. doi: 10.1161/STR.0b013e318296aeca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, Rao-Melacini P, Zhang X, Pais P, Agapay S, Lopez-Jaramillo P, Damasceno A, Langhorne P, McQueen MJ, Rosengren A, Dehghan M, Hankey GJ, Dans AL, Elsayed A, Avezum A, Mondo C, Diener HC, Ryglewicz D, Czlonkowska A, Pogosova N, Weimar C, Iqbal R, Diaz R, Yusoff K, Yusufali A, Oguz A, Wang X, Penaherrera E, Lanas F, Ogah OS, Ogunniyi A, Iversen HK, Malaga G, Rumboldt Z, Oveisgharan S, Al HF, Magazi D, Nilanont Y, Ferguson J, Pare G, Yusuf S. Global and regional effects of potentially modifiable risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet. 2016 Jul 15; doi: 10.1016/S0140-6736(16)30506-2. [DOI] [PubMed] [Google Scholar]
- 17.Peprah E, Wiley K, Troyer J, Adebamowo SN, Adu D, Mayosi BM, Ramsay M, Motala AA, Adebamowo C, Ovbiagele B, Owolabi M. Building a Platform to Enable NCD Research to Address Population Health in Africa: CVD Working Group Discussion at the Sixth H3Africa Consortium Meeting in Zambia. Glob Heart. 2016 Mar;11(1):165–70. doi: 10.1016/j.gheart.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akinyemi RO, Ovbiagele B, Akpalu A, Jenkins C, Sagoe K, Owolabi L, Sarfo F, Obiako R, Gebreziabher M, Melikam E, Warth S, Arulogun O, Lackland D, Ogunniyi A, Tiwari H, Kalaria RN, Arnett D, Owolabi MO. Stroke genomics in people of African ancestry: charting new paths. Cardiovasc J Afr. 2015 Mar;26(2 Suppl 1):S39–S49. doi: 10.5830/CVJA-2015-039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akinyemi RO, Owolabi MO, Oyeniyi T, Ovbiagele B, Arnett DK, Tiwari HK, Walker R, Ogunniyi A, Kalaria RN. Neurogenomics in Africa: Perspectives, progress, possibilities and priorities. J Neurol Sci. 2016 Jul 15;366:213–23. doi: 10.1016/j.jns.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chauhan G, Debette S. Genetic Risk Factors for Ischemic and Hemorrhagic Stroke. Curr Cardiol Rep. 2016 Dec;18(12):124. doi: 10.1007/s11886-016-0804-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holliday EG, Maguire JM, Evans TJ, Koblar SA, Jannes J, Sturm JW, Hankey GJ, Baker R, Golledge J, Parsons MW, Malik R, McEvoy M, Biros E, Lewis MD, Lincz LF, Peel R, Oldmeadow C, Smith W, Moscato P, Barlera S, Bevan S, Bis JC, Boerwinkle E, Boncoraglio GB, Brott TG, Brown RD, Jr, Cheng YC, Cole JW, Cotlarciuc I, Devan WJ, Fornage M, Furie KL, Gretarsdottir S, Gschwendtner A, Ikram MA, Longstreth WT, Jr, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Parati EA, Psaty BM, Sharma P, Stefansson K, Thorleifsson G, Thorsteinsdottir U, Traylor M, Verhaaren BF, Wiggins KL, Worrall BB, Sudlow C, Rothwell PM, Farrall M, Dichgans M, Rosand J, Markus HS, Scott RJ, Levi C, Attia J. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012 Oct;44(10):1147–51. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holliday EG, Traylor M, Malik R, Bevan S, Falcone G, Hopewell JC, Cheng YC, Cotlarciuc I, Bis JC, Boerwinkle E, Boncoraglio GB, Clarke R, Cole JW, Fornage M, Furie KL, Ikram MA, Jannes J, Kittner SJ, Lincz LF, Maguire JM, Meschia JF, Mosley TH, Nalls MA, Oldmeadow C, Parati EA, Psaty BM, Rothwell PM, Seshadri S, Scott RJ, Sharma P, Sudlow C, Wiggins KL, Worrall BB, Rosand J, Mitchell BD, Dichgans M, Markus HS, Levi C, Attia J, Wray NR. Genetic overlap between diagnostic subtypes of ischemic stroke. Stroke. 2015 Mar;46(3):615–9. doi: 10.1161/STROKEAHA.114.007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzourio C, Arima H, Harrap S, Anderson C, Godin O, Woodward M, Neal B, Bousser MG, Chalmers J, Cambien F, MacMahon S. APOE genotype, ethnicity, and the risk of cerebral hemorrhage. Neurology. 2008 Apr 15;70(16):1322–8. doi: 10.1212/01.wnl.0000308819.43401.87. [DOI] [PubMed] [Google Scholar]
- 24.Carty CL, Keene KL, Cheng YC, Meschia JF, Chen WM, Nalls M, Bis JC, Kittner SJ, Rich SS, Tajuddin S, Zonderman AB, Evans MK, Langefeld CD, Gottesman R, Mosley TH, Shahar E, Woo D, Yaffe K, Liu Y, Sale MM, Dichgans M, Malik R, Longstreth WT, Jr, Mitchell BD, Psaty BM, Kooperberg C, Reiner A, Worrall BB, Fornage M. Meta-Analysis of Genome-Wide Association Studies Identifies Genetic Risk Factors for Stroke in African Americans. Stroke. 2015 Aug;46(8):2063–8. doi: 10.1161/STROKEAHA.115.009044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, Anderson CD, Brouwers HB, Valant V, Battey TW, Radmanesh F, Raffeld MR, Baedorf-Kassis S, Deka R, Woo JG, Martin LJ, Haverbusch M, Moomaw CJ, Sun G, Broderick JP, Flaherty ML, Martini SR, Kleindorfer DO, Kissela B, Comeau ME, Jagiella JM, Schmidt H, Freudenberger P, Pichler A, Enzinger C, Hansen BM, Norrving B, Jimenez-Conde J, Giralt-Steinhauer E, Elosua R, Cuadrado-Godia E, Soriano C, Roquer J, Kraft P, Ayres AM, Schwab K, McCauley JL, Pera J, Urbanik A, Rost NS, Goldstein JN, Viswanathan A, Stogerer EM, Tirschwell DL, Selim M, Brown DL, Silliman SL, Worrall BB, Meschia JF, Kidwell CS, Montaner J, Fernandez-Cadenas I, Delgado P, Malik R, Dichgans M, Greenberg SM, Rothwell PM, Lindgren A, Slowik A, Schmidt R, Langefeld CD, Rosand J. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. 2014 Apr 3;94(4):511–21. doi: 10.1016/j.ajhg.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpenter AM, Singh IP, Gandhi CD, Prestigiacomo CJ. Genetic risk factors for spontaneous intracerebral haemorrhage. Nat Rev Neurol. 2016 Jan;12(1):40–9. doi: 10.1038/nrneurol.2015.226. [DOI] [PubMed] [Google Scholar]
- 27.Devan WJ, Falcone GJ, Anderson CD, Jagiella JM, Schmidt H, Hansen BM, Jimenez-Conde J, Giralt-Steinhauer E, Cuadrado-Godia E, Soriano C, Ayres AM, Schwab K, Kassis SB, Valant V, Pera J, Urbanik A, Viswanathan A, Rost NS, Goldstein JN, Freudenberger P, Stogerer EM, Norrving B, Tirschwell DL, Selim M, Brown DL, Silliman SL, Worrall BB, Meschia JF, Kidwell CS, Montaner J, Fernandez-Cadenas I, Delgado P, Greenberg SM, Roquer J, Lindgren A, Slowik A, Schmidt R, Woo D, Rosand J, Biffi A. Heritability estimates identify a substantial genetic contribution to risk and outcome of intracerebral hemorrhage. Stroke. 2013 Jun;44(6):1578–83. doi: 10.1161/STROKEAHA.111.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, Zhao W, Abrantes P, Amouyel P, Attia JR, Battey TW, Berger K, Boncoraglio GB, Chauhan G, Cheng YC, Chen WM, Clarke R, Cotlarciuc I, Debette S, Falcone GJ, Ferro JM, Gamble DM, Ilinca A, Kittner SJ, Kourkoulis CE, Lemmens R, Levi CR, Lichtner P, Lindgren A, Liu J, Meschia JF, Mitchell BD, Oliveira SA, Pera J, Reiner AP, Rothwell PM, Sharma P, Slowik A, Sudlow CL, Tatlisumak T, Thijs V, Vicente AM, Woo D, Seshadri S, Saleheen D, Rosand J, Markus HS, Worrall BB, Dichgans M. Low-frequency and common genetic variation in ischemic stroke: The METASTROKE collaboration. Neurology. 2016 Mar 29;86(13):1217–26. doi: 10.1212/WNL.0000000000002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015 Oct 15;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barry SL. All Africans under the Skin. In: Wolzak E, editor. National Geographic Program: The Genographic Project. 2017. [Google Scholar]
- 31.Morris K. Charles Rotimi: engaging Africa in human genomic research. Lancet. 2010 Oct 23;376(9750):1383. doi: 10.1016/S0140-6736(10)61943-5. [DOI] [PubMed] [Google Scholar]
- 32.Rotimi CN, Jorde LB. Ancestry and disease in the age of genomic medicine. N Engl J Med. 2010 Oct 14;363(16):1551–8. doi: 10.1056/NEJMra0911564. [DOI] [PubMed] [Google Scholar]
- 33.Rotimi CN, Tekola-Ayele F, Baker JL, Shriner D. The African diaspora: history, adaptation and health. Curr Opin Genet Dev. 2016 Dec;41:77–84. doi: 10.1016/j.gde.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN. Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci Rep. 2014 Aug 13;4:6055. doi: 10.1038/srep06055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009 May 22;324(5930):1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010 Aug 13;329(5993):841–5. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker R, Whiting D, Unwin N, Mugusi F, Swai M. Stroke incidence in rural and urban Tanzania: a prospective, community-based study. Lancet Neurology. 2010;9:786–92. doi: 10.1016/S1474-4422(10)70144-7. [DOI] [PubMed] [Google Scholar]
- 38.Ezejimofor MC, Uthman OA, Maduka O, Ezeabasili AC, Onwuchekwa AC, Ezejimofor BC, Asuquo E, Chen YF, Stranges S, Kandala NB. Stroke survivors in Nigeria: A door-to-door prevalence survey from the Niger Delta region. J Neurol Sci. 2017 Jan 15;372:262–9. doi: 10.1016/j.jns.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 39.Khedr EM, Fawi G, Abdela M, Mohammed TA, Ahmed MA, El-Fetoh NA, Zaki AF. Prevalence of Ischemic and Hemorrhagic Strokes in Qena Governorate, Egypt: Community-based Study. J Stroke Cerebrovasc Dis. 2014 Aug;23(7):1843–8. doi: 10.1016/j.jstrokecerebrovasdis.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Agyemang C, Attah-Adjepong G, Owusu-Dabo E, De-Graft AA, Addo J, Edusei AK, Nkum BC, Ogedegbe G. Stroke in Ashanti region of Ghana. Ghana Med J. 2012 Jun;46(2 Suppl):12–7. [PMC free article] [PubMed] [Google Scholar]
- 41.O’Donnell MJ, Xavier D, Liu L, Zhang H, Chin SL, Rao-Melacini P, Rangarajan S, Islam S, Pais P, McQueen MJ, Mondo C, Damasceno A, Lopez-Jaramillo P, Hankey GJ, Dans AL, Yusoff K, Truelsen T, Diener HC, Sacco RL, Ryglewicz D, Czlonkowska A, Weimar C, Wang X, Yusuf S. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. 2010 Jul 10;376(9735):112–23. doi: 10.1016/S0140-6736(10)60834-3. [DOI] [PubMed] [Google Scholar]
- 42.Walker RW, Jusabani A, Aris E, Gray WK, Whiting D, Kabadi G, Mugusi F, Swai M, Alberti G, Unwin N. Post-stroke case fatality within an incident population in rural Tanzania. J Neurol Neurosurg Psychiatry. 2011 Sep;82(9):1001–5. doi: 10.1136/jnnp.2010.231944. [DOI] [PubMed] [Google Scholar]
- 43.Johnson W, Onuma O, Owolabi M, Sachdev S. Stroke: a global response is needed. Bull World Health Organ. 2016 Sep 1;94(9):634–634A. doi: 10.2471/BLT.16.181636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owolabi M, Sarfo F, Howard VJ, Irvin MR, Gebregziabher M, Akinyemi R, Bennett A, Armstrong K, Tiwari HK, Akpalu A, Wahab KW, Owolabi L, Fawale B, Komolafe M, Obiako R, Adebayo P, Manly JM, Ogbole G, Melikam E, Laryea R, Saulson R, Jenkins C, Arnett DK, Lackland DT, Ovbiagele B, Howard G. Stroke in Indigenous Africans, African Americans, and European Americans: Interplay of Racial and Geographic Factors. Stroke. 2017 May;48(5):1169–75. doi: 10.1161/STROKEAHA.116.015937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klag MJ, Whelton PK, Randall BL, Neaton JD, Brancati FL, Stamler J. End-stage renal disease in African-American and white men. 16-year MRFIT findings. JAMA. 1997 Apr 23;277(16):1293–8. [PubMed] [Google Scholar]
- 46.Rao M, Balakrishnan VS. The genetic basis of kidney disease risk in African Americans: MYH9 as a new candidate gene. Am J Kidney Dis. 2009 Apr;53(4):579–83. doi: 10.1053/j.ajkd.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Asimit JL, Hatzikotoulas K, McCarthy M, Morris AP, Zeggini E. Trans-ethnic study design approaches for fine-mapping. Eur J Hum Genet. 2016 Aug;24(9):1330–6. doi: 10.1038/ejhg.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.H3Africa Consortium. H3Africa Consortium Array Available Soon. New cost-effective chip offers unprecedented content for African genomic research. 2017 Feb 5;2017 [Google Scholar]
- 49.Rotimi C, Abayomi A, Abimiku A, Adabayeri VM, Adebamowo C, Adebiyi E, Ademola AD, Adeyemo A, Adu D, Affolabi D, Agongo G, Ajayi S, karolo-Anthony S, Akinyemi R, Akpalu A, Alberts M, Alonso BO, Alzohairy AM, Ameni G, Amodu O, Anabwani G, Andersen K, Arogundade F, Arulogun O, Asogun D, Bakare R, Balde N, Baniecki ML, Beiswanger C, Benkahla A, Bethke L, Boehnke M, Boima V, Brandful J, Brooks AI, Brosius FC, Brown C, Bucheton B, Burke DT, Burnett BG, Carrington-Lawrence S, Carstens N, Chisi J, Christoffels A, Cooper R, Cordell H, Crowther N, Croxton T, de VJ, Derr L, Donkor P, Doumbia S, Duncanson A, Ekem I, El SA, Engel ME, Enyaru JC, Everett D, Fadlelmola FM, Fakunle E, Fischbeck KH, Fischer A, Folarin O, Gamieldien J, Garry RF, Gaseitsiwe S, Gbadegesin R, Ghansah A, Giovanni M, Goesbeck P, Gomez-Olive FX, Grant DS, Grewal R, Guyer M, Hanchard NA, Happi CT, Hazelhurst S, Hennig BJ, Hertz-C, Fowler, Hide W, Hilderbrandt F, Hugo-Hamman C, Ibrahim ME, James R, Jaufeerally-Fakim Y, Jenkins C, Jentsch U, Jiang PP, Joloba M, Jongeneel V, Joubert F, Kader M, Kahn K, Kaleebu P, Kapiga SH, Kassim SK, Kasvosve I, Kayondo J, Keavney B, Kekitiinwa A, Khan SH, Kimmel P, King MC, Kleta R, Koffi M, Kopp J, Kretzler M, Kumuthini J, Kyobe S, Kyobutungi C, Lackland DT, Lacourciere KA, Landoure G, Lawlor R, Lehner T, Lesosky M, Levitt N, Littler K, Lombard Z, Loring JF, Lyantagaye S, Macleod A, Madden EB, Mahomva CR, Makani J, Mamven M, Marape M, Mardon G, Marshall P, Martin DP, Masiga D, Mason R, Mate-Kole M, Matovu E, Mayige M, Mayosi BM, Mbanya JC, McCurdy SA, McCarthy MI, McIlleron H, Mc’Ligeyo SO, Merle C, Mocumbi AO, Mondo C, Moran JV, Motala A, Moxey-Mims M, Mpoloka WS, Msefula CL, Mthiyane T, Mulder N, Mulugeta G, Mumba D, Musuku J, Nagdee M, Nash O, Ndiaye D, Nguyen AQ, Nicol M, Nkomazana O, Norris S, Nsangi B, Nyarko A, Nyirenda M, Obe E, Obiakor R, Oduro A, Ofori-Acquah SF, Ogah O, Ogendo S, Ohene-Frempong K, Ojo A, Olanrewaju T, Oli J, Osafo C, Ouwe MO-B, Ovbiagele B, Owen A, Owolabi MO, Owolabi L, Owusu-Dabo E, Pare G, Parekh R, Patterton HG, Penno MB, Peterson J, Pieper R, Plange-Rhule J, Pollak M, Puzak J, Ramesar RS, Ramsay M, Rasooly R, Reddy S, Sabeti PC, Sagoe K, Salako T, Samassekou O, Sandhu MS, Sankoh O, Sarfo FS, Sarr M, Shaboodien G, Sidibe I, Simo G, Simuunza M, Smeeth L, Sobngwi E, Soodyall H, Sorgho H, Sow BO, Srinivasan S, Stein DJ, Susser ES, Swanepoel C, Tangwa G, Tareila A, Tastan BO, Tayo B, Tiffin N, Tinto H, Tobin E, Tollman SM, Traore M, Treadwell MJ, Troyer J, Tsimako-Johnstone M, Tukei V, Ulasi I, Ulenga N, van RB, Wachinou AP, Waddy SP, Wade A, Wayengera M, Whitworth J, Wideroff L, Winkler CA, Winnicki S, Wonkam A, Yewondwos M, sen T, Yozwiak N, Zar H. Research capacity. Enabling the genomic revolution in Africa. Science. 2014 Jun 20;344(6190):1346–8. doi: 10.1126/science.1251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016 Oct 12;538(7624):161–4. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandhu MS, et al. The Genome Diversity in Africa Project (GDAP) 2017 [Google Scholar]
- 52.Gurdasani D, Carstensen T, Fatumo S, Franklin CS, Wheeler E, Tachmazidou I, Huang J, Karabarinde A, Asiki G. The UG2G initiative: A study of disease susceptibility in 7000 individuals from Uganda using whole genome sequencing and genotyping approaches. 2017 [Google Scholar]
- 53.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, Ritchie GR, Xue Y, Asimit J, Nsubuga RN, Young EH, Pomilla C, Kivinen K, Rockett K, Kamali A, Doumatey AP, Asiki G, Seeley J, Sisay-Joof F, Jallow M, Tollman S, Mekonnen E, Ekong R, Oljira T, Bradman N, Bojang K, Ramsay M, Adeyemo A, Bekele E, Motala A, Norris SA, Pirie F, Kaleebu P, Kwiatkowski D, Tyler-Smith C, Rotimi C, Zeggini E, Sandhu MS. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015 Jan 15;517(7534):327–32. doi: 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clarke L, Zheng-Bradley X, Smith R, Kulesha E, Xiao C, Toneva I, Vaughan B, Preuss D, Leinonen R, Shumway M, Sherry S, Flicek P. The 1000 Genomes Project: data management and community access. Nat Methods. 2012 Apr 27;9(5):459–62. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de VJ, Slabbert M, Pepper MS. Ethical, legal and social issues in the context of the planning stages of the Southern African Human Genome Programme. Med Law. 2012 Mar;31(1):119–52. [PubMed] [Google Scholar]
- 57.Guo MH, Nandakumar SK, Ulirsch JC, Zekavat SM, Buenrostro JD, Natarajan P, Salem RM, Chiarle R, Mitt M, Kals M, Parn K, Fischer K, Milani L, Magi R, Palta P, Gabriel SB, Metspalu A, Lander ES, Kathiresan S, Hirschhorn JN, Esko T, Sankaran VG. Comprehensive population-based genome sequencing provides insight into hematopoietic regulatory mechanisms. Proc Natl Acad Sci U S A. 2017 Jan 17;114(3):E327–E336. doi: 10.1073/pnas.1619052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pengelly RJ, Tapper W, Gibson J, Knut M, Tearle R, Collins A, Ennis S. Whole genome sequences are required to fully resolve the linkage disequilibrium structure of human populations. BMC Genomics. 2015 Sep 3;16:666. doi: 10.1186/s12864-015-1854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Nimwegen KJ, van Soest RA, Veltman JA, Nelen MR, van der Wilt GJ, Vissers LE, Grutters JP. Is the $1000 Genome as Near as We Think? A Cost Analysis of Next-Generation Sequencing. Clin Chem. 2016 Nov;62(11):1458–64. doi: 10.1373/clinchem.2016.258632. [DOI] [PubMed] [Google Scholar]
- 60.Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, Vakkalanka R, Barenboim M, Sei Y, Weinberger DR, Kleinman JE. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc Natl Acad Sci U S A. 2009 Sep 15;106(37):15873–8. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, Manak JR, Berman BP, Henderson BE, Frenkel B, Haiman CA, Freedman M, Tanay A, Coetzee GA. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009 Aug;5(8):e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liang J, Le TH, Edwards DRV, Tayo BO, Gaulton KJ, Smith JA, Lu Y, Jensen RA, Chen G, Yanek LR, Schwander K, Tajuddin SM, Sofer T, Kim W, Kayima J, McKenzie CA, Fox E, Nalls MA, Young JH, Sun YV, Lane JM, Cechova S, Zhou J, Tang H, Fornage M, Musani SK, Wang H, Lee J, Adeyemo A, Dreisbach AW, Forrester T, Chu PL, Cappola A, Evans MK, Morrison AC, Martin LW, Wiggins KL, Hui Q, Zhao W, Jackson RD, Ware EB, Faul JD, Reiner AP, Bray M, Denny JC, Mosley TH, Palmas W, Guo X, Papanicolaou GJ, Penman AD, Polak JF, Rice K, Taylor KD, Boerwinkle E, Bottinger EP, Liu K, Risch N, Hunt SC, Kooperberg C, Zonderman AB, Laurie CC, Becker DM, Cai J, Loos RJF, Psaty BM, Weir DR, Kardia SLR, Arnett DK, Won S, Edwards TL, Redline S, Cooper RS, Rao DC, Rotter JI, Rotimi C, Levy D, Chakravarti A, Zhu X, Franceschini N. Single-trait and multi-trait genome-wide association analyses identify novel loci for blood pressure in African-ancestry populations. PLoS Genet. 2017 May;13(5):e1006728. doi: 10.1371/journal.pgen.1006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He DX, Gu F, Gao F, Hao JJ, Gong D, Gu XT, Mao AQ, Jin J, Fu L, Ma X. Genome-wide profiles of methylation, microRNAs, and gene expression in chemoresistant breast cancer. Sci Rep. 2016 Apr 20;6:24706. doi: 10.1038/srep24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell CJ, Getnet D, Kim MS, Manda SS, Kumar P, Huang TC, Pinto SM, Nirujogi RS, Iwasaki M, Shaw PG, Wu X, Zhong J, Chaerkady R, Marimuthu A, Muthusamy B, Sahasrabuddhe NA, Raju R, Bowman C, Danilova L, Cutler J, Kelkar DS, Drake CG, Prasad TS, Marchionni L, Murakami PN, Scott AF, Shi L, Thierry-Mieg J, Thierry-Mieg D, Irizarry R, Cope L, Ishihama Y, Wang C, Gowda H, Pandey A. A multi-omic analysis of human naive CD4+ T cells. BMC Syst Biol. 2015 Nov 6;9:75. doi: 10.1186/s12918-015-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S, Yin YA, Jiang X, Sahni N, Yi S. Multi-OMICs and Genome Editing Perspectives on Liver Cancer Signaling Networks. Biomed Res Int. 2016;2016:6186281. doi: 10.1155/2016/6186281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arneson D, Shu L, Tsai B, Barrere-Cain R, Sun C, Yang X. Multidimensional Integrative Genomics Approaches to Dissecting Cardiovascular Disease. Front Cardiovasc Med. 2017;4:8. doi: 10.3389/fcvm.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphries CE, Kohli MA, Nathanson L, Whitehead P, Beecham G, Martin E, Mash DC, Pericak-Vance MA, Gilbert J. Integrated whole transcriptome and DNA methylation analysis identifies gene networks specific to late-onset Alzheimer’s disease. J Alzheimers Dis. 2015;44(3):977–87. doi: 10.3233/JAD-141989. [DOI] [PubMed] [Google Scholar]
- 68.Hannon E, Dempster E, Viana J, Burrage J, Smith AR, Macdonald R, St CD, Mustard C, Breen G, Therman S, Kaprio J, Toulopoulou T, Hulshoff Pol HE, Bohlken MM, Kahn RS, Nenadic I, Hultman CM, Murray RM, Collier DA, Bass N, Gurling H, McQuillin A, Schalkwyk L, Mill J. An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 2016 Aug 30;17(1):176. doi: 10.1186/s13059-016-1041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grayson DR, Guidotti A. Merging data from genetic and epigenetic approaches to better understand autistic spectrum disorder. Epigenomics. 2016 Jan;8(1):85–104. doi: 10.2217/epi.15.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu H, Tang Y, Liu DZ, Ran R, Ander BP, Apperson M, Liu XS, Khoury JC, Gregg JP, Pancioli A, Jauch EC, Wagner KR, Verro P, Broderick JP, Sharp FR. Gene expression in peripheral blood differs after cardioembolic compared with large-vessel atherosclerotic stroke: biomarkers for the etiology of ischemic stroke. J Cereb Blood Flow Metab. 2008 Jul;28(7):1320–8. doi: 10.1038/jcbfm.2008.22. [DOI] [PubMed] [Google Scholar]
- 71.Yu L, Wang J, Wang S, Zhang D, Zhao Y, Wang R, Zhao J. DNA methylation regulates gene expression in intracranial aneurysms. World Neurosurg. 2017 Apr 19; doi: 10.1016/j.wneu.2017.04.064. [DOI] [PubMed] [Google Scholar]
- 72.Zhou S, Zhang Y, Wang L, Zhang Z, Cai B, Liu K, Zhang H, Dai M, Sun L, Xu X, Cai H, Liu X, Lu G, Xu G. CDKN2B methylation is associated with carotid artery calcification in ischemic stroke patients. J Transl Med. 2016 Dec 1;14(1):333. doi: 10.1186/s12967-016-1093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gallego-Fabrega C, Carrera C, Reny JL, Fontana P, Slowik A, Pera J, Pezzini A, Serrano-Heras G, Segura T, Marti-Fabregas J, Muino E, Cullell N, Montaner J, Krupinski J, Fernandez-Cadenas I. TRAF3 Epigenetic Regulation Is Associated With Vascular Recurrence in Patients With Ischemic Stroke. Stroke. 2016 May;47(5):1180–6. doi: 10.1161/STROKEAHA.115.012237. [DOI] [PubMed] [Google Scholar]
- 74.Dichgans M, Markus HS. Genetic association studies in stroke: methodological issues and proposed standard criteria. Stroke. 2005 Sep;36(9):2027–31. doi: 10.1161/01.STR.0000177498.21594.9e. [DOI] [PubMed] [Google Scholar]
- 75.Woo D, Debette S, Anderson C. 20th Workshop of the International Stroke Genetics Consortium, November 3–4, 2016, Milan, Italy: 2016.036 ISGC research priorities. Neurol Genet. 2017 Mar;3(1 Suppl 1):S12–S18. doi: 10.1212/NXG.0000000000000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Akpalu A, Sarfo FS, Ovbiagele B, Akinyemi R, Gebregziabher M, Obiako R, Owolabi L, Sagoe K, Jenkins C, Arulogun O, Adamu S, Appiah LT, Adadey MA, Agyekum F, Quansah JA, Mensah YB, Adeoye AM, Singh A, Tosin AO, Ohifemen O, Sani AA, Tabi-Ajayi E, Phillip IO, Isah SY, Tabari NA, Mande A, Agunloye AM, Ogbole GI, Akinyemi JO, Akpa OM, Laryea R, Melikam SE, Adinku D, Uvere E, Burkett NS, Adekunle GF, Kehinde SI, Azuh PC, Dambatta AH, Ishaq NA, Arnett D, Tiwari HK, Lackland D, Owolabi M. Phenotyping Stroke in Sub-Saharan Africa: Stroke Investigative Research and Education Network (SIREN) Phenomics Protocol. Neuroepidemiology. 2015;45(2):73–82. doi: 10.1159/000437372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sarfo FS, Gebregziabher M, Ovbiagele B, Akinyemi R, Owolabi L, Obiako R, Armstrong K, Arulogun O, Akpalu A, Melikam S, Saulson R, Jenkins C, Owolabi M. Validation of the 8-item questionnaire for verifying stroke free status with and without pictograms in three West African languages. eNeurologicalSci. 2016 Jun;3:75–9. doi: 10.1016/j.ensci.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sarfo F, Gebregziabher M, Ovbiagele B, Akinyemi R, Owolabi L, Obiako R, Akpa O, Armstrong K, Akpalu A, Adamu S, Obese V, Boa-Antwi N, Appiah L, Arulogun O, Mensah Y, Adeoye A, Tosin A, Adeleye O, Tabi-Ajayi E, Phillip I, Sani A, Isah S, Tabari N, Mande A, Agunloye A, Ogbole G, Akinyemi J, Laryea R, Melikam S, Uvere E, Adekunle G, Kehinde S, Azuh P, Dambatta A, Ishaq N, Saulson R, Arnett D, Tiwari H, Jenkins C, Lackland D, Owolabi M. Multilingual Validation of the Questionnaire for Verifying Stroke-Free Status in West Africa. Stroke. 2016 Jan 1;47(1):167–72. doi: 10.1161/STROKEAHA.115.010374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owolabi M, Salaam K, Ogunniyi A, Ogbole G, Akinyemi R, Mongkolwat P, Omisore A, Agunloye A, Makanjuola A, Efidi R, Odo J, Akpalu A, Sarfo F, Owolabi L, Obiako OR, Wahab K, Adebayo PB, Komolafe A, Osaigbovo G, Sunmonu T, Olowoyo P, Chukwuonye I, Obiabo Y, Ibinaiye P, Dambatta A, Mensah Y, Abdul S, Olabinrin E, Ikubor J, Oyinloye O, Odunlami F, Akpa O, Melikam S, Saulson R, Ovbiagele B SIREN; AIM on Clear Canvas Enriched Stroke phenotyping Software (ACCESS) NG/PT/NC/2016/2007. Nigeria patent. 2016
- 80.Owolabi M, Ogbole G, Akinyemi R, Salaam K, Akpa O, Mongkolwat P, Omisore A, Agunloye A, Efidi R, Odo J, Makanjuola A, Akpalu A, Sarfo F, Owolabi L, Obiako R, Wahab K, Sanya E, Adebayo P, Komolafe M, Adeoye AM, Akinyemi J, Osaigbovo G, Sunmonu T, Olowoyo P, Chukwuonye I, Obiabo Y, Ibinaiye P, Dambatta A, Mensah Y, Abdul S, Olabinri E, Ikubor J, Oyinloye O, Odunlami F, Melikam E, Saulson R, Kolo P, Ogunniyi A, Ovbiagele B. Development and Reliability of a User-Friendly Multicenter Phenotyping Application for Hemorrhagic and Ischemic Stroke. J Stroke Cerebrovasc Dis. 2017 Jul 28; doi: 10.1016/j.jstrokecerebrovasdis.2017.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Owolabi MO, Akinyemi R, Arnett D, Ovbiagele B, Irvin M, Tiwari H SIREN. Defining the genetic architecture of stroke in people of black ancestry. Oral presentation. 2015 [Google Scholar]
- 82.Owolabi MO, Akinyemi R, Arnett D, Tiwari H, Ovbiagele B SIREN. Is Genetics Important for Stroke in Africa? Invited Oral presentation. 2016 [Google Scholar]
- 83.Akinyemi R, Arnett DK, Tiwari HK, Ovbiagele B, Sarfo F, Srinivasasainagendra V, Irvin MR, Adeoye A, Perry RT, Akpalu A, Jenkins C, Owolabi L, Obiako RO, Wahab K, Sanya E, Komolafe A, Fawale M, Adebayo P, Osaigbovo G, Sunmonu T, Olowoyo P, Chukwuonye I, Obiabo Y, Akpa O, Melikam ES, Saulson R, Kalaria RN, Ogunniyi A, Owolabi MO, SIREN Interleukin-6 (IL-6) rs1800796 and cyclin dependent kinase inhibitor (CDKN2A/CDKN2B) rs2383207 are associated with ischemic stroke in indigenous West African Men. J Neurol Sci. 2017;379:229–35. doi: 10.1016/j.jns.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, Mahajan A, Locke A, Rayner NW, Robertson N, Scott RA, Prokopenko I, Scott LJ, Green T, Sparso T, Thuillier D, Yengo L, Grallert H, Wahl S, Franberg M, Strawbridge RJ, Kestler H, Chheda H, Eisele L, Gustafsson S, Steinthorsdottir V, Thorleifsson G, Qi L, Karssen LC, van Leeuwen EM, Willems SM, Li M, Chen H, Fuchsberger C, Kwan P, Ma C, Linderman M, Lu Y, Thomsen SK, Rundle JK, Beer NL, van de Bunt M, Chalisey A, Kang HM, Voight BF, Abecasis GR, Almgren P, Baldassarre D, Balkau B, Benediktsson R, Bluher M, Boeing H, Bonnycastle LL, Bottinger EP, Burtt NP, Carey J, Charpentier G, Chines PS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Doney AS, Dorkhan M, Edkins S, Eriksson JG, Esko T, Eury E, Fadista J, Flannick J, Fontanillas P, Fox C, Franks PW, Gertow K, Gieger C, Gigante B, Gottesman O, Grant GB, Grarup N, Groves CJ, Hassinen M, Have CT, Herder C, Holmen OL, Hreidarsson AB, Humphries SE, Hunter DJ, Jackson AU, Jonsson A, Jorgensen ME, Jorgensen T, Kao WH, Kerrison ND, Kinnunen L, Klopp N, Kong A, Kovacs P, Kraft P, Kravic J, Langford C, Leander K, Liang L, Lichtner P, Lindgren CM, Lindholm E, Linneberg A, Liu CT, Lobbens S, Luan J, Lyssenko V, Mannisto S, McLeod O, Meyer J, Mihailov E, Mirza G, Muhleisen TW, Muller-Nurasyid M, Navarro C, Nothen MM, Oskolkov NN, Owen KR, Palli D, Pechlivanis S, Peltonen L, Perry JR, Platou CG, Roden M, Ruderfer D, Rybin D, van der Schouw YT, Sennblad B, Sigurethsson G, Stancakova A, Steinbach G, Storm P, Strauch K, Stringham HM, Sun Q, Thorand B, Tikkanen E, Tonjes A, Trakalo J, Tremoli E, Tuomi T, Wennauer R, Wiltshire S, Wood AR, Zeggini E, Dunham I, Birney E, Pasquali L, Ferrer J, Loos RJ, Dupuis J, Florez JC, Boerwinkle E, Pankow JS, van DC, Sijbrands E, Meigs JB, Hu FB, Thorsteinsdottir U, Stefansson K, Lakka TA, Rauramaa R, Stumvoll M, Pedersen NL, Lind L, Keinanen-Kiukaanniemi SM, Korpi-Hyovalti E, Saaristo TE, Saltevo J, Kuusisto J, Laakso M, Metspalu A, Erbel R, Jocke KH, Moebus S, Ripatti S, Salomaa V, Ingelsson E, Boehm BO, Bergman RN, Collins FS, Mohlke KL, Koistinen H, Tuomilehto J, Hveem K, Njolstad I, Deloukas P, Donnelly PJ, Frayling TM, Hattersley AT, de FU, Hamsten A, Illig T, Peters A, Cauchi S, Sladek R, Froguel P, Hansen T, Pedersen O, Morris AD, Palmer CN, Kathiresan S, Melander O, Nilsson PM, Groop LC, Barroso I, Langenberg C, Wareham NJ, O’Callaghan CA, Gloyn AL, Altshuler D, Boehnke M, Teslovich TM, McCarthy MI, Morris AP. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015 Dec;47(12):1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, van Dam RM, Danesh J, Das D, de FU, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, van DC, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, Hattersley AT, Hayward C, Hedman AK, Herder C, Hofman A, Holmen OL, Hovingh K, Hreidarsson AB, Hu C, Hu FB, Hui J, Humphries SE, Hunt SE, Hunter DJ, Hveem K, Hydrie ZI, Ikegami H, Illig T, Ingelsson E, Islam M, Isomaa B, Jackson AU, Jafar T, James A, Jia W, Jockel KH, Jonsson A, Jowett JB, Kadowaki T, Kang HM, Kanoni S, Kao WH, Kathiresan S, Kato N, Katulanda P, Keinanen-Kiukaanniemi KM, Kelly AM, Khan H, Khaw KT, Khor CC, Kim HL, Kim S, Kim YJ, Kinnunen L, Klopp N, Kong A, Korpi-Hyovalti E, Kowlessur S, Kraft P, Kravic J, Kristensen MM, Krithika S, Kumar A, Kumate J, Kuusisto J, Kwak SH, Laakso M, Lagou V, Lakka TA, Langenberg C, Langford C, Lawrence R, Leander K, Lee JM, Lee NR, Li M, Li X, Li Y, Liang J, Liju S, Lim WY, Lind L, Lindgren CM, Lindholm E, Liu CT, Liu JJ, Lobbens S, Long J, Loos RJ, Lu W, Luan J, Lyssenko V, Ma RC, Maeda S, Magi R, Mannisto S, Matthews DR, Meigs JB, Melander O, Metspalu A, Meyer J, Mirza G, Mihailov E, Moebus S, Mohan V, Mohlke KL, Morris AD, Muhleisen TW, Muller-Nurasyid M, Musk B, Nakamura J, Nakashima E, Navarro P, Ng PK, Nica AC, Nilsson PM, Njolstad I, Nothen MM, Ohnaka K, Ong TH, Owen KR, Palmer CN, Pankow JS, Park KS, Parkin M, Pechlivanis S, Pedersen NL, Peltonen L, Perry JR, Peters A, Pinidiyapathirage JM, Platou CG, Potter S, Price JF, Qi L, Radha V, Rallidis L, Rasheed A, Rathman W, Rauramaa R, Raychaudhuri S, Rayner NW, Rees SD, Rehnberg E, Ripatti S, Robertson N, Roden M, Rossin EJ, Rudan I, Rybin D, Saaristo TE, Salomaa V, Saltevo J, Samuel M, Sanghera DK, Saramies J, Scott J, Scott LJ, Scott RA, Segre AV, Sehmi J, Sennblad B, Shah N, Shah S, Shera AS. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014 Mar;46(3):234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pulit SL SiGN. Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol. 2015 Dec 18; doi: 10.1016/S1474-4422(15)00338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim K, Bang SY, Lee HS, Cho SK, Choi CB, Sung YK, Kim TH, Jun JB, Yoo DH, Kang YM, Kim SK, Suh CH, Shim SC, Lee SS, Lee J, Chung WT, Choe JY, Shin HD, Lee JY, Han BG, Nath SK, Eyre S, Bowes J, Pappas DA, Kremer JM, Gonzalez-Gay MA, Rodriguez-Rodriguez L, Arlestig L, Okada Y, Diogo D, Liao KP, Karlson EW, Raychaudhuri S, Rantapaa-Dahlqvist S, Martin J, Klareskog L, Padyukov L, Gregersen PK, Worthington J, Greenberg JD, Plenge RM, Bae SC. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann Rheum Dis. 2015 Mar;74(3):e13. doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matarin M, Brown WM, Scholz S, Simon-Sanchez J, Fung HC, Hernandez D, Gibbs JR, De Vrieze FW, Crews C, Britton A, Langefeld CD, Brott TG, Brown RD, Jr, Worrall BB, Frankel M, Silliman S, Case LD, Singleton A, Hardy JA, Rich SS, Meschia JF. A genome-wide genotyping study in patients with ischaemic stroke: initial analysis and data release. Lancet Neurol. 2007 May;6(5):414–20. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, Kostulas K, Kuhlenbaumer G, Bevan S, Jonsdottir T, Bjarnason H, Saemundsdottir J, Palsson S, Arnar DO, Holm H, Thorgeirsson G, Valdimarsson EM, Sveinbjornsdottir S, Gieger C, Berger K, Wichmann HE, Hillert J, Markus H, Gulcher JR, Ringelstein EB, Kong A, Dichgans M, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008 Oct;64(4):402–9. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 90.Yamada Y, Fuku N, Tanaka M, Aoyagi Y, Sawabe M, Metoki N, Yoshida H, Satoh K, Kato K, Watanabe S, Nozawa Y, Hasegawa A, Kojima T. Identification of CELSR1 as a susceptibility gene for ischemic stroke in Japanese individuals by a genome-wide association study. Atherosclerosis. 2009 Nov;207(1):144–9. doi: 10.1016/j.atherosclerosis.2009.03.038. [DOI] [PubMed] [Google Scholar]
- 91.Schurks M, Buring JE, Ridker PM, Chasman DI, Kurth T. Genetic determinants of cardiovascular events among women with migraine: a genome-wide association study. PLoS One. 2011;6(7):e22106. doi: 10.1371/journal.pone.0022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hata J, Kubo M, Kiyohara Y. Genome-wide association study for ischemic stroke based on the Hisayama study. Nihon Eiseigaku Zasshi. 2011 Jan;66(1):47–52. doi: 10.1265/jjh.66.47. [DOI] [PubMed] [Google Scholar]
- 93.Bilguvar K, Yasuno K, Niemela M, Ruigrok YM, von Und Zu FM, van Duijn CM, van den Berg LH, Mane S, Mason CE, Choi M, Gaal E, Bayri Y, Kolb L, Arlier Z, Ravuri S, Ronkainen A, Tajima A, Laakso A, Hata A, Kasuya H, Koivisto T, Rinne J, Ohman J, Breteler MM, Wijmenga C, State MW, Rinkel GJ, Hernesniemi J, Jaaskelainen JE, Palotie A, Inoue I, Lifton RP, Gunel M. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008 Dec;40(12):1472–7. doi: 10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al. Genome-wide association studies of stroke. N Engl J Med. 2009;360(17):1718–28. doi: 10.1056/NEJMoa0900094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009 Aug;41(8):876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, Jackson CA, Traylor M, Strange A, Su Z, Band G, Syme PD, Malik R, Pera J, Norrving B, Lemmens R, Freeman C, Schanz R, James T, Poole D, Murphy L, Segal H, Cortellini L, Cheng YC, Woo D, Nalls MA, Muller-Myhsok B, Meisinger C, Seedorf U, Ross-Adams H, Boonen S, Wloch-Kopec D, Valant V, Slark J, Furie K, Delavaran H, Langford C, Deloukas P, Edkins S, Hunt S, Gray E, Dronov S, Peltonen L, Gretarsdottir S, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boncoraglio GB, Parati EA, Attia J, Holliday E, Levi C, Franzosi MG, Goel A, Helgadottir A, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Duncanson A, Jankowski J, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Worrall BB, Kittner SJ, Mitchell BD, Kissela B, Meschia JF, Thijs V, Lindgren A, Macleod MJ, Slowik A, Walters M, Rosand J, Sharma P, Farrall M, Sudlow CL, Rothwell PM, Dichgans M, Donnelly P, Markus HS. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012 Feb 5;44(3):328–33. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, Nalls MA, Longstreth W, Wiggins KL, Yadav S, Parati EA, DeStefano AL, Worrall BB, Kittner SJ, Khan MS, Reiner AP, Helgadottir A, Achterberg S, Fernandez-Cadenas I, Abboud S, Schmidt R, Walters M, Chen WM, Ringelstein EB, O’Donnell M, Ho WK, Pera J, Lemmens R, Norrving B, Higgins P, Benn M, Sale M, Kuhlenbaumer G, Doney AS, Vicente AM, Delavaran H, Algra A, Davies G, Oliveira SA, Palmer CN, Deary I, Schmidt H, Pandolfo M, Montaner J, Carty C, de Bakker PI, Kostulas K, Ferro JM, van Zuydam NR, Valdimarsson E, Nordestgaard BG, Lindgren A, Thijs V, Slowik A, Saleheen D, Pare G, Berger K, Thorleifsson G, Hofman A, Mosley TH, Mitchell BD, Furie K, Clarke R, Levi C, Seshadri S, Gschwendtner A, Boncoraglio GB, Sharma P, Bis JC, Gretarsdottir S, Psaty BM, Rothwell PM, Rosand J, Meschia JF, Stefansson K, Dichgans M, Markus HS. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012 Nov;11(11):951–62. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Foroud T, Koller DL, Lai D, Sauerbeck L, Anderson C, Ko N, Deka R, Mosley TH, Fornage M, Woo D, Moomaw CJ, Hornung R, Huston J, Meissner I, Bailey-Wilson JE, Langefeld C, Rouleau G, Connolly ES, Worrall BB, Kleindorfer D, Flaherty ML, Martini S, Mackey J, de los Rios La Rosa, Brown RD, Jr, Broderick JP. Genome-wide association study of intracranial aneurysms confirms role of Anril and SOX17 in disease risk. Stroke. 2012 Nov;43(11):2846–52. doi: 10.1161/STROKEAHA.112.656397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Arning A, Hiersche M, Witten A, Kurlemann G, Kurnik K, Manner D, Stoll M, Nowak-Gottl U. A genome-wide association study identifies a gene network of ADAMTS genes in the predisposition to pediatric stroke. Blood. 2012 Dec 20;120(26):5231–6. doi: 10.1182/blood-2012-07-442038. [DOI] [PubMed] [Google Scholar]
- 100.Williams FM, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, Traylor M, Bevan S, Dichgans M, Rothwell PM, Sudlow C, Farrall M, Silander K, Kaunisto M, Wagner P, Saarela O, Kuulasmaa K, Virtamo J, Salomaa V, Amouyel P, Arveiler D, Ferrieres J, Wiklund PG, Ikram MA, Hofman A, Boncoraglio GB, Parati EA, Helgadottir A, Gretarsdottir S, Thorsteinsdottir U, Thorleifsson G, Stefansson K, Seshadri S, Destefano A, Gschwendtner A, Psaty B, Longstreth W, Mitchell BD, Cheng YC, Clarke R, Ferrario M, Bis JC, Levi C, Attia J, Holliday EG, Scott RJ, Fornage M, Sharma P, Furie KL, Rosand J, Nalls M, Meschia J, Mosely TH, Evans A, Palotie A, Markus HS, Grant PJ, Spector TD. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. 2013 Jan;73(1):16–31. doi: 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Traylor M, Makela KM, Kilarski LL, Holliday EG, Devan WJ, Nalls MA, Wiggins KL, Zhao W, Cheng YC, Achterberg S, Malik R, Sudlow C, Bevan S, Raitoharju E, Oksala N, Thijs V, Lemmens R, Lindgren A, Slowik A, Maguire JM, Walters M, Algra A, Sharma P, Attia JR, Boncoraglio GB, Rothwell PM, de Bakker PI, Bis JC, Saleheen D, Kittner SJ, Mitchell BD, Rosand J, Meschia JF, Levi C, Dichgans M, Lehtimaki T, Lewis CM, Markus HS. A novel MMP12 locus is associated with large artery atherosclerotic stroke using a genome-wide age-at-onset informed approach. PLoS Genet. 2014 Jul;10(7):e1004469. doi: 10.1371/journal.pgen.1004469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Williams SR, Yang Q, Chen F, Liu X, Keene KL, Jacques P, Chen WM, Weinstein G, Hsu FC, Beiser A, Wang L, Bookman E, Doheny KF, Wolf PA, Zilka M, Selhub J, Nelson S, Gogarten SM, Worrall BB, Seshadri S, Sale MM. Genome-wide meta-analysis of homocysteine and methionine metabolism identifies five one carbon metabolism loci and a novel association of ALDH1L1 with ischemic stroke. PLoS Genet. 2014 Mar;10(3):e1004214. doi: 10.1371/journal.pgen.1004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Foroud T, Lai D, Koller D, Van’t Hof F, Kurki MI, Anderson CS, Brown RD, Jr, Connolly ES, Eriksson JG, Flaherty M, Fornage M, von Und Zu FM, Gaal EI, Laakso A, Hernesniemi J, Huston J, Jaaskelainen JE, Kiemeney LA, Kivisaari R, Kleindorfer D, Ko N, Lehto H, Mackey J, Meissner I, Moomaw CJ, Mosley TH, Moskala M, Niemela M, Palotie A, Pera J, Rinkel G, Ripke S, Rouleau G, Ruigrok Y, Sauerbeck L, Slowik A, Vermeulen SH, Woo D, Worrall BB, Broderick J. Genome-wide association study of intracranial aneurysm identifies a new association on chromosome 7. Stroke. 2014 Nov;45(11):3194–9. doi: 10.1161/STROKEAHA.114.006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cotlarciuc I, Malik R, Holliday EG, Ahmadi KR, Pare G, Psaty BM, Fornage M, Hasan N, Rinne PE, Ikram MA, Markus HS, Rosand J, Mitchell BD, Kittner SJ, Meschia JF, van Meurs JB, Uitterlinden AG, Worrall BB, Dichgans M, Sharma P. Effect of genetic variants associated with plasma homocysteine levels on stroke risk. Stroke. 2014 Jul;45(7):1920–4. doi: 10.1161/STROKEAHA.114.005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rannikmae K, Davies G, Thomson PA, Bevan S, Devan WJ, Falcone GJ, Traylor M, Anderson CD, Battey TW, Radmanesh F, Deka R, Woo JG, Martin LJ, Jimenez-Conde J, Selim M, Brown DL, Silliman SL, Kidwell CS, Montaner J, Langefeld CD, Slowik A, Hansen BM, Lindgren AG, Meschia JF, Fornage M, Bis JC, Debette S, Ikram MA, Longstreth WT, Schmidt R, Zhang CR, Yang Q, Sharma P, Kittner SJ, Mitchell BD, Holliday EG, Levi CR, Attia J, Rothwell PM, Poole DL, Boncoraglio GB, Psaty BM, Malik R, Rost N, Worrall BB, Dichgans M, Van AT, Woo D, Markus HS, Seshadri S, Rosand J, Sudlow CL. Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology. 2015 Mar 3;84(9):918–26. doi: 10.1212/WNL.0000000000001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Anderson CD, Falcone GJ, Phuah CL, Radmanesh F, Brouwers HB, Battey TW, Biffi A, Peloso GM, Liu DJ, Ayres AM, Goldstein JN, Viswanathan A, Greenberg SM, Selim M, Meschia JF, Brown DL, Worrall BB, Silliman SL, Tirschwell DL, Flaherty ML, Kraft P, Jagiella JM, Schmidt H, Hansen BM, Jimenez-Conde J, Giralt-Steinhauer E, Elosua R, Cuadrado-Godia E, Soriano C, van Nieuwenhuizen KM, Klijn CJ, Rannikmae K, Samarasekera N, Al-Shahi SR, Sudlow CL, Deary IJ, Morotti A, Pezzini A, Pera J, Urbanik A, Pichler A, Enzinger C, Norrving B, Montaner J, Fernandez-Cadenas I, Delgado P, Roquer J, Lindgren A, Slowik A, Schmidt R, Kidwell CS, Kittner SJ, Waddy SP, Langefeld CD, Abecasis G, Willer CJ, Kathiresan S, Woo D, Rosand J. Genetic variants in CETP increase risk of intracerebral hemorrhage. Ann Neurol. 2016 Nov;80(5):730–40. doi: 10.1002/ana.24780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chauhan G, Arnold CR, Chu AY, Fornage M, et al. Identification of additional risk loci for stroke and small vessel disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2016 Jun;15(7):695–707. doi: 10.1016/S1474-4422(16)00102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cheng YC, ST, GA, HW, TM, AP, HE, MR, XH, KS, CJ, OJ, DJ, RA, ZW, ES, G-GC, Kamatani Y, LM, LD, TV, MT, TT, PA, PE, NB, BS, RP, SC, SA, LA, WM, Jannes J, SJ, CD, DK, LC, KS, BJ, FM, MT, HJ, SK, M-NM, GC, WM, PA, MC, Ikram MA, LWJ, MJ, SS, SP, WB, JC, LC, DM, BG, MH, DS, RA, SD, MB WTCCC-2 Consortium. Genome-Wide Association Analysis of Young-Onset Stroke Identifies a Locus on Chromosome 10q25 Near HABP2. Stroke. 2016;47(2):307–16. doi: 10.1161/STROKEAHA.115.011328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Traylor M, Zhang CR, Adib-Samii P, Devan WJ, Parsons OE, Lanfranconi S, Gregory S, Cloonan L, Falcone GJ, Radmanesh F, Fitzpatrick K, Kanakis A, Barrick TR, Moynihan B, Lewis CM, Boncoraglio GB, Lemmens R, Thijs V, Sudlow C, Wardlaw J, Rothwell PM, Meschia JF, Worrall BB, Levi C, Bevan S, Furie KL, Dichgans M, Rosand J, Markus HS, Rost N. Genome-wide meta-analysis of cerebral white matter hyperintensities in patients with stroke. Neurology. 2016 Jan 12;86(2):146–53. doi: 10.1212/WNL.0000000000002263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Malik R, Dau T, Gonik M, Sivakumar A, Deredge DJ, Edeleva EV, Gotzfried J, van der Laan SW, Pasterkamp G, Beaufort N, Seixas S, Bevan S, Lincz LF, Holliday EG, Burgess AI, Rannikmae K, Minnerup J, Kriebel J, Waldenberger M, Muller-Nurasyid M, Lichtner P, Saleheen D, Rothwell PM, Levi C, Attia J, Sudlow CL, Braun D, Markus HS, Wintrode PL, Berger K, Jenne DE, Dichgans M. Common coding variant in SERPINA1 increases the risk for large artery stroke. Proc Natl Acad Sci U S A. 2017 Apr 4;114(14):3613–8. doi: 10.1073/pnas.1616301114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Traylor M, Malik R, Nalls MA, Cotlarciuc I, Radmanesh F, Thorleifsson G, Hanscombe KB, Langefeld C, Saleheen D, Rost NS, Yet I, Spector TD, Bell JT, Hannon E, Mill J, Chauhan G, Debette S, Bis JC, Longstreth WT, Jr, Ikram MA, Launer LJ, Seshadri S, Hamilton-Bruce MA, Jimenez-Conde J, Cole JW, Schmidt R, Slowik A, Lemmens R, Lindgren A, Melander O, Grewal RP, Sacco RL, Rundek T, Rexrode K, Arnett DK, Johnson JA, Benavente OR, Wasssertheil-Smoller S, Lee JM, Pulit SL, Wong Q, Rich SS, de Bakker PI, McArdle PF, Woo D, Anderson CD, Xu H, Heitsch L, Fornage M, Jern C, Stefansson K, Thorsteinsdottir U, Gretarsdottir S, Lewis CM, Sharma P, Sudlow CL, Rothwell PM, Boncoraglio GB, Thijs V, Levi C, Meschia JF, Rosand J, Kittner SJ, Mitchell BD, Dichgans M, Worrall BB, Markus HS. Genetic variation at 16q24.2 is associated with small vessel stroke. Ann Neurol. 2017 Mar;81(3):383–94. doi: 10.1002/ana.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]