Abstract
Many studies have investigated the diagnostic role of circulating microRNAs (miRNAs) in patients with lung cancer; however, the results still remain inconclusive. An updated system review and meta-analysis was necessary to give a comprehensive evaluation of diagnostic role of circulating miRNAs in lung cancer. Eligible studies were searched in electronical databases. The sensitivity and specificity were used to plot the summary receiver operator characteristic (SROC) curve and calculate the area under the curve (AUC). The between-study heterogeneity was evaluated by Q test and I2 statistics. Subgroup analyses and meta-regression were further performed to explore the potential sources of heterogeneity. A total of 134 studies from 65 articles (6,919 patients with lung cancer and 7,064 controls) were included for analysis. Overall analysis showed that circulating miRNAs had a good diagnostic performance in lung cancers, with a sensitivity of 0.83, a specificity of 0.84, and an AUC of 0.90. Subgroup analysis suggested that combined miRNAs and Caucasian populations may yield relatively higher diagnostic performance. In addition, we found serum might serve as an ideal material to detecting miRNA as good diagnostic performance. We also found the diagnostic role of miRNAs in early stage lung cancer was still relatively high (the sensitivity, specificity and an AUC of stage I/II was 0.81, 0.82 and 0.88; and for stage I, it was 0.80, 0.81, and 0.88). We also identified a panel of miRNAs such as miR-21-5p, miR-223-3p, miR-155-5p and miR-126-3p might serve as potential biomarkers for lung cancer. As a result, circulating miRNAs, particularly the combination of multiple miRNAs, may serve as promising biomarkers for the diagnosis of lung cancer.
Keywords: lung cancer, circulating microRNAs, diagnostic value, meta-analysis
INTRODUCTION
Currently, the mortality rate of lung cancer is still the highest among all cancers in both men and women [1]. The data indicated that an estimated 733,300 new lung cancer cases and 610,200 lung cancer deaths would occur in China in 2015 [2]. According to National Center for Health Statistics in 2017, there were an estimated 222,500 new cases of lung cancer diagnosed, and 155,870 died from lung cancer in the United States [3]. Lung cancer can be divided into two major forms: non-small cell lung cancer (NSCLC) (about 85% of all lung cancers) and small cell lung cancer (SCLC) (about 15%) [4]. Despite advances in early detection and standard treatment, approximately two thirds of NSCLC cases were diagnosed at locally advanced (27.6%) or metastatic (38.1%) disease as the typically asymptomatic at early stages [5]. Thus, the earlier detection of lung cancer would be great meaningful to improve the prognosis of this lethal disease.
Computed tomography (CT), especially low-dose CT is currently widely used as the screening method for lung cancer. But some concerns may exist, such as high false-positive rates, potential over-diagnosis, excessive cost and radiation risk [6]. Recent years, more attentions have been paid in circulating microRNAs (miRNAs) in the field of biomarker discovery for cancer. MiRNAs are short (typically 18-25 nucleotides), single-stranded and highly conserved non-coding RNAs which could negatively regulate gene expression at post-transcriptional level by binding the 3′-untranslated region of target mRNAs, resulting in either mRNA degradation or translational repression [7]. Passively leaked or actively transported from cells, circulating miRNAs could be stably detected in blood and have been used as biomarkers for diagnosis, prognosis or monitoring curative effect in various cancers including lung cancer [7–9].
The diagnostic role of single and various sets of circulating miRNAs in lung cancer have been investigated by many studies. However, the results from individual studies were inconsistent, and the miRNA signatures identified from individual studies were different from each other. Therefore, we conducted this systematic review and meta-analysis to further evaluate the clinical applicability of circulating miRNAs as biomarkers for the diagnosis of lung cancer.
RESULTS
Eligible studies
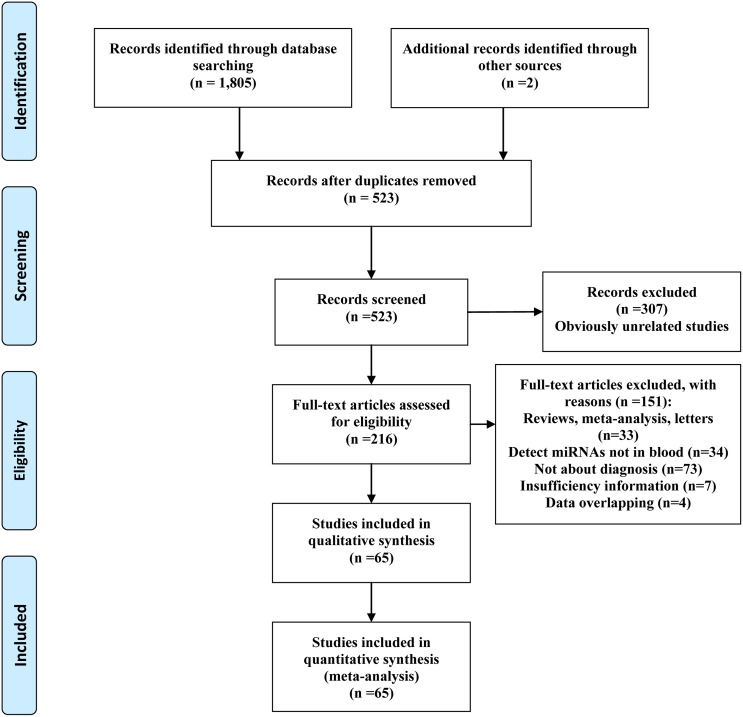
The present work followed the guidelines for systematic reviews and meta-analyses (PRISMA) [10]. 523 articles were identified from initial screen. After reviewing the titles and abstracts, 307 articles were excluded because they obviously did not meet our selection criteria. The remaining 216 articles were further checked by screening the full texts. Finally, a total of 134 studies from 65 articles including 13,983 samples (6,919 patients with lung cancer and 7,064 controls) were qualified for our analysis [11–75]. The process of article selection is summarized in Figure 1. The main characteristics of the included articles are listed in Supplementary Table 1. The qualities of the selected studies according to QUADAS-2 guidelines are showed in Supplementary Figure 1.
Figure 1. Flow diagram of the selection procedure of studies.
Pooled diagnostic accuracy of miRNAs
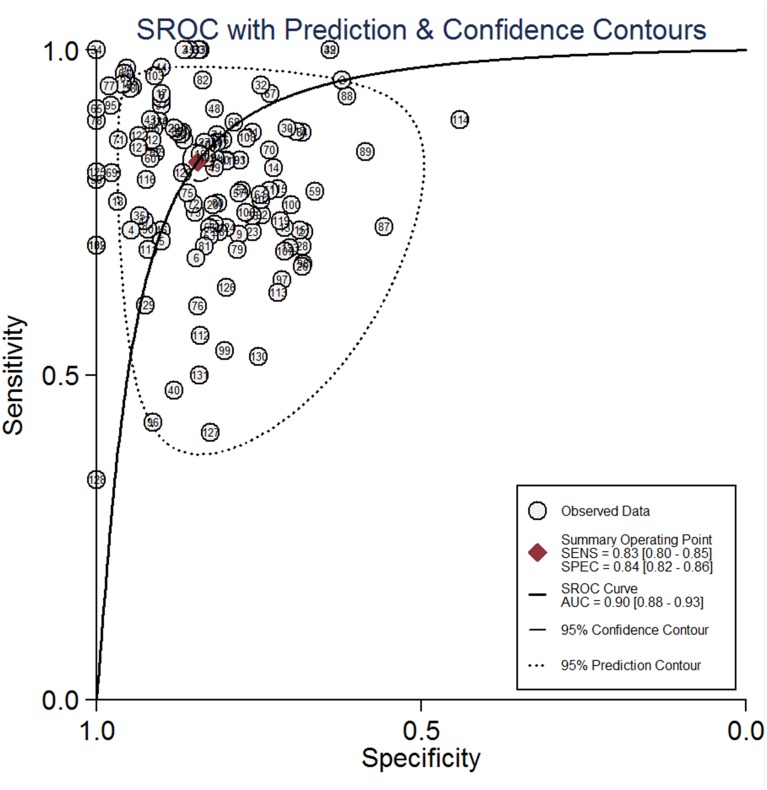
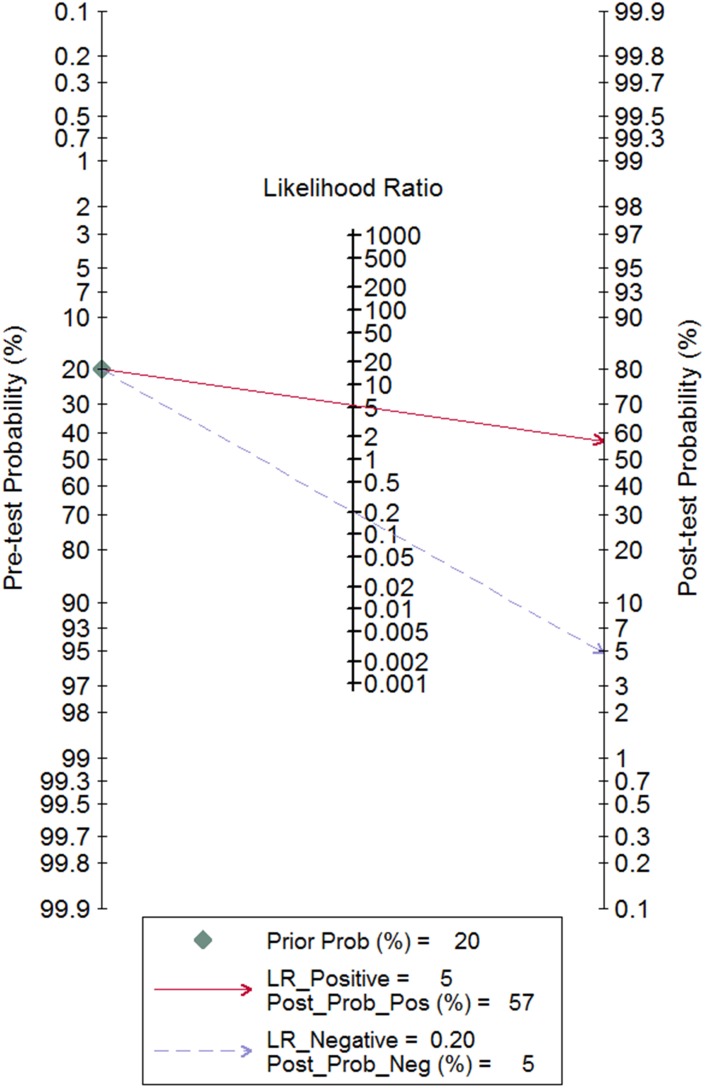
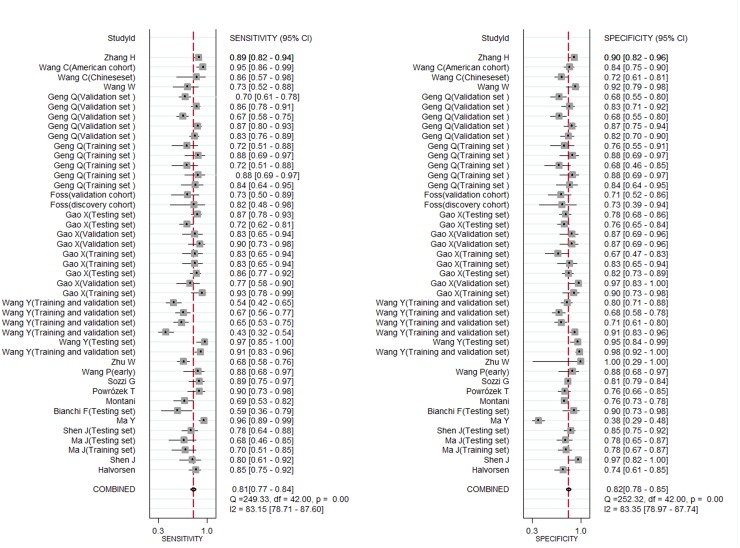
All 134 studies were available for analysis the overall predictive accuracy of miRNAs for detecting lung cancer. The random effects model was applied. The pooled sensitivity was 0.83 (95% CI: 0.80–0.85), the specificity was 0.84 (95% CI: 0.82–0.86), the pooled positive likelihood ratio (PLR) was 5.3(95% CI: 4.7–6.0), the negative likelihood ratio (NLR) was 0.20 (95% CI: 0.18–0.23) and the diagnostic odds ratio (DOR) was 26(95% CI: 21–33) (Table 1). The corresponding summary receiver operator characteristic (SROC) curve is shown in Figure 2. The area under the curve (AUC) value was 0.90 (95% CI: 0.88–0.93) in the overall SROC curve. The Fagan diagram in Figure 3 illustrates the post-test probabilities of circulating miRNAs in lung cancer diagnosis. Our analysis suggested circulating miRNAs may yield a relatively high diagnostic accuracy for detecting lung cancer patients.
Table 1. The main results of meta-analysis.
| Analysis | No. of studies | SEN (95%CI) | SPE (95%CI) | PLR (95%CI) | NLR (95%CI) | DOR (95%CI) | AUC (95%CI) |
|---|---|---|---|---|---|---|---|
| Overall | 134 | 0.83(0.80,0.85),I2=89.3%,p<0.001 | 0.84(0.82,0.86),I2=80.58%,p<0.001 | 5.3(4.7,6.0) | 0.20(0.18,0.23) | 26(21,33) | 0.90(0.88,0.93) |
| Subgroup 1: race | |||||||
| Asian | 86 | 0.81(0.78,0.84),I2=90.67%,p<0.001 | 0.84(0.82,0.86),I2=80.61%,p<0.001 | 5.1(4.4,5.9) | 0.22(0.19,0.26) | 23(18,30) | 0.90(0.87,0.92) |
| Caucasian | 39 | 0.88(0.83,0.91),I2=84.53%,p<0.001 | 0.86(0.82,0.89),I2=85.38%,p<0.001 | 6.2(4.7,8.1) | 0.14(0.11,0.20) | 43(26,68) | 0.93(0.91,0.95) |
| Mixed | 9 | 0.76(0.72,0.79),I2=0.0%,p=0.68 | 0.82(0.79,0.85),I2=0.0%,p=0.59 | 4.3(3.6,5.2) | 0.29(0.25,0.34) | 15(11,20) | 0.86(0.83,0.89) |
| Subgroup2: miRNA profiling | |||||||
| Single miRNA | 98 | 0.79(0.76,0.82),I2=88.46%,p<0.001 | 0.78(0.76,0.81),I2=81.27%,p<0.001 | 3.7(3.3,4.1) | 0.27(0.23,0.31) | 14(11,16) | 0.85(0.82,0.88) |
| Multiple miRNA | 67 | 0.87(0.85,0.89),I2=75.37%,p<0.001 | 0.87(0.85,0.89),I2=81.77%,p<0.001 | 6.9(5.8,8.2) | 0.15(0.12,0.18) | 47(34,64) | 0.94(0.91,0.95) |
| Subgroup3: specimen | |||||||
| Serum | 77 | 0.84(0.81,0.87),I2=91.62%,p<0.001 | 0.84(0.81,0.86),I2=82.12%,p<0.001 | 5.2(4.5,6.1) | 0.19(0.15,0.23) | 28(21,38) | 0.91(0.88,0.93) |
| Plasma | 41 | 0.79(0.75,0.82),I2=84.08%,p<0.001 | 0.85(0.82,0.88),I2=80.03%,p<0.001 | 5.3(4.2,6.8) | 0.25(0.20,0.30) | 22(15,32) | 0.89(0.86,0.91) |
| PBMCs/Neutrophils | 7 | 0.80(0.75,0.84),I2=29.99%,p=0.20 | 0.79(0.75,0.83),I2=0.0%,p=0.54 | 3.8(3.1,4.7) | 0.26(0.21,0.32) | 15(11,21) | 0.85(0.82,0.88) |
| Peripheral blood | 9 | 0.89(0.82,0.94),I2=91.38%,p<0.001 | 0.90(0.81,0.95),I2=89.42%,p<0.001 | 8.5(4.3,16.9) | 0.12(0.06,0.22) | 72(22,236) | 0.95(0.93,0.97) |
| Subgroup4: source of control | |||||||
| Healthy control | 110 | 0.83(0.80,0.85),I2=90.03%,p<0.001 | 0.84(0.82,0.86),I2=81.27%,p<0.001 | 5.3(4.6,6.1) | 0.21(0.18,0.24) | 25(20,33) | 0.90(0.87,0.93) |
| Cancer-free control | 24 | 0.83(0.78,0.87),I2=80.12%,p<0.001 | 0.86(0.82,0.89),I2=85.09%,p<0.001 | 6.0(4.6,7.8) | 0.20(0.15,0.26) | 30(18,48) | 0.91(0.88,0.93) |
| BPD | 9 | 0.77(0.67,0.85),I2=85.15%,p<0.001 | 0.87(0.83,0.90),I2=10.23%,p<0.001 | 5.8(4.2,8.0) | 0.26(0.18,0.39) | 22(11,43) | 0.89(0.86,0.92) |
| Subgroup5: histology type | |||||||
| AD>50% | 74 | 0.83(0.79,0.86), I2=90.98%,p<0.001 | 0.85(0.83,0.88),I2=83.31%,p<0.001 | 5.6(4.7,6.7) | 0.20(0.17,0.25) | 27(20,38) | 0.91(0.88,0.93) |
| AD<50% | 48 | 0.82(0.79,0.85),I2=85.09%,p<0.001 | 0.84(0.81,0.87),I2=78.36,p<0.001 | 5.1(4.2,6.2) | 0.21(0.18,0.26) | 24(17,34) | 0.90(0.87,0.92) |
| NSCLC | 122 | 0.83(0.80,0.85), I2=89.51%,p<0.001 | 0.84(0.82,0.86), I2=78.42%,p<0.001 | 5.2(4.5,5.8) | 0.21(0.18,0.24) | 25(20,31) | 0.90(0.87,0.92) |
| Lung cancer(mixed) | 10 | 0.87(0.80,0.92), I2=81.47%,p<0.001 | 0.85(0.77,0.95) I2=92.35%,p<0.001 | 6.0(3.8,9.5) | 0.15(0.10,0.23) | 40(20,78) | 0.93(0.90,0.95) |
| Subgroup6: stage | |||||||
| I/II>0.6 | 68 | 0.84(0.80,0.87),I2=91.70%,p<0.001 | 0.86(0.83,0.88),I2=82.26%,p<0.001 | 6.0(5.0,7.2) | 0.19(0.15,0.24) | 32(22,45) | 0.92(0.89,0.94) |
| I/II<0.4 | 57 | 0.82(0.79,0.84),I2=84.74%,p<0.001 | 0.83(0.80,0.86),I2=78.01%,p<0.001 | 4.8(4.1,5.7) | 0.22(0.19,,0.26) | 22(16,29) | 0.89(0.86,0.92) |
| Stage I/II patients | 43 | 0.81(0.77,0.84),I2=83.15%,p<0.001 | 0.82(0.78,0.85),I2=83.35%,p<0.001 | 4.5(3.7,5.5) | 0.23(0.19,0.28) | 19(14,27) | 0.88(0.85,0.91) |
| Stage I patients | 20 | 0.80(0.75,0.84),I2=71.63%,p<0.001 | 0.81(0.76,0.86),I2=78.62%,p<0.001 | 4.3(3.2,5.8) | 0.25(0.19,0.32) | 18(10,30) | 0.88(0.84,0.90) |
| Subgroup7: sample size | |||||||
| >150 | 50 | 0.80(0.76,0.84),I2=92.78%,p<0.001 | 0.85(0.82,0.87),I2=84.64%,p<0.001 | 5.3(4.4,6.5) | 0.23(0.19,0.28) | 23(16,33) | 0.90(0.87,0.92) |
| <150 | 84 | 0.84(0.81,0.87),I2=83.53%,p<0.001 | 0.84(0.81,0.86), I2=75.76%,p<0.001 | 5.3(4.5,6.2) | 0.18(0.15,0.22) | 28(21,38) | 0.91(0.88,0.93) |
| Subgroup8: publication year | |||||||
| >2015 | 80 | 0.82(0.79,0.85),I2=90.35%,p<0.001 | 0.85(0.83,0.88),I2=85.43%,p<0.001 | 5.6(4.7,6.7) | 0.21(0.18,0.25) | 27(20,36) | 0.91(0.88,0.93) |
| <2015 | 54 | 0.84(0.80,0.88),I2=86.81%, p<0.001 | 0.83(0.80,0.85),I2=64.91%,p<0.001 | 4.9(4.1,5.7) | 0.19(0.15,0.24) | 26(18,36) | 0.90(0.87,0.92) |
| Subgroup9: specific miRNA | |||||||
| hsa-miR-21-5p | 13 | 0.71(0.64,0.78),I2=82.09%, p<0.001 | 0.77(0.69,0.83),I2=77.44%, p<0.001 | 3.0(2.3,4.0) | 0.38(0.30,0.48) | 8(5,12) | 0.80(0.77,0.84) |
| hsa-miR-155-5p | 6 | 0.81(0.67,0.90),I2=84.93%, p<0.001 | 0.74(0.67,0.81),I2=50.45%, p=0.07 | 3.2(2.3,4.3) | 0.26(0.14,0.46) | 12(5,27) | 0.81(0.77,0.84) |
| hsa-miR-145-5p | 6 | 0.74(0.61,0.83),I2=82.86%, p<0.001 | 0.69(0.56,0.79),I2=81.53%, p<0.001 | 2.4(1.5,3.7) | 0.38(0.23,0.63) | 6(2,15) | 0.77(0.73,0.81) |
SEN=sensitivity, SPE= specificity, PLR= positive likelihood ratio, NLR= negative likelihood ratio, DOR= diagnostic odds ratio, AUC = area under the curve, CI= confidence interval, PBMCs =peripheral blood mononuclear cells, BPD= benign pulmonary disease, AD= adenocarcinoma.
Figure 2. The summary receiver operator characteristic (SROC) curves of circulating miRNAs test for the diagnosis of lung cancer patients in overall population.
Figure 3. Fagan diagram evaluating the positive likelihood ratio (PLR) and negative likelihood ratio (NLR).
Subgroup analyses
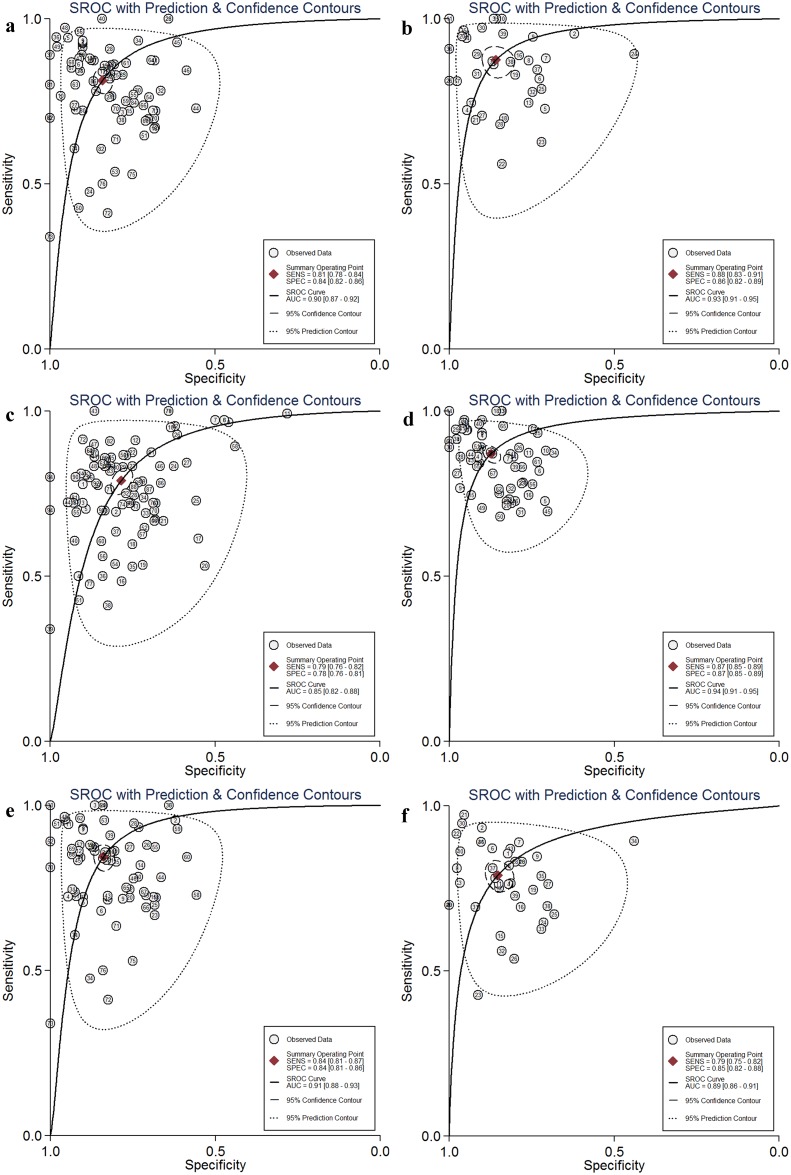
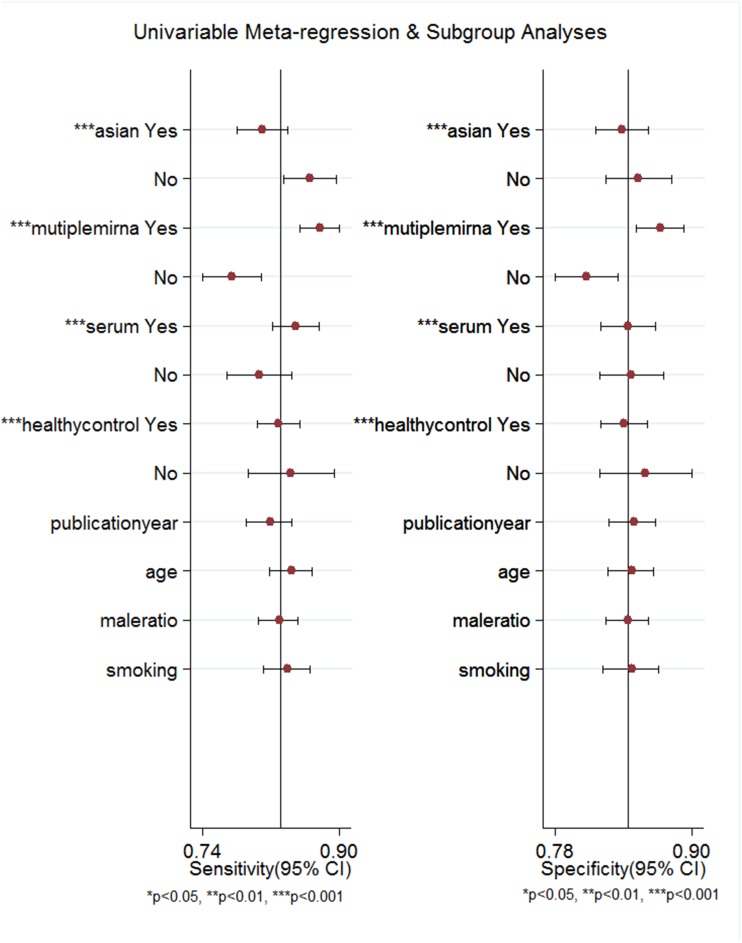
Various subgroup analyses (race, miRNA profiling, specimen, TNM stage, histology type, source of control, sample size and publication year) were also done. The main results of subgroup analyses are summarized in Table 1 and Figure 4.
Figure 4. Subgroup analysis of the summary receiver operator characteristic (SROC) curves of the miRNA test for the diagnosis of lung cancer patients.
(a) Asian population, (b) Caucasian, (c) single miRNA, (d) multiple miRNA, (e) serum, (f) plasma).
Race
In the subgroup analysis according to race, It seemed the circulating miRNAs may yield higher diagnostic performance in Caucasian than in Asian and mixed population (SEN: 0.88 vs 0.81 vs 0.76, SPE: 0.86 vs 0.84 vs 0.82, PLR: 6.2 vs 5.1 vs 4.3, NLR: 0.14 vs 0.22 vs 0.29, DOR: 43 vs 23 vs 15, and AUC: 0.93 vs 0.90 vs 0.86 for Caucasian, Asian and mixed population respectively) (Table 1). Figure 4a and 4b illustrate the SROC curves of diagnostic role of circulating miRNAs in Asian and Caucasian population respectively.
MiRNA profiling
We found combined miRNAs may lead higher diagnostic accuracy when compared with single miRNAs (SEN: 0.87 vs 0.79, SPE: 0.87 vs 0.78, PLR: 6.9 vs 3.7, NLR: 0.15 vs 0.27, DOR: 47 vs 14, and AUC: 0.94 vs 0.85 for multiple (n≥2) and single (n=1) miRNAs respectively) (Table 1 and Figure 4c and 4d).
Specimen
The specimen used in our included studies could be categorized as serum, plasma, peripheral blood mononuclear cells (PBMCs)/neutrophils and peripheral blood. 77 studies applied miRNAs detection in serum, which was the most widely used material. 41 studies applied plasma to detect miRNAs. The diagnostic performance of miRNAs in serum and plasma were good, with sensitivity of 0.84 and 0.79, specificity of 0.84 and 0.85, PLR of 5.2 and 5.3, NLR of 0.19 and 0.25, DOR of 28 and 22, and AUC of 0.91and 0.89 (Figure 4e and 4f). Detecting miRNAs in PBMCs/neutrophils and peripheral blood also had good performance to distinguish lung cancer patients from controls (Table 1).
TNM stage
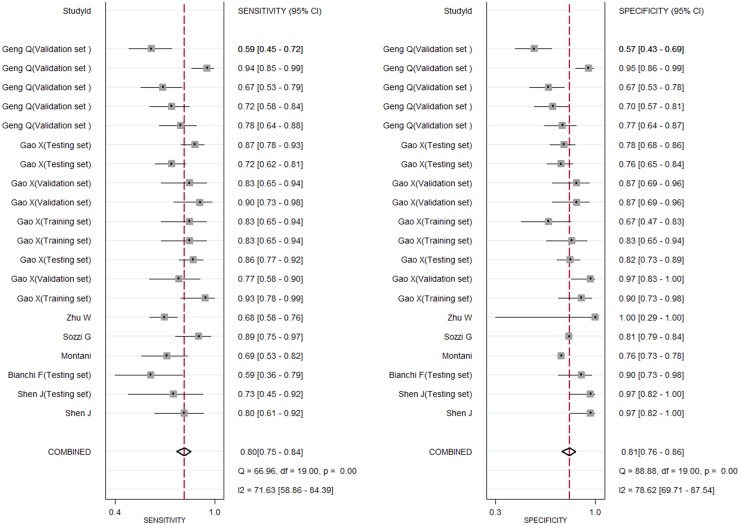
Firstly, we divided studies into two groups according to the percentage of stage I/II in overall lung cancer patients and 0.6 was selected as cutoff value. We found circulating miRNAs in patients with stage I/II≥0.6 and <0.4 all yielded good diagnostic performance. In addition, we further explored the diagnostic role of miRNAs in early stage lung cancer. We found the diagnostic accuracy of stage I/II and an additional stage I group were just similar to overall stage lung cancer (for stage I/II patients, the sensitivity, specificity, PLR, NLR, DOR and AUC were 0.81, 0.82, 4.5, 0.23, 19 and 0.88 respectively; for stage I patients, they were 0.80, 0.81, 4.3, 0.25, 18 and 0.88 respectively) (Figures 5 and 6). This suggested circulating miRNAs could distinguish all stage lung cancer from controls correctly (Table 1).
Figure 5. Forest plots of sensitivity and specifcity for circulating miRNAs in the diagnosis of stage I/II lung cancer.
Figure 6. Forest plots of sensitivity and specifcity for circulating miRNAs in the diagnosis of stage I lung cancer.
Source of controls
The controls of 110 studies were from healthy population, and 24 studies included cancer-free population as controls. Among 24 studies, nine were benign lung disease (BPD). Studies with healthy controls had a pooled sensitivity of 0.83, specificity of 0.84, PLR of 5.3, NLR of 0.21, DOR of 25 and the AUC of 0.90. For studies with cancer-free controls, the pooled sensitivity was 0.83, specificity was 0.86, PLR was 6.0, NLR was 0.20, DOR was 30 and the AUC was 0.91. In BPD controls, the sensitivity was 0.77, specificity was 0.87, PLR was 5.8, NLR was 0.26, DOR was 22 and the AUC was 0.89. Circulating miRNAs could not only screen lung cancer from healthy population, but also distinguish lung cancer from cancer-free patients (Table 1).
Histology type, sample size and publication year
74 studies investigated mostly (≥50%) with adenocarcinoma (AD). We performed subgroup analysis according to the percentage of AD≥50% and <50%, we found both groups had similar diagnostic accuracy. Only two studies evaluated the SCLC, as a result, we failed to conduct subgroup analysis in SCLC. The subgroup analysis was done in NSCLC and lung cancer (mixed type). The result suggested the diagnostic role of miRNAs in both groups was good (SEN: 0.83 vs 0.87, SPE: 0.84 vs 0.85, PLR: 5.2 vs 6.0, NLR: 0.21 vs 0.15, DOR: 25 vs 40, and AUC: 0.90 vs 0.93 for NSCLC and lung cancer, respectively) (Table 1).
The diagnostic accuracy was similar in the subgroup analyses in sample size and publication year (Table 1).
Meta-regression analyses and publication bias
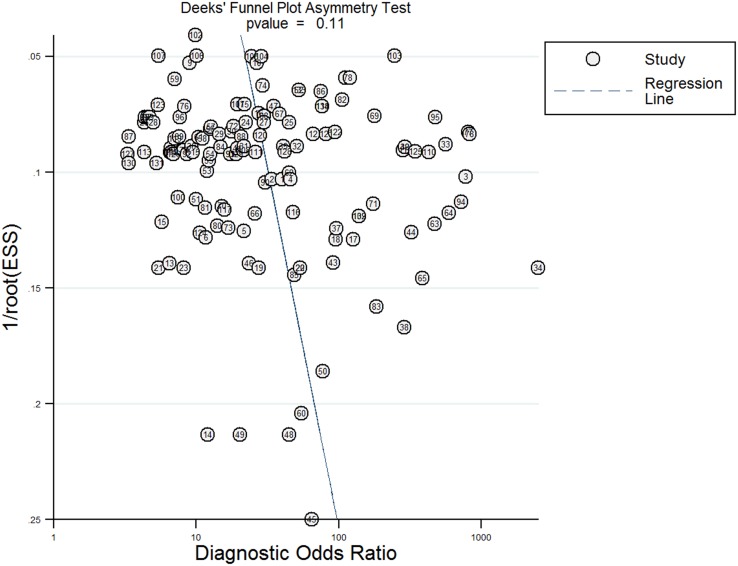
Meta-regression analyses were performed to analyze the potential sources of inter-study heterogeneity. The analysis suggested race (P<0.001), miRNA profiling (P<0.001), specimen (P<0.001), and the source of control (P<0.001) maybe the main sources of heterogeneity (Figure 7). As shown in Figure 8, no significant publication bias was detected by Deeks’funnel plot asymmetry test (P=0.11).
Figure 7. Forest plots of multivariable meta-regression analyses for sensitivity and specificity.
Figure 8. Deeks’ linear regression test of funnel plot asymmetry.
Potential miRNAs as circulating diagnostic biomarkers identified from literature
To identify potential circulating miRNA biomarkers from literature, differentially expressed circulating miRNAs in a consistent direction of change reported by at least two studies were sorted. Table 2 lists the potential miRNAs fulfilled the criteria above. A total of 42 miRNAs were reported by at least two studies in a consistent direction. Except three miRNAs have conflict results (hsa-miR-145-5p, hsa-miR-486-5p and hsa-miR-125a-5p), other 39 miRNAs have aconsistent results. Among 39 miRNAs, 20 miRNAs were upregulated, and the reaming 19 miRNAs were downregulated. Hsa-miR-21-5p was reported upregulated by 13 studies, followed by hsa-miR-223-3p, which was reported by seven studies as upregulated miRNA. Six studies reported two miRNAs (hsa-miR-155-5p and hsa-miR-126-3p); five studies reported two miRNAs (hsa-miR-210-5p and hsa-miR-20a-5p); four studies reported two miRNAs (hsa-miR-182-5p and hsa-miR-148b-3p); and three studies reported five miRNAs (hsa-miR-205-5p, hsa-miR-25-5p, hsa-miR-19b-3p, hsa-let-7a and hsa-let-7d). Also, we tried to examine the individual specificity and sensitivity of these specific miRNAs by meta-analysis. Only three miRNAs (miR-21-5p, miR-155-5p and miR-145-5p) were available for analysis, the analysis showed the diagnostic performance of these three miRNAs were good (SEN: 0.71 vs 0.81 vs 0.74, SPE: 0.77 vs 0.74 vs 0.69, PLR: 3.0 vs 3.2 vs 2.4, NLR: 0.38 vs 0.26 vs 0.38, DOR: 8 vs 12 vs 6, and AUC: 0.80 vs 0.81 vs 0.77 for miR-21-5p, miR-155-5p and miR-145-5p, respectively) (Table 1).
Table 2. The differentially expressed miRNAs with a consistent direction reported in at least two studies.
| miRNAs | Accession | Mature sequence | Source | Direction of expression | Studies | Reference |
|---|---|---|---|---|---|---|
| hsa-miR-21-5p | MIMAT0000076 | uagcuuaucagacugauguuga | Literature | ↑ | 13 | 11,19,25,32, 40,47,48,53,60, 64,66,72,74 |
| hsa-miR-223-3p | MIMAT0000280 | cguguauuugacaagcugaguu | Microarray,literature | ↑ | 7 | 12,13,19,34, 39,46,69 |
| hsa-miR-155-5p | MIMAT0000646 | uuaaugcuaaucgugauaggggu | Literature | ↑ | 6 | 11,17,19,46, 53,70 |
| hsa-miR-210-5p | MIMAT0026475 | agccccugcccaccgcacacug | Literature | ↑ | 5 | 33,47,48,58,72 |
| hsa-miR-20a-5p | MIMAT0000075 | uaaagugcuuauagugcagguag | Microarray,literature | ↑ | 5 | 13,15,19,46,69 |
| hsa-miR-182-5p | MIMAT0000259 | uuuggcaaugguagaacucacacu | Literature | ↑ | 4 | 11,47,48,72 |
| hsa-miR-145-5p | MIMAT0000437 | guccaguuuucccaggaaucccu | Microarray,literature | ↑ | 4 | 13,19,57,69 |
| ↓ | 1 | 53 | ||||
| hsa-miR-205-5p | MIMAT0000266 | uccuucauuccaccggagucug | Microarray,literature | ↑ | 3 | 21,25,67 |
| hsa-miR-25-5p | MIMAT0004498 | aggcggagacuugggcaauug | Microarray | ↑ | 3 | 13,24,67 |
| hsa-miR-19b-3p | MIMAT0000074 | ugugcaaauccaugcaaaacuga | Microarray | ↑ | 3 | 35,37,74 |
| hsa-miR-197-5p | MIMAT0022691 | cggguagagagggcagugggagg | Literature | ↑ | 2 | 11,70 |
| hsa-miR-29a-3p | MIMAT0000086 | acugauuucuuuugguguucag | Microarray | ↑ | 2 | 12,39 |
| hsa-miR-140-5p | MIMAT0000431 | cagugguuuuacccuaugguag | Microarray | ↑ | 2 | 12,39 |
| hsa-miR-221-3p | MIMAT0000278 | agcuacauugucugcuggguuuc | Microarray | ↑ | 2 | 13,74 |
| hsa-miR-222-3p | MIMAT0000279 | agcuacaucuggcuacugggu | Microarray | ↑ | 2 | 13,34 |
| hsa-miR-574-5p | MIMAT0004795 | ugagugugugugugugagugugu | Microarray | ↑ | 2 | 16,24 |
| hsa-miR-324-3p | MIMAT0000762 | acugccccaggugcugcugg | Microarray | ↑ | 2 | 18,24 |
| hsa-miR-200b-3p | MIMAT0000318 | uaauacugccugguaaugauga | Microarray | ↑ | 2 | 21,41 |
| hsa-miR-125b-5p | MIMAT0000423 | ucccugagacccuaacuuguga | Microarray,literature | ↑ | 2 | 21,38 |
| hsa-miR-1244 | MIMAT0005896 | aaguaguugguuuguaugagaugguu | Microarray | ↑ | 2 | 56,76 |
| hsa-miR-183-5p | MIMAT0000261 | uauggcacugguagaauucacu | Literature | ↑ | 2 | 67,72 |
| hsa-miR-126-3p | MIMAT0000445 | ucguaccgugaguaauaaugcg | Microarray,literature | ↓ | 6 | 12,24,46,47, 56,72 |
| hsa-miR-148b-3p | MIMAT0000759 | ucagugcaucacagaacuuugu | Microarray,literature | ↓ | 4 | 12,29,39,64 |
| hsa-miR-486-5p | MIMAT0002177 | uccuguacugagcugccccgag | Microarray | ↓ | 4 | 12,40,47,48 |
| Literature | ↓ | 2 | 31,58 | |||
| hsa-miR-125a-5p | MIMAT0000443 | ucccugagacccuuuaaccuguga | Literature | ↓ | 2 | 56,73 |
| Literature | ↑ | 1 | 57 | |||
| hsa-let-7a | MIMAT0000062 | ugagguaguagguuguauaguu | Microarray,literature | ↓ | 3 | 12,23,24 |
| hsa-let-7d | MIMAT0000065 | agagguaguagguugcauaguu | Microarray | ↓ | 3 | 12,24,39 |
| hsa-miR-328-5p | MIMAT0026486 | gggggggcaggaggggcucaggg | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-191-5p | MIMAT0000440 | caacggaaucccaaaagcagcug | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-92a-3p | MIMAT0000092 | uauugcacuugucccggccugu | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-484 | MIMAT0002174 | ucaggcucaguccccucccgau | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-22-3p | MIMAT0000077 | aagcugccaguugaagaacugu | Microarray | ↓ | 2 | 12,24 |
| hsa-miR-331-3p | MIMAT0000760 | gccccugggccuauccuagaa | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-30c | MIMAT0000244 | uguaaacauccuacacucucagc | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-98-5p | MIMAT0000096 | ugagguaguaaguuguauuguu | Microarray | ↓ | 2 | 12,24 |
| hsa-miR-374a | MIMAT0000727 | uuauaauacaaccugauaagug | Microarray | ↓ | 2 | 12,39 |
| hsa-miR-30b | MIMAT0000420 | uguaaacauccuacacucagcu | Microarray | ↓ | 2 | 12,39 |
| hsa-let-7c | MIMAT0000064 | ugagguaguagguuguaugguu | Literature | ↓ | 2 | 14,24 |
| hsa-let-7e | MIMAT0000066 | ugagguaggagguuguauaguu | Microarray | ↓ | 2 | 24,73 |
| hsa-let-7f | MIMAT0000067 | ugagguaguagauuguauaguu | Microarray,literature | ↓ | 2 | 24,46 |
| hsa-miR-195 | MIMAT0000461 | uagcagcacagaaauauuggc | Microarray,literature | ↓ | 2 | 24,50 |
| hsa-miR-29b-3p | MIMAT0000100 | uagcaccauuugaaaucaguguu | Literature | ↓ | 2 | 35,37 |
DISCUSSION
The development of suitable noninvasive biomarkers is important for the diagnosis of lung cancer. The pooled data in this meta-analysis showed circulating miRNAs had a sensitivity of 0.83 (95% CI 0.80-0.85) and specificity of 0.84 (95% CI 0.82-0.86), with corresponding PLR 5.3 (95% CI: 4.7-6.0) and NLR 0.20 (95% CI: 0.18-0.23). The pooled DOR was 26 (95% CI: 21-33) and the AUC was 0.90 (95% CI: 0.88-0.93). In various subgroup analyses, we found the diagnostic accuracy in each subgroup was very high, suggesting miRNA might be potential biomarker to discriminate lung cancer from controls.
Previous meta-analyses had explored the diagnostic role of miRNA in lung cancer [76–78]. However, our study showed the following advancements when compared with previous works. Firstly, as blood is very common and effective in lung cancer screening and to avoid potential bias caused by specimen, we only focused on circulating miRNAs, but previous works focused on not only blood but also sputum and exhaled breath condensate. It was our first time to comprehensively evaluate the diagnostic role of circulating miRNAs in lung cancer. Secondly, our analysis included 65 articles including 134 studies (6,919 patients with lung cancer and 7,064 controls). The sample size included in the studies by Chen [76], Guo [77] and Wang et al. [78] were 2,623, 3,801 and 3,703 respectively. Our sample size was larger than previous works; as a result, our analysis was more robust and reliable. Besides, the pooled data in this meta-analysis suggested the diagnostic role of circulating miRNA was higher than most recent study by Chen [76], Guo [77] and Wang et al. [78] (with sensitivity of 0.78, 0.76 and 0.78; specificity of 0.80, 0.77 and 0.80; AUC of 0.86, 0.83 and 0.86 in blood-based subgroups in the studies by Chen, Guo and Wang et al. respectively). This was because we included more studies than previous studies. At last, various subgroup analyses such as race, miRNA profiling, specimen, source of control, histology type, stage, sample size and publication year were done in our analysis to provide more information and give a comprehensive insight on the diagnostic role on circulating miRNAs in lung cancer. All these subgroup analyses suggested circulating miRNAs had good diagnostic performance.
The results of subgroup analyses in our study may give some useful information to clinical practice. We found combined miRNAs could yield higher diagnostic performance than single miRNAs. Single miRNAs with good diagnostic performance should be combined to yield higher diagnostic efficacy [49]. Serum was widely used in detecting miRNAs, and miRNAs in serum had good diagnostic value in the diagnosis of lung cancer. Serum was widely used in clinical detection such as blood biochemical tests and protein marker detection. As a result, detecting miRNAs in serum was more convenience for clinical practice [79, 80]. Also, combined miRNAs with serum protein markers, such as cytokeratin 19 fragment (CYFRA21-1), carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), cancer-associated antigen (CA) 125 and CA 19-9 maybe more effective in the diagnosis of lung cancer [58, 81]. We also conducted subgroup analysis based on the source of controls; the source of controls could be mainly divided into healthy population and cancer-free controls. In cancer-free controls, we could also extract a subgroup of BPD (such as pneumonia and benign pulmonary nodules). We found the diagnostic accuracy of miRNAs in these subgroups were similar, suggesting circulating miRNAs could not only serve as screening biomarkers but also could serve as biomarkers to distinguish lung cancer from BPD. Also, the diagnostic value of miRNAs in early stage lung cancer (stage I and stage I/II) is also good when compared with overall patients, suggesting circulating miRNAs were ideal biomarkers in all stage of lung cancers.
The circulating miRNAs may yield higher diagnostic performance in Caucasian than in Asian and mixed population, the racial disparities was found. These differences could be attributed to environmental, diet and life-style [82]. All these changes have been shown to affect the expression of miRNAs. For example, these factors could be affected the expression of miRNAs by methylation of CpG islands in their promoters [82, 83]. Another possible reason was the number of studies included for analysis was different in each groups. In Asian subgroups, 86 studies were included for analysis, but in Caucasian and mixed populations, only 39 and nine studies were included. The studies included for analysis in Asian population was generally two times more than Caucasian population and even more than mixed population.
We think the major obstacle was no uniform circulating miRNA biomarkers used in each study. Most studies selected miRNAs by microarray [12, 13, 18, 21, 22, 24, 26–28, 34–37, 39, 41, 42, 46–49, 52, 55, 59, 61, 64, 67, 74]. Some identified miRNAs by literature [11, 14–16, 19, 23, 25, 29–33, 40, 43–45, 50, 51, 53, 54, 56–58, 60, 62, 63, 65, 66, 68–73, 75, 84]. To give an insight to future studies, we listed the potential miRNAs reported by literature that could serve as circulating biomarkers. We found some upregulated miRNAs such as hsa-miR-21-5p, hsa-miR-223-3p, hsa-miR-155-5p, hsa-miR-210-5p, hsa-miR-205-5p, hsa-miR-25-5p and hsa-miR-19b-3p; some downregulated miRNAs such as hsa-miR-126-3p, hsa-miR-148b-3p, hsa-let-7a and hsa-let-7d, might be served as potential biomarkers. However, we also found some miRNAs such as hsa-miR-486-5p and hsa-miR-125a-5p were reported in inconsistent direction. Four studies found that hsa-miR-486-5p was reduced in peripheral blood of lung cancer patients [12, 40, 47, 48], while others two reported that it was up-regulation in lung cancer patients [31, 58]. The studies by Wang et al. [56] and Zhu et al. [73] reported that hsa-miR-125a-5p was downregulated in serum of lung cancer patients, but another study [57] found it was upregulated in serum. The contradictory results may be caused by inconsistent cutoff values applied by different studies and the potential bias existed in sample selection in each study. These miRNAs with conflicting results should be validated rigorously to verify whether they were suitable to serve as diagnostic biomarkers in lung cancer. In our study, we identified several miRNAs that might serve as potential biomarkers, it would be helpful for further studies to validate them; also, more works should be done in identifying more useful miRNAs and uniform their cutoff values to make them more feasible and effective in clinical practice.
We think the major limitation of our study was high heterogeneity exist in our analysis. Meta-regression analysis was done to explore the potential sources of heterogeneity between included studies. We found race, miRNA profiling, specimen, and the source of control might be the major cause of heterogeneity. Except that, no uniform cutoff values and certain miRNAs were applied in each study. The methodologies for an accurate absolute quantification of miRNAs and selected certain miRNAs limited the cross-comparison between studies performed by different laboratories.
In summary, the present study suggests that circulating miRNAs could serve as non-invasive diagnostic biomarkers for all stage lung cancers. They could not only screen lung cancer from healthy population, but also have a role in distinguishing lung cancer from cancer-free patients. Particularly, combinations of miRNAs are more complete indicators than individual miRNAs. Further subgroup analysis also indicated that serum might serve as the ideal sample specimen for the detecting miRNAs in the diagnosis of lung cancers. We also identified a panel of miRNAs such as miR-21-5p, miR-223-3p, miR-155-5p and miR-126-3p that might serve as potential biomarkers for lung cancer. However, the clinical application of miRNA profiling for lung cancer detection still needs further validation by future studies.
MATERIALS AND METHODS
Literature search
A comprehensive literature search was conducted in the databases of Medline, PubMed, EMBASE and Web of Science. The last search time was May 31, 2017. The following terms and combinations were used to identify studies: “microRNA”, “miRNA”, “lung cancer” and “lung neoplasm”. Furthermore, references of retrieved articles and reviews were manually screened for additional studies.
Inclusion and exclusion criteria
The inclusion criteria were applied to identify the eligible studies: (1) human-based investigations; (2) articles with full texts published in English and Chinese; (3) all lung cancer cases should be confirmed by pathology; (4) miRNA expression level was detected in blood (serum, plasma, peripheral whole blood and leukocyte in peripheral blood); (5) studies regarding the diagnostic potential of circulating miRNAs and lung cancer and provided sufficient data to extract true positive (TP), false positive (FP), true negative (TN), and false negative (FN). The exclusion criteria were as the follows: (1) publications unrelated to the diagnostic values of circulating miRNAs for lung cancer; (2) studies with duplicate data reported in other studies; (3) letters, editorials, case reports or reviews.
Data extraction and quality assessment
The following baseline characteristics and data were extracted: the name of first author, year of publication, country, ethnicity, and sample size; baseline characteristics of participants (age, the percentage of male, smoking status, histology, and stage of lung cancer cases, the source of control, sample species and microRNA profiling). Also, data required for diagnostic meta-analysis (TP, FP, TN and FN) were also extracted from included studies. The quality of each eligible study was assessed by the revised Quality Assessment of Diagnostic Accuracy Studies tool [85]. The data extraction and examination, quality assessments were conducted mainly by two investigators independently. Disagreements between the investigators were resolved by discussion among all authors until reach a consensus.
Extraction of potential circulating miRNAs as diagnostic biomarkers
To identify potential circulating miRNA biomarkers from literature, differentially expressed circulating miRNAs in a consistent direction of change reported by at least two studies were sorted. These miRNAs were analyzed individually or as one of the miRNAs panels. Some studies reported the TP, FP, TN and FN, but some studies did not report them. To evaluate the diagnostic accuracy of these specific miRNAs, the pooled sensitivity and specificity were performed if the data were available for analysis.
Statistical analysis
STATA 12.0 software (StataCorp, College Station, TX, USA) is used to perform all statistical analyses. We used the bivariate random-effects meta-analysis model to calculate the pooled sensitivity (SEN) [TP/(TP+FN)], specificity (SPE) [TN/(TN+FP)], positive likelihood ratio (PLR) [(sensitivity/(1−sensitivity)], negative likelihood ratio (NLR) [(1−specificity)/specificity)] and diagnostic odds ratio (DOR) [PLR/NLR] with their corresponding 95 % confidence intervals (CIs). The summary receiver operator characteristic (SROC) curve was plotted based on the sensitivity and specificity (sensitivity as the vertical axis, specificity as the horizontal axis). The area under the curve (AUC) was also calculated to evaluate the diagnostic accuracy of miRNA in discriminating lung cancer patients from controls [86, 87]. The Q test and I2 test was conducted to analyze the heterogeneity between studies. A P value less than 0.10 for Q test or I2 more than 50% indicated that there is substantial between-study heterogeneity [88]. To further explore the potential sources of heterogeneity, subgroup analyses and meta-regression were performed according to the characteristics of the included studies. Publication bias was assessed by using Deek's funnel plot asymmetry test (P<0.10 indicating statistically significant) [89].
SUPPLEMENTARY MATERIALS FIGURE AND TABLE
Abbreviations
- AD
adenocarcinoma
- AUC
area under the curve
- BPD
benign lung disease
- CI
confidence intervals
- CT
computed tomography
- DOR
diagnostic odds ratio
- FN
false negative
- FP
false positive
- NLR
negative likelihood ratio
- NSCLC
non-small-cell lung cancer
- PBMCs
peripheral blood mononuclear cells
- PLR
positive likelihood ratio
- PRISMA
preferred reporting items for meta-analyses
- SCLC
small cell lung cancer
- SEN
sensitivity
- SPE
specifcity
- SROC
summary receiver operating characteristic
- TN
true negative
- TP
true positive
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This work was supported by the PHD Innovation Fund of Kunming Medical University (No.2017B005), the Project of Basic Applied Research in Yunnan Province (2016FB145), the National Natural Science Foundation of China (No. 81460441), the Internal Institution Project of Ministry of Health of Yunnan Province (No.2017NS200) and the Health Science and Technology Project of Yunnan Province (No.2014NS001).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. http://doi.org/10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–32. doi: 10.3322/caac.21338. http://doi.org/10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. http://doi.org/10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. http://doi.org/10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgensztern D, Ng SH, Gao F, Govindan R. Trends in stage distribution for patients with non-small cell lung cancer: a National Cancer Database survey. J Thorac Oncol. 2010;5:29–33. doi: 10.1097/JTO.0b013e3181c5920c. http://doi.org/10.1097/JTO.0b013e3181c5920c. [DOI] [PubMed] [Google Scholar]
- 6.van der Aalst CM, Ten Haaf K, de Koning HJ. Lung cancer screening: latest developments and unanswered questions. Lancet Respir Med. 2016;4:749–61. doi: 10.1016/S2213-2600(16)30200-4. http://doi.org/10.1016/S2213-2600(16)30200-4. [DOI] [PubMed] [Google Scholar]
- 7.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–58. doi: 10.1016/S1470-2045(12)70073-6. http://doi.org/10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 8.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. http://doi.org/10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: 10.1016/j.tcb.2011.12.001. http://doi.org/10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. http://doi.org/10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys. 2013;67:875–84. doi: 10.1007/s12013-013-9575-y. http://doi.org/10.1007/s12013-013-9575-y. [DOI] [PubMed] [Google Scholar]
- 12.Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’olio V, Bernard L, Pelosi G, Maisonneuve P, Veronesi G, Di Fiore PP. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. http://doi.org/10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, et al. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer. 2012;130:1620–8. doi: 10.1002/ijc.26177. http://doi.org/10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- 14.Dou H, Wang Y, Su G, Zhao S. Decreased plasma let-7c and miR-152 as noninvasive biomarker for non-small-cell lung cancer. Int J Clin Exp Med. 2015;8:9291–8. [PMC free article] [PubMed] [Google Scholar]
- 15.Fan L, Qi H, Teng J, Su B, Chen H, Wang C, Xia Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016;37:7777–84. doi: 10.1007/s13277-015-4608-3. http://doi.org/10.1007/s13277-015-4608-3. [DOI] [PubMed] [Google Scholar]
- 16.Foss KM, Sima C, Ugolini D, Neri M, Allen KE, Weiss GJ. miR-1254 and miR-574-5p: serum-based microRNA biomarkers for early-stage non-small cell lung cancer. J Thorac Oncol. 2011;6:482–8. doi: 10.1097/JTO.0b013e318208c785. http://doi.org/10.1097/JTO.0b013e318208c785. [DOI] [PubMed] [Google Scholar]
- 17.Gao F, Chang J, Wang H, Zhang G. Potential diagnostic value of miR-155 in serum from lung adenocarcinoma patients. Oncol Rep. 2014;31:351–7. doi: 10.3892/or.2013.2830. http://doi.org/10.3892/or.2013.2830. [DOI] [PubMed] [Google Scholar]
- 18.Gao X, Wang Y, Zhao H, Wei F, Zhang X, Su Y, Wang C, Li H, Ren X. Plasma miR-324-3p and miR-1285 as diagnostic and prognostic biomarkers for early stage lung squamous cell carcinoma. Oncotarget. 2016;7:59664–75. doi: 10.18632/oncotarget.11198. http://doi.org/10.18632/oncotarget.11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15:149. doi: 10.1186/s12931-014-0149-3. http://doi.org/10.1186/s12931-014-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Zhang Y, Zhang Y, Shi Y, Xi J, Fan H, Xu S. Decreased expression of miR-204 in plasma is associated with a poor prognosis in patients with non-small cell lung cancer. Int J Mol Med. 2015;36:1720–6. doi: 10.3892/ijmm.2015.2388. http://doi.org/10.3892/ijmm.2015.2388. [DOI] [PubMed] [Google Scholar]
- 21.Halvorsen AR, Bjaanaes M, LeBlanc M, Holm AM, Bolstad N, Rubio L, Penalver JC, Cervera J, Mojarrieta JC, Lopez-Guerrero JA, Brustugun OT, Helland A. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget. 2016;7:37250–9. doi: 10.18632/oncotarget.9363. http://doi.org/10.18632/oncotarget.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennessey PT, Sanford T, Choudhary A, Mydlarz WW, Brown D, Adai AT, Ochs MF, Ahrendt SA, Mambo E, Califano JA. Serum microRNA biomarkers for detection of non-small cell lung cancer. PLoS One. 2012;7:e32307. doi: 10.1371/journal.pone.0032307. http://doi.org/10.1371/journal.pone.0032307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong HC, Kim EK, Lee JH, Lee JM, Yoo HN, Kim JK. Aberrant expression of let-7a miRNA in the blood of non-small cell lung cancer patients. Mol Med Rep. 2011;4:383–7. doi: 10.3892/mmr.2011.430. http://doi.org/10.3892/mmr.2011.430. [DOI] [PubMed] [Google Scholar]
- 24.Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, Scheffler M, Huwer H, Lenhof HP, Meese E. miRNAs in lung cancer - studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer. 2009;9:353. doi: 10.1186/1471-2407-9-353. http://doi.org/10.1186/1471-2407-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le HB, Zhu WY, Chen DD, He JY, Huang YY, Liu XG, Zhang YK. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med Oncol. 2012;29:3190–7. doi: 10.1007/s12032-012-0303-z. http://doi.org/10.1007/s12032-012-0303-z. [DOI] [PubMed] [Google Scholar]
- 26.Leidinger P, Brefort T, Backes C, Krapp M, Galata V, Beier M, Kohlhaas J, Huwer H, Meese E, Keller A. High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer. Oncotarget. 2016;7:4611–23. doi: 10.18632/oncotarget.6566. http://doi.org/10.18632/oncotarget.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leidinger P, Backes C, Dahmke IN, Galata V, Huwer H, Stehle I, Bals R, Keller A, Meese E. What makes a blood cell based miRNA expression pattern disease specific?--a miRNome analysis of blood cell subsets in lung cancer patients and healthy controls. Oncotarget. 2014;5:9484–97. doi: 10.18632/oncotarget.2419. http://doi.org/10.18632/oncotarget.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leidinger P, Keller A, Borries A, Huwer H, Rohling M, Huebers J, Lenhof HP, Meese E. Specific peripheral miRNA profiles for distinguishing lung cancer from COPD. Lung Cancer. 2011;74:41–7. doi: 10.1016/j.lungcan.2011.02.003. http://doi.org/10.1016/j.lungcan.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Chen YY, Li SQ, Huang C, Qin YZ. Expression of miR-148/152 family as potential biomarkers in non-small-cell lung cancer. Med Sci Monit. 2015;21:1155–61. doi: 10.12659/MSM.892940. http://doi.org/10.12659/MSM.892940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Zhang Q, Wu L, Jia C, Shi F, Li S, Peng A, Zhang G, Song X, Wang C. Serum miR-499 as a novel diagnostic and prognostic biomarker in non-small cell lung cancer. Oncol Rep. 2014;31:1961–7. doi: 10.3892/or.2014.3029. http://doi.org/10.3892/or.2014.3029. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Wang Y, Zhang Q, Tang L, Liu X, Dai Y, Xiao L, Huang S, Chen L, Guo Z, Lu J, Yuan K. MicroRNA-486 as a biomarker for early diagnosis and recurrence of non-small cell lung cancer. PLoS One. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. http://doi.org/10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Li W, Ouyang Q, Hu S, Tang J. Detection of lung cancer with blood microRNA-21 expression levels in Chinese population. Oncol Lett. 2011;2:991–4. doi: 10.3892/ol.2011.351. http://doi.org/10.3892/ol.2011.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li ZH, Zhang H, Yang ZG, Wen GQ, Cui YB, Shao GG. Prognostic significance of serum microRNA-210 levels in nonsmall-cell lung cancer. J Int Med Res. 2013;41:1437–44. doi: 10.1177/0300060513497560. http://doi.org/10.1177/0300060513497560. [DOI] [PubMed] [Google Scholar]
- 34.Lv S, Xue J, Wu C, Wang L, Wu J, Xu S, Liang X, Lou J. Identification of a panel of serum microRNAs as biomarkers for early detection of lung adenocarcinoma. J Cancer. 2017;8:48–56. doi: 10.7150/jca.16644. http://doi.org/10.7150/jca.16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Lin Y, Zhan M, Mann DL, Stass SA, Jiang F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest. 2015;95:1197–206. doi: 10.1038/labinvest.2015.88. http://doi.org/10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J, Li N, Lin Y, Gupta C, Jiang F. Circulating neutrophil microRNAs as biomarkers for the detection of lung cancer. Biomark Cancer. 2016;8:1–7. doi: 10.4137/BIC.S37333. http://doi.org/10.4137/BIC.S37333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma J, Guarnera MA, Zhou W, Fang H, Jiang F. A prediction model based on biomarkers and clinical characteristics for detection of lung cancer in pulmonary nodules. Transl Oncol. 2017;10:40–5. doi: 10.1016/j.tranon.2016.11.001. http://doi.org/10.1016/j.tranon.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol. 2012;138:2045–50. doi: 10.1007/s00432-012-1285-0. http://doi.org/10.1007/s00432-012-1285-0. [DOI] [PubMed] [Google Scholar]
- 39.Montani F, Marzi MJ, Dezi F, Dama E, Carletti RM, Bonizzi G, Bertolotti R, Bellomi M, Rampinelli C, Maisonneuve P, Spaggiari L, Veronesi G, Nicassio F, et al. miR-Test: a blood test for lung cancer early detection. J Natl Cancer Inst. 2015;107:djv063. doi: 10.1093/jnci/djv063. http://doi.org/10.1093/jnci/djv063. [DOI] [PubMed] [Google Scholar]
- 40.Mozzoni P, Banda I, Goldoni M, Corradi M, Tiseo M, Acampa O, Balestra V, Ampollini L, Casalini A, Carbognani P, Mutti A. Plasma and EBC microRNAs as early biomarkers of non-small-cell lung cancer. Biomarkers. 2013;18:679–86. doi: 10.3109/1354750X.2013.845610. http://doi.org/10.3109/1354750X.2013.845610. [DOI] [PubMed] [Google Scholar]
- 41.Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, Ramnath N, Gotoh N, Beer DG, Chen G. A novel serum 4-microRNA signature for lung cancer detection. Sci Rep. 2015;5:12464. doi: 10.1038/srep12464. http://doi.org/10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patnaik SK, Yendamuri S, Kannisto E, Kucharczuk JC, Singhal S, Vachani A. MicroRNA expression profiles of whole blood in lung adenocarcinoma. PLoS One. 2012;7:e46045. doi: 10.1371/journal.pone.0046045. http://doi.org/10.1371/journal.pone.0046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peng H, Wang J, Li J, Zhao M, Huang SK, Gu YY, Li Y, Sun XJ, Yang L, Luo Q, Huang CZ. A circulating non-coding RNA panel as an early detection predictor of non-small cell lung cancer. Life Sci. 2016;151:235–42. doi: 10.1016/j.lfs.2016.03.002. http://doi.org/10.1016/j.lfs.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 44.Powrozek T, Krawczyk P, Kowalski DM, Kuznar-Kaminska B, Winiarczyk K, Olszyna-Serementa M, Batura-Gabryel H, Milanowski J. Application of plasma circulating microRNA-448, 506, 4316, and 4478 analysis for non-invasive diagnosis of lung cancer. Tumour Biol. 2016;37:2049–55. doi: 10.1007/s13277-015-3971-4. http://doi.org/10.1007/s13277-015-3971-4. [DOI] [PubMed] [Google Scholar]
- 45.Powrozek T, Krawczyk P, Kowalski DM, Winiarczyk K, Olszyna-Serementa M, Milanowski J. Plasma circulating microRNA-944 and microRNA-3662 as potential histologic type-specific early lung cancer biomarkers. Transl Res. 2015;166:315–23. doi: 10.1016/j.trsl.2015.05.009. http://doi.org/10.1016/j.trsl.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH, Brest P, Hofman P. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS One. 2013;8:e54596. doi: 10.1371/journal.pone.0054596. http://doi.org/10.1371/journal.pone.0054596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL, Jiang Z, Fang H, Katz RL, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–87. doi: 10.1038/labinvest.2010.194. http://doi.org/10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen J, Liu Z, Todd NW, Zhang H, Liao J, Yu L, Guarnera MA, Li R, Cai L, Zhan M, Jiang F. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. http://doi.org/10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sozzi G, Boeri M, Rossi M, Verri C, Suatoni P, Bravi F, Roz L, Conte D, Grassi M, Sverzellati N, Marchiano A, Negri E, La Vecchia C, et al. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J Clin Oncol. 2014;32:768–73. doi: 10.1200/JCO.2013.50.4357. http://doi.org/10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su K, Zhang T, Wang Y, Hao G. Diagnostic and prognostic value of plasma microRNA-195 in patients with non-small cell lung cancer. World J Surg Oncol. 2016;14:224. doi: 10.1186/s12957-016-0980-8. http://doi.org/10.1186/s12957-016-0980-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Sun L, Chen Y, Su Q, Tang X, Liang Y, Che G, Luo F. Increased plasma miRNA-30a as a biomarker for non-small cell lung cancer. Med Sci Monit. 2016;22:647–55. doi: 10.12659/MSM.897330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tai MC, Yanagisawa K, Nakatochi M, Hotta N, Hosono Y, Kawaguchi K, Naito M, Taniguchi H, Wakai K, Yokoi K, Takahashi T. Blood-borne miRNA profile-based diagnostic classifier for lung adenocarcinoma. Sci Rep. 2016;6:31389. doi: 10.1038/srep31389. http://doi.org/10.1038/srep31389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A, Jiao W, Wang Y. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22:540–8. doi: 10.1097/CEJ.0b013e32835f3be9. http://doi.org/10.1097/CEJ.0b013e32835f3be9. [DOI] [PubMed] [Google Scholar]
- 54.Ulivi P, Foschi G, Mengozzi M, Scarpi E, Silvestrini R, Amadori D, Zoli W. Peripheral blood miR-328 expression as a potential biomarker for the early diagnosis of NSCLC. Int J Mol Sci. 2013;14:10332–42. doi: 10.3390/ijms140510332. http://doi.org/10.3390/ijms140510332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang C, Ding M, Xia M, Chen S, Van Le A, Soto-Gil R, Shen Y, Wang N, Wang J, Gu W, Wang X, Zhang Y, Zen K, et al. A five-miRNA panel identified from a multicentric case-control study serves as a novel diagnostic tool for ethnically diverse non-small-cell lung cancer patients. EBioMedicine. 2015;2:1377–85. doi: 10.1016/j.ebiom.2015.07.034. http://doi.org/10.1016/j.ebiom.2015.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang P, Yang D, Zhang H, Wei X, Ma T, Cheng Z, Hong Q, Hu J, Zhuo H, Song Y, Jia C, Jing F, Jin Q, et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin Lung Cancer. 2015;16:313–9. doi: 10.1016/j.cllc.2014.12.006. http://doi.org/10.1016/j.cllc.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Wang RJ, Zheng YH, Wang P, Zhang JZ. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8:765–71. [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Zhi X, Zhang Y, An G, Feng G. Role of plasma MicroRNAs in the early diagnosis of non-small-cell lung cancers: a case-control study. J Thorac Dis. 2016;8:1645–52. doi: 10.21037/jtd.2016.06.21. http://doi.org/10.21037/jtd.2016.06.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Zhao H, Gao X, Wei F, Zhang X, Su Y, Wang C, Li H, Ren X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070–86. doi: 10.18632/oncotarget.8429. http://doi.org/10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J, Liu LK, Gao W, Zhu CJ, Liu YQ, Cheng T, Shu YQ. Reduction of plasma microRNA-21 is associated with chemotherapeutic response in patients with non-small cell lung cancer. Chin J Cancer Res. 2011;23:123–8. doi: 10.1007/s11670-011-0123-2. http://doi.org/10.1007/s11670-011-0123-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wozniak MB, Scelo G, Muller DC, Mukeria A, Zaridze D, Brennan P. Circulating microRNAs as non-invasive biomarkers for early detection of non-small-cell lung cancer. PLoS One. 2015;10:e0125026. doi: 10.1371/journal.pone.0125026. http://doi.org/10.1371/journal.pone.0125026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan HJ, Ma JY, Wang L, Gu W. Expression and significance of circulating microRNA-31 in lung cancer patients. Med Sci Monit. 2015;21:722–6. doi: 10.12659/MSM.893213. http://doi.org/10.12659/MSM.893213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang YL, Xu LP, Zhuo FL, Wang TY. Prognostic value of microRNA-10b overexpression in peripheral blood mononuclear cells of nonsmall-cell lung cancer patients. Tumour Biol. 2015;36:7069–75. doi: 10.1007/s13277-015-3366-6. http://doi.org/10.1007/s13277-015-3366-6. [DOI] [PubMed] [Google Scholar]
- 64.Yang JS, Li BJ, Lu HW, Chen Y, Lu C, Zhu RX, Liu SH, Yi QT, Li J, Song CH. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015;36:3035–42. doi: 10.1007/s13277-014-2938-1. http://doi.org/10.1007/s13277-014-2938-1. [DOI] [PubMed] [Google Scholar]
- 65.Yu Y, Zuo J, Tan Q, Zar Thin K, Li P, Zhu M, Yu M, Fu Z, Liang C, Tu J. Plasma miR-92a-2 as a biomarker for small cell lung cancer. Cancer Biomark. 2017;18:319–27. doi: 10.3233/CBM-160254. http://doi.org/10.3233/CBM-160254. [DOI] [PubMed] [Google Scholar]
- 66.Zhao W, Zhao JJ, Zhang L, Xu QF, Zhao YM, Shi XY, Xu AG. Serum miR-21 level: a potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int J Clin Exp Med. 2015;8:14759–63. [PMC free article] [PubMed] [Google Scholar]
- 67.Zaporozhchenko IA, Morozkin ES, Skvortsova TE, Ponomaryova AA, Rykova EY, Cherdyntseva NV, Polovnikov ES, Pashkovskaya OA, Pokushalov EA, Vlassov VV, Laktionov PP. Plasma miR-19b and miR-183 as potential biomarkers of lung cancer. PLoS One. 2016;11:e0165261. doi: 10.1371/journal.pone.0165261. http://doi.org/10.1371/journal.pone.0165261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zeng XL, Zhang SY, Zheng JF, Yuan H, Wang Y. Altered miR-143 and miR-150 expressions in peripheral blood mononuclear cells for diagnosis of non-small cell lung cancer. Chin Med J (Engl) 2013;126:4510–6. [PubMed] [Google Scholar]
- 69.Zhang H, Mao F, Shen T, Luo Q, Ding Z, Qian L, Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13:669–76. doi: 10.3892/ol.2016.5462. http://doi.org/10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575. [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou C, Chen Z, Dong J, Li J, Shi X, Sun N, Luo M, Zhou F, Tan F, He J. Combination of serum miRNAs with Cyfra21-1 for the diagnosis of non-small cell lung cancer. Cancer Lett. 2015;367:138–46. doi: 10.1016/j.canlet.2015.07.015. http://doi.org/10.1016/j.canlet.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 72.Zhu W, Zhou K, Zha Y, Chen D, He J, Ma H, Liu X, Le H Zhang Y. Diagnostic value of serum miR-182, miR-183, miR-210, and miR-126 levels in patients with early-stage non-small cell lung cancer. PLoS One. 2016;11:e0153046. doi: 10.1371/journal.pone.0153046. http://doi.org/10.1371/journal.pone.0153046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu WY, Luo B, An JY, He JY, Chen DD, Xu LY, Huang YY, Liu XG, Le HB Zhang YK. Differential expression of miR-125a-5p and let-7e predicts the progression and prognosis of non-small cell lung cancer. Cancer Invest. 2014;32:394–401. doi: 10.3109/07357907.2014.922569. http://doi.org/10.3109/07357907.2014.922569. [DOI] [PubMed] [Google Scholar]
- 74.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, Cheng W, Wang F, Qi LW, Chen Y, Huang Z, Wang T, Zhu D, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–25. doi: 10.18632/oncotarget.14311. http://doi.org/10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W, Li W, Ding M, Yuan H, Yang J, Meng W, Jin E, Wang X, Ma S. Identification of miRNAs as non-invasive biomarkers for early diagnosis of lung cancers. Tumour Biol. 2016 doi: 10.1007/s13277-016-5442-y. http://doi.org/10.1007/s13277-016-5442-y. [DOI] [PubMed] [Google Scholar]
- 76.Chen L, Jin H. MicroRNAs as novel biomarkers in the diagnosis of non-small cell lung cancer: a meta-analysis based on 20 studies. Tumour Biol. 2014;35:9119–29. doi: 10.1007/s13277-014-2188-2. http://doi.org/10.1007/s13277-014-2188-2. [DOI] [PubMed] [Google Scholar]
- 77.Guo Z, Zhao C, Wang Z. MicroRNAs as ideal biomarkers for the diagnosis of lung cancer. Tumour Biol. 2014;35:10395–407. doi: 10.1007/s13277-014-2330-1. http://doi.org/10.1007/s13277-014-2330-1. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, Wu S, Zhao L, Zhao J, Liu J, Wang Z. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology. 2015;20:56–65. doi: 10.1111/resp.12444. http://doi.org/10.1111/resp.12444. [DOI] [PubMed] [Google Scholar]
- 79.Nakamura H, Nishimura T. History, molecular features, and clinical importance of conventional serum biomarkers in lung cancer. Surg Today. 2017;47:1037–59. doi: 10.1007/s00595-017-1477-y. http://doi.org/10.1007/s00595-017-1477-y. [DOI] [PubMed] [Google Scholar]
- 80.Armand-Labit V, Pradines A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol Concepts. 2017;8:61–81. doi: 10.1515/bmc-2017-0002. http://doi.org/10.1515/bmc-2017-0002. [DOI] [PubMed] [Google Scholar]
- 81.Li X, Zhang Q, Jin X, Cao L. Combining serum miRNAs, CEA, and CYFRA21-1 with imaging and clinical features to distinguish benign and malignant pulmonary nodules: a pilot study : Xianfeng Li et al.: Combining biomarker, imaging, and clinical features to distinguish pulmonary nodules. World J Surg Oncol. 2017;15:107. doi: 10.1186/s12957-017-1171-y. http://doi.org/10.1186/s12957-017-1171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahmad A, Azim S, Zubair H, Khan MA, Singh S, Carter JE, Rocconi RP, Singh AP. Epigenetic basis of cancer health disparities: looking beyond genetic differences. Biochim Biophys Acta. 2017;1868:16–28. doi: 10.1016/j.bbcan.2017.01.001. http://doi.org/10.1016/j.bbcan.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jones J, Grizzle W, Wang H, Yates C. MicroRNAs that affect prostate cancer: emphasis on prostate cancer in African Americans. Biotech Histochem. 2013;88:410–24. doi: 10.3109/10520295.2013.807069. http://doi.org/10.3109/10520295.2013.807069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gou WF, Sun HZ, Zhao S, Niu ZF, Mao XY, Takano Y, Zheng HC. Downregulated inhibitor of growth 3 (ING3) expression during colorectal carcinogenesis. Indian J Med Res. 2014;139:561–7. [PMC free article] [PubMed] [Google Scholar]
- 85.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM. QUADAS-2 Group. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. http://doi.org/10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 86.Deeks JJ. Systematic reviews in health care: systematic reviews of evaluations of diagnostic and screening tests. BMJ. 2001;323:157–62. doi: 10.1136/bmj.323.7305.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56:1129–35. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 88.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. http://doi.org/10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–93. doi: 10.1016/j.jclinepi.2005.01.016. http://doi.org/10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.