ABSTRACT
Salmonella enterica serovar Typhimurium is a leading cause of foodborne disease worldwide. Severe infections result from the ability of S. Typhimurium to survive within host immune cells, despite being exposed to various host antimicrobial factors. SodCI, a copper-zinc-cofactored superoxide dismutase, is required to defend against phagocytic superoxide. SodCII, an additional periplasmic superoxide dismutase, although produced during infection, does not function in the host. Previous studies suggested that CueP, a periplasmic copper binding protein, facilitates acquisition of copper by SodCII. CopA and GolT, both inner membrane ATPases that pump copper from the cytoplasm to the periplasm, are a source of copper for CueP. Using in vitro SOD assays, we found that SodCI can also utilize CueP to acquire copper. However, both SodCI and SodCII have a significant fraction of activity independent of CueP and cytoplasmic copper export. We utilized a series of mouse competition assays to address the in vivo role of CueP-mediated SodC activation. A copA golT cueP triple mutant was equally as competitive as the wild type, suggesting that sufficient SodCI is active to defend against phagocytic superoxide independent of CueP and cytoplasmic copper export. We also confirmed that a strain containing a modified SodCII, which is capable of complementing a sodCI deletion, was fully virulent in a copA golT cueP background competed against the wild type. These competitions also address the potential impact of cytoplasmic copper toxicity within the phagosome. Our data suggest that Salmonella does not encounter inhibitory concentrations of copper during systemic infection.
IMPORTANCE Salmonella is a leading cause of gastrointestinal disease worldwide. In severe cases, Salmonella can cause life-threatening systemic infections, particularly in very young children, the elderly, or people who are immunocompromised. To cause disease, Salmonella must survive the hostile environment inside host immune cells, a location in which most bacteria are killed. Our work examines how one particular metal, copper, is acquired by Salmonella to activate a protein important for survival within immune cells. At high levels, copper itself can inhibit Salmonella. Using a strain of Salmonella that cannot detoxify intracellular copper, we also addressed the in vivo role of copper as an antimicrobial agent.
KEYWORDS: copper efflux, Salmonella, SodC
INTRODUCTION
Salmonella enterica serovar Typhimurium is a leading cause of bacterial foodborne illness and is capable of causing life-threatening systemic disease (1). During extraintestinal infection, Salmonella resides inside host macrophages in a modified phagosome called the Salmonella-containing vacuole (SCV) (2–4). SodCI is important for Salmonella survival during systemic infection. Located in the periplasm, SodCI specifically defends against extracellular superoxide generated by the oxidative burst in macrophages (5). In the absence of SodCI, S. Typhimurium is approximately 10-fold attenuated (5–7). SodCI converts two molecules of superoxide into hydrogen peroxide and water, with the reaction being catalyzed by a copper atom in the active site; a zinc atom is also present for structural purposes (8). SodCI is exported to the periplasm via the Sec pathway, where the protein then folds and acquires metal cofactors (6).
Salmonella expresses a second Cu/Zn periplasmic superoxide dismutase (SOD), SodCII. Despite these superoxide dismutases being 60% identical at the amino acid level, our lab and others have previously shown that Salmonella strains lacking SodCII are fully virulent (6, 9–12). Two important properties that affect the contributions of SodCI and SodCII to virulence were identified. First, SodCI noncovalently binds peptidoglycan, whereas SodCII is released into the milieu upon partial disruption of the outer membrane, such as that caused by antimicrobial peptides (13). Second, SodCI is protease resistant, whereas SodCII is protease sensitive; proteases are an additional defense mechanism utilized by phagocytes. Modifying SodCII to remain tethered in the periplasm or protease resistant allows the enzyme to actively defend against superoxide during infection (13–15). SodCI has a higher affinity for its metal cofactors than SodCII (16). Importantly, this difference in metal affinity does not influence the differential function of SodCI and SodCII in vivo (6, 14, 16).
Copper is an essential transition metal used in a variety of biochemical reactions, including electron transfer, oxidation-reduction reactions, and free-radical scavenging (17). However, accumulation of intracellular copper is toxic. This creates a delicate balance between acquiring sufficient copper and avoiding deleterious side effects (18). ATP7A and ATP7B are human P1B-type ATPases that transport copper across membranes throughout the body. Humans with mutations in ATP7A or ATP7B have Menkes' and Wilson's disease, respectively, both of which are debilitating genetic disorders caused by inability to appropriately traffic copper (19–21). Bacteria produce homologous P-type ATPases, which are necessary to prevent the accumulation of cytoplasmic copper (22). Based on in vitro experiments and studies in mammalian cells, it was originally hypothesized that cytoplasmic copper toxicity resulted from DNA damage mediated by Fenton-like reactions of Cu1+ with hydrogen peroxide, generating highly reactive hydroxyl radicals (23–25). Macomber et al. (26) demonstrated that copper does not catalyze formation of oxidative DNA damage in the cytoplasm but actually protects against iron-mediated Fenton reactions by competing with iron for binding. They went on to identify certain dehydratases as the primary cytoplasmic target of copper toxicity in Escherichia coli (26). The partially exposed nature of the iron-sulfur cluster in this enzyme family allows interaction with copper, which can displace iron, disrupting activity. The susceptible class of enzymes includes dihydroxy-acid dehydratase and isopropylmalate isomerase, which are involved in branched-chain amino acid synthesis, fumarase A, and 6-phosphogluconate dehydratase (27).
Copper toxicity necessitates that bacteria contain cytoplasmic copper detoxification systems. In Salmonella, two transcriptional regulators, CueR and GolS, detect intracellular copper. CueR regulates copA, cueO, and cueP (28–30). CopA is an inner membrane P-type ATPase capable of pumping copper from the cytoplasm into the periplasm. CueP is a periplasmic copper binding protein, and CueO is a periplasmic multi-copper-ion oxidase. GolS positively regulates golT and golB (31, 32). GolT is also a P-type ATPase, and GolB is a cytoplasmic copper binding protein (Fig. 1). CopA and GolT are functionally redundant in export of copper from the cytoplasm; single mutations in copA or golT do not confer significant growth defects in copper-supplemented medium in vitro, where a copA golT double mutant is growth inhibited (32). The periplasmic copper binding protein CueP binds copper exported by either CopA or GolT. Periplasmic copper can also be oxidized to Cu2+ by CueO, potentially hindering copper from crossing the inner membrane and/or preventing Cu+1 from reacting with hydrogen peroxide to generate hydroxyl radicals (33, 34). Previous reports suggest that a Salmonella cueO mutant is attenuated for survival in the spleen and liver after oral infection, based on reduced CFU recovery relative to that for the wild type (29). We have previously reported that a cueO mutant is not significantly attenuated in an intraperitoneal (i.p.) competition assay with the wild type (35).
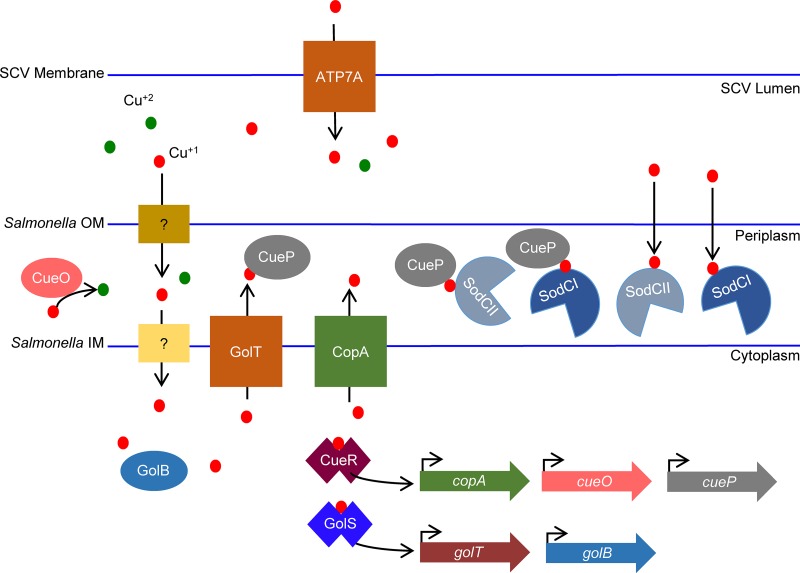
FIG 1.
Schematic of copper trafficking in S. Typhimurium. Copper gains entry to the periplasm via nonspecific porins or transporters. In the periplasm, Cu1+ can be oxidized by CueO to Cu2+. Cu1+ can cross the inner membrane via unspecified mechanisms. CueR and GolS are transcriptional regulators that bind Cu and activate target gene expression. CopA and GolT are inner membrane P-type ATPases that pump copper back to the periplasm, where it can be bound by CueP. CueP can deliver copper to SodCI and SodCII. SodCI and SodCII are also able to acquire copper via CueP-independent mechanisms. During infection, phagocytes upregulate expression of a copper-specific transporter, ATP7A, that potentially pumps copper into the phagosome.
The fate of periplasmic copper in S. Typhimurium, after being bound by CueP, is unclear. In addition to copper binding, it was suggested that under high copper concentrations, CueP confers additional protection to the cell by reducing Cu2+ to Cu0, presumably blocking copper-mediated Fenton reactions in the periplasm (36). DsbC and DsbG are periplasmic disulfide bond isomerases (37). DsbC has been shown to be required to protect the periplasm from copper-mediated nonnative disulfide bond formation in E. coli (38). In addition, work in Salmonella suggests that DsbC contributes to maintaining CueP in a reduced state (39). Osman et al. have reported that, in vitro, CueP facilitates copper acquisition by SodCII in the periplasm (40). In the absence of CueP or both CopA and GolT, SodCII was produced but not completely metalated. The requirement of CopA or GolT implies that the copper source for CueP originates from the cytoplasm. Due to limitations in the experimental setup, SodCI was not tested.
During systemic infection, phagocytes alter normal copper trafficking and increase expression of transporters that import copper into the phagosome (41). Due to the antimicrobial properties of copper, it is appealing to presume that the host has evolved to use copper as an additional defense mechanism. One specific copper transporter, ATP7A, is actively recruited to phagosomes in response to bacterial infections. Macrophages with a mutation in this transporter have increased susceptibility to infection by E. coli (41). Recently, Ladomersky et al. (42) provided data supporting a role for ATP7A in resistance to Salmonella infection by increasing the concentration of copper in the Salmonella-containing vacuole (SCV). The overall impact of increased copper concentrations in vivo along with the role of cytoplasmic copper export in the ability of SodCI and SodCII to acquire copper in the SCV is unclear.
In this study, we first addressed the contribution of CueP to metalation of SodCI in vitro. Although more significant for SodCII, our data demonstrate that CueP and cytoplasmic copper export can also contribute to activation of SodCI. However, both enzymes can acquire copper independent of the copper efflux systems. We then moved to an in vivo infection model to determine if cytoplasmic copper detoxification was important for Salmonella survival during systemic infection and if SodCI or SodCII requires CueP-mediated activation to be fully virulent. SodCI and SodCII are sufficiently active independent of CueP or CopA and GolT. Moreover, our data suggest that S. Typhimurium does not experience significant cytoplasmic copper stress in the Salmonella-containing vacuole.
RESULTS
CopA or GolT is required to protect the cytoplasm from copper stress.
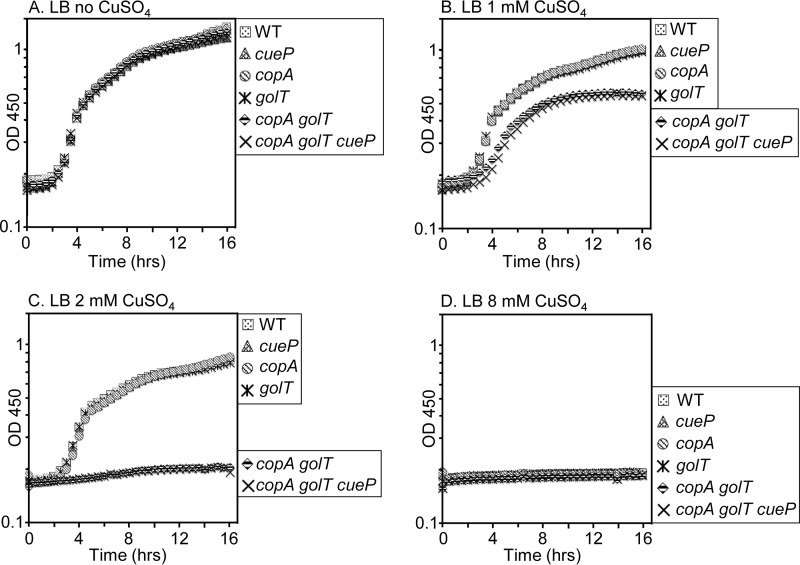
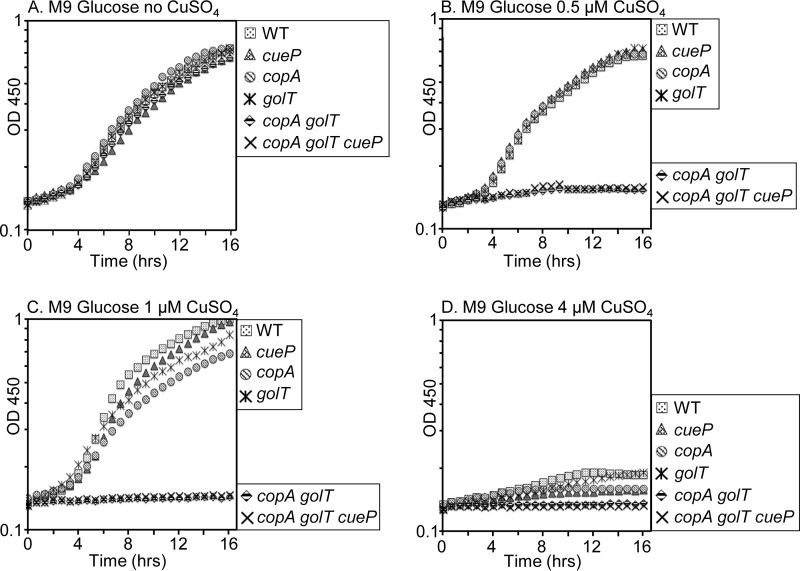
To test the requirement for CopA, GolT, and CueP in copper metalation of SodCI and SodCII, we first generated gene deletions of copA, golT, and cueP (43) and then constructed a copA golT double mutant and a copA golT cueP triple mutant. To confirm that the copA and golT mutants were defective in copper export, growth assays were performed under increasing concentrations of copper sulfate (CuSO4); accumulation of copper in the cytoplasm is expected to inhibit growth (27). Comparing the optical density at 450 nm during overnight growth in rich medium (lysogeny broth [LB]) without added CuSO4, all mutants grew at similar rates and reached comparable endpoint optical densities (ODs) (Fig. 2A). With the addition of 1 mM CuSO4, the copA golT double mutant and copA golT cueP triple mutant demonstrated a growth defect, reaching a final OD approximately 50% lower than that of any single mutant or the wild type (Fig. 2B). At 2 mM CuSO4, the copA golT and copA golT cueP mutants did not grow (Fig. 2C); increasing the concentration to 8 mM CuSO4 inhibited growth of all strains (Fig. 2D). This is in agreement with previous reports that copA or golT export systems are redundant in Salmonella (32). Metabolites in rich media are able to bind and sequester copper away from cells. We repeated the growth assay in M9 minimal medium and showed that even at 0.5 μM CuSO4, a copA golT mutant exhibited a growth defect (Fig. 3B). Interestingly, at 1 μM CuSO4, the copA and golT single mutants appear to have a slight growth defect compared to the cueP mutant and the wild type, whereas the double and triple mutants do not grow (Fig. 3C). At 4 μM CuSO4 all six strains were severely inhibited for growth (Fig. 3D). A cueP mutant in these assays has no detectable phenotype, implying that the growth defect is due to cytoplasmic accumulation of copper. These experiments demonstrate that either CopA or GolT is sufficient to protect the cytoplasm from copper stress. At increasing concentrations of copper, 8 mM in rich medium and 4 μM in minimal medium, even wild-type Salmonella demonstrates a significant growth defect.
FIG 2.
Growth curves of the wild type (WT) or mutants grown in increasing concentrations of copper sulfate (CuSO4). Overnight LB cultures were diluted to an OD of 0.01 in LB with 0.2% glucose and grown highly aerated at 37°C until reaching an OD of 0.2. Each strain was then inoculated 1:50 into a 96-well plate containing a total of 250 μl LB with the indicated concentration of CuSO4. The OD450 at 30-min intervals is plotted. The data are representative of three independent experiments. The strains used were 14028, JS2089, JS2087, JS2086, JS2091, and JS2092.
FIG 3.
Growth curves of the wild type or mutants grown in increasing concentrations of copper sulfate. Strains were grown overnight in LB, washed with M9 glucose minimal medium, diluted to an OD of 0.01 in M9 glucose, and grown highly aerated at 37°C until reaching an OD of 0.2. Each strain was then inoculated 1:50 into a 96-well plate containing a total of 250 μl M9 glucose with the indicated concentration of CuSO4. The OD450 at 30-min intervals is plotted. The data are representative of three independent experiments. The strains used were 14028, JS2089, JS2087, JS2086, JS2091, and JS2092.
Cytoplasmic copper detoxification and CueP can contribute to metalation of SodCI and SodCII.
Next we addressed the impact of the cytoplasmic copper detoxification system on the ability of SodCI and SodCII to acquire copper as a cofactor. Both enzymes are exported to the periplasm as unfolded polypeptides via the Sec pathway (11, 44). Presumably, both enzymes then acquire copper and zinc cofactors within the periplasm. Since copper is required for SodC activity, we can use enzyme activity as a proxy for metalation status. By adding exogenous copper, we can activate any apoenzyme to determine the total amount of enzyme present in each sample (6, 14). If CueP and CopA or GolT contribute to metalation of the SodC enzymes, we would expect that in the absence of these proteins, a reduced fraction of the total enzyme would be active before remetalation. We used the xanthine oxidase and cytochrome c method to assay superoxide dismutase activity (45).
To simplify the experiment, the SOD assays were done in a strain deleted for the two cytoplasmic superoxide dismutase genes, sodA and sodB, along with both sodCI and sodCII. The SOD gene of interest was then cloned into the expression vector pWSK29 (46) and transformed into the SOD-null strain, resulting in an ∼10-fold increase in SodC activity (44). Overexpression of SOD should exacerbate the potential need for CueP and cytoplasmic copper export to metalate the enzyme, while also facilitating the detection of differences in SOD activity.
We first attempted to confirm previously published results suggesting that CueP contributes to SodCII activity by functioning as a copper chaperone (40). SodCII was isolated by osmotic shock in these experiments (6, 47). Osmotic shock partially disrupts the outer membrane, resulting in release of SodCII and other periplasmic proteins. Initially, we compared the activity of SodCII in a wild-type background to that of SodCII in a copA golT cueP triple mutant background. Data are presented as the fraction of active SOD in any given extract out of the total amount of SOD, as determined by remetalating any apoenzyme by addition of exogenous copper. Periplasmic extracts from the control strain, containing an empty vector, did not show any detectable SOD activity in a wild-type or triple mutant background (data not shown).
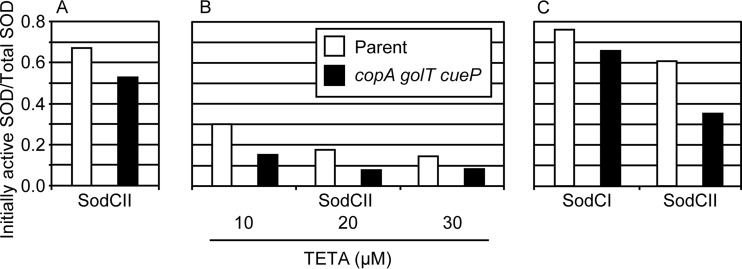
The data presented in Fig. 4A are representative of three independent experiments. Consistent with previous data (6, 14), only a fraction of SodCII is metalated. This fraction of active SodCII enzyme was consistently reduced in a triple mutant background compared to in the background with the export system intact. While it is clear from our data that cytoplasmic copper export and CueP can contribute to activation of SodCII, we did note variability in both the fraction of SodCII that was initially active and the relative contribution of the copper export pathway to activation of the enzyme. The SodCII activity was reduced 20 to 50% in the export mutant background compared to the parent strain background across all experiments. To try to further exacerbate the phenotype, we added triethylenetetramine (TETA), a copper-specific chelator, to the growth medium. We anticipated that the contribution of CueP-mediated copper delivery to SodCII might increase as the concentration of available copper became more limited. We compared the fractions of initial SodCII activity in medium containing increasing concentrations of TETA (Fig. 4B). As the concentration of TETA increased, the initial fractions of active SOD in the wild-type and triple mutant backgrounds both decreased. Surprisingly, the chelator did not enhance the importance of CueP, CopA, and GolT. In each case, the mutant background had approximately half the activity of the wild-type background.
FIG 4.
CueP can contribute to activation of SodCI and SodCII. SOD assays were performed using the xanthine oxidase-cytochrome c method. Cultures were grown overnight in 10 ml LB with TETA when indicated. Strains were devoid of all native SOD and expressed only the listed SodC from a pWSK29 vector. The osmotic shockates (A and B) or whole-cell extracts (C) were remetalated with 45 μM CuSO4. Data shown are the initial SOD activity divided by the SOD activity after remetalation and are representative of three independent experiments. The strains used were JS2093 and JS2094 containing either pMR101 or pMR102.
Determining the metalation status of SodCI was more challenging because SodCI is not released by osmotic shock but remains tethered within the periplasm (10, 11). To assay SodCI, we used glass beads to mechanically disrupt cells to obtain whole-cell extracts. As a control to ensure that this method of isolation does not alter the metalation status, we also assayed SodCII using the same lysis protocol. In whole-cell extracts, we again saw that SodCII showed a reduced fraction of initial activity in a copA golT cueP mutant background compared to the wild-type background (Fig. 4C). Similar to our results for SodCII, SodCI was generally less active in a copA golT cueP background than in the wild-type background. A higher percentage of SodCI than of SodCII is initially active in both the wild-type and the triple mutant backgrounds, consistent with previous data demonstrating that SodCI has higher affinity for metal cofactors than SodCII (6, 14, 16). Overall, these data suggest that under certain conditions, the copper detoxification pathway can also contribute to activation of SodCI. However, it is apparent that SodCI and SodCII do not absolutely require CueP and cytoplasmic copper export for activation; we can activate the enzyme simply by adding copper to an extract. In addition, strains lacking copA, golT, and cueP still have significant fractions of SodCI and SodCII that are active, although we did consistently observe a decrease in activity in the absence of the copper systems.
CopA, GolT, and CueP are not required for SodCI or SodCII activity in vivo.
The results above suggest that, while not absolutely required, CueP can facilitate metalation of SodCI. We wanted to investigate if this supply pathway was important for SodCI activity during systemic infection. Previous work in our lab and others studying SodCI and SodCII found that only SodCI is important for Salmonella survival during systemic infection (6, 12). To determine the effect of copA, golT, or cueP mutations in vivo, we turned to the competition assay, a powerful tool to compare virulence of two strains. A sodCI mutant is 7- to 10-fold attenuated in a competition assay with the wild type (5). Mice were infected by the intraperitoneal route, bypassing intestinal invasion and directly testing the ability of each strain to survive systemically. Competition assays were done in 6- to 8-week-old BALB/c mice using an inoculum of ∼500 bacteria mixed in a 1:1 ratio.
If CueP-mediated SodCI activation is important for defense against phagocytic superoxide, we would expect mutants with mutations in cueP or copA and golT to be attenuated relative to the wild type in a competition assay. This set of competition assays also allowed us to address whether an inhibitory amount of copper stress exists in the Salmonella-containing vacuole. Neither a cueP mutant nor a copA golT double mutant had any virulence defect compared to the wild type (Table 1). These results suggest that a sufficient amount of SodCI is able to obtain copper during systemic infection independent of the cytoplasmic copper detoxification pathway and CueP. To further probe the system, we tested the triple mutant against the wild type. Similarly to the single and double mutants, the triple mutant competed equally with the wild type.
TABLE 1.
Competition assays of copper-trafficking mutants
| Relevant genotype of S. Typhimurium straina: |
Mouse strainb | Median CI | No. of mice | P valuec | Fold attenuationd | |
|---|---|---|---|---|---|---|
| A | B | |||||
| ΔcueP | WT | BALB/c | 1.16 | 4 | NS | — |
| ΔcopA ΔgolT | WT | BALB/c | 1.00 | 8 | NS | — |
| ΔcueP ΔcopA ΔgolT | WT | BALB/c | 3.14 | 4 | NS | — |
| ΔcueP ΔcopA ΔgolT | WT | C3H | 2.40 | 6 | NS | — |
| ΔcueP ΔcopA ΔgolT | WT | C57 | 4.30 | 5 | NS | — |
The strains used were 14028, JS2089, JS2090, and JS2092.
Mouse genetic background: BALB/c, BALB/cAnNHsd; C3H, C3H/HeNHsd; C57, C57BL/6J.
By Student's t test comparing CI versus inoculum. NS, not significant.
Reciprocal of median CI, if significant. —, CI not significant.
Different mouse genotypes are known to have different susceptibilities to Salmonella infection (48). We repeated the competition with the copA golT cueP triple mutant versus the wild type in C3H (NRAMP1/Slc11a1 wild-type) and C57BL/6J mice. The results were similar to those of the BALB/c competition; the triple mutant was equally virulent as the wild type. From these data, we conclude that SodCI is able to acquire sufficient copper to defend against the oxidative burst independent of CueP, CopA, and GolT. The in vitro growth assays demonstrate that a copA golT mutant has a growth defect relative to the wild type in the presence of 1 mM copper in rich medium and 0.5 μM in minimal glucose medium. Based on these results, we can conclude that Salmonella does not encounter high enough concentrations of copper in the SCV for a copA golT mutant to be attenuated.
To more directly test if SodCI enzymatic activity is reduced in the absence of CueP and cytoplasmic copper export, we determined the phenotype of a sodCI copA golT cueP strain competed against the copA golT cueP parent strain. The sodCI copA golT cueP strain was 8-fold attenuated, indistinguishable from the phenotype in a wild-type background (Table 2), showing that the enzyme has acquired sufficient copper.
TABLE 2.
Competition assays of ΔsodCI in copper-trafficking mutant backgrounds
| Relevant genotype of S. Typhimurium straina: |
Median CI | No. of mice | P valueb | Fold attenuationc | |
|---|---|---|---|---|---|
| A | B | ||||
| ΔsodCI ΔcueP | ΔcueP | 0.17 | 7 | 0.001 | 6.0 |
| ΔsodCI ΔcopA ΔgolT | ΔcopA ΔgolT | 0.23 | 4 | 0.034 | 4.4 |
| ΔsodCI ΔcopA ΔgolT ΔcueP | ΔcopA ΔgolT ΔcueP | 0.13d | 7 | 0.005 | 7.7 |
| ΔsodCI | WT | 0.15d | 4 | <0.005 | 6.7 |
The strains used were 14028, JS2096, JS192, JS2095, JS2091, JS2097, and JS2092.
By Student's t test comparing CI versus inoculum.
Reciprocal of median CI, if significant.
Not significantly different from one another; P = 0.47.
The data above suggest that Salmonella has sufficient SodCI activity during systemic infection independent of cytoplasmic copper export. SodCI is reported to have higher affinity for metal cofactors than SodCII (Fig. 4C) (5, 10, 13). We have previously generated a hybrid SodCII protein that is protease resistant but still released by osmotic shock. Protease-resistant SodCII can complement SodCI in vivo yet has metal affinities similar to those of wild-type SodCII (14). Starting with a strain background that has deletions of sodCI and sodCII, the protease-resistant sodCII hybrid gene was integrated into the sodCII locus as described previously (14). The strain expressing the protease-resistant SodCII was approximately 5-fold more virulent than the sodCI sodCII double mutant strain, confirming the ability of this SodC to defend against phagocytic superoxide. A cueP mutant in the protease-resistant SodCII background was equally competitive in a competition assay against the SodCII protease-resistant strain (Table 3). This confirms that during systemic infection, CueP is not required to activate sufficient protease-resistant SodCII to functionally defend against phagocytic superoxide.
TABLE 3.
Competition assays addressing the importance of CueP in defense against oxidative stress
| Relevant genotype of S. Typhimurium straina: |
Median CI | No. of mice | P valueb | Fold attenuationc | |
|---|---|---|---|---|---|
| A | B | ||||
| ΔsodCI ΔsodCII | sodCIIres+ ΔsodCI ΔsodCII | 0.21 | 3 | 0.0085 | 4.8 |
| ΔcueP sodCIIres+ | sodCIIres+ | 0.86 | 3 | NS | — |
| ΔcueP ΔsodCI ΔsodCII | ΔsodCI ΔsodCII | 0.64 | 5 | NS | — |
| ΔdsbC ΔdsbG | WT | 1.60 | 5 | NS | — |
| ΔsodCI ΔdsbC ΔdsbG | ΔdsbC ΔdsbG | 0.14 | 5 | <0.0005 | 7.1 |
The strains used were 14028, JS2100, JS1176, JS2101, JS2102, JS2098, and JS2099.
By Student's t test comparing CI versus inoculum; NS, not significant.
Reciprocal of median CI, if significant. —, CI not significant.
We can also use the competition assay to address the claim that CueP can directly scavenge superoxide (36). If CueP alone can protect against superoxide during infection, we would expect a cueP sodC strain to be attenuated relative to a sodC strain. A cueP mutant in a sodCI sodCII background was equally virulent as the parent strain (Table 3). This suggests that CueP does not have a direct role in protecting Salmonella against phagocytic superoxide. If copper is causing periplasmic stress due to inappropriate disulfide bond formation (38), we would expect a dsbC dsbG double mutant to be attenuated relative to the wild type. We find that the double mutant competes equally with the wild type (Table 3). Moreover, if dsbC were required to maintain CueP activity, which is required to activate SodC, we would expect a sodCI mutant in a dsbC dsbG background to compete equally with a dsbC dsbG strain. However, we see that a sodCI dsbC dsbG strain is 7-fold attenuated relative to the parent, the same phenotype we see when SodCI is fully functional (Table 3).
DISCUSSION
We addressed the role of CueP and cytoplasmic copper export in Salmonella survival during systemic infection. First, we asked if cytoplasmic copper export and CueP-mediated copper delivery are required to activate sufficient SodC to defend Salmonella against the oxidative burst. Second, we determined if Salmonella experiences sufficient copper stress in the phagosome to cause growth inhibition and affect virulence.
Our in vitro data suggest that the copper detoxification pathway in Salmonella can contribute to metalation of both SodCI and SodCII. The phenotype is apparent only under conditions when copper is limited in the medium. If we grow the cultures with addition of exogenous copper, we no longer see any effect of CueP in SodC activation (data not shown). The ability to activate the enzyme in cell extracts simply by providing exogenous copper, even in strains lacking cueP, confirms that CueP-mediated delivery is not absolutely required for activation.
Our competition assay results show that sufficient SodC is active to defend Salmonella against phagocytic superoxide in strains lacking copA, golT, and cueP. This result is consistent across multiple mouse genotypes. Based on previous work, we can infer that far more SodC is produced than is necessary to defend Salmonella against superoxide generated within the phagosome (14). This implies that even a reduced fraction of active SodC in a copA golT cueP mutant background can be sufficient to protect Salmonella against phagocytic superoxide. Our assays are not sensitive enough to determine if the fraction of active SodC is also reduced in vivo in a copA golT cueP mutant. Our data also do not support CueP having any significant contribution to direct scavenging of reactive oxygen species (36). A cueP mutant competes equally with the wild type in a competition assay. In agreement, we do not see a phenotype for a dsbC mutant, and DsbC is reportedly necessary to keep CueP in a reduced state (39).
Copper has long been used as an antimicrobial agent (49), and many bacterial pathogens can be inhibited in vitro by addition of exogenous copper. The immune system has apparently evolved to take advantage of the toxic effects of copper. Notably, the ATP7A copper transporter is trafficked to the phagosome in E. coli-infected macrophages, increasing the concentration of copper in the vacuole (41, 42). Thus, bacterial copper efflux systems are often important for pathogenesis. For examples, in Neisseria gonorrhoeae, a copA mutation decreases intracellular survival in human cervical epithelial cells (50). Streptococcus pneumoniae copA mutants have decreased virulence in pulmonary, intravenous, and intraperitoneal infection models (51), while a copper efflux mutant of Mycobacterium tuberculosis shows decreased survival in guinea pigs (52).
Results on the role of copper in Salmonella Typhimurium virulence have been somewhat variable. Previous experiments in Salmonella Typhimurium have identified a phenotype for a copA golT double mutant in RAW264.7 cells (32). Similarly, Ladomersky et al. (42) reported that activated mouse peritoneal macrophages can kill S. Typhimurium and that this killing activity is partially dependent on ATP7A. Moreover, a copA golT mutant was more sensitive to macrophage killing, and this increased sensitivity was also dependent on ATP7A (42). In their study, a copA golT mutant of S. Typhimurium strain SL1344 was ∼2-fold attenuated in an i.p. competition assay in C57BL/6J mice but equally virulent in mice lacking ATP7A in myeloid cells. In contrast, Osman et al. (32) reported no difference in bacterial CFU between SL1344 and the copA golT mutant in tissues of orally infected C57BL/6J mice. We also observed no phenotype conferred by deletion of copA, golT, and cueP in S. Typhimurium strain 14028 in i.p. competition assays in BALB/c, C57BL/6J, or C3H/HeN mice.
Our data suggest that the level of copper exposure in the SCV does not exceed amounts necessary to cause significant cytoplasmic stress. However, copper could still be playing an important role during Salmonella infection, potentially explaining the subtle differences observed in the various studies described above. There could be differences in the relative role of copper in different immune cells. For example, previous work found that neutrophils and inflammatory monocytes are able to kill Salmonella in vivo and that within different subsets of macrophages, survival can be variable (53). The Salmonella organisms that we recovered from infected mice are from those cells in which Salmonella survived. Our data suggest that in this case, copper toxicity does not significantly impact the results. However, in cells where Salmonella is effectively controlled, copper may have a more significant contribution. This could explain the phenotypes observed in in vitro activated mouse-derived macrophages, which are efficient at killing Salmonella. Also, the Atp7alysMcre mouse model created by Ladomersky et al. (42) is missing ATP7A in all macrophages and monocytes, impacting a simple interpretation of the results.
In those macrophages where Salmonella does survive, alterations in vesicular trafficking caused by the SPI2 type three secretion system (TTSS) effectors could be reducing delivery of the ATP7A copper transporter to the SCV, analogous to the role of the TTSS effectors in altering delivery of reactive oxygen or nitrogen species to the SCV (54, 55). Whereas ATP7A colocalizes with E. coli-containing phagosomes (41, 42), infection of bone marrow-derived macrophages with Salmonella resulted in formation of distinct copper-containing intracellular vesicles, which were dependent on copper uptake by the cell (56). However, these vesicles were distinct from the SCV or lysosomes. Perhaps a more detailed analysis of specific types of immune cells or infection models will further clarify the role of copper in Salmonella infection.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains and plasmids are listed in Table 4. All Salmonella strains used in this study are derived from Salmonella enterica serovar Typhimurium strain 14028 (American Type Culture Collection). Lambda Red-mediated recombination was used for allelic replacement of genes with antibiotic resistance cassettes (57). Primers for lambda Red and any PCRs were purchased from IDT Inc. Endpoints for each deletion are listed in Table 4. The resulting constructs were moved into an otherwise wild-type background via P22 HT105/1 int-201-mediated transduction (58) and confirmed via PCR. The temperature-sensitive plasmid pCP20 carrying FLP recombinase (59) was used to remove antibiotic resistance cassettes containing FLP recombination target (FRT) sites when necessary.
TABLE 4.
Bacterial strains and plasmids
| Strain or plasmid | Genotypea | Deletion endpoints (bp)b | Source or referencec |
|---|---|---|---|
| Strain | |||
| 14028 | Wild type | ATCC | |
| JS192 | ΔsodCI-1::Aph | 9 | |
| JS1176 | ΔsodCII::sodCII1–133 sodCI136–157 (sodCIIres) ΔsodCI-108::Cm | 14 | |
| JS2085 | ΔdsbC105::Kn | 3222825–3223540 | |
| JS2086 | ΔgolT1::Kn | 399253–401541 | |
| JS2087 | ΔcopA1::Cm | 558639–561140 | |
| JS2088 | ΔdsbG106::Cm | 669962–670696 | |
| JS2089 | ΔcueP2::Cm | 3850516–3851055 | |
| JS2090 | ΔgolT1::Kn ΔcopA1::Cm | ||
| JS2091 | ΔgolT1 ΔcopA1 | ||
| JS2092 | ΔgolT1 ΔcopA1 ΔcueP2::Cm | ||
| JS2093 | ΔsodA101 ΔsodB102 ΔsodCI-105 ΔsodCII-103 | ||
| JS2094 | ΔsodA101 ΔsodB102 ΔsodCI-105 ΔsodCII-103 ΔcueP2 ΔgolT1 ΔcopA1::Kn | ||
| JS2095 | ΔgolT1 ΔcopA1 ΔsodCI-105::Cm | ||
| JS2096 | ΔcueP2::Cm ΔsodCI-1::Aph | ||
| JS2097 | ΔgolT1 ΔcopA1 ΔcueP2::Cm ΔsodCI-1::Aph | ||
| JS2098 | ΔdsbC105 ΔdsbG106::Cm | ||
| JS2099 | ΔdsbC105 ΔdsbG106::Cm ΔsodCI-1::Aph | ||
| JS2100 | ΔsodCI-105 ΔsodCII-103 | ||
| JS2101 | ΔcueP2::Kn ΔsodCII::sodCII1–133 sodCI136–157 (sodCIIres) ΔsodCI-108::Cm | ||
| JS2102 | ΔsodCI-105 ΔsodCII-103 ΔcueP2::Cm | ||
| Plasmids | |||
| pMR101 | pWSK29::sodCI+ | ||
| pMR102 | pWSK29::sodCII+ |
All Salmonella strains are isogenic derivatives of S. enterica serovar Typhimurium strain 14028. Subscript numbers in hybrid genes indicate the amino acids of the mature protein encoded.
Base pairs that are deleted (inclusive), defined by the S. Typhimurium 14028 genome sequence (NC_016856.1).
The source is this study unless otherwise specified. ATCC, American Type Culture Collection.
Growth conditions.
Bacteria were grown in lysogeny broth (LB) broth at 37°C with shaking for 16 to 18 h, with the exception of strains containing pCP20 or pKD46, which were grown at 30°C. As necessary, antibiotics were used at the following concentration: chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; and ampicillin, 50 μg/ml. For growth curves, 2-ml overnight cultures were grown in LB, diluted to an OD of 0.01 in 3 ml LB with 0.2% glucose or M9 minimal medium with 0.4% glucose, and grown with shaking at 37°C in a 50-ml baffled flask until reaching an OD of 0.2. Each strain was inoculated 1:50 into a 96-well plate containing 250 μl LB (no glucose) or M9 glucose with various concentrations of CuSO4. the OD at 450 nm (OD450) was recorded every 15 min for 16 h using an ELx808IU absorbance reader (BioTek), with the temperature set to 37°C and a 15-s mixing prior to each OD450 reading.
SOD assays.
Biochemical analysis of SOD enzymes was done using periplasmic extracts or whole-cell lysates. Periplasmic extracts were obtained by osmotic shock (14). To ensure that the measured SOD activity was specific to the SOD of interest, strains devoid of all four native SODs (SodA, SodB, SodCI, and SodCII) were transformed with pWKS29-derived plasmids encoding the specific SOD of interest or an empty vector. To study the impact of cytoplasmic copper export in this strain background, copA, golT, and cueP deletions were individually introduced via P22 transduction. Overnight 10-ml cultures were grown in LB with ampicillin. The copper-specific chelator triethylenetetramine (TETA) was added as indicated. Cultures were harvested via centrifugation at 4°C and then washed three times in an equal volume of cold 50 mM potassium phosphate buffer (pH 7.4). After the third wash, cells were resuspended in 5 ml of plasmolysis buffer (50 mM Tris, 2.5 mM EDTA, 20% [wt/vol] sucrose, pH 7.4) and allowed to sit at room temperature for 15 min before centrifugation for 5 min at 7,800 rpm. Cells were resuspended in 2.5 ml cold deionized water. After a 15-min incubation on ice, cells were recentrifuged, and the supernatant collected was considered the osmotic shockate. For experiments requiring whole-cell extracts, overnight cultures were washed twice in 50 mM potassium phosphate buffer (pH 7.4) and resuspended in 600 μl of the same buffer. Approximately 200 μl of glass disruptor beads (0.1 mm; USA Scientific) was added to the cells, and the mixture was placed in a Digital Disruptor Genie at 4°C. Cells were agitated at 3,000 rpm for 2 min, followed by a 2-min rest on ice. The cycle was repeated 3 additional times. The lysate was then centrifuged for 15 min at 4°C and 14,000 rpm. The supernatant was collected, transferred to a new 1.5-ml microcentrifuge, tube and spun a second time at 14,000 rpm. The resulting supernatant was considered the whole-cell extract.
Superoxide dismutase activity was determined using the xanthine oxidase-cytochrome c method (45). The protein concentration for osmotic shock or whole-cell lysates was determined using a Bradford assay (Bio-Rad). To determine the total amount of SOD present in a given extract, 45 μM copper sulfate was added to 100 μl of the initial extract and allowed to gently mix at room temperature for 30 min. The amount of SOD activity after remetalation was considered the total SOD activity. Dividing the initial activity by the total activity gives the initial fraction of active SOD. We previously determined that addition of copper sulfate to extracts does not affect the SOD assay, and thus we did not require dialysis to remove excess copper.
Competition assays.
All animal work was reviewed and approved by the University of Illinois IACUC and performed under protocol 15214. Competition experiments were done in female 6- to 8-week-old mice. BALB/cAnNHsd and C3H/HeNHsd mice were purchased from Envigo (formerly Harlan), and C57BL/6J mice were purchased from The Jackson Laboratory. Strains for each competition were grown overnight in LB (16 h), mixed together in a 1:1 ratio, and then diluted to a target inoculum of ∼500 CFU in 200 μl sterile 0.15 M NaCl. Infections were done via the intraperitoneal route. An aliquot of each inoculum was spread on LB agar to enumerate the total inoculum and replica plated to the appropriate selective medium to calculate the input ratio for each competition. At 4 to 5 days postinfection, animals were sacrificed, and their spleens were removed and homogenized. Dilutions of spleen homogenate were plated on LB and replica plated to appropriate selective medium to calculate the output ratio for each competition. The competitive index (CI) was calculated as (percent strain A recovered/percent strain B recovered)/(percent strain A inoculated/percent strain B inoculated). Statistical comparison of individual competitions was done using the Student t test.
ACKNOWLEDGMENTS
This study was supported by NIH grants AI111455 and AI123381 to J.M.S.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank James Imlay for helpful discussions.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol 11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holden DW. 2002. Trafficking of the Salmonella vacuole in macrophages. Traffic 3:161–169. doi: 10.1034/j.1600-0854.2002.030301.x. [DOI] [PubMed] [Google Scholar]
- 4.Kuhle V, Hensel M. 2004. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci 61:2812–2826. doi: 10.1007/s00018-004-4248-z. [DOI] [PubMed] [Google Scholar]
- 5.Craig M, Slauch JM. 2009. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4:e4975. doi: 10.1371/journal.pone.0004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krishnakumar R, Craig M, Imlay JA, Slauch JM. 2004. Differences in enzymatic properties allow SodCI but not SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium strain 14028. J Bacteriol 186:5230–5238. doi: 10.1128/JB.186.16.5230-5238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golubeva YA, Slauch JM. 2006. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutase SodCI is a member of the PhoPQ regulon and is induced in macrophages. J Bacteriol 188:7853–7861. doi: 10.1128/JB.00706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pesce A, Battistoni A, Stroppolo ME, Polizio F, Nardini M, Kroll JS, Langford PR, O'Neill P, Sette M, Desideri A, Bolognesi M. 2000. Functional and crystallographic characterization of Salmonella typhimurium Cu,Zn superoxide dismutase coded by the sodCI virulence gene. J Mol Biol 302:465–478. doi: 10.1006/jmbi.2000.4074. [DOI] [PubMed] [Google Scholar]
- 9.Ho TD, Slauch JM. 2001. Characterization of grvA, an antivirulence gene on the Gifsy-2 phage in Salmonella enterica serovar Typhimurium. J Bacteriol 183:611–620. doi: 10.1128/JB.183.2.611-620.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farrant JL, Sansone A, Canvin JR, Pallen MJ, Langford PR, Wallis TS, Dougan G, Kroll JS. 1997. Bacterial copper- and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol Microbiol 25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 11.Fang FC, DeGroote MA, Foster JW, Baumler AJ, Ochsner U, Testerman T, Bearson S, Giard JC, Xu Y, Campbell G, Laessig T. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc Natl Acad Sci U S A 96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slauch JM. 2011. How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol 80:580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim B, Richards SM, Gunn JS, Slauch JM. 2010. Protecting from antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 192:2140–2149. doi: 10.1128/JB.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rushing MD, Slauch JM. 2011. Either periplasmic tethering or protease resistance is sufficient to allow a SodC to protect Salmonella enterica serovar Typhimurium from phagocytic superoxide. Mol Microbiol 82:952–963. doi: 10.1111/j.1365-2958.2011.07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tidhar A, Rushing MD, Kim B, Slauch JM. 2015. Periplasmic superoxide dismutase SodCI of Salmonella binds peptidoglycan to remain tethered within the periplasm. Mol Microbiol 97:832–843. doi: 10.1111/mmi.13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ammendola S, Pasquali P, Pacello F, Rotilio G, Castor M, Libby SJ, Figueroa-Bossi N, Bossi L, Fang FC, Battistoni A. 2008. Regulatory and structural differences in the Cu,Zn-superoxide dismutases of Salmonella enterica and their significance for virulence. J Biol Chem 283:13688–13699. doi: 10.1074/jbc.M710499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Festa RA, Thiele DJ. 2011. Copper: an essential metal in biology. Curr Biol 21:R877-83. doi: 10.1016/j.cub.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ladomersky E, Petris MJ. 2015. Copper tolerance and virulence in bacteria. Metallomics 7:957–964. doi: 10.1039/C4MT00327F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bull PC, Thomas GR, Rommens JM, Forbes JR, Cox DW. 1993. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet 5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 20.Tanzi RE, Petrukhin K, Chernov I, Pellequer JL, Wasco W, Ross B, Romano DM, Parano E, Pavone L, Brzustowicz LM, Devoto M, Peppercorn J, Bush AI, Sternlieb I, Pirastu M, Gusella JF, Evgrafow O, Penchaszadeh GK, Honig B, Edelman IS, Soares MB, Scheinberg IH, Gilliam TC. 1993. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet 5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 21.Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY. 2007. Function and regulation of human copper-transporting ATPases. Physiol Rev 87:1011–1046. doi: 10.1152/physrev.00004.2006. [DOI] [PubMed] [Google Scholar]
- 22.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A 97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunther MR, Hanna PM, Mason RP, Cohen MS. 1995. Hydroxyl radical formation from cuprous ion and hydrogen peroxide: a spin-trapping study. Arch Biochem Biophys 316:515–522. doi: 10.1006/abbi.1995.1068. [DOI] [PubMed] [Google Scholar]
- 24.Halliwell B, Gutteridge JM. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol 186:1–85. doi: 10.1016/0076-6879(90)86093-B. [DOI] [PubMed] [Google Scholar]
- 25.Saleha Banu B, Ishaq M, Danadevi K, Padmavathi P, Ahuja YR. 2004. DNA damage in leukocytes of mice treated with copper sulfate. Food Chem Toxicol 42:1931–1936. doi: 10.1016/j.fct.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espariz M, Checa SK, Audero ME, Pontel LB, Soncini FC. 2007. Dissecting the Salmonella response to copper. Microbiology 153:2989–2997. doi: 10.1099/mic.0.2007/006536-0. [DOI] [PubMed] [Google Scholar]
- 29.Achard ME, Tree JJ, Holden JA, Simpfendorfer KR, Wijburg OL, Strugnell RA, Schembri MA, Sweet MJ, Jennings MP, McEwan AG. 2010. The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infect Immun 78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontel LB, Soncini FC. 2009. Alternative periplasmic copper-resistance mechanisms in Gram negative bacteria. Mol Microbiol 73:212–225. doi: 10.1111/j.1365-2958.2009.06763.x. [DOI] [PubMed] [Google Scholar]
- 31.Checa SK, Espariz M, Audero ME, Botta PE, Spinelli SV, Soncini FC. 2007. Bacterial sensing of and resistance to gold salts. Mol Microbiol 63:1307–1318. doi: 10.1111/j.1365-2958.2007.05590.x. [DOI] [PubMed] [Google Scholar]
- 32.Osman D, Waldron KJ, Denton H, Taylor CM, Grant AJ, Mastroeni P, Robinson NJ, Cavet JS. 2010. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. J Biol Chem 285:25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 34.Chaturvedi KS, Henderson JP. 2014. Pathogenic adaptations to host-derived antibacterial copper. Front Cell Infect Microbiol 4:3. doi: 10.3389/fcimb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Craig M, Sadik AY, Golubeva YA, Tidhar A, Slauch JM. 2013. Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89:887–902. doi: 10.1111/mmi.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon BY, Yeom JH, Kim JS, Um SH, Jo I, Lee K, Kim YH, Ha NC. 2014. Direct ROS scavenging activity of CueP from Salmonella enterica serovar Typhimurium. Mol Cells 37:100–108. doi: 10.14348/molcells.2014.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. 2009. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol 7:215–225. doi: 10.1038/nrmicro2087. [DOI] [PubMed] [Google Scholar]
- 38.Hiniker A, Collet JF, Bardwell JC. 2005. Copper stress causes an in vivo requirement for the Escherichia coli disulfide isomerase DsbC. J Biol Chem 280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- 39.Yoon BY, Kim JS, Um SH, Jo I, Yoo JW, Lee K, Kim YH, Ha NC. 2014. Periplasmic disulfide isomerase DsbC is involved in the reduction of copper binding protein CueP from Salmonella enterica serovar Typhimurium. Biochem Biophys Res Commun 446:971–976. doi: 10.1016/j.bbrc.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 40.Osman D, Patterson CJ, Bailey K, Fisher K, Robinson NJ, Rigby SE, Cavet JS. 2013. The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol Microbiol 87:466–477. doi: 10.1111/mmi.12107. [DOI] [PubMed] [Google Scholar]
- 41.White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ladomersky E, Khan A, Shanbhag V, Cavet JS, Chan J, Weisman GA, Petris MJ. 26 June 2017. Host and pathogen copper-transporting P-type ATPases function antagonistically during Salmonella infection. Infect Immun doi: 10.1128/IAI.00351-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnakumar R, Kim B, Mollo EA, Imlay JA, Slauch JM. 2007. Structural properties of periplasmic SodCI that correlate with virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 189:4343–4352. doi: 10.1128/JB.00010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCord JM, Fridovich I. 1969. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem 244:6049–6055. [PubMed] [Google Scholar]
- 46.Wang RF, Kushner SR. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195–199. doi: 10.1016/0378-1119(91)90366-J. [DOI] [PubMed] [Google Scholar]
- 47.Imlay KR, Imlay JA. 1996. Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J Bacteriol 178:2564–2571. doi: 10.1128/jb.178.9.2564-2571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy MF, Malo D. 2002. Genetic regulation of host responses to Salmonella infection in mice. Genes Immun 3:381–393. doi: 10.1038/sj.gene.6363924. [DOI] [PubMed] [Google Scholar]
- 49.Borkow G, Gabbay J. 2005. Copper as a biocidal tool. Curr Med Chem 12:2163–2175. doi: 10.2174/0929867054637617. [DOI] [PubMed] [Google Scholar]
- 50.Djoko KY, Franiek JA, Edwards JL, Falsetta ML, Kidd SP, Potter AJ, Chen NH, Apicella MA, Jennings MP, McEwan AG. 2012. Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infect Immun 80:1065–1071. doi: 10.1128/IAI.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson MD, Kehl-Fie TE, Klein R, Kelly J, Burnham C, Mann B, Rosch JW. 2015. Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance. Infect Immun 83:1684–1694. doi: 10.1128/IAI.03015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolschendorf F, Ackart D, Shrestha TB, Hascall-Dove L, Nolan S, Lamichhane G, Wang Y, Bossmann SH, Basaraba RJ, Niederweis M. 2011. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 108:1621–1626. doi: 10.1073/pnas.1009261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burton NA, Schurmann N, Casse O, Steeb AK, Claudi B, Zankl J, Schmidt A, Bumann D. 2014. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15:72–83. doi: 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 54.Vazquez-Torres A, Xu Y, Jones-Carson J, Holden DW, Lucia SM, Dinauer MC, Mastroeni P, Fang FC. 2000. Salmonella pathogenicity island 2-dependent evasion of the phagocyte NADPH oxidase. Science 287:1655–1658. doi: 10.1126/science.287.5458.1655. [DOI] [PubMed] [Google Scholar]
- 55.Chakravortty D, Hansen-Wester I, Hensel M. 2002. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med 195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achard ME, Stafford SL, Bokil NJ, Chartres J, Bernhardt PV, Schembri MA, Sweet MJ, McEwan AG. 2012. Copper redistribution in murine macrophages in response to Salmonella infection. Biochem J 444:51–57. doi: 10.1042/BJ20112180. [DOI] [PubMed] [Google Scholar]
- 57.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maloy SR, Stewart VJ, Taylor RK. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 59.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]