Abstract
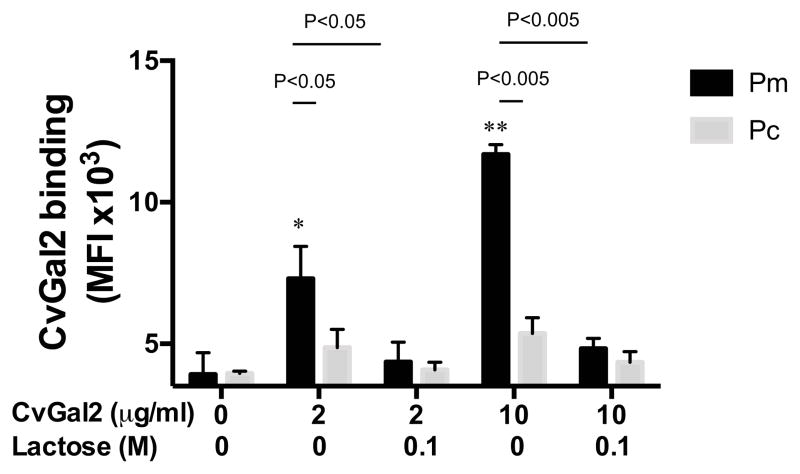
Galectins are highly conserved lectins that are key to multiple biological functions, including pathogen recognition and regulation of immune responses. We previously reported that CvGal1, a galectin expressed in phagocytic cells (hemocytes) of the eastern oyster (Crassostrea virginica), is “hijacked” by the parasite Perkinsus marinus to enter the host, where it causes systemic infection and death. A screening of an oyster hemocyte cDNA library revealed a novel galectin, which we designated CvGal2, with four tandemly arrayed carbohydrate recognition domains (CRDs). A phylogentic analysis of the CvGal2 CRDs suggests close relationships with homologous CRDs from CvGal1. A glycan array analysis, however, revealed that unlike CvGal1 that preferentially binds to the blood group A tetrasaccharide, CvGal2 recognizes both blood group A and B tetrasaccharides and related structures, suggesting that CvGal2 has broader binding specificity. Further, SPR analysis demonstrated significant differences in the binding kinetics of CvGal1 and CvGal2, and structural modeling revealed substantial differences in their interactions with the oligosaccharide ligands. CvGal2 is homogeneously distributed in the hemocyte cytoplasm, is released to the extracellular space, and binds to the hemocyte surface. CvGal2 binds to P. marinus trophozoites in a dose-dependent and β-galactoside-specific manner. Strikingly, negligible binding of CvGal2 was observed for P. chesapeaki, a sympatric parasite species mostly prevalent in the clams Mya arenaria and Macoma balthica. The differential recognition of Perkinsus species by the oyster galectins is consistent with their relative prevalence in oyster and clam species, and supports their role in facilitating parasite entry and infectivity in a host-preferential manner.
Graphical Abstract
Invertebrates display effective innate immunity for defense against microbial infection (1, 2). Potentially pathogenic viruses, bacteria, fungi and eukaryotic parasites are identified by a battery of soluble and cell-associated recognition factors, several of which have been structurally and functionally conserved along the lineages leading to the vertebrates (3, 4). Among them, a diversified lectin repertoire mediates the binding interactions with potential pathogens, resulting in agglutination, immobilization and opsonization, leading to phagocytosis or encapsulation (5, 6). However, a variety of microbial pathogens and parasites overcome the immune mechanisms of the host and establish successful infections that may lead to chronic or acute disease (7–10). Among these, the protozoan parasite Perkinsus marinus causes “Dermo” disease in the eastern oyster Crassostrea virginica and is responsible for catastrophic losses in both native and farmed oyster populations, with a significant impact on the integrity of the estuarine environment (8–12). Another Perkinsus species, P. chesapeaki (= P. andrewsi), which is sympatric with P. marinus along most of its distribution range, preferentially infects clams (13–16). Although P. chesapeaki can also infect the eastern oyster (17, 18) its pathogenicity for bivalves remains to be confirmed (19).
Perkinsus marinus trophozoites are phagocytosed by the oyster hemocytes present in various tissues exposed to the environment, such as the gills, gut, mantle, and palps (20, 21). In a previous study (22, 23) we identified in C. virginica a galectin of unique structure, which we designated CvGal [CvGal1 in a subsequent report (22, 23), and thereafter in this text] that facilitates parasite entry into the oyster hemocytes. Galectins are an evolutionarily conserved family of β-galactoside-binding lectins, members of which have been identified in most eukaryotic organisms, from fungi to mammals, the latter usually displaying a complex galectin repertoire (24, 25). By binding to endogenous carbohydrate moieties, galectins exert not only diverse regulatory effects on early developmental processes (26, 27) but also as a tight homeostatic control of both innate and adaptive immune responses, including acute and allergic inflammation (28), neutrophil adhesion and motility (29), macrophage activation (30) and development, activation, and apoptosis of B and T cells among others (31–33). In addition, by binding exogenous glycans on the surface of potentially pathogenic microbes, parasites, and fungi, galectins can function as pattern recognition receptors (PRRs) in innate immunity (6). This apparent paradox in recognition of “self” and “non-self” ligands underscores the significant gaps in our knowledge about the structural and biophysical aspects of the interactions of galectins with endogenous and microbial carbohydrate moieties (34).
CvGal1 is expressed in the oyster hemocytes and released to the extracellular environment where it recognizes carbohydrate moieties on both the hemocyte cells surface and P. marinus trophozoites, promoting their phagocytosis. The phagocytosed trophozoites survive intracellular killing and proliferate. As the infected hemocytes migrate into the internal milieu of the oyster, they provide not only the means of parasite uptake and entry, but also an environment favorable for parasite proliferation and dissemination (22, 35).
To further elucidate the structural and functional diversity of the galectin repertoire of the eastern oyster and its potential role(s) in parasite recognition and host entry, we screened the oyster cDNA library to identify the proteins containing potential carbohydrate recognition domains (CRD) that display the galectin sequence motif. This screening yielded a novel galectin, which we designated CvGal2 (C. virginica galectin 2). In this study, we analyzed the gene organization, and phylogenetic, structural, biochemical and functional aspects of CvGal2, including its binding to P. marinus and P. chesapeaki. The results concerning the CvGal2-mediated differential recognition of these two Perkinsus spp. suggest that the oyster galectins facilitate parasite entry and infectivity in a host-preferential manner.
EXPERIMENTAL PROCEDURES
Reagents
Carbohydrates (monosaccharides, oligosaccharides, and glycoproteins, at the highest purity available), Kanamycin, PMSF, lysozyme, and β-mercaptoethanol (2-ME) were purchased from Sigma-Aldrich (St. Louis, MO). Neoglycoproteins were obtained from Accurate Chemical & Scientific Corporation (Westbury, NY) [type 2 blood group A tetrasaccharide-BSA, 20 tetrasaccharide units/BSA polypeptide], V-labs Inc. (Covington, LA) [blood group A trisaccharide-BSA, 19 trisaccharide units/BSA polypeptide; and N-acetylgalactosamine-BSA, 28 GalNAc units/BSA polypeptide], or Sigma-Aldrich [GalNAc-BSA, 19 GalNAc units/BSA polypeptide]. Other neoglycoproteins (GlcNAc-BSA, 35 GlcNAc units/BSA polypeptide and Gal-BSA, 23 Gal units/BSA polypeptide) were a generous gift from Yuan Chuan Lee (Department of Biology, Johns Hopkins University, Baltimore, MD). The HRP-labeled goat anti-rabbit IgG was obtained from Bio-Rad (Hercules, CA). The Alexa 488-labeled goat anti-rabbit IgG was obtained from Invitrogen (Eugene, OR). Protein electrophoresis reagents were purchased from Bio-Rad (Hercules, CA). Restriction enzymes were acquired from New England Biolabs (Ipswich, MA). Revert Aid First Strand cDNA Synthesis Kit and DreamTaq PCR Master Mix were from Thermo Fisher Scientific (Pittsburgh PA). Protease inhibitor cocktail was purchased from CalBiochem (La Jolla, CA). DNase was purchase from Novagen (Billerica, MA).
Oyster hemocytes and tissues
Adult eastern oysters (C. virginica), averaging 40–50 g each, were obtained from a local commercial firm, their shells notched, and the animals acclimated in 20-liter tanks as reported elsewhere (22, 23). Hemolymph was drawn through the shell notch by puncture of the adductor muscle using 18-gauge needle, and the hemolymph was collected in plastic tubes kept on wet ice for the preparation of hemocyte suspensions as previously described (22, 23). Briefly, hemocytes from pooled hemolymph samples from 6 to 12 oysters, were collected by centrifugation (800 × g for 10 min at 4 °C), washed three times in ice cold HEPES-buffered saline (HBS; 10 mM HEPES, 430 mM NaCl, 10 mM CaCl2, pH 7.3), and used for protein or RNA extraction as described below, or suspended in HBS containing glutaraldehyde (10 mM), incubated on wet ice for 10 min, washed three times in HBS, and stored at 4 °C until use. For collection of pallial fluid and various tissues, oysters were shucked open, the pallial fluid was collected, and the suspended cells were separated by centrifugation at 3000 rpm for 5 min at 4°C. The soft parts of the oyster were dissected and the selected tissues (mantle, gill, palp, rectum, digestive glands, heart, muscle, pallial fluid) were collected on ice for the preparation of cell extracts and RNA extraction. The dissected tissues, pallial fluid cell pellet, and hemocytes freshly isolated as described above were lysed by sonication in PBS containing protease inhibitors cocktail (Calbiochem), and the clear supernatant was collected after centrifugation at 3000 rpm for 10 min at 4 °C. For isolation of total RNA the freshly isolated hemocytes, pallial fluid cell pellet, and the aforementioned tissues were extracted by using TRIzole reagent (Invitrogen) as previously described (36). RNA was quantified in a Nanodrop Bioanalyzer at 260/280 nm.
Parasite trophozoites
Clonal cultures of P. marinus and P. chesapeaki strains both established in our laboratory [P. marinus strain PRA 240 (37), P. chesapeaki (=P. andrewsi) (14)], and obtained from ATCC were propagated as reported earlier (38). Cultures in log phase of growth were harvested and fixed with PFA as previously described (22, 23) for staining and flow cytometry analysis.
Analysis of CvGal2 transcripts and genomic sequences
Primers for PCR amplification
Primers used for all PCR are shown in Table 1 and order from Sigma-Aldrich. Several primers were designed based on cDNA sequence of CvGal2, whereas the others were designed based on the clone sequences obtained by genomic PCR described below.
Table 1.
Primers used for CvGal2 genomic DNA analysis
| Primer | Sequence | mRNA position | DNA position | Mark* | Seg. |
|---|---|---|---|---|---|
| CA1-F | CGGTTCAAGTCAAAGGCGAT | 1296–1315 | Exon 10 | amp | 5th |
| CA3-R | CCCTTCTGATGGTCGTGGAT | 289–270 | Exon 4 | amp | 1st |
| CA4-F | CATTTCAACGTTCGGAGGGG | 1493–1512 | Exon 12 | amp | 6th |
| CA4-R | AAGTTGCAGTCTCCGTCCAT | 1737–1718 | Exon 13 | amp | 5th |
| CA5-F | CTCCGTGTGGTTTTCTTCGC | 24–43 | Exon 1 | amp | 1st |
| CA5-R | ATTCGTATATTATCATAGTGCAGC | 1922–1945 | Exon 13 | amp | 6th |
| CA7-F | TTGTAACACTCGCACTTGGG | Intron1 | seq | 1st | |
| CA7-R | TTTTCCTGCCTCTCTGATCC | 112–93 | Exon 2 | seq | 1st |
| CA8-F | TTCACCCGATGCATGGTAGT | Intron 3 | amp | 2nd | |
| CA9-F | ACAACCAGACATGCATGACT | Intron 3 | seq | 2nd | |
| CA9-R | ATGTTCCTGGCCGTATTCCT | 1381–1400 | Exon 11 | amp | 4th |
| CA10-F | CTCGCCATCTATCCACGTCT | Intron 1 | seq | 1st | |
| CA10-R | ACCTTCCGTTTTTCCTGCCT | 101–118 | Exon 2-Intron 2 | seq | 1st |
| CA11-F | GGGCGGGAAAGTACCTCATC | 468–487 | Exon 4 | amp | 3rd |
| CA11-R | CTAACCACGGTCCTCTGTGC | 702–683 | Exon 6 | amp | 2nd |
| CA12-F | GGCGAGGATGCAATGA | 938–953 | Exon 8 | amp | 4th |
| CA12-R | CCGCGGATTGAAGTGAAGA | 1094–1075 | Exon 9 | amp | 3rd |
| CA14-R | GATGTTGTTCCGGTTGTCATTG | 658–637 | Exon 6 | seq | 3rd |
| CA15-R | CGGATTTCCGCGGTGTGTAA | 849–868 | Exon 7 | seq | 3rd |
| CA16-R | TTTCAGTGGGTTTACCAAGG | Intron 4 | seq | 3rd |
amp: for segment amplification; seq: for sequence
Identification of CvGal2 transcripts
For RT-PCR, first-strand cDNA was synthesized from 1 μg of total RNA using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). First-strand cDNA for 3′- and 5′-RACE (rapid amplification of cDNA ends) was synthesized from 5 μg of hemocytes’ total RNA using GeneRacer kit (Invitrogen). Each PCR amplification described below was conducted using a MyCycler thermal cycler (Bio-Rad) in a total volume of 20 μl with 0.5 U of Ex-TaqDNA polymerase (Takara), 800 μM dNTP, and the forward and reverse primers (500 nM each) indicated in Table 1.
Genomic PCR
Genomic DNA was purified using DNeasy 96 Blood and Tissue kit (Qiagen) according to manufacturer’s protocol. PCR amplification was conducted using 100 ng of gemonic DNA in a total volume of 20 μl with forward and reverse primers (400 nM each) using DreamTaq PCR Master Mix (Thermo Scientific). At the first round, fragments 1–3 were amplified by PCR with primer sets of CA5F and CA3R, CA1F and CA2R, CA4F and CA5R, respectively. At the second round, the fragment 2 was amplified with primers CA8F and CA9R (Table 1).
DNA sequencing
PCR amplicons were isolated and purified with Gel Extraction kit (Qiagen), and cloned into pGEM-T vector using pGEM-T Easy Vector System (Promega). The positive clones were picked and plasmids were extracted with Mini-Prep Kit (Qiagen) for sequencing. DNA sequencing was conducted from both directions using dye termination reactions, and analyzed on an Applied Biosystems model 373 Stretch sequencer. Sequence assembly was performed manually. Calculations of theoretical molecular mass and pI from the deduced amino acid sequence were performed with ProtParam (www.Expasy.org).
Phylogenic analysis
Outgroup sequences for both 2- and 4-CRD galectins were selected from the bivalves Argopecten irradians and Pinctada fucata, and the gastropod snail Biomphalaria glabrata. Additional 2-CRD galectins were collected from Crassostrea gigas, and three other bivalve species. In the C. gigas genome several identical or nearly identical sequences were found. All of these sequences were aligned against the CvGal1 sequence and the two most complete sequences (EKC37204.1, EKC31760.1, both with 4 CRDs) were selected for further analysis. The CRDs from the tandem CRD galectin sequences were identified according to the domain predictions of blastp, and used to create single CRD alignments. Thus, for each gene the four (for 4-CRD genes) or two (for 2-CRD genes) CRDs formed individual alignment rows in the alignment and phylogenetic tree. Alignments were constructed using clustal Omega software with the –full-iter option (39). Phylogenetic analysis was performed using RAxML with the optimal JTT model with gamma site to site rate variation and 100 rapid bootstrap replicates (40).
Recombinant expression of CvGal2 and production of anti-CvGal2 antibodies
Construction of an expression vector for CvGal2
A DNA fragment encoding the complete CvGal2 ORF was amplified by PCR with 5′-CAT ATG CGG ATC AGA GAG GCA GG and 3′-CTC GAG TTA GAT AAC AAC TTC CGG TTC G. The amplicon was subcloned into pGEM-T vector, and a plasmid with the correct sequence was digested with NdeI and XhoI (both from New England Biolabs), purified after 1% agarose gel electrophoresis, and subcloned into pET30a(+) vector (Novagen).
Expression and purification of recombinant CvGal2 (rCvGal2)
The CvGal2 construct was transformed into E. coli Rosetta (DE3) pLysS competent cells (Novagen). The expression of rCvGal2 was induced by 0.1 mM isopropyl β-D-thiogalactoside at 23 °C for 16 h in 3 liters of LB medium containing 15 μg/ml kanamycin and 34 μg/ml chloramphenicol. The soluble proteins were extracted with BugBuster (Novagen) containing 1 mM PMSF, 1 mM protease inhibitor cocktail, 10 μg/ml of DNase, 10 μg/ml of lysozyme, and 0.07% of 2-ME, contained most of the recombinant CvGal2 (approximately 80%). This fraction was loaded onto a column packed with 4 ml of lactose-Sepharose (Sigma-Aldrich). After washing the column thoroughly with PBS containing 10 mM β-mercaptoethanol (PBS/2-ME), the rCvGal2 was eluted with 0.1 M lactose in PBS/2-ME.
Protein determinations
Protein concentrations were estimated with the Protein Assay kit I (Bio-Rad) in 96-well flat-bottom plates as described by the manufacturer in the microassay format, using crystalline BSA as a standard. After 5 min of combining protein and dye, the reactions were read at 595 nm on SpectraMax340 Plate Reader (Molecular Devices; Sunnyvale, CA) controlled by SoftmaxPro software, version 1.
Production of anti-rCvGal2 antibodies
Anti-CvGal2 antisera were raised in New Zealand White rabbits following Tasumi and Vasta (2007) (22). The IgG was purified on Protein A-Sepharose, followed by immuno-affinity purification of the specific Igs on the immobilized rCvGal2 as previously described (22). Cross-reactive antibodies were removed by passing the purified anti-CvGal1 and anti-CvGal2 antibodies through the immobilized rCvGal2 and rCvGal1 respectively, and the specific and unbound antibody fractions were collected and tested for specificity in Western blot.
Analysis of CvGal2 expression
RT-PCR amplification
First-strand cDNA synthesis and PCR amplification was conducted as described above. For detection of CvGal2 expression levels, CA3F and CA3R primers were used (Table 1); for β-actin, ACTIN_F and ACTIN_R primers were used. PCR products were separated in 1.5% agarose gels, and DNA bands were visualized by staining with ethidium bromide.
Western blot
Cells were lysed with ice cold 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% v/v Triton X, protease inhibitor (1:100), 0.1 mM PMSF. Concentration of proteins in lysates was determined by standard BCA assay (Bio-Rad). Equivalent levels of protein from different tissues of oyster were subjected to electrophoresis on 4–15% mini-protean TGX precast gels (Bio-Rad, #456-1084) and transferred onto PVDF membrane (PRC #88518). The membranes were blocked in 5% nonfat milk and probed with primary antibodies at 1:3000 dilutions. Next, membranes were washed in PBS-T followed by incubation with HRP-linked secondary rabbit polyclonal antibody (1:3000) and the bands were visualized with enhanced chemiluminescence (ECL) reagents following exposure of membranes to x-ray film (Kodak, Rochester, NY).
Immunofluorescence analysis of attached hemocytes
Two hundred microliters of hemolymph were added to each well of poly-D-lysine-coated eight-well chamber slides (Biocoat; BD Biosciences) and incubated at room temperature for 1 h. Each well was gently washed with HBS three times, and the attached cells were fixed with 200 μl of 2% PFA in HBS for 30 min. After washing with HBS for 5 min three times, half of the wells were treated with 200 μl of 0.05% Triton X-100 in HBS for 10 min at room temperature, whereas the remaining wells were incubated with buffer only. After washing all wells with HBS three times, blocking was conducted with 1% BSA in HBS for 1 h at room temperature. The fixed cells were stained with anti-CvGal2 (and anti-CvGal1for comparative purposes), or pre-immune rabbit IgG for 1 h at room temperature, followed by incubation with FITC-labeled anti-rabbit IgG for 1 h at room temperature. The slides were mounted with ProLong Antifade Kit (Invitrogen) for observation by fluorescence microscopy (Carl Zeiss Microscopy GmbH, Germany). The images were captured and analyzed with ZEN 2011 software (Carl Zeiss Microscopy GmbH).
Characterization of the carbohydrate specificity of CvGal2
Enzymatic treatment of glycoproteins
Porcine stomach mucin (PSM; 30 μg/ml, final concentration) was incubated with PNGase F (25 × 103 U/ml as final concentration in G7 reaction buffer, pH7.5), O-glycosidase (2 x106 U/ml in G7, pH7.5), α-N-acetylgalactosaminidase (103 U/ml in G7, pH7.5), α-L-fucosidase (1 U/ml in G6, pH5.5), β1,3-galactosidase (500 U/ml in G6, pH5.5), or buffer only (G6 or G7 as control) at 37 °C for 48 h. To prepare desialylated glycoproteins, fetuin, PSM, ovine submaxillary mucin (OSM), or bovine submaxillary mucin (BSM) (10 μg/ml in Tris buffer pH5.5) were incubated with neuraminidase (10 mU/ml as final concentration) at 37 °C overnight. The neuraminidase-treated glycoproteins were boiled for 10 min to deactivate the enzyme before use. The untreated (fetuin, PSM, OSM, and BSM), and desialylated (asialofetuin, asialo-PSM, asialo-OSM, and asialo-BSM) glycoproteins as well as glycosidase-treated hemocyte extracts were diluted in PBS and used for coating ninety six-well plates for binding and binding-inhibition assays, as described below.
Solid phase binding, binding-inhibition, and competitive inhibition assays
Solid phase lectin binding assays were carried out as described elsewhere (22, 41). Briefly, ninety six-well microtiter plates were coated with authentic untreated or enzyme-treated glycoproteins or neoglycoproteins (20 μg/ml in PBS) for 3 h at 37 °C. The plates were washed with 0.1% Tween in PBS (PBST) and blocked overnight with 3% BSA in PBS at 4 °C. The plates were washed 3 times with cold PBS/2-ME and stored at 4 °C. For binding studies, rCvGal2 (100 μl; 0.5 μg/ml in PBS/2-ME) were dispensed into the coated plates and incubated for 1 h at 4 °C. For binding-inhibition studies, rCvGal2 (0.2 μg/ml) was pre-incubated with serial dilutions of PSM, asialo-PSM, OSM, asialo-OSM, BSM, or asialo-BSM (1:3 serial dilutions starting at 10 μg/ml) for 1 h at 4 °C in PBS/2-ME. The rCvGal-inhibitor mixtures were delivered into the coated plates and incubated for 1 h at 4 °C. Controls were the substitutions of the glycan inhibitor solution by PBS, and substitution of purified lectin by PBS. After three washes with PBS/2-ME, the plates were fixed with 2% formaldehyde in PBS for 30 min at 37 °C. The wells were washed twice with 100 mM glycine in PBS and three times with PBST. Binding of rCvGal2 was detected by purified rabbit anti-CvGal2 IgG, followed by an HRP-conjugated anti-rabbit antibody. The plates were developed with TMB substrate for 5 min, and the reaction was stopped by adding 1 M HCl. Optical densities at 450 nm were measured on SpectraMax340 Plate Reader (Molecular Devices) controlled by SoftmaxPro software, version 1. All experiments were carried out in triplicate and repeated at least twice.
Glycan array analysis
Glycan array analysis was carried out at the Core H of the Consortium for Functional Glycomics (CFG) at Emory University, on the version 5.0 of the array printed with 611 glycans in replicates of 6. The rCvGal2 (1 mg/ml) was dialyzed against PBS/2-ME, and diluted to 20 and 200 μg/ml before adding onto array for analysis. The bound rCvGal2 was detected with rabbit anti-CvGal2 purified IgG, followed by a goat anti-rabbit IgG labeled with AlexaFluor 488 (Invitrogen).
Comparative homology modeling
The structure of CvGal2 was modeled using the toad (Bufo arenarum) ovary galectin-1 (BaGal1) as template (PDBid 1GAN; (42)) The four CRDs of CvGal2 (A: 1–144, B: 145–26, C:287–419, and D:420–556) showed sequence identities with 1GAN and CvGal1 in the lower twenties (Table 2). The initial automatic alignment between template and target CRDs was performed using the program Molecular Operating Enviroment (Moe v 2013.8001; Chemical Computing Group Inc) and after inspection was adjusted to maintain the galectin’s archetypical recognition of the galactoside, which using Bgal1 numbering are Trp69—Gal, (Asn62, Glu72)—O6-Gal, (Arg49, His45, Asn47)—O4-Gal, and (Arg49, Glu72)—O4- or O3-GlcNAc in the core type 1 and 2 respectively (Sup. Fig. 1A).
Table 2.
Pairwise percentage identity between the BaGal1 (template), CvGal1 and CvGal2 (target)
| BaGal1 | CvGal1 | CvGal2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chains | A | B | C | D | A | B | C | D | ||
|
|
|
|||||||||
| BaGal1 | 100 | 18.6 | 22.8 | 18.8 | 22.8 | 19.9 | 23.7 | 21.6 | 21.5 | |
| CvGAl1 | A | 20.1 | 100 | 33.1 | 32.6 | 37.5 | 45.2 | 34.1 | 23.7 | 29.2 |
| B | 23.1 | 31 | 100 | 31.2 | 30.9 | 30.1 | 40 | 22.3 | 29.2 | |
| C | 19.4 | 31 | 31.6 | 100 | 35.3 | 32.9 | 31.1 | 41 | 29.2 | |
| D | 23.1 | 35.2 | 30.9 | 34.8 | 100 | 30.8 | 27.4 | 25.9 | 38.9 | |
| CvGal2 | A | 21.6 | 45.5 | 32.4 | 34.8 | 33.1 | 100 | 27.4 | 23 | 27.8 |
| B | 23.9 | 31.7 | 39.7 | 30.4 | 27.2 | 25.3 | 100 | 25.9 | 28.5 | |
| C | 22.4 | 22.8 | 22.8 | 41.3 | 26.5 | 21.9 | 26.7 | 100 | 27.8 | |
| D | 23.1 | 29 | 30.9 | 30.4 | 41.2 | 27.4 | 30.4 | 28.8 | 100 | |
Initial structural models based on the alignment were generated with MOE homology modeling routines and manually adjusted afterwards by minimization to improve locally the interaction with the modeled ligands. An Amber12HT force field (Amber12 parametrization (43) 2012 and EHT (44) was used for all energy calculations in MOE. The antigens were built and optimized using the Glycam.org server with their glycam force field.
Surface plasmon resonance (SPR) measurements
Surface plasmon resonance measurements were carried on a Biacore T100 (GE Healthcare) instrument at 25 °C. rCvGal2 and neoglycoproteins (blood group A tetrasaccharide-BSA, blood group A trisaccharide-BSA, GalNAc-BSA, Gal-BSA and GlcNAc-BSA) were immobilized on individual CM5 chips in duplicate cells, until reaching 3000 response units by using amine coupling kit provided by the manufacturer. A reference channel was immobilized with ethanolamine. Binding analyses were performed by injecting a solution of analytes (PSM, asialofetuin, and oligosaccharides) over four cells at 2-fold increasing concentration in 10 mM HEPES, pH 7.4 containing 150 mM NaCl and 0.005% surfactant P20 at a flow rate of 20 μL/min for 2 min and allowed to dissociate for another 5 min. The surface was regenerated after each cycle by injecting 3M MgCl2 (aq.) solution for 3 min at a flow rate of 30 μL/min. Data were collected at the rate of 10 Hz. T-100 Biacore evaluation software was utilized to subtract the appropriate blank reference, and to fit the sensorgram globally by applying a 1:1 Langmuir model. Graphpad prism software was used to deduce EC50 values for analytes PSM and ASF. Mass transfer effects were checked by the tc values displayed by the T-100 biacore evaluation software. No significant mass transportation effects were observed.
Analysis of interactions between CvGal2, oyster hemocytes, and Perkinsus trophozoites
Enzymatic treatment of hemocyte extracts and intact hemocytes
Hemocyte extracts (100 μg/ml) were incubated with PNGase F (25 × 103 U/ml as final concentration in G7 reaction buffer, pH7.5), O-glycosidase (2 x106 U/ml in G7, pH7.5), α-N-acetylgalactosaminidase (103 U/ml in G7, pH7.5), α-L-fucosidase (1 U/ml in G6, pH5.5), β1,3-galactosidase (500 U/ml in G6, pH5.5), or buffer only (G6 or G7 as control) at 37 °C for 48 h. Fixed intact hemocytes were incubated with PNGase F (25×103 units/ml), O-glycosidase (2×106 units/ml), α-N-acetylgalactosaminidase (103 U/ml), or buffer only at 37 °C for 72 h.
Solid phase binding of CvGal2 to hemocyte extracts and competitive inhibition assays
Solid phase binding assays to examine binding of CvGal2 to hemocyte extracts were carried out as described above for glycoproteins, following Ahmed et al (2002)(41). Briefly, ninety six-well microtiter plates were coated with untreated or enzyme-treated hemocyte extracts (0.5 μg/ml in PBS) for 3 h at 37 °C. The plates were washed with PBST and blocked overnight with 3% BSA in PBS at 4 °C. The plates were washed 3 times with cold PBS/2-ME and stored at 4 °C. For binding studies, rCvGal2 (100 μl; 0.5 μg/ml in PBS/2-ME) was dispensed into the coated plates and incubated for 1 h at 4 °C. For competitive binding studies, the wells were incubated with either anti-A or anti-B blood group antibodies (100 μl; 0.5 μg/ml in PBS) for 1 h at 4 °C, and after three washes with PBS/2-ME, the rCvGal2 (100 μl; 0.5 μg/ml in PBS/2-ME) was dispensed and incubated as above. After three washes with PBS/2-ME, the plates were fixed with 2% formaldehyde in PBS for 30 min at 37 °C. The wells were washed twice with 100 mM glycine in PBS and three times with PBST. Binding of rCvGal2 was detected by purified rabbit anti-CvGal2 IgG, followed by an HRP-conjugated anti-rabbit antibody. The plates were developed with TMB substrate for 5 min, and the reaction was stopped by adding 1 M HCl. Optical densities at 450 nm were measured on SpectraMax340 Plate Reader (Molecular Devices) controlled by SoftmaxPro software, version 1. All experiments were carried out in triplicate and repeated at least twice.
Flow cytometry analysis of galectin binding to oyster hemocytes and Perkinsus spp
Fixed hemocytes or fixed P. marinus trophozoites were incubated with 3% BSA in HBS 1 h, and subsequently incubated with 0–100 μg/ml of rCvGal1 or rCvGal2 for 1 h, both at room temperature. The cells were washed three times, and stained with 100 μl of rabbit anti-CvGal1 or anti-CvGal2 IgG respectively for 1 h, followed by incubation with Alexa 488-labeled anti-rabbit IgG for 1 h, both at room temperature. After three washes in HBS, the stained cells were analyzed with Accuri C6 flow cytometer. For competition experiments, cells were pre-incubated with of 0–100 μg/ml of rCvGal1 or rCvGal2, anti-A, or anti-B antibody for 30 min before adding 2 μg/ml of rCvGal2 or rCvGal1 respectively, followed by antibody detection.
Statistical analyses
Numerical results were quantified using Image J software. Comparison of two groups was performed by Student’s t-test for the comparison of non-paired samples. All results with p<0.05 were considered statistically significant.
RESULTS AND DISCUSSION
The roles of galectins in the recognition of carbohydrate moieties on the surface of viruses, bacteria, and eukaryotic parasites as a first step of the innate immune response has been firmly established in recent years [Reviewed in (6)]. Recent studies have corroborated their roles not only in recognition of potential pathogens, but also their key functions as effector factors in microbial clearance from the internal milieu (45–48). However, some eukaryotic parasites have adapted to subvert the roles of lectins in recognition and defense, to facilitate their infectivity for the hosts (22, 23, 49) or vectors (50). Among these, the parasite P. marinus causes Dermo disease in the eastern oyster C. virginica, and has been responsible for the progressive decline of native oyster populations along the Gulf and Atlantic coast of North America, with a virtual collapse of the shellfisheries in certain areas such as the Chesapeake Bay, and associated detrimental effects on water quality and the integrity of the estuarine ecosystem (8–12). In recent years, not only the oyster/P. marinus system has generated significant interest due to the economical and environmental impact of Dermo disease, but it has also emerged as a very useful model for addressing the lectin-mediated mechanisms involved in host recognition and entry (51, 52). Further, this model system has also been useful for elucidating strategies for intracellular survival of biomedically relevant parasites (53, 54).
In previous studies we identified in the eastern oyster hemolymph a diverse lectin repertoire with prominent specificities for galactose and N-acetylated hexosamines (55, 56). Our further studies identified the galactose-binding lectin as a novel galectin with a unique domain organization and binding properties, that we initially designated CvGal (22) [CvGal1 in subsequent reports (23, 57)]. CvGal1 displays a unique four-CRD organization that showed stronger binding to galactosamine and N-acetylgalactosamine than to D-galactose, particularly blood group A oligosaccharides (22, 23, 57). Infectious challenge induces transient CvGal1 expression, translocation to the hemocyte periphery, secretion, and binding to the hemocyte glycocalyx, where it can function as a PRR. A significant fraction of the secreted CvGal1 remains free in extracellular space as a soluble protein in plasma, mucus, or pallial fluid, where it can function as soluble opsonin. We showed that CvGal1 recognizes microalgae, and proposed that by cross-linking of sugars on their glycocalyx with the hemocyte surface has a key role in feeding by mediating hemocyte uptake and intracellular digestion of the phytoplankton (22). CvGal1 also recognizes a wide variety of environmental bacteria, some of which are well-established pathogens, and therefore would play a key role in immune recognition and defense, by phagocytosis and intracellular oxidative killing of the potentially infectious microbes (22). Most notably, however, is that P. marinus trophozoites are strongly recognized by CvGal1, and the immune/feeding functions of CvGal1 are subverted to become a parasite receptor that facilitates passive host entry by phagocytosis (22). The observation that pre-exposure of the oyster hemocytes to a specific anti-CvGal1 antibody only partially reduced phagocytosis even at the highest antibody concentrations tested (22), was intriguing and led us to propose the existence of additional hemocyte receptor(s) for Perkinsus entry (22).
As mammals express a very diverse and complex galectin repertoire comprised by over a dozen distinct galectin species (5), we hypothesized that additional oyster galectins might also function as hemocyte receptors for P. marinus uptake. A screening of an oyster hemocyte cDNA library yielded only one distinct galectin transcript sequence, which we designated CvGal2. In this study, we proceeded to characterize the molecular, structural, and binding properties of CvGal2 with the goal of gaining further understanding of the functional aspects of the oyster’s galectins as hemocyte receptors for P. marinus infection.
Gene structure and CRD organization of CvGal2
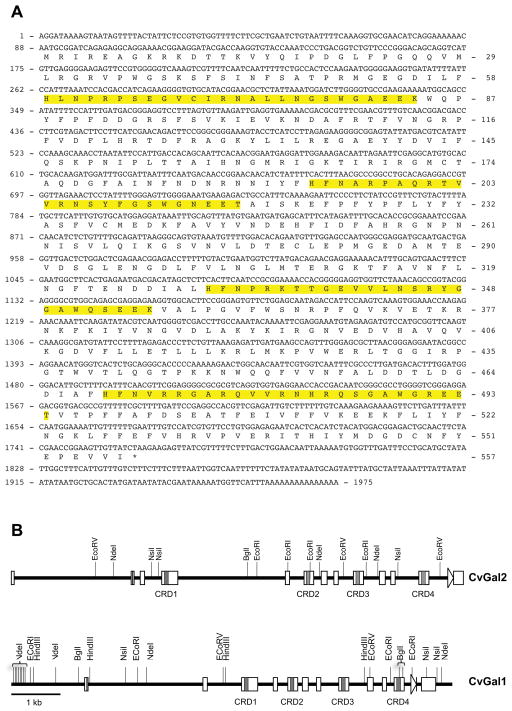
Using degenerate PCR primers, we identified in the eastern oyster a novel galectin cDNA sequence, which we named CvGal2, that spanned 1975 nt (Fig. 1A) containing a 1674 nt open reading frame. This sequence encoded a protein of 557 amino acid residues, that included 4 potential CRDs, with a calculated molecular mass of 64.1 kDa and a predicted isoelectric point of 8.11. This sequence was verified by amplifying the full-length coding region, which yielded a product of identical sequence, including the 5′- and 3′-UTRs, of 88 and 213 nt, respectively. To determine the structure of the gene that encode for CvGal2, we used a PCR amplification approach on genomic DNA with primers designed from the cDNA sequence, and that enabled the identification of the intron sequences (Table 1). The CvGal2 gene sequence (~9 kb), obtained by the alignment of multiple sequences of overlapping PCR products, is composed of 13 exons (lengths in downstream order: 58, 60, 89, 328, 86, 202, 120, 89, 205, 120, 89, 205, and 114 nt) separated by 12 introns (2.382, 0.125, 0.351, 2.189, 0.295, 0.153, 0.138, 0.308, 0.324, 0.115, 0.482, and 0.389 kbp), a structure similar to that of CvGal1 (Fig. 1B). None of the introns is present within the regions encoding the individual CRDs.
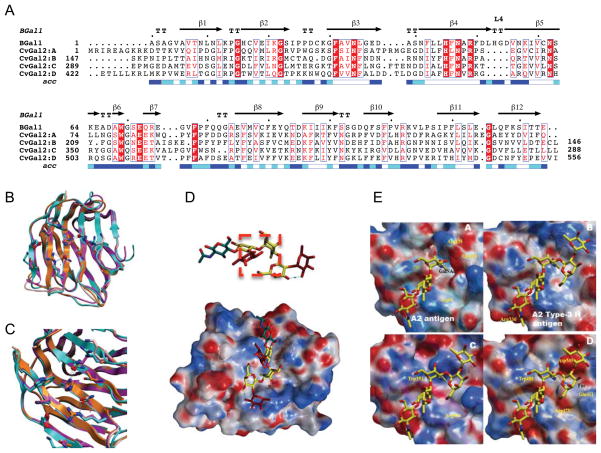
Figure 1. CvGal2 gene.
(A) Nucleotide sequence of cDNA and deduced amino acid sequence of CvGal2. Numbers on the left and the right indicate the nucleotide and amino acid positions, respectively. Asterisks mark the stop codon. The CRD domains are highlighted. (B) Genomic structure of CvGal2 and CvGal1 for comparison. Line indicates the position of start codon and arrow indicates the stop codon, with open boxes showing exons. Shaped area indicates the CRD regions listed below.
The oyster galectin CvGal2 carries four tandemly arrayed CRDs, which are similar in sequence, but not identical. Thus, CvGal2 shares a similar gene structure and CRD organization with CvGal1. The number and organization of the CRDs in the oyster galectins CvGal1 and CvGal2 represent a unique feature in the galectin family that suggests unique binding properties. Since the affinity of interactions between lectins and their carbohydrate ligands is relatively weak, the simultaneous binding of multiple CRDs to multivalent ligands can significantly increase the avidity of the interaction (58). In lectins carrying a single CRD, such as the mannose binding lectin, high avidity for the target is achieved by the association of peptide subunits into oligomeric structures in which multiple CRDs simultaneously interact with ligands (59, 60). The presence of tandemly-arrayed CRDs encoded within a single polypeptide such as in the oyster galectins would lead to the cooperative binding independent of oligomer formation, and may be critical for the recognition, agglutination, and immobilization of microbial pathogens (22, 61). Further, the binding to hemocyte glycans of the multiple CRDs in the oyster galectins would facilitate crosslinking and lattice formation at the hemocyte surface, which may trigger signaling pathways leading to phagocytosis and cell activation as described for other lectins (62).
Phylogenetic analysis of CvGal2
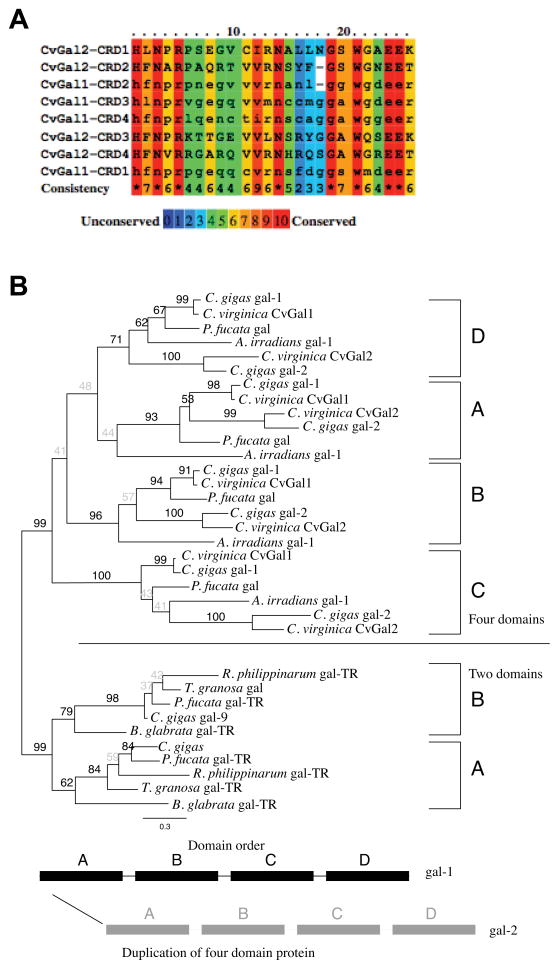
The CvGal2 gene encodes four tandemly-arrayed CRD structures, and thus is similarly organized as the gene encoding for CvGal1 (Fig. 1). Alignment of the CRD sequences from CvGal2 and CvGal1, revealed high conservation of the primary structure of domains responsible for ligand binding, particularly of the amino acid residues critical β-galactoside recognition (Fig. 2A). Therefore, we analyzed the phylogenic relationships of CvGal2 to CvGal1, and to all available galectin sequences from other bivalve mollusk species, using the galectins from the Atlantic bay scallop Argopecten irradians, the pearl oyster Pinctada fucata, and the planorbid freshwater gastropod Biomphalaria glabrata as the outgroups. The Pacific oyster C. gigas genome contains at least five duplicate partial galectin sequences randomly named, which converge to two genes (in this study named gal-1 and gal-2) with high identity to CvGal1 and CvGal2, respectively (Sup. Table 2). These two genes shared by the two Crassostrea species (C. virginica and C. gigas) each contained four tandemly repeated CRDs, here labeled A – D based on their order from the N-terminus to the C-terminus. Each of the domains formed a single monophyletic clade in the phylogeny with lowest bootstrap support for the first domain (A) (<60%), 71 % support for the fourth domain (D), and high support for domains B (96%) and C (100%) (Fig. 2B). The two C. gigas galectins, gal-1 and gal-2 were distinct so that within the clades corresponding to the individual CRD domains gal-1 and gal-2 from the two Crassostrea species formed two distinct well-supported groups (Fig. 2B). However, the individual domains from the two outgroup species, A. irradians and P. fucata, had different relationships within each domain to the C. gigas gal-1 and gal-2, albeit mostly with poor bootstrap support. Finally, there was a distinct division between the 2- and 4-CRD galectins with 99% bootstrap support. Within the 2-CRD galectins, again two distinct clades were formed, corresponding to the first and second domains with 62% and 79% bootstrap support, respectively. While the branching order of the species was not consistent between the first (A) and second (B) domains, B. glabrata was a consistent, well supported outgroup.
Figure 2. Phylogenetic analysis of CvGal2.
(A) Alignment of 4 CRDs of CvGal2 with those of CvGal1. The color codes for the amino acid conservation are illustrated below. (B) Phylogenetic analysis of the individual CRDs of CvGal2 with those of galectins from various mollusk species. Four-CRDs galectins include: C. virginica CvGal1 (ABG75998.1), C. gigas gal-1 (EKC37204.1), C. gigas gal-2 (EKC31760.1), Pinctada fucata gal (ACO36044.1), Argopecten irradians (ACS72240); two CRDs include: C. gigas gal-9 (EKC40501.1), P.fucata gal-TR (ADU60337.1), Ruditapes philippinarum gal-TR (ACA09732.1), Tegillarca granosa gal (AGH06131.1), Biomphalaria glabrata gal-TR (ABQ09359.1).
The distance-based tree was consistent with phylogenetic analysis of galectins reported elsewhere (22, 63, 64). However, the phylogenic analysis of the individual CRDs of CvGal2 and CvGal1, and galectins from other bivalve species, revealed that each CRD formed a single monophyletic clade. Thus, the results suggest that within the genus Crassostrea, the duplication of the 4-CRD galectins into gal-1 and gal-2 (or CvGal1 and CvGal2) likely occurred prior to speciation into C. gigas and C. virginica, because the gal-1 and gal-2 were consistently distinct in the CRD phylogeny. Likely, the most recent common ancestor of the two species contained the two 4-CRD genes and one 2-CRD gene. Furthermore, the 4-CRD and 2-CRD galectins do not appear to be the result of simple duplication or reduction. The reason is that between the two possible scenarios, one involving the duplication (or reduction) of an entire multidomain gene and the second involving duplication of individual domains within a gene, based on the phylogenetic analysis the former must be ruled out. In other words, under the first scenario a 2-CRD gene could be duplicated into a 4-CRD gene. However, the individual domain phylogeny shows the 4-CRD galectins are not interleaved with the 2-CRD galectins, so that the 2- and 4-CRD genes are distinct, and not the product of simple duplication (or reduction).
Tissue expression and distribution of CvGal2
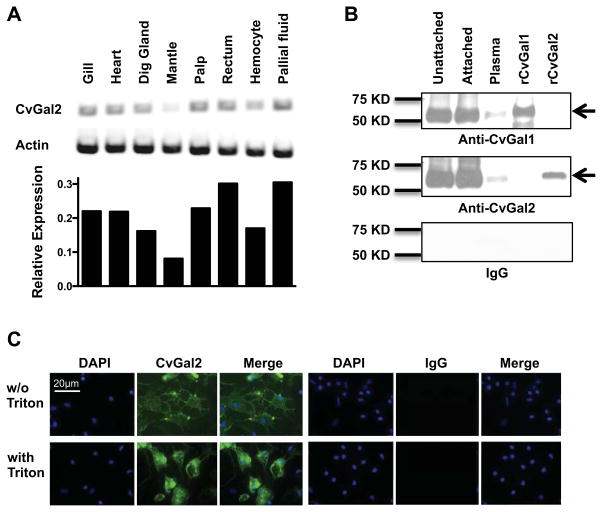
To examine the spatial distribution of CvGal2 transcripts in oyster tissues, we used RT-PCR with primers based on the CvGal2 cDNA sequence. CvGal2 transcripts were present in all tissues tested, which included gill, heart, digestive gland, mantle, palp, rectum, hemocytes, and cells present in the pallial fluid (Fig. 3A). To examine the presence and subcellular localization of the mature CvGal2 protein, we cloned and expressed the recombinant CvGal2 and raised an antiserum against the rCvGal2. Because the amino acid sequences of CvGal1 and CvGal2 are similar (albeit not identical), we further passed the anti-CvGal2 antibody through an rCvGal1 affinity column to absorb any potential cross-reactive antibodies against CvGal1 and reciprocally, the anti-CvGal1 antibody through an rCvGal2 affinity column. The specificity of these anti-rCvGal1 and anti-rCvGal2 antibodies was verified by Western blot, testing crude hemocyte extracts and plasma, with the purified rCvGal1 and rCvGal2 as specificity controls. Single bands of the expected electrophoretic mobility were detected in crude extracts from circulating unattached hemocytes, hemocytes that had attached to a plastic surface, and the supernatant plasma from the latter, as well as the purified rCvGal1 and rCvGal2 controls, respectively (Fig. 3B). No signal was observed in the pre-immune IgG controls (Fig. 3B). The results confirmed the specificity of the antibodies for the authentic CvGal1 and CvGal2, supported the RT-PCR results indicating that the CvGal2 is transcribed in the hemocyte, and revealed that CvGal2 is secreted into extracellular space. Subsequently, we examined by immunofluorescence the subcellular distribution of CvGal2 in attached hemocytes, by comparing the fluorescence signal distribution in intact cells with those that had been permeabilized by Triton-X100 (Fig. 3C). In addition to highly fluorescent sparse granular structures, the CvGal2 signal was detected on the plasma membrane of the intact attached hemocytes, although it was weaker than the mostly homogeneous stain observed in the cytoplasm of the permeabilized hemocytes (Fig. 3C). No fluorescence was observed in the pre-immune IgG controls (Fig. 3C).
Figure 3. Expression of CvGal2 in Eastern oyster tissues.
(A) mRNA expression of CvGal2 in different tissues of the eastern oyster was assessed by RT-PCR. Relative expression levels relative to actin are shown. (B) CvGal1 and CvGal2 protein in unattached hemocytes, attached hemocytes, or plasma was detected by Western blot. Recombinant CvGal1 (rCvGal1) and CvGal2 (rCvGal2) were included as specificity controls for the antibody, and a membrane exposed to pre-immune IgG is included as a control for IgG specificity. (C) Hemocytes were allowed to attach for 1 hour, and the presence of CvGal2 on cell surface (w/o Triton) or total (with Triton) was revealed by fluorescence microscopy after antibody staining. Images from cells stained with pre-immune IgG (IgG) are included as controls for IgG specificity. Scale bar: 20μm.
Although the CvGal2 transcripts were detected in all oyster tissues tested, these are abundant in either circulating or resident hemocytes. Thus, it is likely that as we observed for CvGal1 (22), the CvGal2 transcripts identified in these tissues actually originate in the infiltrating hemocytes. The P. marinus trophozoites enter the oyster through filter-feeding and initially become in contact with the gills, palps, gut or mantle, where they are phagocytosed by resident hemocytes (65) and survive intracellular oxidative stress by their effective anti-oxidative mechanisms (53, 54, 66, 67). The trophozoites proliferate in phagosome-like structures, eventually lysing the host hemocytes, and causing systemic infection and death of the oyster, which by releasing large numbers of parasites into the water column spreads the infection to neighboring oysters (20, 35). Thus, the expression of CvGal2, as well as CvGal1 (22, 23), is associated with the cells and tissues that have been proposed as the entry sites for P. marinus trophozoites (20, 35, 37). Further, although CvGal2 is abundant in the hemocyte cytoplasm, it is also secreted into the extracellular space and binds to the hemocyte membrane glycocalyx. Because CvGal2 lacks a signal peptide, its presence in the extracellular space suggests that the unconventional secretion mechanism reported for mammalian galectins may also be involved in CvGal2 secretion (68). In contrast with CvGal1, however, which was only expressed in about 30% of the hemocytes and is mostly associated with granular cells, CvGal2 is homogeneously distributed in all hemocytes examined, both granulocytes and hyalinocytes, suggesting a broader array of biological activities such as crosslinking of potential microbial pathogens, cell adhesion, and activation of hemocyte signaling pathways (69–72).
Glycan array analysis
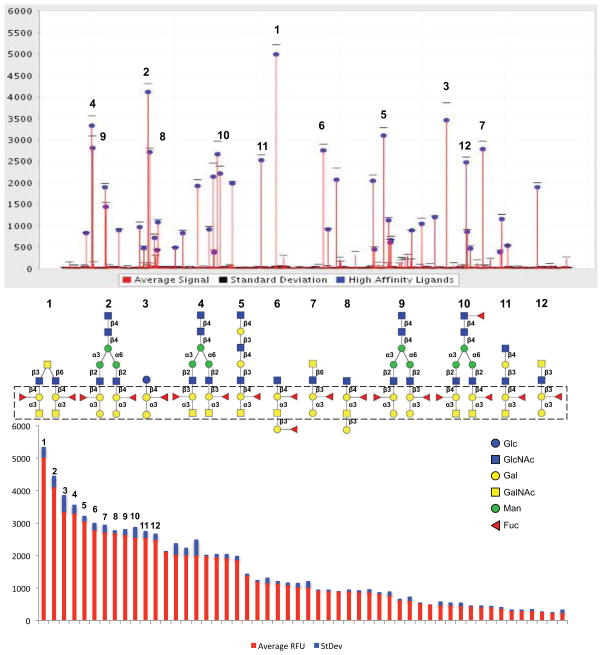
The glycan array binding profile of rCvGal2 (20 and 200 μg/ml) (http://www.functionalglycomics.org/glycomics/publicdata/home.jsp) revealed that CvGal2 recognizes with high affinity carbohydrates containing non-reducing terminal Gal and GalNAc (Fig. 4; Sup. Table 1). The strongest binder in the glycan array was the complex bi-antennary structure GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-6(GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-3)GalNAc (Fig. 4A, 1), followed by Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-2Manα1-6(Galα1-3(Fucα1-2)Galβ1-4GlcNAcβ1-2Manα1-3)Manβ1-4GlcNAcβ1-4GlcNAc (Fig. 4A, 2), Galα1-3(Fucα1-2)Galβ1-4Glcβ (Fig. 4A, 3), and 6 mono-antennary and 3 bi-antennary glycans with core structure (Gal/GalNAc)α1-3(Fucα1-2)Galβ1-4/3GlcNAc (Fig. 4A, 4–12). This suggested that the anomeric linkage β1-4 or β1-3 of the subterminal Gal to the GlcNAc affects the binding, and that CvGal2 prefers type-2 backbone structures Galβ1-4GlcNAc in N-acetyllactosamine and galactose. In addition, the Fuc linked in α1-2 to the subterminal Gal was present in all preferred ligands. Thus, rCvGal2 recognizes with high specificity array elements displaying blood group A/B type 2 tetrasaccharides, followed by the type 1 structures (Fig. 4). This is in contrast with the clear preference for blood group A over blood group B oligosaccharides we had observed in a similar analysis on CvGal1 (23, 57). As in CvGal1, however, the Fuc linked in α1-2 to the subterminal Gal appeared to further strengthen the binding. Thus, unlike most described galectins that recognize oligosaccharides exhibiting galactosyl units at the non-reducing end, particularly N-acetyllactosamine moieties (45, 73–75), this study together with previous reports from our laboratory (22, 23, 57) reveal that the oyster galectins CvGal1 and CvGal2 display specificity for ABH blood groups. The sequence alignment of CvGal2 CRDs with those of vertebrates and bivalve species showed that two amino acid residues (His53 and Asp55), that interact with the nitrogen of the N-acetyl group, are missing in all four CvGal2 CRDs. In the C. elegans galectin Lec-6, the absence of these two residues confers distinct carbohydrate specificity relative to that of the mammalian prototype galectins (41). A few other galectins, from fungi to mammals, share similar binding properties for blood group oligosaccharides. The galectin CGL2 from inky cap mushroom Coprinus cinereus, is specific for blood group A moieties (76, 77) whereas the mammalian galectins 2, 3, 4 and 8 can recognize A and B oligosaccharides (74, 75).
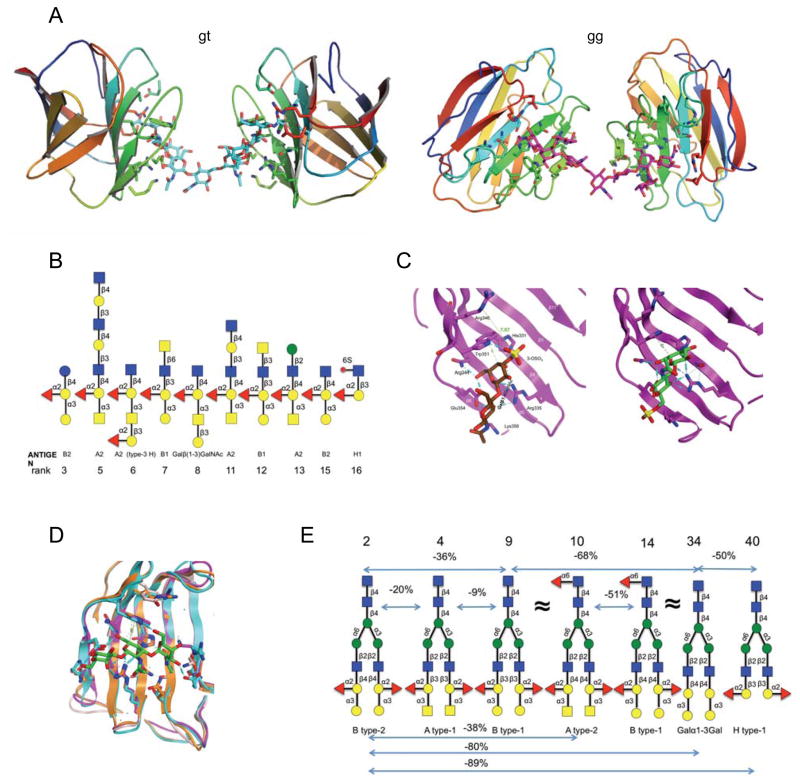
Figure 4. Glycan array analysis of CvGal2 specificity.
Recombinant CvGal2 was subjected to glycan array analysis (Consortium for Functional Glycomics). The fluorescent signals along the glycan array are shown with the top-ranked signals labeled (top panel), and rearranged by signal strength as high to low from left to right (bottom panel). The twelve best ranked glycan CvGal2 ligands are shown with their core structures boxed (middle panel).
Comparative modeling
To rationalize the structural basis for the binding properties of CvGal2 described above, we modeled its structure using the Bufo arenarum galectin-1 (BaGal1) as template [PDBid 1GAN; (42)] and analyzed the interactions of the CRDs with the carbohydrate structures identified in the glycan array as CvGal2 ligands. The four CRDs of CvGal2 (A: 1–144, B: 145–286, C:287–419, and D:420–556) show sequence identities with BaGal1 and CvGal1 in the lower twenties, ranging from 19% to 23% (Table 2). Large sequence variations, insertions and deletions, with respect to the template were accommodated in permissive regions of the template structure such as solvent exposed loops (Fig. 5A). Modeling the CvGal2 structure revealed interesting features in the quaternary structure of this galectin. The sequence alignment of CRDs of CvGal2 with those in the BaGal1 template revealed longer N-terminal regions of CRD A and D that may form an additional N-terminal β-strand, homologous to the first β-strand of Coprinus cinereus galectin-2 CGL2 (77). This extra strand places the N- and C-terminals of the CRD beside each other at one side of the β-sandwich opposite to the binding site. These additional strands could interfere with the typical homodimerization by an extended β-sheet observed in galectin-1. Instead, the intervening stretches between CRDs suggest a quaternary arrangement in which the A and D CRDs are displayed in the opposite side of B and C.
Figure 5. Models of individual CvGal2 CRDs.
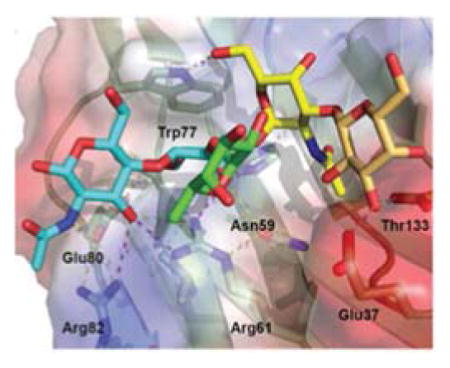
A) Alignment with toad galectin1 (BaGal1) as template [PDBid 1GAN; (42)]. B) Overlay of the four CRDs (CRD1: CRD2: CRD3: CRD4) based on the model. C) Residues of the glycoside recognition site and other in β3 and β11 with possible glycan interactions. D) Accessible surface of the CvGal2:A colored by electrostatic potential A2 type-3 antigen [Fuc α1-2Galβ1-3GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAc] in this galectins extended site. E) Electrostatic surface of the four CvGal2’s CRD models with a A2 antigen (CRD:A) and A2 type-3 antigen (CRDs:B-D) bound.
The model of the CRDs (A–D) also provided the structural basis of the carbohydrate specificity revealed by the glycan array and biochemical analyses. The modeling alignment between BaGal1 and the four CRDs of CvGal2 preserves the interactions that recognize the β-galactoside ligand: Trp69—Gal, (Asn62, Glu72)—O6-Gal, (Arg49, His45, Asn47)—O4-Gal, and (Arg49, Glu72)—O4- and O3-GlcNAc in the core type 1 and 2, respectively (Fig. 5B). This alignment identifies a shorter sequence between strands β4 and β5 that results in a smaller loop 4 compared to that in proto type galectins (ie. galectin-1) such as BaGal1 (42). In contrast, loop 3 (except in the B CRD) is longer than in BaGal1. At loop 4, a conserved histidine residue (His 53 in BaGal1, Fig. S1) makes an apolar contact with the C2 and O2 atoms of the disaccharide’s Gal moiety. This histidine residue –highly conserved in most galectin-1s– is missing in CvGal1 (23) and CvGal2 (Fig. 5A), and also in CGL2–a galectin of fungal origin (77). The main feature in the alignment between template and targets is that like CvGal1 (23) and CGL2 (77), CvGal2 shows a short loop 4. In addition β3 and β11 strands show specificity-relevant modifications (Fig. 5C).
Recognition of the α(1-2)fucose of H type 2 structures
The 2′-fucosyl moiety may loosely accommodate in the space left over by the shortening of loop 4 and only contacting with its 6-methyl group the aliphatic side of the galactoside-binding arginine. Although it is tempting to associate the small size of the loop 4 with the recognition of H antigens, human galectin-1 (hGal-1)–which have a long loop 4– recognizes galactosides better than α(1-2)fucosylated H antigens (Sup. Table 1). The docking simulation of an H-antigen in hGal-1 is shown in Fig. S1B. The required Fuc conformation (Fig. S1B) is not compatible with the α(1-3) linked pyranose of A and B oligosaccharides and explain the lack of affinity of hGal-1 for these moieties (see hGal-1 in Sup. Table 1). This short loop opens the site to easily accommodate simultaneously the 2′-fucosyl group and the α(1-3)Gal[NAc] of A and B oligosaccharides, with an apparent ‘gain’ of affinity with respect to hGal-1. hGal-1 –with a long loop 4– recognizes well H epitopes (Sup. Table 1) but with a conformation of the α(1-2)fucosyl not achievable by A or B antigens. Loop 4 in the conformation observed in galectin-1 (galectin-3 also has a long loop, but in a different conformation) selects against A and B moieties (hGal-1 in Sup. Table 1 and Fig. S1B). No direct polar interactions with protein groups are predicted with the Fuc (Fig. 5D). Water-mediated interactions with Fuc can explain the observed high affinity for the H containing groups observed (Sup. Table 1).
Recognition of the α(1,3)galactose of blood-group A and B antigens
The 6-OH of the α(1,3) linked galactose may H-bond the 5-NH group of the binding site tryptophan in CvGal2 (Trp69 equivalent in BaGal1). The tryptophan 5-NH at the galactoside binding-site –A:79, B:212, C:351, and D:489 in the respective CRDs– H-bonds to the hydroxymethyl OH group of the α3-linked pyranose in a gt (O6 gauge to O5 and trans to C4) conformation. The 2-NAc group of the α3-linked GalNAc of A oligosaccharides docks in an hydrophobic pocket at the strand β3 – on top of A:Ser48, B:Ala180, C:Ala315, and D:Val453 (Fig. 5E). A similar coordination was observed in the complex of CGL2 with an A2 oligosaccharide (PDBid: 1UFL) (77). Two strategically placed small residues on the N-terminus of β3 (Fig. 5B) —[G/A/S/T]F[A/S]hN (h stands for hydrophobic) — open a pocket to docks the methyl-group of the 2-NAc moiety. Variations in sequences, like in CvGal2:D that shows bulkier residues (Q; and a V), may disrupt this recognition.
Recognition of bi-antennary glycans, sulfated moieties, and additional carbohydrate units at the reducing or non-reducing ends
CvGal2 strongly recognizes –up to order of magnitude higher than the corresponding core glycoside– epitopes displaying blood group A and B antigens in the glycan array (Sup. Table 1), which closely agrees with the predicted model. Due to the multivalency associated with this 4-CRD lectin, several bi-antennary glycans appear in the first ten ranked epitopes (Fig. 4). Figure 6A shows how two CRDs –from the same or different CvGal2 molecules– may recognize any of the two lowest energy structures (gt and gg) of the O-GalNAc extended core 2 ranked first for the glycan array (Fig. 4). CvGal2 binds core type-1 –Galβ(1,3)GlcNAc– and core type-2 –Galβ(1-4)GlcNAc. Despite the predominance of core 2 epitopes in the array (48.5% of the array glycans) a significant number (seven) of core 1 structures (only 12% of all glycans in the array), are among the first twenty ligands ranked, suggesting a preference for core 1 (p-value for a random selection = 0.0005) (Fig. 6B).
Figure 6. Model of glycan recognition by CvGal2.
A) Multiple recognition of the first ranked bi-antennary glycan (gt and gi: two lowest energy structures of the O-GalNAc extended core 2 recogination by two CRDs. B) Glycans with common core structure on the strong binding list. C) Sulfation of 3-OH (c1) or 6-OH (c2) group of non-reducing end galactose. D) Model of the recognition of A type-4 epitope by CvGal’s CRDS. Ligth blue: Orange: Magenta and pink. E) Selected biantennary N-Glycans of complex-type. CvGal2 recognizes A and B of type 1 and 2 epitopes. The negative percentages are the evaluation of the % change in the fluorescent signal ((Fj−Fi)/Fi×100%). α(1-3)galatosylation and α(1-2) fucosylation provides apparent ten- and five-fold affinity enhancement, respectively.
CvGal2 recognizes sulfated core glycosides 20-fold stronger than non-sulfated ones (Sup. Table 1). Sulfation of 3-OH at the non-reducing end of core 2 or of 6-OH at the reducing-end of core 1 galactoside is recognized for the CvGal2 (Sup. Table 1). Our model suggest that 3-OH sulfation at the galactose is recognized by the NH5 group of the binding side Trp by forming an H-bond with a sulfate O (Fig. 6C). However, the glycan array results from different galectins (Sup. Table 1) suggest that such interaction alone is not sufficient to explain the galectins’ binding-pattern –all of them share the same Trp. A comparison between glycan array profiles of galectins in Sup. Table 1 shows that the presence or absence of a positively charged residue at the C-terminal of strand β5 divides strong binders (CvGal2 B and C; CGL2, and hGal1) from weak binders (CvGal1 and Gal-3). Sulfation of the 6-OH group at the reducing end of glycosides type 1 are better recognized by CvGal2 than by the other galectins in Sup. Table 1 – apparently smaller (Thr in B and D) or more flexible positively charged residue (Lys, in the CDRs A and C) accommodate better the substitution than the more rigid and bulkier Arg of the other galectins.
Interestingly, CvGal2 also recognizes strongly A type-4 and Forssman glycotopes (Sup. Table 1), although it would appear physically impossible to accommodate the α(1-4)galactosylation at the non-reducing end of the galactoside, without affecting its recognition. A plausible explanation of the observed affinity is the shift in recognition of a carbohydrate moiety at the non-reducing end, to the GalNAcα(1-3)GalNAcβ(1,3)Galα1-4 segment of the glycotope (Fig. 6D). In this alternative interaction, the third Gal moiety takes the place of the glycoside Glc[NAc] only loosing an H-bond to the binding site Glu (Glu – 4OH of the GlcNAc in the core type 2).
The CvGal2 CRD cannot recognize extensions from the galactoside reducing end (Fig. 5D). Thus, with the exception of (1–6) linkages to the glucosamine that may fold-back to contact the proteins, no affinity trends for CvGal2 were detected for additions at the reducing end (Fig. 6B and Sup. Table 1). Hydroxyls at 3 and 4 positions of the β(1,3)-linked Gal of H type-3 precursors can be recognized by polar residues at β11 —A: R135-E136; B:K269 C:K402; D:D545 (Fig. 5C). The β11 residues of the CvGal2 CRD A may select for non-substituted 3-OH group – H type-3 oligosaccharides (Fig. 5E, A) – while CRDs B, C and D can accommodate additional terminal carbohydrates (Fig. 5E, B–D). Binding to the array oligosaccharides with short linkers like Sp0 (a two carbons chain), however, shows different results than the slightly longer Sp8 linker (Sup. Table 1), suggesting interference with the recognition of the glycoside’s Glc[NAc] moiety at the reducing end. To analyze the effect of α(1-3)galactosylation and α(1-2)fucosylation at the non-reducing end independent of linkage effects, we compared a selection of bi-antennary complex-type N-glycans from the glycan-array elements (Fig. 6E). This comparison among glycans of the same basic structure at the non-reducing end would exclude any contributions from different backbone and linkages to the interaction, isolating the recognition of particular features of the glycotope. CvGal2 recognizes strongly A and B oligosaccharides of type 1 and 2, with a weaker preference for B moieties. Lack of α(1-2)fucosylation or α(1-3)galactosylation weaken this recognition 5- and 10-fold, respectively (Fig. 6E).
The structural model of CvGal2 enabled a comprehensive interpretation of the glycan array binding profile, including its preference for blood group oligosaccharides, the relative high prevalence of bi-antennary structures, and the recognition of sulfated moieties and Forssman antigens. While the archetypical galectin carbohydrate-binding site recognizes core disaccharides Galβ(1-[3/4])Glc [or GlcNAc], the glycan array results indicated that CvGal2 binds to oligosaccharides displaying blood group A and B oligosaccharides, in agreement with the structural model. The modeling revealed that like in CvGal1 (23) and CGL2 (77), the shorter loop 4 and a longer loop 3 in CvGal2 relative to the typical proto type galectins (23, 42) provides the structural basis for the preference for blood group oligosaccharides. The recognition of the N-acetyl group at the non-reducing GalNAc in blood group A is established by a pocket formed on the N-terminus that docks the methyl-group of the N-acetyl moiety, a coordination similar to that observed in the CGL2-A2 antigen complex (PDBid 1ULF) (77). The shorter loop 4 opens the site to accommodate simultaneously the 2′-fucosyl group and the α(1-3)Gal[NAc] of A and B antigens, with an apparent ‘gain’ of affinity with respect to the mammalian prototype galectins such as human galectin-1 (hGal-1) that possess the typical long loop 4 (42). In this regard, the four CvGal2 CRDs (A–D) show sequence variations that suggest differences in their fine specificity for A, B and H oligosaccharides. Recognition of the latter, however, can not be attributed solely to the shorter loop 4 since hGal-1 recognizes α(1-2)fucosylated H antigens better than galactosides (74). However, in contrast with CvGal2, hGal-1 recognizes H epitopes with a conformation of the α(1-2)fucosyl that is not achievable by A or B antigens (74).
The presence of several core 1 bi-antennary glycans within the top ranked bound oligosaccharides strongly suggest that the potential multivalency of the CvGal2 may contribute by crosslinking the O-GalNAc in extended core 1 (and possibly core 2) structures. The model also revealed the structural basis for the strong binding of CvGal2 to sulfated oligosaccharides, as not only established by the H-bond between the sulfate O and the NH5 group of the Trp common to other galectins, but also by the smaller or more flexible positively charged residues in the CvGal2 CRDs that can better accommodate the sulfation of the 6OH group at the reducing end of the type 1 structures. Finally, the recognition by CvGal2 of α(1-4)galactosylation at the non-reducing end of the oligosaccharide revealed by the binding to A type-4 and Forssman glycotopes in the glycan array, which initially appears as structurally incompatible with the CvGal2 model, can be explained by a shift in the recognition of the non-reducing end moiety to the GalNAcα(1-3)GalNAcβ(1,3)Galα1-4 segment of the glycotope, in which the third Gal unit takes the place of the Glc[NAc].
Binding of CvGal2 to neoglycoproteins and natural glycoproteins in a solid phase assay, and binding affinity measurements (SPR)
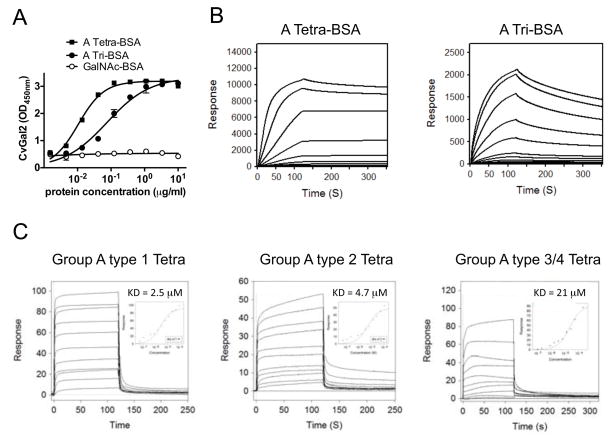
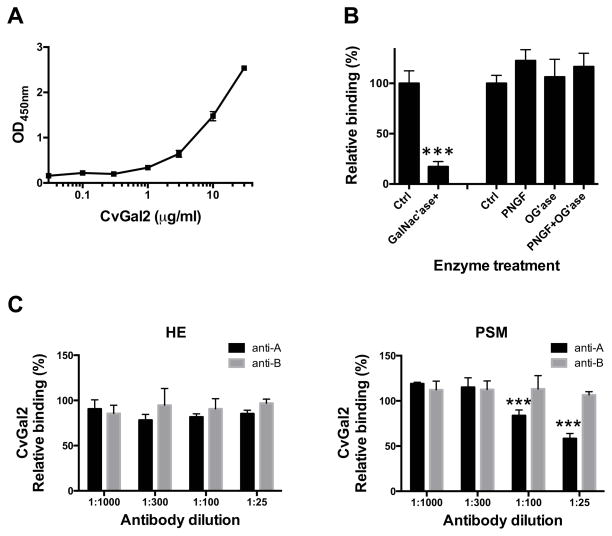
To quantitatively assess the binding of CvGal2 to blood group oligosaccharides we evaluated the binding of CvGal2 to neoglycoproteins that display blood group A tetrasaccharide, the A trisaccharide, or GalNAc. CvGal2 effectively bound to the blood group A tetrasaccharide-BSA, with lower affinity to the A trisaccharide-BSA, and showed no binding to GalNAc-BSA (Fig. 7A). SPR analysis revealed that the KDs for CvGal2 binding to blood group A tetrasaccharide-BSA (20 units/BSA) and trisaccharide-BSA (19 units/BSA) are 4.8 nM and 60 nM, respectively (Fig. 7B). We further assessed by SPR the binding affinity of CvGal2 with various blood group A/B/H oligosaccharides. The results revealed that CvGal2 bound effectively (KD = 2.5 to 21μM) to three types (type 1, 2, and 3/4) of blood group A tetrasaccharides. BSA linked tetra- and trisaccharide bind in nanomolar range due to avidity resulting from multivalent sites (19 or 20 saccharide units/BSA) of neoglycoproteins.
Figure 7. Recognition of blood group A oligosaccharide by CvGal2.
(A) Blood group A tetrasaccharide-BSA (A Tetra-BSA), blood group A trisaccharide-BSA (A Tri-BSA), or N-Acetylgalactosamine-BSA (GalNAc-BSA) were delivered at the concentrations indicated (serial dilution starting from 10 μg/ml; 100 μl/well) into 96-well plates and the binding of CvGal2 (0.2 μg/ml) was assessed by ELISA. Data show optical density at 450 nm (OD450nm) in triplicates with standard error (SEM). (B) Neoglycoproteins were immobilized up to 1000 response units on CM5 chips, and the binding of rCvGal2 was assessed by SPR, flowing through rCvGal2 as the analyte. The SPR sensorgrams were recorded with 2-fold serial dilutions of the analyte starting from 100μg/ml. Sensorgrams for the binding of rCvGal2 onto blood group A tetrasaccharide-BSA (A Tetra-BSA) and trisaccharide-BSA (A Tri-BSA) are shown. Negligible responses were observed on sensorgrams for GalNAc-BSA (not shown). Biacore T100 evaluation software was used to deduce KD values. (C) SPR was performed with immobilized rCvGal2 and the indicated oligosaccharides as analyte. SPR sensorgrams were recorded with 2-fold serial dilutions starting from 100 μg/ml. SPR sensorgram are shown with inset as 1:1 steady state fitting curve.
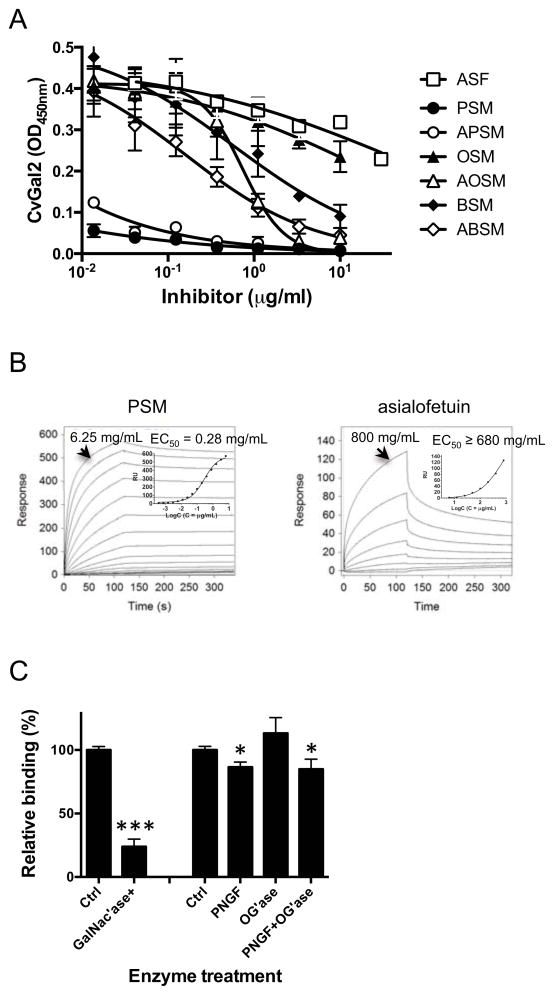
We further evaluated by a solid phase assay the inhibition of the binding of CvGal2 to asialofetuin by glycoproteins that display some of the oligosaccharides identified as CvGal2 ligands in the glycan array (Fig. 8A). These include glycoproteins that contain abundant blood group oligosaccharides, like PSM (78), and the desialylated forms of PSM and fetuin that are rich in non-reducing terminal galactose (79). Asialo-PSM (IC50<0.01 μg/ml) and PSM (IC50< 0.01 μg/ml) behaved as the strongest inhibitors, at least >40-fold better than any other glycoprotein tested. These were followed by asialo-BSM (IC50: 0.037 to 0.708 μg/ml), BSM (0.089 to 3.997), asialo-OSM (0.571 to 0.949), and OSM (IC50: 5.13 to 44.79 μg/ml), all stronger than asialo-fetuin (5.86 to 348.2 ug/ml) itself (Table 3).
Figure 8. Binding of CvGal2 to natural glycoproteins.
A) Binding of rCvGal2 to asialofetuin (20 μg/ml) in the presence of serial dilution of asialofetuin (ASF), PSM, asialo-PSM (APSM), OSM, asialo-OSM (AOSM), BSM, or asialo-BSM (ABSM) were measured in solid-based binding assay. Data show optical density at 450 nm (OD450nm) in triplicates with standard error (SEM). IC50 were calculated (Table 3). (B) The binding of CvGal2 to natural glycoproteins was measured in SPR with immobilized rCvGal2, using PSM (left) or asialo-fetuin (right) as analytes. The sensorgrams were recorded with 2-fold serial dilutions starting at the highest concentration lindicated. SPR sensorgram was shown with inset as the 1:1 binding curve. Graphpad Prism software was used to deduce EC50 values. (C) PSM (50 ng/ml) coated wells were treated with neuraminidase (Ctrl) plus N-acetylgalactosidase (GalNac’ase), PNGase F (PNGF), O-glycosidase (OG’ase), or both. Binding of rCvGal2 to treated wells was measured in a solid-phase binding assay. Relative binding to untreated wells was calculated, and the mean with SEM from triplicates is shown. *: p<0.05; ***: p<0.001 vs. control
Table 3.
Inhibition of CvGal2 binding to asialofetuin by glycoproteins
| Inhibitor | IC50 (95%CI, μg/ml) |
|---|---|
| Asialofetuin | 5.856 to 348.2 |
| PSM | <0.01 * |
| asialo-PSM | <0.01 * |
| OSM | 5.131 to 44.79 |
| asialo-OSM | 0.571 to 0.949 |
| BSM | 0.089 to 3.997 |
| asialo-BSM | 0.037 to 0.708 |
Too low to be calculated accurately (see figure 8A).
We subsequently assessed by SPR the binding kinetics of rCvGal2 to the glycoproteins identified above as CvGal2 ligands, such as PSM and asialofetuin. The results showed that in agreement with the binding-inhibition studies, PSM behaved as the strongest binding ligand with an EC50 value of 0.28 μg/ml (Fig. 8B), while for asialofetuin the binding of CvGal2 was significantly weaker (EC50 ≥ 680μg/μl) (Fig. 8B). Next we evaluated the binding ligands on PSM by removing GalNAc with enzyme. The results showed that 70% reduction of CvGal2 binding to GalNAcase-treated PSM than to the untreated control. On the other hand, binding was reduced by PNGase F treatment significantly but not completely (Fig. 8C).
These results not only corroborated the glycan array analysis and validated the structural models, but also provided information about the specificity of CvGal2 for blood group oligosaccharide moieties that are covalently bound to polypetide scaffolds and presented in distinct displays, revealing notable differences with the binding properties of CvGal1. For example, the recognition of blood group A tetra- and trisaccharides on neoglycoproteins by CvGal2 observed in solid phase binding assays and SPR analysis, was in sharp contrast with CvGal1, which only recognizes the tetrasaccharide (23). The binding affinity of CvGal2 for the types 1, 2 and 3/4 blood group A oligosaccharides also confirmed the above results from the glycan array analysis and the structural modeling. Furthermore, binding-inhibition and SPR measurements confirmed that glycoproteins that display abundant ABH blood group oligosaccharides and non-reducing terminal galactose, like PSM and asialo-PSM (78, 79), are the preferred ligands for CvGal2. While fetuin displays abundant sialylated N-acetyllactosamines [Siaα2-3Galβ1-4GlcNAcβ] (79), PSM is rich in oligosaccharides that display non-reducing terminal GalNAc and L-Fuc, namely blood group H [Fucα1-2Galβ1-4GlcNAcβ] and A [GalNAcα1-3(Fucα1-2)Galβ1-4GlcNAcβ] moieties (78). The similar binding of CvGal2 to intact and asialo PSM, and significantly weaker binding to asialofetuin indicates that CvGal2 only recognizes relatively weakly those subterminal sugars, such as galactose, that are exposed by the cleavage of non-reducing terminal sialic acids. In contrast, CvGal1 showed significantly stronger binding to asialo-PSM than to PSM, and effectively recognized asialofetuin, suggesting that subterminal sugars unmasked by sialidases also contribute to the binding of CvGal1 to asialo-PSM and asialofetuin (23). The comparative assessment of CvGal2 binding to untreated PSM and PSM that had been treated with PNGase F and O-glycosidase not only verified that the CvGal2 binding to PSM is carbohydrate-mediated, but also revealed that the oligosaccharide ligands for CvGal2 in PSM are N-linked. This result identifies a significant difference in the binding properties of CvGal1 and CvGal2, since the PSM ligands recognized by the former are both N- and O-linked (23). Furthermore, pretreatment of PSM with GalNAc’ase clearly showed that GalNAc at the non-reducing end is required for the binding of CvGal2 to the PSM’s blood group A oligosaccharides, in agreement with the glycan array and modeling studies.
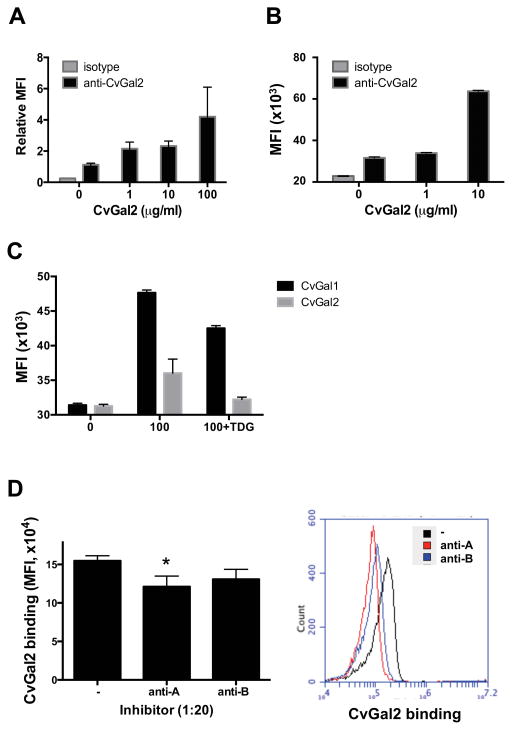
Binding of rCvGal2 to hemocyte surface glycans
Based on the results above, we tested whether CvGal2 recognizes glycans on the surface of hemocytes, as we had previously observed for CvGal1 (22, 23). First, we used ELISA to evaluate the presence of CvGal2 ligands in hemocyte extracts, and the results showed that CvGal2 binds to the hemocyte extract in a dose-dependent manner (Fig. 9A). Pre-treatment of hemocyte extract with GalNAcase decreased the binding over 80% (Fig. 9B), whereas no effect was observed with either PNGase F or O-glycosidase treatment.
Figure 9. Recombinant CvGal2 binds to hemocyte extract.
(A) Hemocyte extract (1 μg/ml) coated wells were incubated with different concentration of rCvGal2 (0 to 30 μg/ml). Binding of rCvGal2 was measured in solid-based binding assay, and mean optical density at 450 nm (OD450nm) in triplicates with SEM was shown. (B) Hemocyte extract (1 μg/ml) coated wells were treated with neuraminidase (Ctrl) plus N-acetylgalactosidase (GalNac’ase), PNGase F (PNGF), O-glycosidase (OG’ase), or both PNGF and OG’ase. Binding of rCvGal2 to treated wells was measured as above. Relative binding to untreated cells was calculated, and the mean from triplicates with SEM is shown. *: p<0.05; ***: p<0.001 vs. control. (C) Hemocyte extract (1 μg/ml) (left panel) or PSM (50 ng/ml) (right panel) coated wells were pre-incubated with different concentration of anti-A or anti-B (1:25 to 1:1000 dilution) before incubation with rCvGal2 (2 μg/ml). Binding of rCvGal2 to treated wells was measured as above. Relative binding to untreated cells was calculated, and the mean from triplicates with SEM is shown. ***: p<0.001 vs. control.
Next, we evaluated by a competitive ELISA if the CvGal2 ligands in the hemocyte extract are recognized by anti-A or anti-B monoclonal antibodies. Surprisingly, neither the anti-A nor anti-B antibodies block the binding of CvGal2 to the hemocyte extract up to 1:25 dilution, while anti-A antibody reduced in 20% and 40% the binding to PSM at 1:100 and 1:25 dilutions, respectively (Fig. 9C). We tested the binding of CvGal2 to intact circulating hemocytes by flow cytometry, and by immunofluoresence the CvGal2 binding to hemocytes attached and spread on a plastic surface. rCvGal2 bound to both the circulating and the attached hemocytes in a dose-dependent (Fig. 10A&B) and carbohydrate-specific (Fig. 10C) manner. In contrast to what we observed with the hemocyte extract, the anti-A antibody reduced the binding of CvGal2 to the intact hemocytes in 20% (p<0.05), while the anti-B antibody only reduced the binding marginally (p=0.10) (Fig. 10D).
Figure 10. Recombinant CvGal2 binds to oyster hemocytes.
(A) Attached hemocytes were incubated with different concentration of rCvGal2 for 1h at RT. The binding of rCvGal2 to attached hemocytes were detected and revealed under fluorescent microscope as described in Materials and Methods. The fluorescent intensity from at least 5 randomly selected views was quantified and normalized to the ones without exogenous rCvGal2 (0). Data are shown as relative mean fluorescent intensity (MFI)±SD. (B) Freshly isolated hemocytes were fixed as unattached and incubated with different concentration of rCvGal2 (0–10 μg/ml) for 1h at RT. The cells were stained with anti-CvGal2 antibody followed by FITC-conjugated secondary antibody; and the binding of recombinant proteins was revealed by flow cytometry. Data show as MFI±95%CI. (C) Fixed hemocytes were incubated with CvGal1 or CvGal2 (100 μg/ml) with or without thiodigalactose (TDG) for binding assay by flow cytometry as above. (D) Fixed cells were pre-incubated with anti-A or anti-B antibodies to outcompete rCvGal2 binding before adding rCvGal2 (2 μg/ml) for binding assay by flow cytometry as above. Data are shown as MFI±95%CI, *: p<0.05; ***: p<0.001 vs. control (−).
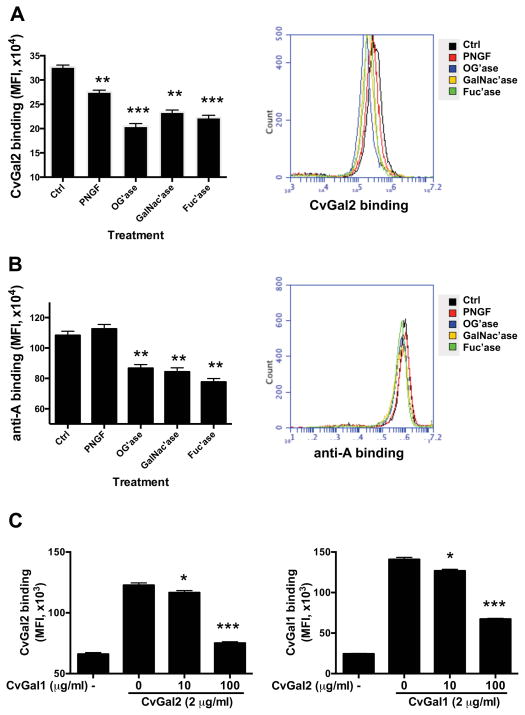
By the use of glycosidase treatments we subsequently investigated the type of glycan structures on hemocytes responsible for CvGal2 binding. Both PNGase F and O-glycosidase reduced the CvGal2 binding 15% (p<0.05) and 40% (p<0.05), respectively, suggesting that the CvGal2 ligands were displayed on both N-linked and O-linked cell surface glycans. GalNAcase and fucosidase treatments reduced the binding at least 30% (Fig. 11A). For comparative purposes, the binding of the anti-A antibody to the glycosidase-treated hemocyte surface was evaluated. PNGase F treatment didn’t reduce the anti-A binding, but O-Glycosidase reduced binding at least 20%, confirming the presence of O-linked blood group A moieties on the hemocyte surface. Both GalNAcase and fucosidase reduced anti-A binding to the hemocytes in 20% (Fig. 11B).
Figure 11. Binding of rCvGal2 and anti-A/B to intact, Gal-ase and GalNAc-ase-treated hemocytes.
(A) Fixed hemocytes were treated with PNGase F (PNGF), O-Glycosidase (OG’ase), N-acetylgalactosidase (GalNac’ase), or Fucosidase (Fuc’ase), and incubated with CvGal2 to determine the binding by flow cytometry. (B) The fixed cells were treated as above and stained with anti-A antibody for flow cytometry analysis. (C) Fixed cells were pre-incubated with rCvGal1 (0–100 μg/ml) or rCvGal2 (0–100 μg/ml) before adding rCvGal2 (2 μg/ml) or rCvgal1 (2 μg/ml) respectively for binding assay by flow cytometry as above. Data are shown as MFI±95%CI. *: p<0.05; **: p<0.01; ***: p<0.001 vs. Ctrl.
Next, we evaluated whether CvGal2 and CvGal1 share the same carbohydrate ligands on the hemocyte surface. Pre-incubation with CvGal1 (100 μg/ml) reduced the CvGal2 binding up to 90%, in a dose-dependent manner (Fig. 11C, left panel). In the reciprocal experiment, pre-incubation with CvGal2 (100 μg/ml) reduced the CvGal1 binding up to 60%, also in a dose-dependent manner (Fig. 11C, right panel). The results suggest that the carbohydrate ligands for CvGal1 and CvGal2 on the hemocyte surface only overlap partially.
The analysis of binding of CvGal2 to oyster hemocyte glycoproteins was consistent with those obtained with purified mammalian glycoproteins in that GalNAc is a key sugar recognized by CvGal2 on the surface of the oyster hemocytes. As the binding of CvGal2 to PSM could be effectively outcompeted by pretreatment with a monoclonal anti-A antibody, the lack of a similar strong inhibitory effect of the same monoclonal with oyster hemocyte extract or intact hemocytes suggest that CvGal2 can recognize not only blood group A oligosaccharides on the surface of hemocytes, but also other distinct N- or O-linked GalNAc-bearing moieties that lack the blood group A structure. Significant reduction in CvGal2 binding by pretreatment of the hemocytes with GalNAc’ase and fucosidase, however, confirm that blood group A oligosaccharides on the surface of hemocytes partially contribute to CvGal2 binding. This observation also points to differences in binding between CvGal1 and CvGal2, since the binding of the former to the hemocyte surface is mostly attributable to blood group A carbohydrate moieties (23). Finally, the reciprocal partial competitive binding to the hemocyte surface of both CvGal1 and CvGal2 clearly shows that although ligands common to both galectins are exposed, these also bind to non-overlapping surface ligands that suggest distinct biological roles for each galectin.
In a previous study we established that CvGal1 is expressed and secreted by a hemocyte subpopulation, most likely granulocytes, and binds to β-integrin and dominin on the hemocyte surface (23, 57). It is possible that CvGal2, which is expressed equally in both granular hemocytes and hyalinocytes, also binds to either one or both CvGal1 ligands, but the results suggest that additional hemocyte sugar moieties are recognized by CvGal2. In either case, the partial sharing of glycan ligands on the hemocyte cell surface, together with the observation that CvGal1 and CvGal2 possess multiple CRDs that exhibit differences in specificity as shown by the modeling study, suggest a complex interplay of opsonic properties for potential pathogens of both galectins together with the effects of their selected CRDs binding to the hemocyte surface, leading to cell activation by synergistic activities or downregulation by competitive inhibition of galectin binding to the hemocyte surface.
Differential recognition of sympatric Perkinsus species by rCvGal2
As CvGal1 binds to P. marinus cell surface and promotes parasite entry (22, 23), we examined the potential binding of CvGal2 to P. marinus and compared it to P. chesapeaki, a sympatric Perkinsus species that is mostly prevalent in clam species. CvGal2 bound strongly to P. marinus in a dose-dependent and carbohydrate-specific manner, (Fig. 12), but only negligible binding to P. chesapeaki was observed. This striking selectivity in recognition properties of CvGal2 for Perkinsus species may have functional implications in parasite infectivity and pathogenicity. Along the mid-Atlantic coast of USA, and particularly in the Chesapeake and Delaware Bays, Perkinsus chesapeaki is a species sympatric with P. marinus (17). Although as mentioned above, P. marinus is prevalent and pathogenic for the eastern oyster (17), P. chesapeaki is mostly prevalent in sympatric clam species, such as Mya arenaria, Macoma balthica, Tagelus plebeius, Mulinia lateralis, Rangia cuneata, Cyrtopleura costata and Mercenaria mercenaria (10, 14, 15, 17–19). Further, the pathogenicity of P. chesapeaki for the eastern oyster has not been rigorously demonstrated, and it remains unclear if the relatively lower prevalences observed in the eastern oyster are due to this species being a less suitable or permissive host for P. chesapeaki as compared to P. marinus (17, 18). As CvGal1 and CvGal2 strongly bind to the P. marinus cell surface and promote parasite entry into the host (22, 23), the negligible binding of CvGal2 to P. chesapeaki may explain its low prevalence relative to P. marinus in the eastern oyster, and perhaps the reduced pathogenicity. The diversified binding specificity and biological functions of CvGal1 and CvGal2 may provide an evolutionary advantage to the oyster against infectious challenge, but P. marinus has co-evolved to subvert their role(s) to gain entry into the host. In this regard, P. chesapeaki would have instead co-evolved with other bivalve hosts, including the aforementioned clam species. Thus, based on our results it is tempting to speculate that host preference and pathogenicity of Perkinsus species for sympatric bivalve species may be the result of a close co-evolutionary process between a parasite species and a bivalve host. This would be determined by the adaptation of the parasite glycocalyx to specific host recognition factors, such as the galectins CvGal1 and CvGal2, that would facilitate parasite entry into the preferred host such as the eastern oyster (6). Current studies in our laboratory are aimed at assessing potential differences in the binding of CvGal1 and CvGal2 to glycotopes on the hemocyte surface, P. marinus trophozoites, phytoplankton food, and microbial pathogens to address these unanswered questions.
Figure 12. Binding of rCvGal1 and rCvgal2 to Perkinsus parasites.
P. marinus (Pm) or P. chespeaky (Pc) were fixed and incubated with 0–10 μg/ml of rCvGal2 (bottom), which was pre-incubated with (0.1) or without (0) 0.1 M of lactose, for 1h at RT. The cells were stained respectively with anti-CvGal2 antibody followed by FITC-conjugated secondary antibody; and the binding of recombinant proteins was revealed by flow cytometry. Data are shown as MFI±95%CI. p<0.05; **: p<0.01; ***: p<0.001 vs. no rCvGal2 (0); Significant differences between two samples are shown in bar with p values.
Acknowledgments
Funding Source Statement: This work was supported by grants IOS-0822257 and IOS-1063729 from the National Science Foundation, and grant 5R01 GM070589 from the National Institutes of Health to GRV, and grant R01 GM080374 from the National Institutes of Health to LXW.
We are grateful to Dr. David Smith and Dr. Jamie Molinaro from the Core H, Consortium for Functional Glycomics, Emory University, Atlanta, GA, for the glycan array analysis of CvGal2.
Abbreviations used in the text
- CvGal2
Crassostrea virginica galectin 2
- BaGal1
Bufo arenarum galectin-1
- hGal-1
human galectin-1
- CGL2
Coprinus cinereus galectin-2
- CRD
carbohydrate recognition domain
- GalNAc
N-acetyl-D-galactosamine
- GlcNAc
N-acetyl-D-glucosamine
- LacNAc
N-acetyllactosamine
- TDG
thiodigalactose
- BSM
bovine submaxillary mucin
- OSM
ovine submaxillary mucin
- PSM
porcine stomach mucin
- SPR
surface plasmon resonance
- PRR
pattern recognition receptors
- 2-ME
β-mercaptoethanol
- PBS/2-ME
PBS containing 10 mM β-mercaptoethanol
- PBST
0.1% Tween in PBS
- HBS
HEPES buffered saline
- RACE
rapid amplification of cDNA ends
- FC
flow cytofluorometry
- PFA
paraformaldehyde
References
- 1.Bachere E, Gueguen Y, Gonzalez M, de Lorgeril J, Garnier J, Romestand B. Insights into the anti-microbial defense of marine invertebrates: the penaeid shrimps and the oyster Crassostrea gigas. Immunological reviews. 2004;198:149–168. doi: 10.1111/j.0105-2896.2004.00115.x. [DOI] [PubMed] [Google Scholar]
- 2.Boehm T. Evolution of vertebrate immunity. Current biology: CB. 2012;22:R722–732. doi: 10.1016/j.cub.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 4.Vasta GR, Ahmed H, Odom EW. Structural and functional diversity of lectin repertoires in invertebrates, protochordates and ectothermic vertebrates. Current opinion in structural biology. 2004;14:617–630. doi: 10.1016/j.sbi.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Vasta GR, Ahmed H. Animal Lectins: A Functional View. CRC Press; 2008. [Google Scholar]
- 6.Vasta GR. Roles of galectins in infection. Nature reviews. Microbiology. 2009;7:424–438. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadevall A, Pirofski LA. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infection and immunity. 2000;68:6511–6518. doi: 10.1128/iai.68.12.6511-6518.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dame R, Bushek D, Allen D, Lewitus A, Edwards D, Koepfler E, Gregory L. Ecosystem response to bivalve density reduction: management implications. Aquat Ecol. 2002;36:51–65. [Google Scholar]
- 9.Harvell CD, Kim K, Burkholder JM, Colwell RR, Epstein PR, Grimes DJ, Hofmann EE, Lipp EK, Osterhaus AD, Overstreet RM, Porter JW, Smith GW, Vasta GR. Emerging marine diseases--climate links and anthropogenic factors. Science. 1999;285:1505–1510. doi: 10.1126/science.285.5433.1505. [DOI] [PubMed] [Google Scholar]
- 10.Dungan CF, Hamilton RM, Hudson KL, McCollough CB, Reece KS. Two epizootic diseases in Chesapeake Bay commercial clams, Mya arenaria and Tagelus plebeius. Dis Aquat Organ. 2002;50:67–78. doi: 10.3354/dao050067. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy VS, Newell RIE, Eble AF. The Eastern Oyster: Crassostrea virginica. 2. University of Maryland Sea Grant Publications; 1996. [Google Scholar]
- 12.Andrews JD. History of Perkinsus marinus, a pathogen of oysters in Chesapeake Bay 1950–1984. Journal of Shellfish Research. 1996;15:13–16. [Google Scholar]
- 13.McLaughlin SM, Tall BD, Shaheen A, Elsayed EE, Faisal M. Zoosporulation of a new Perkinsus species isolated from the gills of the softshell clam Mya arenaria. Parasite. 2000;7:115–122. doi: 10.1051/parasite/2000072115. [DOI] [PubMed] [Google Scholar]
- 14.Coss CA, Robledo JA, Ruiz GM, Vasta GR. Description of Perkinsus andrewsi n. sp isolated from the Baltic clam (Macoma balthica) by characterization of the ribosomal RNA locus, and development of a species-specific PCR-based diagnostic assay. The Journal of eukaryotic microbiology. 2001;48:52–61. doi: 10.1111/j.1550-7408.2001.tb00415.x. [DOI] [PubMed] [Google Scholar]
- 15.Burreson EM, Reece KS, Dungan CF. Molecular, morphological, and experimental evidence support the synonymy of Perkinsus chesapeaki and Perkinsus andrewsi. The Journal of eukaryotic microbiology. 2005;52:258–270. doi: 10.1111/j.1550-7408.2005.05-00035.x. [DOI] [PubMed] [Google Scholar]
- 16.Coss CA, Robledo JA, Vasta GR. Fine structure of clonally propagated in vitro life stages of a Perkinsus sp. isolated from the Baltic clam Macoma balthica. The Journal of eukaryotic microbiology. 2001;48:38–51. doi: 10.1111/j.1550-7408.2001.tb00414.x. [DOI] [PubMed] [Google Scholar]
- 17.Pecher WT, Alavi MR, Schott EJ, Fernandez-Robledo JA, Roth L, Berg ST, Vasta GR. Assessment of the northern distribution range of selected Perkinsus species in eastern oysters (Crassostrea virginica) and hard clams (Mercenaria mercenaria) with the use of PCR-based detection assays. The Journal of parasitology. 2008;94:410–422. doi: 10.1645/GE-1282.1. [DOI] [PubMed] [Google Scholar]
- 18.Reece KS, Dungan CF, Burreson EM. Molecular epizootiology of Perkinsus marinus and P. chesapeaki infections among wild oysters and clams in Chesapeake Bay, USA. Dis Aquat Organ. 2008;82:237–248. doi: 10.3354/dao01997. [DOI] [PubMed] [Google Scholar]
- 19.Bushek D, Landau B, Scarpa E. Perkinsus chesapeaki in stout razor clams Tagelus plebeius from Delaware Bay. Dis Aquat Organ. 2008;78:243–247. doi: 10.3354/dao01871. [DOI] [PubMed] [Google Scholar]
- 20.Bushek D, Ford SE, Chintala MM. Comparison of in vitro-cultured and wild-type Perkinsus marinus. III. Fecal elimination and its role in transmission. Dis Aquat Organ. 2002;51:217–225. doi: 10.3354/dao051217. [DOI] [PubMed] [Google Scholar]
- 21.Ford SE, Chintala MM, Bushek D. Comparison of in vitro-cultured and wild-type Perkinsus marinus. I. Pathogen virulence. Dis Aquat Organ. 2002;51:187–201. doi: 10.3354/dao051187. [DOI] [PubMed] [Google Scholar]
- 22.Tasumi S, Vasta GR. A galectin of unique domain organization from hemocytes of the Eastern oyster (Crassostrea virginica) is a receptor for the protistan parasite Perkinsus marinus. J Immunol. 2007;179:3086–3098. doi: 10.4049/jimmunol.179.5.3086. [DOI] [PubMed] [Google Scholar]
- 23.Feng C, Ghosh A, Amin MN, Giomarelli B, Shridhar S, Banerjee A, Fernandez-Robledo JA, Bianchet MA, Wang LX, Wilson IB, Vasta GR. The galectin CvGal1 from the eastern oyster (Crassostrea virginica) binds to blood group A oligosaccharides on the hemocyte surface. The Journal of biological chemistry. 2013;288:24394–24409. doi: 10.1074/jbc.M113.476531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper DNW. Galectinomics: finding themes in complexity. Biochimica et biophysica acta. 2002;1572:209–231. doi: 10.1016/s0304-4165(02)00310-0. [DOI] [PubMed] [Google Scholar]
- 25.Vasta GR, Quesenberry M, Ahmed H, O’Leary N. C-type lectins and galectins mediate innate and adaptive immune functions: their roles in the complement activation pathway. Dev Comp Immunol. 1999;23:401–420. doi: 10.1016/s0145-305x(99)00020-8. [DOI] [PubMed] [Google Scholar]
- 26.Liao DI, Kapadia G, Ahmed H, Vasta GR, Herzberg O. Structure of S-lectin, a developmentally regulated vertebrate beta-galactoside-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:1428–1432. doi: 10.1073/pnas.91.4.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed H, Du S-J, Vasta GR. Knockdown of a galectin-1-like protein in zebrafish (Danio rerio) causes defects in skeletal muscle development. Glycoconj J. 2009;26:277–283. doi: 10.1007/s10719-008-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu FT. Galectins: a new family of regulators of inflammation. Clin Immunol. 2000;97:79–88. doi: 10.1006/clim.2000.4912. [DOI] [PubMed] [Google Scholar]
- 29.Sato S, Ouellet N, Pelletier I, Simard M, Rancourt A, Bergeron MG. Role of galectin-3 as an adhesion molecule for neutrophil extravasation during streptococcal pneumonia. J Immunol. 2002;168:1813–1822. doi: 10.4049/jimmunol.168.4.1813. [DOI] [PubMed] [Google Scholar]
- 30.Barrionuevo P, Beigier-Bompadre M, Ilarregui JM, Toscano MA, Bianco GA, Isturiz MA, Rabinovich GA. A novel function for galectin-1 at the crossroad of innate and adaptive immunity: galectin-1 regulates monocyte/macrophage physiology through a nonapoptotic ERK-dependent pathway. J Immunol. 2007;178:436–445. doi: 10.4049/jimmunol.178.1.436. [DOI] [PubMed] [Google Scholar]
- 31.Rabinovich GA, Rubinstein N, Toscano MA. Role of galectins in inflammatory and immunomodulatory processes. Biochimica et biophysica acta. 2002;1572:274–284. doi: 10.1016/s0304-4165(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 32.Novelli F, Allione A, Wells V, Forni G, Mallucci L. Negative cell cycle control of human T cells by beta-galactoside binding protein (beta GBP): induction of programmed cell death in leukaemic cells. J Cell Physiol. 1999;178:102–108. doi: 10.1002/(SICI)1097-4652(199901)178:1<102::AID-JCP13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 33.Perillo NL, Pace KE, Seilhamer JJ, Baum LG. Apoptosis of T cells mediated by galectin-1. Nature. 1995;378:736–739. doi: 10.1038/378736a0. [DOI] [PubMed] [Google Scholar]
- 34.Vasta GR, Ahmed H, Nita-Lazar M, Banerjee A, Pasek M, Shridhar S, Guha P, Fernandez-Robledo JA. Galectins as self/non-self recognition receptors in innate and adaptive immunity: an unresolved paradox. Frontiers in immunology. 2012;3:199. doi: 10.3389/fimmu.2012.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu F-LE. Laboratory investigations of susceptibility, infectivity, and transmission of Perkinsus marinus in oysters. Journal of Shellfish Research. 1996;15:57–66. [Google Scholar]
- 36.Feng C, Zhang L, Nguyen C, Vogel SN, Goldblum SE, Blackwelder WC, Cross AS. Neuraminidase reprograms lung tissue and potentiates lipopolysaccharide-induced acute lung injury in mice. J Immunol. 2013;191:4828–4837. doi: 10.4049/jimmunol.1202673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandez-Robledo JA, Lin Z, Vasta GR. Transfection of the protozoan parasite Perkinsus marinus. Molecular and Biochemical Parasitology. 2008;157:44–53. doi: 10.1016/j.molbiopara.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Gauthier JD, Vasta GR. In vitro culture of the Eastern parasite Perkinsus marinus: optimization of the methodology. Journal of Invertebrate Pathology. 1995;66:156–168. [Google Scholar]
- 39.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed H, Bianchet MA, Amzel LM, Hirabayashi J, Kasai K, Giga-Hama Y, Tohda H, Vasta GR. Novel carbohydrate specificity of the 16-kDa galectin from Caenorhabditis elegans: binding to blood group precursor oligosaccharides (type 1, type 2, Talpha, and Tbeta) and gangliosides. Glycobiology. 2002;12:451–461. doi: 10.1093/glycob/cwf052. [DOI] [PubMed] [Google Scholar]
- 42.Bianchet MA, Ahmed H, Vasta GR, Amzel LM. Soluble beta-galactosyl-binding lectin (galectin) from toad ovary: crystallographic studies of two protein-sugar complexes. Proteins. 2000;40:378–388. doi: 10.1002/1097-0134(20000815)40:3<378::aid-prot40>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Case DA, et al. AMBER 12. University of California; San Francisco: 2012. [Google Scholar]
- 44.Gerber PR, Muller K. MAB, a generally applicable molecular force field for structure modelling in medicinal chemistry. Journal of computer-aided molecular design. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- 45.Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, Smith DF, Cummings RD. Innate immune lectins kill bacteria expressing blood group antigen. Nature medicine. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levroney EL, Aguilar HC, Fulcher JA, Kohatsu L, Pace KE, Pang M, Gurney KB, Baum LG, Lee B. Novel innate immune functions for galectin-1: galectin-1 inhibits cell fusion by Nipah virus envelope glycoproteins and augments dendritic cell secretion of proinflammatory cytokines. J Immunol. 2005;175:413–420. doi: 10.4049/jimmunol.175.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toledo KA, Fermino ML, del Andrade CC, Riul TB, Alves RT, Muller VD, Russo RR, Stowell SR, Cummings RD, Aquino VH, Dias-Baruffi M. Galectin-1 Exerts Inhibitory Effects during DENV-1 Infection. PloS one. 2014;9:e112474. doi: 10.1371/journal.pone.0112474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X-W, Xu J-D, Zhao X-F, Vasta GR, Wang J-X. A shrimp C-type lectin inhibits proliferation of the hemolymph microbiota by maintaining the expression of antimicrobial peptides. The Journal of biological chemistry. 2014;289:11779–11790. doi: 10.1074/jbc.M114.552307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okumura CY, Baum LG, Johnson PJ. Galectin-1 on cervical epithelial cells is a receptor for the sexually transmitted human parasite Trichomonas vaginalis. Cell Microbiol. 2008;10:2078–2090. doi: 10.1111/j.1462-5822.2008.01190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kamhawi S, Ramalho-Ortigao M, Pham VM, Kumar S, Lawyer PG, Turco SJ, Barillas-Mury C, Sacks DL, Valenzuela JG. A role for insect galectins in parasite survival. Cell. 2004;119:329–341. doi: 10.1016/j.cell.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 51.Hager KM, Carruthers VB. MARveling at parasite invasion. Trends in parasitology. 2008;24:51–54. doi: 10.1016/j.pt.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petri WA, Jr, Haque R, Mann BJ. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite. Entamoeba histolytica, Annual review of microbiology. 2002;56:39–64. doi: 10.1146/annurev.micro.56.012302.160959. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Robledo JA, Schott EJ, Vasta GR. Perkinsus marinus superoxide dismutase 2 (PmSOD2) localizes to single-membrane subcellular compartments. Biochemical and biophysical research communications. 2008;375:215–219. doi: 10.1016/j.bbrc.2008.07.162. [DOI] [PubMed] [Google Scholar]
- 54.Schott EJ, Vasta GR. The PmSOD1 gene of the protistan parasite Perkinsus marinus complements the sod2Δ mutant of Saccharomyces cerevisiae, and directs an iron superoxide dismutase to mitochondria. Mol Biochem Parasitol. 2003;126:81–92. doi: 10.1016/s0166-6851(02)00271-2. [DOI] [PubMed] [Google Scholar]
- 55.Vasta GR, Sullivan JT, Cheng TC, Marchalonis JJ, Warr GW. A cell membrane-associated lectin of the oyster hemocyte. Journal of Invertebrate Pathology. 1982;40:367–377. [Google Scholar]
- 56.Vasta GR, Cheng TC, Marchalonis JJ. A lectin on the hemocyte membrane of the oyster (Crassostrea virginica) Cellular immunology. 1984;88:475–488. doi: 10.1016/0008-8749(84)90179-5. [DOI] [PubMed] [Google Scholar]
- 57.Kurz S, Jin C, Hykollari A, Gregorich D, Giomarelli B, Vasta GR, Wilson IB, Paschinger K. Hemocytes and plasma of the eastern oyster (Crassostrea virginica) display a diverse repertoire of sulfated and blood group A-modified N-glycans. The Journal of biological chemistry. 2013;288:24410–24428. doi: 10.1074/jbc.M113.478933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dam TK, Brewer CF. Effects of clustered epitopes in multivalent ligand-receptor interactions. Biochemistry. 2008;47:8470–8476. doi: 10.1021/bi801208b. [DOI] [PubMed] [Google Scholar]
- 59.Ng KK, Drickamer K, Weis WI. Structural analysis of monosaccharide recognition by rat liver mannose-binding protein. The Journal of biological chemistry. 1996;271:663–674. doi: 10.1074/jbc.271.2.663. [DOI] [PubMed] [Google Scholar]
- 60.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu Rev Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 61.Davicino RC, Elicabe RJ, Di Genaro MS, Rabinovich GA. Coupling pathogen recognition to innate immunity through glycan-dependent mechanisms. Int Immunopharmacol. 2011;11:1457–1463. doi: 10.1016/j.intimp.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 62.Vasta GR, Ahmed H, Bianchet MA, Fernandez-Robledo JA, Amzel LM. Diversity in recognition of glycans by F-type lectins and galectins: molecular, structural, and biophysical aspects. Ann N Y Acad Sci. 2012;1253:14–26. doi: 10.1111/j.1749-6632.2012.06698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Houzelstein D, Goncalves IR, Fadden AJ, Sidhu SS, Cooper DNW, Drickamer K, Leffler H, Poirier F. Phylogenetic analysis of the vertebrate galectin family. Mol Biol Evol. 2004;21:1177–1187. doi: 10.1093/molbev/msh082. [DOI] [PubMed] [Google Scholar]
- 64.Lensch M, Lohr M, Russwurm R, Vidal M, Kaltner H, Andre S, Gabius H-J. Unique sequence and expression profiles of rat galectins-5 and -9 as a result of species-specific gene divergence. Int J Biochem Cell Biol. 2006;38:1741–1758. doi: 10.1016/j.biocel.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Allam B, Ashton-Alcox KA, Ford SE. Flow cytometric comparison of haemocytes from three species of bivalve molluscs. Fish & shellfish immunology. 2002;13:141–158. doi: 10.1006/fsim.2001.0389. [DOI] [PubMed] [Google Scholar]
- 66.Lin Z, Fernandez-Robledo JA, Cellier MF, Vasta GR. The natural resistance-associated macrophage protein from the protozoan parasite Perkinsus marinus mediates iron uptake. Biochemistry. 2011;50:6340–6355. doi: 10.1021/bi200343h. [DOI] [PubMed] [Google Scholar]
- 67.Schott EJ, Pecher WT, Okafor F, Vasta GR. The protistan parasite Perkinsus marinus is resistant to selected reactive oxygen species. Exp Parasitol. 2003;105:232–240. doi: 10.1016/j.exppara.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Hughes RC. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochimica et biophysica acta. 1999;1473:172–185. doi: 10.1016/s0304-4165(99)00177-4. [DOI] [PubMed] [Google Scholar]
- 69.Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 70.Camby I, Le Mercier M, Lefranc F, Kiss R. Galectin-1: a small protein with major functions. Glycobiology. 2006;16:157. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]
- 71.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126:855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 72.Zick Y, Eisenstein M, Goren RA, Hadari YR, Levy Y, Ronen D. Role of galectin-8 as a modulator of cell adhesion and cell growth. Glycoconj J. 2004;19:517–526. doi: 10.1023/B:GLYC.0000014081.55445.af. [DOI] [PubMed] [Google Scholar]
- 73.Di Lella S, Sundblad V, Cerliani JP, Guardia CM, Estrin DA, Vasta GR, Rabinovich GA. When galectins recognize glycans: from biochemistry to physiology and back again. Biochemistry. 2011;50:7842–7857. doi: 10.1021/bi201121m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, Leffler H, Smith DF, Cummings RD. Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. The Journal of biological chemistry. 2008;283:10109–10123. doi: 10.1074/jbc.M709545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stowell SR, Arthur CM, Slanina KA, Horton JR, Smith DF, Cummings RD. Dimeric Galectin-8 induces phosphatidylserine exposure in leukocytes through polylactosamine recognition by the C-terminal domain. The Journal of biological chemistry. 2008;283:20547–20559. doi: 10.1074/jbc.M802495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cooper DN, Boulianne RP, Charlton S, Farrell EM, Sucher A, Lu BC. Fungal galectins, sequence and specificity of two isolectins from Coprinus cinereus. The Journal of biological chemistry. 1997;272:1514–1521. doi: 10.1074/jbc.272.3.1514. [DOI] [PubMed] [Google Scholar]
- 77.Walser PJ, Haebel PW, Kunzler M, Sargent D, Kues U, Aebi M, Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure. 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Thomsson KA, Karlsson H, Hansson GC. Sequencing of sulfated oligosaccharides from mucins by liquid chromatography and electrospray ionization tandem mass spectrometry. Analytical chemistry. 2000;72:4543–4549. doi: 10.1021/ac000631b. [DOI] [PubMed] [Google Scholar]
- 79.Baenziger JU, Fiete D. Structure of the complex oligosaccharides of fetuin. The Journal of biological chemistry. 1979;254:789–795. [PubMed] [Google Scholar]