Computational simulations are used to probe potential mechanisms through which nuclear dynein organizes forces in an anisotropic manner to promote centrosome separation. Two mechanisms are key: one relies on steric interactions between microtubules and centrosomes and the other on the initial position of centrosomes in the cell.
Abstract
Centrosome separation along the surface of the nucleus at the onset of mitosis is critical for bipolar spindle assembly. Dynein anchored on the nuclear envelope is known to be important for centrosome separation, but it is unclear how nuclear dynein forces are organized in an anisotropic manner to promote the movement of centrosomes away from each other. Here we use computational simulations of Caenorhabditis elegans embryos to address this fundamental question, testing three potential mechanisms by which nuclear dynein may act. First, our analysis shows that expansion of the nuclear volume per se does not generate nuclear dynein–driven separation forces. Second, we find that steric interactions between microtubules and centrosomes contribute to robust onset of nuclear dynein–mediated centrosome separation. Third, we find that the initial position of centrosomes, between the male pronucleus and cell cortex at the embryo posterior, is a key determinant in organizing microtubule aster asymmetry to power nuclear dynein–dependent separation. Overall our work reveals that accurate initial centrosome position, together with steric interactions, ensures proper anisotropic organization of nuclear dynein forces to separate centrosomes, thus ensuring robust bipolar spindle assembly.
INTRODUCTION
How organelles are positioned within the confines of the cell is of paramount importance for proper cell physiology. One striking illustration of such importance is the process of centrosome separation that occurs at the onset of mitosis in animal cells (reviewed in Tanenbaum and Medema, 2010). Initially, the two centrosomes present at this stage of the cell cycle are positioned close to one another, near the outer nuclear envelope. During prophase, the two centrosomes separate along the nuclear surface, reaching opposite positions on the nucleus, thus ensuring efficient bipolar spindle assembly and faithful chromosome segregation. How centrosomes separate in an accurate and robust manner remains incompletely understood.
Microtubules and the minus end–directed dynein motor complex (hereafter referred to as dynein for simplicity) are important for centrosome separation in a range of metazoan organisms (reviewed in Tanenbaum and Medema, 2010). For instance, human cells partially depleted of kinesin-5 rely on dynein at the nuclear envelope for efficient centrosome separation (Raaijmakers et al., 2012). In one-cell Caenorhabditis elegans embryos, dynein is essential for centrosome separation (Gönczy et al., 1999), a requirement that is exerted through two distinct protein pools: dynein anchored at the cell cortex and dynein anchored at the nuclear envelope (De Simone et al., 2016).
Some degree of anisotropy in the organization of dynein-dependent forces is fundamental for centrosome separation to occur. Indeed, if dynein motors are distributed homogeneously and microtubules grow uniformly in all directions, forces exerted on the microtubules asters will balance each other on average, resulting in a null net force. Whereas stochastic forces, including those exerted by dynein motors, could result in slight diffusion-like separation of the two centrosomes, mechanisms that impart robust anisotropy to the system are likely to be required to ensure efficient centrosome separation. For cortical dynein, flows of the actomyosin network that occur in a directional manner and translocate the cortically bound motor protein provide a source of anisotropy in one-cell C. elegans embryos (De Simone et al., 2016). By contrast, the mechanisms ensuring anisotropic organization of nuclear dynein forces are not well understood in the worm or in other organisms.
The fact that the contribution of nuclear dynein can be clearly separated from that of cortical dynein in the one-cell C. elegans embryo provides a favorable setting to dissect the underlying mechanisms. Here we use computational simulations with a previously developed model of centrosome separation to probe three mechanisms by which nuclear dynein forces might contribute to anisotropic organization of forces to separate centrosomes.
RESULTS AND DISCUSSION
In the one-cell C. elegans embryo, as in most metazoan species, centrioles are contributed to the embryo strictly by the sperm. As a result, centrosomes in the newly fertilized worm zygote are associated with the male pronucleus, confined between it and the cell cortex at the future posterior of the embryo (Supplemental Video S1). Centrosome separation begins in that location and then continues as the two microtubule asters grow in size and move away from the posterior cortex, together with the associated male pronucleus (Supplemental Video S1).
In previous work (De Simone et al., 2016), we developed a comprehensive computational model of centrosome separation in the one-cell C. elegans embryo that relies on parameters determined experimentally or estimated within a sensible range (Supplemental Table S1). Of importance, this model has been challenged and validated with experimental data, both from the wild-type and a variety of perturbation conditions, including depletion of select pools of dynein (De Simone et al., 2016). Of particular importance, simulations and experiments concurred to show that centrosome separation requires the combined action of cortical and nuclear dynein, with the process failing entirely in embryos lacking both components (De Simone et al., 2016).
Here we use this computational model to investigate mechanisms by which nuclear dynein might contribute to centrosome separation. To focus strictly on these mechanisms, we simulated embryos lacking cortical dynein, such as upon depletion of the cortical dynein-anchoring components GOA-1 and GPA-16 (Nguyen-Ngoc et al., 2007), as well as lacking the female pronucleus, a condition not accessible experimentally but one that can be readily simulated (Figure 1, A–C, and Supplemental Video S2). How could nuclear dynein and microtubules organize forces in an anisotropic manner to promote centrosome separation? We considered and challenged computationally three possible mechanisms underlying such anisotropy (Figure 1D).
FIGURE 1:
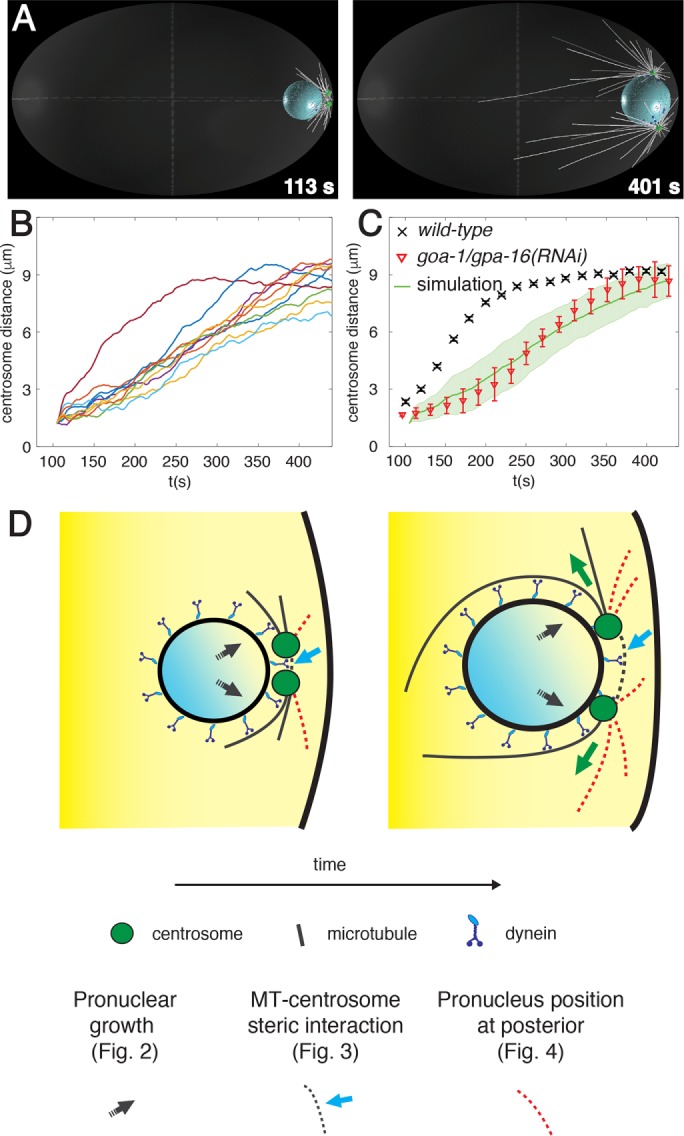
Nuclear dynein–driven centrosome separation in the one-cell C. elegans embryo. (A) Snapshots from computer simulation of centrosome separation driven by dynein at the nuclear envelope. Here and thereafter, male pronucleus (blue sphere), nuclear dynein (blue dots), centrosomes (green disks), microtubules (white lines), and cortex (light gray ellipse in transparence) are depicted. For visual clarity, only bound motors and one-fourth of microtubules are shown. All simulated embryos are 50 µm long. For consistency with previous work (De Simone et al., 2016), the beginning of the simulations in all cases corresponds to time = 104 s of the synchronized experimental data set in De Simone et al. (2016), where more information can be found as well. See also Supplemental Video S2. (B, C) Quantification of centrosome separation driven by nuclear dynein in computer simulations. Centrosome distance curves (n = 10) as a function of time (B), as well as their averages with SD (C), compared with experimental data of centrosome separation in one-cell embryos expressing the centrosomal marker GFP::TAC-1 and depleted of cortical dynein using goa-1/gpa-16(RNAi) (n = 20; data from De Simone et al., 2016). (D) Potential mechanisms organizing nuclear dynein forces anisotropically and thus favoring centrosome movement away from each other (left, onset of separation; right, during separation). Black arrow: growth of the male pronucleus (Figure 2); cyan arrow and gray dashed lines: steric interactions between microtubules emanating from one centrosome and the other centrosome (Figure 3); red dashed lines: position of centrosomes confined between the male pronucleus and the posterior cortex (Figure 4).
The first potential mechanism stems from the growth of the male pronucleus that is concomitant with centrosome separation (Figure 1D, black arrows; De Simone et al., 2016). Because centrosomes are anchored to the nuclear envelope via dynein (Malone et al., 2003), the expansion of the nuclear volume could conceivably move the two centrosomes apart from one another, thus resulting in an increased distance between them. To investigate this possibility, we simulated centrosome dynamics in a situation in which the male pronucleus does not increase in size. As shown in Figure 2, A–C, as well as in Supplemental Table S2, which also reports statistical analysis of the entire data set, we found that centrosome separation velocity is slightly slower and the final separation distance slightly smaller in this case than in the control condition. We conclude that centrosomes separate less efficiently on a small male pronucleus.
FIGURE 2:
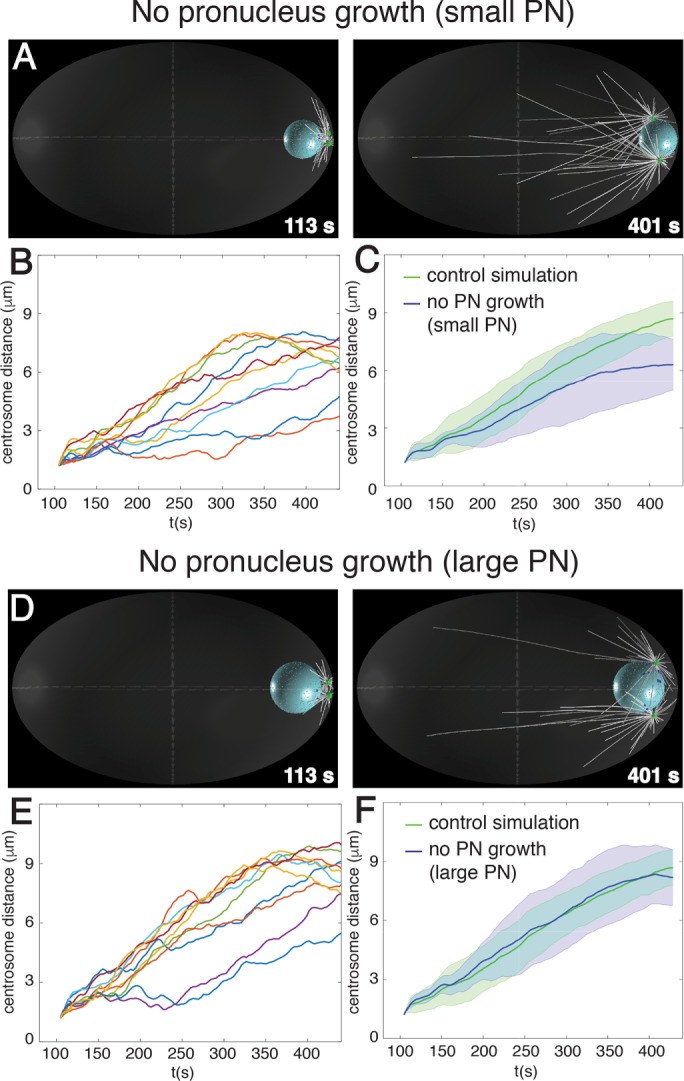
Nuclear expansion does not generate nuclear dynein–driven separation forces. Snapshots (A, D) and quantification (B, C, E, F) from computer simulation of centrosome separation in which the male pronucleus has a constant small size (A–C) or a constant large size (D–F). Individual centrosome distance curves (n = 10) as a function of time (B, E), as well as their averages with SD (C, F, blue). Here and hereafter, centrosome separation in control simulations (see Figure 1B) is shown for comparison (green).
Next we addressed whether nuclear growth contributes to centrosome separation by exerting an actual force onto the two anchored centrosomes or instead merely by allowing the system to reach a larger maximum distance between them. To distinguish between these possibilities, we repeated the simulation with a male pronucleus also of constant volume but with the final, large, size. We found in this case that the pace and maximal extent of centrosome separation are similar as those in the control condition (Figure 2, D–F, and Supplemental Table S2). We conclude that growth of the male pronucleus contributes to centrosome separation merely by increasing the maximal distance that can be achieved between the two centrosomes.
We considered a second potential mechanism contributing to anisotropy of forces during centrosome separation: steric interaction between microtubules and centrosomes (Figure 1D, cyan arrow and gray dashed lines). Microtubules originating from one centrosome and directed toward the other one eventually collide with it, thus starting to push on it through microtubule polymerization forces. It is clear that such pushing forces cannot separate centrosomes on their own because centrosomes do not separate in embryos depleted of both cortical and nuclear dynein but with intact microtubules (Gönczy et al., 1999; De Simone et al., 2016). Nevertheless, such pushing forces might trigger catastrophe of these very microtubules, resulting in their depletion from the region between centrosomes and thereby to a slight asymmetry of microtubule asters, as observed experimentally in reconstitution assays (Janson et al., 2003). To investigate computationally the contribution of steric interactions between microtubules and centrosomes in the one-cell C. elegans embryo, we simulated centrosome separation in their absence and found that centrosome separation velocity is reduced to a slight but significant extent (Figure 3, A–C, Supplemental Table S2). Of interest, in addition, we noted that the process becomes less robust in this condition, being strongly impaired in a fraction of the simulations (Figure 3B, three bottommost traces), resulting in a large SD from the average centrosome separation curve (Figure 3C).
FIGURE 3:
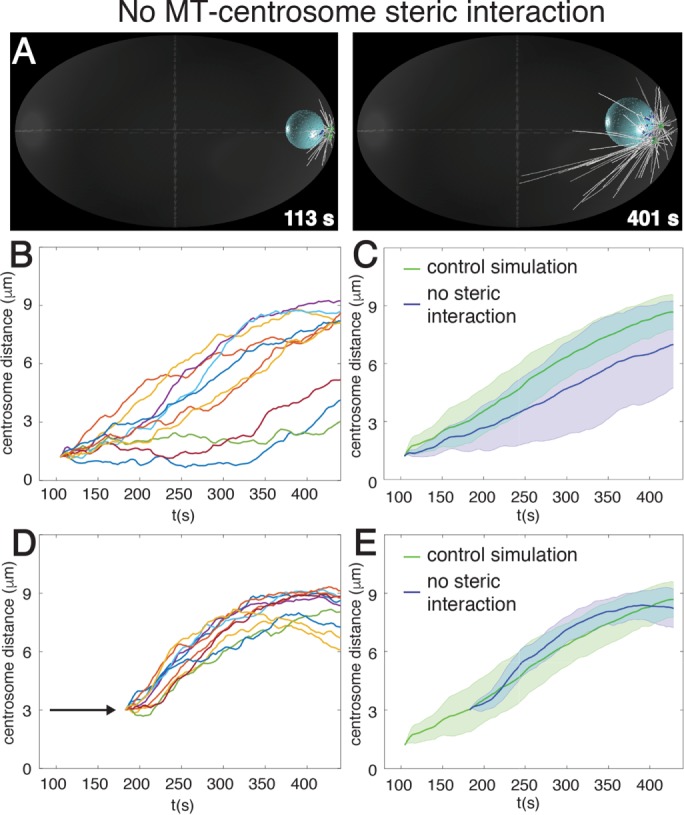
Steric interaction between microtubules and centrosomes promotes centrosome separation onset. Snapshots (A) and quantification (B, C) from computer simulation of centrosome separation in which microtubules do not interact sterically with centrosomes. Centrosome distance curves (n = 10) as a function of time (B), as well as their averages with SD (C, blue). (D, E) Quantification of centrosome separation in computer simulations in which microtubules do not interact sterically with centrosomes, as in A–C, but where centrosomes are positioned 3 µm from each other at the beginning of the simulation. Centrosome distance curves (n = 10) as a function of time for individual simulations (D), as well as their averages with SD (E, blue).
Simple geometrical considerations indicate that if microtubules grow at the same average pace in every direction, then the fraction of microtubules within each aster that is oriented toward the other one decreases quadratically with the distance between centrosomes. Therefore, we reasoned that steric interactions between microtubules and centrosomes might contribute to the force imbalance at the onset of centrosome separation when centrosomes are close to each other but become negligible thereafter. To test this idea, we performed simulations again without steric interactions between microtubules and centrosomes but now positioning centrosomes 3 µm away from one another at the beginning of the simulation in order to move past the normal onset stage. Strikingly, we found that centrosome separation proceeds in an efficient and robust manner in this case, just like in the control condition (Figure 3, D and E, and Supplemental Table S2). Overall we conclude that steric interactions between microtubules and centrosomes contribute to the robustness of centrosome separation at the onset of the process but become dispensable thereafter.
A third potential mechanism that we considered is rooted in the initial position of the two centrosomes, which are confined between the male pronucleus and the cell cortex at the very posterior of the embryo (Figure 1D, red dashed microtubules). Owing to this spatial constraint, posterior-directed microtubules will encounter the cell cortex and bend or undergo catastrophes more readily than those directed anteriorly, which are free to polymerize toward the embryo center. Because these longer microtubules offer more binding sites to nuclear dynein motors than the shorter ones, they will exert stronger forces than those present between the two centrosomes; such a slight asymmetry of microtubule asters should favor centrosome separation. To test this hypothesis, we simulated centrosome dynamics when placing the male pronucleus and associated centrosomes in the cell center, where all microtubules can grow to become long. Of importance, we found that centrosome separation is severely compromised in this case (Figure 4, A–C, and Supplemental Table S2). Furthermore, we simulated intermediate distances between the posterior and the center and found that the pace of centrosome separation decreases rapidly once the male pronucleus and associated centrosomes are located away from the posteriormost position (Figure 4D).
FIGURE 4:
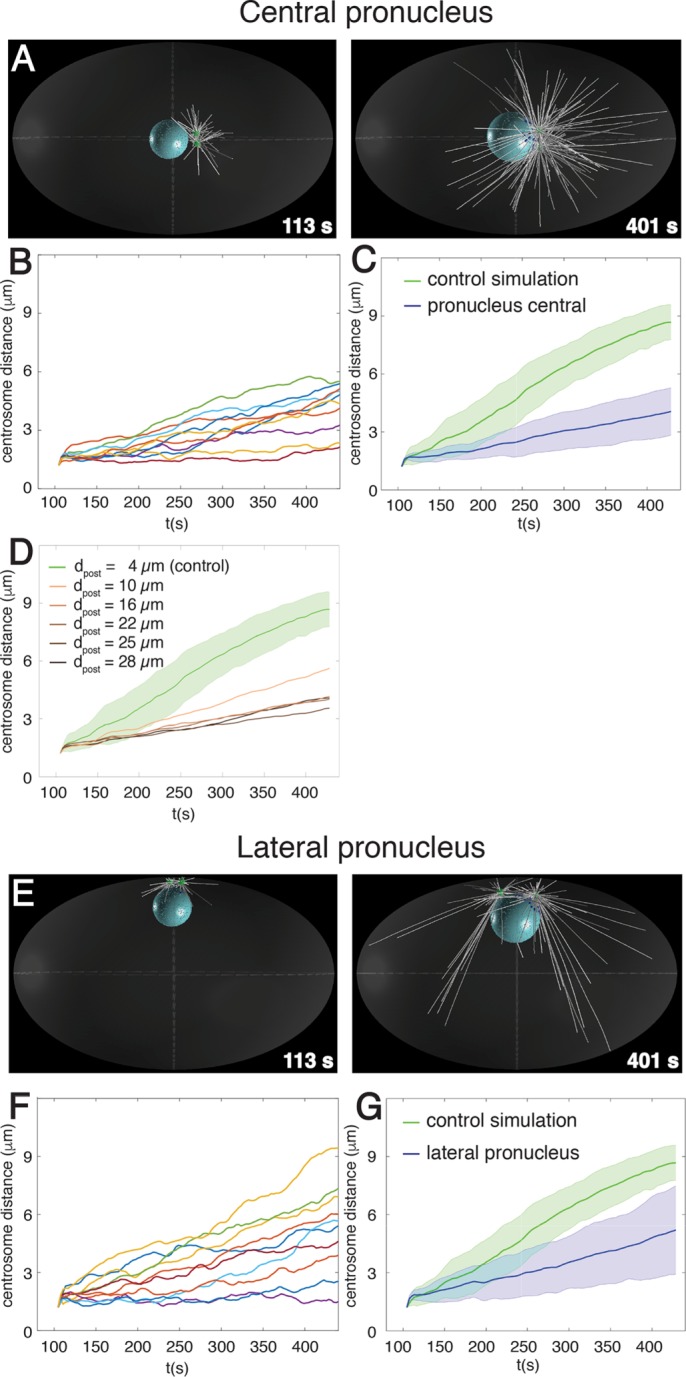
Position of pronucleus–centrosomes complex organizes nuclear dynein forces to favor centrosome separation. Snapshots (A) and quantification (B, C) from computer simulation of centrosome separation in which the male pronucleus and associated centrosomes are located initially in the center of the embryo. Centrosome distance curves (n = 10) as a function of time (B), as well as their averages with SD (C, blue). (D) Quantification of centrosome separation in computer simulations in which the male pronucleus and associated centrosomes are located initially in the indicated positions along the anterior–posterior embryonic axis; dpost is the distance of the center of the male pronucleus from the posterior cortex (4 µm in control condition). Average simulated centrosome distance curves as a function of time (n = 10 in each case; for visual clarity, SDs are not shown). (E–G) Snapshots (E) and quantification (F, G) from computer simulation of centrosome separation in which the male pronucleus and associated centrosomes are located initially laterally. Centrosome distance curves (n = 10) as a function of time (F), as well as their averages with SD (G, blue).
Next we set out to explore whether the particular curvature of the cell cortex present at the posterior pole is important or whether confining the centrosomes between the male pronucleus and the cortex may suffice, regardless of the actual curvature. To this end, we conducted another set of simulations in which the male pronucleus and associated centrosomes are placed next to the lateral cortex, which exhibits a smaller curvature than the posterior cortex. As reported in Figure 4, E–G, and Supplemental Table S2, we found that centrosome separation is significantly compromised in this case compared with the control condition (Figure 4, E–G, and Supplemental Table S2). Overall these findings indicate that spatial confinement of centrosomes at the posteriormost location observed in the wild type is critical for efficient centrosome separation.
Do the two anisotropy-generating mechanisms described so far suffice to explain centrosome separation powered by nuclear dynein and microtubules? To address this question, we simulated centrosome dynamics in embryos lacking these two mechanisms. Strikingly, we found that centrosome separation is essentially abolished when steric interaction does not occur and the male pronucleus plus the associated centrosomes are positioned in the cell center (Figure 5, Supplemental Table S2, and Supplemental Video S2). Overall we conclude that two distinct and partially redundant mechanisms together generate anisotropic nuclear dynein forces, thus ensuring robust centrosome separation in one-cell C. elegans embryos.
FIGURE 5:
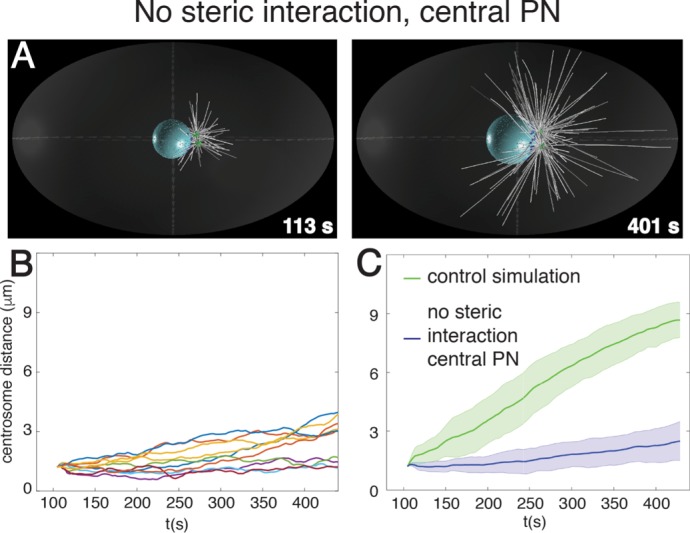
Two mechanisms together organize nuclear dynein forces to drive centrosome separation. Snapshots (A) and quantification (B, C) of centrosome separation in simulations in which microtubules do not interact sterically with centrosomes and the male pronucleus plus associated centrosomes are located in the center of the embryo. Individual centrosome distance curves (n = 10) as a function of time (B), as well as their averages with SD (C, blue). See also Supplemental Video S2.
In conclusion, computational simulations enabled us to readily probe the contribution of three mechanisms that could have potentially organized nuclear dynein forces to separate centrosomes. Computational modeling offers the potential to investigate the behavior of complex systems in a rigorous manner based on the theory of cytoskeletal mechanics and thus reach conclusions that would often be difficult to derive using intuitive reasoning alone. Although a wealth of mutant and RNA interference conditions is available in C. elegans to analyze cell division processes, computational simulations enable one to investigate theoretically potential mechanisms that can be difficult to dissect experimentally. Moreover, whereas experimental analysis of the consequences of gene inactivation is well suited to address whether a feature is necessary for a given process, computational simulations are optimal for testing whether it is also sufficient. In the case of centrosome separation, our computer simulations indicate that two features, namely steric interactions between microtubules and centrosomes, as well as spatial confinement of centrosomes, are together sufficient to explain how anisotropic forces can be exerted by nuclear dynein to separate centrosomes. These findings imply that in embryos with a single centrosome, for instance, after fertilization by zyg-1 or sas-5 mutant sperm (O’Connell et al., 2001; Delattre et al., 2004), movements of this single centrosome should be analogous to those taking place when both centrosomes are present. However, because a measure of centrosome distance is not available in embryos with a single centrosome, it is difficult to distinguish centrosome movements due to forces that would normally drive separation from those due to other forces, including stochastic forces and those that bring centrosome to the cell center.
Analogous strategies to those uncovered here may be used in other systems to separate centrosomes. Thus, in Drosophila embryos, centrosomes are also positioned between the cortex and the nucleus, being attached to the nuclear envelope using dynein (Robinson et al., 1999; Cytrynbaum et al., 2005). Moreover, nuclear dynein can drive centrosome separation in mammalian cells derived by directed evolution after kinesin-5 impairment (Raaijmakers et al., 2012). Perhaps also in that case steric interactions between microtubules and centrosomes, as well as nuclear position within the cell, are critical to ensure anisotropy and robust centrosome separation. An additional mechanism by which microtubule aster asymmetry can be generated has been proposed to be at play in centrosome movement in fly, frog, and fish embryos (Cytrynbaum et al., 2005; Wühr et al., 2010). In this mechanism, the presence at centrosomes of a negative regulator of microtubules, such as a protein of the kinesin-13 depolymerizing motor family (reviewed in Ems-McClung and Walczak, 2010), could preferentially affect microtubules between the two centrosomes, the separation of which would thereby be fostered.
Given the importance of accurate centrosome separation for proper spindle assembly and faithful chromosome segregation, a combination of partially redundant mechanisms, as revealed here through computational simulations, might have been indeed favored during evolution.
MATERIALS AND METHODS
Computational simulations
Computer simulations were performed using the open source project Cytosim (Nedelec and Foethke, 2007; available at https://github.com/nedelec/cytosim), as described in detail previously (De Simone et al., 2016). In brief, the motion of elastic fibers, that is, microtubules, and solids, that is, centrosomes and male pronucleus, in a viscous fluid is simulated by overdamped Langevin equations. All stochastic events (motor binding, catastrophes, nucleation) are generated as first-order random events. The cell boundary, a 50 × 30 × 30 μm ellipsoid, is considered to exert soft confinement forces on microtubules, centrosomes, and male pronucleus. Similarly, a soft steric interaction applies to microtubules and male pronucleus, as well as microtubules and centrosomes (unless stated otherwise), preventing these objects from overlapping. Interactions between microtubules are not considered.
Microtubules are flexible fibers nucleated by centrosomes that switch from growing to shrinking phases with constant probabilities. Centrosomes are spheres 1 μm in diameter covered by microtubule nucleation sites that, when empty, nucleate a microtubule with a constant probability.
The male pronucleus is a sphere that grows in radius at a constant rate. Dynein motors are distributed on the surface of the male pronucleus with a constant density. Unbound motors can bind microtubules within their binding range with a certain rate. Each microtubule can bind multiple dynein motors simultaneously. Dynein motors have a base fixed on the pronucleus and can bind to microtubules that are within their binding range. When a motor is bound to a microtubule, its attachment point moves along the microtubule length with a linear force velocity relationship. A linear elastic force is exerted between the base and the attachment point of a bound motor.
At the start of the simulation, centrosomes are located at the posterior side of the male pronucleus and are separated by 1.2 μm, unless stated otherwise. No microtubules are polymerized, and thus all dynein motors are unbound.
The simulation parameters are the same as those used in De Simone et al. (2016) and are also provided in Supplemental Table S1 here.
Statistical analysis of centrosome separation in different conditions
To quantitatively compare centrosome separation in different simulations, we determined the following three signature features of centrosome separation from the simulated centrosome distance curves: onset time, separation velocity, and plateau distance. To this end, we pooled the centrosome distance curves as a function of time for each simulation condition and fitted them with the effective model
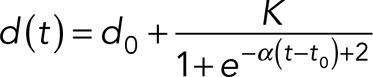 |
where t is time, d(t) is the centrosome distance, d0 is the initial centrosome separation, α is the separation rate, and K + d0 is the plateau distance. t0 corresponds to centrosome separation onset time, defined as the intercept on d = d0 of the tangent in the inflection point for which d = d0 +(K/2). Parameters from different conditions were compared using the zi test.
Supplementary Material
Acknowledgments
We are very grateful to Antoine Spahr and Coralie Busso for help in conducting exploratory experiments related to this work. We thank Niccolò Banterle, Simon Blanchoud, and François Nédélec for comments on the manuscript. We thank François Nédélec also for help in developing the computational model and Niccolò Banterle for fruitful discussions. This work was supported by the Swiss National Science Foundation (3100A0-122500/1 and 31003A_155942). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-12-0823) on July 12, 2017.
REFERENCES
- Cytrynbaum EN, Sommi P, Brust-Mascher I, Scholey JM, Mogilner A. Early spindle assembly in Drosophila embryos: role of force balance involving cytoskeletal dynamics and nuclear mechanics. Mol Biol Cell. 2005;16:4967–4981. doi: 10.1091/mbc.E05-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Simone A, Nédélec F, Gönczy P. Dynein transmits polarized actomyosin cortical flows to promote centrosome separation. Cell Rep. 2016;14:2250–2262. doi: 10.1016/j.celrep.2016.01.077. [DOI] [PubMed] [Google Scholar]
- Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gönczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Walczak CE. Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin Cell Dev Biol. 2010;21:276–282. doi: 10.1016/j.semcdb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;147:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, De Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J Cell Biol. 2003;161:1029–1034. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/s0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- Nedelec F, Foethke D. Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys. 2007;9:427. [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gönczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RGHP, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Wühr M, Tan ES, Parker SK, Detrich HW, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol. 2010;20:2040–2045. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


