Abstract
The biosynthetic potential of marine-sediment-derived Gram-negative bacteria is poorly understood. Sampling of California near-shore marine environments afforded isolation of numerous Gram-negative bacteria in the Proteobacteria and Bacteriodetes phyla, which were grown in the laboratory to provide extracts whose metabolites were identified by comparative analyses of LC-mass spectrometry and MSn data. Overall, we developed an assemblage of seven bacterial strains grown in five different media types designed to coax out unique secondary metabolite production as a function of varying culture conditions. The changes in metabolite production patterns were tracked using the GNPS MS2 fragmentation pattern analysis tool. A variety of nitrogen-rich metabolites were visualized from the different strains grown in different media, and strikingly, all of the strains examined produced the same new, proton-atom-deficient compound, 1-methyl-4-methylthio-β-carboline (1), C13H12N2S. Scaleup liquid culture of Achromobacter spanius (order: Burkholderiales; class: Betaproteobacteria) provided material for the final structure elucidation. The methods successfully combined in this work should stimulate future studies of molecules from marine-derived Gram-negative bacteria.
Graphical abstract
Harnessing the biosynthetic machinery of marine-derived bacteria should open the door to expand on the treasure trove of bioactive molecules obtained over the past 40+ years from terrestrial strains.1 An insightful progress report by Williams2 highlighted that from 1997 to 2008 marine natural products-driven research on Gram-negative nonphotosynthetic bacteria accounted for only 16% of 659 compounds reported, illustrating that research on such taxa appears underexplored. A similar situation is shown in literature published during 2014.3 A total of 164 new marine compounds were described from four heterotrophic bacterial phyla consisting of 71.3% from Gram-positive Actinobacteria vs 15.9% from Gram-positive Firmicutes, 10.4% from Gram-negative Proteobacteria, 1.2% from Gram-negative Bacteriodetes, and 1.2% via eDNA workup of a sediment sample. Similarly, we discussed in a 2014 review4 that the chemical study of marine-derived Gram-negative bacteria is underdeveloped. This was followed by our 2105 report describing strategies to use California near-shore environments as a source of a Gram-negative bacterium that in culture afforded kailuin cyclic depsipeptides.5 Further underscoring new opportunities is a 2015 Jensen–Fenical6 team publication describing a unique collection of 20 Gram-negative marine-derived strains from the Bacteroidetes and Proteobacteria phyla, which produced aryl alkaloids. Another relevant recent finding was the disclosure of salimyxins A/B from a cultured Gram-negative (phylum: Proteobacteria) strain,7 isolated from a Santa Barbara, California, beach sediment. Adding to this record was the isolation of the tunicate-derived cytotoxin didemnin B from cultures of the Gram-negative Tistrella (phylum: Proteobacteria), isolated by two independent research groups during the study of a marine sediment from Japan and a Red Sea water column sample.8
Described herein are results on the further mining of a UC Santa Cruz library of Gram-negative bacterial strains for chemical diversity. Our collection now includes samples representing more than 100 operational taxonomic units (OTUs), and the work flow involves a three-step process. First, we employ low-nutrient, long-incubation isolation methods to maximize the variety of isolatable Gram-negative strains. This is followed up by taxonomic identification to limit redundancy and de-emphasize exploration of common Gram-negative rods of the Proteobacteria, especially Vibrio, Pseudoalteromonas, and Pseudomonas. Second, each unique strain is grown in a panel of five liquid media (see Table S1) with the idea that diversity in primary metabolism and environmental stress may trigger unique secondary metabolite production. Finally, extracts from each strain and media condition are analyzed using the GNPS MS/MS9 networking strategy introduced in 2012 by Dorrestein–Bandeira.9a By tagging each extract’s mass spectra with its strain and media attributes, especially when accurate (experimental error ±0.003 amu) MS1 and MS2 data are obtained, we can explore dereplication–identification of metabolites produced in a strain- or media-dependent manner. Reported below are results from a campaign to chemically mine Gram-negative bacteria driven by the characterization (a) of known molecules rich in nitrogen atoms, (b) of compounds possessing new structural diversity, and (c) of substances whose scaffolds can potentially answer questions about the symbiotic relationships driving marine microbial heterocycle biosynthesis.
RESULTS AND DISCUSSION
In this project we sought new approaches to gain an efficient understanding of the biosynthetic potential of new sediment-derived Gram-negative strains being added to our collection. Many strains were identified as distinct OTUs10 by 16S rRNA and included (a) 54 cultivable Gram-negative bacteria obtained at 41 sites in the Monterey Bay, California, and (b) 96 Gram-negative bacterial strains responsive to culturing from sampling at 66 beaches in Humboldt, California. Overall, a subset of 80 strains from this cohort were characterized as members of the phyla Proteobacteria (P) and Bacteriodetes (B) as follows: Monterey Bay strains P = 21, B = 18 vs Humboldt strains P = 36, B = 5. Signature metabolites from marine-derived members of both phyla are poorly understood. Also, the relative presence of many samples from the Bacteriodetes of the Monterey Bay was unexpected and presented an opportunity for additional discovery. We needed a chemodiversity prioritization filter to rank this library of >150 samples, and the strategy of genome mining provided one possible pathway. Eventually, two methods were chosen to shift from a time-consuming trial- and-error analysis, and these involved (a) aggressive use of MSn data processed by the GNPS tools9 for comparative metabolomics analysis and/or (b) in silico analysis of draft genome sequencing data using tools such as antiSMASH11 to assess biosynthetic richness via the presence of putative secondary metabolite gene clusters. We employed the former strategy since our strains were not well represented among currently sequenced microbial genomes in public repositories and large-scale sequencing efforts could not be justified without motivating chemistry. In addition, successful use of bioinformatics tools to pinpoint specific molecules through gene cluster analysis of Gram-negative marine-derived bacteria has been modest.12
The kailuin cyclic depsipeptide mixtures5 biosynthesized from P. halotolerans (Gram-negative strain M128SB283Ax), obtained from Monterey Bay, California, coastal littoral zone sediments, provided an excellent test-bed for additional proof of concept studies. Overall, five samples, along with two Photobacterium halotolerans (strains M128SB283Ax and M132NC031Ax: phylum Proteobacteria, class γ-proteobacteria), were chosen for MS-based metabolomics studies. These strains are (i) Proteobacteria Achromobacter spanius (class β-Proteobacteria), Pseudomonas benzenivorans (class γ-Proteobacteria), and Pseudoaltermonas elyakovii (class γ-Proteobacteria) and (ii) Bacteriodetes Pontibacter korlensis (class Cytophaga) and Cellulophaga baltica (class Flavobacteria). The criteria for assembling this set included (a) their confirmed 16s rRNA sequences substantiating their classification as Gram-negative bacteria, (b) the availability of multiple extracts from cultures grown in our five standard media, and (c) the ability to reculture each sample from a cryopreserved sample. Aside from scant work on P. halotolerans5 and P. elyakovii,13 there are no reports on the biosynthetic products from these other four species.
The process to assess the biosynthetic richness of each strain involved metabolite profiling by mass spectrometry molecular networking,9 and final profiles created for three samples are shown in Figure 1. There has been intense interest by the natural products community in using the tool called Global Natural Product Social Molecular Networking (GNPS).9b Most studies have involved comparing data collected against benchmark compounds housed in individual laboratories or those in shared libraries (described on the Web site http://gnps.ucsd.edu/). Our implementation of this strategy was different. It involved searching for comparative uniqueness among metabolites potentially biosynthesized from our collections by subjecting each strain to culturing in the standard media types (summarized in Table S1). It is important to note that three of these (coded: M1, M3, M4) are made up with “seawater salts” and a “trace elements solution”. Alternatively, two others (coded: M2, M5) are constituted with DI-H2O only.
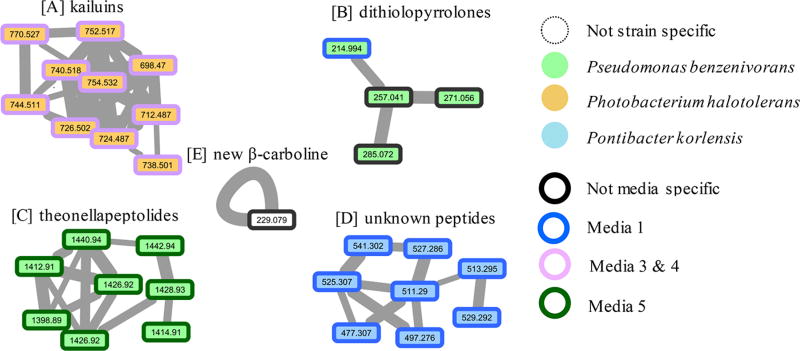
Figure 1.
Examples of molecular networks visualized after analysis of accurate MS2 data via the Global Natural Products Social Molecular Networking (GNPS; http://gnps.ucsd.edu) tools. The networks were obtained from analyses of crude extracts from near-shore-derived Gram-negative bacteria grown in five different media (see Table S1). Annotations depict variation in metabolite production based on strain and media conditions. Line thickness between nodes describes the similarity of the linked parent masses’ MS2 spectra. Panels illustrate trends as follows: (A) P. halotolerans yields kailuins in both media 3 and 4 (e.g., kailuin D, [M + H]+ m/z 752.517); (B) P. benzenivorans yields some dithiolopyrrolones, particularly in medium 1 (e.g., holomycin, [M + H]+ m/z 214.994); (C) P. benzenivorans yields theonellapeptolides in medium 5 (e.g., theonellapeptolide Ib, [M + Na]+ m/z 1440.94); (D) P. korlensis yields polypeptides in medium 1; and (E) six strains produce 1-methyl-4-methylthio-β-carboline (1) ([M + H]+ m/z 229.079) in all media including those shown here and Achromobacter spanius, Pseudoaltermonas elyakovii, and Cellulophaga baltica (see Figure S14).
As noted above, each of the seven strains were cultured in the five standard liquid media (Table S1), which provided a large array of individual samples plus five media “blanks”. We obtained high-accuracy MS1 (±0.003 amu) data coupled with medium-accuracy MS2 fragmentation data for the 10 most intense ions from each primary scan and used these spectra as input for mass spectral networking via the GNPS tool. A sample of the more distinctive networks obtained is represented by the interconnected nodes depicted in panels A–D of Figure 1. In contrast to the complexity of these networks the result shown in panel E contained only one node with an m/z 229.079. This type of network is difficult to recognize among the hundreds of other single-node clusters that are commonly obtained in GNPS network outputs. We flagged this mass as potentially important based on its high peak area in strain M125SB302Ax and found it was present as a very minor component in the extracts of all strains listed above based on extracted ion chromatograms of the original spectra. However, the low relative ion intensity prevented its selection for MS2 in three strains, precluding its inclusion in the GNPS network for those strains (Figure 1). The types of metabolites present in A– D of Figure 1 will be discussed first followed by an explanation of the unusual circumstance and molecular structure of the [M + H]+ m/z 229 compound produced by all the strains in multiple media types.
Analyzing the MS2 network results obtained for P. halotolerans (strain M128SB283Ax) seemed a logical first step because we had rigorously explored this strain under various culture conditions and found it reproducibly afforded kailuins B–E, G, and H, which could be isolated in good yields.5 The compounds identified from semipreparative HPLC-based isolations were nearly identical to those observed using the network data (see panel A) for P. halotolerans grown in both media M3 and M4. Alternatively, these compounds were not observed when this strain was grown in media M1, M2, and M5. Specifically, the six kailuin compounds (B, C, D, E, G, H) could be correlated to four of the 10 nodes visualized in panel A containing accurate (±0.003 amu) MS1 data including kailuins B and C ([M + H]+ m/z 726.502), kailuin D ([M + H]+ m/z 752.517), kailuins E and H ([M + H]+ m/z 754.532), and kailuin G ([M + H]+ m/z 740.518). In addition, six other analogues, not purified during our isolation work, could be inferred as obtainable via the [M + H]+ MS1 data in other network nodes: kailuin D·18 ([M + H]+ m/z 770.527), kailuin C (or B) ·18 ([M + H]+ m/z 744.511), tetradehydro kailuin G ([M + H]+ m/z 736.501), didehydro kailuin C (or B) ([M + H]+ m/z 724.487), kailuin A ([M + H]+ m/z 698.470), and unknown kailuin Z ([M + H]+ m/z 712.487).
The next action was to decipher the profiles shown in Figure 1, panels B–D. The sample P. benzenivorans produced different compounds dependent on the media type. Revealed in panel B is that holomycin ([M + H]+ m/z 214.994, C7H7N2O2S2)14a is produced in M1, and its presence in the culture broths was verified by isolation and comparison of its NMR data to that in the literature. Different, yet related heterocycles are produced in M1 and M4, including a trio of xenorhabdin15 −CH2− homologues [new: [M + H]+ m/z 257.041 (C10H13N2O2S2); xeno-1: [M + H]+ m/z 271.056 (C11H15N2O2S2); and xeno-2: [M + H]+ m/z 285.072 (C12H17N2O2S2)]. Compounds of this dithiolopyrrolone class have been isolated from a number of Gram-positive and Gram-negative bacteria, particularly from Actinobacteria and γ-Proteobacteria.14b There are additional sulfur-containing heterocycles under study from the large-scale culturing of this strain, and they will be described elsewhere. By contrast, culturing this strain in medium M5 produced a very different cluster of compounds visualized in the eight molecular network nodes depicted in panel C. The metabolite with [M + Na]+ m/z 1440.94 can be proposed to be theonellapeptolide Ie (C71H127N13O16), which we previously isolated from Indo-Pacific Theonella sponges.16 However, this observation must be regarded as very preliminary because our repeated attempts to isolate these compounds from large-scale reculturing have, to date, been unsuccessful. Panel D depicts the elaboration of undescribed peptides by P. korlensis when grown in medium M1, also reproducibly produced during large-scale culturing. The molecule with [M + H]+ m/z 511.2906 has a formula of C28H39N4O5 and can be sequenced as Val-Phe-Val-Phe, justified by accurate MS2 fragmentation data. Surprisingly, this compound is not in the literature, and the same is evident for the other compounds visualized in the network of panel D, which also have acyclic peptide-type MS2 fragmentation patterns.
An unusual circumstance is associated with the simple network pattern of Figure 1 panel E, because the single m/z node associated with an [M + H]+ m/z 229.079 was seen from the culturing of several distinct strains. This new compound, eventually determined to be 1-methyl-4-methylthio-β-carboline (1), was always observed (by LC-MS or direct isolation) as a minor component from seven strains (three shown in Figure 1 and four in Figure S14), each grown in multiple media (see Experimental Section for the strain list). Also shown in Figure S16 is that for the medium 4 control (no inoculation with a Gram-negative strain) a peak corresponding to m/z 229.079 is not observed. The nonselective strain/media outcome observed here was in sharp contrast to the patterns discussed above for Figure 1, panels A–D, wherein each metabolite network varied as a function of the bacterium and culture medium. Further illustrating the latter situation is that the kailuins (Figure 1 panel A) were observed only from P. halotolerans (strain M128SB238Ax) cultured in either media M3 or M4. The steps in the isolation and purification of 0.9 mg of 1 from a 20 L liquid culture (M4) of A. spanius (strain M125SB302Ax) are shown in Figure S14.
An initial indication that the characterization of compound 1, with isotopic mass = 228.0721 and subsequently deduced molecular formula C13H12N2S, would not be a straightforward dereplication came from two observations. First, 71 hits were obtained from a Dictionary of Natural Products CHEMnet-BASE17 search for compounds with masses differing from the isotopic mass of 1 by ±0.007 amu. Second, none of these known molecules fit the NMR data for 1 shown in Table 1. Establishing the correct molecular formula of 1 required several steps. Inspecting the accurate mass [M + H]+ cluster m/z 229.0786 (100%), 230.0816 (14.6%), and 231.0741 (4.69%) (Figure S1) required one S atom (enhanced intensity of the [M + H + 2]+ peak). A ChemCalc18 search for formulas within ±0.003 amu of the experimental [M + H]+ m/z yielded seven possibilities; three were ruled out based on unsaturation number (U#) misfits, and two lacking an S atom were dropped. The remaining two [M + H]+ formulas were (a) C13H13N2S (U# = 9) and (b) C8H13N4O2S (U# = 5), and only the former was consistent with the 2D NMR formula of C13H11 (Table 1).
Table 1.
Experimental NMR Data (125/600 MHz, DMSO-d6, 25°C) for 1-Methyl-4-methylthio-β-carboline (1)
| position | δC, typea | δH, J (Hz) | gHMBC correlations | 1JCH (Hz)b | NOEc |
|---|---|---|---|---|---|
| 1 | 139.6, C | ||||
| 3 | 134.8, CH | 8.11, s | 1, 4, 4a, 4b, 9a, | 180 | 4′ |
| 4 | 125.2, C | ||||
| 4a | 124.3, C | ||||
| 4b | 120.9, C | ||||
| 5 | 123.4, CH | 8.37, d (J = 7.9) | 4a, 4b, 7, 8, 8a | 160 | 4′, 6 |
| 6 | 119.5, CH | 7.28, ddd (J = 7.8, 7.0, 0.7) | 4b, 5, 7, 8, 8a | 161 | 5, 7 |
| 7 | 127.5, CH | 7.55, ddd (J = 8.1, 7.0, 1.2) | 4b, 5, 6, 8a | 159 | 6 |
| 8 | 111.9, CH | 7.63, d (J = 8.1) | 4b, 5, 6 | 163 | NH |
| 8a | 140.2, C | ||||
| NH | 11.75, s | 4a, 4b, 8a, 9a | 1′, 8 | ||
| 9a | 133.8, C | ||||
| 1′ | 20.1, CH3 | 2.74, s | 1, 3, 4, 9a | 127 | NH |
| 4′ | 15.2, CH3 | 2.65, s | 3,e 4 | 141 | 3, d 5e,d |
Carbon type determined by gHSQC.
Values measured by 1JCH breakthrough in gHMBC spectrum.
Determined by 2D NOESY.
Data obtained by 1D NOE.
Weak correlation.
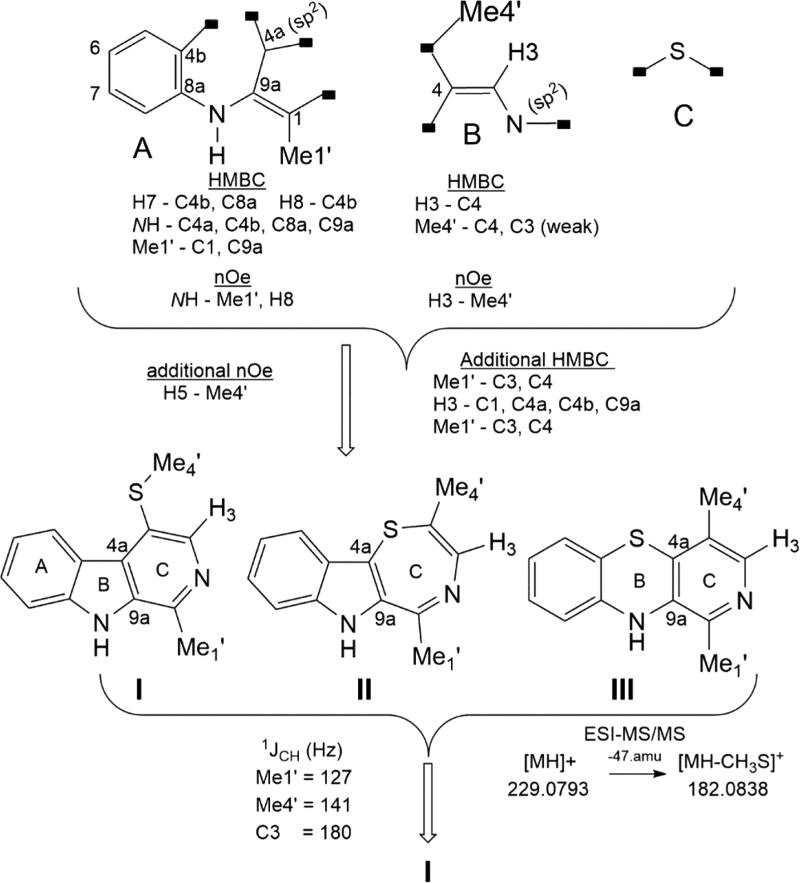
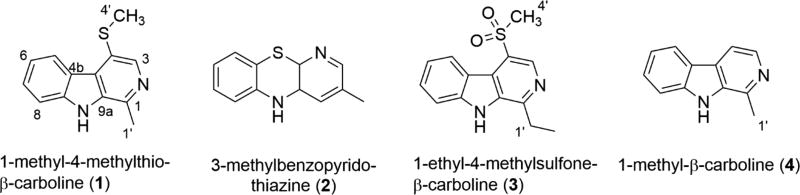
Key insights to guide the next steps of the structure determination came from additional hits observed during literature searches for compounds with similarity to 1 using atom counts of C10–14N2S. The hit list included (a) C12H10N2S (U# = 9), a synthetic compound we named 3-methylbenzopyridothiazine (2),19 and (b) C14H14N2SO2 (U# = 9), a bryzoan metabolite 1-ethyl-4-methylsulfone-β-carboline (3).20 Also relevant to this set is 1-methyl-β-carboline (aka harman) (4),21 a plant metabolite. It was tempting to assume that 1 possessed a tricyclic structure analogous to that present in 2 or 3 (see Tables 1 and S2 for NMR data). Three substructures, A–C shown in Figure 3, were assembled. The 1H NMR data revealed an o-disubstituted benzene ring, which could be expanded to partial structure A based on the four sets of HMBC and NOE correlations observed. The atom constellation in partial structure B was justified by three sets of HMBC and NOE data. Using the additional NOE and HMBC data shown in Figure 3 allowed joining A–C in three different ways as complete structures I–III. However, further progress to distinguish among them was complicated because several 2D NMR correlations observed appeared due to either 4JH−C or 5JH−C couplings. Another difficulty was that the atom count for the B/C rings in each possibility fits into a well-known general limitation in structure elucidation involving densely functionalized molecules, having a relatively low count of H’s, large U#, many carbons without attached protons, and several heteroatoms. The so-called Crews rule22 is often cited in discussing difficulties in assigning a final structure in such situations based only on 2D NMR data. We first commented on this circumstance in 200723 and must now briefly revisit the conclusion that the Crews rule is obsolete24 in view of new 2D NMR experiments such as the inverted 1JCC 1,n-ADEQUATE.25 We disagree with that idea because this and other 2D NMR data on hydrogen-deficient compounds will not always provide an unequivocal outcome and the entries collected in Figure S1526 provide case examples, especially for counts of H/(C + Z) < 0.45.
Figure 3.
Rationale used to propose structure I for the compound with an [M + H]+ m/z 229.0786 of formula C13H12N2S. This information flow includes substructures A–C, merged working structures I–III, and a final structure I.
Success in distinguishing among the three candidate structures I–III (ratio H/C + Z = 0.67) hinged on collection and interpretation of the additional 1JCH and MSn information outlined in Figure 3. All structures were in agreement with the vinyl C-3 1JCH = 180 Hz (Table 1) shown vs those of the standard values (Chart S1) for R2C=C(H)Y: (i) 159–160 Hz (Y = C), (ii) 178–182 Hz (Y = N), (iii) 189 Hz (Y = S). Significantly, only I was consistent with the CH3-1′ 1JCH = 127 Hz (requiring a C−CH3) and CH3-4′ 1JCH = 141 (requiring an S-CH3) based on standard values (Chart S1). Similarly, the ESIMS2 fragment at m/z 182.0838 (Figure S1), via loss of CH3S•, was in accord with structure I. Finally, the 13C NMR shifts of the indole ring atoms of 1 and 4 were nearly identical (Table S1). Thus, the working structure I was deduced to fit that of 1-methyl-4-methylthio-β-carboline (1). Because thioethers can oxidize to sulfoxides and sulfones, the extracted ion chromatograms of the extract (Figure S14) were examined for these potential analogues of 1. As shown in Figure S17, there was no evidence of a sulfoxide analogue (m/z 245.07), and only a trace amount seemed evident for a sulfone analogue (m/z 261.07) of 1.
Overall, this study illustrates the potential of near-shore California environments as a source of unusual Gram-negative bacteria especially from the less commonly encountered Bacteroidetes phylum. We continue to use a baiting strategy often employed to isolate terrestrial myxobacteria, consisting of E. coli-streaked WCX agar plates made with NaCl-containing SWS (see Table S1) and recommend this to be effective in building collections of the unusual marine-derived Gram-negative bacteria strains. Re-examining the production of kailuin cyclic depsipeptides from P. halotolerans and tracking their appearance via accurate mass spectrometry data, especially by MS2 networking, has provided a powerful connection between our previous isolation studies and an informatics-driven prioritization scheme exemplified in Figure 1. Extending this strategy to explore extracts or semipurified fractions from various strains afforded insights about their potential to biosynthetically express a variety of heteroatom-rich molecules. A serendipitous outcome of this study is that all seven strains produce 1-methyl-4-methylthio-β-carboline (1), which was unexpectedly difficult to characterize because of the paucity of H atoms directly attached to the B/C-rings. The biological function of 1, ubiquitously produced by strains in our lab, remains unknown; however this relatively simple compound could operate as a signaling molecule similar to that observed for the salinipostins recently discovered from marine-derived Gram-positive bacteria27 or as an agent to deter pathogens analogous to the indole-containing phytoalexin camalexin.28 The close structural relationship between 1-methyl-4-methylthio-β-carboline (1) reported here from Gram-negative bacteria and 1-ethyl-4-methylsulfone-β-carboline (3) obtained from a bryozoan20 is parallel to the recent report on the isolation of bromotyrosine-derived alkaloids from cultures of marine sponge-derived Gram-negative bacterium, which are also constituents of Verongida sponges.29 The outcomes achieved herein provide encouragement that our ongoing work with sediments collected from near-shore environments or sponges obtained from Indo-Pacific coral reefs will afford chemically productive Gram-negative bacteria.
EXPERIMENTAL SECTION
General Experimental Procedures
UV spectra were measured with a Thermo Ultimate 3000 DAD. IR spectra were measured with a PerkinElmer Spectrum One FTIR spectrometer. All NMR experiments were run on a Varian Unity spectrometer (500 MHz for 1H), a Varian UNITY INOVA spectrometer (600 MHz for 1H) equipped with a 5 mm triple resonance (HCN) cold probe, or a Bruker spectrometer (800 MHz for 1H) outfitted with a 5 mm triple resonance (HCN) inverse cold probe. Residual solvent shifts for DMSO-d6 or CD3OD were defined as δH 2.50 or 3.31, respectively, for proton spectra and δC 39.52 or 49.00 for carbon spectra in accordance with reference spectra. Accurate mass measurements for molecular formula determinations were obtained on a Thermo Velos Pro Orbitrap ESI-FTMS. Prefractionation HPLC was performed using a Phenomenex Gemini-NX 5 µm C18 (50 × 21.1 mm) column. Semipreparative HPLC fractions were generated using a Phenomenex Luna 5 µm C18 (250 × 10 mm) column. Analytical LC-MS analysis was performed on samples at a concentration of approximately 0.5–10 mg/mL, using a reversed-phase 150 × 4.60 mm 5 µm C18 Phenomenex Luna column in conjunction with a 4.0 × 3.0 mm C18 (octadecyl) guard column and cartridge (holder part number: KJ0-4282; cartridge part number: AJ0-4287, Phenomenex, Inc.).
Biological Material
The following bacterial strains used in this study were all isolated from sediment samples obtained at California state beaches during 2012–2013. The strain M125SB302Ax was isolated from a backshore sediment collection at Zmudowski State Beach, Monterey County. The strain M132NC031Ax was isolated from a water collection at Kellogg Beach, Del Norte County. Strains M135NC101Ax, M135NC362Ax, and M135NC397Ax were each isolated from backshore sediments collected respectively at Enderts, Crab County Park, Del Norte County, and Black Sands Beach, Humboldt County. The strain M138NC113Bx was isolated from a foreshore sediment collection at Wilson Creek Beach, Del Norte County. Isolation and characterization of strain M128SB283Ax was reported previously.5 Each strain was isolated on SWS-WCX30 agar medium containing 750 mL of seawater salts, 250 mL of DI H2O, 1 g of CaCl2·H2O, and 15 g of agar per liter with 50 mg/mL cycloheximide (see Table S1 for SWS recipe). The pH of the isolation medium was adjusted to 7.2–7.5 with NaOH prior to agar being added and autoclaved for 45 min at 120 °C. To select for growth of Gram-negative bacteria, methods originally designed for the isolation of myxobacteria were adapted for use in this work.31 This involved streaking the isolation plate with E. coli prior to the addition of sediment, enabling observation of “predatory” and/or proteolytic behavior.32
Nucleic Acid Extraction, Sequencing, and Strain Identification
Genomic DNA extractions were performed using the GeneElute Bacterial Genomic DNA kit as per instructions (Sigma #NA2110-1KT). The DNA was quantified by NanoDrop (Thermo Scientific) and stored at −20 °C. The 16S rRNA genes were amplified from genomic DNA with primers corresponding to E. coli 16S rRNA gene sequences: 27F (AGAGTTTGATCCTGGCTCAG) and 1492R (ACGGCTACCTTGTTACGACTT) (IDT DNA). Each PCR mixture contained 50–100 ng of genomic DNA, 0.5 mM of each primer, and Taq Master Mix (2.5U Taq polymerase, Qiagen). The PCR program consisted of 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension step at 72 °C for 5 min. Amplification products were examined by agarose gel electrophoresis and purified when sequenced at the University of California Berkeley Sequencing Facility. The resulting sequences were queried at the NCBI’s refseq database using BLAST.33
Strain Identifications
The individual bacterial strains (see Figure 1, Figure S9) were provisionally identified based on comparison of their 16S rRNA to the RefSeq database, where each strain’s rRNA sequence had >98% identity to a reference organism. Strain codes are followed by their GenBank accession numbers and provisional ID: M125SB302Ax (KT354561), Achromobacter spanius; M132NC031Ax (KT354559), Photobacterium halotolerans; M135NC101Ax (KT354535), Pseudomonas benzenivorans; M135NC362Ax (KT354518), Pontibacter korlensis; M135NC397Ax (KT354516), Cellulophagia baltica; M138NCNC113Bx (KT354558), Pseudoalteromonas elyakovii. Strain M128SB283Ax was described previously.5 All strains used in this study are maintained as cryopreserved glycerol stocks at the University of California, Santa Cruz.
Culture Conditions
One-liter volumes of five media types (M1, M3–M5, Table S1) were placed in 2 L flasks and adjusted to pH 6.8 before autoclaving at 120 °C for 45 min. Each liter was inoculated with 1 mL of a saturated overnight culture and cultured at 25 °C for 3 weeks with shaking at 150 rpm before extraction with equal volumes of EtOAc. For production of 1-methyl-4-methylthio-β-carboline (1), strain M125SB302Ax was grown in medium M4 (Table S1) as stated above at a 20 L scale.
Mass Spectral GNPS Networking
All MS2 similarity networks were generated using the GNPS server at http://gnps.ucsd.edu9 and visualized in Cytoscape at http://www.cytoscape.org. UHPLC-MS data were acquired using a Dionex Ultimate 3000 UHPLC paired with a Thermo Velos Pro Oribtrap. The UHPLC was run with a Phenomenex Kinetex 1.7 µm, 2.1 × 100 mm column equipped with guard cartridge (part no. AJ0-9000, AJ0-8782) maintained at 35 °C during analysis. The MS was operated in data-dependent MS/MS mode in which the top 10 most intense ions were selected for fragmentation. The first scan was performed by FTMS (resolution 30 000, m/z range 110–2000), while MS2 spectra were recorded in the “rapid” ion trap mode after collision-induced dissociation (performed at a normalized collision energy of 35%). All data were acquired as centroid spectra. The ubiquitous plasticizers n-BBS ([M + H]+ m/z 214.0896) and diisooctyl phthalate ([M + H]+ m/z 391.2843) were used as internal standards with the detector’s lock mass feature. In GNPS, the default parameters (as of January 2017) were used except the following: precursor ion mass tolerance of ±0.01 amu; fragment ion mass tolerance of ±0.2 amu; minimum matched fragment ions. The group and attribute files needed for network visualization were generated via a Java program available from UCSC (https://mnpr.chem.ucsc.edu/programs/GAparse/) that assigns attributes based on file names. The primary attributes generated relevant to this work are strain, media type, and media controls, which allowed us to remove media components from consideration while observing trends in strain- and media-specific metabolite production.
Extraction and Isolation
An overview of the general method is shown in Figure S14. A 20 L amount of fermentation broth from strain M125SB302Ax, grown in medium M4 (Table S1), as stated above, was extracted with EtOAc, which was dried by rotary evaporation. A portion of the organic extract (192 mg of 319.7 mg total) was prefractionated by HPLC using methods reported previously.5 Analytical HPLC fractions were generated using a Phenomenex Luna 5 µm C18 (150 × 4.60 mm) column. Prefraction F3 (2.8 mg) was subjected to analytical HPLC using MeCN/H2O (with 0.1% formic acid) at 1 mL/min, UV absorbance detection at 310 nm, and the following step gradient: 10% MeCN for 3.0 min, linear gradient to 82.5% MeCN over 17 min, linear gradient to 100% MeCN over 0.10 min, hold at 100% MeCN for 15 min. Scale-up isolation yields (see Figure S9) were 0.4 mg of F3H1 (tR = 5.2 min), 0.2 mg of F3H2 (tR = 7.4 min), 0.4 mg of F3H3 (tR = 11.7 min), 0.9 mg of F3H4 (1-methyl-4-methylthio-β-carboline (1)) (tR = 14.8 min), 0.1 mg of F3H5 (tR = 15.1 to 20.0 min), and 0.6 mg of F3H6 (tR = 20.0 to 35 min).
1-Methyl-4-methylthio-β-carboline (1): off-white film; UV (MeCN/H2O) λmax 256, 294, 379 nm (Figure S12); IR (film) νmax 3152, 2924, 1620, 1455, 1389, 1324, 739 cm−1 (Figure S13); 13C and 1H NMR data, Table 1; ESITOFMS m/z [M + H]+ 229.0787 (calcd for C13H13N2S, 229.0799).
Supplementary Material
Figure 2.
Proposed structure for compound 1 alongside similar tricyclic alkaloids.
Acknowledgments
This work was supported by grants from the NIH R01 CA47135 (P.C.) and Diversity Research Supplements R01 CA47135 (-23S1 in support of P.S. and -25S1 in support of G.N.) and NIH R25 GM104552 (fellowship to D.C.). We also acknowledge funding from NSF CHE1427922 for purchase of the Thermo Velos Pro electrospray ionization hybrid ion traporbitrap mass spectrometer and NIH 1S10OD018455-01 for the acquisition of an 800 MHz NMR spectrometer and helium cryoprobe. Thanks also to a reviewer for insightful comments and pointing out the results in ref 20.
Footnotes
ASSOCIATED CONTENT
- Media recipes for microbial cultures, copies of NMR (including 2D) spectroscopic data for 1, ESI-FTMS data for 1, UV data for 1, IR data for 1, examples of relevant reference literature data for many compounds, selected LC-MS traces for media controls, and selected LC-MS extracted ion chromatograms (PDF)
The authors declare no competing financial interest.
NOTE ADDED AFTER ASAP PUBLICATION
This paper was published ASAP on August 4, 2017, with an error in the Supporting Information. The corrected version was reposted on August 8, 2017.
References
- 1.Cragg GM, Newman DJ. Biochim. Biophys. Acta, Gen. Subj. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams PG. Trends Biotechnol. 2009;27:45–52. doi: 10.1016/j.tibtech.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Blunt JW, Copp BR, Keyzers RA, Munro MHG, Prinsep MR. Nat. Prod. Rep. 2016;33:382–43. doi: 10.1039/c5np00156k. [DOI] [PubMed] [Google Scholar]
- 4.Still PC, Johnson TA, Theodore CM, Loveridge ST, Crews P. J. Nat. Prod. 2014;77:690–702. doi: 10.1021/np500041x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theodore CM, Lorig-Roach N, Still PC, Johnson TA, Draskovic M, Schwochert JA, Naphen CN, Crews MS, Barker SA, Valeriote FA, Lokey RS, Crews P. J. Nat. Prod. 2015;78:441–452. doi: 10.1021/np500840n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi EJ, Nam S−J, Paul L, Beatty D, Kauffman CA, Jensen PJ, Fenical W. Chem. Biol. 2015;22:1270–1279. doi: 10.1016/j.chembiol.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 7.Felder S, Kehraus S, Neu E, Bierbaum G, Schäberle TF, König GM. ChemBioChem. 2013;22:1363–71. doi: 10.1002/cbic.201300268. [DOI] [PubMed] [Google Scholar]
- 8.(a) Xu Y, Kersten RD, Nam S-J, Lu L, Al-Suwailem AM, Zheng H, Fenical W, Dorrestein PC, Moore BS, Qian P-Y. J. Am. Chem. Soc. 2012;134:8625–8632. doi: 10.1021/ja301735a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tsukimoto M, Nagaoka M, Shishido Y, Fujimoto J, Nishisaka F, Matsumoto S, Harunari E, Imada C, Matsuzaki T. J. Nat. Prod. 2011;74:2329–2331. doi: 10.1021/np200543z. [DOI] [PubMed] [Google Scholar]
- 9.(a) Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, van der Voort M, Pogliano K, Gross H, Raaijmakers JM, Moore BS, Laskin J, Bandeira N, Dorrestein PC. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1743–E1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crüsemann M, O’Neill EC, Larson CB, Melnik AV, Floros DJ, da Silva RR, Jensen PR, Dorrestein PC, Moore BS. J. Nat. Prod. 2017;80:588–597. doi: 10.1021/acs.jnatprod.6b00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bálint M, Bahram M, Eren AM, Faust K, Fuhrman JA, Lindahl B, O’Hara RB, Öpik M, Sogin ML, Unterseher M, Tedersoo L. FEMS Microbiol. Rev. 2016;40:686–700. doi: 10.1093/femsre/fuw017. [DOI] [PubMed] [Google Scholar]
- 11.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Muller R, Wohlleben W, Breitling R, Takano E, Medema MH. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machado H, Sonnenschein EC, Melchiorsen J, Gram L. BMC Genomics. 2015;16:158–170. doi: 10.1186/s12864-015-1365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SJ, Kim BG, Park HJ, Yim JH. Prep. Biochem. Biotechnol. 2016;46:261–266. doi: 10.1080/10826068.2015.1015568. [DOI] [PubMed] [Google Scholar]
- 14.(a) Liras P. Appl. Microbiol. Biotechnol. 2014;98:1023–1030. doi: 10.1007/s00253-013-5410-z. [DOI] [PubMed] [Google Scholar]; (b) Qin Z, Huang S, Yu Y, Deng H. Mar. Drugs. 2013;11:3970–3997. doi: 10.3390/md11103970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInerney BV, Gregson RP, Lacey MJ, Akhurst RJ, Lyons GR, Rhodes SH, Smith DRJ, Engelhardt LM, White AH. J. Nat. Prod. 1991;54:774–784. doi: 10.1021/np50075a005. [DOI] [PubMed] [Google Scholar]
- 16.Clark DP, Carroll J, Naylor S, Crews P. J. Org. Chem. 1998;63:8757–8764. [Google Scholar]
- 17. [accessed Mar 28, 2017]; http://dnp.chemnetbase.com.
- 18.(a) Patiny L, Borel A. J. Chem. Inf. Model. 2013;53:1223–1228. doi: 10.1021/ci300563h. [DOI] [PubMed] [Google Scholar]; (b) [accessed Mar 28, 2017]; http://www.chemcalc.org.
- 19.Hu W, Zhang S. J. Org. Chem. 2015;80:6128–6132. doi: 10.1021/acs.joc.5b00568. [DOI] [PubMed] [Google Scholar]
- 20.Prinsep MR, Blunt JW, Munro MHG. J. Nat. Prod. 1991;54:1068–1076. doi: 10.1021/np50076a023. [DOI] [PubMed] [Google Scholar]
- 21.Koike K, Sakamoto Y, Ohmoto T. Org. Magn. Reson. 1984;22:471–473. [Google Scholar]
- 22.Molinski TF, Morinaka BI. Tetrahedron. 2012;68:9301–9478. doi: 10.1016/j.tet.2011.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(a) White KN, Amagata T, Oliver AG, Tenney K, Wenzel PJ, Crews P. J. Org. Chem. 2008;73:8719–8722. doi: 10.1021/jo800960w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Crews P. Lecture “Discovering and Characterizing Provocative Small Molecules from Simple Marine Organisms” at Center for Marine Natural Products and Drug Discovery Symposium. Seoul Korea: 2007. [Google Scholar]
- 24.Senior MM, Williamson RT, Martin GE. J. Nat. Prod. 2013;76:2088–2093. doi: 10.1021/np400562u. [DOI] [PubMed] [Google Scholar]
- 25.Buevich AV, Williamson RT, Martin GE. J. Nat. Prod. 2013;77:1942–1947. doi: 10.1021/np500445s. [DOI] [PubMed] [Google Scholar]
- 26.(a) Brastianos HC, Vottero E, Patrick BO, Van Soest R, Matainaho T, Mauk AG, Andersen RJ. J. Am. Chem. Soc. 2006;128:16046–16047. doi: 10.1021/ja067211+. [DOI] [PubMed] [Google Scholar]; (b) Hughes CC, Prieto-Davo A, Jensen PR, Fenical W. Org. Lett. 2008;10:629–631. doi: 10.1021/ol702952n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Inman WD, O’Neill-Johnson M, Crews Phillip. J. Am. Chem. Soc. 1990;112:1–4. [Google Scholar]; (d) Tan C, Liu Z, Chen S, Huang X, Cui H, Long Y, Lu Y, She Z. Sci. Rep. 2016;6:36609. doi: 10.1038/srep36609. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Moquin-Pattey, Guyot M. Tetrahedron. 1989;45:3445–3450. [Google Scholar]; (f) Ren Z, Hao Y, Hu X. Org. Lett. 2016;18:4958–4961. doi: 10.1021/acs.orglett.6b02424. [DOI] [PubMed] [Google Scholar]; (g) Schmitz FJ, DeGuzman FS, Hossain MB, Van der Helm D. J. Org. Chem. 1991;56:804–808. [Google Scholar]; (h) White KN, Amagata T, Oliver AG, Tenney K, Wenzel PJ, Crews P. J. Org. Chem. 2008;73(22):8719–8722. doi: 10.1021/jo800960w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Hughes CC, MacMillan JB, Gaudencio SP, Jensen PR, Fenical W. Angew. Chem., Int. Ed. 2009;48:725–727. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Ribaucourt A, Hodgson DM. Org. Lett. 2016;18:4364–4367. doi: 10.1021/acs.orglett.6b02120. [DOI] [PubMed] [Google Scholar]; (k) Choi EJ, Nam S-J, Paul L, Beatty D, Kauffman CA, Jensen PR, Fenical W. Chem. Biol. 2015;22(9):1270–1279. doi: 10.1016/j.chembiol.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 27.Schulze CJ, Navarro G, Ebert D, DeRisi J, Linington RG. J. Org. Chem. 2015;80:1312–1320. doi: 10.1021/jo5024409. [DOI] [PubMed] [Google Scholar]
- 28.Jimenez LD, Ayer WA, Tewari JP. Phytoprotection. 1997;78:99–103. [Google Scholar]
- 29.Nicacio KJ, Ioca LP, Froes AM, Leomil L, Appolinario LR, Thompson CC, Thompson FL, Ferreira AG, Williams DE, Andersen RJ, Eustaquio AS, Berlinck RGS. J. Nat. Prod. 2017;80:235–240. doi: 10.1021/acs.jnatprod.6b00838. [DOI] [PubMed] [Google Scholar]
- 30.Reichenbach H, Dworkin M. In: The Prokaryotes. 2. Balows A, Truëper HG, Dworkin M, Harder W, Schleifer K-H, editors. Springer-Verlag; Berlin: 1992. pp. 3416–3487. [Google Scholar]
- 31.Iizuka T, Jojima Y, Fudou R, Yamanaka S. FEMS Microbiol. Lett. 1998;169:317–322. doi: 10.1111/j.1574-6968.1998.tb13335.x. [DOI] [PubMed] [Google Scholar]
- 32.Li Y-Z, Hu W, Zhang Y-Q, Qiu Z-J, Zhang Y, Wu B-H. J. Microbiol. Methods. 2002;50:205–209. doi: 10.1016/s0167-7012(02)00029-5. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.