Abstract
Cytochrome P450 2D6 (CYP2D6) participates in the metabolism of approximately 20–25% of prescribed drugs. Genetic polymorphisms influence the expression and/or activity of CYP2D6, and inter-individual differences in drug activation and elimination caused by CYP2D6 genetic variants were reported. However, little is known about the potential modulation of CYP2D6 expression by microRNAs (miRNAs). In the current study, by using in silico prediction of the stabilities of miRNA/mRNA complexes, we screened 38 miRNA candidates that may interact with the transcript of CYP2D6. An inverse correlation between the expression of miRNA hsa-miR-370-3p and the expression of CYP2D6 was observed in human liver tissue samples. Electrophoretic mobility shift assays confirmed that hsa-miR-370-3p was able to directly bind to its cognate target within the coding region of the CYP2D6 transcript. The transfection of hsa-miR-370-3p mimics into the HepG2CYP2D6 cell line, a genetically modified cell line that overexpresses exogenous CYP2D6, was able to suppress the expression of CYP2D6 significantly at both mRNA and protein levels. The transfection of hsa-miR-370-3p mimics was also able to inhibit endogenous mRNA expression and/or protein production of CYP2D6 in HepaRG cells. Furthermore, in HepaRG, HepG2, and Huh7 cells, dexamethasone-induced expression of CYP2D6 was inhibited by hsa-miR-370-3p mimics. To investigate whether the miRNA mediated suppression is caused by inhibiting protein translation or promoting mRNA degradation, an actinomycin D assay was used to measure the stability of CYP2D6 transcripts. The results indicated that hsa-miR-370-3p mimics facilitated significantly the degradation of CYP2D6 mRNA. In addition, proteomics analyses of proteins isolated from the miRNA/mRNA/protein complex suggested that a group of multifunctional proteins facilitated the interaction between hsa-miR-370-3p and CYP2D6, thereby promoting mRNA degradation.
Keywords: CYP2D6, hsa-miR-370-3p, Drug metabolizing enzymes, Epigenetics, Pharmacogenomics
1. Introduction
Cytochrome P450 (CYP) 2D6 is a very important drug metabolizing enzyme (DME). Despite its low abundance (CYP2D6 consists only 2% of the total CYPs in liver), CYP2D6 is involved in metabolizing approximately 20–25% of all drugs [1]. The substrate spectrum for this enzyme covers a broad range of drugs, including the anticancer drug tamoxifen, anti-psychotropic drugs (perphenazine, thioridazine, haloperidol, and nortriptyline), and anti-cardiovascular disease drugs (antiarrhythmics, antiemetics, and opioids) [1]. The inter-individual variability in the enzyme activity/expression of CYP2D6 is substantial, accounting for the different responses to certain drugs by different individuals [2,3]. There are several factors contributing to the inter-individual variability in the activity/expression of CYPs, including genetic variants, environmental factors, and epigenetic regulations [4]. A variety of CYP2D6 genetic polymorphisms has been reported in populations with different ethnicities and geographic regions [5]. Currently, more than 100 allelic variants have been identified in the CYP2D6 gene, suggesting that genetic variation is the major factor responsible for the inter-individual variation in the activity/expression of the enzyme (http://www.cypalleles.ki.se/cyp2d6.htm). CYP2D6*2, *3, *4,*5, *6,*10 and *41 are more frequently observed in Caucasian populations, and CYP2D6*10 and *36 are more prevalent in Asians [6].
Environmental factors, pathophysiological factors, and drug-drug interactions also affect the activity/expression of P450 enzymes. Several clinical studies indicated that CYP2D6 is inducible by corticosteroids, based on the observation that the clearance of CYP2D6 substrates (e.g. metoprolol and dextromethorphan) increases significantly during pregnancy [7–9]. Recently, it was confirmed that both endogenous and exogenous corticosteroids can induce CYP2D6 in human primary hepatocytes and HepaRG cells [10].
In addition to genetic polymorphisms and environmental influences, epigenetic modification is another important mechanism that perturbs the expression of various DMEs [11]. MicroRNAs (miRNAs) are a group of small noncoding RNAs with 20–24 nucleotides that act as important regulators in the gene expression network. Currently, more than 2500 miRNAs are known in humans (http://www.mirbase.org/, version 21). CYP1B1 was the first DME identified to be regulated by a miRNA, which was demonstrated by the pioneering study showing that human CYPs can be modulated post-transcriptionally by miRNAs [12]. Thereafter, several important human DME genes have been identified to be regulated by miRNAs, such as CYP3A4 by miR-27b, and CYP2E1 by miR-378 [13,14]. In our previous studies, we demonstrated that hsa-miR-29a-3p, hsa-miR-128-3p, and hsa-miR-25-3p regulated CYP2C19, CYP2C9, ALDH5A1 and CYP2B6, respectively, by suppressing their expression [15–18]. However, the effects of miRNAs on the expression of CYP2D6 have not been studied.
In the present study, a combination of in silico, in vitro and proteomics approaches was used to systematically investigate the interaction between miRNAs and CYP2D6. As a result, hsa-mir-370-3p was found to suppress CYP2D6 by recruiting miRNA/mRNA binding proteins and nucleases into the hsa-miR-370-3p target site at the CYP2D6 coding region, thus facilitating the degradation of CYP2D6 transcripts.
2. Materials and methods
2.1. Cell culture
HepaRG cells were obtained from Life Technologies (Carlsbad, CA) and maintained in Williams’ Medium E (without L-Glutamine and Phenol Red, Life Technologies) containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Lawrenceville, GA). Human hepatic HepG2 and Huh7 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in DMEM medium (Life Technologies) containing 10% FBS. The HepG2CYP2D6 cell line, a genetically modified cell line that overexpresses exogenous CYP2D6, and HepG2EV cell line, a genetically modified cell line with an empty vector for CYP2D6 (as a negative control), have been established and described previously [19]. Both cell lines were maintained in DMEM medium containing 10% FBS and 10 μg/ml Blasticidin S (Life Technologies). The passage number for each cell line was less than 12 when the experiments were performed.
2.2. Reagents and chemicals
Thiazolyl blue tetrazolium bromide (MTT), actinomycin D, and dexamethasone were obtained from Sigma-Aldrich (St. Louis, MO). Actinomycin D was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich), and dexamethasone was prepared in methanol (Sigma-Aldrich).
2.3. In silico analyses
The miRNAs that potentially bind to their target sites within the full length of CYP2D6 transcript were screened through the four public databases, including microRNA.org (http://www.microrna.org/), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_prediction.html), TargetScan (Release 6.2, http://www.targets-can.org), and miRTar.human (http://mirtar.mbc.nctu.edu.tw/human/).
The RNAhybrid program (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid) was used to calculate the minimum free energy of hybridization for candidate miRNAs with their putative binding sites detected within the CYP2D6 mRNA sequence (NM_000106).
2.4. Correlation analysis of CYP2D6 mRNA and miRNAs in human liver tissues
The miRNA and mRNA expression levels were obtained from The Cancer Genome Atlas (TCGA) public database (https://cancer-genome.nih.gov/), which includes 49 pairs of liver tumor tissue samples and adjacent non-tumor liver tissue samples. The correlation between the expression of candidate miRNAs and the expression of CYP2D6 mRNA in human non-tumor liver tissue samples was estimated by Pearson correlation analysis.
2.5. RNA electrophoretic mobility shift assay
RNA electrophoretic mobility shift assay (EMSA) was performed according to the protocol provided by the LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific, Waltham, MA), which was described in our previously studies [15–17]. Briefly, 200 nmol IRDye1800-dye labeled hsa-miR-370-3p and/or cy5.5™ labeled dye-hsa-miR-370-3p TAR (targeting sequences in CYP2D6 coding region) oligonucleotides (Intergrated DNA Technologies, Coralville, Iowa) were mixed in the 1 × REMSA Binding Buffer supplemented with 5% glycerol, 200 mM KCl, and 100 mM MgCl2. The binding specificity was confirmed in each case by the addition of a 50-fold molar excess of unlabeled oligonucleotides (cold-hsa-miR-370-3p) into the reaction system 5 min before the addition of dye-labeled probes. To test the RNA:protein interactions, 2 μg of HepaRG cell cytoplasmic extract was added to reaction mixtures and 1 μg of tRNA was added as a nonspecific competitor RNA. After incubation at 25 °C for 20 min to allow the formation of hybridizations, the reaction mixture was separated on a 12% PAGE by electrophoresis (100V, 150 min) at 4 °C and then detected with an Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). Oligonucleotide sequence information for EAMS is listed in Table 1.
Table 1.
Primers used in qRT-PCR assays and oligonucleotides used in EMSA.
| RT Primers & Oligonucleotides | Sequences | Application |
|---|---|---|
| CYP2D6 | RT-F: 5′-TAGTGGTGGCTGACCTGTTCTCT-3′ RT-R: 5′-TCGTCGATCTCCTGTTGGACA-3′ |
RT-PCR |
| GAPDH | RT-F: 5′-GAAATCCCATCACCATCTTCCAGG-3′ RT-R: 5′-GAGCCCCAGCCTTCTCCATG-3′ |
RT-PCR |
| hsa-miR-370-3p | RT-F: 5′-GCCTGCTGGGGTGGAACCTGGT-3′ | RT-PCR |
| U6 | RT-F: 5′-CTCGCTTCGGCAGCACA-3′ RT-R: 5′-AACGCTTCACGAATTTGCGT-3′ |
RT-PCR |
| dye-miR-370-3p | /5IRD800CWN/rGrC rCrUrG rCrUrG rGrGrG rUrGrG rArArC rCrUrG rGrU | EMSA |
| dye-miR-370-mut | /5IRD800CWN/rGrC rCrUrA rCrUrA rGrArG rUrGrG rArArC rCrUrG rGrU | EMSA |
| cold-miR-370-3p | rGrCrC rUrGrC rUrGrG rGrGrU rGrGrA rArCrC rUrGrG rU | EMSA |
| dye-miR370-Tar | /5Cy55/mCmU mUmCmC mUmGmC mCmUmU mUmCmU mCmAmG mCmAmG mGmC | EMSA |
2.6. Exogenous CYP2D6 expression assays
The HepG2CYP2D6 cells were constructed using the lentiviral expression vector pLV-EF1α-MCS-IRES-Bsd (Biosettia, San Diego, CA) as described in our previous study [19]. Cells were seeded in 24-well plates at a density of 2 × 105 cells per well and incubated overnight without antibiotics. miRNAs (Intergrated DNA Technologies) including miRNA mimic negative control, hsa-miR-370-3p mimic, hsa-miR-370-3p mimic-mutant, miRNA inhibitor negative control, or hsa-miR-370-3p inhibitor were mixed with lipofectamine 2000 (Thermo Scientific) and transfected into cells at a final concentration of 50 nmol/L. Cells were collected 48 h after transfection to perform reverse-transcription PCR (RT-PCR) and Western blot analyses, respectively. Each independent experiment was performed in triplicate.
2.7. CYP2D6 induction
Cells were seeded in 24-wells plates and dexamethasone was added to the DMEM medium at final concentrations of 1 μM, 2 μM, or 4 μM for a period of 72 h. Then qRT-PCR, Western blot, and CYP2D6 activity analyses were performed to detect CYP2D6 mRNA expression, protein expression, and enzyme activity. Cells incubated with 0.9% methanol (the solvent for dexamethasone) was used as a vehicle control. Each experiment was carried out three times.
2.8. RNA extraction and qRT-PCR
Total RNA was extracted from HepaRG, HepG2, HepG2CYP2D6, and Huh7 cells using the miRNeasy Mini Kit (Qiagen, Valencia, CA). The first-strand cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) or a miScript® II RT Kit (Qiagen). qRT-PCR was performed on an ABI Prism7900 Sequence Detection System (Applied Biosystems) based on the instructions provided with the QuantiFast SYBR1 Green RT-PCR Kit (Qiagen). A negative control (with primers and SYBR green mixture but without cDNA) was performed to monitor the potential contamination. The relative expression levels of CYP2D6 and hsa-miR-370-3p were calculated using the 2−ΔΔCt method and normalized to the expression level of the housekeeping gene, GAPDH and the U6 small nuclear RNA, respectively. Primers for qRT-PCR were obtained from Interagrated DNA Technologies and the sequences are provided in Table 1.
2.9. Western blot analysis
Protein extracts from HepaRG, HepG2, HepG2CYP2D6, and Huh7 cells were prepared using RIPA Lysis buffer with protease inhibitors (Life Technologies) and quantified using Pierce BCA Protein assay kit (Thermo Scientific). Equal amounts of the total proteins from different experimental groups were separated on a 10% polyacrylamide gel and then transferred to a 0.45-μm polyvinylidene difluoride membrane (Millipore, Bedford, MA). Western blot was performed following the instructions from the Odyssey™ Western Blotting Kit (LI-COR Biosciences). The rabbit-anti-human CYP2D6 polyclonal primary antibody and the mouse GAPDH monoclonal primary antibody were purchased from GeneTex (Irvine, CA) and Abcam (Cambridge, MA), respectively. The protein bands were visualized after the scanning of fluorescence signal and the image analyses were performed with the Odyssey CLx Infrared Imaging System.
2.10. Assays for CYP2D6 enzyme activity
Enzymatic activities of CYP2D6 in HepG2 cells and Huh7 cells treated with or without dexamethasone were determined using P450-Glo™ assays (Promega, Madison, WI) according to the manufacturer’s instructions. Following a 72 h period, cells were incubated with fresh culture medium containing the corresponding CYP2D6 substrate (Luciferin-ME EGE) at 37 °C for 3 h. Then 75 μl of culture medium was transferred to a white 96-well plate, and 75 μl of luciferin detection reagent was added in each well. The luminescence was measured with a BioTek Cytation 5 cell imaging multimode reader. HepG2CYP2D6 cells were used as a positive control. The MTT assay was performed using the same cells, to evaluate the cell viability, and the luminescence was normalized with the MTT assay data. Each experiment was conducted in triplicate.
2.11. mRNA decay measurements
The stability of the CYP2D6 mRNA was assessed by adding 5 μg/ml actinomycin D into the cell culture medium to inhibit mRNA transcription. At the indicated time points, the relative amount of specific mRNA remaining in each sample could be correlated with mRNA degradation. Total RNA was extracted at 0, 4, 8, and 24 h after actinomycin D treatment, and the endogenous CYP2D6 mRNA levels were analyzed by qRT-PCR. Since the mRNA levels for GAPDH were not changed after actinomycin D treatment, the GAPDH gene was used as a reference gene, and the ratio of CYP2D6 and GAPDH in each sample was calculated [20].
2.12. Mass spectrometry analysis of miRNA-protein complex
The gel band representing the mRNA/miRNA/protein complex was cut from the native PAGE gel, and fixed in 50% methanol and 10% acetic acid. It was then washed with 50 mM NH4HCO3 (pH 8.0), followed by washing with Milli-Q water. Next, the gel band was cut into smaller pieces and then dried using a speed vacuum. The samples were digested by trypsin (Promega) overnight at 37 °C. The resulted peptides were extracted by sonication in 80% acetonitrile/0.1% trifluoroacetic acid, and then lyophilized. The extracted peptides were analyzed by a reversed-phase nanoflow LC-tandem mass spectrometry (RP nanoLC–MS/MS) using a method similar to that described previously [21]. Briefly, peptides were re-dissolved in 0.1% formic acid and injected onto a 180 μm i.d. × 20 mm C18 trap column (100 Å, 5 μm) and separated on a 75 μm i.d. × 150 mm BEH C18 column (130 Å, 1.7 μm, Waters, Milford, MA), which was coupled to an Orbitrap mass spectrometer (LTQ-Orbitrap Elite, Thermo Electron, San Jose, CA). Peptide separation was performed at a flow rate of 0.5 μL/min using a step gradient of 2–42% solvent B (0.1% formic acid in acetonitrile) for 40 min and 42–98% solvent B for 10 min. Both solvents A (0.1% formic acid in water) and B were delivered by a nanoAcquity UPLC system (Waters, Milford, MA). The mass spectrometer was operated in a data dependent mode in which each full MS scan (acquired in the Orbitrap analyzer) was followed by 15 MS/MS scans (acquired in the ion trap) where the 15 most abundant peptide molecular ions were dynamically selected from the prior MS scan for collision-induced dissociation (CID) using a normalized collision energy of 35%. The raw MS/MS data were searched using the SEQUEST HT running under Proteome Discoverer (version 1.4, Thermo Scientific) against a human UniProt proteome database (http://www.uniprot.org) for the identification of peptides and proteins within the gel band.
2.13. Constructing a miRNA/mRNA/protein interaction network
The miRNA/mRNA/protein interaction network for analyzing the miRNA/mRNA/protein complex was built up as following steps. All proteins identified by mass spectrometry were subjected to protein function annotation, using information from a variety of databases. Briefly, the RNA-protein interaction data were downloaded from the RAID 2.0 database (http://www.rna-society.org/raid/index.html), and then miRNA-binding proteins and mRNA-binding proteins were selected from the interaction data. An RNA degradation-related protein was considered if the protein is related to the term “RNA degradation” in the KEGG PATHWAY database (http://www.genome.jp/kegg/pathway.html). Nucleases were extracted by querying the keyword “Nuclease” in the UniProt database (http://www.uniprot.org/). Interaction data of miRNA-binding proteins, mRNA-binding proteins, RNA degradation related proteins and nucleases were retrieved from the InBio_Map database (https://www.intomics.com/inbio/map/#home). The miRNA-protein interaction network was assembled using Cytoscape (version 3.4) software (http://www.cytoscape.org/).
2.14. Statistics
All statistical analyses were performed using SPSS software, version 17.0. Pearson’s correlation analysis was used to analyze the correlation between CYP2D6 mRNA levels and miRNA hsa-miR-370-3p levels in the original data of human liver tissues published in the TCGA database. Two tailed Student’s t-test (for comparing data from two groups) or one way analysis of variance (ANOVA) (for comparing data from multiple groups) was used to compare the differences in CYP2D6 and hsa-miR-370-3p RNA levels, CYP2D6 protein levels, or enzyme activities between subgroups. P < 0.05 was considered statistically significant.
3. Results
3.1. Screening and identification of miRNAs potentially regulate CYP2D6
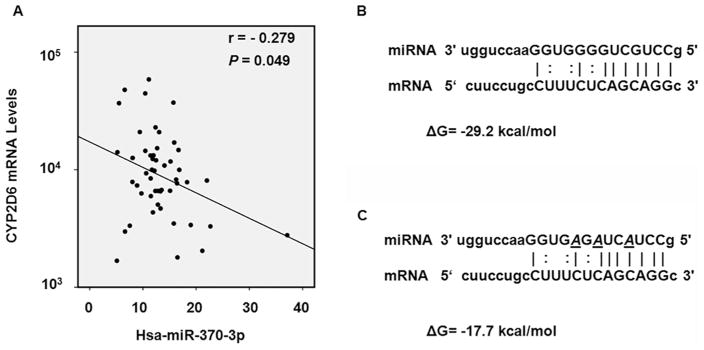
Three databases including microRNA.org, PITA, and TargetScan were used to predict potential miRNAs that bind to the 3′-UTR of CYP2D6 mRNA; the database miRTar.human was used to predict binding sites of miRNAs in 3′-UTR, 5′-UTR, and the coding region of the gene. The screening results from all four databases did not provide any putative miRNA biding sites within the 3′-UTR of CYP2D6. However, results from the miRTar.human database suggested several miRNAs may target CYP2D6 in the coding region of the CYP2D6 transcript. Then the free energy of hybridization between candidate miRNAs and their cognate targets was calculated, and the correlation between the expression of miRNA and the expression of CYP2D6 in human liver tissues was analyzed (Table 2). As shown in Table 2 and Fig. 1, the free energy of hybridization between hsa-370-3p and its cognate target at the CYP2D6 coding region was −29 kcal/mol; and the expression of hsa-370-3p was inversely correlated with the expression of CYP2D6 mRNA in human liver tissues (r = −0.279, P < 0.05). In our previous studies, we have demonstrated that a minimum free energy of hybridization lower than −20 kcal/mol was sufficient for the formation of miRNA/mRNA complex [15–18]. Based on these criteria, the hsa-miR-370-3p was selected for further analyses.
Table 2.
MicroRNAs targeting CYP2D6 coding region predicted by miRTar.Human.
| Name | Correlation | Free energy* | Position# |
|---|---|---|---|
| hsa-miR-37-3p | −0.279 | −29.2 | 4012–4131 |
| hsa-miR-372-3p | −0.213 | −24.8 | 4185–4206 |
| hsa-miR-542-5p | −0.187 | −21.8 | 2098–2551 |
| hsa-miR-1306-3p | −0.174 | −27.4 | 157–174 |
| hsa-miR-1262 | −0.168 | −21.2 | 1096–1116 |
| hsa-miR-1292-5p | −0.168 | −30.8 | 1121–1142 |
| hsa-miR-214-3p | −0.157 | −22.4 | 211–232 |
| hsa-miR-665 | −0.154 | −28.1 | 4248–4268 |
| hsa-miR-94 | −0.151 | −27.9 | 1803–1824 |
| hsa-miR-382-5p | −0.145 | −26.3 | 1749–17700 |
| hsa-miR-23a-5p | −0.138 | −22.7 | 2702–2913 |
| hsa-miR-744-5p | −0.131 | −30.0 | 4285–4306 |
| hsa-miR-125a-3p | −0.130 | −24.4 | 1984–2005 |
| hsa-miR-105-3p | −0.121 | −25.1 | 3010–3031 |
| hsa-miR-582-3p | −0.118 | −21.2 | 252–975 |
| hsa-miR-323a-5p | −0.118 | −24.7 | 2965–2986 |
| hsa-miR-103a-2-5p | −0.109 | −26.3 | 3931–3951 |
| hsa-miR-629-3p | −0.058 | −25.4 | 3929–3949 |
| hsa-miR-1226-3p | −0.040 | −27.5 | 1048–1069 |
| hsa-miR-135b-3p | −0.033 | −30.1 | 3308–3329 |
| hsa-miR-3191-3p | −0.030 | −24.2 | 3355–3376 |
| hsa-miR-23b-5p | −0.025 | −25.6 | 2702–2913 |
| hsa-miR-942-5p | −0.016 | −26.2 | 2688–2899 |
| hsa-miR-194-3p | −0.007 | −25.0 | 4205–4226 |
| hsa-miR-32-3p | 0.002 | −21.1 | 2564–2585 |
| hsa-miR-664a-5p | 0.016 | −30.8 | 4325–4346 |
| hsa-miR-1291 | 0.026 | −24.1 | 3973–3993 |
| hsa-miR-766-3p | 0.042 | −24.2 | 1774–1795 |
| hsa-miR-1266-5p | 0.048 | −24.2 | 2706–2917 |
| hsa-miR-3065-3p | 0.052 | −28.5 | 3905–3926 |
| hsa-miR-328-3p | 0.056 | −23.7 | 3977–3994 |
| hsa-miR-3194-5p | 0.057 | −27.5 | 2561–258 |
| hsa-miR-671-5p | 0.064 | −27.1 | 2583–2607 |
| hsa-miR-3157-5p | 0.083 | −26.4 | 2937–2958 |
| hsa-miR-185-3p | 0.088 | −29.4 | 2647–2668 |
| hsa-miR-34a-5p | 0.121 | −23.2 | 192–213 |
| hsa-miR-454-5p | 0.122 | −24.2 | 3262–3283 |
| hsa-miR-2114-5p | 0.139 | −24.7 | 2053–2071 |
Calculated by the RNAhybrid program (http://bibiserv2.cebitec.uni-bielefeld.de/rnahybrid).
Position is defined as the nucleotide position at the CYP2D6 coding region in NM_000106, and the first nucleotide at the transcription start site is counted as position 1.
Fig. 1.
In silico analyses of the interaction between hsa-miRNA-370-3p and CYP2D6 mRNA. (A) According to the TCGA public database, the level of hsa-miRNA-370-3p was negatively correlated with the CYP2D6 mRNA level in non-tumor human liver tissues (r = −0.279 P = 0.049). The data points represent the relative expression levels of CYP2D6 mRNA and hsa-mir-370-3p, and the unit was defined as RPKM (Reads Per Kilobase Million). (B) Predicted hybrid complexes formed by hsa-miR-370-3p and its targeted sequence within the coding region of wild-type CYP2D6 mRNA. The free energy of hybridization is −29.2 kcal/mol. (C) A mutated hsa-miR-370-3p sequence was designed to include three altered nucleotides (italic, underlined). The predicted free energy of hybridization of the mutated hsa-miR-370-3p sequence is −17.7 kcal/mol.
3.2. hsa-miR-370-3p interacted with its target in CYP2D6 directly
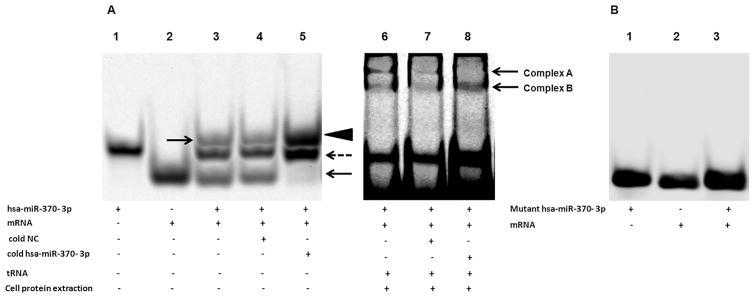
RNA EMSA was performed to explore whether or not hsa-miR-370-3p binds to its target within the CYP2D6 coding region directly. As shown in Fig. 2 A, when hsa-miR-370-3p interacted with its cognate mRNA to form a duplex, the duplex ran much slower than the free oligonucleotides during electrophoresis (lane 3, the duplex was indicated by an arrow). In the competition assays, adding 50-fold molar excess of unlabeled negative control oligonucleotide to the mixture did not influence the complexes formation (lane 4). In the presence of 50-fold molar excess of cold hsa-miR-370-3p, a large amount of labeled mRNA interacted with cold miRNA to form a bigger complex with a higher density (lane 5, the bigger duplex was indicated by an arrowhead), while the density of the band representing the free labeled mRNA target was significantly decreased (lane 5, free un-interacted mRNA was indicated by an arrow). On the other hand, the free hsa-miR-370-3p band became much stronger, because the competition between cold hsa-miR-370-3p and labeled hsa-miR-370-3p made more labeled hsa-miR-370-3p un-reacted (lane 5, indicated by a dotted arrow). When 3 nucleotides in the hsa-miR-370-3p sequence were mutated, the free energy of hybridization between the mutated miRNA and its targeted mRNA was reduced to −17.7 kcal/mol (Fig. 1C), which is not favorable to form a stable miRNA/mRNA complex. This was confirmed by the RNA EMSA assay, as the mutant hsa-miR-370-3p failed to interact with its targeted sequence (Fig. 2B). These results indicated that hsa-miR-370-3p interacted with its target in the CYP2D6 coding region directly and specifically. When cytoplasmic extract from HepaRG cells was added to the mixture, two miRNA/mRNA/protein complexes were formed (Fig. 2A, lanes 6, 7, and 8, complexes A and B). Complex A was competitively inhibited by 50-fold molar excess of cold hsa-miR-370-3p (lane 8), suggesting a specific interaction among mRNA, miRNA and proteins. When the Ago1, Ago2, Ago3, or Ago4-specific antibody was added respectively, no miRNA/mRNA/protein/antibody super-shift band was detected (data not shown). Therefore, the miRNA/mRNA/protein complex A was collected to perform mass spectrometry analysis, and the results are described in Section 3.7.
Fig. 2.
RNA electrophoretic mobility shift assays (EMSAs) demonstrated that the hsa-miR-370-3p oligonucleotides interacted directly with CYP2D6 mRNA oligonucleotides. (A) Hsa-miR-370-3p and/or CYP2D6 mRNA oligonucleotides were incubated in the mixture to form miRNA/mRNA hybrids. Lane 1 = free miRNA, Lane 2 = free mRNA, Lane 3 = miRNA + mRNA (arrow indicates miRNA/mRNA hybridization), Lane 4 = miRNA + mRNA + cold negative control (NC, cold NC does not affect hybridization), Lane 5 = miRNA + mRNA + cold hsa-miR-370-3p (hybridization was inhibited, arrowhead indicates the miRNA/mRNA hybridization, dotted arrow indicates the free miRNA, arrow indicates the free mRNA). Lane 6 to lane 8 represent the RNA:protein interactions. After cells cytoplasmic extract was added to reaction mixtures, there was a miRNA/mRNA/protein complex formation (complexes A and B). Complex A was competitively inhibited by 50-fold molar excess of cold hsa-miR-370-3p (lane 8). (B) Mutated hsa-miR-370-3p failed to form miRNA/mRNA hybridization with CYP2D6 mRNA oligonucleotides.
3.3. hsa-miR-370-3p suppressed the exogenous expression of CYP2D6 in HepG2CYP2D6 cells
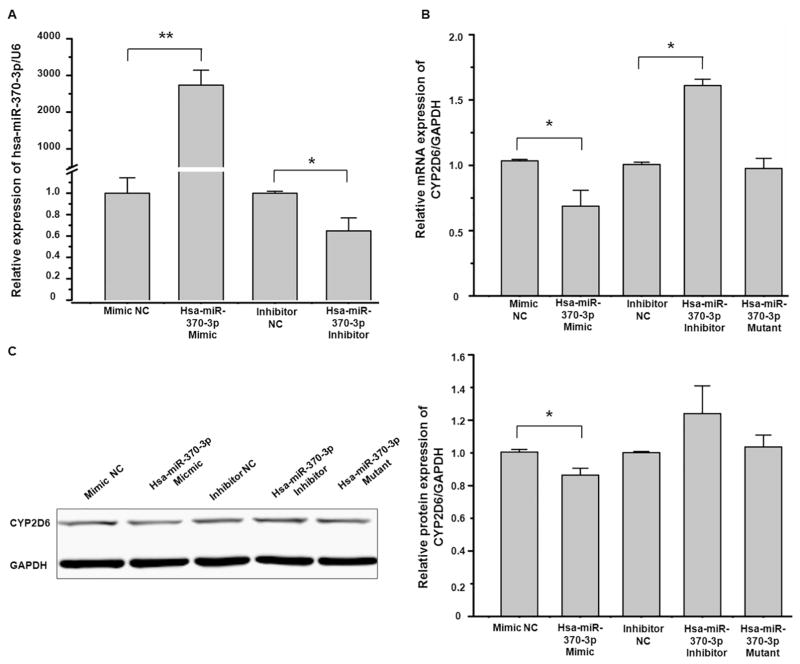
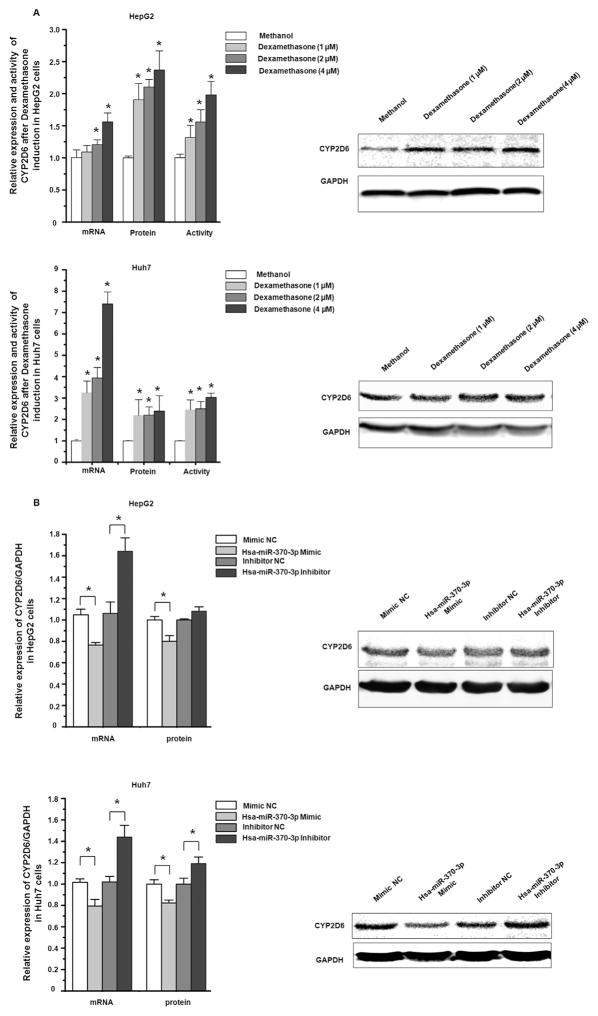
We used HepG2CYP2D6 cells to investigate whether hsa-miR-370-3p could regulate the expression of CYP2D6. As shown in Fig. 3, the transfection of hsa-miR-370-3p mimic drastically increased the intracellular level of hsa-miR-370-3p (Fig. 3A), and suppressed the expression of CYP2D6 at mRNA and protein levels (Fig. 3B and C), whereas the transfection of hsa-miR-370-3p inhibitor decreased the hsa-miR-370-3p level (Fig. 3A) and increased the abundance of CYP2D6 mRNA and protein (Fig. 3B and C). Hsa-miR-370-3p-mutant did not show any effect on the expression of CYP2D6 (Fig. 3B and C). In Fig. 3 C, although the difference of CYP2D6 protein expression between the hsa-miR-370-3p inhibitor treatment group and the inhibitor NC treatment group was statistically insignificantly (P value > 0.05), the trend of the CYP2D6 protein level was upward in the hsa-miR-370-3p inhibitor treatment group.
Fig. 3.
Hsa-miR-370-3p suppressed exogenous CYP2D6 expression. HepG2CYP2D6 cells were transiently transfected using 50 nmol/L mimic negative control, hsa-miR-370-3p mimic, hsa-miR-370-3p mimic-mutant, miRNA inhibitor negative control, or hsa-miR-370-3p inhibitor, respectively. (A) The transfection of hsa-miR-370-3p mimic increased the hsa-miR-370-3p level in HepG2CYP2D6 cells apparently, whereas hsa-miR-370-3p inhibitor decreased the hsa-miR-370-3p level. (B) The expression of CYP2D6 mRNA was detected by qRT-PCR. The hsa-miR-370-3p mimic suppressed exogenous CYP2D6 expression, whereas the hsa-miR-370-3p inhibitor increased CYP2D6 expression. The hsa-miR-370-3p-mutant did not show any effect on CYP2D6 expression. (C) Western blot analysis showed that CYP2D6 protein levels were decreased in HepG2CYP2D6 cells transfected with the hsa-miR-370-3p mimic, and were increased in cells transfected with the hsa-miR-370-3p inhibitor. The hsa-miR-370-3p-mutant did not show any effect on CYP2D6 protein expression. Data shown as mean ± SD (n = 3). * P < 0.05 vs. Negative Control. ** P < 0.01 vs. Negative Control.
3.4. hsa-miR-370-3p suppressed endogenous expression of CYP2D6 in HepaRG cells
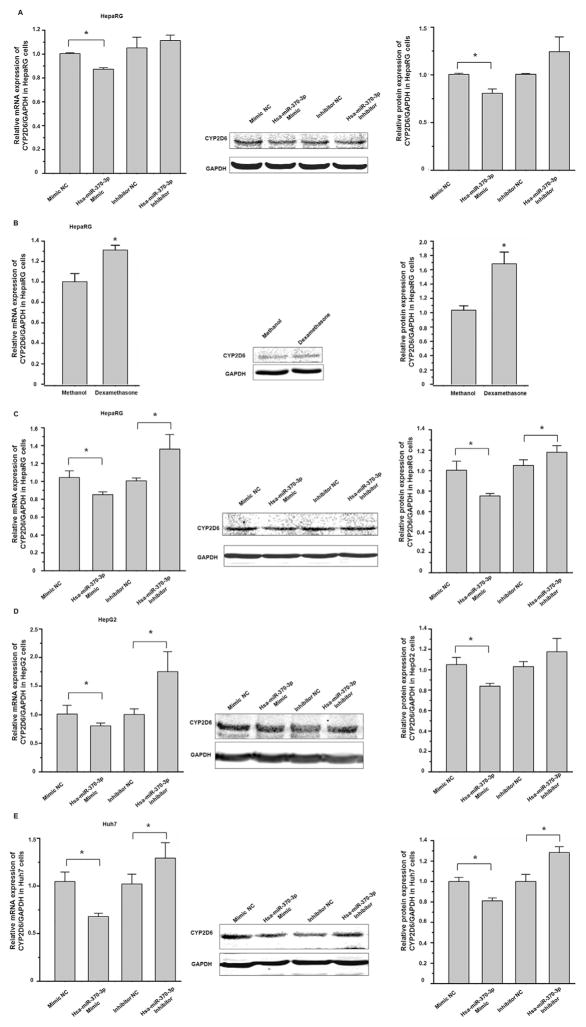
HepaRG, a terminally differentiated hepatic cell line, expresses many DMEs at comparable levels to those in primary human hepatocytes [22]. As shown in Fig. 4A, the transfection of hsa-miR-370-3p mimic into HepaRG cells suppressed the endogenous expression of CYP2D6 at both mRNA and protein levels. We used dexamethasone to induce CYP2D6 in HepaRG cells, to investigate whether the transfection of the hsa-miR-370-3p mimic can suppress the induction of CYP2D6. As shown in Fig. 4B, after dexamethasone incubation, the expression of CYP2D6 increased 1.5-fold and 1.68-fold at mRNA and protein levels, respectively. When hsa-miR-370-3p mimic was transfected into HepaRG cells prior to dexamethasone treatment, the induction of CYP2D6 at both mRNA and protein levels was attenuated (85% of that in NC treated cells at mRNA level, and 75% of that in NC treated cells at protein level. P < 0.05, Fig. 4C).
Fig. 4.
Hsa-miR-370-3p suppressed endogenous CYP2D6 expression in HepaRG, HepG2, and Huh7 cells. Cells were transiently transfected using 50 nmol/L mimic negative control, hsa-miR-370-3p mimic, miRNA inhibitor negative control, and hsa-miR-370-3p inhibitor, respectively. (A) The hsa-miR-370-3p mimic suppressed endogenous CYP2D6 expression in HepaRG cells. (B) Treatment with dexamethasone led to increased levels of CYP2D6 mRNA and protein in HepaRG cells. (C) MiRNAs transfection of HepaRG cells prior to dexamethasone treatment showed that the induction of CYP2D6 by dexamethasone could be attenuated by hsa-miR-370-3p mimic, whereas hsa-miR-370-3p inhibitor up-regulated the CYP2D6 level. In hepatocarcinoma cells, the hsa-miR-370-3p mimic suppressed endogenous CYP2D6 levels, whereas hsa-miR-370-3p inhibitor increased the CYP2D6 levels in HepG2 cells (D) and Huh7 cells (E), respectively. Data shown as mean ± SD (n = 3). *P < 0.05 vs. Negative Control.
3.5. hsa-miR-370-3p attenuated the induction of CYP2D6 by dexamethasone in hepatic HepG2 and Huh7 cells
Hepatoma cell lines HepG2 and Huh7 are commonly used as in vitro cell models in pharmacological and toxicological studies owing to the inducibility of CYPs. Because the expression of CYPs in cultured cells may be influenced by the cell passages, we tested the effects of cell culture passages on the expression of CYP2D6 using HepG2 cells (with passage numbers of 3–7), and the Huh7 cells (with passage numbers of 6–9). The qRT-PCR results from independent experiments showed that the differences in the expression of CYP2D6 mRNA in HepG2 cells and in Huh7 cells with different passages were not statistically significant (P > 0.05, data not shown). Then we examined the effects of hsa-miR-370-3p on the expression of CYP2D6 in both HepG2 and Huh7 cell lines. Although the endogenous expression level of CYP2D6 was relatively low in these cells, we still detected the suppression effect of hsa-miR-370-3p mimic on the expression of CYP2D6 at mRNA and protein levels (Fig. 4D & E). We next studied the induction of CYP2D6 by dexamethasone in these cells, and explored the effect of hsa-miR-370-3p on CYP2D6 expression. As shown in Fig. 5 A, dexamethasone upregulated the mRNA, protein, and activity levels of CYP2D6 in a concentration-dependent manner, in both HepG2 and Huh7 cells. When miRNAs was transfected into cells prior to dexamethasone treatment, hsa-miR-370-3p mimic attenuated the dexamethasone-induced expression of CYP2D6 in both HepG2 (reduced by 24% at mRNA level and 20% at protein level, compared to controls) and Huh7 cells (reduced by 21% at mRNA level and 20% at protein level, all P < 0.05, Fig. 5B).
Fig. 5.
Hsa-miR-370-3p inhibited drug-induced CYP2D6 expression in HepG2 and Huh7 cells. (A) Cells were seeded in 24-wells plates and dexamethasone was added to the DMEM medium at final concentrations of 1 μM, 2 μM, or 4 μM for a period of 72 h. After dexamethasone treatment, the CYP2D6 expression in HepG2 and Huh7 cells were upregulated in both mRNA and protein levels. The enzyme activity was also increased in a concentration-dependent pattern. (B) MiRNAs transfection of HepG2 and Huh7 cells prior to dexamethasone treatment showed that the induction of CYP2D6 by dexamethasone could be attenuated by hsa-miR-370-3p mimic, whereas hsa-miR-370-3p inhibitor increased the CYP2D6 levels. Data shown as mean ± SD (n = 3). *P < 0.05 vs. Negative Control.
3.6. Hsa-miR-370-3p decreased the stability of CYP2D6 transcript
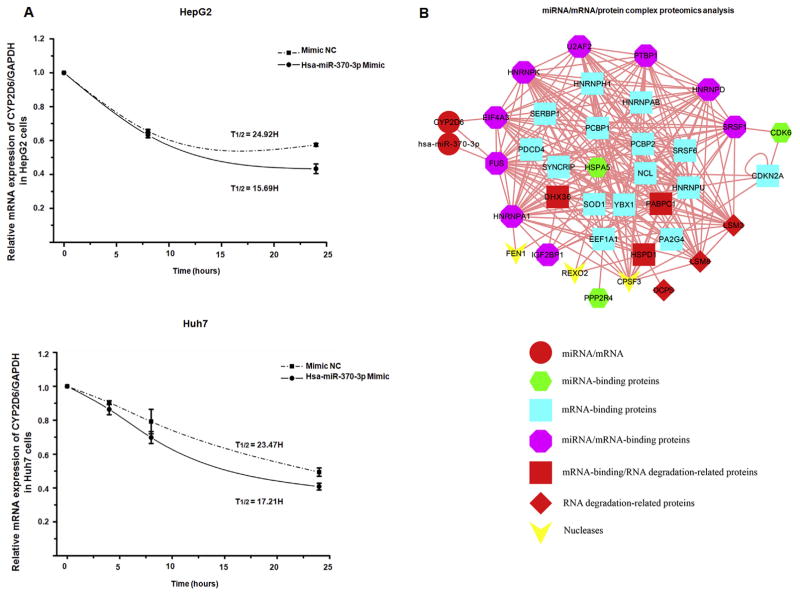
To investigate whether hsa-miR-370-3p plays a role in decreasing CYP2D6 mRNA stability, we used actinomycin D to inhibit mRNA transcription and then measured the half-life of the CYP2D6 transcript in HepG2 and Huh7 cells. As shown in Fig. 6A, the calculated half-life of the CYP2D6 transcript in HepG2 cells transfected with the hsa-miR-370-3p mimic was 16-h, compared to a half-life of 25-h in the negative control miRNA-transfected cells. Similar results were obtained in Huh7 cells, with a 17-h half-life of CYP2D6 transcript in hsa-miR-370-3p mimic transfected cells compared to a 23-h half-life of CYP2D6 transcripts in negative control cells. These results suggested that hsa-miR-370-3p facilitated the degradation of endogenous CYP2D6 transcripts and resulted in a suppression of CYP2D6.
Fig. 6.
Hsa-miR-370-3p inhibited CYP2D6 expression via mRNA degradation in HepG2 and Huh7 cells. (A) HepG2 and Huh7 cells were incubated with actinomycin D to inhibit transcription, and CYP2D6 mRNA expression was quantified by qRT-PCR at 0, 4, 8, and 24 h after treatment. The half-life of CYP2D6 mRNA in HepG2 cells transfected with hsa-miR-370-3p mimic was 15.96-h, and that of the mimic NC treated cells was 24.92-h. The half-life of CYP2D6 mRNA in Huh7 cells transfected with hsa-miR-370-3p mimic was 14.34-h, and that of the mimic NC treated cells was 23.47-h. Data shown as mean ± SD (n = 3). (B) The interaction complex of miRNA/mRNA/protein was separated on 12% PAGE. The band representing the complex was excised and subjected to protein identification using mass spectrometry. The identified proteins were used to assemble a miRNA/mRNA/protein network. Three nucleases and six RNA degradation-related proteins participate in this network, indicating degradation of CYP2D6 mRNA.
3.7. Proteomics analysis of the miRNA/mRNA/protein complex
Mass spectrometry analysis of proteins from the isolated miRNA/mRNA/protein complex revealed that, among identified proteins, RNA binding proteins and proteins associated with RNA degradation were highly enriched. Specifically, identified proteins including miRNA-binding proteins, mRNA-binding proteins, nucleases and mRNA degradation-related proteins showed higher abundance. To analyze specific molecular interactions within the complex, mRNA/miRNA/protein interaction networks were built up using bioinformatics approaches for the mass spectrometry identified proteins (Fig. 6B). The interaction network suggested that FUS, a protein component of heterogeneous nuclear ribonucleoprotein complex with many biological functions, directly interacts with the hsa-miR-370-3p/CYP2D6-mRNA duplex, and the eukaryotic translation initiation factor EIF4A3 directly interacts with CYP2D6 mRNA. Many other miRNA and/or mRNA binding proteins may directly or indirectly interact with FUS and EIF4A3. The protein FUS belongs to the FET family of RNA-binding proteins that have been implicated in many cellular processes, including regulation of gene expression, maintenance of genomic integrity and mRNA/microRNA processing [23,24]. In addition, three nucleases and six RNA degradation-related proteins are involved in the formation of miRNA/mRNA/protein complex, indicating their roles in the degradation of CYP2D6 mRNA. Although our data only identified two proteins that could directly interact with hsa-miR-370-3p and/or CYP2D6 mRNA, there is a possibility that other proteins may play the same role, which requires further research to determine.
4. Discussions
Accumulating evidence has indicated that miRNAs are important components involved in post-transcriptional regulation of many genes, including DMEs [12]. Several CYP genes have been shown to be regulated by miRNAs binding their 3′-UTR, including CYP2B6, CYP3A4, CYP1A2, and CYP1B1 [11,12,17]. Besides miRNA binding to 3′-UTRs, the binding of miRNAs to their targets within mRNA coding regions can also modulate gene expression. It has been reported that miR-602 and miR-608 regulated the expression of sonic hedgehog (SHH) by targeting their binding sites located in the coding region of the SHH mRNA [25]. In our previous study, we also demonstrated that hsa-miR-29a-3p modulates CYP2C19 by targeting a coding region [15]. As for CYP2D6, the full length of 3′-UTR only contains 75 nucleotides, which dramatically decreases the chance to be targeted by miRNAs. Previous in silico and in vitro experiments showed that several miRNAs, such as hsa-miR-34a and hsa-miR-449a, can regulate CYP2D6 indirectly via suppression of the expression of hepatic nuclear factor 4α, a transcriptional factor that influences the expression and activity of CYP2D6 [26]. In the present study, although the correlation between CYP2D6 mRNA and hsa-miR-370-3p (analysis of data derived from the TCGA database) was weak, we systematically validated roles of miRNAs in the regulation of CYP2D6 using multilayer experiments, including in silico, in vitro and proteomics approaches, and demonstrated that CYP2D6 can be modulated by hsa-miRNA-370-3p directly with the binding site within the coding region.
It is known that mature miRNAs incorporate into the RNA-induced silencing complex (RISC) that recognizes the 3′-UTR of the targeted mRNA, thus causing mRNA cleavage [27]. The key components of the RISC complex are the Argonaute proteins, including Ago1, Ago2, Ago3, and Ago4. Additional proteins also have been reported to be associated with the RISC-complex, such as Vasa intronic gene (VIG) protein, Fragile X-related protein and the Gemin3 [28,29]. Our proteomics analysis of the miRNA/mRNA/protein complex indicated that many proteins other than Ago family members were involved in the formation of the CYP2D6 miRNA/mRNA/protein complex. Among the identified proteins, FUS and EIF4A3 may have a close relationship with CYP2D6 transcripts, by directly interacting with hsa-miR-370-3p/CYP2D6-mRNA duplex or with CYP2D6 mRNA (Fig. 6 B). EIF4A3, a core component of the splicing-dependent multiprotein exon junction complex (EJC), is essential for nonsense-mediated mRNA decay (NMD) [30]. FUS is involved in multiple facets of DNA/RNA metabolism, including maintenance of genomic stability, and regulation of gene expression by modulation of transcription and RNA metabolism [23,24]. Interacting with protein TDP-43, FUS protein is a multifunctional protein whose roles in mRNA processing are largely unknown. It is notable, however, that FUS can shuttle between nucleus and cytoplasm and ultimately contribute to the spatiotemporal fates of mRNA, specifically impacting mRNA stability, translocation, translation and decay [31]. Although our results in the present study indicated that EIF4A3 and FUS are involved in the CYP2D6 mRNA decay process, the precise mechanism underlying hsa-miR-370-3p mediated CYP2D6 regulation is still unclear, and more biochemical studies are warranted.
Alternatively, miRNAs can exert their ability on targeted mRNAs silencing via a slicer-independent mechanism [32]. For example, a miRNA can significantly down-regulate the protein production with little change in the mRNA level, suggesting that the mRNA is silenced by the miRNA through repressing the protein translation [33]. In this study, hsa-miR-370-3p decreased CYP2D6 mRNA level in parallel with the protein level (Fig. 4 D, 20% in mRNA decrease and 17% in protein decrease in HepG2 cells), suggesting that hsa-miR-370-3p suppressed the CYP2D6 via mRNA degradation, which is consistent with our mRNA stability data. Although specific functions of the proteins in the miRNA/mRNA/protein complex remain unclear, the current study provides insight on the mechanism of miRNAs in the suppression of CYP2D6 by targeting its transcripts at the coding region thus facilitating mRNA degradation.
Cytochrome P450 enzymes are involved in the activation and/or detoxification of xenobiotics; therefore, a change in the expression of CYPs in hepatocytes could influence the clearance and toxicity of drugs. It was reported that concurrent use of CYP2D6 substrates or inhibitors could influence cardiovascular patients’ therapeutic responses, in some cases even result in adverse effects. For example, when acute myocardial infarction patients were treated with metoprolol (a substrate for CYP2D6) and co-treated with paroxetine (an inhibitor of CYP2D6), paroxetine inhibited the metabolism of metoprolol and increased its concentration maximum (Cmax), resulting in enhanced therapeutic effects and/or adverse effects, such as severe dizziness, fainting and heart failure [34]. In anti-cancer therapy, many studies have demonstrated that altered expression of DMEs can influence the sensitivity and toxicity of drugs in target cells, and the expression of DMEs plays a pivotal role in chemotherapy sensitivity, resistance, and/or carcinogenesis [35,36]. The enzyme activity of CYP2D6 significantly impacts the anti-estrogen effect of tamoxifen in cancer therapy, because it is the most important CYP in the conversion of tamoxifen to 4-OH tamoxifen [37]. Consequently, patients who are poor CYP2D6 metabolizers have shorter relapse-free and disease-free survival [38]. A group of clinical data reported that the expression level of hsa-miR-370-3p in breast tumor tissues was negatively correlated to overall survival status of patients [39]. In view of our results, it is reasonable to speculate that hsa-miR-370-3p could suppress the expression of CYP2D6, resulting in a decreased therapeutic effect of tamoxifen. Therefore, any drug that can increase the expression of hsa-miR-370-3p may cause an impaired therapeutic response of tamoxifen, thus impacting the process of miRNA mediated drug-drug interaction that affects the efficacy of tamoxifen.
Dexamethasone, a potent exogenous corticosteroid that is widely used in anti-inflammation, immunosuppression, and anti-cancer therapeutics, is an inducer of CYP2D6 [10]. In our study, using HepaRG cells, the transfection of hsa-miR-370-3p mimic blocked dexamethasone-induced CYP2D6 induction, whereas the hsa-miR-370-3p inhibitor increased the expression of CYP2D6 (Fig. 4C), suggesting that hsa-miR-370-3p might be a potential mediator in drug-drug interactions through inhibition or induction of CYP2D6.
In summary, we demonstrated that hsa-miR-370-3p suppresses CYP2D6 expression by targeting a specific sequence located within the coding region of CYP2D6 for its mRNA degradation. Proteomics approach suggested that several multifunctional proteins are involved in the CYP2D6 transcript decay process. Our results may provide insight regarding a novel epigenetic mechanism for regulating CYP2D6 gene expression.
Acknowledgments
This study was supported and funded by the National Center for Toxicological Research (Project E0731311 and E0757801), U.S. Food and Drug Administration.
Abbreviations
- CYP
cytochrome P450
- miRNA
microRNA
- DMEs
drug metabolizing enzymes
- Ago
argonaute RISC catalytic component
- RISC
RNA-induced silence complex
- EMSA
electrophoretic mobility shift assay
Footnotes
Author contributions
Participated in Study design: Zeng L., Chen Y., Tong W., Yu D. and Ning B.
Conducted experiments: Zeng L., Chen Y., Knox B., Wang Y., Yu D, Yu L., Ren Z., Chen S., and Wu Y.
Performed data analysis: Zeng L., Yu D., Yu L., Guo L., Shi T., Chen J., Liu D., Huang K., Tong W., and Ning B.
Wrote or contributed to the writing of the manuscript: Zeng L., Chen S., Ren Z., Guo L., Yu L., Chen J., Yu D., Knox B., Tong W., and Ning B.
Disclaimer
The information in these materials is not a formal dissemination of the U.S. Food and Drug Administration.
References
- 1.Rendic S. Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002;34(1–2):83–448. doi: 10.1081/dmr-120001392. [DOI] [PubMed] [Google Scholar]
- 2.Daly AK. Polymorphic variants of cytochrome P450: relevance to cancer and other diseases. Adv Pharmacol. 2015;74:85–111. doi: 10.1016/bs.apha.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Teh LK, Bertilsson L. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet. 2012;27(1):55–67. doi: 10.2133/dmpk.dmpk-11-rv-121. [DOI] [PubMed] [Google Scholar]
- 4.Tracy TS, Chaudhry AS, Prasad B, Thummel KE, Schuetz EG, Zhong XB, et al. Interindividual variability in cytochrome P450-mediated drug metabolism. Drug Metab Dispos. 2016;44(3):343–351. doi: 10.1124/dmd.115.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5(1):6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 6.Bradford LD. CYP2D6 allele frequency in European Caucasians Asians, Africans and their descendants. Pharmacogenomics. 2002;3(2):229–243. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 7.Tracy TS, Venkataramanan R, Glover DD, Caritis SN H. National, Institute for Child, U. Human Development Network of Maternal-Fetal-Medicine. Temporal changes in drug metabolism (CYP1A2, CYP2D6 and CYP3A Activity) during pregnancy. Am J Obstet Gynecol. 2005;192(2):633–639. doi: 10.1016/j.ajog.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Ryu RJ, Eyal S, Easterling TR, Caritis SN, Venkataraman R, Hankins G, et al. Pharmacokinetics of metoprolol during pregnancy and lactation. J Clin Pharmacol. 2016;56(5):581–589. doi: 10.1002/jcph.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadelius M, Darj E, Frenne G, Rane A. Induction of CYP2D6 in pregnancy. Clin Pharmacol Ther. 1997;62(4):400–407. doi: 10.1016/S0009-9236(97)90118-1. [DOI] [PubMed] [Google Scholar]
- 10.Farooq M, Kelly EJ, Unadkat JD. CYP2D6 is inducible by endogenous and exogenous corticosteroids. Drug Metab Dispos. 2016;44(5):750–757. doi: 10.1124/dmd.115.069229. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Koturbash I, Tolleson WH, Guo L, Yu D, Chen S, Hong H, et al. MicroRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark Med. 2015;9(11):1153–1176. doi: 10.2217/bmm.15.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res. 2006;66(18):9090–9098. doi: 10.1158/0008-5472.CAN-06-1403. [DOI] [PubMed] [Google Scholar]
- 13.Pan YZ, Gao W, Yu AM. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab Dispos. 2009;37(10):2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohri T, Nakajima M, Fukami T, Takamiya M, Aoki Y, Yokoi T. Human CYP2E1 is regulated by miR-378. Biochem Pharmacol. 2010;79(7):1045–1052. doi: 10.1016/j.bcp.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Yu D, Green B, Tolleson WH, Jin Y, Mei N, Guo Y, et al. MicroRNA hsa-miR-29a-3p modulates CYP2C19 in human liver cells. Biochem Pharmacol. 2015;98(1):215–223. doi: 10.1016/j.bcp.2015.08.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, et al. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y, Yu D, Tolleson WH, Knox B, Wang Y, Chen S, et al. MicroRNA hsa-miR-25-3p suppresses the expression and drug induction of CYP2B6 in human hepatocytes. Biochem Pharmacol. 2016;113:88–96. doi: 10.1016/j.bcp.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu D, Tolleson WH, Knox B, Jin Y, Guo L, Guo Y, et al. Modulation of ALDH5A1 and SLC22A7 by microRNA hsa-miR-29a-3p in human liver cells. Biochem Pharmacol. 2015;98(4):671–680. doi: 10.1016/j.bcp.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xuan J, Chen S, Ning B, Tolleson WH, Guo L. Development of HepG2-derived cells expressing cytochrome P450s for assessing metabolism-associated drug-induced liver toxicity. Chem Biol Interact. 2016;255:63–73. doi: 10.1016/j.cbi.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leclerc GJ, Leclerc GM, Barredo JC. Real-time RT-PCR analysis of mRNA decay: half-life of Beta-actin mRNA in human leukemia CCRF-CEM and Nalm-6 cell lines. Cancer Cell Int. 2002;2(1):1. doi: 10.1186/1475-2867-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen QR, Yu LR, Tsang P, Wei JS, Song YK, Cheuk A, et al. Systematic proteome analysis identifies transcription factor YY1 as a direct target of miR-34a. J Proteome Res. 2011;10(2):479–487. doi: 10.1021/pr1006697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanebratt KP, Andersson TB. HepaRG cells as an in vitro model for evaluation of cytochrome P450 induction in humans. Drug Metab Dispos. 2008;36(1):137–145. doi: 10.1124/dmd.107.017418. [DOI] [PubMed] [Google Scholar]
- 23.Masuda A, Takeda J, Ohno K. FUS-mediated regulation of alternative RNA processing in neurons: insights from global transcriptome analysis. Wiley Interdiscip Rev RNA. 2016;7(3):330–340. doi: 10.1002/wrna.1338. [DOI] [PubMed] [Google Scholar]
- 24.Masuda A, Takeda J, Okuno T, Okamoto T, Ohkawara B, Ito M, et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015;29(10):1045–1057. doi: 10.1101/gad.255737.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis Rheumatol. 2015;67(2):423–434. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramamoorthy A, Li L, Gaedigk A, Bradford LD, Benson EA, Flockhart DA, et al. In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4alpha expression. Drug Metab Dispos. 2012;40(4):726–733. doi: 10.1124/dmd.111.040329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, et al. MiRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16(19):2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibuya T, Tange TO, Sonenberg N, Moore MJ. EIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11(4):346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- 31.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21(16):1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goryachkina K, Burbello A, Boldueva S, Babak S, Bergman U, Bertilsson L. Inhibition of metoprolol metabolism and potentiation of its effects by paroxetine in routinely treated patients with acute myocardial infarction (AMI) Eur J Clin Pharmacol. 2008;64(3):275–282. doi: 10.1007/s00228-007-0404-3. [DOI] [PubMed] [Google Scholar]
- 35.Khlifi R, Messaoud O, Rebai A, Hamza-Chaffai A. Polymorphisms in the human cytochrome P450 and arylamine N-acetyltransferase: susceptibility to head and neck cancers. Biomed Res Int. 2013;2013:582768. doi: 10.1155/2013/582768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 enzymes: novel options for cancer therapeutics. Mol Cancer Ther. 2004;3(3):363–371. [PubMed] [Google Scholar]
- 37.Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, et al. Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst. 2003;95(23):1758–1764. doi: 10.1093/jnci/djg108. [DOI] [PubMed] [Google Scholar]
- 38.Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101(1):113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 39.Sim J, Ahn H, Abdul R, Kim H, Yi KJ, Chung YM, et al. High MicroRNA-370 expression correlates with tumor progression and poor prognosis in breast cancer. J Breast Cancer. 2015;18(4):323–328. doi: 10.4048/jbc.2015.18.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]