Abstract
Imaging and chromosome conformation capture studies have revealed several layers of chromosome organization, including segregation into megabase-large active and inactive compartments, and partitioning into sub-megabase domains (TADs). Yet, it remains unclear how these layers of organization form, interact with one another and impact genome functions. Here, we show that deletion of the cohesin-loading factor Nipbl, in mouse liver, leads to a dramatic reorganization of chromosomal folding. TADs and associated peaks vanish globally, even in the absence of transcriptional changes. In contrast, compartmental segregation is preserved and even reinforced. Strikingly, the disappearance of TADs unmasks a finer compartment structure that accurately reflects the underlying epigenetic landscape. These observations demonstrate that the 3D organization of the genome results from the interplay of two independent mechanisms: 1) cohesin-independent segregation of the genome into fine-scale compartments, defined by chromatin state; 2) cohesin-dependent formation of TADs, possibly by loop extrusion, which contributes to guide distant enhancers to their target genes.
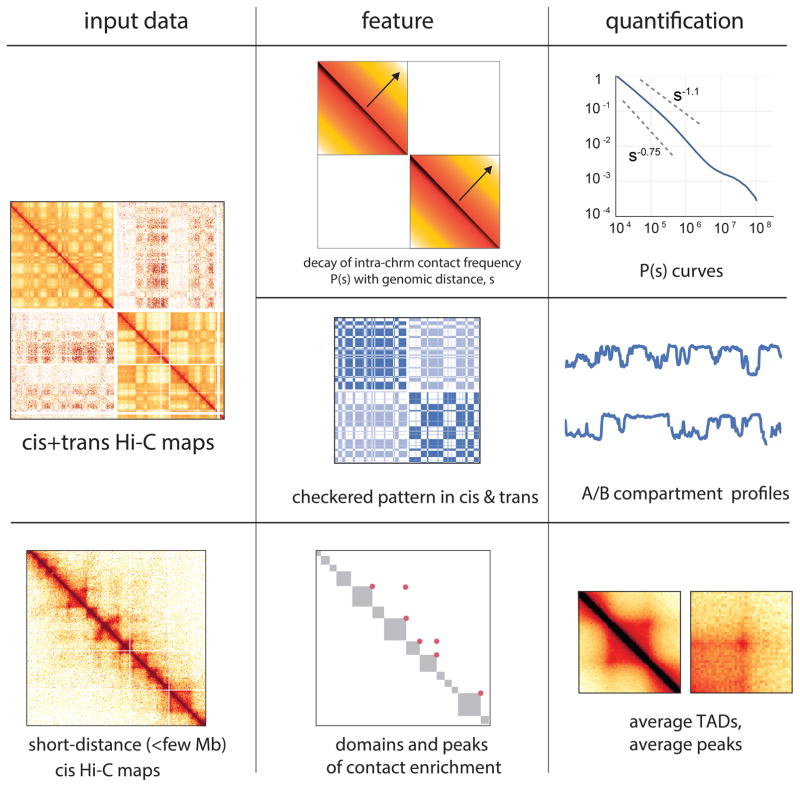
The three-dimensional organization of chromosomes is tightly related to their biological function 1–3. Hi-C maps have revealed key features of the 3D organization of metazoan chromosomes, including compartmentalization, TADs, and interaction peaks 4,5(Extended Data Figure 1). Compartmentalization is visible as a characteristic checkerboard pattern of contact enrichment both in cis and trans between megabase-sized genomic intervals of the same type, reflecting spatial segregation of transcriptionally active (type A) and inactive (type B) chromatin6. TADs appear as squares of enriched contact frequency with s harp boundaries; they usually span hundreds of kilobases and do not necessarily exhibit any checkering 7,8. TADs are thought to contribute to gene expression, notably by promoting or preventing interactions between promoters and distant regulatory elements 9–11. Finally, peaks (often termed loops 12) are visible as focal enrichments in contact frequency between pairs of loci, often at the corner of TAD squares. Despite a lack of supporting mechanistic experiments, compartments, TADs and peaks are typically assumed to constitute hierarchical levels of chromosome folding. Yet, the connection between them remains poorly understood.
Architectural proteins, notably cohesin and CTCF, are believed to play crucial roles for interphase chromatin organization 13. Cohesin and CTCF co-localize at TAD boundaries and the bases of Hi-C peaks 7,12, but their roles are not fully defined. The recently proposed loop extrusion model 14,15 yields predictions that are consistent with experiments. In this model, TADs emerge from the progressive extrusion of chromatin loops by a protein complex (e.g. cohesin) until it dissociates from chromatin or reaches a boundary element (e.g. CTCF sites). Recent experimental manipulation of CTCF motifs 15–17 and CTCF depletion 18 support the proposed role of CTCFs as boundary elements, but cohesin’s role in interphase chromatin organization and the process of loop extrusion remains ambiguous, as experimental depletions of cohesin have shown limited impact on chromatin organisation 19–21
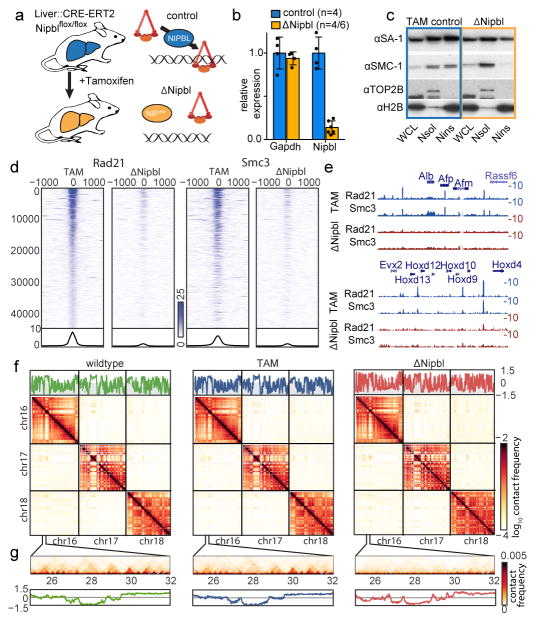
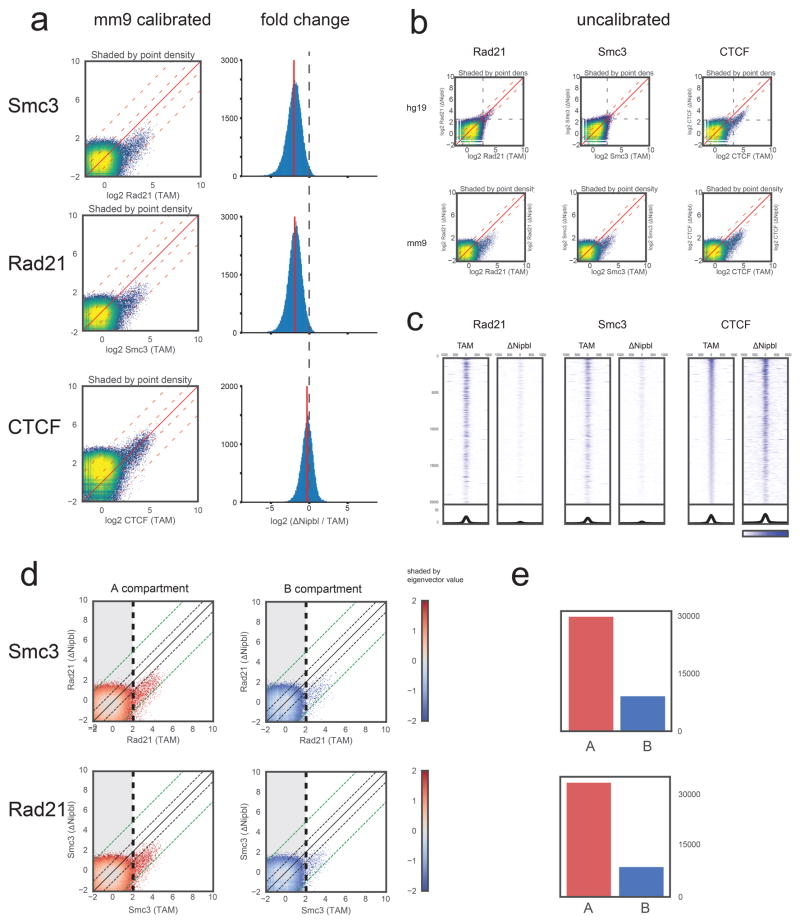
To determine the role of cohesin in interphase chromatin, we deleted NIPBL/SCC2, the factor which is necessary for cohesin loading on chromatin22. Since the turnover of chromosome-bound cohesin is ~20 minutes23,24, constant loading is required for its presence on DNA. We achieved efficient deletion of Nipbl in non-dividing hepatocytes by using a liver-specific, tamoxifen-inducible Cre driver (Fig. 1a), which circumvents the lethality of Nipbl +/− mice and the essentiality of cohesin in dividing cells (Extended Data Figure 2). Ten days after tamoxifen injection, Nipbl expression was dramatically reduced (Fig. 1b) and led to a displacement of cohesin from the chromatin fraction to the soluble nuclear fraction, indicating loss of cohesin from chromosomes (Fig. 1c). This strong depletion of chromatin-bound cohesin is also observed by calibrated ChIP-seq 25 for RAD21 and SMC3 both genome-wide (Extended Data Figure 3) and at WT cohesin peaks (Fig. 1d–e). A conservative estimate indicates at least a 4- to 6-fold decrease in chromatin-associated cohesin. No particular pathological signs compared to control animals (mock-injected Nipblflox/flox; Ttr-cre/Esr1 animals or tamoxifen-injected Nipbl+/+; Ttr-cre/Esr1 animals) were observed in the liver. Hepatocytes showed no sign of cell death or proliferation (Extended Data Figure 2).
Figure 1. Overview of the experimental design.
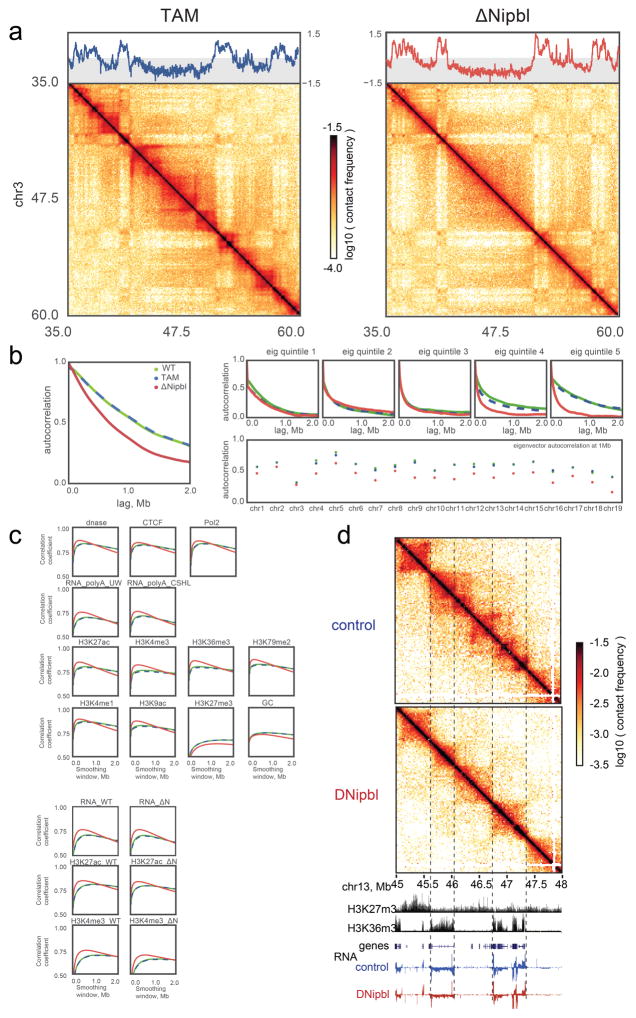
(a) S We deleted a conditional allele of Nipbl (Extended Data Fig. 2) in adult hepatocytes using a liver-specific driver for the CRE-ERT2 fusion protein after injection of Tamoxifen. In absence of Nipbl, cohesin (represented by a triangular ring) is not loaded on chromatin. (b) Expression of Nipbl and Gapdh (control) by RT-qPCR in control (n=4) and ΔNipbl (n=6 for Nipbl; 4 for Gapdh) hepatocytes. Mean normalized gene expression (using Pgk1 as internal control, and WT expression level set as 1) is displayed as mean and s.d and compared using unpaired two-sided t-test (95% CI control=[0.7033–1.297]; mutant=[0.05731–0.2198] for Nipbl expression). (c) Western blots of hepatocyte protein extracts (WCL: whole cell lysate, Nsol (nuclear extract, soluble fraction) Nins (insoluble, chromatin fraction)) showed displacement of cohesin structural subunits (SA-1, SMC1) from the chromatin-bound fraction. H2B and TOP2B distribution serves as controls for loading and enrichment of two nuclear fractions. Experiment repeated on two biologically independent samples per condition. See Supplementary Data 1 for gel data source. (d) Stacked heatmaps of calibrated ChIP-seq signal (for Rad21 and SMC3) at WT Rad21 peaks ranked by fold change over input in the TAM control condition. (e) ChIP-seq tracks for Rad21 and SMC3 over representative genomic regions. (f–g) Hi-C maps at 20kb resolution of WT (left), TAM control (middle) and ΔNipbl cells (right). Top – cis compartment tracks. Middle – cis and trans contact maps of chr16–18. (g) An example of short-range Hi-C map at chr16:25–32Mb with compartment tracks.
To assess the consequence of Nipbl depletion and cohesin loss on chromosome organization, we performed tethered chromatin conformation capture 26 (referred below as Hi-C) on purified hepatocytes from wildtype (WT), tamoxifen control (TAM) and ΔNipbl animals (Fig. 1f). For each of these three conditions, two biological replicates were generated; 5 out of 6 replicates produced >25 million interactions at >10kb separation, on par with other primary tissue Hi-C (27, Supplementary Tables 1–2). The contact maps obtained from each biological replicate showed extensive similarities (Supplementary Data 2), allowing us to pool the two replicate datasets to generate Hi-C maps for the three different conditions. We compared Hi-C maps for ΔNipbl and control cells by examining compartments, TADs, peaks, and global scaling of the contact probability P(s) 28(Extended Data Figure 1).
Disappearance of TADs and peaks
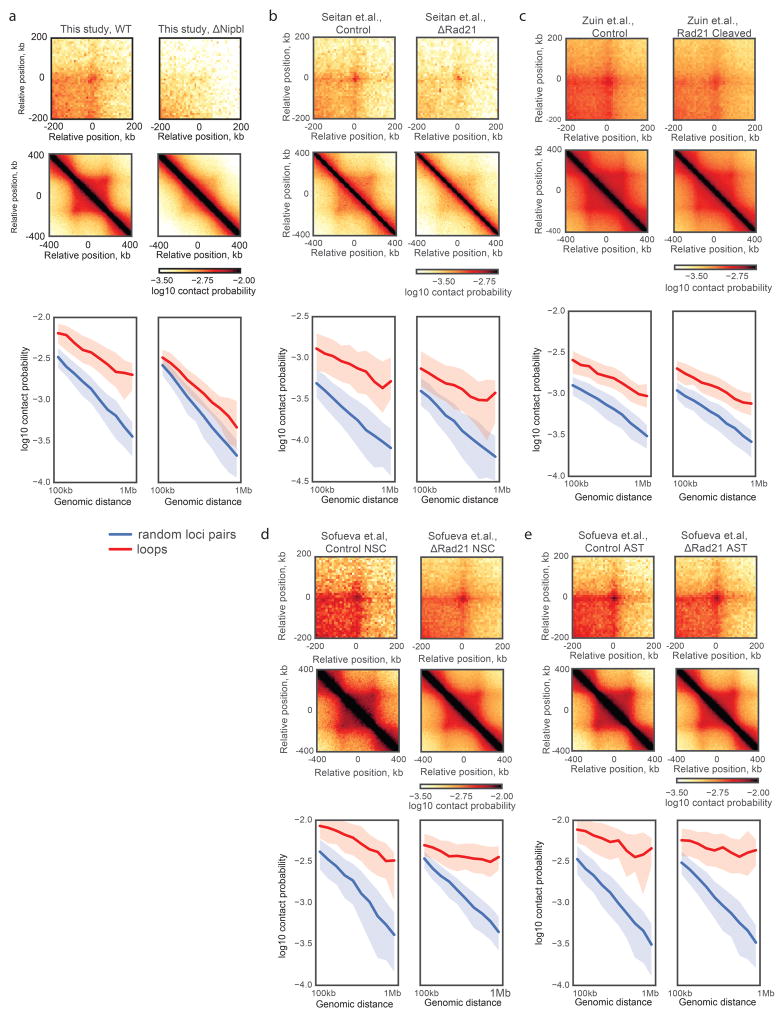
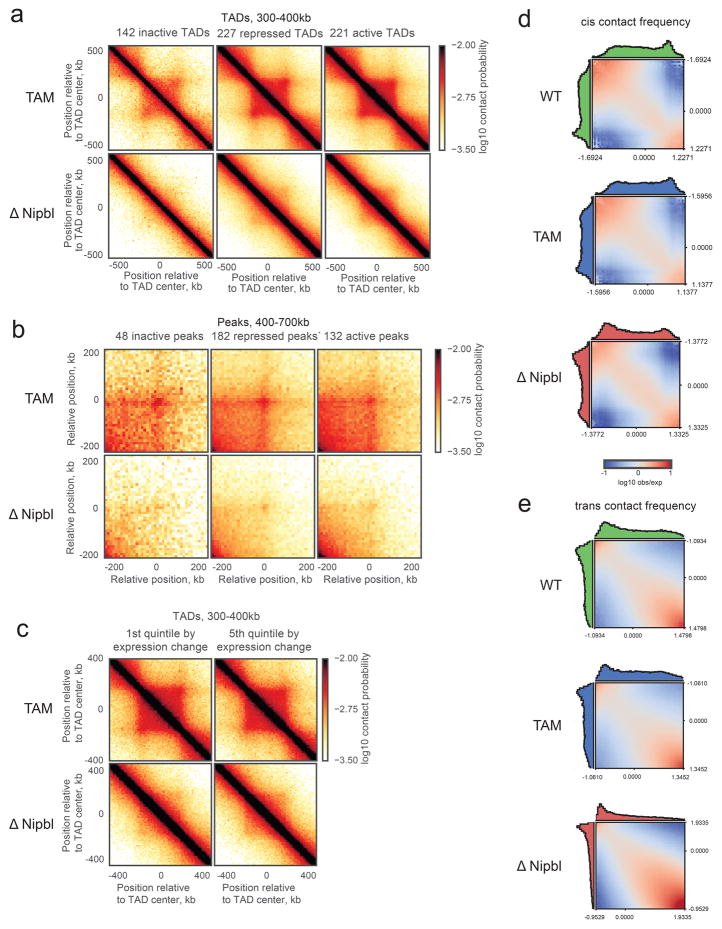
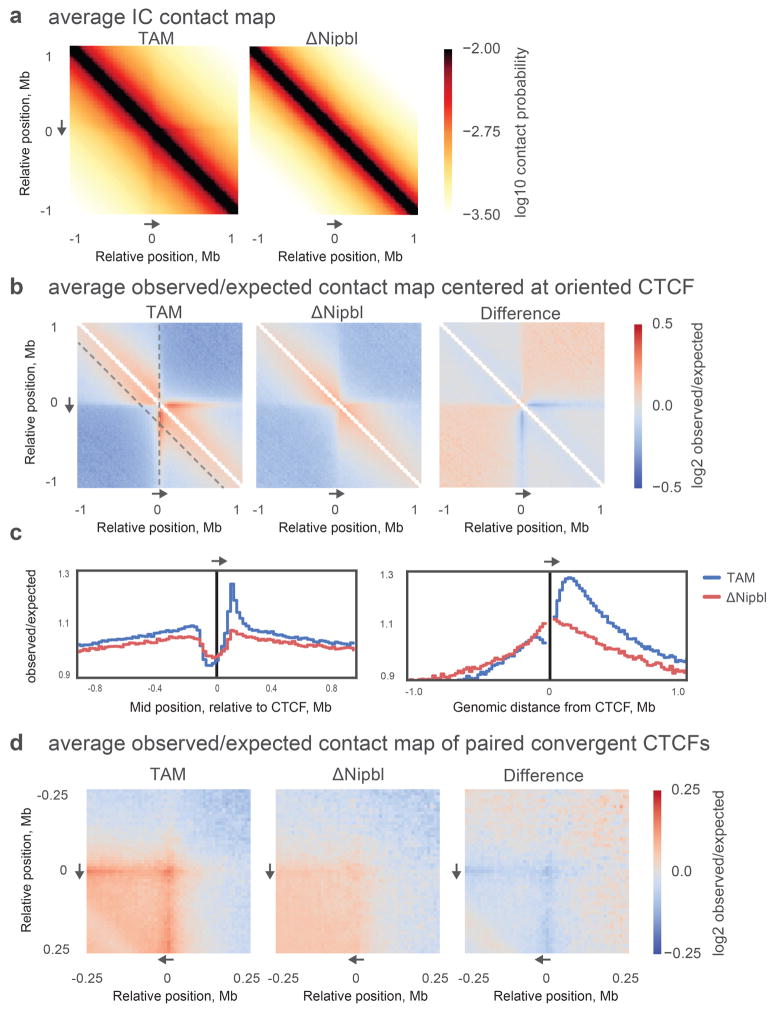
Our Hi-C maps reveal a striking effect of Nipbl deletion on genome organization (Fig. 1f–g), contrasting with the very mild changes reported in previous cohesin depletion experiments (Extended Data Figure 4) 19–21. Compared to WT and TAM control samples, ΔNipbl cells show genome-wide disappearance of local TAD patterns (Fig. 1g) but persistence of A/B compartmentalization (Fig. 1f). Disappearance of TADs in ΔNipbl is widespread and evident in individual maps (Fig. 2a), as well as on the composite map constructed by averaging the ΔNipbl Hi-C map around locations of domain boundaries detected in WT maps (Fig. 2c, Extended Data Figure 5). Some local organization is retained in regions with higher activity (A-compartment), where cohesin is about 3-fold more abundant than in inactive regions (B-compartment) in TAM (Extended Data Figures 5, 2e). We show below that these structures are not residual or new TADs, but unmasked small compartments. TADs-associated peaks of contact enrichment disappear as well in ΔNipbl maps, showing up to 4-fold reduction in contact frequency (Fig. 2d and Extended Data 4a), notably between convergent CTCF sites. Insulation and directionality of the contact footprint of CTCF sites also disappear upon Nipbl deletion (Extended Data Figure 6). However, we observe no effect of Nipbl deletion on CTCF occupancy, demonstrating that the loss of TADs and CTCF contact in ΔNipbl is not due to loss or changes in CTCF occupancy (Extended Data Figure 3), This further strengthens the very distinct roles of CTCF and cohesin in shaping chromosome architecture14,18.
Figure 2. Nipbl deletion leads to disappearance of TADs and peaks from Hi-C contact maps.
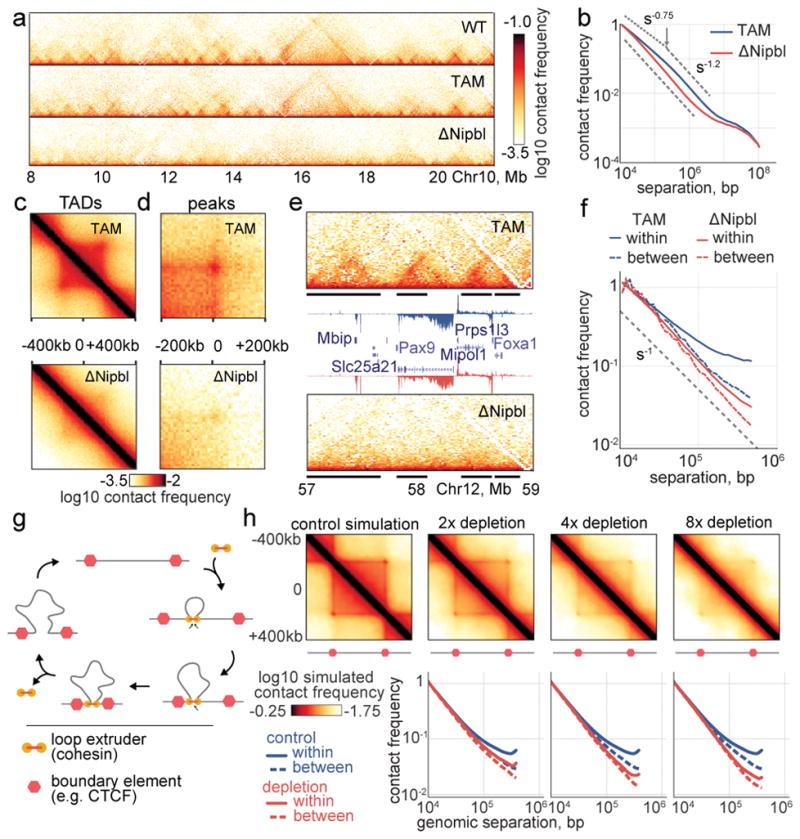
(a) The Hi-C map for chr10:8–21Mb illustrating loss of TADs and peaks. (b) Genome-wide P(s) curves in TAM control and ΔNipbl, normalized to unity at s=10kb. (c, d) The average Hi-C map of (c) 564 TADs of length 300–400kb and (d) 102 peaks of separation 500–600kb. (e) The Hi-C map of an example 2Mb region chr12:57–59Mb (top – TAM control, bottom – ΔNipbl) with expression tracks (middle – annotated genes and RPM normalized RNA-seq; sense above axis, antisense below; TAM in blue, ΔNipbl in red). Black bars show WT TADs. Hi-C and RNA-seq experiments were repeated independently on two and four biological replicates, respectively, with similar results. (f) P(s) curves plotted separately for contacts formed within or between TADs of size 300–500kb. (g) The cartoon representation of the loop extrusion model of cohesin action 14. In this model, cohesins form cis-loops by first binding to adjacent loci on chromosomes (top and right diagrams). After binding, cohesins translocate along the fiber in both directions, effectively extruding a loop (bottom diagrams). Extrusion halts when cohesins reach boundary elements. Extruded loops disassemble when cohesins unbind from the chromosome (left diagram). (h) Polymer simulations of loop extrusion reproduce the effects of cohesin depletion. Top row – average maps of TADs formed on contact maps in polymer simulations of loop extrusion. Left-to-right: the impact of sequential cohesin depletion on the contact map of a TAD in simulations. Bottom row – P(s) calculated separately within and between TADs.
Importantly, these changes cannot be attributed to altered gene expression in ΔNipbl cells, as TAD patterns vanished equally in regions with unchanged expression (Fig. 2e) or with up- and down-regulated transcription (Extended Data Figure 5c). This major reorganization of chromatin architecture is also reflected in the curves of contact frequency P(s) as a function of genomic distance s 3,29. In control samples, like in other mammalian cells 14,15, P(s) curve has two regimes: a more shallow decay for s<200Kb (P(s)~s−0.7), and a steeper scaling for 200Kb < s < 3Mb (P(s)~s−1.2). Loss of chromatin-associated cohesin leads to disappearance of the first regime, producing a single decay of contact probability across the whole range (Fig. 2b). This observation suggests that the first scaling regime reflects the compaction of the genome associated with TADs. We confirmed this by calculating P(s) separately within and between WT TAD intervals: in controls, P(s) within TADs decreases more slowly than P(s) between TADs. In ΔNipbl, the within P(s) curve collapses to the between P(s) curve, indicating that the characteristic enrichment of contacts within TADs is lost and that chromatin folding becomes more uniform and decompacted (Fig. 2f), which is consistent with decompaction observed by imaging upon Nipbl reduction 30.
Next, we simulated the effects of Nipbl depletion in our model of loop extrusion, by reducing the number of extruding cohesins (Fig. 2g). For each simulated concentration of extruding cohesins, we calculated Hi-C maps and P(s) within and between TADs. In simulations, 8-fold depletion is required to achieve agreement with our experimental data, manifested by (i) noticeable disappearance of TAD and corner peak enrichments; (ii) loss of the P(s)~s−0.7 regime in the scaling; (iii) decompaction of chromatin (Fig. 2h). Together, these analyses and the observed effects of Nipbl deletion indicate that cohesin plays a central role in the local compaction of chromosomes, and support that this effect is mediated by the production of dynamic populations of extruded chromatin loops between boundary elements, which forms TAD and corner peak patterns in interphase Hi-C maps 14,15.
Enhanced and finer compartmentalization
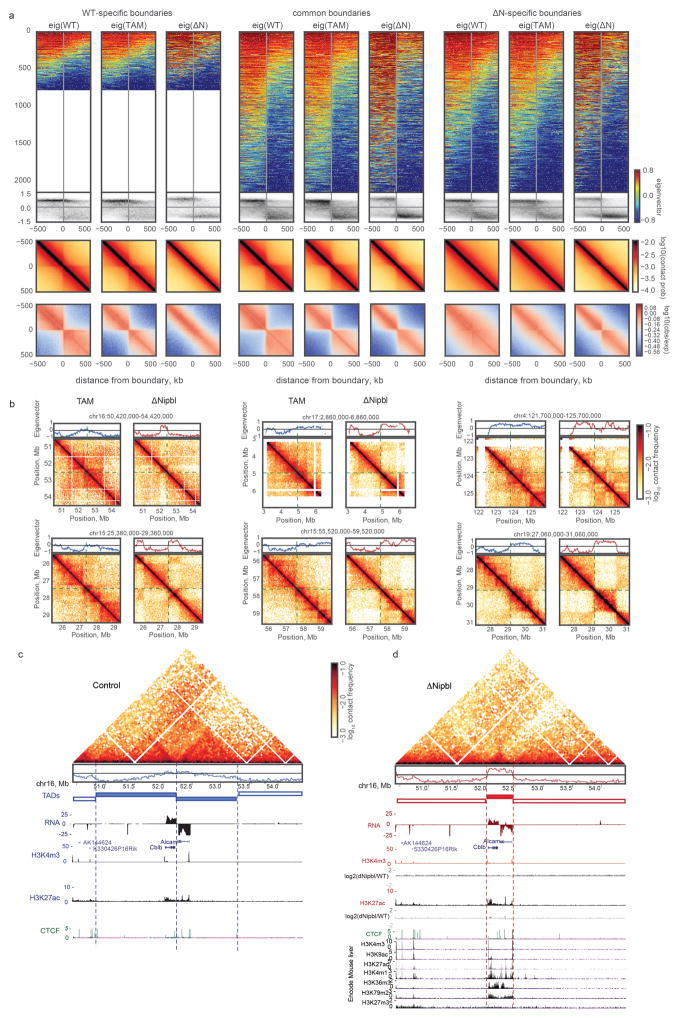
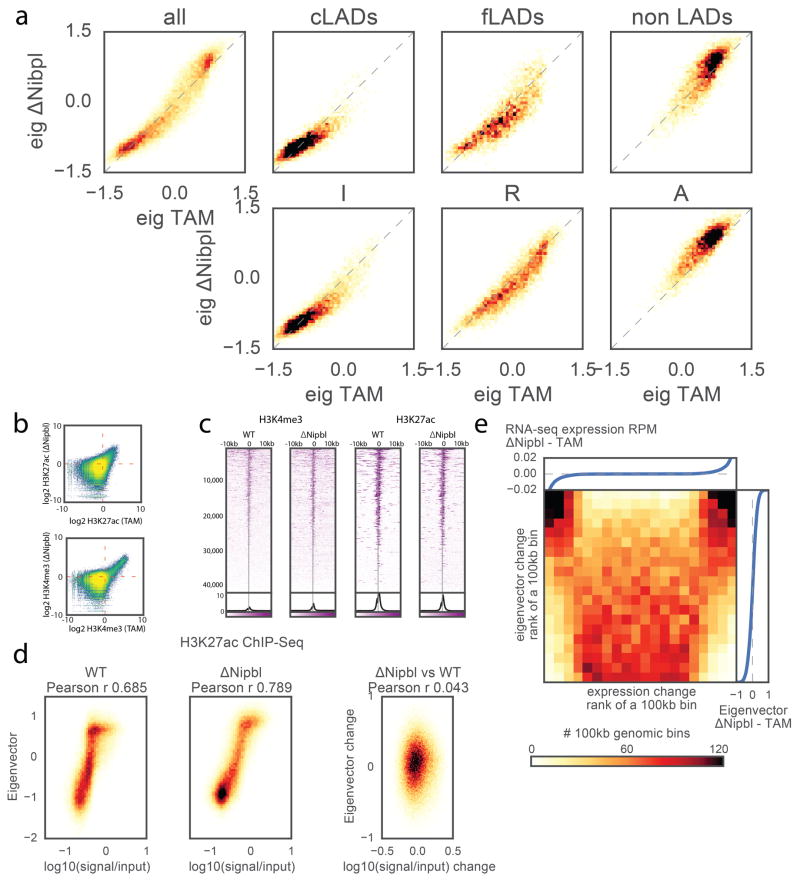
We next examined compartmentalization of chromatin in ΔNipbl cells. As noted earlier, in contrast to the drastic loss of TADs, compartmentalization still exists (Fig. 1) and is enhanced (~1.8 fold) (Extended Data Figure 5d,e, Supplementary Data 2,3). Moreover, closer examination of Hi-C data and compartment tracks reveals the emergence of series of shorter compartmental intervals in ΔNipbl cells with small B-like regions appearing inside A-regions (Fig. 3a,b, Extended Data Figure 7a). This finer compartmentalization is reflected in the shorter autocorrelation length of the compartment track (150Kb in ΔNipbl vs ~500Kb in WT and TAM (Extended Data Figure 7b). It explains most of the remaining or new domains and boundaries seen in ΔNipbl Hi-C maps (Extended Data Figures 8a). These emerging B-like regions possess the hallmarks of compartmentalization: (i) they are visible as local depressions in the ΔNipbl compartment track (Fig. 3b) and (ii) they show preferential interactions with other B-regions both in far cis- and in trans-chromosomal maps (Fig. 3b). Since their diagonal squares lack both corner peaks and enriched borders, and exhibit mutual checkering, we conclude that these intervals do not represent newly formed TADs. In contrast, predominantly B-type regions of the genome, do not show a similar fragmentation in ΔNipbl cells despite a complete loss of TAD patterns (Extended Data Figure 7a,b). Finally, Nipbl depletion reveals that WT TADs can span regions of opposing compartment type (Extended Data Figure 8b–d). Taken together, our observations defy the common notion of TADs simply being the building blocks of larger compartmental segments. Instead, TADs and compartments represent two independent, potentially antagonistic types of chromosomal organization.
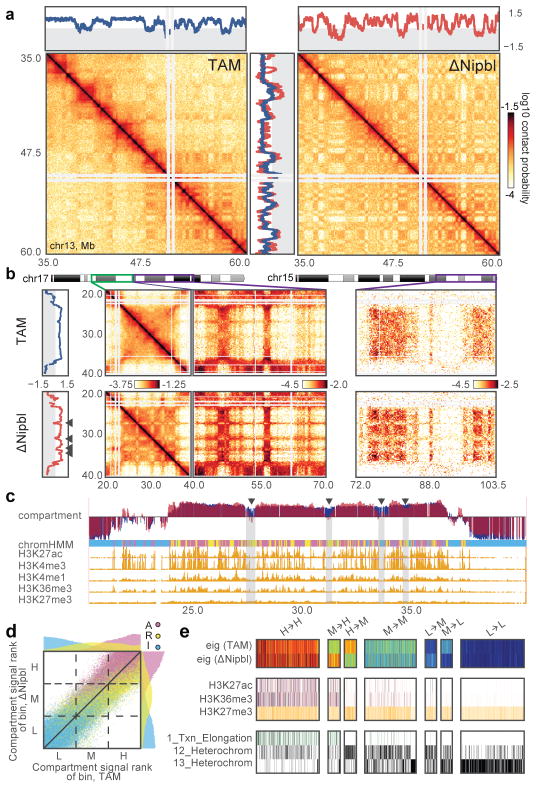
Figure 3. Nipbl deletion leads to activity-dependent alteration of compartment structure.
(a) An example region (chr13:35–60Mb) showing changes in compartmentalization. Top and central panels – 20kb cis compartment tracks. (b) Hi-C maps of an example region at 200kb with predominantly positive (type A) compartment signal in TAM control (top) showing fragmentation upon Nipbl deletion (bottom), manifested in the alternating contact patterns of short- (<10Mb, middle left) and long-range cis (middle right) and trans contact maps (right panel). In (a–b), the two Hi-C replicates of each condition show similar results. (c) The loci experiencing a local drop are depleted in epigenetic marks of activity. Top to bottom: compartment track in TAM (blue) and ΔNipbl (red); simplified ChromHMM state assignments: active (magenta) / repressed (yellow)/inert (cyan); ENCODE activity-related histone ChIP-seq for adult mouse liver cells. In (b–c), arrowheads indicate local drops in compartment signal. (d) Rank correlation of 20kb compartment tracks (n=113,372 20kb genomic bins) in TAM and ΔNipbl, colored by simplified ChromHMM state. Top and right margins – histograms of compartment signal ranks split by simplified ChromHMM state in ΔNipbl (right) and TAM (top). The dashed lines show the tercile borders, splitting bins into equal-sized groups of low (L), middle (M) and high (H) compartment signal. (e) Epigenetic profiles of bins transitioning between compartment signal terciles upon Nibpl deletion. Top to bottom: compartment track in WT and ΔNipbl; ENCODE histone marks; ChromHMM states characteristic of active, repressed and inert chromatin. The bins that transitioned from the middle to the high tercile are enriched in activity marks, while bins transitioning from the high to the middle tercile were depleted in those marks.
Strikingly, we found that the compartmentalization profile of the ΔNipbl Hi-C map reflects local transcriptional activity and chromatin state better than that of the WT Hi-C map (Fig. 3c, Extended Data Figure 7c–d, 8c–d). The compartment track of ΔNipbl cells shows a stronger correlation with tracks of activity-associated epigenetic marks, e.g. H3K27ac, H3K4me3, DNase hypersensitivity, transcription factor binding etc., smoothed over a wide range of window sizes (Extended Data Figure 7c). To understand the relationship between epigenetic state and the change in compartment status, we compared the compartment tracks to the mouse liver chromatin state segmentation (ChromHMM31) simplified into three state categories: Active, Repressed and Inert (see Methods). While inert regions are relatively unaffected by Nipbl deletion, regions of repressed and active chromatin further diverge in their compartment status (Fig. 3c,d), producing local peaks in the compartment track in active regions, and local B-like depressions in repressed regions (Fig. 3c). Furthermore, regions of facultative lamin-B1 association 32 are enriched in regions showing a reduction in compartment signal (from A to B-like), while those showing lamin-B1 association across different mouse cell lines are primarily B-type in both WT and the mutant (Extended Data Figure 9a, Methods). Importantly, these changes in compartmentalization cannot be attributed to changes in expression or in the activity marks (H3K27ac, H3K4me3) that are largely unperturbed in the mutant at the scales relevant to compartmentalization (Extended Data Figures 8d,9b–e). In summary, absence of cohesin enhances the compartmentalization of active and inactive chromatin: (i) A and B regions, as detected by Hi-C, form fewer contacts between one another, (ii) a finer compartment division emerges, (iii) this fine compartment structure corresponds better to the local functional states of the genome, even when considering those observed in WT. The fact that we observe no effect on activity marks indicates that cohesin and TADs do not play a role in the maintenance of epigenetic state, though this does not rule out possible roles in its establishment. A recent study has observed a similar preservation of epigenetic state upon CTCF depletion 18.
Altogether, these results indicate that chromatin has an intrinsic tendency to form small-scale, specific compartments that reflect co-segregation based on the local epigenetic landscape and transcriptional activity, and that Nipbl and cohesin activity interfere with those clear subdivisions by bringing together and mixing loci with opposing states.
Transcriptional changes upon TAD loss
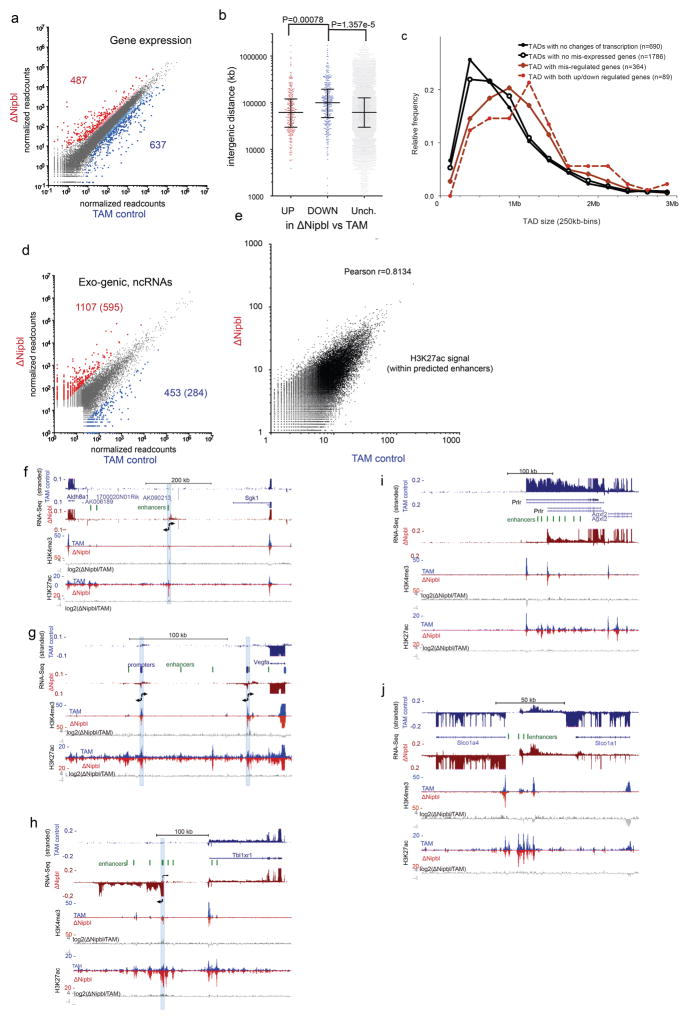
We next examined the effect of Nipbl deletion and disappearance of TADs on transcription. About a thousand genes are significantly mis-expressed (637 down-regulated, median fold-change=0.32, 487 up-regulated, median fold-change=3.15, with DESeq2 tools) upon Nipbl deletion (Fig. 4a and Extended Data Figure 10a–e). Gene ontology enrichment analysis does not give very strong indication of preferential effect on a biological function (Supplementary Table 4), reflecting possibly indirect and adaptive transcriptional changes.
Figure 4. Transcriptional changes in Nipbl mutants reveal possible enhancer-promoter miscommunication in absence of TADs.
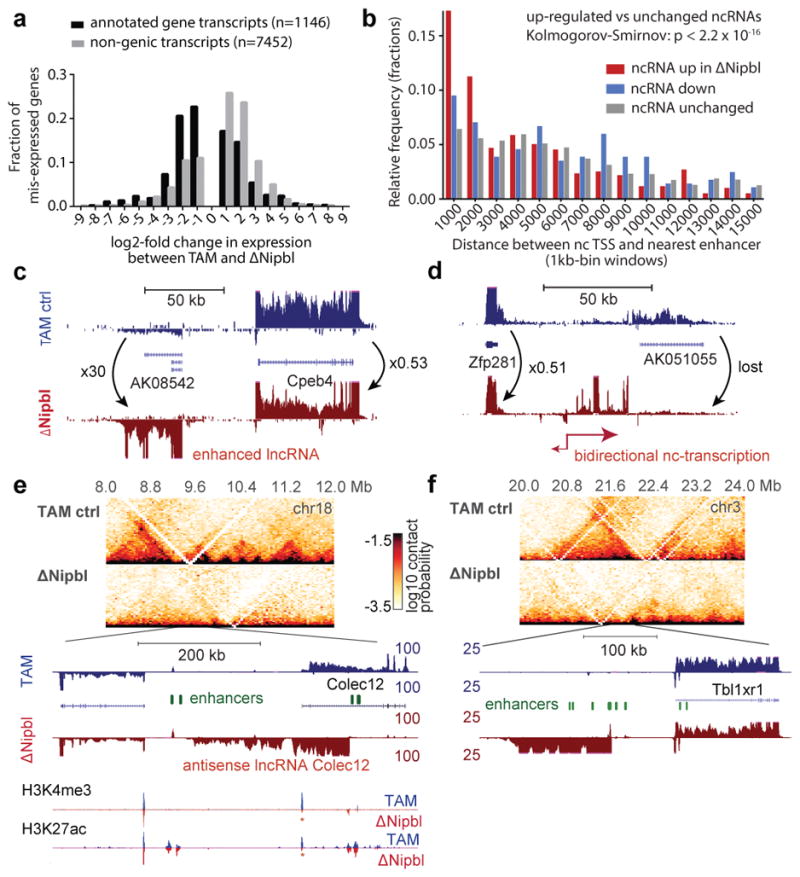
(a) Distribution of fold-changes for genes (black, n=1146)) and exo-genic transcripts (light grey, n=7452) showing at least a two-fold change in expression. (b–c) Examples of transcriptional changes, with TAM control stranded RNA-seq tracks in blue, and ΔNipbl in red. Four replicates per condition, each confirming the reported changes, were combined. (d) Distribution of distances between the transcriptional start of the ncRNAs (up log2(foldchange)>3, n=595; down log2(foldchange)<−3, n=284; unchanged −0.5<log2(foldchange)<0.5, n=2238) and the nearest enhancer. (e,f) Switches in transcription at the Colec12 and Tbl1xr1 loci. Top panels show 40kb Hi-C maps of the 4Mb neighborhood. (e) RNA-seq shows loss of Colec12 transcripts, replaced by antisense transcripts initiated from an intronic enhancer. H3K4me3 and H3K27ac profiles show no changes at distal enhancers (green ovals), while peaks at the Colec12 promoter disappear (red asterisk). (f) Exo-genic transcription emerges in ΔNipbl upstream of the Tbl1xr1 gene on chr3.
While H3K27ac (and H3K4me3) peaks at promoters of affected genes change in coherence with expression changes, distal peaks (marking active distant enhancers) are mostly unaffected (Extended Data Figure 10f–i), indicating that while transcriptional changes did occur, the regulatory potential of the cells was mostly unperturbed. There is so far no reliable way to identify a priori genes for which distal regulatory interactions are essential. However, we noticed that down-regulated genes were surrounded by a larger intergenic space (defined by the distance separating the TSS of their immediate neighbors) than up-regulated or unaffected ones (Extended Data Figure 10b) and that transcriptional changes are concentrated within regions that form larger TADs in WT. This characteristic genomic context of transcriptional alterations is consistent with defective long-range regulatory interactions in ΔNipbl cells.
Closer examination of RNA-Seq tracks revealed a widespread up-regulation of exogenic (intergenic or antisense intragenic) transcription in ΔNipbl cells (Fig. 4a). Interestingly, while we found more genes down-regulated than up-regulated, the trend was opposite in exogenic regions. Using a conservative approach (see Methods), we found 1107 non-genic transcripts or transcribed regions, which showed at least an 8-fold enhanced transcription in ΔNipbl cells; among these, 232 corresponded to non-coding RNAs, which were not or barely detected in control samples, and often not annotated (Extended Data Figure 10d). The new transcription is often bi-directional, (Fig. 4c–d, Extended Data Figure 10f–j) and occurs either at small pre-exiting H3K4me3 peaks, corresponding likely to poised promoters, or at active H3K27ac enhancers (Fig. 4b,e,f, Extended Data Figure 10f–j). We saw several examples of reciprocal expression changes (i.e. down-regulation of a gene being followed by up-regulation of an adjacent gene or of a new non-coding transcripts) (Fig. 4c–d, Extended Data Figure 10f–j), but often, new non-coding transcription arises without measurable impact on surrounding genes. While the chromatin profile suggests that enhancers retain their normal activity and therefore regulatory potential, this rise in intergenic transcription initiated in the vicinity of distal regulatory elements suggests that Nipbl loss impairs enhancer communication: with a reduced range of contact due to absence of TADs, some enhancers may not reach their target promoters and instead transfer their activity on nearby alternative, sometimes cryptic, targets (including themselves).
Cohesin is central to chromosome folding
Overall, our findings provide insights into the mechanisms that organize the interphase genome in 3D and their relation to gene expression. Our data shows the essential role of cohesin in the formation of TADs. It is possible that the cohesin depletion in previous studies that did not report such drastic effects 19–21 was insufficient to achieve substantial loss of TADs. Our simulations suggest that TADs would still be pronounced at 2-fold cohesin depletion, requiring ~8-fold depletion for loss of TADs, which is close to what we observed in ΔNipbl samples. It is however also possible that NIPBL may affect cohesin activity at several levels, i.e. not only as a loader but also by facilitating ATP hydrolysis and loop extrusion 33, which could further impact TAD formation. This could also account for why a recent deletion of Scc4/Mau2, a co-factor of NIPBL, showed a milder effect on TAD formation34.
While our manuscript was under review, several preprints reported direct perturbations to cohesin, producing results consistent with this study and thus providing additional support that loss of cohesin is responsible for the effects we observe. Single-nucleus Hi-C for cohesin (Rad21/Scc1) knockout zygotes demonstrated complete loss of TADs and associated peaks 35. Emergence of fine compartmentalization, however, was not reported likely due to the limited number of nuclei. Two other preprints reporting degron-mediated depletion of Rad21/Scc1 in human cell lines observed concordant effects on TADs, peaks, and compartmentalization to our study 36,37. Rao et al. also identified intriguing emergent “loop” cliques enriched in super-enhancers upon cohesin depletion37. We also observed such features in our primary hepatocytes using a new Hi-C browser 38, though we note that in both cases these patterns do not resemble the sharp cis-only focal patterns of CTCF-associated Hi-C peaks commonly referred to as loops 12. Rather, these larger patches appear to be enhanced compartmental interactions between very active regions that line up well with actively transcribed genes 38.
Two independent folding principles
Our results challenge the classic picture of genome architecture in which TADs/peaks and compartments represent well-defined hierarchically folded entities, with individual TADs combining to form compartmental regions. Remarkably, the changes we observed demonstrate that there are at least two mechanisms of independent origin whose overlapping action organizes mammalian interphase chromatin (Fig. 5). First, the global spatial compartmentalization of active and inactive regions of the genome is achieved by a cohesin-independent mechanism, which acts globally and across scales, including scales smaller than previously appreciated. This compartmentalization is however blurred by the action of a second, cohesin-dependent, mechanism that compacts chromatin locally, independently of its status. Noteworthy, the independence of the two mechanisms is supported the absence of compartments, despite of the existence of TADs, in maternal zygotic pronuclei 39.
Figure 5. Two independent but overlapping modes of chromosomal 3D organization.
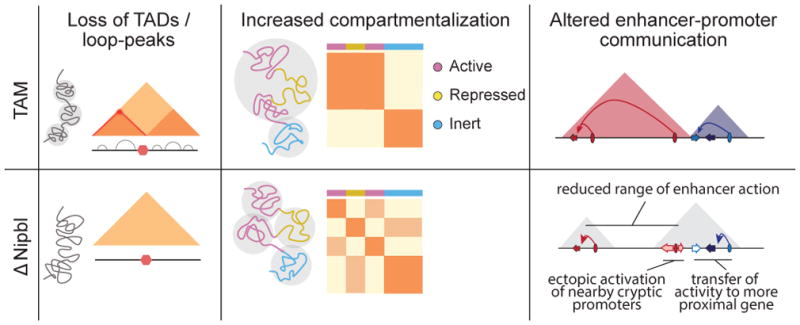
TADs (colored triangles) and Hi-C peaks disappear upon Nipbl deletion (left), unmasking a stronger and finer compartmentalization (middle) that is visible as a fragmented checkered pattern in the mutant Hi-C map relative to that of the WT and whose alternating member regions more faithfully track transcriptional activity. The resulting reduction of contact range (right) thwarts distant enhancers (ovals) from acting on their normal target genes (arrows, with colored ones indicating active genes, white ones inactive), leading them to act instead on neighboring genes or cryptic promoters located in their vicinity. The active units make up new compartmental regions (grey triangle).
The co-existence of two processes with different modes and scales of action can help explain the difficulties in the field in delineating and unambiguously classifying the different features of Hi-C maps, leading to a confusing plethora of denominations and definitions 7,8,12,40–42. Our data demonstrates clearly the existence of local, cohesin-dependent, self-interacting domains identifiable as TADs. The experimental ability to now distinguish these two modes of chromosome organization provides avenues to dissect the process(es) governing their formation and maintenance, as well as to characterize their relationship to gene expression. In this respect, the ability of cohesin to bring regions of different activities together may play an essential role in initiating changes in gene expression driven by distant enhancers, while compartmentalization may maintain existing regulatory interactions.
On-line Methods
Experimental procedures
Generation of Nipblflox/flox mice
The Nipbl locus was targeted by introduction of two loxP sites flanking exon 18 via homologous recombination in E14 mouse embryonic stem cells. Individual ESC clones were screened for successful recombination by Southern blotting deploying two unique probes hybridizing 5′ and 3′ to the integration site, respectively. Cells of a successful clone were injected into mouse blastocysts and resultant chimera were bred to C57BL/6J mice. Offspring were analyzed for successful germ line transmission by PCR and Southern blotting. Nipblflox/+ mice were maintained on C57BL/6J genetic background.
For acute deletion of the floxed exon, we used either a constitutive ubiquitous CRE-driver (Hprt::Cre 43), a limb-specific CRE-driver (Prx1::Cre 44) or an inducible, liver-specific Cre allele (Ttr-cre/Esr1 45.
Mice were genotyped by PCR using specifc primer pairs (details available on request).
All lines were maintained by breeding with C57Bl/6J mice. Mouse experiments were conducted in accordance with the principles and guidelines in place at European Molecular Biology Laboratory, as defined and overseen by its Institutional Animal Care and Use Committee, in accordance with the European Directive 2010/63/EU. The experimental protocols followed were reviewed and approved by the EMBL Institutional Animal Care and Use Committee under the project “Function of cohesin and condensing complexes”.
Generation and preparation of Nipbl−/− adult primary hepatocytes
To inactivate Nipbl in adult mouse hepatocytes, Nipblflox/+ mice were crossed with mice carrying an inducible, liver-specific Cre allele (Ttr-cre/Esr1 45. Resultant double heterozygous mice were back-crossed to Nipblflox/flox. For experiments we used animals homozygous for the floxed Nipbl allele either carrying one or no copy of the inducible, liver-specific Cre allele as sample (Ttr-cre/Esr1+/wt ; Nipblflox/flox ) or control (Ttr-cre/Esr1wt/wt ; Nipblflox/flox ), respectively.
12 week old mice were injected with 1mg Tamoxifen (100μl of 10mg/ml Tamoxifen in corn oil) on 5 consecutive days. After keeping these mice for another 5 days without injection, they were sacrificed and the hepatocytes were harvested. Until this time point, mice displayed no abnormal behavior, weight loss or any other obvious physiological changes. This was also the case, when we kept mice for additional 4 days without injection to test for any adverse effects immediately after our experimental time point.
The liver was dissected and the left lateral lobe was prepped for a two-step perfusion adapted from 46,47. First, the liver is perfused with an EDTA-containing buffer removing Ca2+ from the tissue. This weakens the integrity of the desmosome, which is then digested during the subsequent perfusion with a Ca2+ rich buffer containing collagenase. The freed hepatocytes were rinsed through a cell strainer and washed four times with ice-cold Ca2+ rich buffer without collagenase. For each wash the cells were spun at low centrifugal force (60g for 1 min), to reduce non-mesenchymal cells and debris, hence, enriching intact hepatocytes in the sample. Part of each sample was fixed with 1% PFA for 10 minutes at room temperature. Fixed and unfixed hepatocytes were aliquoted and frozen in liN2 for later use.
Nipbl RNA levels and activity
Unfixed hepatocyte aliquots were thawed and RNA was prepared with Qiagen RNeasy Kit. cDNA was generated using NEB ProtoScript® First Strand cDNA Synthesis Kit with random primer mix. RT-qPCR was performed with Applied SYBR Green PCR Master Mix and following primers: Nipbl-qPCR_F TCCCCAGTATGACCCTGTTT, Nipbl-qPCR_R AGAACATTTAGCCCGTTTGG, Gapdh-qPCR_F CTCCCACTCTTCCACCTTCG, Gapdh-qPCR_R CCACCACCCTGTTGCTGTAG, RTqPCR_Pgk1_Fwd TGGTATACCTGCTGGCTGGATGG and RTqPCR_Pgk1_Rev GACCCACAGCCTCGGCATATTTC.
For Western blots unfixed hepatocyte aliquots were lysed and fractionated with a Subcellular Protein Fractionation Kit (ThermoFisher). The blots were probed with antibodies against cohesin subunits SA-1 and SMC1 (a courtesy of Ana Losada, CNIO, Madrid) and Topo IIβ (611492, BD Biosciences) and Histone H2B (07-371, Millipore) as control for nuclear soluble and nuclear insoluble fractions, respectively.
Immunohistochemistry on liver
Slices of adult livers were collected and fixed in 4% PFA overnight. After dehydration, the tissues were embedded in paraffin and sectioned at 6μm. The sections were deparaffinized with xylene, rehydrated and antigens were retrieved by boiling in citrate buffer. The sections were blocked in 10% FBS and incubated with primary antibodies (αphospho-Histone H3, 06-570 Millipore; αcleaved-caspase-3, #9661 Cell Signaling) at 4°C overnight. Primary antibodies were detected with goat anti-rabbit IgG Alexa Fluor® 568 secondary antibody (A-11011, Invitrogen) and counter stained with DAPI. Images were acquired using confocal microscopy.
RNA-seq libraries and sequencing
RNA integrity was tested with Bioanalyzer (Agilent RNA Nano Kit) and ribosomal RNA was removed using Ribo-Zero rRNA Removal Kit (Illumina) prior to library preparation. Strand-specific libraries were prepared with NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®. After amplification and size selection with Agencourt AMPure XP beads (Beckmann Coulter) their size-distributions were determined with Bioanalyzer. Equimolar pools of libraries were sequenced with Illumina HiSeq2000 (50bp, single end). We retrieved on average 25 mio reads per sample, of which 19 mio reads were uniquely mapped to the reference genome (NCBI37/mm9).
ChIP-seq libraries and sequencing
For histone ChIP-Seq, fixed aliquots of hepatocytes were hypotonically lysed and sonicated in 1% SDS/TE. An aliquot of each sample was reverse cross-linked in order to determine chromatin concentration and sonication efficiency. 20μg chromatin per sample was diluted in RIPA and incubated with 1.5μg of either αH3k4me3 antibody (C15410003-50, Diagenode) or αH3K27Ac antibody (ab4729, Abcam) at 4°C, overnight. The antibodies were retrieved with Dynabeads (IgA, Invitrogen) and bound chromatin was washed and eluted. After reverse cross-linking, the amount of ChIPped and input DNA was determined with Qubit (Thermo Fisher). The libraries were prepared with NEBNext® ChIP-Seq Library Prep Kit for Illumina®. After amplification and size selection with E-Gel® SizeSelect™ (Thermo Fisher) their size-distributions were determined with Bioanalyzer. Equimolar pools of libraries were sequenced with Illumina HiSeq2000 (50bp, single end). We retrieved on average 20 mio reads per sample, of which 15 mio reads were uniquely mapped to the reference genome (NCBI37/mm9).
For cohesin subunits and CTCF ChIP-Seq, cell lysis, isolation of nuclei and shearing of chromatin were performed according to ref. 52. Briefly, cross-linked samples were thawed on ice and incubated in lysis buffer (5mM PIPES pH 8.0, 85mM KCl, 0.5% IGEPAL CA-630) supplemented with complete, EDTA-free Protease Inhibitor Cocktail (Sigma) for 45 minutes prior to transfer to shearing cuvette (Covaris-520130). Lysis was performed in a Covaris E220 sonicator for 5 minutes according to the following parameters: Peak power = 75W, Duty factor = 2%, 200 cycles/burst, temperature = 4C. Following lysis, nuclei were pelleted in a chilled 4C centrifuge at 1000g for 5 minutes and resuspended in shearing buffer (10mM TrisHCl pH 8.0, 0.1% SDS, 1mM) EDTA supplemented with complete, EDTA-free Protease Inhibitor Cocktail (Sigma) and transferred to a clean shearing cuvette. Chromatin was sheared in a Covaris E220 sonicator for 17 minutes with the following settings: Peak power = 140W, Duty factor = 5%, 200 cycles/burst, temperature = 4C. An aliquot of 1% of the total chromatin lysate was reverse cross-linked and subsequently used to validate chromatin fragment size distribution, concentration and for the construction of an input control library. The antibodies used (Anti-Rad21: Abcam ab992; Anti-CTCF: Millipore 07-729; anti-SMC3: Abcam ab9263) were raised against epitotes fully conserved between the mouse and human proteins. Each antibody was incubated overnight with 100μl of Dynabeads M-280 sheep anti-rabbit. Chromatin Immunoprecipitations were performed successively (Rad21, followed by CTCF, followed by SMC3) using unbound chromatin from the first IP to perform the next one. As IP efficiency is very low for this protein, we did not observe a change of signal when comparing sequential to parallel IP performed on control chromatin. Antibody-bound beads were washed 2x in ice cold PBS and incubated with 75ug of sheared mouse hepatocyte chromatin pooled with 75ug HEK293 human chromatin (as internal control and calibration) in RIPA buffer (50mM TrisHCl, pH 8.0, 0.15M NaCl, 1% Triton X-100 and 0.1% sodium deoxycholate) overnight at 4C on a rotating wheel and then subsequently washed 2x each in RIPA, RIPA-500 (50mM TrisHCl, pH 8.0, 0.5M NaCl, 1% Triton X-100 and 0.1% sodium deoxycholate), LiCl buffer (50mM TrisHCl, pH 8.0, 1mM EDTA, 1% IGEPAL CA-630, 0.7% sodium deoxcholate and 0.5% LiCl), and TE for 5 min per wash on a rotating wheel. Beads were resuspended in TE with 50ug/mL RNaseA and incubated for 30 min at 37C, followed by reverse cross-linking with 0.5 mg/ml Proteinase K in TE supplemented with 0.5% SDS overnight at 65C in a shaking thermomixer at 1200rpm. DNA was purified with the QIAquick nucleotide removal kit (Qiagen 28304). Multiplexed libraries were prepared according to the NEBNext ChIP-seq library preparation protocol and amplified with 15 PCR cycles using Thermo Phusion HF Mastermix (Fisher 10402678). Sequencing was performed on a NextSeq500 sequencer using a HighOutput Kit v2 75 (Illumina FC-404-2005).
HEK293 cells which chromatin is used as internal control for the IP were not authenticated, nor screened for mycoplasma contamination.
Tethered Chromatin Capture (TCC)
Roughly 100 mio fixed hepatocytes per sample were processed according to Kalhor et al. 26 using HindIII. Libraries were PCR-amplified (12 cycles) and size selected with E-Gel® SizeSelect™ (Thermo Fisher). Equimolar pools of libraries were sequenced with Illumina HiSeq2000 (50bp, paired end). We retrieved between 100 and 150 mio paired reads per sample, of which ~40% had both sides uniquely mapped to the reference genome (NCBI37/mm9).
Computational analysis
Preparation of Hi-C maps
We mapped the sequence of Hi-C molecules to reference mouse genome assembly mm9 using Bowtie 2.2.8 and the iterative mapping strategy, as described in 48 and implemented in the hiclib library for Python (publicly available at https://bitbucket.org/mirnylab/hiclib). Upon filtering PCR duplicates and reads mapped to multiple or zero locations, we aggregated the reads pairs into 20kb and 100kb genomic bins to produce Hi-C contact matrices.
Filtering of Hi-C maps
Low-coverage bins were excluded from further analysis using the MAD-max (maximum allowed median absolute deviation) filter on genomic coverage. MAD-max is a robust heuristic filtering method based on the empirical observation that the total number of contacts per each genomic bin follows a lognormal distribution. This shape of a distribution is expected when different factors (GC content, mappability, restriction fragment density, etc) affect the visibility of a genomic bin in a multiplicative fashion. The MAD-max procedure has three steps:
Normalize the numbers of contacts at each genomic bin via dividing them by their corresponding chromosome-wide medians. This step accounts for a possible variation in chromosome copy numbers, e.g. in chrX.
Fit the distribution of normalized contact numbers with a log normal distribution. To make this step more robust against skew caused by heavy tails, we find the center of the distribution as the median of log(normalized contact numbers), and the width as the Median Absolute Deviation from this median, or, MAD.
Filter out the low-coverage genomic bins, which are 3 standard deviations below the center of the distribution. The exact conversion ratio between MAD and standard deviations is 1.4826 and we approximate 3.0 standard deviations with 5.0 MADs.
Our MAD-max filtering procedure removed between 1.4% to 2.2% of non-zero bins in the six experimental replicates processed at 20kb resolution and between 1.8% to 3.0% at 100kb resolution.
To remove the short-range Hi-C artifacts - unligated and self-ligated Hi-C molecules - we removed contacts mapping to the same or adjacent genomic bins. The filtered 20kb and 100kb contacts matrices were then normalized using the iterative correction procedure (IC) 48, such that the genome-wide sum of contact probability for each row/column equals 1.0. Observed/expected contact maps were obtained by dividing each diagonal of a contact map by its average value over non-filtered genomic bins.
The same procedure was used to analyze other existing Hi-C datasets on cohesin-depleted cells 19–21.
Compartment analysis via eigenvector decomposition
The compartment structure of Hi-C maps was detected using a modified procedure from 48. In short, compartment tracks (i.e. the values of compartment signal across all genomic bins) were quantified as the dominant eigenvector of the observed/expected 20kb and 100kb cis contacts maps upon subtraction of 1.0 and rescaling by the magnitude of the square root of its eigenvalue, as implemented in hiclib. We refer to continuous genomic regions of relatively uniform compartment signal as compartment intervals. We introduce the genome-wide measure of a Hi-C map compartmentalization as , where AA is the average contact enrichment between pairs of loci with a strong compartment A signal (those with eigenvector values from the top 20% of the genome-wide range of eigenvector values), BB is the average contact enrichment between pairs of loci with a strong compartment B signal (those with eigenvector values from the bottom 20% of the genome-wide range of eigenvector values) and AB is the average contact depletion between pairs of loci with a strong compartment A and B signal.
Domain detection (Lavaburst) and peak coordinates
To identify domains, we used a segmentation algorithm very similar to 49, which divides the genome into domains in such a way as to maximize a global domain scoring function. We used two different scoring functions: one was the corner score function from 12 and the other was based on network modularity 50, which is a metric widely used to detect communities in networks. The modularity score for a domain spanning genomic bins a to b inclusively is given by
where A is the contact matrix and N is the corresponding matrix of a penalizing background model. The resolution parameter controls the strength of the penalty and therefore the characteristic size of the domains identified.
By restricting the solution space to contiguous segmentations, both calculating domain scores and finding the highest scoring segmentation can be reduced to O(n^2) dynamic programming algorithms. Optimal segmentation, in particular, becomes the well-known max-sum algorithm on a weighted directed acyclic graph 51. Furthermore, one can marginalize over the space of all possible segmentations to obtain a linear genomic track of boundary scores for a given domain scoring function, also in O(n^2) time. The implementation of these and related algorithms is provided in the lavaburst package (https://github.com/nezar-compbio/lavaburst).
The domain calls used in composite heatmaps were computed using the corner score on 20kb resolution matrices. To robustly call insulating boundaries across different conditions, we exploit the multi-resolution nature of the modularity score and compute the average marginal boundary scores on 20kb WT and ΔNipbl contact matrices sweeping over a range of gamma values to obtain a 1D boundary (i.e., insulation) track. Short intervals representing insulating loci were called by thresholding on the boundary score, and the common and unique loci to each condition were determined by interval intersection.
To characterize the structure of known peaks in our data, we used the list of peaks detected in Hi-C maps for CH12-LX mouse cell line in 12.
Analysis of epigenetic features
To study the influence of the epigenetic chromatin state on genomic architecture, we used the ChromHMM genomic state annotation from 52. Briefly, the authors trained the ChromHMM genomic segmentation model on H3K4me1, H3K4me3, H3K36me3, H3K27me3, H3K27ac, CTCF and RNA polymerase II ENCODE tracks for mouse liver. This model assigned to each genomic loci one of 15 possible epigenetic states. For our analyses, we further grouped these 15 states into three: Active (states 1–11, 14 and 15 characterized by presence of PolII), Repressed (states 10–12, characterized by high H3K27me3 and low H3K27ac emission probabilities) and Inert (state 13, lack of any signal).
To find the average footprint of a chromatin-bound CTCF, we detected all occurrences of the M1 CTCF motif 53 in the mm9 genome (filtered by p-value > 0.0001), intersected with CTCF binding peaks in ENCODE adult mouse liver dataset 54. This procedure yielded 27840 peaks.
To study how lamina association affects genome compartmentalization, we used the dataset of Lamin-B1-binding loci from 55 containing data from four mouse cell lines: embryonic stem cells (ESC), multipotent neural precursor cells (NPC), terminally differentiated astrocytes (AC) and embryonic fibroblasts (MEF). We then selected 100kb genomic bins with more than 60 locations probed for lamin-B1 binding. We called the bins as constitutive LAD (cLAD) or non-LAD bins if >90% of probed locations were bound to lamina or not bound to lamina across all four tested cell lines, respectively. Finally, bins were called facultative LADs if for more than 90% of probes showed lamina binding in some cell lines, but not in others.
RNA-Seq
We mapped the RNA-seq data to mm9 reference mouse genome assembly and GENCODE vM1 transcriptome 56 using STAR v.2.5.0a 57 and scripts from the ENCODE RNA-Seq pipeline [https://github.com/ENCODE-DCC/long-rna-seq-pipeline]. To obtain the tracks of local transcription, we aggregated the uniquely mapped reads into RPM-normalized bigWig files using the built-in STAR functionality. To find differentially expressed genes, we aggregated the read counts at the gene level using HTSeq 58 with the “union” option and called DE genes with DESeq2 59.
For identification of exogenic transcripts, we aligned reads on the mm9 genome using HISAT2 60 with the option to authorize splicing events. From these alignments, we count the number of reads in sliding windows of 1kb (with a step of 600nt) taking each strand of the genome as separate. We use a gaussian mixture model (2 distributions) to find a cutoff value for readcounts differentiating “expressed” regions from noise. We choose a cutoff of 15 reads according to the best fitted gaussian distributions. To avoid overestimating the number of exogenic transcripts, we merged adjacent expressed windows (on the same strand) as composite transcripts. We used spliced reads to merge expressed windows that were not directly adjacent, but linked by spliced reads, with a cutoff of 7 splice reads. The resulting merged and linked windows defined the region corresponding to the exogenic transcription unit, and we took its 5′ end (using the strand information, and after trimming the 5′end of all nucleotides with no coverage) as transcriptional start site.
ChIP-Seq
We mapped the sequences obtained in ChIP-seq experiments using workflows based on the ENCODE 3 pipeline [https://github.com/ENCODE-DCC/chip-seq-pipeline].
For the CTCF and cohesin (Rad21 and Smc3) ChIP experiments, we aligned the reads from the pooled experimental and calibration (human spike-in) samples using bwa mem to a combined reference consisting of the mm9 and hg19 genome assemblies. For each condition, the reads that mapped uniquely to one of the two assemblies were bucketed into two separate datasets. The remainder of the ChIP-seq processing pipeline was applied to both the hg19-mapped and mm9-mapped reads in order to generate raw signal tracks. Because the proportion of human spike-in DNA was the same in both conditions, we used the difference in hg19 read coverage between the TAM control and ΔNipbl conditions to “calibrate” the raw mm9 signal pileups in order to make the binding profiles quantitatively comparable. Briefly, for each factor (CTCF, Rad21, Smc3), we computed genome-wide binned coverage at 200bp of the hg19-mapped reads. Because the background signal that makes up most of the genome-wide track exhibits different signal-to-noise characteristics than the binding peaks whose signal we wish to compare (as can be surmised from Extended Data Figure 6b), we used the ratio of the top 0.2% of 200bp bins in TAM vs ΔNipbl to rescale the mm9 signal in the ΔNipbl condition in order to be compared with the mm9 signal in the TAM condition. Peak calling was performed using MACS2 on each of the mm9 datasets using alignments of mm9-mapped input DNA as background.
GO enrichment analysis
GO enrichment analysis was performed on significantly differentially expressed genes (DESeq2 adjusted p-value < 0.5) using the online Functional Annotation Tool from DAVID version 6.8 (http://david.ncifcrf.gov), selecting annotation terms from the GOTERM_BP_DIRECT, GOTERM_MF_DIRECT, and GOTERM_CC_DIRECT categories and using default parameters. For the background gene list, we took the subset of gene IDs reported by DESeq2 that were expressed in either the TAM control condition or the ΔNipbl condition. ED Table 1 displays the list of enriched GO terms with a Benjamini-Hochberg p-value < 0.05.
Simulations of loop extrusion
To investigate the impact of NIPBL depletion on TADs and associated peaks in the context of loop extrusion, we performed simulations that couple the 1D dynamics of loop extrusion by LEFs with 3D polymer dynamics, as previously 14. We then generated contact maps and calculated P(s), also as previously described.
We considered a system of 42 consecutive TADs of 400kb, where impermeable BEs were placed between each pair of neighboring TADs. We modeled the chromatin fiber as a series of 1kb monomers (~6 nucleosomes, ~20nm), such that each TAD was 400 monomers. Polymer connectivity, stiffness, and excluded volume were implemented as previously described 14. Extruded loops held by LEFs were implemented by connecting the two monomers held by the two heads of each LEF with a harmonic bond, as previously 14,61.
Three-dimensional polymer dynamics were implemented using OpenMM, a high-performance GPU-assisted molecular dynamics API 62,63. Simulations were initialized from a compact polymer conformation, as described in 64, created on a cubic lattice a box of the size (PBC box – 2). Prior to a block of simulations, LEF dynamics were advanced by 500,000 steps. To allow the polymer fiber to equilibrate with this set of LEF-imposed bonds, simulations were then advanced for 400 blocks of simulations. After that, 2000 blocks (ie. blocks of 1D+3D dynamics) of simulations were performed and their conformations were recorded. After that, LEF dynamics were advanced by 500,000 steps, and the process was repeated, until 5000 conformations for each parameter set were obtained.
For the control simulation, we considered LEFs with 200kb processivity and 400kb separation. To investigate the effect of depleting the amount of bound cohesin, we then increased LEF separation by 2-fold, 4-fold, and 8-fold. All simulations used a stiffness of 2, density of 0.2, 3D-to-1D-steps of 2500. To generate contact maps from simulated conformations, a capture radius of 6 was used.
For display, simulated contact maps were first binned using 10 monomer (10kb) bins. Then, the map was normalized such the average value of the first diagonal equals one, and log10 transformed. Finally, the color-scale was clipped to show 1.5 logarithmic orders of magnitude as its dynamic range.
To calculate P(s) plots from simulated data, a contact map with 1 monomer resolution was used. For each diagonal of the contact map, we determined which regions within 400-monomer TADs and which were between TADs. We then averaged the values in logarithmically-spaced bins of increasing distance with a step of 1.1. Similar to experimental data, P(s) curves were then vertically shifted such that P(s) at 10kb was equal to 1.
Extended Data
Extended Data Figure 1. Overview of various features of chromosomal architecture detected and quantified in Hi-C contact maps.
Top row – intra-chromosomal maps show the decay of contact frequency with genomic distance, which can be quantified with the curves of contact frequency P(s) vs genomic separation s. Middle row – both intra- and inter-chromosomal maps display a checkered pattern caused by compartmentalization of the genome. This pattern can be quantified by a continuous genomic track obtained via eigenvector decomposition of either cis or trans maps. Bottom row – intra-chromosomal maps at short genomic distance scales reveal domains of enriched contact frequency, which appear as bright squares along the main diagonal, and peaks which appear as bright dots connecting two loci. Both can be detected and quantified using specialized algorithms.
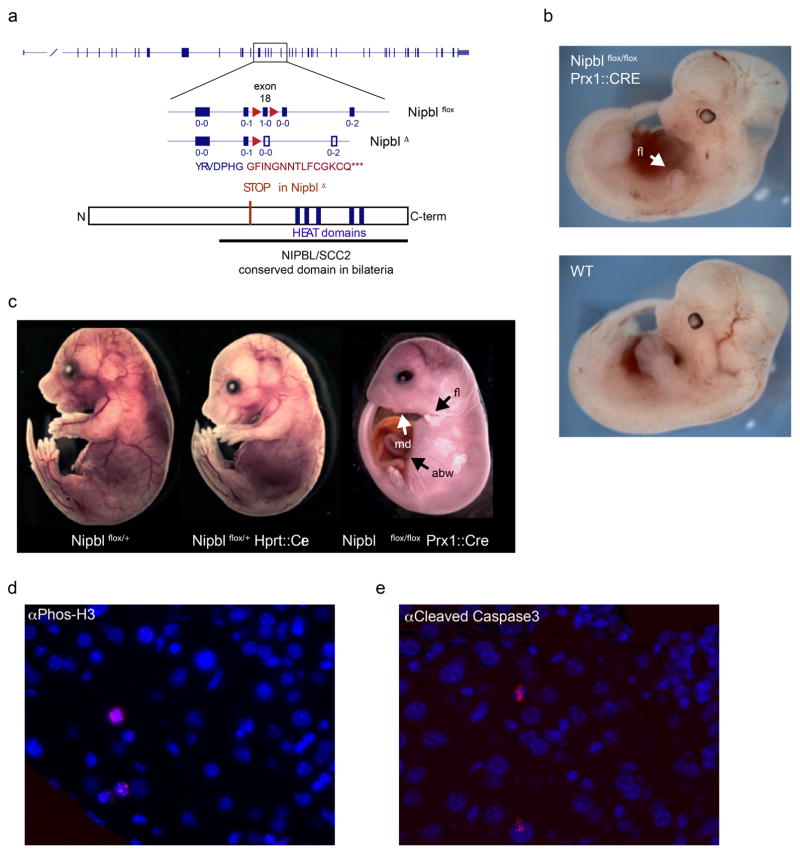
Extended Data Figure 2. Conditional inactivation of Nipbl in mice.
(a) Schematic representation of the conditional allele, with loxP sites (red triangles) flanking exon 18. The reading frame of each exon is indicated below the corresponding square, as “x-x”. Deletion of exon 18 leads to a frame-shift introducing a premature stop codon (indicated by amino acids in red). The resulting protein lacks the critical HEAT domains conserved in NIPBL/SCC2 proteins. (b–c) E12 embryos (b) and E18 fetuses (c) carrying the conditional Nipbl allele (Nipblflox) and either ubiquitous (Hprt:Cre 43) or limb-specific (Prx1::Cre 44) Cre recombinase drivers. Structures expressing Cre are rapidly lost in Nipblflox/flox animals. Heterozygous Nipblflox/+ animals are grossly morphologically normal, but die soon after birth, as reported for other Nipbl loss of function alleles 65. fl=forelimb; md:mandibule; abw=abdominal wall. (d–e) Histochemical staining of liver section of adult ΔNipbl hepatocytes (Nipblflox/flox; Ttr::CreERT2; 10 days after Tamoxifen injection) for a proliferation marker (Phos-H3) (d) and apoptosis (cleaved Caspase3) (e) (both showed in red). Nuclei are stained with DAPI (blue). Staining were performed once.
Extended Data Figure 3. Calibrated ChIP-seq for CTCF and cohesin (Rad21 and Smc3).
(a) Left: Comparison of calibrated mouse ChIP-seq data. For each factor (CTCF, Rad21, Smc3), we used the ratio of the top 0.2% of 200bp bins (~29,000 points) in the TAM hg19 fraction vs the ΔNipbl hg19 fraction to rescale the mm9 signal in the ΔNipbl condition in order to be compared with the mm9 signal in the TAM condition (see Methods). The scatter plot heatmaps in the left column are shaded by point density. Right: Log2 fold difference ratio between ΔNipbl and TAM for ChIP-seq peaks (points in with ChIP in TAM>2.0). While the calibrated CTCF binding signal stays relatively constant between conditions (within the 2-fold envelope), most of the calibrated cohesin binding signal drops by ~2–8 fold in ΔNipbl, with a mean depletion rate of 3.7 fold. (b) Uncalibrated ChIP-seq reads used for the calibration human cell (hg19, left) and mouse (mm9, right). Processed mapped reads were converted to genome-wide signal tracks binned at 200bp resolution. Scatter plots of these genomic tracks (TAM vs ΔNipbl) are shown as heatmaps shaded by point density. For cohesin subunits, the hg19 signal has a similar profile in both conditions, but the ΔNipbl signal is diminished in the uncalibrated mm9 fraction. The uncalibrated mm9 signal appears to go down in ΔNipbl for CTCF as well; however, a similar diminishing effect is seen in the hg19 data. (c) Stacked heatmaps of calibrated ChIP-seq signal at the top 20,000 CTCF binding sites (peaks with an assigned CTCF motif), ranked by fold change over input in the TAM control condition. (d) Scatter plots of calibrated ChIP-seq tracks as in (b), but split into two groups by compartment type A (compartment eigenvector>0) and B (eigenvector assignment < 0). The shading is colored by the eigenvector signal. Black and green dashed diagonal lines demarcate the 2-fold and 8-fold envelopes, respectively. (e) Total cohesin occupancy. Bar plot showing the number of ChIP-seq 200bp points in the non-shaded area of the scatter plots (bins with TAM signal > 2) representing high confidence binding signal in WT. While cohesin binding is more than 3-fold more prevalent in A-compartment regions, the scatter plots show that both A and B regions respond equally to Nipbl deletion.
Extended Data Figure 4. Deletion of Nipbl in this study leads to a more robust disappearance of TADs and associated peaks as compared to previous techniques.
(a) genetic deletion of Nipbl in hepatocytes, this study. (b) deletion of Rad21 in thymocytes 20. (c) proteolytic cleavage of RAD21 in HEK293T cells 19 (d–e) deletion of Rad21 in NSCs and ASTs 21. For each dataset: left column, top panels – the average Hi-C map around 102 peaks with size range 500–600kb 12 in WT and ΔNipbl contact maps; middle panels – the average Hi-C map of TADs called in each dataset; bottom panels – the relative contact probability between pairs of peak loci vs genomic distance, compared to randomly selected pairs of loci. The thick line shows the median contact probability; the shading shows the envelope between the 25th and 75th percentiles of contact probability at each genomic separation. Note significant changes in the contact probability at peaks (red line) upon Nipbl depletion (a) and little changes in other studies (b–e). All studies used comparable Hi-C sequencing depth (Supplementary Table 3)
Extended Data Figure 5. Residual structures are observed in active and repressed regions of the genome after Nipbl deletion.
For each TAD, an activity was assigned based on the dominant simplified ChromHMM state category. (a) The average contact map of 300–400kb TADs in inert, repressed and active regions of the genome. (b) The average contact map of 300–700kb peaks in inert, repressed and active regions of the genome. (c) The average contact maps of most upregulated 20% (left) and most downregulated 20% (right) of 300–400kb TADs. (d) Compartmentalization saddle plots: average interaction frequencies between pairs of loci (100kb bins) arranged by their compartment signal (eigenvector value). The interaction frequencies in cis (top row) are computed for observed/expected contact maps. Notice enrichment of AA and depletion of AB interactions in ΔNipbl cells. Histograms along the axes show the distributions of eigenvector values.
Extended Data Figure 6. The average Hi-C contact footprint of CTCF sites.
CTCF peaks were detected in our ChIP-Seq data from TAM control cells and supported by an underlying CTCF binding motif occurrence. (a) Average iteratively corrected 20kb-resolution contact map of around ~42,000 CTCF peaks in TAM and ΔNipbl cells. Individual contributing snippets to the composite heatmap were oriented such that the CTCF motif points in the direction shown by the grey arrow. (b) Average 20kb-resolution contact map normalized by the expected contact frequency at a given genomic separation. (c) Average observed over expected contact frequency curves along “slices” of the composite heatmaps depicted by dashed grey lines in (b): left panel – the insulation profile at 200kb separation (diagonal dashed line on (b)), right panel – the “virtual 4C” curve (vertical dashed line on (b)) of the composite heatmap. (d) Average 10kb-resolution observed over expected contact map centered on ~11,000 pairs of CTCF ChIP-seq peaks with convergent motif orientations separated by 200 +/− 10kb.
Extended Data Figure 7. Fragmentation upon Nipbl deletion in smaller alternating regions of A and B compartment-type is activity-dependent.
(a) Example region (chr3:35–60Mb) illustrating lack of compartment fragmentation in predominantly B-type regions yet robust disappearance of TADs. Top – compartment eigenvector, Bottom – contact matrix snapshot. (b) Autocorrelation of eigenvector tracks reveals genome-wide fragmentation of active compartments. Left – the genome-wide Spearman coefficient of correlation of the 20kb cis eigenvector values (n=113,372) of pairs of loci as a function of their genomic separation (autocorrelation). Top right – eigenvector correlation of locus pairs split by quintile of the eigenvector value of the upstream locus. Bottom right – chromosome-wide values of eigenvector correlation of locus pairs separated by 1Mb. (c) Spearman coefficient of correlation between the smoothed histone and TF ChIP-seq and RNA-seq tracks and the 20kb cis eigenvectors (n=113,372) as a function of the smoothing window size. Left group of panels – ENCODE data, right – data from this study. First and second rows – histone marks, third row – RNA-seq tracks, fourth row – miscellaneous tracks (DNase hypersensitivity, CTCF and PolII ChIP-seq and GC content). ΔNipbl eigenvectors show an increased correlation with tracks associated with transcriptional activity yet a decreased correlation with the repression-associated track of H3K27me3 and GC content. (d) An example of a large WT A-type compartment region (chr13:45–48Mb). ΔNipbl compartment transitions highlighted by black dashed lines. TAD boundaries in the WT are shifted or lost and replaced by compartment transitions in ΔNipbl cells. Histone ChIP-seq tracks 66 and stranded RNA-seq tracks (blue: TAM hepatocytes, red; ΔNipbl cells) highlight that WT/TAM TADs do not strictly follow the underlying chromatin activities, whereas the new checkered pattern in ΔNipbl cells delineated bydashed lines correspond precisely to active versus inactive chromatin domains. In (a) and (d), both replicates of each condition show similar results.
Extended Data Figure 8. TADs and compartments constitute independent layers of genome organization.
(a) The residual contact-insulating boundaries in ΔNipbl are associated with compartment transitions. The first group of columns considers the boundaries detected in WT cells only, the second pair considers boundaries detected both in WT and ΔNipbl cells, the last pair considers boundaries detected in ΔNipbl only. For each group, the first, second and third columns display data (eigenvectors and Hi-C) from WT, TAM and ΔNipbl cells, respectively. Within each column: the top row – a stack of eigenvector tracks in a +/− 500kb window around boundaries, oriented such that the left-half of the window has greater average signal value and sorted by the average WT eigenvector value in the window. The second row – density histogram of eigenvector values as a function of the distance to the boundary. The third and fourth row – the boundary-centered average contact probability and observed/expected contact ratio, respectively. The density histograms show that common and ΔNipbl-specific boundaries correspond to sharp transitions of compartment signals in ΔNipbl cells, in contrast to the more diffuse signal at these positions in WT and TAM cells. (b) Boundaries of former TADs and new compartment domains do not coincide. Examples of TADs detected in WT cells, which contain cross sharp compartment transitions revealed by ΔNipbl contact maps. Left column – TAM control data, right column – ΔNipbl data. Top of each figure – local compartment signal in the corresponding cell type. The contact maps are centered at the sharp compartment transition in ΔNipbl. These examples illustrate that that chromatin-bound cohesins can locally interfere with genome compartmentalization. (c, d) New compartments do not respect TAD boundaries but the underlying chromatin domains. A large region (chr16:50420000–54420000) adopts a very different 3D organization in control (c) (in blue) and ΔNipbl cells (d) (in red). Hi-C data are shown, as well as the eigenvector values in the two conditions. RNA-Seq tracks showed minimal changes of expression (Alcam expression is reduced by 2-fold in ΔNipbl cells) and chromatin states. ChIP-Seq tracks for H3K27ac and H3K4me3 are shown in the two conditions, with log2 ratio tracks under the ΔNipbl (d) panel. Encode tracks (corresponding to WT liver cells) are shown in the grey boxed area. The new structure adopted in ΔNipbl cells put together the two active genes which are normally in different TADs in the same domain, corresponding to the active chromatin linear domain. In (b–d), both replicates of each condition show similar result.
Extended Data Figure 9. Eigenvector change upon Nipbl deletion is activity-dependent and uncorrelated with changes in gene expression or epigenetic marks.
(a) Correlation of cis eigenvector values of 100kb genomic bins before and after Nipbl deletion, split by the functional state of chromatin. Top row, left to right: genome-wide relationship; bins showing constitutive lamin-B1 association across 4 mouse cell types (cLADs); bins showing variable (facultative) lamin-B1 association (fLADs); binds not showing any association (non LADs). Bottom row: bins assigned the Inert ChromHMM simplified state; bins assigned the Repressed state; bins assigned the Active state. (b) Scatter plot of genome-wide ChIP-seq signal binned at 200bp in WT vs ΔNipbl. Top – H3K27ac, bottom – H3K4me3. (c) Stacked heatmap of histone ChIP-seq signal over input +/− 10kb around putative TSS sites sorted by total H3K27ac signal in WT and oriented by TSS strand. From left to right: H3K4me3 in WT and ΔNipbl, followed by H3K27ac in WT and ΔNipbl. (d) ChIP-Seq signal for histone marks of activity vs eigenvector value of 20kb bins, top row – H3K27ac, bottom row – H3K4me3. Left column – WT cells, middle column – ΔNipbl cells, right column – correlation of changes in both signals upon Nipbl deletion. (e) The change in the compartment structure upon Nipbl deletion cannot be attributed to the sign of the local expression change. The heatmap shows the number of 100kb genomic bins as a function of the ranks of expression change and the eigenvector change. The attached plots show the correspondence between the values of expression change (top) or eigenvector change (right) and their ranks.
Extended Data Figure 10. Expression changes in ΔNipbl hepatocytes.
(a) Changes in gene expression between TAM controls and ΔNipbl liver cells (four replicates for each condition) analyzed with DESeq2 59. Genes with significant changes in gene expression (FDR > 0.05) are colored in red (up-regulated, n=487) or blue (down-regulated, n=637), with larger dots corresponding to gene with a fold-change > 3. (b) Intergenic distances for the different categories of disregulated genes (with fold-change >3; up-regulated=268; down-regulated=350; unchanged=15055). Statistical differences determined by an unpaired two-tailed t-test. The differences between means were 50020.40 (CI 95% = 27723.85–72316.95) and 52185 (CI 95%=21824–82547) for comparison between down-regulated vs unchanged, and down-regulated vs up-regulated, respectively (c) Size distribution of the TADs observed in WT (lost in ΔNipbl) depending on the degree alteration of their transcriptional states. The size of TAD with transcriptional changes (red) is significantly larger than those that do not show transcriptional alterations (black) (Kolmogorov-Smirnov, P=4.095e-08) (d) Change in transcription in non-genic intervals (including inter-genic and antisense within gene bodies). Gene expression was calculated as the normalized number of read within intervals defined by merging adjacent 1kb windows showing readcounts over background (see Methods). The numbers of non-coding transcription up-regulated (in red) or down-regulated (in blue) in ΔNipbl compared to the TAM control is given (P-value <0.01 using a two-tailed t-test, four replicates per condition, fold-change higher than 8), with the second number indicating the high-confidence events (labelled with coloured dots, expression value over an arbitrary threshold of 30 reads) which constitute the list used for subsequent analyses. (e) Comparison of control and ΔNipbl H3K27ac normalized signals within predicted liver enhancer elements (n=51850; readcounts within +/− 500bp of predicted enhancer peak) 66. (f–i) Examples of transcriptional changes upon Nipbl deletion. Stranded RNA-Seq and ChIP-Seq tracks (H3K4me3, H3K27ac) are shown for control (blue) and ΔNipbl (red) samples. Comparison of the chromatin profiles are shown with log2(ΔNipbl/TAM) tracks for H3K4me3 and H3K27ac (in grey). Active enhancers (peaks of high H3K27ac, H3K4me1 66, low H3K4me3) are depicted as green ovals. (f) chr10:21,090,000–21,781,000. Bidirectional transcription (position labeled with a blue bar) arises from an isolated enhancer in ΔNipbl cells. (g) chr17:45,945,000–46,176000. Bidirectional transcription (position labeled with a blue bar) arises from two cryptic promoters (H3K4me3 peaks, no/weak transcription in TAM) downstream of Vegfa. (h) chr3:21,712,500–22,126,240. A new transcript from a cryptic promoter 100 kb upstream of Tbl1xr1. H3K27ac signal is enhanced at peaks surrounding the activated cryptic promoter. (i) chr15:9,873,000–10,354,700. Promoter switch for Prlr, from an upstream promoter to a more downstream one surrounded by active enhancers. (j) chr6:141,743,961–141,904,692. Downregulation of Slco1a1 and concomitant up-regulation of Slco1a4 and non-coding intergenic transcripts (arrowheads). Distance of Slco1a4 promoter to intergenic enhancers is less than 10kb, compared to 80 kb for Slco1a1.
Supplementary Material
Acknowledgments
This work would not have been possible without the important contribution of the members of the EMBL Laboratory Animal Resources Facility, particularly Silke Feller, for animal welfare and husbandry. We thank members of the Mirny and the Spitz labs, John Marioni (EMBL/EBI)) for many productive discussions and helpful suggestions. We greatly thank Heather Marlow (IP) for her irreplaceable help and expertise in generating the cohesin and CTCF ChIP-Seq data. We thank the EMBL Genomics Core Facility and Pasteur Biomics for expert assistance and support in sequencing the different genomic libraries. We thank Ana Losada (CNIO) for generously providing antibodies. W.S and A.P were supported by an EMBL Interdisciplinary Postdoc (EIPOD) Fellowship under Marie Curie Actions COFUND. The work in the Mirny lab is supported by R01 GM114190, U54 DK107980 from the NIH, and 1504942 from the NSF. The collaboration is also partially supported by MIT-France MISTI Fund. The work in the Spitz lab was supported by EMBL, the Pasteur Institute and the Deutsche Forschungsgesellschaft (DFG grant: SP 1331/3-1) and included funding from the European Commission’s Seventh Framework Programme through the Collaborative Research Project RADIANT (Grant Agreement no: 305626, to W.H.).
Footnotes
Author Contributions
W.S. and F.S. conceived the study and designed the experiments, with input and advice from C.H. Experimental data was generated by W.S., with the help of A.P for TCC, who also carried out preliminary analyses of the TCC datasets, with advice from W.H. N.A. and A.G. performed computational analysis of Hi-C, RNA-seq, ChIP-seq and other relevant datasets. Y.L-M and N.A.F. contributed to analysis of transcription data. G.F. performed computer simulations of cohesin depletion and assisted with data analysis and paper writing. L.M. provided advice on data analysis and simulations. W.S., N.A., A.G., L.M. and F.S. wrote the paper with input from the other authors.
The authors declare no competing financial interests.
Code availability.
The software developed to process the data in our study is available at https://github.com/mirnylab/.
Data availability statement
The reference and accession number to published data used and analysed in this work are indicated in Supplementary Table 3. The Hi-C, ChIP-seq and RNA-seq data generated and analysed are available in the GEO repository, under accession number GSE93431. Links to HiGlass visualizations of Hi-C, ChIP-seq and RNA-seq data obtained in this study are available at http://mirnylab.mit.edu/projects/nipbl and at http://higlass.io.
References
- 1.Bickmore WA, van Steensel B. Genome Architecture: Domain Organization of Interphase Chromosomes. Cell. 2013;152:1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods? Bioessays. 2013;35:818–828. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupiáñez DG, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symmons O, et al. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev Cell. 2016;39:529–543. doi: 10.1016/j.devcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014 doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genomics Hum Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 14.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Reports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seitan VC, et al. Cohesin-based chromatin interactions enable regulated gene expression within pre-existing architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sofueva S, et al. Cohesin-mediated interactions organize chromosomal domain architecture. EMBO J. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 23.Tedeschi A, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlich D, Koch B, Dupeux F, Peters JM, Ellenberg J. Live-cell imaging reveals a stable cohesin-chromatin interaction after but not before DNA replication. Curr Biol. 2006;16:1571–1578. doi: 10.1016/j.cub.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 25.Hu B, et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 2015;43:e132. doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt AD, et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Reports. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lajoie BR, Dekker J, Kaplan N. The Hitchhiker's guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naumova N, et al. Organization of the Mitotic Chromosome. Science. 2013 doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nolen LD, Boyle S, Ansari M, Pritchard E, Bickmore WA. Regional chromatin decompaction in Cornelia de Lange syndrome associated with NIPBL disruption can be uncoupled from cohesin and CTCF. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rhodes J, Mazza D, Nasmyth K, Uphoff S. Scc2/Nipbl Hops Between Chromosomal Cohesin Rings After Loading. biorxiv.org. 2017 doi: 10.1101/136754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haarhuis JHI, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gassler J, et al. A Mechanism of Cohesin-Dependent Loop Extrusion Organizes Zygotic Genome Architecture. bioRxiv. 2017;177766 doi: 10.1101/177766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wutz G, et al. CTCF, WAPL and PDS5 proteins control the formation of TADs and loops by cohesin. biorxiv.org. 2017;177444 doi: 10.1101/177444. [DOI] [Google Scholar]
- 37.Rao S, et al. Cohesin Loss Eliminates All Loop Domains, Leading To Links Among Superenhancers And Downregulation Of Nearby Genes. bioRxiv. 2017;139782 doi: 10.1101/139782. [DOI] [Google Scholar]
- 38.Kerpedjiev P, et al. HiGlass: Web-based Visual Comparison And Exploration Of Genome Interaction Maps. bioRxiv. 2017;121889 doi: 10.1101/121889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flyamer IM, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips-Cremins JE, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fraser J, et al. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol. 2015;11:852. doi: 10.15252/msb.20156492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sexton T, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Tang SHE, Silva FJ, Tsark WMK, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 44.Logan M, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 45.Tannour-Louet M, Porteu A, Vaulont S, Kahn A, Vasseur-Cognet M. A tamoxifen-inducible chimeric Cre recombinase specifically effective in the fetal and adult mouse liver. Hepatology. 2002;35:1072–1081. doi: 10.1053/jhep.2002.33164. [DOI] [PubMed] [Google Scholar]
- 46.Li WC, Ralphs KL, Tosh D. Isolation and culture of adult mouse hepatocytes. Methods Mol Biol. 2010;633:185–196. doi: 10.1007/978-1-59745-019-5_13. [DOI] [PubMed] [Google Scholar]
- 47.Gonçalves LA, Vigário AM, Penha-Gonçalves C. Improved isolation of murine hepatocytes for in vitro malaria liver stage studies. Malar J. 2007;6:169. doi: 10.1186/1475-2875-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imakaev M, et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filippova D, Patro R, Duggal G, Kingsford C. Identification of alternative topological domains in chromatin. Algorithms Mol Biol. 2014;9:14. doi: 10.1186/1748-7188-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bishop CM. Pattern recognition. Machine Learning. 2006;128 [Google Scholar]
- 52.Bogu GK, et al. Chromatin and RNA Maps Reveal Regulatory Long Noncoding RNAs in Mouse. Mol Cell Biol. 2015;36:809–819. doi: 10.1128/MCB.00955-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt D, et al. Waves of Retrotransposon Expansion Remodel Genome Organization and CTCF Binding in Multiple Mammalian Lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yue F, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016;5 doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eastman P, et al. OpenMM 4: A Reusable, Extensible, Hardware Independent Library for High Performance Molecular Simulation. J Chem Theory Comput. 2013;9:461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eastman P, Pande VS. Efficient nonbonded interactions for molecular dynamics on a graphics processing unit. J Comput Chem. 2010;31:1268–1272. doi: 10.1002/jcc.21413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imakaev MV, Tchourine KM, Nechaev SK, Mirny LA. Effects of topological constraints on globular polymers. Soft Matter. 2015;11:665–671. doi: 10.1039/c4sm02099e. [DOI] [PubMed] [Google Scholar]
- 65.Kawauchi S, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/−) mouse, a model of Cornelia de Lange Syndrome. PLoS Genet. 2009;5:e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.