Abstract
Interindividual differences in hypolipidemic effect of simvastatin are observed in clinic, and metabolomic studies have uncovered the association between variations of bacterial-derived metabolites and therapeutic outcomes. In current study, we investigated the gut microbial-involved mechanisms underlying the different responses to simvastatin. Male C57BL/6J mice were given high-fat/cholesterol diet (HFD) 8 weeks and then orally administered simvastatin (20 mg/kg, once a day) for 4 weeks with or without antibiotic (100 mg/kg Imipenem : Cilastatin Sodium). We observed simvastatin reduced the levels of serum TC, LDL, HDL and TG in HFD-fed mice, but this effect was attenuated by antibiotic which altered gut microbiota composition. Subsequent metabolomic study indicated that gut microbiota modulation changed the serum metabolic and bile acid profiles in simvastatin-treated mice. Moreover, our results showed that simvastatin stimulated the expression of hepatic CYP7A1, CYP7B1 and Farnesoid X receptor (FXR) in HFD-fed mice, which were impaired by gut microbiota modulation. In summary, our results revealed that the hypolipidemic effect of simvastatin was correlated with the composition of gut microbiota, and the attenuated hypolipidemic effect of simvastatin by gut microbiota modulation was associated with the suppression on hepatic CYP7A1, CYP7B1 and FXR proteins that regulate bile acids synthesis from cholesterol.
Keywords: Simvastatin, hypolipidemic effect, antibiotic, gut microbiota, metabolomics
Introduction
Simvastatin (SV) is one of the most widely used statins for reducing LDL-cholesterol (LDL-c) and preventing cardiovascular disease (CVD) by inhibiting HMG-CoA reductase 1–3. In clinic, obvious variations in therapeutic benefits and LDL-c reduction upon SV therapy have been observed among patients, in which some “good” responders showed over 60% reduction in LDL-c, whereas those “poor” responders only had less than 10% reduction in LDL-c 4. However, the mechanisms underlying the interindividual variation in efficacy of SV are poorly known. Kaddurah-Daouk R et al. have conducted a series of investigations for deciphering the metabolic association with efficacy of SV between good and poor responders by using metabolomic approach5, 6. They found that several panels of metabolites were differently altered in good and poor responders upon SV therapy such as lipids, fatty acids, amino acids and gut microbiota-derived secondary bile acids. Moreover, they observed a strong correlation between the degree of LDL reduction and baseline levels of several secondary bile acids, including lithocholic acid (LCA), taurolithocholic acid (TLCA) and glycolithocholic acid (GLCA) which are produced by gut microbiota7. In addition, the different responses to SV therapy are also associated with the higher baseline level of coprostanol in patients, which is derived from hydroxylation of cholesterol by gut microbiota7. Accordingly, this evidence highlights the involvement of gut microbiota in affecting individual responses to SV.
It has been well recognized that gut microbiota plays critical roles in development of various diseases8, as well as influencing both the pharmacokinetics and pharmacodynamics of many drugs 9, 10. In vitro experiment coupled with metabolomics indicates that SV is degraded into various metabolites by human intestinal microbiota 11, while a similar study uncovered that the biotransformation of orally administered lovastatin could be altered in antibiotic-treated rats compared to normal rats 12. As a result, we speculated that the different responses towards SV therapy were due to the differences in gut microbiota.
In current study, we compared the hypolipidemic effect of SV in high-fat/cholesterol-diet (HFD) fed mice with or without vancomycin treatment, as well as their metabolic impacts and expression of genes involved in cholesterol metabolism pathway in liver. Our results showed that gut microbiota modulation with antibiotic attenuated the hypolipidemic effect of SV in HFD fed mice. Moreover, gut microbiota modulation resulted in obvious alteration of serum metabolic profiles in SV-treated mice including a panel of phospholipids and bile acids, as well as the expression of hepatic CYP7a1 and CYP7a1 genes and their protein. Our current results indicated that the different responses to SV therapy was, at least partially, due to the variation of gut microbiota.
Material and Methods
Chemicals
Simvastatin (SV) was purchased from MSD Pharmaceutical Company Limited (Hangzhou, China). TIENAM (Imipenem:Cilastatin Sodium=1:1) was obtained from Merck Sharp & Dohme Corp. All the primary and secondary antibodies for the western blot analysis were purchased from Cell Signaling Technology (USA). TRIzol reagent used for total RNA extraction was purchased from Sigma-Aldrich (USA). High capacity cDNA reverse transcription kit and SYBR Master Mix were purchased from Applied Biosystems (USA); RIPA Lysis Buffer was purchased from Beyotime company (China). SuperSignal West Pico Chemiluminescent Subatrate was purchased from Thermo Fisher Scientific (USA). The derivatization reagent for GC-MS analysis, N,O-Bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) was purchased from Regis (Morton Grove,IL, USA).
Animal experiments
Sixty male C57BL/6J mice (4-week-old) were purchased from Shanghai Laboratory Animal Center (Shanghai, China) and housed in a light-controlled room kept at a temperature of 23±3°C and a relative humidity of 50±5% with free access to water and a normal standard chow diet. All the mice were acclimatized for 1 week prior to the experiments. Seven mice were randomly selected and fed with normal chow diet as the control group (Con, n=7), while the rest 53 mice were fed with high-fat/cholesterol-diet (HFD) for 8 weeks. Then, the mice were divided into four groups as follows: HFD group (HFD, n=8), antibiotic-treated group (AB, n=10), simvastatin-treated group (SV, n=13) and combined antibiotic and simvastatin treatment group (AB_SV, n=12). The mice in AB and AB_SV groups were orally administrated with TIENAM (100 mg/kg) once a day during experiment according to the reference 13. The mice in SV and AB_SV groups were treated with SV (20 mg/kg•day) by gavage. After another four weeks treatment, all the animals were sacrificed and the samples were collected and stored at −80°C for subsequent analysis.
Analysis of serum TC, TG, LDL, HDL and liver histology
The serum TC, TG, LDL, and HDL were analyzed with enzymatic assay kits according the manufacture’s instruction. For liver histological analysis, the liver tissue was first fixed in 10 % (volume/volume) formaldehyde, embedded in paraffin, and stained with hematoxylin-eosin (HE) according to the standard protocol.
Fecal DNA extraction and gut microbiota analysis
Fecal samples were collected from cecum of mice during sacrifice and frozen at −80°C until use. Bacterial genomic DNA was extracted with 100mg fecal samples using a fast DNA stool mini kit (QIAGEN, Germany) following the manufacturer’s instructions. The extracted DNA samples were used as template for amplification of the V3 region of 16S rRNA gene. The PCR amplification, pyrosequencing of PCR amplicons and quality control were performed on Ion PGM™ System according to reference 14. The acquired valid and representative sequences of each sample were against Greengenes database using nearest alignment space termination algorithm15, and constructed a neighbor-joining tree with ARB 16. Operational taxonomic units (OTUs) were delineated at 97% similarity level with Mothur software. The representative sequence of each OTU was selected with the most abundance and subjected to RDP classifier for taxonomical assignment with a bootstrap cutoff of 60% 17, 18. Alpha diversity was assessed with Rarefaction analysis and the Shannon-Wiener index with QIIME 19. Weighted Fast UniFrac principal coordinate analysis (PCoA) was performed with the phylogenetic tree constructed by each OTU generated with QIIME 19.
Serum metabolic profiling with GC-MS and UPLC-QTOF-MS
Serum samples stored at −80 °C were thawed and vortexed for 5 s at room temperature. To a tube containing 10 μL of internal standard (0.1 mg/mL dulcitol), 30 μL serum sample was added and vortexed for 5 s. Subsequently, 120 μL of ice-cold methanol/chloroform (3:1) was added, the resulting mixture was vortexed for 30 s and placed at −20 °C for 20 min before centrifugation at 16 000 g and 4 °C for 15 min. Quality control (QC) sample pooled from representative serum samples of mice in every group were prepared and analyzed with the same procedure as that for the experiment samples in each batch. The supernatant was utilized for metabolic profiling with GC-MS and UPLC-QTOF-MS, respectively. The process of instrumental analysis and data preprocessing was described as the following:
GC-MS analysis and data preprocessing
One vial containing 90 μL of supernatant of each sample was dried under a gentle nitrogen stream, and 30 μL of 20 mg/mL methoxylamine hydrochloride in pyridine was subsequently added. The resultant mixture was vortexed vigorously for 30 s and incubated at 37 °C for 90 min. Thirty microliters of BSTFA (with 1% TMCS) was added into the mixture, which was derivatized at 70 °C for 60 min prior to injection. At the same time, a blank derivatization sample (using deionized water instead of serum sample) was prepared in order to remove the background noise produced during sample preparation and GC/MS analysis. The derivatized serum samples were analyzed on an Agilent 7890A gas chromatography system coupled to an Agilent 5975C MSD system with inert Triple-Axis Detector (Agilent, CA). A HP-5MS fused-silica capillary column (30 m × 0.25 mm × 0.25 μm, Agilent J&W Scientific, Folsom, CA, USA) was utilized to separate the derivatives. Helium was used as the carrier gas at a constant flow rate of 1 mL/min through the column. The injection volume was 1 μL, and the solvent delay time was 5 min. The initial oven temperature was held at 80 °C for 2 min, ramped to 300 °C at a rate of 10 °C/min, and finally held at 300 °C for 6 min. The temperature of the injector, transfer line, and ion source (electron impact) was set to 250, 290, and 230 °C, respectively. The collision energy was 70 eV. Mass data was acquired in a full-scan mode (m/z 50–600). The samples were analyzed in a random sequence.
The peak picking, alignment, deconvolution, and further processing of raw GC-MS data were referred to the previous published protocols 20. The final data was exported as a peak table file, including observations (sample name), variables (rt_mz), and peak abundances. The data was normalized against total peak abundances before performing univariate and multivariate statistics.
UPLC-QTOF-MS analysis and data preprocessing
Another one tube containing 10 μL supernatant from each sample was dried and reconstituted in 100 μL acetonitrile/water (1:1, v/v) containing 1 μg/mL L-phenylalanine-13C9,15N as internal standard prior to UPLC-QTOF-MS analysis. Chromatographic separation was performed on a Waters Acquity UPLC system with a BEH C18 column (2.1mm × 100 mm, 1.7 μm) at a flow rate of 0.4 mL/min and 50 °C column temperature. The injection volume was 2 μL. The mobile phases consisted of water (phase A) and methanol (phase B), both with 0.1% formic acid (v/v). A linear gradient elution was performed with the following program: 0–0.5 min, 1% B; 1.5 min, 40% B; 5 min, 80% B; 9.3 min, 100% B; 12 min, 100% B; 12.01 min, 1% B and held to 14 min.
The eluents were analyzed on a hybrid quadrupole time-of-light mass spectrometer (Triple TOF 4600 system, AB Sciex, Comcord, ON, Canada) equipped with a DuoSpray ion source in positive ion mode. The pressures of nebulizer gas (GS1), heater gas (GS2) and curtain gas (CUR) were set to 50 psi, 50 psi, and 45 psi, respectively. Ionization voltage was set to 5000 V and spray temperature was 550 °C. A typical information dependent acquisition comprising the acquisition of a survey TOF MS spectrum and then a MS/MS experiment was applied in the analysis. The TOF MS scan was operated under the high resolution settings with a range of 50 – 1000 m/z and an accumulation time of 250 ms. The declustering potential (DP) and collision energy (CE) were set at 60 V and 10 eV, respectively, in the positive ion mode. In the second experiment, up to 10 candidate precursors per scan cycle were fragmented in collision-induced dissociation (CID) by a CE setting at 45 ± 15 eV, and the data were collected at a range of 50 – 1000 m/z with 10 ms accumulation time for the products of each precursor. The software for controlling instrument and collecting data was Analyst TF 1.6 (AB Sciex, Comcord, ON, Canada).
The raw data of UPLC-QTOF-MS were transformed to mzXML format (ProteoWizard) and then processed by XCMS and CAMERA packages in R software platform. In XCMS package, the peak picking (method= centWave, ppm=15, peakwidth=c(5,20), snthresh=10), alignment (bw =6 and 3 for the first and second grouping, respectively), and retention time correction (method=obiwarp) were conducted. In CAMERA package, the annotations of isotope peak, adducts, and fragments were performed with default parameters. The final data was exported as a peak table file, including observations (sample name), variables (rt_mz), and peak abundances. The data was normalized against total peak abundances before performing univariate and multivariate statistics.
Statistical analysis and identification of differential variables
For multivariate statistical analysis, the normalized data from GC-MS and UPLC-QTOF-MS were imported to Simca-P software (version 11.0), where the data were preprocessed by UV scaling and mean centering before performing PCA, and PLS-DA. The model quality is described by the R2X or R2Y and Q2 values. R2X (PCA) or R2Y (PLS-DA) is defined as the proportion of variance in the data explained by the models and indicates the goodness of fit. Q2 is defined as the proportion of variance in the data predictable by the model and indicates the predictability of current model, calculated by cross-validation procedure. In order to avoid model over-fitting, a default 7-round cross-validation in Simca-P software was performed throughout to determine the optimal number of principal components. For univariate statistical analysis, the Student’s t test was conducted on the normalized data. The variables with VIP>1 values of PLS-DA model and p<0.05 values of Student’s t test were identified as potential differential metabolites. Fold change was calculated as with the normalized peak intensity.
Structural identification of metabolites
For GC-MS data, the AMDIS software was applied to deconvolute mass spectra from raw GC-MS data, and the purified mass spectra were automatically matched with an in-house standard library including retention time and mass spectra, Golm Metabolome Database, and Agilent Fiehn GC/MS Metabolomics RTL Library (matching similarity larger than 70%). For UPLC-QTOF-MS data, the accurate m/z of precursors and product ions were matched against Metlin, MassBank, LipidBlast databases and in-house standard library including retention time, accurate precursors, and product ions.
Analysis of serum bile acids with UPLC-MS/MS
Ten μL of isotope labeled internal standards (125 ng/mL in 50 % aqueous methanol) and 90 μL of methanol/acetonitrile (5:3, v/v) were added to 25 μL of thawed serum sample. The mixture was vortexed for 1 min and placed at 4 °C for 30 min prior to centrifugation at 16000 g for 15 min (4 °C). The supernatant was dried under nitrogen stream and reconstituted in 50 μL of aqueous methanol (v/v). The re-dissolved solution was vortexed for 2 min and centrifuged at 16000 g for 15 min (4 °C). The supernatant was used for detecting serum bile acids with UPLC-MS/MS. Chromatographic separation was performed on an Agilent Poroshell 120 SB-C18 column (2.7μm, 2.1mm×50mm) with a flow of 0.4 mL/min and 40 °C column temperature. The mobile phases consisted of water (phase A) and methanol (phase B), both with 10 mM ammonium acetate and 0.012% formic acid (v/v),. A linear gradient elution was used with the following program: 0–0.5 min, 35%B; 2.5min, 65%B; 6.7min, 80%B; 6.71min, 100%B; 8 min, 100%B; 8.01min, 35%B and held to 10min. The eluents were detected by an AB Sciex TripleQuad 5500 (Comcord, ON, Canada) equipped with electrospray ionization (ESI) source in negative ion mode. The pressures of nebulizer gas (GS1), heater gas (GS2), curtain gas (CUR), and collision gas (CAD) were set to 50, 50, 30, and 8 psi, respectively. Ionization voltage was set to −4500 V and spray temperature was 550 °C. The precursors were fragmented and monitored in Multiple Reaction Monitoring (MRM) mode. The dwelling time was 10 ms for all transitions. The raw data were processed in Analyst 1.5.2 (AB Sciex, Comcord, ON, Canada). The relative quantification was obtained by normalized the peak area against internal standard (Cholic acid-D4).
Hepatic gene expression analysis with RT2 Profiler PCR Array
To test the hepatic gene expression involved in cholesterol metabolism pathway, commercial RT2 Profiler PCR Arrays were used for quantitation of 84 targeted genes involved in cholesterol metabolism pathway (Cat no. PAHS-080Z, QIAGEN, Germany). Briefly, the total RNA was extracted with RNeasy Mini Kit (Cat no. 74104, QIAGEN, Germany) and 1μg of total RNA was subjected to first strand cDNA synthesis with RT2 HT First Strand Kit (Cat no. 330411, QIAGEN, Germany) according to the manufacturer’s instructions. The synthesized cDNA samples were analyzed with RT2 Profiler PCR Arrays in triplicate on Bio-rad Connect (Bio-rad, USA).
Western blot
The expression of several hepatic proteins was analyzed with western blot. Briefly, the total proteins were extracted from about 20 mg liver tissues with 500 μL of RIPA Lysis Buffer (Beyotime, China) according to well-established protocols. The protein concentrations were determined with Pierce Coomassie Protein Assay kit (Thermo Scientific). A total of 20 μg protein was loaded into each lane and separated by 10% SDS-PAGE gel, and then transferred to a PVDF membrane (Bio-rad, USA). The membrane was then blocked with 10% milk at room temperature. The membranes were incubated with primary antibodies (1:1000 dilution, Cell Signaling Technology, USA) overnight at 4°C. Then, the membranes were washed with TBST (20 mM Tris-HCL, 137mM NaCl, and 0.1% Tween20, pH7.5) three times at 10min interval. The HRP-conjugated secondary antibodies were then incubated for 1h at room temperature. The membranes were exposed with SuperSignal West Pico Chemiluminescent Subatrate (Thermo Fisher Scientific, USA) and the resulting bands were quantified by using Amersham Imager 600 system (General Electric Company, USA). GAPDH was used as a control.
Results
Antibiotic treatment attenuated the hypolipidemic effect of SV in mice
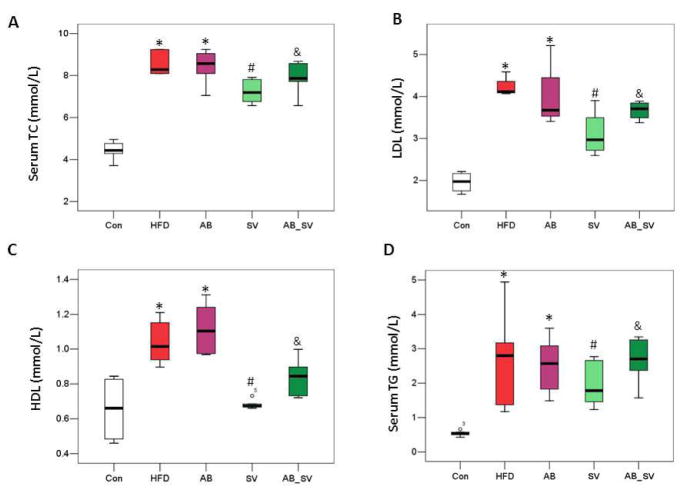
To test whether the different responses to SV treatment was associated with the differences in gut microbiota, mice were treated with SV for 8 weeks with or without AB intervention. The serum levels of TC, LDL, HDL and TG were significantly elevated in both HFD and AB groups indicating that AB did not affect the lipids metabolism per se. SV treatment significantly reduced the levels of TC, LDL, HDL and TG, while the concomitant administration of AB attenuated the hypolipidemic effect of SV (Fig 1), indicating that the hypolipidemic effect of SV was affected by gut microbiota modulation.
Figure 1.
Antibiotic treatment attenuated the hypolipidemic effect of SV. A-D: Serum TC, LDL, HDL and TG levels among groups. *P<0.05 vs Con group; #P<0.05 vs HFD group; &P<0.05 vs SV group. Data are mean ± S.D. Comparisons between groups were analyzed with two-tailed Student’s t-test. Con: control group; HFD: high-fat/cholesterol diet group; AB: antibiotic group; SV: simvastatin (20 mg/kg) group; AB_SV: simvastatin (20 mg/kg) in the presence of antibiotic treatment.
Antibiotic treatment altered the composition of gut microbiota
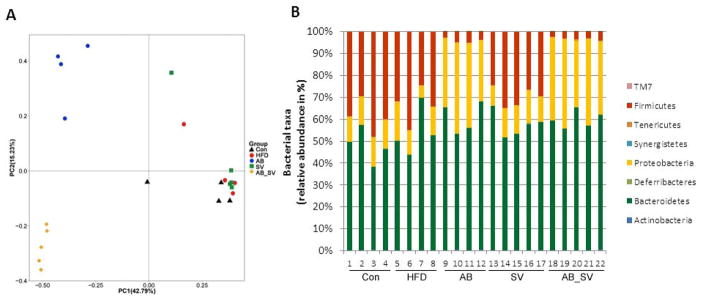
We next detected the compositional changes of gut microbiota by AB administration by using pyrosequencing on the V3 region of 16S rDNA of bacteria. A total of 263732 valid reads were obtained from 22 fecal samples after normalization to the sample with minimal number of valid reads, and averagely each sample had 11987±55 reads for following analysis. Then, OTUs were binned with acquired valid reads at 97% similarity level against the Greengene database 15. The Rarefaction and Shannon-Wiener curves showed that most of the diversities of bacteria in samples have been covered in the current sequencing depth (data not shown). A total of 39966 OTUs were used for phyla analysis with RDP classifier at a bootstrap cutoff of 60%. The most abundant phyla included Bacteroidetes (21108 OTUs, contributing to 52.81% of all OTUs), Firmicutes (9042 OTUs, contributing to 22.62% of all OTUs), Proteobacteria (7732 OTUs, contributing to 19.35% of all OTUs) and Actinobacteria (82 OTUs, contributing to 0.21% of all OTUs). Then, PCoA was performed to compare the differences of OTUs abundance among the five groups. Samples from AB and AB_SV groups were distinctly separated from Con, HFD and SV groups along the PC1 (42.79%), which accounted for the largest proportion of total variation. Interestingly, AB_SV group was also separated from AB group by PC2 (15.23%) (Fig 2A). Thus, AB treatment significantly altered the composition of gut microbiota, and the compositional differences of gut microbiota may exist between AB and AB_SV groups suggesting that SV treatment may also exert some impacts on gut microbiota. HFD feeding resulted in the increase in abundance of Bacteroidetes, decrease of Firmicutes and Actinobacteria, while Proteobacteria kept stable. In AB and AB_SV group, Firmicutes were greatly depleted, whereas Bacteroidetes and Proteobacteria were increased similarly. However, Actinobacteria and Tenericutes were differently altered in AB and AB_SV groups. SV treatment mainly increased the abundance of Bacteroidetes, but reduced Firmicutes and Actinobacteria (Fig 2B). Therefore, the composition of gut microbiota was dramatically altered by AB administration.
Figure 2.
Antibiotic treatment altered the composition of gut microbiota. A: Weighted UniFrac PCoA plot based on OTU abundance of each mouse. Each point in the plot represents the gut microbiota of one mouse. B: The relative taxonomic abundance at the phylum level of gut microbiota among groups.
Serum metabolic profiles among groups
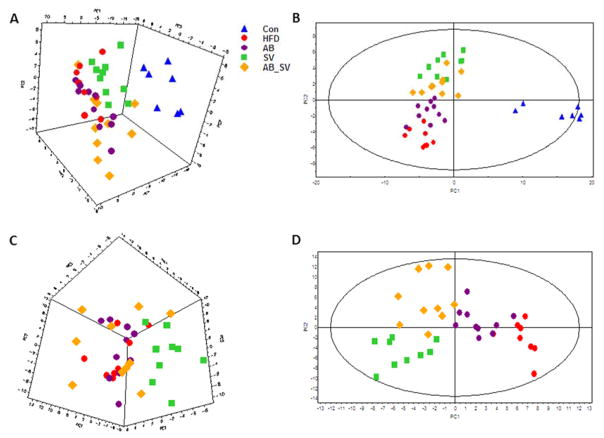
To describe the metabolic impacts of gut microbiota modulation on SV-treated mice, serum metabolic profiling was performed by using combined GC-MS and UPLC-QTOF-MS metabolomic approach. First, the global metabolic profiles were compared with unsupervised PCA, which incorporated the identified 182 serum metabolites (including 99 from GC-MS and 83 from UPLC-QTOF-MS). It showed that samples from control group were distinctly separated from all the rest groups, whereas HFD and AB groups were almost clustered together. Most samples from SV and AB_SV groups were clearly separated (Fig 3A). Then, a supervised PLS-DA model was constructed, in which control group was separated from the rest four groups by PC1, while SV and AB_SV groups were separated from both HFD and AB groups by PC2 (Fig 3B). The quality of the PCA and PLS-DA models were described in Table S1.
Figure 3.
Serum metabolic profiles among groups. A-B: The 3D PCA score plot and PLS-DA model of all of the five groups based on the identified metabolites in serum with combined GC-MS and UPLC-QTOF-MS. C-D: The 3D PCA score plot and PLS-DA model of the serum metabolites of the four groups, except control group.
To further characterize the metabolic differences between SV and AB_SV groups, we constructed a PCA model with the four HFD feeding groups, in which SV group was clearly separated with the rest three groups, and the latter three groups were hardly divided (Fig 3C). The following PLS-DA model showed that both HFD and AB groups were divided with SV and AB_SV groups by PC1, whereas SV and AB_SV were separated by PC2 (Fig 3D). Accordingly, these results indicated that HFD feeding resulted in transparent alterations in serum metabolic profile, and AB per se did not change it. Moreover, SV treatment dramatically altered the metabolic profile of HFD feeding mice, but not in AB_SV group.
To investigate the metabolic involvement in therapeutic effect of SV, the key differential metabolites were determined with the criteria of both VIP>1 in PLS-DA model and P<0.05 in Student’s t test between control and HFD groups, and were also significantly restored by SV treatment compared to HFD group. A total of 26 key differential metabolites were selected and most of them were increased, except for the decrease of glycolic acid, glycerol and lysoPC (20:4) in HFD group. The increased differential metabolites in HFD group mainly belonged to monoglyceride (MG) including MG(18:1), MG(18:2), and MG(20:5), lysophospholipids (LysoPCs) including LysoPC(16:1), LysoPC(18:0), LysoPC(18:1), LysoPC(18:2) and LysoPC(20:4), phospholipids (PCs) including PC(36:7), PC(38:7), PC(32:2), PC(40:8), PC(34:3), PC(36:5), PC(32:1), and PC(36:3), and phosphatidylethanolamines (PEs) including PE(38:4), PE(36:2) and PE(36:1), as well as cholesterol, phenylalanine, glycerol-3-phosphate, alanine, ribose-5-phosphate. Moreover, most of these metabolites were differently altered in SV and AB_SV groups, except for the lysoPCs that were changed with similar extent in both SV and AB_SV groups (Table 1).
Table 1.
Serum differential metabolites based on combined GC-MS and UPLC-QTOFMS
| Metabolites | RT_Min | Mz | P value
|
Fold Change (vs Con)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HFD vs Con | AB vs HFD | SV vs HFD | AB_SV vs HFD | HFD | AB | SV | AB_SV | |||
| Glycolic acidGC | 5.34 | 177 | 0.000 | 0.840 | 0.030 | 0.889 | 0.59 | 0.59 | 0.71 | 0.58 |
| AlanineGC | 5.66 | 116 | 0.001 | 0.737 | 0.018 | 0.598 | 1.87 | 1.80 | 1.43 | 1.73 |
| GlycerolGC | 7.96 | 205 | 0.009 | 0.346 | 0.003 | 0.000 | 0.72 | 0.82 | 1.15 | 1.17 |
| Glycerol-3-phosphateGC | 13.96 | 357 | 0.008 | 0.017 | 0.028 | 0.049 | 1.28 | 1.12 | 1.07 | 1.06 |
| Ribose-5-hosphateGC | 17.48 | 315 | 0.004 | 0.635 | 0.003 | 0.224 | 2.41 | 2.61 | 1.38 | 1.95 |
| CholesterolGC | 25.09 | 329 | 0.000 | 0.720 | 0.000 | 0.102 | 2.36 | 2.29 | 1.34 | 2.00 |
| PhenylalanineLC | 1.88 | 166.0857 | 0.002 | 0.066 | 0.020 | 0.081 | 1.54 | 1.37 | 1.25 | 1.35 |
| MG(18:1)GC | 22.38 | 397 | 0.000 | 0.985 | 0.000 | 0.081 | 5.23 | 5.22 | 2.82 | 4.34 |
| MG(20:5) LC | 5.12 | 399.2492 | 0.000 | 0.077 | 0.009 | 0.040 | 5.55 | 3.96 | 3.12 | 3.75 |
| MG(18:2) LC | 7.58 | 377.2648 | 0.000 | 0.612 | 0.002 | 0.116 | 4.26 | 3.99 | 2.77 | 3.56 |
| LysoPC(16:1) LC | 6.8 | 494.3225 | 0.000 | 0.005 | 0.007 | 0.001 | 2.07 | 1.53 | 1.35 | 1.24 |
| LysoPC(20:4) LC | 7.04 | 544.3374 | 0.022 | 0.946 | 0.033 | 0.046 | 0.67 | 0.66 | 0.52 | 0.55 |
| LysoPC(18:2) LC | 7.06 | 542.3206 | 0.000 | 0.008 | 0.001 | 0.000 | 4.23 | 3.26 | 2.85 | 2.72 |
| LysoPC(18:1) LC | 7.55 | 522.3541 | 0.007 | 0.038 | 0.001 | 0.000 | 1.78 | 1.45 | 1.14 | 1.18 |
| LysoPC(18:0) LC | 8.12 | 524.3696 | 0.000 | 0.408 | 0.038 | 0.069 | 1.71 | 1.60 | 1.40 | 1.53 |
| PC(36:7) LC | 10.11 | 776.517 | 0.000 | 0.757 | 0.001 | 0.835 | 2.62 | 2.79 | 1.61 | 2.53 |
| PC(38:7) LC | 10.12 | 826.5335 | 0.000 | 0.520 | 0.003 | 0.992 | 2.46 | 2.65 | 1.57 | 2.46 |
| PC(32:2) LC | 10.16 | 752.5183 | 0.000 | 0.895 | 0.002 | 0.186 | 6.23 | 6.15 | 3.84 | 5.17 |
| PC(40:8) LC | 10.21 | 852.5493 | 0.000 | 0.702 | 0.020 | 0.759 | 2.14 | 2.07 | 1.65 | 2.25 |
| PC(34:3) LC | 10.23 | 778.5343 | 0.000 | 0.951 | 0.007 | 0.384 | 4.11 | 4.09 | 2.83 | 3.68 |
| PC(36:5) LC | 10.26 | 802.5341 | 0.000 | 0.202 | 0.001 | 0.596 | 2.14 | 2.40 | 1.36 | 2.00 |
| PC(32:1) LC | 10.43 | 754.5338 | 0.005 | 0.446 | 0.007 | 0.235 | 1.49 | 1.59 | 0.97 | 1.28 |
| PC(36:3) LC | 10.57 | 806.5656 | 0.004 | 0.718 | 0.030 | 0.542 | 1.71 | 1.77 | 1.32 | 1.92 |
| PE(38:4) LC | 10.29 | 790.5348 | 0.001 | 0.505 | 0.020 | 0.479 | 1.99 | 2.15 | 1.37 | 2.25 |
| PE(36:2) LC | 10.34 | 744.5518 | 0.000 | 0.951 | 0.038 | 0.947 | 6.00 | 6.03 | 4.63 | 6.06 |
| PE(36:1) LC | 10.61 | 768.5499 | 0.001 | 0.698 | 0.045 | 0.721 | 2.04 | 2.14 | 1.56 | 2.16 |
In addition to the measurement of serum TC with biochemical kit, we found that the relative concentrations of serum free cholesterol were also significantly increased in HFD group and greatly reduced in SV, but not in AB_SV group detected by GC-MS. Altogether, the serum metabolic profile indicated that HFD resulted in dramatically metabolic alteration, which was effectively restored by SV, but not by AB_SV suggesting the metabolic and gut microbial involvement in mediating hypolipidemic efficacy of SV.
Serum bile acid profiles among groups
Since bile acids are derived from cholesterol and metabolized by gut microbiota, the variation of serum cholesterol level is associated with the alteration of bile acid metabolism 21. To investigate the involvement of bile acid metabolism in hypolipidemic effect of SV with or without gut microbiota modulation, we measured the levels of 16 bile acids in serum samples from different groups with UPLC-MS/MS. The 16 detected bile acids include 7 unconjugated- (CA, CDCA, DCA, LCA, HDCA, 7-MDCA, and UDCA), 5 tauro-conjugated- (TDCA, TCA, TUDCA, THDCA, and TCDCA) and 4 glyco-conjugated (GUDCA, GCDCA, GDCA, and GCA) bile acids. Both CA and CDCA are the primary bile acids of mammalian animals. The relative concentrations of CA, TCA and GCA were dramatically reduced in HFD group compared to control group, in which only TCA was further depleted by SV treatment. However, gut microbiota modulation did not affect their concentrations either alone or combined with SV. In contrary, increased levels of CDCA and TCDCA were observed in both HFD and AB groups, whereas GCDCA was similar among the 5 groups. TCDCA was obviously deceased either in SV or AB_SV groups with similar extent, however, CDCA was only reduced by SV treatment, but not AB_SV. DCA is derived from CA by gut microbiota, and could be metabolized into either GDCA or TDCA. HFD feeding greatly depleted TDCA, but not DCA and GDCA. SV treatment reduced DCA levels compared to HFD group, whereas GDCA was comparable among the five groups. UDCA is a kind of hydrophilic bile acid which is used as a cholagogue and choleretic agent in clinic. In our current study, we observed that both UDCA, and its conjugated forms, GUDCA and TUDCA were dramatically up-regulated in HFD and AB groups, and significantly reduced in SV group, but not in AB_SV group. LCA is derived from CDCA by gut microbiota. We observed that the relative concentration of LCA was increased in HFD group compared to control group, and was further up-regulated in SV group, but was greatly depleted in either AB or AB_SV group. In addition, HFD feeding increased the concentration of serum HDCA and 7-MDCA, which were significantly decreased by either AB or SV treatment (Table 2).
Table 2.
Serum bile acid profiles among groups
| Bile acids | RT_min | Parent ion (mz) | Product ion (mz) | Mean±SD
|
Fold Change (vs Con)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Con | HFD | AB | SV | AB_SV | HFD | AB | SV | AB_SV | ||||
| TUDCA | 2.96 | 498.3 | 80 | 0.0805±0.0998 | 5.2828±6.9763 | 4.7295±5.0536* | 1.0151±0.4211 | 1.8002±0.9833& | 65.61 | 58.74 | 12.61 | 22.36 |
| GUDCA | 3.04 | 448.3 | 74 | 0.0017±0.0008 | 0.0046±0.0044 | 0.0042±0.0019* | 0.0029±0.0007 | 0.0043±0.0027 | 2.65 | 2.38 | 1.67 | 2.44 |
| THDCA | 3.07 | 498.3 | 80 | 0.1073±0.1149 | 0.182±0.1491 | 0.0886±0.0614 | 0.1136±0.1383 | 0.0673±0.0388 | 1.70 | 0.83 | 1.06 | 0.63 |
| TCA | 3.31 | 514.3 | 80 | 1.873±1.2593 | 0.5804±0.8325* | 0.2201±0.3018** | 0.1143±0.0843 | 0.2158±0.1544 | 0.31 | 0.12 | 0.06 | 0.12 |
| 7-MDCA | 3.43 | 405.3 | 405.3 | 0.265±0.112 | 2.7288±2.1263* | 0.5026±0.2601* | 0.7531±0.5404## | 0.579±0.4514 | 10.30 | 1.90 | 2.84 | 2.18 |
| GCA | 3.45 | 464.3 | 74 | 0.0051±0.0021 | 0.0009±0.0005*** | 0.001±0.0011*** | 0.0012±0.0009 | 0.0011±0.0006 | 0.18 | 0.20 | 0.24 | 0.22 |
| TCDCA | 3.97 | 498.3 | 80 | 0.197±0.1831 | 1.5298±1.2291* | 1.1785±0.6616** | 0.8986±0.681 | 0.8904±0.4683 | 7.76 | 5.98 | 4.56 | 4.52 |
| UDCA | 4.05 | 391 | 391 | 0.0406±0.0118 | 13.22±4.331*** | 10.8291±3.3802*** | 8.4961±4.6281# | 9.9859±4.6574 | 325.56 | 266.68 | 209.23 | 245.91 |
| GCDCA | 4.13 | 448.3 | 74 | 0.0057±0.0022 | 0.0052±0.0017 | 0.0059±0.0013 | 0.0057±0.0023 | 0.0076±0.0039 | 0.90 | 1.03 | 1.00 | 1.33 |
| TDCA | 4.15 | 498.3 | 80 | 0.5944±0.1754 | 0.0408±0.0419*** | 0.0067±0.0117*** | 0.0583±0.0866 | 0.0042±0.0053 | 0.07 | 0.01 | 0.10 | 0.01 |
| HDCA | 4.37 | 391 | 391 | 0.0997±0.0883 | 1.1284±0.6062** | 0.4921±0.2867** | 0.3741±0.2657## | 0.4804±0.2867 | 11.32 | 4.94 | 3.75 | 4.82 |
| GDCA | 4.37 | 448.3 | 74 | 0.0013±0.0009 | 0.0009±0.0005 | 0.001±0.0005 | 0.0009±0.0004 | 0.0009±0.0007 | 0.69 | 0.79 | 0.66 | 0.68 |
| CA | 4.61 | 407.3 | 407.3 | 2.5382±1.0373 | 0.7663±0.66** | 0.4553±0.5234*** | 0.4928±0.4746 | 0.9349±0.8208 | 0.30 | 0.18 | 0.19 | 0.37 |
| CDCA | 5.73 | 391 | 391 | 0.0502±0.0222 | 1.6084±1.1658** | 1.0426±0.6207** | 0.481±0.3377## | 0.9791±0.5268& | 32.06 | 20.78 | 9.59 | 19.52 |
| DCA | 5.92 | 391 | 391 | 0.8958±0.3654 | 0.9238±0.591 | 0.5648±0.2739* | 0.4138±0.2587# | 0.6737±0.6199 | 1.03 | 0.63 | 0.46 | 0.75 |
| LCA | 7.03 | 375.3 | 375.3 | 0.0956±0.0412 | 0.6162±0.1859*** | 0.1849±0.3937 | 1.1976±0.7897 | 0.0897±0.0624& | 6.45 | 1.93 | 12.53 | 0.94 |
P<0.05,
P<0.01,
P<0.001 vs Con group;
P<0.05,
P<0.01,
P<0.001 vs HFD group;
P<0.05 vs SV group.
Comparisons between groups were analyzed with two-tailed Student’s t-test. Con: control group; HFD: high-fat/cholesterol diet group; AB: antibiotic group; SV: simvastatin (20 mg/kg) group; AB_SV: simvastatin (20 mg/kg) in the presence of antibiotic treatment.
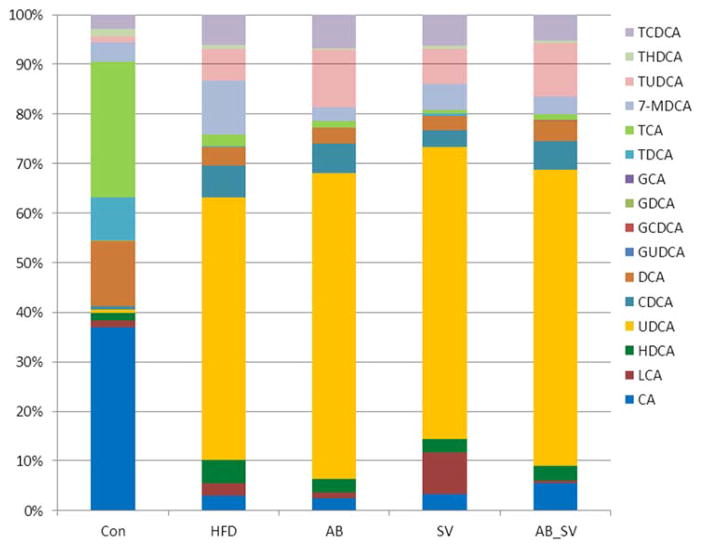
Meanwhile, the relative abundance of the detected 16 bile acids was further analyzed. Generally, HFD feeding resulted in obvious changes of detected bile acids in abundance, and minor changes were observed in either AB or AB_SV group compared to HFD group. We found that the bile acids in control group mainly consisted of CA, DCA, TCA, and TDCA, all of which were obviously reduced in HFD group. The relative abundance of UDCA was over 50% in HFD feeding groups either in presence of AB, SV or not, while LCA was particularly increased by SV treatment (Figure 4). Altogether, these results indicated that HFD feeding dramatically changed the serum bile acid profiles, which were differently altered in SV and AB_SV groups.
Figure 4.
The relative abundance of detected serum bile acids among groups. The relative abundance of 16 bile acids was compared among groups which were detected by UPLC-MS/MS.
Expression analysis of hepatic genes in cholesterol metabolism pathways
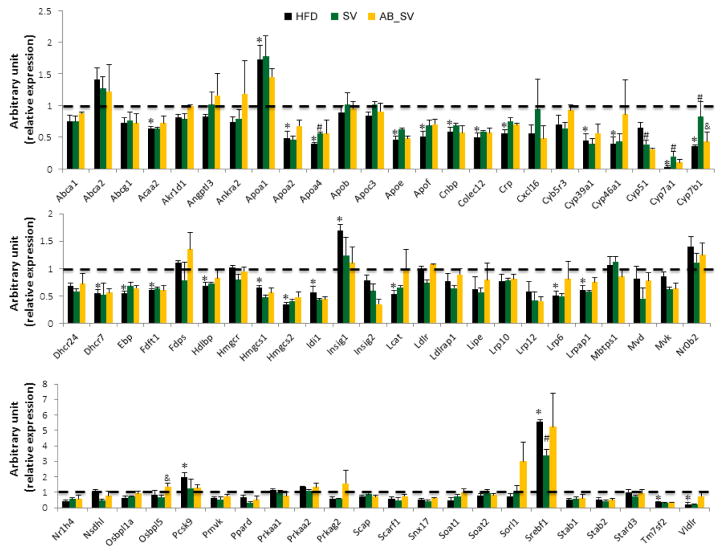
Given the observed differences in hypolipidemic effect and serum bile acids profile between SV and AB_SV groups, we wondered whether the differences in hypolipidemic effect between SV and AB_SV groups were due to the transcriptional regulation on genes involved in cholesterol metabolism pathways. We analyzed the expression of 84 genes that are critically involved in cholesterol metabolism pathway with commercial RT2 Profiler PCR Array, among which 14 genes were excluded because of extremely low signaling in all samples. Since AB alone did not affect the levels of serum lipids, the gene expression analysis was specifically focused on samples from Con, HFD, SV and AB_SV groups. The transcriptional expression of 28 genes was significantly changed by HFD feeding in comparison with Con group, however, only a very small part of the differently expressed genes were reversed by SV treatment such as Apoa4, Cyp7a1, Cyp7b1, Cyp51 and Srebf1 compared to HFD group. Among these significantly altered genes, only Cyp7b1 was statistically restored in AB_SV group compared to SV group, while Cyp7a1 was also restored by AB_SV, but without statistical significance (Fig 5). Therefore, the transcriptional data suggested that the attenuated hypolipidemic effect in AB_SV group was probably associated with the process of bile acids synthesis.
Figure 5.
Summary of hepatic gene expression involved in cholesterol metabolism. The expression analysis of genes involved in cholesterol metabolism pathway by using RT2 Profiler Array. Data are means±S.E.M of triplicates for each group. *indicates P<0.05 compared to Control group, #P<0.05 compared to HFD group, &P<0.05 compared to SV group with two-tailed Student’s t test.
Gut microbiota modulation decreased SV-induced protein expression in regulating bile acids synthesis
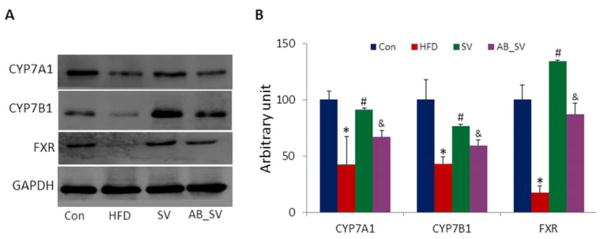
Given the critical roles of bile acids synthesis in affecting cholesterol level, as well as the observed transcriptional variations of Cyp7a1 and Cyp7b1 genes among different groups, we further measured their expression at protein level. Our results showed that the expression of these two proteins were significantly down-regulated by HFD feeding. SV treatment obviously up-regulated the expression of both CYP7A1 and CYP7B1 proteins, and this effect was attenuated by gut microbiota modulation (Fig 6A–B). Farnesoid X Receptor (FXR) is one of the intracellular ligand-activated nuclear receptors that plays critical roles in maintaining cholesterol, triglyceride, glucose and bile acid homeostasis 21. To test whether the attenuated hypolipidemic effect of AB_SV was associated with the regulation on FXR, we measured the expression of FXR protein among groups. We found that HFD feeding resulted in dramatic suppression on FXR protein expression, and was significantly up-regulated in SV group. However, the SV-induced up-regulation of FXR was obviously attenuated in AB_SV group (Fig 6A–B). Altogether, our current results indicated that the attenuated hypolipidemic effect of SV by gut microbiota modulation was associated with the suppression on CYP7A1, CYP7B1 and FXR protein expression, which regulate the bile acid synthesis from cholesterol.
Figure 6.
Expression of hepatic proteins with western blot. A: The representative bands of hepatic CYP7A1, CYP7B1 and FXR proteins detected with western blot (n=4). B: The statistical results of protein expression. Data are means±S.E.M. *indicates P<0.05 compared to Control group, #P<0.05 compared to HFD group, &P<0.05 compared to SV group with two-tailed Student’s t test.
Discussion
Our current results showed that gut microbiota modulation by antibiotic could attenuate the hypolipidemic effect of SV in HFD feeding mice, and this effect was accompanied by obvious alterations of serum metabolic and bile acids profiles. Moreover, we found that the attenuated hypolipidemic effect in AB_SV mice was associated with the suppression on CYP7A1, CYP7B1 and FXR proteins that played critical role in bile acids synthesis.
Statins are widely used lipid-lowering drugs by inhibiting HMG-CoA reductase, while their LDL-C lowering effect varied greatly in clinic 4. Although it is well recognized that the efficacy of statins could be affected by genetic factor 22, only a small proportion of therapeutic variance could be explained by genetic polymorphism 23. Recent publications show that different responses to SV therapy correlates with the baseline variations of several kinds of metabolites such as amino acids 5, phospholipid metabolites 6 and some secondary bacterial-derived bile acids 7. Meanwhile, SV is metabolized by anaerobic bacteria in human fecal suspension into several SV-derived metabolites step by step 11. These results highlight the potential involvement of gut microbiota in affecting the hypolipidemic effect of SV. Our results showed that oral administration of antibiotic not only obviously altered the composition of gut microbiota, but attenuated the hypolipidemic effect of SV as well, demonstrating that different responses to hypolipidemic effect of SV was associated with the differences in gut microbiota. We observed that the bacterial abundance of Gram-positive Firmicutes phylum was greatly reduced, while Gram-negative Proteobacteria phylum was increased by antibiotic administration. Nevertheless, such imbalance between Gram-positive and negative bacteria in AB-treated mice led to the uncertainty of whether the attenuated lipid-lowering effect of SV was due to the reduced Gram-positive bacteria or the increased Gram-negative bacteria individually or jointly.
Although the metabolic profiles of different responses to SV treatment have been extensively investigated, these studies are mainly focused on identifying differential metabolites either at pre- or post-dose of SV treatment that correlate with therapeutic outcomes 5–7. In our current study, we adopted combined GC-MS and UPLC-QTOF-MS-based metabolomic approach to analyze the metabolic profiles among different groups. Our results indicated that the metabolic profile of SV-treated mice was distinctly different from the rest groups, and the metabolic impact of SV was also altered by gut microbiota modulation. Among the identified 26 differential metabolites, some were consistent with the previous observation in human subjects treated with SV. For example, the decreased free cholesterol in current SV-treated mice was also observed in human subjects who were given SV (40mg/d) for six weeks 5. In addition to suppression of HMG-CoA reductase, SV can also up-regulate triacyglycerol lipase activity 24, which may lead to the increase of glycerol in human blood. Consistently, we found that the reduced glycerol by HFD feeding was restored in both SV and AB_SV groups.
Phospholipids PCs and PEs are critical for lipoprotein membrane structure and functions 25, 26. Obvious reduction of PCs and PEs are observed in good responders upon SV treatment 6. Consistently, our results showed that the increased PCs, PEs, and MGs in HFD group were significantly decreased in SV, but not in AB_SV group, suggesting that gut microbiota modulation altered the metabolism of phospholipids. LysoPCs are formed by hydrolysis of PCs by the enzyme of phospholipase A2 27, or lecithin:cholesterol acyltransferase (Lcat) which is secreted from liver 28. Interestingly, the alterations of LysoPCs were very similar, as well as the mRNA expression of hepatic Lcat gene in both SV and AB_SV groups in our current study. In addition, study indicates SV can inhibit the de novo synthesis of PCs by decreasing phosphocholine cytidylyltransferase activity leading to the decrease of plasma lipids 29. Accordingly, we postulated that gut microbiota modulation might impair the suppression of PCs synthesis by SV in the context of HFD feeding.
There are complex interplays between bile acids and cholesterol metabolism, which are also co-metabolized by gut microbiota 21. Meanwhile, primary bile acids (CA and CDCA) and SV are commonly metabolized by CYP3A4 30, 31, as well as the shared transporters such as multidrug resistance gene 1 (MDR1), P-glycoprotein, multidrug resistance-associated protein 2 (MRP2), and organic anion-transporting polypeptide 1B1 32, 33. Therefore, the variations in hypolipidemic effect of SV are associated with bile acids metabolism, and some bacterial-derived secondary bile acids are predictive for the extent of LDL reduction in SV- treated patients 7. We speculated that the attenuated hypolipidemic effect of SV by gut microbiota modulation might be associated with the variations in bile acids metabolism. Our results showed that the profile of the 16 analyzed bile acids in serum varied obviously among groups. LCA is derived from CDCA by intestinal bacteria of Clostridium, a genus of Gram-positive bacteria 34, which is identified as a marker for good response to SV treatment 7. We found that the increased LCA by HFD feeding was almost doubled in SV group, but sustained at lower concentration in AB and AB_SV groups suggesting that the Clostridium bacteria-derived LCA by HFD feeding was impaired by antibiotic. Interestingly, the relative concentrations of UDCA, GUDCA and TUDCA were increased by HFD feeding, and significantly reduced by SV treatment. Although a close association was observed between pretreatment concentrations of UDCA, GUDCA and LDL reduction in SV-treated patients 7, it was unclear about the roles of reduced UDCA, GUDCA and TUDCA in SV-treated mice. As a result, our results indicated that the attenuated hypolipidemic effect of SV by gut microbiota modulation accompanied with variations of serum bile acids, which implied that the impact of gut microbiota modulation on hypolipidemic effect of SV might be associated with the alterations in bile acids metabolism. CYP7A1 is a critical rate-limiting enzyme for bile acid synthesis from cholesterol that is encoded by Cyp7a1 gene 35, 36. The enhanced enzymatic production of 7-hydroxycholesterol catalyzed by CYP7A1 has been observed in SV-treated macrophages 37. Our results showed that the expression of hepatic Cyp7a1 gene and its protein was dramatically suppressed by HFD feeding, and greatly stimulated by SV treatment, however, this effect was impaired in AB_SV group. Similar result was also observed in CYP7b1 gene and its protein, which catalyzes the “alternative” pathway of bile acid synthesis 38. FXR is an important nuclear receptor for regulating bile acid homeostasis via modulation of a series of targeted genes such as Cyp7a1, Cyp8b1, Bsep, Ntcp and so on39. On the other hand, FXR can be activated by either free or conjugated bile acids such as CDCA, LCA and DCA. It is observed that mice lacking Fxr gene have elevated LDL and total triglycerides in blood when they are fed with high-cholesterol diet 40. Moreover, activation of FXR by agonist or bile acids will lead to reduction of HDL in wide-type mice, and decrease of LDL, HDL and triglycerides in hypercholesterolemia mice 41. Our current results indicated that hepatic FXR was stimulated by SV in the context of HFD feeding, and this effect was impaired by antibiotic treatment. It is reported that the activation of FXR by statins is associated with their diversified actions 42, however, little is known about the relationship between the hypolipidemic effect of statins and activation of hepatic FXR so far. Our current results revealed that the hypolipidemic effect of SV might be associated with the stimulation of hepatic FXR and FXR-regulated targeted genes, which could be impaired by gut microbiota modulation. Further studies are warranted to investigate the roles of activated hepatic FXR in mediating the hypolipidemic effect of SV, as well as the gut microbial contribution in affecting FXR activation and the downstream biological significance.
Supplementary Material
Table S1. Summary of the PCA and PLS-DA Modelsa
Acknowledgments
Funding Sources
This work was financially supported by Shanghai Pujiang Program (14PJD031) from the Science and Technology Commission of Shanghai Municipality, Shanghai Creative Research Fund (ZYX-CXYJ-017) of Higher Education, Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning from Shanghai Municipal Education Commission, and National Natural Science Foundation of China ( No. 81673662).
We would like to thank Dr. Xianfu Gao (Shanghai Prof Leader Biotech Co, Ltd) for his assistance with the metabolomic experiment.
Footnotes
Notes: The authors declare no competing financial interest.
Accession number of sequences
The sequence information of 16S rDNA from fecal samples has been uploaded to the Sequence Read Archive database under the accession number SRP093219.
Author Contributions
XY. He, NN. Zheng & JJ. He conducted the animal experiment, sample analysis and data mining. Can Liu helped in animal experiment and sample analysis. Jing Feng helped in sample analysis. Wei Jia and Houkai Li designed the study and wrote the manuscript. All authors have approved the final version of the manuscript.
References
- 1.Grundy SM, Cleeman JI, Merz CN, Brewer HB, Jr, Clark LT, Hunninghake DB, Pasternak RC, Smith SC, Jr, Stone NJ National Heart L, Blood I, American College of Cardiology F, American Heart A. Implications of recent clinical trials for the National Cholesterol Education Program Adult Treatment Panel III guidelines. Circulation. 2004;110:227–239. doi: 10.1161/01.CIR.0000133317.49796.0E. [DOI] [PubMed] [Google Scholar]
- 2.Zhou Z, Rahme E, Pilote L. Are statins created equal? Evidence from randomized trials of pravastatin, simvastatin, and atorvastatin for cardiovascular disease prevention. Am Heart J. 2006;151:273–281. doi: 10.1016/j.ahj.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, Kirby A, Sourjina T, Peto R, Collins R, Simes R Cholesterol Treatment Trialists, C. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, Rotter JI, Nickerson DA, Yang H, Saad M, Krauss RM. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–850. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 5.Trupp M, Zhu H, Wikoff WR, Baillie RA, Zeng ZB, Karp PD, Fiehn O, Krauss RM, Kaddurah-Daouk R. Metabolomics reveals amino acids contribute to variation in response to simvastatin treatment. PLoS One. 2012;7:e38386. doi: 10.1371/journal.pone.0038386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaddurah-Daouk R, Baillie RA, Zhu H, Zeng ZB, Wiest MM, Nguyen UT, Watkins SM, Krauss RM. Lipidomic analysis of variation in response to simvastatin in the Cholesterol and Pharmacogenetics Study. Metabolomics. 2010;6:191–201. doi: 10.1007/s11306-010-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaddurah-Daouk R, Baillie RA, Zhu H, Zeng ZB, Wiest MM, Nguyen UT, Wojnoonski K, Watkins SM, Trupp M, Krauss RM. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One. 2011;6:e25482. doi: 10.1371/journal.pone.0025482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 9.Li H, He J, Jia W. The influence of gut microbiota on drug metabolism and toxicity. Expert Opin Drug Metab Toxicol. 2016;12:31–40. doi: 10.1517/17425255.2016.1121234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DH. Gut Microbiota-Mediated Drug-Antibiotic Interactions. Drug Metab Dispos. 2015;43:1581–1589. doi: 10.1124/dmd.115.063867. [DOI] [PubMed] [Google Scholar]
- 11.Aura AM, Mattila I, Hyotylainen T, Gopalacharyulu P, Bounsaythip C, Oresic M, Oksman-Caldentey KM. Drug metabolome of the simvastatin formed by human intestinal microbiota in vitro. Mol Biosyst. 2011;7:437–446. doi: 10.1039/c0mb00023j. [DOI] [PubMed] [Google Scholar]
- 12.Yoo DH, Kim IS, Van Le TK, Jung IH, Yoo HH, Kim DH. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos. 2014;42:1508–1513. doi: 10.1124/dmd.114.058354. [DOI] [PubMed] [Google Scholar]
- 13.Zheng XJ, Zhao AH, Xie GX, Chi Y, Zhao LJ, Li HK, Wang CR, Bao YQ, Jia WP, Luther M, Su MM, Nicholson JK, Jia W. Melamine-Induced Renal Toxicity Is Mediated by the Gut Microbiota. Science Translational Medicine. 2013:5. doi: 10.1126/scitranslmed.3005114. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Zhao Y, Zhang M, Pang X, Xu J, Kang C, Li M, Zhang C, Zhang Z, Zhang Y, Li X, Ning G, Zhao L. Structural changes of gut microbiota during berberine-mediated prevention of obesity and insulin resistance in high-fat diet-fed rats. PLoS One. 2012;7:e42529. doi: 10.1371/journal.pone.0042529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Pujos-Guillot E, Sebedio JL. Development of a quantitative metabolomic approach to study clinical human fecal water metabolome based on trimethylsilylation derivatization and GC/MS analysis. Anal Chem. 2010;82:6447–6456. doi: 10.1021/ac1006552. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT, Ridker PM, Chatterjee A, Rotter JI, Nickerson DA, Stephens M, Krauss RM. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5:e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangravite LM, Wilke RA, Zhang J, Krauss RM. Pharmacogenomics of statin response. Curr Opin Mol Ther. 2008;10:555–561. [PubMed] [Google Scholar]
- 24.Piorunska-Stolzmann M, Piorunska-Mikolajczak A, Mikolajczyk Z. Effect of simvastatin on trioleylglycerol hydrolysis and transacylation with cholesterol in serum of outpatients with coronary heart disease. Drugs Exp Clin Res. 2003;29:37–43. [PubMed] [Google Scholar]
- 25.Lagace TA, Ridgway ND. The role of phospholipids in the biological activity and structure of the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2499–2510. doi: 10.1016/j.bbamcr.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney G, Nazir D, Clarke C, Goettsche G. Ethanolamine and choline phospholipids in nascent very-low-density lipoprotein particles. Clin Invest Med. 1996;19:243–250. [PubMed] [Google Scholar]
- 27.Carlquist JF, Muhlestein JB, Anderson JL. Lipoprotein-associated phospholipase A2: a new biomarker for cardiovascular risk assessment and potential therapeutic target. Expert Rev Mol Diagn. 2007;7:511–517. doi: 10.1586/14737159.7.5.511. [DOI] [PubMed] [Google Scholar]
- 28.Soupene E, Borja MS, Borda M, Larkin SK, Kuypers FA. Featured Article: Alterations of lecithin cholesterol acyltransferase activity and apolipoprotein A-I functionality in human sickle blood. Exp Biol Med (Maywood) 2016;241:1933–1942. doi: 10.1177/1535370216657447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagita T, Yamamoto K, Ishida S, Sonda K, Morito F, Saku K, Sakai T. Effects of simvastatin, a cholesterol synthesis inhibitor, on phosphatidylcholine synthesis in HepG2 cells. Clin Ther. 1994;16:200–208. [PubMed] [Google Scholar]
- 30.Deo AK, Bandiera SM. Identification of human hepatic cytochrome p450 enzymes involved in the biotransformation of cholic and chenodeoxycholic acid. Drug Metab Dispos. 2008;36:1983–1991. doi: 10.1124/dmd.108.022194. [DOI] [PubMed] [Google Scholar]
- 31.Shitara Y, Sugiyama Y. Pharmacokinetic and pharmacodynamic alterations of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase inhibitors: drug-drug interactions and interindividual differences in transporter and metabolic enzyme functions. Pharmacol Ther. 2006;112:71–105. doi: 10.1016/j.pharmthera.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Holtzman CW, Wiggins BS, Spinler SA. Role of P-glycoprotein in statin drug interactions. Pharmacotherapy. 2006;26:1601–1607. doi: 10.1592/phco.26.11.1601. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Mireles RJ, Campbell SD, Lin J, Mills JB, Xu JJ, Smolarek TA. Differential interaction of 3-hydroxy-3-methylglutaryl-coa reductase inhibitors with ABCB1, ABCC2, and OATP1B1. Drug Metab Dispos. 2005;33:537–546. doi: 10.1124/dmd.104.002477. [DOI] [PubMed] [Google Scholar]
- 34.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25:1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen JC, Cali JJ, Jelinek DF, Mehrabian M, Sparkes RS, Lusis AJ, Russell DW, Hobbs HH. Cloning of the human cholesterol 7 alpha-hydroxylase gene (CYP7) and localization to chromosome 8q11-q12. Genomics. 1992;14:153–161. doi: 10.1016/s0888-7543(05)80298-8. [DOI] [PubMed] [Google Scholar]
- 37.Xu X, Zhang A, Halquist MS, Yuan X, Henderson SC, Dewey WL, Li PL, Li N, Zhang F. Simvastatin promotes NPC1-mediated free cholesterol efflux from lysosomes through CYP7A1/LXRalpha signalling pathway in oxLDL-loaded macrophages. J Cell Mol Med. 2016 doi: 10.1111/jcmm.12970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren S, Marques D, Redford K, Hylemon PB, Gil G, Vlahcevic ZR, Pandak WM. Regulation of oxysterol 7alpha-hydroxylase (CYP7B1) in the rat. Metabolism. 2003;52:636–642. doi: 10.1053/meta.2003.50106. [DOI] [PubMed] [Google Scholar]
- 39.Claudel T, Staels B, Kuipers F. The Farnesoid X receptor: a molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol. 2005;25:2020–2030. doi: 10.1161/01.ATV.0000178994.21828.a7. [DOI] [PubMed] [Google Scholar]
- 40.Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howe K, Sanat F, Thumser AE, Coleman T, Plant N. The statin class of HMG-CoA reductase inhibitors demonstrate differential activation of the nuclear receptors PXR, CAR and FXR, as well as their downstream target genes. Xenobiotica. 2011;41:519–529. doi: 10.3109/00498254.2011.569773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of the PCA and PLS-DA Modelsa