Abstract
Autosomal recessive polycystic kidney disease (ARPKD), usually considered to be a genetically homogeneous disease caused by mutations in PKHD1, has been associated with ciliary dysfunction. Here, we describe mutations in the DAZ interacting protein 1-like (DZIP1L) gene in patients with ARPKD, findings we have further validated by loss-of-function studies in mice and zebrafish. DZIP1L localizes to centrioles and at the distal end of basal bodies, and interacts with septin2, a protein implicated in maintenance of the periciliary diffusion barrier at the ciliary transition zone. Consistent with a defect in the diffusion barrier, we found that the ciliary membrane translocation of the PKD proteins, polycystin-1 and −2, is compromised in DZIP1L mutant cells. Together, these data provide the first conclusive evidence that ARPKD is not a homogeneous disorder, and establishes DZIP1L as a second gene involved in its pathogenesis.
Keywords: Autosomal recessive polycystic kidney disease, DZIP1L, cilia, transition zone, septin2, polycystin
INTRODUCTION
Polycystic kidney disease (PKD) is the most common form of heritable kidney disease. In contrast to the later onset of autosomal dominant PKD (ADPKD), the autosomal recessive form (ARPKD) manifests perinatally or in childhood with enlarged kidneys1, and arterial hypertension in most surviving children2. Hepatic ductal plate malformation (DPM) can be detected histologically, but often only manifests clinically with increasing age.
In addition to ADPKD and ARPKD, cystic kidney disease is a feature of some syndromic ciliopathies, which arise from dysfunctional primary cilia3. Cilia are microtubule-based organelles that project from the surface of most vertebrate cells, and function in chemo- and mechano-sensation and fluid transport. The ciliary axoneme is anchored by the centriole-derived basal body, and is ensheathed by the ciliary membrane. Located between the basal body and axoneme, the transition zone forms a diffusion barrier that modulates the selective passage of proteins to and from the cilium4–6.
PKHD1, encoding fibrocystin (FPC)/polyductin, is widely believed to be the only ARPKD gene7. Like the two ADPKD proteins, polycystin-1 and −2 (PC1 and 2), FPC has been shown to localize to cilia8,9 and associate with the PC1/2 complex, which has long been thought to regulate calcium (Ca2+) signalling in response to urine flow10–12. However, the role of PC1/2 complex in Ca2+ signaling and the theory that primary cilia are Ca2+-responsive mechanosensors has recently been refuted13–15. Thus, the precise mechanisms underpinning PKD remain elusive.
Here, we report the identification of a second ARPKD locus, with homozygous mutations in the cilia-related gene DZIP1L in seven patients from four families. We further validated these findings in mouse and zebrafish models. We show that DZIP1L localizes to centrioles and at the distal end of the basal body, and interacts with septin2 (SEPT2), a protein involved in maintenance of the diffusion barrier at the transition zone. Consistent with these data, we found that in DZIP1L deficient primary cilia, PC1 and PC2 show altered distribution along the ciliary membrane.
RESULTS
DZIP1L is a new ARPKD gene
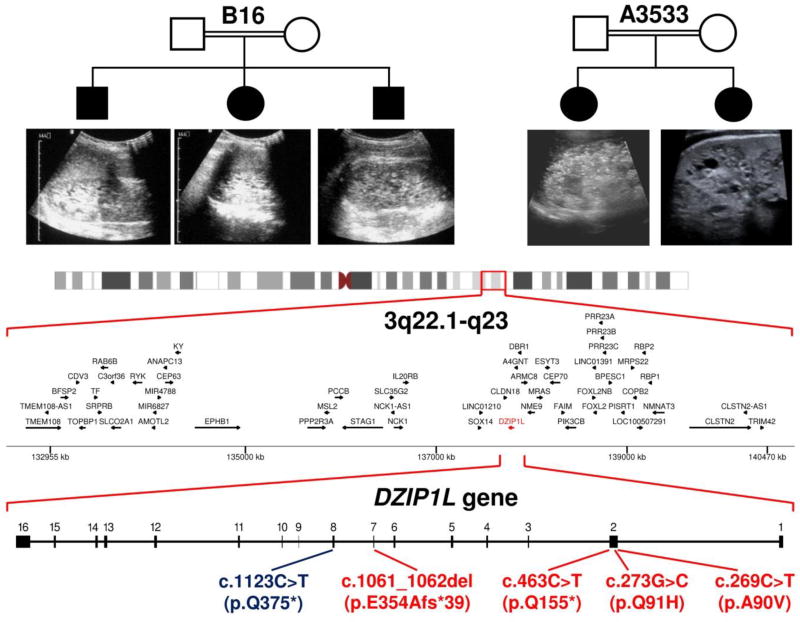
In two unrelated consanguineous multiplex pedigrees (B16 and A3533) with a total of five children affected by ARPKD, mutations in known PKD genes were excluded by linkage analysis and sequencing (Fig. 1; data not shown). Genome-wide SNP analysis in DNA from parents and all affected children revealed a single overlapping homozygous peak, suggesting a new disease locus on chromosome 3q22, in a 7.5Mb region of genomic DNA (Fig. 1). By whole-exome sequencing (WES) and subsequent Sanger sequencing, we identified different homozygous missense DZIP1L mutations that segregated with disease in each family (Family B16: c.269C>T (p.Ala90Val); Family A3533: c.273G>C, (p.Gln91His); Accession number: NM_173543.2) (Fig. 1, Supplementary Fig. 1 and Supplementary Table 1). Interestingly, these mutations affect adjacent evolutionarily conserved residues of DZIP1L, and are bioinformatically predicted to be pathogenic (Table 1). No further convincing variants were identified in the candidate region in these pedigrees. All three affected individuals in family B16 showed enlarged polycystic kidneys with reduced cortico-medullary differentiation, and developed arterial hypertension early during childhood. A comparable clinical course was observed in the two affected girls in family A3533 (Table 1).
Figure 1.
Mapping of a new ARPKD locus on 3q22.1-q23 and identification of DZIP1L mutations. Genome-wide SNP analysis performed in two unrelated consanguineous multiplex pedigrees (shown at top) with a total of five children affected by ARPKD, resulted in identification of a single overlapping 7.5 Mb region of homozygosity on chromosome 3q22.1-q23. By various sequencing approaches, we identified different homozygous DZIP1L mutations in these and other consanguineous families with ARPKD (see text for details). In parallel, we identified an ENU-induced recessive mouse model (see Fig. 2) bearing the homozygous Dzip1l nonsense mutation c.1123C>T (p.Gln375*) (in blue).
Table 1.
Mutations of DZIP1L in patients with ARPKD.
| Family -Individual |
Gender | Ethnic origin |
Mutation | Protein Change |
Exon/ Intron (state) |
Continuously conserved to |
Polyphen-2/ MutTast/SIFT |
ExAC | Parental consang. |
Phenotype |
|---|---|---|---|---|---|---|---|---|---|---|
| B16-1 | male | Turkey | c.269C>T | p.Ala90Val | 2 (hom) | Xenopus tropicalis | 0.99/DC/dam | 0/5/116158 | yes | ARPKD, enlarged polycystic kidneys with increased echogenicity (kidney volumes > 97th percentile), diminished CMD, pepper-salt pattern. Arterial hypertension since early toddler age. ESRD at age 18, RTX at age 26. |
| B16-2 | female | Turkey | c.269C>T | p.Ala90Val | 2 (hom) | Xenopus tropicalis | 0.99/DC/dam | 0/5/116158 | yes | ARPKD, kidney morphology like elder brother with polycystic kidneys of increased echogenicity and volumes constantly > 97. percentile (at age 11: left kidney 175 ml, right 190 ml; at age 20: left 225 ml, right 217 ml). Arterial hypertension since early toddler age. ESRD 26 y, RTX 27 y. |
| B16-3 | male | Turkey | c.269C>T | p.Ala90Val | 2 (hom) | Xenopus tropicalis | 0.99/DC/dam | 0/5/116158 | yes | Characteristic ARPKD phenotype with hyperechogenic kidneys with lack of CMD. Kidney volumes at age 3: left 75th p., right 97th p.; age 7: left 97 ml (90th p.), right 112 ml (97th p.); age 14: left 167 ml, right 240 ml (both > 97th p.) Hepatosplenomegaly. Arterial hypertension since early toddler age. Normal kidney function at age 15 y (serum creatinine 0.9 mg/dl, creatinine clearance 112 ml/min/1.73 m2). |
| A3533-1 | female | Arab | c.273G>C | p.Gln91His | 2 (hom) | Danio rerio | 1.00/DC/dam | 0/0/116158 | yes | ARPKD, enlarged polycystic kidneys with increased echogenicity. Arterial hypertension first diagnosed at age 8. ESRD and RTX 12 y. |
| A3533-2 | female | Arab | c.273G>C | p.Gln91His | 2 (hom) | Danio rerio | 1.00/DC/dam | 0/0/116158 | yes | ARPKD, enlarged polycystic kidneys with increased echogenicity. Arterial hypertension first diagnosed at age 3. Kidney function at age 9 y: Serum creatinine 0.8 mg/dl, creatinine clearance 94 ml/min/1.73 m2, slightly increased cystatin C 1.4 mg/l, cystatin-GFR 67 ml/min/1.73 m2. |
| B155 | male | Palestine | c.463C>T | p.Gln155* | 2 (hom) | Danio rerio | NA/DC/NA | 0/0/116158 | yes | First diagnosed with arterial hypertension and massively, symmetrically enlarged kidneys with hyperechogenicity with poor CMD at 8 months of age. ESRD 18 y, RTX 20 y. |
| B8031 | female | Egypt | c.1061_1062del | p.Glu354Alafs*39 | 7 (hom) | Danio rerio | NA/NA/NA | 0/0/116158 | yes | Characteristic ARPKD phenotype with bilaterally enlarged kidneys of increased echogenicity, twinkling phenomenon/calcification spots, diminished CMD, normal renal function at 13 y. Kidney volumes 90th-97th p. (at age 8: 146 ml; age 10: 163 ml; age 13: 195 ml). |
DZIP1L cDNA mutations are numbered according to human cDNA reference sequence NM_173543.2. +1 corresponds to the A of ATG start translation codon in exon 2.
Abbreviations: ARPKD, autosomal recessive polycystic kidneys; CMD, cortico-medullary differentiation; het, heterozygous; hom, homozygous; DC, ExAC, Exome Aggregation Consortium; MutTast, Mutation Taster; disease causing (MutTast); SIFT, sorting intolerant from tolerant; dam, damaging (SIFT); ESRD, end-stage renal disease; NA, not applicable; RTX, renal transplantation; TOP, termination of pregnancy; y, years.
Next, we analysed DZIP1L by Sanger sequencing in 218 unrelated patients with suspected ARPKD or sporadic PKD, the majority of whom had tested negative for mutations in known cystoprotein genes. A further 1330 patients with a PKD phenotype (n=525) or a nephronopthisis (NPHP)-related complex ciliopathy (n=805) were analyzed by next-generation sequencing (NGS), mainly targeted multi-gene panel testing16. Thereby, we identified different homozygous protein truncating DZIP1L mutations (c.463C>T; (p.Gln155*) and c.1061_1062del; (p.Glu354Alafs*39)) in two additional unrelated consanguineous pedigrees, B155 and B8031, with hallmarks of ARPKD (Fig. 1, Table 1, Supplementary Fig. 1).
DZIP1L encodes a 767 amino acid (aa) protein containing a C2H2-type zinc finger motif (aa 166–189) and a series of coiled-coil domains (aa 205–406) (Supplementary Fig. 2), and previous overexpression studies showed localization to the ciliary base17. In zebrafish and mice, we and others have implicated the paralogue Dzip1 in hedgehog (HH) signalling and ciliogenesis18–22, but there is little known about Dzip1l function in any organism.
A Dzip1l loss-of-function mouse model
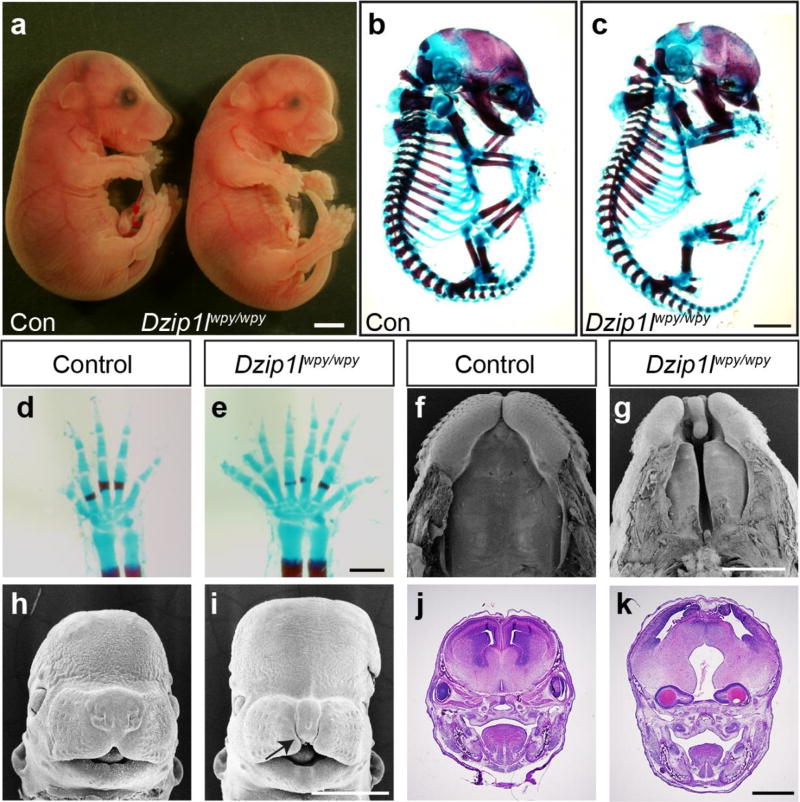
In a recessive N-ethyl-N-nitrosourea (ENU) mutagenesis screen23, we identified a mouse mutant (warpy or wpy) with widespread embryonic dysmorphologies. Linkage analysis mapped the mutation to a 64 Mb region on chromosome 9, and subsequent WES identified 8 homozygous mutations only one of which, a nonsense mutation in the region encoding the coiled-coil domains of Dzip1l (c.1123C>T; p.Gln375*; Accession number: NM_028258.4), was within the linkage interval (Supplementary Fig. 3a). On a C57BL/6:C3H mixed or a pure C57BL/6 background, homozygous Dzip1lwpy/wpy mutants displayed a range of phenotypes, including highly penetrant polydactyly of all four limbs and craniofacial defects (Fig. 2), including full or partial cleft palate and bilateral cleft lip (Fig. 2f–i). Gross eye abnormalities were evident in 67% (n=30/45) of embryos scored from 13.5 days post coitum (dpc) on, in some cases manifesting as internalised eyes (Fig. 2j,k). Homozygous C57BL/6:C3H mutant embryos were recovered at roughly the expected Mendelian frequency at mid-gestation (23.3% at 10.5dpc, n=66/283), but although some embryos were recovered up to 18.5dpc, viability was decreased at later stages.
Figure 2.
Dzip1lwpy/wpy mutant embryos show multiple dysmorphic features. (a) Control (left) and mutant (right) embryos at 17.5dpc. (b–e) Skeletal preps at 17.5dpc highlight defects in the craniofacial complex and polydactyly in Dzip1lwpy/wpy embryos (c,e) relative to controls (b,d). Dzip1lwpy/wpy mutant embryos consistently show some degree of cleft palate (f,g; arrowhead in g) and bilateral cleft lip (f-i; arrows in g,i). Scanning electron micrographs of 15.5dpc embryos shown with lower jaw removed in f,g; 13.5dpc embryo heads in h,i. (j,k) In a subset of embryos (n=30/45 scored) the eyes appeared abnormal, and in severe cases were buried within the skull (arrow in k). Images shown are of frontal sections through 15.5dpc embryonic heads. Note control embryos shown in (b,f,j) are heterozygous embryos that show no obvious phenotypic differences from wild-type controls, as shown in (a,d,h). The limbs shown in (d,e) are from different embryos than those shown in (b,c). Scale bars in a,b,c = 2mm; d,e = 0.5mm; f-k = 1mm. All data presented are from embryos on a C57BL/6:C3H mixed background.
These phenotypes, in particular polydactyly, suggested a defect in HH signalling, a pathway regulated by the primary cilium24. Accordingly, we saw expansion of Hoxd13 and gremlin (Grem1) expression into the anterior fore- and hindlimb bud of 11.5dpc Dzip1lwpy/wpy embryos (n=3; Supplementary Fig. 3b; hindlimb data not shown), suggesting a decrease in the repressor form of the GLI3 transcription factor that mediates HH signalling in the limb25. Consistent with cilia and GLI3 regulating HH signalling downstream of ligand, the expression of sonic hedgehog (Shh) appeared unaltered in mutant limbs. Interestingly, while patched1 (Ptch1) was not consistently altered, another universal downstream marker of HH signalling, Gli1, showed subtle but consistent ectopic anterior expression (Supplementary Fig.3b). We next assessed the ability of Dzip1lwpy/wpy mouse embryonic fibroblasts (MEFs) to respond to the HH agonist SAG, and showed an attenuated response relative to wild-type MEFs, as measured by qRT-PCR for Gli1 (Supplementary Fig.3d). In contrast to the limb and MEFs, we saw no evidence of altered HH signalling across the dorsal-ventral axis of the neural tube in Dzip1lwpy/wpy embryos, as assessed by staining for markers of SHH-dependent neuronal populations, regardless of the anterior-posterior level of the sections (n=2 for Shh, 3 for other markers; Supplementary Fig. 3c). Our data suggest tissue-specific effects on HH signalling in Dzip1lwpy/wpy embryos, with gene expression changes in the limb consistent with polydactyly.
Dzip1lwpy/wpy mutants develop cystic kidney disease
On a mixed C57BL/6:C3H background, Dzip1lwpy/wpy embryos display cystic kidneys from 15.5dpc (data not shown). To analyse the postnatal progression of this phenotype, we crossed the mice to an outbred CD1 background, and after 4 generations, were able to recover live mice. These mice still displayed limb defects, albeit less severe than the inbred embryos, but gross facial clefting phenotypes were rare. This suggests that some of the dysmorphologies seen in the Dzip1lwpy/wpy embryos, but not in ARPKD patients, may be due to genetic background effects. Homozygous mutants were born at the expected Mendelian frequency (25.2%, n=30/119), but showed a generalised failure to thrive, and all were sacrificed by postnatal day (P)21.
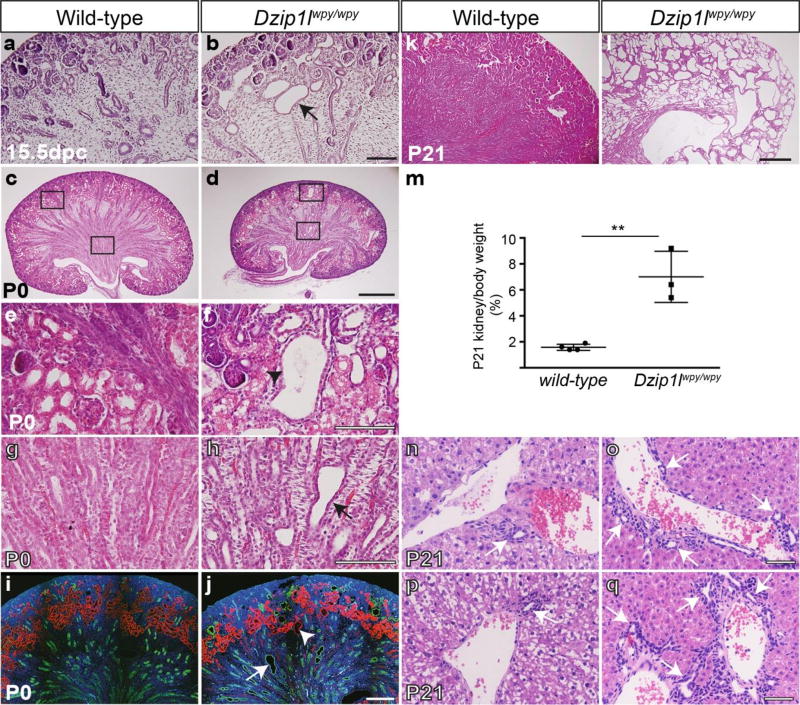
Histological analysis of kidney sections following at least four crosses to CD1 revealed a highly penetrant progressive cystic kidney phenotype (Fig. 3). Late in embryogenesis, the most obvious cysts were located toward the renal cortex (Fig. 3a,b; n=4 for 15.5dpc). At P0 (n=6) we observed marked dilatations throughout the medullary region that stained positive for the collecting duct marker Aquaporin2 (Aqp2) (Fig. 3c–f,h,j). Some more cortical cysts stained positive for a proximal tubule marker (LTL-biotin) (Fig. 3e,f,i). The phenotype increased in severity with age, and P21 kidneys were massively cystic and larger than wild-type when corrected for body weight (Fig. 3k–m; n=3). Consistent with ARPKD in humans, we also observed subtle signs of DPM (excess of bile ducts) in the livers of Dzip1lwpy/wpy mice at P21 (n=2) (Fig. 3n–q and Supplementary Fig. 4). Hepatic fibrosis was not detected, likely due to the young age of these mice.
Figure 3.
Dzip1lwpy/wpy mice display progressive cystic kidney disease and hepatic defects following at least 4 crosses to a CD1 background. (a,b) Following H&E staining, cortical cysts were evident from 15.5dpc in Dzip1lwpy/wpy kidneys (arrow in b; n=4 independent embryos). (c,d,g,h) At P0, both cortical cysts (arrowhead in d,g) and medullary dilatations (arrow in d,h) were observed (n=6). (g,h) Show high power images of the regions marked in (d). (e,f,i,j) Immunofluorescence analysis of sections from P0 kidneys stained with Aqp2 (collecting ducts; green), and LTL-biotin (proximal tubules; magenta). (i,j) Show high power images of the proximal tubule cyst (i) and collecting duct dilatation (j) marked with an arrowhead and arrow respectively in (f). (k,l) By P21, cysts extend throughout the kidney, and (m) when corrected for body weight Dzip1lwpy/wpy kidneys are heavier than wild-type (n= 3 mutant, 4 wild-type embryos). Statistical analysis based on unpaired t-test with Welch’s correction to account for unequal variances, *p=0.05. Error bars show SEM. A significant difference was confirmed with a Student’s t-test on log transformed data, where the variances are equalized (not shown). (n–q) H&E staining of P21 liver sections. Arrows mark an excess of bile ducts, which nearly circumferentially surround portal vein branches in P21 Dzip1lwpy/wpy mutant (o,q) compared to wild-type (n,p) mice, indicating DPM (n=2). Scale bars in a,b = 100µm; c-f = 200µm, k,l = 500µm and g-j and n-q = 50µm.
Inactivation of zebrafish dzip1l causes phenotypes consistent with ciliary dysfunction
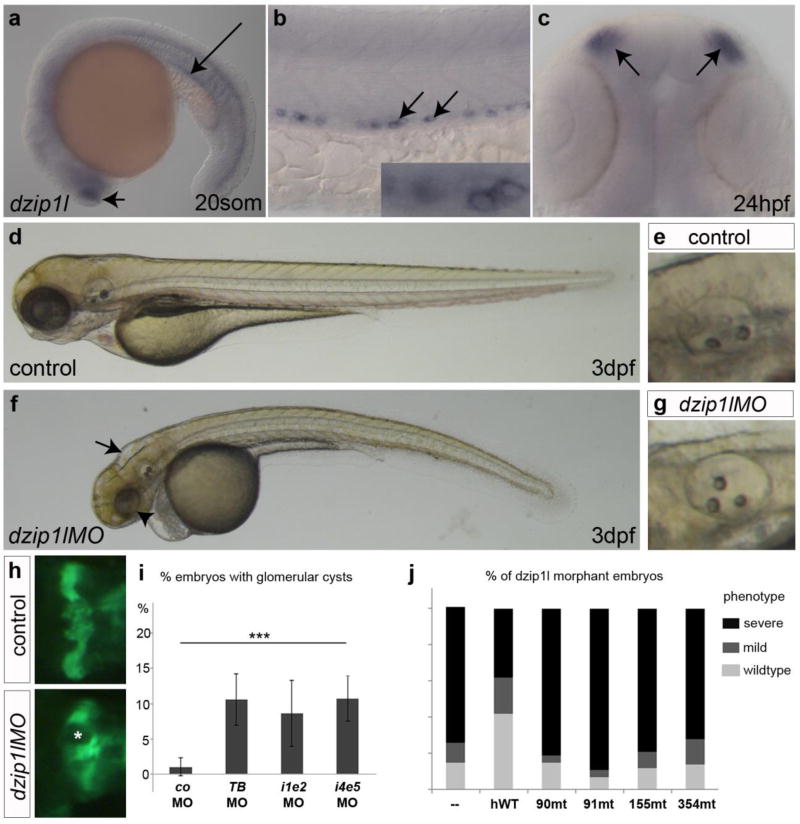
The single orthologue of DZIP1L in the zebrafish genome (Accession number: NM_0010372277.1; Supplementary Fig. 2) is maternally expressed (Supplementary Fig. 5a), and 1 day post-fertilization (dpf) embryos express fairly ubiquitous low levels of dzip1l, with a more discrete accumulation in the embryonic kidneys - the pronephric ducts (Supplementary Fig. 6a,b). Unlike the solitary primary cilium in mammalian kidney cells, zebrafish pronephric duct cells harbour motile cilia, with the mid-section of each duct containing cells bearing mono-motile cilia interspersed with multiciliated cells (MCCs). dzip1l expression was observed in the mid-section of the pronephric ducts in a pattern consistent with the MCCs (Supplementary Fig. 6b), and in the nasal placodes, another tissue populated by MCCs (Supplementary Fig. 6c).
We next used four different antisense morpholino oligonucleotides (one translation blocking and three splice inhibitory) to knockdown dzip1l function, with all four producing largely identical cilia-related phenotypes. From 2 dpf on, dzip1l morphants presented a slight body axis curvature, hydrocephalus and defects in otolith formation in the inner ear (Supplementary Fig. 6d–g). Otolith defects were observed in 12–15% of translation blocking (n=258) and splice inhibitory morpholino (n=584) injected embryos, but not in controls (n=195) (Supplementary Fig. 6e,g,i). Despite the ciliary dysfunction phenotypes, no obvious alterations in Hh signalling were detected in 1–2 dpf dzip1l morphant embryos by in situ hybridization for ptc1, eng1 and nkx2.2b expression (data not shown).
To investigate defects in glomerular and pronephric tubules, we injected the dzip1l translation blocking morpholino into Tg(wt1b::GFP) transgenic embryos that express GFP in the pronephros26. Approximately 10% of morphant embryos displayed glomerular cysts (n=100) (Supplementary Fig. 6h,i). Specificity of the dzip1l morphant phenotypes was demonstrated by co-injection of embryos with the translation blocking morpholino and wild-type dzip1l mRNA. 85% of the morphants showed severe morphological defects, compared to 36% of the mRNA co-injected embryos (Supplementary Fig. 6j). mRNA rescue was confirmed with a second (splice blocking) morpholino (data not shown). RT-PCR analysis confirmed mis-splicing (Supplementary Fig. 5b–d).
We next generated a mutant dzip1l allele using CRISPR/Cas9 technology, targeting exons 2 and 13 with two separate guide RNAs to induce a large deletion (Supplementary Fig. 5e,f). Embryos homozygous for this deletion showed highly penetrant defects in otolith deposition, but no additional morphological abnormalities (Supplementary Fig. 5g). Since dzip1l is expressed maternally, we raised zygotic mutants to adulthood, and bred them to obtain maternal-zygotic mutant embryos. Like the zygotic mutants, maternal-zygotic dzip1l deficient embryos exhibited defective otolith deposition, but no other overt morphological changes (data not shown).
DZIP1L localizes to centrioles and to the distal end of basal bodies
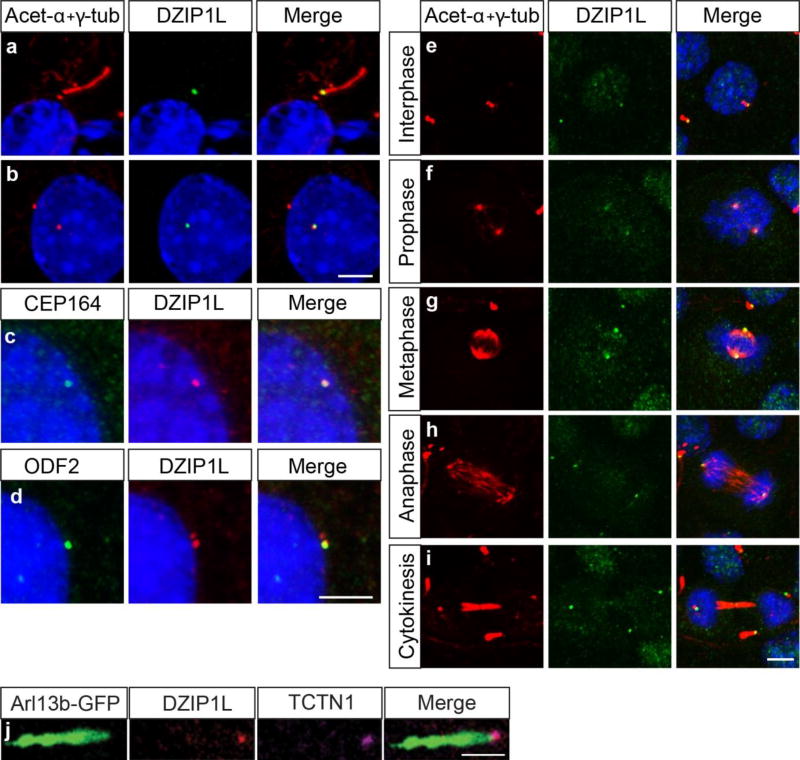
To investigate the subcellular localization of endogenous DZIP1L we used two commercial antibodies, one raised to the C-terminus of DZIP1L (Sigma-Aldrich), and one to the entire protein (Abnova). Both antibodies stained centrioles and basal bodies in mouse kidney inner medullary collecting duct (IMCD3) cells, MEFs and human dermal fibroblasts (Fig. 4a–i and Supplementary Fig. 7a,c,f and data not shown). Furthermore, DZIP1L remained localized to the centrioles throughout the cell cycle, even when cilia are disassembled during mitosis (Fig. 4e–i). The mother centriole, from which the basal body is derived, acquires sub-distal and distal appendages, with distal appendages or transition fibres anchoring the basal body to the ciliary membrane at the transition zone27. DZIP1L staining overlaps with both sub-distal (ODF2) and distal (CEP164) appendage markers in IMCD3 cells (Fig. 4c,d). This suggests that DZIP1L localizes to the distal end of the mother centriole, possibly at the site where the transition zone forms. Consistent with this, DZIP1L co-localizes with the transition zone protein tectonic1 (TCTN1) in ciliated human fibroblasts (Fig. 4j). This was confirmed in retinal pigment epithelium (RPE-1) cells by superresolution microscopy (3D-SIM) that improved resolution (by a factor of 2 in all dimensions; to give ∼120 nm resolution laterally, xy), and revealed that DZIP1L and TCTN1 indeed localize in close proximity of each other (Fig. 4k).
Figure 4.
DZIP1L localization overlaps with basal body, centrosome and transition zone markers. (a,b) IF staining for DZIP1L (green) and acetylated-α-tubulin and γ-tubulin (both in magenta) in serum starved IMCD3 kidney cells. DZIP1L staining overlaps with the basal body in ciliated cells (a) and the centrosome in non-ciliated cells (b). (c–d) Co-localization of DZIP1L (magenta) with the distal (CEP164, green in c) and subdistal (ODF2, green in d) appendage proteins. (e–i) DZIP1L (green) tracks with the centrioles (magenta) throughout all stages of the cell cycle in IMCD3 cells. (j) TCTN1 (magenta) and DZIP1L (cyan) co-localize at the transition zone in human dermal fibroblasts. ARL13B-GFP was used to mark the ciliary membrane around the axoneme (green in j). (k) 3D-SIM superresolution microscopy on RPE-1 (human retinal pigment epithelial) cells confirms both DZIP1L (magenta) and TCTN1 (cyan) are closely associated at the transition zone. Cilium labeled with ARL13B-GFP (green). Nuclei are stained with DAPI (blue). Scale bars in a-I = 5µm; j = 1µm; k = 500 nm. DZIP1L stained with Sigma C-terminal antibody, except in panels c,d,j,k where the Abnova antibody was used.
As expected, the C-terminal DZIP1L antibody (Sigma) did not detect specific staining in mutant Dzip1lwpy/wpy MEFs or human fibroblasts from patient B155 (p.Gln155*), both of which harbour protein-truncating mutations (Supplementary Fig. 7a,b,f,g). However, some residual staining was detected with the Abnova antibody raised to the entire DZIP1L protein, in both mutant cell types (Supplementary Fig. 7c,d and data not shown). Consistent with this, qRT-PCR showed reduced but not absent Dzip1l expression in Dzip1lwpy/wpy MEFs, with no compensatory change in Dzip1 (Supplementary Fig. 7e). Immunoblotting with the Abnova antibody revealed a band at the predicted size of DZIP1L in control but not B155 human cells, confirming a lack of full-length protein in mutant cells (Supplementary Fig. 7h). While we cannot rule out the possibility that the truncated mouse and human DZIP1L proteins retain some function, both would lack significant functional domains.
Cilia number is largely unaffected in Dzip1l mutant tissue
In early passage cultures, serum-starved B155 mutant human fibroblasts displayed no obvious difference to controls in the percentage of ciliated cells, cilia morphology or localization of a number of markers of cilia and basal bodies (ARL13B, acetylated-α-tubulin, CEP164, DZIP1, and TCTN1; data not shown). However, in MEFs we consistently observed a decrease in the percentage of ciliated cells in serum-starved Dzip1lwpy/wpy versus control cultures (Supplementary Fig. 8a). Interestingly, in a number of embryonic mouse tissues, including the limb bud mesenchyme, we saw no difference in the relative proportion of cilia between mutant and wild-type embryos (n=3; Supplementary Fig. 8b). The only location we saw evidence of fewer cilia was in the collecting duct dilatations in the Dzip1lwpy/wpy kidney (data not shown), but this is likely to be an effect of the dilatations rather than a cause.
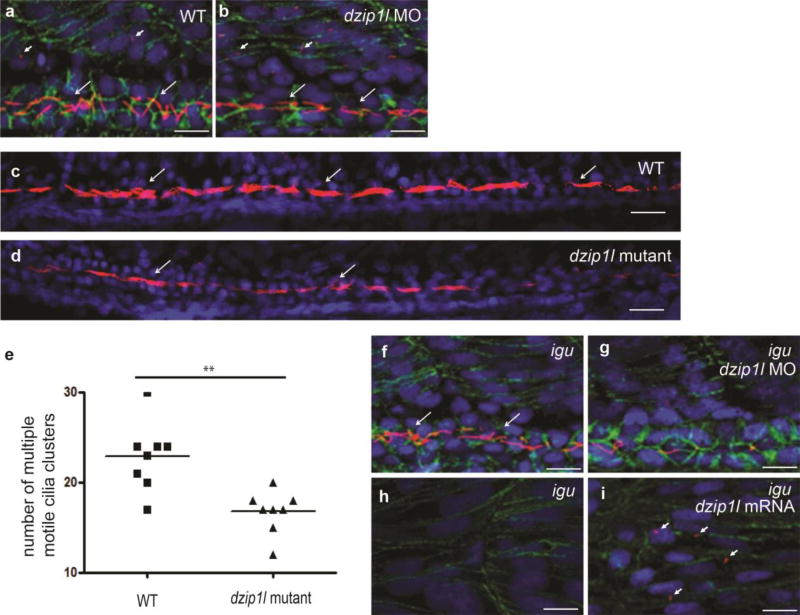
We also saw no obvious primary cilia defects in dzip1l deficient zebrafish embryos (Fig. 5a,b; data not shown). However, consistent with prominent expression of dzip1l in MCCs in kidney tubules, numbers of cilia bundles were consistently reduced in morphant and zygotic and maternal-zygotic dzip1l mutant embryos (Fig. 5c–e; data not shown).
Figure 5.
Dzip1l is required for ciliary differentiation in the zebrafish kidney tubule. (a) Primary cilia (short arrows; myotome) and mono motile cilia (long arrows; pronephric duct) in a 24 hpf wild-type embryo. (b) No obvious difference in primary (short arrows) and mono motile cilia (long arrows) differentiation in a dzip1l splice morpholino (e5i5) injected embryo at 24 hpf (n=10). (c) MCC motile cilia bundles (arrows) in the pronephric duct of a wild-type embryo at 48 hpf (arrows). (d) MCC motile cilia bundles (arrows) are reduced in a 48 hpf maternal-zygotic dzip1l mutant zebrafish embryo (n=8). (e) Quantification of MCC cilia bundles in wild-type and maternal-zygotic dzip1l mutant zebrafish embryos. 8 embryos from each group were analyzed. **p=0.0017 based on unpaired Student’s t-test. Error bars show SEM. In all panels, cilia were stained with anti-acetylated tubulin antibody (magenta) and nuclei with DAPI (blue); in panels a,b,f-i cell membranes were stained with β-catenin antibodies (green). Scale bars in a,b,f-i = 10µm; c,d = 100µm.
DZIP1L associates with the ciliary transition zone protein SEPT2
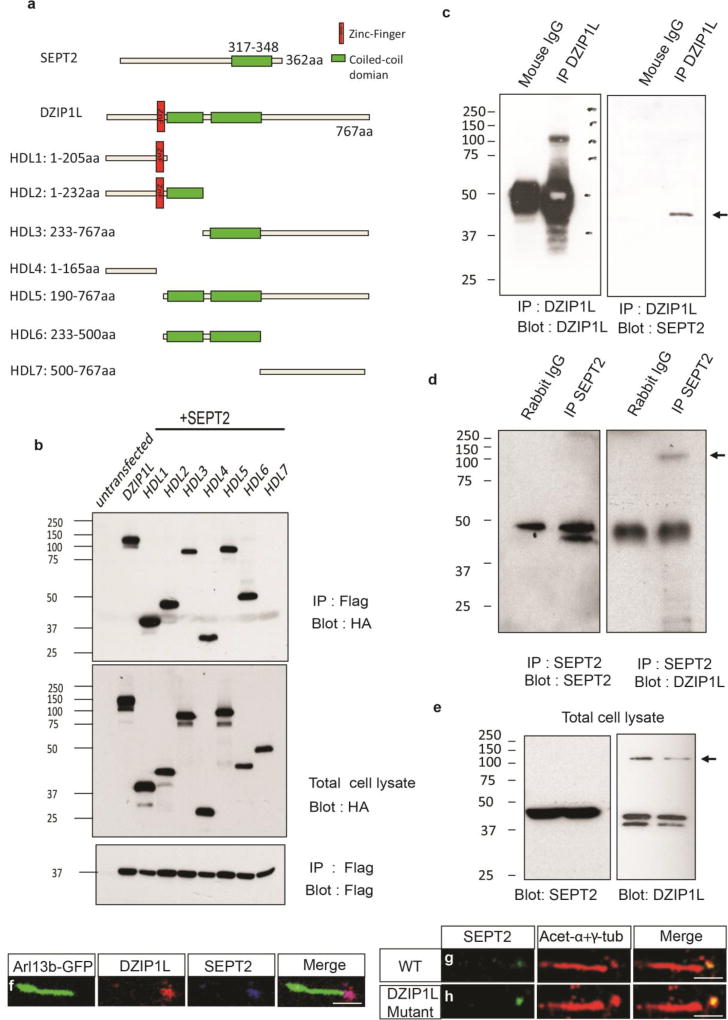
To further explore a function for DZIP1L at the cilium, we used human DZIP1L as bait to screen for interacting proteins in yeast two-hybrid libraries of highly ciliated human tissues (testis, lung and brain). From all three libraries, SEPT2 was identified as a high confidence interactor of DZIP1L (Supplementary Table 2). Septins are cytoskeletal GTPases, and SEPT2 functions as part of the periciliary diffusion barrier at the transition zone28,29. The interaction was verified by co-immunoprecipitation of epitope-tagged DZIP1L and SEPT2 (Fig. 6a) from transfected HEK293T cells (Fig. 6b), and by co-IP of the endogenous proteins from RPE-1 cells (Fig. 6c–e). Moreover, immunostaining of human dermal fibroblasts showed co-localization of DZIP1L and SEPT2 at the ciliary base (Fig. 6f). In support of the specificity of the interaction, we were unable to detect interaction in co-IP experiments between transfected SEPT2 and several other transition zone (NPHS1, MKS1, B9D1) and basal body appendage (CEP89) proteins. Likewise, we failed to detect any interaction between these proteins and DZIP1L (Supplementary Fig. 9).
Figure 6.
DZIP1L associates with the ciliary transition zone protein SEPT2. (a) Schematic diagram of SEPT2, DZIP1L and a series of DZIP1L deletion mutants. (b) Interaction between SEPT2 (N-terminal FLAG) and DZIP1L and various DZIP1L deletion mutants (C-terminal HA) as determined by IP following transfection in HEK293T cells. (c,d) Endogenous interaction between DZIP1L and SEPT2 in RPE-1 cells. IP was performed with either the anti-DZIP1L antibody (Abnova; c) or the anti-SEPT2 antibody (d). (e) Expression of DZIP1L and SEPT2 in total cell lysate. (f) Co-localization of DZIP1L (cyan) and SEPT2 (magenta) in human dermal fibroblasts. ARL13B-GFP was used to mark the ciliary axoneme (green). (g,h) SEPT2 (green) localization to the transition zone was unaffected in DZIP1L mutant dermal fibroblasts from individual B155 (p.Gln155*). Ciliary axonemes and basal bodies were labelled with anti-acetylated tubulin and anti-γ-tubulin antibodies (magenta), respectively. Scale bars in f-h = 1µm. All co-IP experiments were repeated at least three times.
To localize the domains in DZIP1L that interact with SEPT2, we generated a series of epitope-tagged N- and C-terminal deletion DZIP1L constructs, and tested their ability to associate with SEPT2 in co-IP assays (Fig. 6a). We found that all, except a C-terminal fragment (from aa 500 to the end of the protein), were able to interact with SEPT2 (Fig. 6b). Intriguingly, even a small fragment consisting of the N-terminal 165 amino acids and lacking the zinc-finger and the coiled coil domains (effectively mimicking the predicted truncation of DZIP1L in patient B155), retained the ability to interact with SEPT2. This may explain why we did not observe any alteration in SEPT2 localization at the transition zone in the B155 DZIP1L mutant human fibroblasts (Fig. 6g,h).
Loss of DZIP1L affects the localization of PKD proteins to the ciliary membrane
The cystic kidney phenotype caused by loss of DZIP1L led us to investigate the ciliary localization of the two ADPKD proteins PC1 and PC2 in DZIP1L mutant cells. Using an N-terminal PC1 monoclonal antibody (7e12)30 (see Supplementary Fig. 10 for validation), we found that in Dzip1lwpy/wpy ciliated MEFs, PC1 was more frequently enriched at the proximal end of the axoneme compared to wild-type MEFs, which frequently showed PC1 staining along the length of the axoneme (Fig. 7a–c). In control human fibroblasts we detected PC1 at the basal body and along the axoneme, whereas there was a reduction in PC1 accumulation along the ciliary membrane of fibroblasts from individual B155 (Fig. 7d–f). PC2 ciliary membrane localization was similarly affected in B155 fibroblasts (polyclonal antibody to aa 349–361; see online methods for details and Supplementary Fig. 11 for validation) (Fig. 7g–i). This suggests that in the absence of correct DZIP1L function, the ciliary membrane distribution of PC1 and PC2 is compromised, possibly reflecting a defect in the barrier function of the transition zone. However, this is unlikely to be a generalized defect since the HH co-receptor smoothened (SMO) localizes normally along the axoneme in response to SAG treatment in Dzip1lwpy/wpy MEFs (Supplementary Fig. 11). While we also analysed the localization of FPC in human fibroblasts, in our hands two independent antibodies revealed prominent staining around the basal body as described previously8,9, but not along the axoneme. This pattern remained unaffected in cells from individual B155 (data not shown).
Figure 7.
Loss of DZIP1L affects the localization of PC1 and PC2 to the ciliary membrane. (a) In most wild-type ciliated MEFs PC1 (green; 7e12 mAb, Abcam) staining was distributed along the length of the axoneme as marked with ARL13B (magenta). (b) By contrast PC1 staining was more often concentrated at the proximal end of the axoneme in Dzip1lwpy/wpy MEFs (c) Quantification of the percentage of ciliated cells with PC1 along the axoneme (n=6, derived from 3 MEF cell lines from independent embryos, each counted in two separate experiments; between 50–185 cells counted per replicate, total 737 Dzip1l+/+ and 649 Dzip1lwpy/wpy cells counted). (d,e) Decreased PC1 (green) localization in the ciliary membrane in human dermal fibroblasts from affected individual B155 (p.Gln155*). ARL13B (magenta) marks the ciliary axoneme. (g,h) Decreased PC2 localization (green) in the ciliary membrane in DZIP1L mutant human dermal fibroblasts. Acetylated-α–tubulin and γ-tubulin (magenta) mark the cilia. (f,i) Quantification of the percentage of ciliated cells with PC1 and PC2 along the axoneme, respectively, in control and DZIP1L mutant human dermal fibroblasts. For human cells, cilia were counted in three experiments, with cells from three independent coverslips counted for each experiment (approximately 100 ciliated cells counted on each coverslip). In all cases, quantification included cilia with or without detectable PC1 or PC2 staining. Statistical analyses based on unpaired Student’s t-test. Error bars show SEM. For PC1 staining on MEFs, ****p<0.0001; PC1 and PC2 staining on human dermal fibroblasts, ***p<0.001. Scale bars in a,b = 2µm; d,e,g,h = 1µm.
DISCUSSION
In clinical genetics, there is a strong demand for early and reliable diagnosis of ARPKD, which is only feasible by molecular genetic analysis. To date, PKHD1 has been thought to be the only gene responsible for ARPKD. We have now identified DZIP1L mutations in ARPKD, thereby describing a second locus for this disease. While the clinical manifestations in our patients were first detected prenatally or during early childhood, none showed perinatal demise, which affects about 30% of ARPKD patients with PKHD1 mutations31,32. Mutations in DZIP1L are a considerably rarer cause of ARPKD than PKHD1 mutations, maybe due in part to the larger size of PKHD1 relative to DZIP1L. Furthermore, our limited data suggest that the region encoding the N-terminus of DZIP1L may be more susceptible to mutations, a concept supported by in silico data that predict pathogenicity scores to decay towards the C-terminus (Supplementary Fig. 12). Although we have not screened large numbers of embryonically lethal patients, in view of homozygous truncating mutations in two of our patients we hypothesize that it is unlikely that DZIP1L mutations play a major role in this cohort. This is consistent with the birth of homozygous mutant outbred mice at expected frequencies. Despite the relatively rare occurrence of pathogenic DZIP1L mutations, ARPKD NGS diagnostic multi-gene panels should nevertheless target this gene.
Ultrasonographic patterns observed in our patients phenocopy PKHD1-linked ARPKD, with characteristic bilaterally enlarged, echogenic kidneys, poor cortico-medullary differentiation and multiple tiny cysts. Macrocysts are uncommon in infants, although they may be present with advanced clinical course. Renal failure is rarely a cause of neonatal demise in ARPKD, but renal function variably deteriorates over time. Most of our patients developed chronic renal failure during school age; end-stage renal disease (ESRD) occurred in four. However, most patients were younger than 20 years at the time of analysis, and the remaining may develop ESRD later in life. For PKHD1-linked ARPKD, 25% of non-perinatal patients required kidney transplantation by age 3233. Most of our patients developed arterial hypertension in the first years of life, which is typical for ARPKD. DPM with congenital hepatic fibrosis is another hallmark of ARPKD. We detected subtle early signs of DPM in Dzip1lwpy/wpy mice, with the absence of more severe liver pathology, including hepatic fibrosis, likely due to the early demise of these mice. We can only speculate from our findings in the mouse that hepatic defects might also be present in our patients.
Consistent with the human phenotype, Dzip1lwpy/wpy mutant mice display an early onset, progressive, cystic kidney phenotype, with cysts arising from the collecting ducts and proximal tubules. While ARPKD usually affects the collecting ducts and distal tubules, individuals may present with involvement of other nephron segments, including proximal tubules34 (and unpublished data, CB). Moreover, a number of Pkhd1 mouse models display proximal tubule cysts, although the phenotype in mice carrying different alleles of Pkhd1 is variable, and kidney involvement generally manifests later than in Dzip1lwpy/wpy mice35,36,37.
Unlike patients with DZIP1L mutations, Dzip1lwpy/wpy mouse embryos display a number of extrarenal dysmorphologies including limb and craniofacial defects. The amelioration of craniofacial phenotypes on crossing to an outbred background suggests that at least some of these differences may be due to genetic background, and indeed many ciliary-related phenotypes are subject to variation based on genetic modifying alleles38. It is not uncommon to see phenotypic differences between human and mouse models. For example, non-renal and hepatobiliary phenotypes have been reported in PKD mouse models, including craniofacial, cardiovascular and skeletal defects in homozygous Pkd1 mutant mice39,40. While we cannot exclude the involvement of DZIP1L mutations in syndromic ciliopathies, pathogenic mutations have not been detected in large cohorts of ciliopathy patients screened to date. However, our data raise the possibility that variation at DZIP1L may genetically interact with other PKD or ciliopathy loci to modify phenotype.
Our zebrafish analysis also supports a role in the kidney, with reduced motile cilia in dzip1l-deficient pronephric tubules. Although the mammalian kidney is populated by primary cilia, this nevertheless suggests a conserved requirement of Dzip1l at cilia for proper kidney function across the vertebrates. It is notable that although dzip1l morphant and mutant embryos exhibited overlapping phenotypes (for example, otolith and motile cilia defects), we failed to observe body curvature and hydrocephalus in either zygotic or maternal-zygotic mutants. These effects could be morpholino-induced artefacts. However, given that all four morpholinos produced these phenotypes, it is possible that a degree of genetic compensation is operating in the mutants, as recently described41,42.
Our data suggest context or tissue-specific roles for DZIP1L in primary ciliogenesis and HH signalling in the mouse, whereas loss of the protein in zebrafish causes less prominent effects on these processes. Previous studies in cultured human cells suggested a redundant role for DZIP1 and DZIP1L in ciliogenesis17, a conclusion supported by our zebrafish in vivo experiments (data not shown). DZIP1 localizes to the basal body, the transition zone, and mother centriole22,43. Our findings reveal a similar localization pattern for DZIP1L, and proteomic studies in Chlamydomonas reinhardtii further support DZIP1L as a transition zone protein44. There is some debate over the precise boundaries between the basal body, transition zone and axoneme6, and some proteins shuttle between these structures. In keeping with a transition zone localization, we found that DZIP1L associates with SEPT2, a component of the transition zone diffusion barrier. While DZIP1L has not been rigorously associated with any of the known transition zone protein complexes, a recent mass spectroscopy study included DZIP1L amongst an extensive list of proteins pulled down by Inturned, a core component of the CPLANE transition zone complex that recruits a subset of intraflagellar transport A (IFT-A) proteins to basal bodies45. Although this raises the possibility that DZIP1L may act in the CPLANE complex, this requires further detailed analysis.
Consistent with a role for DZIP1L at the transition zone, ADPKD proteins PC1 and PC2 showed altered distribution along the ciliary membrane in the absence of correct DZIP1L function. In the renal tubules, PC1 and PC2 act in a ciliary membrane complex, and FPC has been shown to bind to and regulate PC212. However, we were not able to detect FPC along the ciliary axoneme to determine if this localization was also compromised by loss of DZIP1L function. In terms of how ciliary mislocalization of the polycystins may contribute to PKD, it has been proposed that the PC1/2 complex suppresses a yet to be identified cilia-dependent cyst formation signal that is inappropriately activated when the PC1/2 complex is unable to properly traffic to cilia46. Whether mislocalization of the polycystins is a major factor contributing to PKD in DZIP1L mutant mice and humans remains to be determined. Nevertheless, our data demonstrate DZIP1L dysfunction as a novel cause of ARPKD, and extend our understanding of the genetic heterogeneity underlying the pathology of this disorder.
(ONLINE) METHODS
Ethics approvals
Human blood samples for DNA extraction and fibroblasts were obtained with written informed consent. All investigations were conducted according to the Declaration of Helsinki, and the study was approved by the Institutional Review Board of the Ethics Committee of the University Freiburg, University of Aachen, University of Michigan, Boston Children`s Hospital and collaborating institutions. All mouse experimentation was performed under approval from the Institutional Review Board of Australian National University and the relevant animal ethics committee of The University of Queensland, and conformed to ethical guidelines. All experiments involving zebrafish were approved by the animal committees of respective institutes (Regierungspräsidium Baden-Württemberg and the Singapore National Advisory Committee on Laboratory Animal Research).
Data availability
For data release, for each of the families the respective IRB-related protocols are being followed. Sequence and variant data will be deposited into dbGaP as a suitable public repository (https://www.ncbi.nlm.nih.gov/gap).
Human haplotype analysis and SNP genome-wide linkage analysis (homozygosity mapping)
Initially, linkage to PKHD1 was excluded by haplotype analysis. Two informative flanking markers were typed in our index pedigree B16. DNA samples were available from parents and all affected individuals. Primers for PCR amplification were as published before and are available on request. Microsatellite markers were analyzed on an ABI PRISM 3130 genetic analyzer (Applied Biosystems, Darmstadt, Germany). Subsequently, we performed genome-wide SNP mapping in the above first-degree consanguineous index family with three affected children (LOD score 2.4; Fig. 1) and family A3533 as reported previously47.
DZIP1L mutation analysis
Mutation analysis was done by direct sequencing for the 15 exons encoding the 767 amino acid (aa) DZIP1L protein (GenBank: NM_173543.2, NP_775814; mutation numbering corresponds to the A of the ATG-translation initiation codon in exon 2). Genomic DNA from an affected individual was amplified by PCR with oligonucleotide primers complementary to flanking intronic sequences. Primers used for PCR and direct sequencing are available on request. Samples were run and analyzed on an ABI PRISM 3130 genetic analyzer (Applied Biosystems).
In addition to conventional Sanger sequencing of DZIP1L in a total of 218 patients with suspected PKD, we performed different next-generation sequencing (NGS) based approaches. First, we used PCR-based 48.48 Access Array microfluidic technology (Fluidigm™) with consecutive NGS. We applied a multiplexing approach allowing PCR-based amplification of 53 amplicons (44 exons) for 48 DNA samples simultaneously in one known and two candidate genes, including DZIP1L. A total of 96 patients with a PKD-related phenotype were analyzed. After two rounds of amplification followed by indexing of all patient-derived products with 96 different 10 bp-barcodes in a subsequent PCR, 2×150 bidirectional sequencing was performed on a MiSeq platform (Illumina™). Second, all exons and adjacent intronic boundaries of a different number of genes (dependent on the version of our customized multi-gene panel, including DZIP1L) known or hypothesized to cause PKD and other ciliopathies were targeted by a custom SeqCap EZ choice sequence capture library (NimbleGen, Madison, Wisconsin, USA) and subsequently sequenced on a Roche 454 GS FLX or an Illumina MiSeq or HiSeq platform (2×150 PE) according to the manufacturer’s protocol. A total of 1234 patients with a polycystic kidney disease phenotype (n=429) or an NPHP-related complex ciliopathy (n=805) were analyzed with an average coverage of 60-fold (GS FLX), 120-fold (MiSeq) or more than 200-fold (HiSeq), respectively. Bioinformatic analysis was performed using the Roche GS Reference Mapper™ software (v2.6), SeqPilot SeqNext moduleTM (v3.5.2, JSI medical systems, Kippenheim, Germany) as well as an in-house bioinformatic pipeline. For all approaches, potential mutations were confirmed by Sanger sequencing and shown to segregate with the phenotype. No further mutation thought to be of pathogenic relevance for the disease phenotype was present among the patients described in this manuscript. Whole-exome sequencing (WES) and mapping of reads was carried out as previously described48,49.
Mouse strains and mutation detection
N-ethyl-N-nitrosourea (ENU) mutagenesis screening was performed at the Australian Phenomics Facility essentially as previously described 23, except that C3H was used as the mapping strain. Phenotypic screening was at 14.5dpc. The initial genome scan involved screening a pool of affected (C57BL/6xC3H) F2 mice and a pool of unaffected F2 mice with a set of ∼100 markers that are polymorphic between C57BL/6 and C3H mice and on average span the genome at 20Mb intervals. The allele specific SNP markers were designed using available software to amplify a subset of validated SNPs. SNP typing was performed using the Amplifluor kit (Chemicon). The individual DNA samples from each of the affected and unaffected F2’s were run with markers that showed possible linkage to the mutation. Following this approach, the wpy mutation was mapped to a 64Mb region on the mouse genome (GRCm38) on Chr 9 between 48804027 (rs3023210) and 112866211 (rs3679771). In this interval there are >900 genes (GRCm38). Eight genes were chosen as potential candidates and subjected to Sanger sequencing of all candidate exons +/−15bp, used an Applied Biosystems 3730xl capillary sequencer, using Big Dye Terminator (BDT) chemistry version 3.1 (Applied Biosytems). The raw trace files were analysed using Lasergene software (DNAstar) against the C57BL/6 mouse reference genome (NCBI). One mutation was identified in intron 19–20 of Kif9 102bp distal to exon 19 (T to C), however was unlikely to be causal. Subsequent whole exome sequencing, as previously described50, was performed on samples enriched for exonic sequences by Nimblegen solution-based capture technologies, using the Illumina GAIIx system employing 100bp end reads. This resulted in the identification of 8 homozygous mutations of which only that in Dzip1l lay within the previously mapped critical interval. The mutation in Dzip1l was confirmed using affecteds and unaffecteds from the mapping screen run with an amplifluor assay designed to the Dzip1l SNV, and also by Sanger sequencing at the Australian Genomic Research Facility. Subsequently, a PCR and restriction enzyme-based genotyping assay was developed (details available on request).
For detailed phenotypic analysis, C57BL/6:C3H mice were subsequently crossed to C57BL/6 wild-type mice through 5 generations (total of 7 wild-type crosses from G0 mutagenised founder), promoting removal of incidental ENU-induced mutations. A pure C57BL/6 line was also maintained by reviving a frozen G1 C57BL/6 individual and crossing through 9 generations with wild-type C57BL/6 mice. The initial analysis of the Dzip1lwpy/wpy phenotype was performed on the C57BL/6:C3H mixed background following at least 2 backcrosses, and was broadly consistent in subsequent generations and also on the C57BL/6 pure background in all generations. All data presented on the outbred CD1 background follows at least 4 crosses to CD1 (in total this line has now been crossed to CD1 through 10 generations). On all background strains the overall phenotype has segregated with the Dzip1l mutation through many meioses (with amelioration of the craniofacial phenotypes on crossing to a CD1 background). In most cases wild-type littermates were used as controls, except in those cases stated where heterozygous embryos were used (Dzip1lwpy/+ heterozygous mice display no discernible morphological differences from wild-type).
Antibodies, stains and primers
Primary antibodies: mouse anti-ARL13B and rabbit-anti-ARL13B (NeuroMab, Clone N295B/66; 1:300 dilution for immunofluorescence (IF) and ProteinTech Group. #17711-1-AP; 1:500 for IF); rabbit-anti-α-tubulin (Sigma-Aldrich T5192; 1:500 for IF); rabbit-anti-acetylated-α–tubulin (Cell Signalling Technology #5335; 1:800 for IF); mouse-anti-acetylated-α–tubulin (Sigma-Aldrich 6-11B-1; #T 6793; 1:500–700 for IF); mouse-anti-γ-tubulin (Sigma-Aldrich T6557; 1:500 for IF), rabbit-anti-DZIP1L (Sigma-Aldrich #HPA030404; 1:500 for IF); mouse-anti-DZIP1L (Abnova-MaxPab H00199221-B01P; 1:500 for IF; 1:1000 for western blot); rabbit-anti-DZIP1 (Proteintech Group, #13779-1-AP; 1:500 for IF); rabbit-anti-SEPT2 (Sigma-Aldrich, #HPA018481;1:2000 for western blot; 1:500 for IF); rabbit-anti-CEP164 (SAB3500022; Sigma), rabbit-anti-TCTN1 (ProteinTech Group, #15004-1-AP; 1:500 for IF); mouse-anti-Polycystin 1 (Abcam, #AB74115; Santa Cruz Biotechnology, # sc-130554; 1:200 for IF); rabbit-anti-Polycystin 2 (MV12; gift from R. Witzgall; 1:500 for IF); rabbit-anti-FPC (PKHD1)9 (hAR-Np and hAR-c2P; gift from G. Wu; 1:10 and 1:20, respectively); rabbit-anti-SMO51 was a kind gift of Rajat Rohatgi (Stanford University; 1:500 for IF); rabbit-anti-IFT88 (Proteintech 13967-1-AP; 1:300 for IF); anti-Aquaporin 2 antibody (Millipore; Cat # AB3274; 1:200 for IF); rabbit-anti-β-catenin antibody (Abcam, #AB6302; 1:500 for IF); goat-anti-HNF3β (M-20) (Santa-Cruz, #sc-6544; 1:50 for IF); goat-anti-SHH(N-19) (Santa-Cruz, #sc-1194; 1:100 for IF); the mouse monoclonal antibodies for Isl-1 (1:200 for IF), Pax6 (1:10 for IF) and Nkx2.2 (1:50 for IF) were obtained from the Developmental Studies Hybridoma Bank (developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242.Bank); biotinylated Lotus Tetragonolobus Lectin (LTL) (Vector laboratories; Cat # B1325; 1:200). Secondary antibodies (all used at 1:500 for IF): Alexa 488 goat anti-rabbit (Invitrogen A-11034); Alexa 488 goat-anti mouse (Invitrogen A-11029); Alexa 555 goat anti-rabbit (Invitrogen A-21428), Alexa 594 goat anti-rabbit (Invitrogen A-11037); Alexa 594 goat anti-mouse (Invitrogen A-11032). Primers used in this study are presented in Supplementary Table 3.
Mouse skeletal preparations, in situ hybridization, scanning electron microscopy and histology
Mouse skeletal preparations52, whole mount in situ hybridization53 and probes used in the limb54 have been described previously. Anti-sense RNA probes were prepared by transcribing linearized cDNA clones with SP6 and T7 polymerases using the digoxigenin labeling mix (Roche). Scanning electron microscopy methods and preparation of mouse tissues for histological analysis have been described previously55. For histological analysis, sections were stained with haematoxylin and eosin (H&E), Elastica van Gieson (EvG) or Silver using standard procedures.
Cell culture
Control and mutant MEFs were isolated from trunks of 13.5dpc littermate embryos by standard methods56 and cultured as primary cell lines for up to 5 passages in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen) containing 10% fetal bovine serum (FBS). Mouse inner medullary collecting duct (IMCD3) cells were grown under the same conditions. Human dermal fibroblasts were derived from skin biopsies from ARPKD affected individual B155 and age- and sex-matched control individuals using standard techniques. Human fibroblasts and HEK293T cells were maintained in DMEM (Sigma, St. Louis, IL, USA) with 4500 mg/L glucose and 10% FBS (HyClone, SH30071.03HI). RPE-1 cells and RPE-1 cells with stable expression of ARL13B-GFP were cultured in ATCC formulated DMEM:F12 Medium (Catalog No. 30–2006) with 10% FBS and 0.01 mg/ml hygromycin B. Cultures of immortalized mouse kidney cells from heterozygous Pkd1flox/− and Pkd2+/− mice as well as Pkd1−/− and Pkd2−/− null mutant cells (derived by inactivating the flox allele with Cre recombinase via transfection in the case of Pkd157 and via spontaneous in-vivo conversion for Pkd258,59) (gifts of S. Somlo) were cultured in DMEM containing 2% fetal bovine serum (FBS), 10,000U/µl IFNγ, 10mg/L Insulin, 6.7µg/L Selenium, 5.5mg/L Transferrin and 1.2ug/L Triiodo-L-thyronine. Established cell lines, human fibroblasts and primary MEFs tested negative for mycoplasma.
IF analysis of mouse and human cells and mouse tissue sections
For IF analysis, cells were grown on glass coverslips to approximately 80%-90% confluency in DMEM + 10% FBS, at which time the medium was replaced with DMEM containing 0.2% FBS for 24h or 0.1% FBS for 48h to induce ciliogenesis. For analysis of SMO the reduced serum media was supplemented with SAG (100nM, Alexis Biochemicals) or diluent alone for 24h. For cell cycle analysis cells were synchronized by serum starvation as above for 24h, the media was replaced with DMEM + 10% FBS and cells were fixed at different timepoints to optimize capture of cells at different stages of the cell cycle. Cells were fixed in either ice-cold methanol for 5 minutes (co-localization experiments involving Abnova anti-DZIP1L and PC2 antibodies) or 4% paraformaldehyde (PFA) for 10 minutes. PFA fixed cells were permeabilised in PBTX (PBS, 0.1% Triton X-100) for 10 minutes. Fixed cells were washed in PBS, and incubated with 2% bovine serum albumin in PBS for 1 hour, followed by 1 hour with primary antibody. After brief washes in PBS, cells were incubated with secondary antibody for 1 hour, and 10 minutes wash with 6-Diamidino-2-Phenylindole (DAPI) to stain nuclei, and mounted on glass slides with Dako Fluorescence mounting medium and imaged on a Zeiss LSM-710 or Olympus FluoView upright laser scanning confocal microscope with a 63x or 100x objectives. For acquisition of 3D structured illumination superresolution microscopy (3D-SIM) images the sample preparation was modified to use high-precision coverslips (#1.5H) and VectaShield H-1000 mounting media (Vector Laboratories) in an effort to minimise spherical aberration. 3D-SIM was performed using a DeltaVision OMX v4 Blaze microscope (GE Healthcare) equipped with 488, 568 and 647 nm lasers, and BGR-FR filter drawer as previously described60.
IF of embryo sections was performed as previously described61. For quantification of cilia in limb sections, three confocal Z-stacks were performed in each of 3 different sections from the limb (9 stacks in total per limb). Given the density of cells in the tissue, and to attempt to account for orientation of cilia in different planes, nuclei were manually counted from the third image of each 5-image stack, while cilia were counted from all 5. Note that the final cilia/nuclei ratio does not necessarily represent the actual percentage of ciliated cells in the tissue, but is a relative value across the samples. The same conclusion was obtained from counting both nuclei and cilia from just the third image of each stack.
Quantitative real time RT-PCR (qRT-PCR)
MEFs were cultured in DMEM containing 10% FBS to 80% confluency, before replacing with DMEM+0.2% FBS for 24h. For SAG treatment experiment, the reduced serum media was supplemented with SAG (100nM, Alexis Biochemicals) or diluent alone for 24h. Total RNA was extracted using TriReagent (Sigma-Aldrich), and first-strand cDNA synthesis was performed using SuperScript III First-Strand Synthesis system (Invitrogen) using oligo dT primers. cDNA was amplified with ABI SYBR Green PCR master mix using transcript specific primers (Supplementary Table 3). Temperature cycling and real-time detection of SYBR Green fluorescence used an Applied Biosystems 7900HT Fast Real-Time PCR system. Technical triplicate reactions were performed for each sample (gene of interest and normalising gene). Relative transcript levels were determined using the 2−ΔCT method.
Zebrafish strains and maintenance
Zebrafish (Danio rerio) were raised under standard conditions at 28°C. TU and AB wild-type embryo strains were used and staged according to Kimmel62. The Tg(wt1b::GFP) strain has been described previously26.
Zebrafish embryo microinjections
Morpholinos were synthesized by Gene Tools and re-suspended in water at a stock concentration of 1mM. Morpholinos were injected into one- to two-cell stage eggs, with each morpholino injected at least four times for each analysis. The following volumes of morpholinos were injected: 1.5nl of translation blocking morpholino (0.25mM working solution), i1e2 morpholino (0.05mM), i4e5 morpholino (0.35mM) and 0.75nl e5i5 morpholino (1mM). For rescue experiments, sense mRNAs of zebrafish and human DZIP1L (in the pSP64T or pCS2 vectors) were in vitro transcribed using the mMessage mMachine kit (Life technologies). For rescue of dzip1l morphants, embryos were co-injected with zebrafish dzip1l mRNA (100ng/µl).
RT-PCR and in situ hybridization with zebrafish embryos
Zebrafish total RNA was isolated by Trizol (Ambion Life Technologies, # 15596018). RT-PCR was carried out using one-step RT-PCR kit (QIAGEN,# 210212). 1µg of total RNA from each experimental group was used to synthesize the 1st strand cDNA. dzip1l specific primers were used to check mis-splicing induced by the morpholinos. actinb1 and ef1α RT-PCR product levels were used as loading control. Whole mount in situ hybridization with the zebrafish dzip1l, ptc1, eng1a and nkx2.2b antisense mRNA probes was performed according to standard protocols.
Whole-mount IF staining of zebrafish embryos and confocal microscopy
Zebrafish embryos were fixed in Dent’s fixative (80% methanol, 20% DMSO) for 3 hours or 4% paraformaldehyde for 2 hours at room temperature, followed by storing in absolute methanol at −20°C. Embryos were then rehydrated in a decreasing methanol:PBS gradient. Blocking buffer (1% (w/v) BSA, 1% DMSO, 0.5% Triton X-100 and 2% sheep serum in PBS) was used to block the embryos for 1 hour. The embryos were then incubated with primary antibodies at 4°C over-night. After washing embryos with PBDT (1% (w/v) BSA, 1% DMSO, 0.5% Triton X-100 in PBS), the embryos were then incubated in fluorophore-conjugated secondary antibodies and DAPI for 3 hours at room temperature. The stained embryos were mounted in 70% glycerol and imaged by confocal microscopy. Image acquisition and analysis was performed using the Olympus FluoView FV10-ASW software and an Olympus FluoView upright confocal microscope.
Generation of dzip1l CRISPR mutants
Design of sgRNAs: sgRNAs for the dzip1l gene were designed using the web tool CHOPCHOP63. Target sites for the sgRNAs were designed by seeking sequences corresponding to the GGN18NGG sequence in the DNA. Two sgRNAs, targeting exon 2 and exon 13, respectively, were designed. To check off-target effects of the sgRNAs, basic local alignment search tool (BLAST) was used and the results were visually analysed. Off-target sequences with no mismatches in the last 15 nt, including the NGG PAM, were discarded. Synthesis of sgRNAs: sgRNAs were synthesized based on a modified two-component system described by Bassett and colleagues64. Briefly, PCR was performed with a forward oligonucleotide encoding the T7 polymerase binding site and the sgRNA target sequence and a common reverse oligonucleotide encoding the remainder of the sgRNA sequence using Phusion polymerase (NEB, M0530S) (see Supplementary Table 3 for the oligonucleotide sequences). In vitro transcription was performed using 250ng of the purified DNA template with MEGAshortscript T7 kit (Ambion, AM1354M) and the sgRNAs were purified as per manufacturer’s instruction. Micro-injection of zebrafish eggs: A mixture of 800ng of the Cas9 protein (Toolgen) and 500ng of each sgRNA were incubated at 37°C for 15 minutes. One-cell stage embryos were injected with 1 nl of the mixture. Genomic DNA extraction and PCR analysis to identify mutations: Genomic DNA extraction from embryos and fins from adult fish were done using the alkaline lysis method65. For embryonic genomic DNA extraction, 8–10 single embryos at 2 dpf were used. To identify the mutations, primers were designed to bind upstream and downstream of the expected double-stranded breaks in exon 2 and exon 13 (Supplementary Table 3). The PCR amplified genomic DNA region from the mutants were TOPO-cloned and sequenced to confirm the deletion.
Yeast two hybrid screen, immunoprecipitation and western blot
Yeast two hybrid screen with human DZIP1L protein (three fragments: aa 1–232, 200–540 and 540–767) was performed by Myriad pharmaceuticals. For immunoprecipitation of endogenous DZIP1L and SEPT2 and vice versa, RPE-1 or HEK293T cells were lysed in 1ml of cell lysis buffer (20mM Tris at PH 8.0, 137mM NaCl, 1%Triton X-100, 2mM EDTA in PBS) with proteinase inhibitor cocktail (Roche, Switzerland). Protein concentrations were estimated by using Bio-Rad protein assay reagent (BioRad Laboratories, USA) according to the manufacturer’s instructions and reading the absorbance at 595 nm with a spectrophotometer. Equal amounts of total protein lysates (0.5mg-10mg) were incubated with DZIP1L or SEPT2 antibodies (2µg) and protein A-agarose (Oncogene Research Products, USA) overnight at 4°C. The immunoprecipitates were washed 5 times with cell lysis buffer, boiled in Laemmili buffer for 5 min and resolved by SDS-PAGE. For western blot analysis, cells were lysed in RIPA buffer (50mM Tris-HCL, pH7.2, 150mM NaCl, 1% Triton x-100, 1% (w/v) deoxycholic acid, 0.1% (w/v) sodium dodecyl sulphate, 0.25mM EDTA with proteinase inhibitor cocktail). The proteins in the lysate were separated by SDS-PAGE. For interaction of SEPT2, different DZIP1L constructs and NPHP1, MKS1, B9D1 and CEP89, transient transfections into HEK293T cells were carried out using JetPrimePolyPlus transfection kit (PolyPlus Transfections) according to the manufacturer’s instructions. Cell lysates were prepared in lysis buffer (20mM Tris-HCl, pH7.4, 150mM NaCl, 1mM EDTA, 0.25% deoxycholate, 1% NP-40 and protease inhibitors). ImmunoCruz IP/WB Optima C beads (Santa cruz Biotechnology INC) were incubated with 2µg of M2 FLAG antibody (F1804; Sigma) for 3 hours at 4°C and subsequently washed in lysis buffer. Control and transiently transfected cell lysates were added to the antibody-beads conjugate, and incubated further overnight for 4°C. Immunoprecipitates were washed in lysis buffer, separated by SDS PAGE, transferred onto PVDF membranes and probed with rabbit anti-HA antibodies (77–5500; Thermo Fisher Scientific) followed by goat-anti rabbit-HRP secondary antibody (Biorad) and chemiluminescence (Super Signal West Pico; Thermo Fisher Scientific).
Alignment of DZIP1L amino-acid sequences
Human, mouse and zebrafish DZIP1L sequence alignment was performed using the MegAlign software (DNASTAR Lasergene 12 Core Suite) following the Clustal W Method.
Statistical analyses
Statistical analyses used Prism version 4.0, 6.0 or 7.0 software (Graphpad). In most cases P-values are based on an unpaired Student’s t-test (see figure legends for details), the mean value is shown and error bars represent standard error of the mean (SEM). In these cases variances are not significantly different. The exception is the analysis in Fig. 3m where both the sample sizes and variances are unequal, and an unpaired t-test was used with Welch’s correction. The significance in this case was confirmed by a Student’s t-test on log transformed data where the variances are equalized (not shown). For animal experiments no specific randomization or routine blinding strategies were employed. Regardless of whether statistical tests were applied, as per standard practice, all animal experiments were performed on multiple embryos/mice/morphants or primary cell lines derived from multiple independent embryos. For both mice and zebrafish the number of individuals used in each experiment is detailed in the text and/or relevant figure legend.
Supplementary Material
Acknowledgments
The authors would like to thank the patients and their families for their cooperation and interest in the study, M. T. Toh and Y. T. Koh for technical assistance, W. I. Goh of the Institute of Medical Biology’s Microscopy Unit for help with superresolution microscopy, J. Lefevre and N. Hamilton for advice on statistical analyses, and C. Cortés, M. Pitt, F. Olsson, L. Zhao, L. Wilkinson and P. Karaith Oliva for assistance, insightful discussion and advice. Thanks also to C. Has for kindly providing control fibroblasts, S. Somlo, M. Ma and K. Dong for Pkd1 and Pkd2 mutant cells, L. Lei for the RPE-1 cell line stably expressing Arl13b–GFP, L. Pelletier for basal body and transition zone protein cDNA clones, and R. Witzgall, G. Wu and R. Rohatgi for antibodies. We would also like to thank staff of the University of Queensland (UQ) QBP animal house for help with mouse husbandry, and the Australian Phenomics Facility for maintaining mice throughout the screen. Confocal microscopy at UQ was carried out in the Institute for Molecular Bioscience Dynamic Imaging Facility for Cancer Biology, developed with the generous support of the Australian Cancer Research Foundation. We also acknowledge the Australian Microscopy & Microanalysis Facility (AMMRF) at the Centre for Microscopy and Microanalysis at UQ. MH, SN, VF and CB are employees of Bioscientia/Sonic Healthcare, CW was a recipient of a University of Queensland Vice-Chancellor’s Senior Research Fellowship and SR is a Senior Principal Investigator with the Institute of Molecular and Cell Biology, Singapore. MHL is a Senior Principal Research Fellow of the Australian National Health and Medical Research Council (NHMRC). FH is the Warren E. Grupe Professor. This work was supported by grants from the German Research Fund (DFG) to KZ and CB, the DFG Collaborative Research Centre (SFB) KIDGEM 1140 and the Federal Ministry of Education and Research (BMBF, 01GM1515C, project 2.3) to CB, the Australian NHMRC to CW (APP1045464), the National Institutes of Health NIH (DK068306) to FH and the Agency for Science, Technology and Research (A*STAR) of Singapore to WH and SR. This paper is dedicated to the memory of Markus Nauck, a friend and colleague, who recently passed away.
Footnotes
Author contributions
LH, PJK, SYT, SV, and SR performed the zebrafish mutant and morphant analysis, protein interaction studies, localization experiments with human fibroblasts and validation of PC antibodies. WH supervised the protein interaction studies. EO, CK and DE performed the zebrafish morpholino analysis. MCRG, GK, VM and ADC performed the mouse analyses, and MCRG and GK performed the localization experiments with mouse cells. RT, PP and ACP performed the screen that identified the mouse mutant, and BW was involved in mapping and identifying the mouse mutation. GDW performed the superresolution microscopy experiments. MHL assisted with design and analysis of mouse kidney experiments. NOB, NH, VF, and SN performed the human mutation analysis. SN, VF, MH, HYG, EAO, FH, and CB carried out the WES data processing and analyses. SN, VF, EW, UV, HYG, KZ, FH, and CB recruited and clinically characterized the study subjects and collected samples. BH performed histologic evaluation of the data. SR, CW, and CB conceived the project, designed and supervised the experiments, analysed and interpreted the data, and wrote the manuscript. All authors reviewed the final manuscript.
Competing financial interest statement
The authors declare that there are no competing financial interests.
References
- 1.Guay-Woodford LM, et al. Consensus expert recommendations for the diagnosis and management of autosomal recessive polycystic kidney disease: report of an international conference. J Pediatr. 2014;165:611–7. doi: 10.1016/j.jpeds.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann C, et al. Clinical consequences of PKHD1 mutations in 164 patients with autosomal-recessive polycystic kidney disease (ARPKD) Kidney Int. 2005;67:829–48. doi: 10.1111/j.1523-1755.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 3.Habbig S, Liebau MC. Ciliopathies - from rare inherited cystic kidney diseases to basic cellular function. Mol Cell Pediatr. 2015;2:8. doi: 10.1186/s40348-015-0019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Gonzalo FR, et al. A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat Genet. 2011;43:776–84. doi: 10.1038/ng.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen VL, et al. Formation of the transition zone by Mks5/Rpgrip1L establishes a ciliary zone of exclusion (CIZE) that compartmentalises ciliary signalling proteins and controls PIP2 ciliary abundance. EMBO J. 2015;34:2537–56. doi: 10.15252/embj.201488044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13:608–18. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guay-Woodford LM. Autosomal recessive polycystic kidney disease: the prototype of the hepato-renal fibrocystic diseases. J Pediatr Genet. 2014;3:89–101. doi: 10.3233/PGE-14092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Luo Y, Wilson PD, Witman GB, Zhou J. The autosomal recessive polycystic kidney disease protein is localized to primary cilia, with concentration in the basal body area. J Am Soc Nephrol. 2004;15:592–602. doi: 10.1097/01.asn.0000113793.12558.1d. [DOI] [PubMed] [Google Scholar]
- 9.Zhang MZ, et al. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc Natl Acad Sci U S A. 2004;101:2311–6. doi: 10.1073/pnas.0400073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, et al. Kinesin-2 mediates physical and functional interactions between polycystin-2 and fibrocystin. Human molecular genetics. 2006;15:3280–3292. doi: 10.1093/hmg/ddl404. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, et al. Fibrocystin/polyductin, found in the same protein complex with polycystin-2, regulates calcium responses in kidney epithelia. Mol Cell Biol. 2007;27:3241–52. doi: 10.1128/MCB.00072-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim I, et al. Fibrocystin/polyductin modulates renal tubular formation by regulating polycystin-2 expression and function. J Am Soc Nephrol. 2008;19:455–68. doi: 10.1681/ASN.2007070770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delling M, DeCaen PG, Doerner JF, Febvay S, Clapham DE. Primary cilia are specialized calcium signalling organelles. Nature. 2013;504:311–4. doi: 10.1038/nature12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaen PG, Delling M, Vien TN, Clapham DE. Direct recording and molecular identification of the calcium channel of primary cilia. Nature. 2013;504:315–8. doi: 10.1038/nature12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delling M, et al. Primary cilia are not calcium-responsive mechanosensors. Nature. 2016;531:656–60. doi: 10.1038/nature17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisenberger T, et al. An efficient and comprehensive strategy for genetic diagnostics of polycystic kidney disease. PLoS One. 2015;10:e0116680. doi: 10.1371/journal.pone.0116680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazer AM, et al. The Zn finger protein Iguana impacts Hedgehog signaling by promoting ciliogenesis. Dev Biol. 2010;337:148–56. doi: 10.1016/j.ydbio.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tay SY, et al. The iguana/DZIP1 protein is a novel component of the ciliogenic pathway essential for axonemal biogenesis. Dev Dyn. 2010;239:527–34. doi: 10.1002/dvdy.22199. [DOI] [PubMed] [Google Scholar]
- 19.Wolff C, et al. iguana encodes a novel zinc-finger protein with coiled-coil domains essential for Hedgehog signal transduction in the zebrafish embryo. Genes Dev. 2004;18:1565–76. doi: 10.1101/gad.296004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekimizu K, et al. The zebrafish iguana locus encodes Dzip1, a novel zinc-finger protein required for proper regulation of Hedgehog signaling. Development. 2004;131:2521–32. doi: 10.1242/dev.01059. [DOI] [PubMed] [Google Scholar]
- 21.Kim HR, Richardson J, van Eeden F, Ingham PW. Gli2a protein localization reveals a role for Iguana/DZIP1 in primary ciliogenesis and a dependence of Hedgehog signal transduction on primary cilia in the zebrafish. BMC Biol. 2010;8:65. doi: 10.1186/1741-7007-8-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Low WC, Liu A, Wang B. Centrosomal protein DZIP1 regulates Hedgehog signaling by promoting cytoplasmic retention of transcription factor GLI3 and affecting ciliogenesis. J Biol Chem. 2013;288:29518–29. doi: 10.1074/jbc.M113.492066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papathanasiou P, et al. A recessive screen for genes regulating hematopoietic stem cells. Blood. 2010;116:5849–58. doi: 10.1182/blood-2010-04-269951. [DOI] [PubMed] [Google Scholar]
- 24.Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–30. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–83. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 26.Perner B, Englert C, Bollig F. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev Biol. 2007;309:87–96. doi: 10.1016/j.ydbio.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol. 2011;193:435–44. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Q, et al. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–9. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chih B, et al. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat Cell Biol. 2012;14:61–72. doi: 10.1038/ncb2410. [DOI] [PubMed] [Google Scholar]
- 30.Ong AC, et al. Polycystin-1 expression in PKD1, early-onset PKD1, and TSC2/PKD1 cystic tissue. Kidney Int. 1999;56:1324–33. doi: 10.1046/j.1523-1755.1999.00659.x. [DOI] [PubMed] [Google Scholar]
- 31.Bergmann C, et al. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1) J Am Soc Nephrol. 2003;14:76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- 32.Bergmann C, et al. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD) Hum Mutat. 2004;23:453–63. doi: 10.1002/humu.20029. [DOI] [PubMed] [Google Scholar]
- 33.Gunay-Aygun M, et al. Correlation of kidney function, volume and imaging findings, and PKHD1 mutations in 73 patients with autosomal recessive polycystic kidney disease. Clin J Am Soc Nephrol. 2010;5:972–84. doi: 10.2215/CJN.07141009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakanishi K, Sweeney WE, Jr, Zerres K, Guay-Woodford LM, Avner ED. Proximal tubular cysts in fetal human autosomal recessive polycystic kidney disease. J Am Soc Nephrol. 2000;11:760–3. doi: 10.1681/ASN.V114760. [DOI] [PubMed] [Google Scholar]
- 35.Williams SS, Cobo-Stark P, James LR, Somlo S, Igarashi P. Kidney cysts, pancreatic cysts, and biliary disease in a mouse model of autosomal recessive polycystic kidney disease. Pediatr Nephrol. 2008;23:733–41. doi: 10.1007/s00467-007-0735-4. [DOI] [PubMed] [Google Scholar]
- 36.Bakeberg JL, et al. Epitope-tagged Pkhd1 tracks the processing, secretion, and localization of fibrocystin. J Am Soc Nephrol. 2011;22:2266–77. doi: 10.1681/ASN.2010111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woollard JR, et al. A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney international. 2007;72:328–336. doi: 10.1038/sj.ki.5002294. [DOI] [PubMed] [Google Scholar]
- 38.Davis EE, Katsanis N. The ciliopathies: a transitional model into systems biology of human genetic disease. Curr Opin Genet Dev. 2012;22:290–303. doi: 10.1016/j.gde.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boulter C, et al. Cardiovascular, skeletal, and renal defects in mice with a targeted disruption of the Pkd1 gene. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:12174–12179. doi: 10.1073/pnas.211191098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kolpakova-Hart E, et al. Growth of cranial synchondroses and sutures requires polycystin-1. Dev Biol. 2008;321:407–19. doi: 10.1016/j.ydbio.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–3. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 42.Blum M, De Robertis EM, Wallingford JB, Niehrs C. Morpholinos: Antisense and Sensibility. Dev Cell. 2015;35:145–9. doi: 10.1016/j.devcel.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, et al. GSK3beta-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol. 2015;13:e1002129. doi: 10.1371/journal.pbio.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diener DR, Lupetti P, Rosenbaum JL. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol. 2015;25:379–84. doi: 10.1016/j.cub.2014.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toriyama M, et al. The ciliopathy-associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat Genet. 2016;48:648–56. doi: 10.1038/ng.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat Genet. 2013;45:1004–12. doi: 10.1038/ng.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun DA, et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet. 2016;48:457–65. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–5. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoff S, et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat Genet. 2013;45:951–6. doi: 10.1038/ng.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews TD, et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. 2012;2:120061. doi: 10.1098/rsob.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–6. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 52.Bruce SJ, et al. Inactivation of Patched1 in the mouse limb has novel inhibitory effects on the chondrogenic program. J Biol Chem. 2010;285:27967–81. doi: 10.1074/jbc.M109.091785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fowles LF, et al. Genomic screen for genes involved in mammalian craniofacial development. Genesis. 2003;35:73–87. doi: 10.1002/gene.10165. [DOI] [PubMed] [Google Scholar]
- 54.Butterfield NC, et al. Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development. 2009;136:3515–24. doi: 10.1242/dev.037507. [DOI] [PubMed] [Google Scholar]
- 55.Ashe A, et al. Mutations in mouse Ift144 model the craniofacial, limb and rib defects in skeletal ciliopathies. Hum Mol Genet. 2012;21:1808–23. doi: 10.1093/hmg/ddr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shibazaki S, et al. Cyst formation and activation of the extracellular regulated kinase pathway after kidney specific inactivation of Pkd1. Hum Mol Genet. 2008;17:1505–16. doi: 10.1093/hmg/ddn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grimm DH, et al. Polycystin-1 distribution is modulated by polycystin-2 expression in mammalian cells. J Biol Chem. 2003;278:36786–93. doi: 10.1074/jbc.M306536200. [DOI] [PubMed] [Google Scholar]
- 59.Wu G, et al. Somatic inactivation of Pkd2 results in polycystic kidney disease. Cell. 1998;93:177–88. doi: 10.1016/s0092-8674(00)81570-6. [DOI] [PubMed] [Google Scholar]
- 60.Xie W, Horn HF, Wright GD. Superresolution Microscopy of the Nuclear Envelope and Associated Proteins. Methods Mol Biol. 2016;1411:83–97. doi: 10.1007/978-1-4939-3530-7_4. [DOI] [PubMed] [Google Scholar]
- 61.Metzis V, et al. Patched1 is required in neural crest cells for the prevention of orofacial clefts. Hum Mol Genet. 2013;22:5026–35. doi: 10.1093/hmg/ddt353. [DOI] [PubMed] [Google Scholar]
- 62.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 63.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–7. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–8. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu C, Zhang Y, Yao S, Wei Y. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One. 2014;9:e98282. doi: 10.1371/journal.pone.0098282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For data release, for each of the families the respective IRB-related protocols are being followed. Sequence and variant data will be deposited into dbGaP as a suitable public repository (https://www.ncbi.nlm.nih.gov/gap).