Abstract
Accumulating evidences suggest that longnon-coding RNAs (lncRNAs) play functional roles in development of different cancers, including cancer initiation and progression. Metastasis associated lung adenocarcinoma transcript 1(MALAT1) is a well-known lncRNA which was previously shown to be a direct target of miR-125b in bladder cancer (BCa) and to promote cancer progression and invasion. However, little is known whether MALAT1 can also target miR-125b. In the present study, using CRISPR-based technologies and qRT-PCR, we show that MALAT1 is capable of suppressing mature miR-125b and increasing the expression of its target genes (Bcl-2 and MMP-13), but has no effect on pri-miR-125b and pre-miR-125b. We observe that the biotin-labeled MALAT1-RNA probe is able to pull down Ago2 and miR-125b and that the negative regulation of miR-125b by MALAT1 is dependent on Ago2. Importantly, the results of flow cytometry assay and transwell assay reveal that the MALAT1-mediated cancer progression is in part due to specific suppression of miR-125b and activation of its two target genes. All together, these data suggest that the “MALAT1-miR-125b-Bcl-2 / MMP-13” axis plays an important role in the progression of BCa, thereby may provide a potential therapeutic strategy for the treatment of human BCa.
Keywords: MALAT1, miR-125b, Ago-2, bladder cancer, CRISPR-based technology.
Introduction
Urothelial bladder cancer is one of the most common malignant tumors in the urinary system, and the prognosis is relatively poor1. There should be a multistep process that involves the activation of proto-oncogenes and the inactivation of tumor suppressor genes in the development and progression of bladder cancer2, 3. The abnormal activities of the downstream gene pathways caused by the expression changes of many non-coding RNAs have been reported to play a key role in this disease 4, 5.
Long non-coding RNAs (lncRNAs) are RNA molecules whose transcripts are more than 200nt in length 6. They do not encode proteins, but play important roles in regulating gene expression through both epigenetic and posttranscriptional mechanisms 6. Although little is known about the carcinogenic mechanisms of lncRNAs, many lncRNAs have been found to be abnormally expressed in bladder cancer 7, including UCA1 (urothelial carcinoma associated) 8, HOTAIR (HOX transcript antisense intergenic RNA) 9 and TINCR (tissue differentiation-inducing non-protein coding RNA) 10.Yet, the biological roles of lncRNAs in bladder cancers are largely unknown. Metastasis associated lung adenocarcinoma transcript 1(MALAT1) is a long non-coding RNA that is upregulated in many cancers, such as breast, pancreatic, lung, colon, prostate and liver cancer 4. Our previous studies have shown that MALAT1inhibits tumor cell apoptosis and promotes cell invasion in bladder cancer 11. However, the underlying mechanism of MALAT1-mediated gene expression having an impact on tumorigenesis is still unclear.
As another type of non-coding RNA, miRNAs (micro RNAs) have also been revealed to play a similar role in tumors 12. Accumulating evidences demonstrated that abnormal expression of miRNAs plays a critical role in regulating the biological behaviors of tumor cells by modulating the mRNA levels of its downstream target genes 12. For example, miR-125b is a very active member of the miRNA family and upregulated in various tumors including leukemia 13, colorectal cancer 14, bladder cancer 15 and lung cancer 16. Previous studies have revealed that miR-125b suppresses cancer cell proliferation and promotes apoptosis by inhibiting the expression of the anti-apoptotic proteins, such as B cell leukemia 2(Bcl-2) 17 and matrix metallopeptidase 13(MMP13) 18.Likewise, our group has indicated that miR-125b functions as a tumor suppressor in bladder cancer by down-regulating MALAT1 15. We showed that the MALAT1 is a direct target of miR-125b through interaction with the putative miR-125b-binding site at MALAT1 RNA.
Intriguingly, some recent studies showed that whereas miRNAs can degrade lncRNAs, at the same time, lncRNAs can also repress miRNA expression19-21. miR-125b negatively regulates MALAT1 possibly through the RNA-induced silencing complex (RISC), suggesting that there may be a reciprocal repression feedback loop between miR-125b and MALAT1.We hypothesized that MALAT1can also suppress miR-125b by acting as a molecular sponge in bladder cancer, which may in part account for MALAT1-mediated cancer progression.
Results
MALAT1 negatively regulates miR-125b
To determine whether MALAT1 can suppress miR-125b expression, we have used the newly-developed technologies "CRISPR Activation" (CRISPRa)22 and "CRISPR Interference" (CRISPRi)23 to up- and down-regulate MALAT1 expression respectively causing for its higher stability and lower off-target effect than RNAi 24, 25.
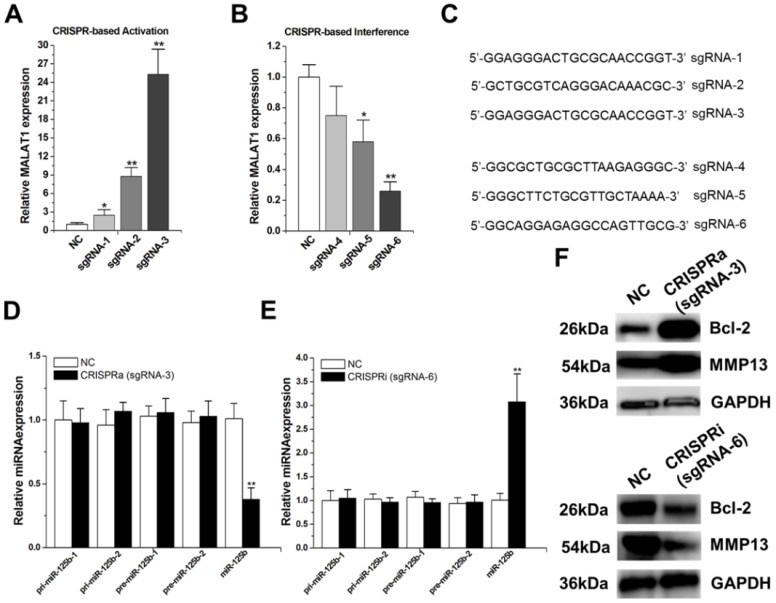
We targeted the dCas9-VP64 protein to the promoter regions of MALAT1 using designed sgRNAs (sgRNA1~3).The expression of these sgRNAs induced various significant increases in MALAT1 expression, and the strongest activation was achieved with sgRNA-3 (Figure 1A).
Figure 1.
MALAT1 negatively regulates miR-125b expression. A) Activation ofMALAT1expressionusing the designed sgRNAs. The error bars indicate the s.d. from five different experiments. NC, negative control. *P < 0.05, relative to the negative control by paired, one-sided t-test. **P < 0.01, relative to the negative control by paired, one-sided t-test. B) Repression ofMALAT1expressionusing the designed sgRNAs. The error bars indicate the s.d. from five different experiments. NC, negative control. *P < 0.05, relative to the negative control by paired, one-sided t-test. **P < 0.01, relative to the negative control by paired, one-sided t-test. C) cDNA sequences of the first 20nt of the sgRNAs. D) Effect of activation of MALAT1on pri-miR-125b, pre-miR-125b and mature miR-125b in T24 cells. **P < 0.01, relative to the negative control by paired, one-sided t-test. E) Effect of repression of MALAT1on pri-miR-125b, pre-miR-125b and mature miR-125b in T24 cells. The error bars indicate the s.d. from five different experiments. **P < 0.01, relative to the negative control by paired, one-sided t-test. F) After activation or repression of MALAT1, the protein expression changes of Bcl-2 and MMP-13 were detected by western-blot. Uncropped blots were presented in Supplementary Figure 1.
We then targeted the dCas9 protein to the promoter regions of MALAT1 using another group of designed sgRNAs (sgRNA4~6).Both sgRNA-5 and sgRNA-6 repressed MALAT1 expression to some extent (Figure 1B), with highest repression observed with sgRNA-6. Sequences of the first 20 nucleotides of these engineered sgRNAs were shown in Figure 1C. We chose sgRNA-3 and sgRNA-6 to do the following experiments.
Firstly, we introduced dCas9-VP64 and sgRNA-3 into T24 cells and found that activation of MALAT1 reduced mature miR-125b level (Figure 1D). Then, we introduced dCas9 and sgRNA-6 into T24 cells and observed that inactivation of MALAT1 increased mature miR-125b level (Figure 1E). To dissect the underlying mechanism of this negative regulation of miR-125b by MALAT1, we examined the effect of activation or inactivation of MALAT1 on the levels ofpri-miR-125b and pre-miR-125b. As shown in Figure 1D and E, MALAT1 has no effect on pri-miR-125b or pre-miR-125b, suggesting a possible post-transcriptional regulation involved. Moreover, our previous study found that MALAT1 contains two direct binding sites for mature miR-125b,5'-ttuggcacgaacacctTCAGGGa-3' (position 3386, R_002819) and 5'-tttatttccagaaagTCAGGGg-3' (position6183, NR_002819), in its 3'UTR 20.Taken together, these findings implicate that MALAT1 may suppress miR-125b by acting as a molecular sponge in bladder cancer.
MALAT1 affects Expression of Endogenous miR-125b Targets
To determine whether MALAT1 affects the expression of endogenous miR-125b targets, we also analysed the impact of MALAT1 on the expression levels of two well known miR-125b targets, “Bcl-2”17 and “MMP13”23. They were detected by western-blot after transfection of CRISPR-encoding plasmid or the negative control, and the results showed that MALAT1 inhibits endogenous miR-125b function, promoting Bcl-2 and MMP13 protein expression (Figure 1F; Supplementary Figure 1).
Association of both miR-125b and MALAT1 with the Ago2-complex
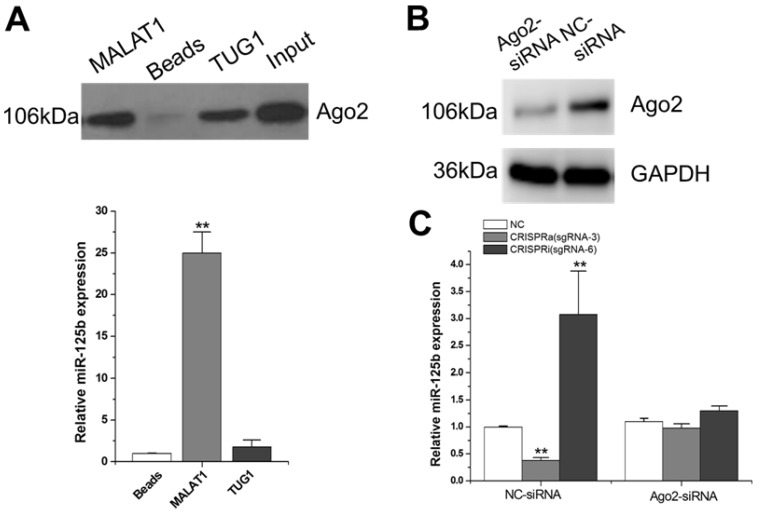
Argonaute 2 (AGO2, EIF2C2) is the only member in AGO family with catalytic activity during the silencing processes of RNA-induced silencing complex (RISC) 26. AGO2 has been demonstrated involved in tumorgenesis through miRNAs-dependent or independent ways 27. Since MALAT1-mediated regulation of miR-125b is likely through a post-transcriptional mechanism, we reasoned that MALAT1 may degrade miR-125b through the Ago2-complex. We synthesized a RNA probe containing partial MALAT1 sequence, labeled it with biotin and then mixed it with T24 cellular extract. After precipitation with streptavidin beads, Ago2 was detected by western blot (Figure 2A; Supplementary Figure 1). The results showed that MALAT1 interacts with Ago2. Interestingly, we also detected a significant amount of miR-125b in this MALAT1-pulled down pellet (Figure 2A). There was no obvious increase inthemiR-125b when TUG1 (another lncRNA studied by our group) RNA probe was used as a negative control.
Figure 2.
Studies on the molecular mechanisms of MALAT1-mediated miR-125b repression. A) Detection of Ago2 using western-blot, and detection of miR-125b using qRT-PCR. The error bars indicate the s.d. from five different experiments. **P < 0.01, relative to the beads group by paired, one-sided t-test. Uncropped blots were presented in Supplementary Figure 1. B) Ago2 siRNA or siRNA negative control was transfected into T24 cells. Then Ago2 protein expression was detected by western-blot. Uncropped blots were presented in Supplementary Figure 1. C) miR-125b expression was detected by qRT-PCR after co-transfection of CRISPR encoding plasmid and the siRNA. The error bars indicate the s.d. from five different experiments. NC, negative control. **P < 0.01, relative to the negative control group by paired, one-sided t-test.
To further determine whether MALAT1 degrades miR-125b through Ago2-mediated RNAi pathway, we used Ago2 siRNA to downregulate Ago2 expression at protein level (Figure 2B; Supplementary Figure 1). Compared to the negative control siRNA, the downregulation effect of miR-125b by MALAT1 was markedly eliminated with the presence of Ago2 siRNA (Figure 2C). All these data implied that MALAT1 can function as a natural miRNA sponge and reduce miR-125b expression through the Ago2-mediated post-transcriptional regulation mechanism.
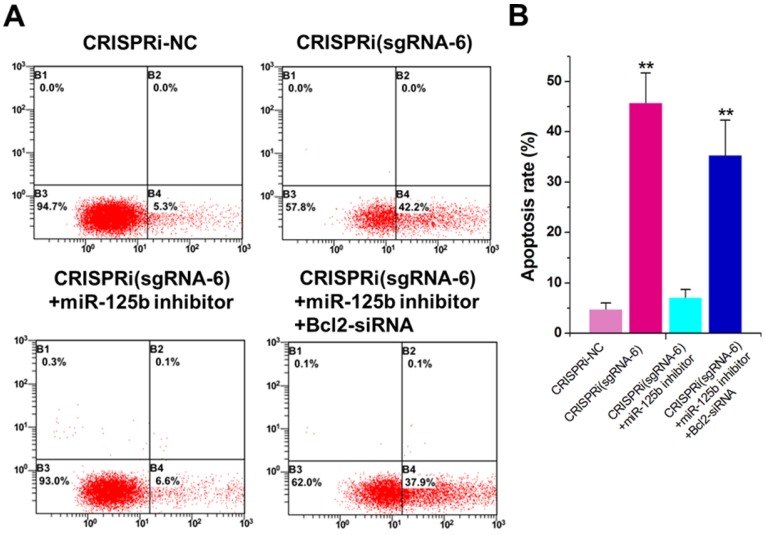
miR-125b inhibitor partly reversed the apoptotic changes caused by CRISPRi-MALAT1
The increased apoptotic rate was observed inCRISPRi (sgRNA-6)-transfected T24 cells by flow cytometry assay (Figure 3A). To determine whether this phenomenon was mediated by up-regulation of miR-125b, we used the miRNA inhibitor and found that inhibitor for miR-125b partly reversed T24 apoptosis induced by CRISPRi (sgRNA-6) (Figure 3A and B). Interestingly, further addition of Bcl-2 siRNA partly recovered T24 apoptosis induced by CRISPRi (sgRNA-6), suggesting that there may be a “MALAT1-miR-125b-Bcl-2”axis in T24 cells. We have also analyzed and confirmed these effects on apoptosis in another bladder cancer cell line 5637 (Supplementary Figure 2).
Figure 3.
miR-125b inhibitor partly reversed the apoptotic changes caused by CRISPRi-MALAT1. A) Cell apoptosis changes were determined using flow cytometry analysis and representative images in T24 cells were shown. B) Cell apoptosis induction observed in T24 cells.The error bars indicate the s.d. from three different experiments. NC, negative control. **P < 0.01, relative to the negative control group by paired, one-sided t-test.
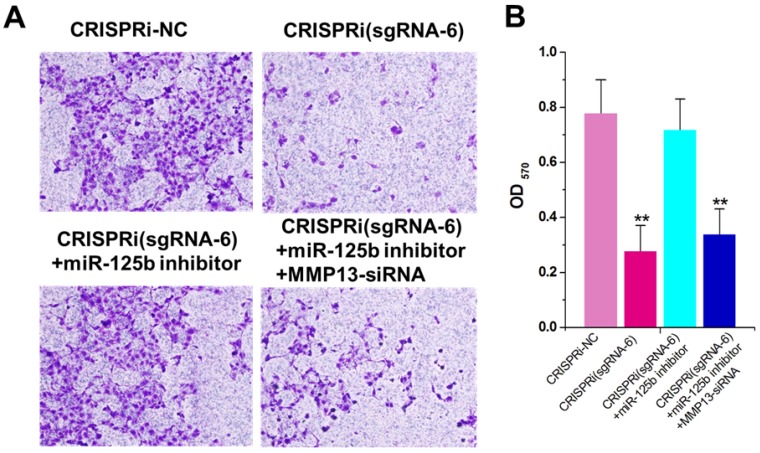
miR-125b inhibitor partly reversed the motility changes caused by CRISPRi-MALAT1
The decreased invasion ability was observed in CRISPRi (sgRNA-6)-transfected T24 cells by transwell assay (Figure 4A). To determine whether this phenomenon was also mediated by up-regulation of miR-125b, we used the miRNA inhibitor again and found that inhibitor for miR-125b partly reversed T24 invasion inhibition induced by CRISPRi (sgRNA-6) (Figure 4A and B). Interestingly, further addition of MMP13 siRNA partly recovered T24 quiescence induced by CRISPRi (sgRNA-6), suggesting that there may be a “MALAT1-miR-125b-MMP13” axis in T24 cells. We have also analyzed and confirmed these effects on invasion in another bladder cancer cell line 5637 (Supplementary Figure 3).
Figure 4.
miR-125b inhibitor partly reversed the motility changes caused by CRISPRi-MALAT1. A) Cell motility changes were determined using transwell assay and representative images in T24 cells were shown. Scale bar, 100μm in each figure. B) Inhibition of cell invasion observed in T24cells.The error bars indicate the s.d. from three different experiments. NC, negative control. **P < 0.01, relative to the negative control group by paired, one-sided t-test.
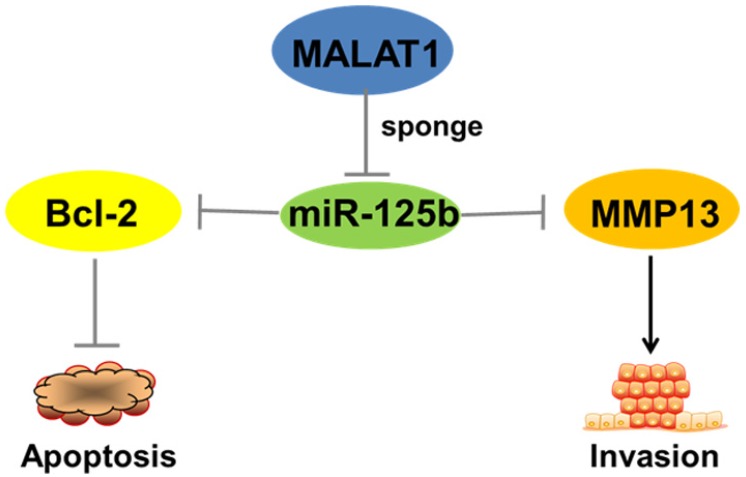
All together, the above results show that MALAT1''sponges''miR-125b in bladder cancer to regulate the expression of Bcl-2 and MMP-13, functional genes that inhibit apoptosis and promote invasion (Figure 5).
Figure 5.
Schematic representation of the circuitry linking MALAT1, miR-125b, Bcl-2, MMP13 and cell apoptosis or invasion. “MALAT1-miR-125b-Bcl-2/MMP13” axis plays a critical role in the complex network of regulatory interactions governing progression of bladder cancer.
Discussion
In our present study, we investigated whether MALAT1 can also suppressmiR-125b in bladder cancer. We have used the newly-developed technologies CRISPRa and CRISPRi. The results implicated that MALAT1 may suppress miR-125b by acting as a molecular sponge in bladder cancer, through a post-transcriptional mechanism. To further determine whether MALAT1 degrades miR-125b through Ago2-mediated RNAi pathway, we used Ago2 siRNA to downregulate Ago2 expression at protein level. All the data implied thatMALAT1 can function as a natural miRNA sponge and reduce miR-125b expression through the Ago2-mediatedpost-transcriptional regulation mechanism. Furthermore, we clearly show that MALAT1 promotes bladder cancer cell invasion and inhibits its apoptosis in part through the suppression of miR-125b. Collectively, these results suggest that miR-125b-MALAT1 interaction plays an important role in the progression of bladder cancer.
MiRNAs and lncRNAs are two new classes of RNAs and constitute an important component of the repertoire of non-coding RNAs5. The miRNAs were initially shown to inhibit expression of specific target mRNA using miRNA binding sites. After that, some lncRNAs were also reported to be the direct targets of various miRNAs. Further evidences reveal that miRNAs target lncRNA in the nucleus in an Ago2-dependent manner 27. Intriguingly, accumulating evidences have shown that lncRNAs can antagonize miRNA function 19-21, 28-30.
During the last few years, many researchers have constructed artificial miRNA sponges 31, which sequester and degrade miRNAs. Interestingly, emerging evidences indicated that lncRNAs can modulate the role and functions of miRNAs, acting as a molecular sponge, such as HOTAIR 32, H19 20, ATB 19, ANRIL 33 and so on. Among them, previous studies have demonstrated MALAT1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206 34. Likewise, our data also found the similar result in bladder cancer cells, suggesting that MALAT1 could function as an effective miRNAs sponge to suppress miR-125b. Furthermore, we observe that the biotin-labeled MALAT1-RNA probe is able to pull down Ago2 and miR-125b and that the negative regulation of miR-125b by MALAT1 is dependent on Ago2, which revealed MALAT1 can function as a natural miRNA sponge through the Ago2-mediated post-transcriptional regulation mechanism.
miR-125b, as a member of miR-125 family, has been reported to be involved in various cancers and other diseases as either repressors or promoters 35. Although dysregulation of miR-125b has been verified in various cancer types, its role in disease is not completely understood 36. Thus, to understand the role of miR-125b in tumorigensis, we should identify various miR-125b target genes. A number of studies demonstrated miR-125b plays crucial roles in many different cellular processes like cell differentiation, apoptosis and invasion by targeting several genes 37, MMP13 18, Bcl-2 17, BAK1 37 and so on. In bladder cancer, it was reported that MMP13 was over-expressed in tumor cells, particularly at the invading edges. Similarly, Bcl-2, an anti-apoptotic protein, plays an important role in apoptosis which is largely regulated by the Bcl-2 family mediated pathway 17. In our experiments, further addition of Bcl-2/MMP13siRNApartly recovered cell apoptosis and quiescence respectively induced by CRISPRi. These data suggest that the “MALAT1-miR-125b-Bcl-2 / MMP-13” axis plays an important role in the progression of bladder cancer, which can explain the interaction between Bcl-2 and MALAT-1, thus providing a new therapeutic strategy for patients.
The present study is the first to demonstrate that there is reciprocal repression between miR-125b and MALAT1, and to verify that "MALAT1-miR-125b-Bcl-2 / MMP-13"axis exerts function in bladder cancer. Additionally, to ensure the precision of this study, we used the CRISPR technology which can achieve up to 100 fold transcriptional repression or activation 38. Taken together, our present study provides a new strategy for cancer research. However, it should be emphasized that there are several limitations in our study. Firstly, our conclusions were only based on the responses of bladder cancer cell lines, which might not reflect processes in the intact organism and should be confirmed in animal models. Secondly, previous studies have demonstrated that MALAT1 can function as a competing endogenous RNA and impact myoblast differentiation through inhibiting miR-133 39. Thus, apart from cancer research, our findings about miR-125b-MALAT1 interaction still needs to be confirmed in other biological processes, such as myogenesis, metabolism regulation, embryonic development and so on. Thirdly, MALAT1 contains binding sites for additional miRNAs, suggesting that MALAT1 may also regulate other miRNAs, such as miR-133, miR-206, etc. Whether there is any crosstalk between the pathways regulated by these miRNAs remains to be investigated.
In summary, our study reveals that there is reciprocal repression between miR-125b and MALAT1 in bladder cancer cell, suggesting that this reciprocal repression is related to the pathway involving the Ago2-mediated post-transcriptional regulation. Furthermore, our work highlights the significance of “MALAT1-miR-125b-Bcl-2 / MMP-13” axis in tumorigenesis including tumor apoptosis and invasion, which adds a new piece of puzzle to the non-coding RNA regulatory networks.
Materials and Methods
Cell lines and cell culture
The bladder cancer cell lineT24 and 5637were purchased from the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, China. Cells were cultured in DMEM medium containing10 % fetal bovine serum and incubated at 37 °C in 5 % CO2.
Plasmids construction
The plasmid vectors pcDNA-dCas9-HA (Plasmid #61355, Addgene) and M-SPn-VP64 (Plasmid #48674, Addgene) were used to transiently express in active Cas9 protein (for CRISPR-based interference) and Cas9-VP64 fusion protein (for CRISPR-based activation) respectively in mammalian cells. The designed cDNA sequence for each sgRNA was designed (http://crispr-era.stanford.edu/), synthesized and inserted into pGPU6/GFP/Neo vector digested with Bam HI / Bbs I.
Cell transfection
T24 or 5637 cells were incubated for 24 h prior to transfection. According to the manufacturer's instructions, the plasmids were transiently transfected into T24 or 5637cells by using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). siRNAs were transfected using RNAfectin reagent (Applied Biological Materials) following the manufacturer's protocol.
RNA extraction and qRT-PCR
At 48 hours after transfection, 1 × 106 cells of T24were collected for extracting total RNA. Total cellular RNA was extracted from different groups with TRIzol reagent (Invitrogen, USA) following the manufacturer's procedures. The concentration and purity of total RNA were measured by using UV spectrophotometer analysis at 260 nm.10mg of total RNA was converted to cDNA using the All-in-One™ miRNA qRT-PCR Detection Kit (Gene-CopoieaInc, Rockville, MD, USA).To specifically detect the expression levels of MALAT1,pri-miR-125b and pre-miR-125b, we used the SYBR Green method with the designed primers. GAPDH or U6 was used as an internal control. The primer sequences were as follows: MALAT1 forward: 5'-AAAGCAAGGTCTCCCCACAAG-3', reverse: 5'-GGTCTGTGCTAGATCAAAAGGCA-3'; GAPDH forward: 5'-CGCTCTCTGCTCCTCCTGTTC-3', reverse: 5'-ATCCGTTGACTCCGACCTTCAC-3'; pri-miR-125b-1 forward: 5'-CGAACAGAAATTGCCTGTCA-3', reverse: 5'-ACCAAATTTCCAGGATGCAA-3'; pri-miR-125b-2 forward: 5'-GAATTCTACCGCATCAAACCA-3', reverse: 5'-TCTGGTGGTAACTGCAGACAA-3'; pre-miR-125b-1 forward: 5'-TGCGCTCCTCTCAGTCCCTGAGA-3', reverse: 5'-GACTCGCAGCTCCCAAGA-3'; pre-miR-125b-2 forward: 5'-ACCAGACTTTTCCTAGTCCCT-3', reverse: 5'-AGGTCCCAAGAGCCTGACTT-3'. Homo sapiens RNAU6 was purchased from GeneCopoiea Inc, Rockville, MD, USA. To detect the relative expression level of mature miR-125b, we use the poly A polymerase-based SYBRGreen method (SBI). Expression fold changes were calculated using 2-ΔΔCtmethods.
Western blot analysis
Cells were grown in 10-cm dishes and homogenized in buffer (10 nMTris-HCl, 5 mM EDTA, 50 nMNaCl, 50 mM sodium fluoride, 30 mM sodium pyrophosphate, 1% Triton-X, 200 µM sodium orthovanadate, 1 mM phenyl methyl sulfonyl fluoride, 1µg/mL pepstatin, 2µg/mL leupeptin and 5µg/mL aprotinin). The protein concentration was determined using the Pierce (Rockford, IL) BCA (bicinchoninic acid) protein assay. Equal amounts of whole protein extract were electrophoresed onto 12% or 15% sodium dodecyl sulfate polyacryl-amide gel electrophoresis gels and then transferred to nitrocellulose membranes (Millipore, Billerica, MA) using a semi-dry transfer cell (Bio-Rad Laboratories, Hercules, CA) at 1 mA/cm2 for 2 h. After transfered to polyvinyl difluoride membranes, samples were blocked in 1% dry milk and incubated over-night with the primary antibodies. Antibodies specific to Bcl-2, MMP13, Ago2 and GAPDH were purchased from Santa Cruz, Dallas, Texas, USA. The levels of proteins were detected with horseradish peroxidase-linked secondary antibodies and revealed using the ECL® System (GE Healthcare, Milan, Italy).
RNA precipitation
To determine whether MALAT1 is associated with the RISC complex, we carried out an RNA precipitation assay using synthesized patialMALAT1 sequence containing miR-125b binding site as a probe and then detected Ago2 from the pellet using western blot and miR-125b using qRT-PCR as described elsewhere12.
Flow cytometry assay
At 48 hours post-transfection, T24 or 5637cells were harvested and resuspended in fixation fluid. 5µl Annexin V - FIFC and 2µl propidium iodide were added to 500 µl cell suspension. Cell apoptosis was then determined by using flow cytometry (EPICS, XL-4, Beckman, CA, USA). In the graphs, the quadrant respectively stands for dead cells, living cells, early apoptotic cells and late apoptotic cells.
Transwell assay
After 24 hours post-transfection, about 1×105 cells with 100 μl serum-free medium were plated into the upper chambers (24-well insert, pore size 8 μm, Corning) which were added with Matrigel (1:8, 50μl/well, BD Bioscience, San Jose, CA, USA). Concurrently, the lower chambers were filled with 500 μl medium containing 10 % fetal bovine serum. Cells were cultured at 37°C in a 5% CO2 atmosphere for 48 hours. Cells under the surface of the lower chamber were washed with 1×PBS, fixed with 4% paraformaldehyde for 20 min, stained with 0.1 % crystal violet for 25 min, and then washed 3 times. Invaded cells were observed under the inverted microscope and imaged. Afterwards, each chamber with the invaded cells was soaked into 1ml 33% acetic acid for 10 min to wash out the crystal violet. Then 100μl of 33% acetic acid was added into per well of 96-well plates, and the absorbance was measured at a wavelength of 570nm using a microplate reader (Bio-Rad, Hercules, CA, USA). Experiments were performed in triplicate.
Statistical analysis
All experimental assays were performed in triplicate. All data were presented as mean± standard deviation (SD). All statistical analyses were executed by using SPSS 20.0 software (IBM, Chicago, IL, USA). P value of less than 0.05 was considered to be statistically significant.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81402103), the Shenzhen Municipal Government of China (ZDSYS201504301722174, JCYJ20150330102720130, GJHZ20150316154912494) and Special Support Funds of Shenzhen for Introduced High-Level Medical Team.
Author contribution
H.X., X.L., Z.C., Y.F. and A.H. performed experiments and data analysis. Y.L. and H.X. designed the project and wrote the paper. W.H., J.L. and Y.L. supervised the project. W.H. and Y.L. provided financial support for the project.
Biography
 Dr. Yuchen Liu's research interests lie at the interface of information science, molecular engineering, and cancer therapy. The current focus is to engineer information directed self-assembly of nucleic acid (DNA/RNA) structures and devices, and to exploit such systems to do useful molecular work, e.g. detecting and destroying cancer cells for diagnosis and therapeutic applications. He has published over 50 SCI papers in this field and he won a gold medal at the Internally Genetically Engineered Machine Competition (iGEM) for his work in genetic technologies in 2015.
Dr. Yuchen Liu's research interests lie at the interface of information science, molecular engineering, and cancer therapy. The current focus is to engineer information directed self-assembly of nucleic acid (DNA/RNA) structures and devices, and to exploit such systems to do useful molecular work, e.g. detecting and destroying cancer cells for diagnosis and therapeutic applications. He has published over 50 SCI papers in this field and he won a gold medal at the Internally Genetically Engineered Machine Competition (iGEM) for his work in genetic technologies in 2015.
References
- 1.Alvarez Kindelan J, Campos Hernandez JP, Lopez Beltran A, Requena Tapia MJ. [The 2004 WHO classification of bladder tumors: a summary and commentary] Actas Urol Esp. 2007;31:978–88. doi: 10.1016/s0210-4806(07)73761-0. [DOI] [PubMed] [Google Scholar]
- 2.Al Hussain TO, Akhtar M. Molecular basis of urinary bladder cancer. Adv Anat Pathol. 2013;20:53–60. doi: 10.1097/PAP.0b013e31827bd0ec. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, Zhan Y, Chen Z, He A, Li J, Wu H. et al. Directing cellular information flow via CRISPR signal conductors. Nature methods. 2016;13:938–44. doi: 10.1038/nmeth.3994. [DOI] [PubMed] [Google Scholar]
- 4.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–8. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 5.Ostankovitch M, Pyle AM. Noncoding RNAs: A story of networks and long-distance relationships. J Mol Biol. 2013;425:3577–81. doi: 10.1016/j.jmb.2013.07.032. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ. et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. 2006;12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- 9.Sun X, Du P, Yuan W, Du Z, Yu M, Yu X. et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Liu Y, He A, Li J, Chen M, Zhan Y. et al. Theophylline controllable RNAi-based genetic switches regulate expression of lncRNA TINCR and malignant phenotypes in bladder cancer cells. Sci Rep. 2016;6:30798. doi: 10.1038/srep30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y, Liu Y, Nie L, Gui Y, Cai Z. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81:209. doi: 10.1016/j.urology.2012.08.044. e1-7. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY. et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/beta-catenin signaling. Molecular cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H. et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin Cancer Res. 2015;21:4234–42. doi: 10.1158/1078-0432.CCR-14-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Y, Liu Y, Zhang H, Wang T, Diao R, Jiang Z, Hsa-miR-125b suppresses bladder cancer development by down-regulating oncogene SIRT7 and oncogenic long noncoding RNA MALAT1. FEBS Lett; 2013. [DOI] [PubMed] [Google Scholar]
- 16.Talekar M, Trivedi M, Shah P, Ouyang Q, Oka A, Gandham S. et al. Combination wt-p53 and MicroRNA-125b Transfection in a Genetically Engineered Lung Cancer Model Using Dual CD44/EGFR-targeting Nanoparticles. Mol Ther. 2016;24:759–69. doi: 10.1038/mt.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao A, Zeng Q, Xie X, Zhou J, Yue W, Li Y. et al. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J Genet Genomics. 2012;39:29–35. doi: 10.1016/j.jgg.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Wu D, Ding J, Wang L, Pan H, Zhou Z, Zhou J. et al. microRNA-125b inhibits cell migration and invasion by targeting matrix metallopeptidase 13 in bladder cancer. Oncol Lett. 2013;5:829–34. doi: 10.3892/ol.2013.1123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Ma CC, Xiong Z, Zhu GN, Wang C, Zong G, Wang HL. et al. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35:90. doi: 10.1186/s13046-016-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L. et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–12. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M. et al. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–68. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–9. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–37. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander ES. The Heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 25.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–6. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Z, Jin H, Qian Q. Argonaute 2: A Novel Rising Star in Cancer Research. J Cancer. 2015;6:877–82. doi: 10.7150/jca.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X. et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer letters. 2016;388:281–91. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhao X, Wang P, Liu J, Zheng J, Liu Y, Chen J. et al. Gas5 Exerts Tumor-suppressive Functions in Human Glioma Cells by Targeting miR-222. Mol Ther. 2015;23:1899–911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S. et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–6. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke J, Yao YL, Zheng J, Wang P, Liu YH, Ma J. et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget. 2015;6:21934–49. doi: 10.18632/oncotarget.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L. et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–92. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang SH, Zhang WJ, Wu XC, Zhang MD, Weng MZ, Zhou D. et al. Long non-coding RNA Malat1 promotes gallbladder cancer development by acting as a molecular sponge to regulate miR-206. Oncotarget. 2016;7:37857–67. doi: 10.18632/oncotarget.9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin H, Sun Y, Wang X, Park J, Zhang Y, Li M. et al. Progress on the relationship between miR-125 family and tumorigenesis. Exp Cell Res. 2015;339:252–60. doi: 10.1016/j.yexcr.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 37.Bousquet M, Nguyen D, Chen C, Shields L, Lodish HF. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica. 2012;97:1713–21. doi: 10.3324/haematol.2011.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jusiak B, Cleto S, Perez-Pinera P, Lu TK. Engineering Synthetic Gene Circuits in Living Cells with CRISPR Technology. Trends Biotechnol. 2016;34:535–47. doi: 10.1016/j.tibtech.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Han X, Yang F, Cao H, Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–64. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.