Abstract
Caseinolytic peptidase B (ClpB) plays a pivotal role in suppressing and reversing protein aggregation. Toxoplasma gondii is an intracellular parasitic protozoan that infects a wide variety of mammals and birds and therefore is exposed to a broad range of living condition. We screened ToxoDB (http://ToxoDB.org) and identified 10 putative T. gondii genes encoding members of the Clp superfamily of caseinolytic proteases and chaperones. Of these, we focused on characterizing the Class I ATP-dependent molecular chaperones TgClpB1, TgClpB2, and TgClpB3. We found that TgClpB1, the most divergent of the five T. gondii Class I Clp ATPases, is cytoplasmic, TgClpB2 is found in the mitochondria of the parasites, and TgClpB3 is a ClpB with novel apicoplast localization. Knockout strains of TgClpB1 and TgClpB2 were established by CRISPR/Cas9 mutagenesis, and their complementing strains were constructed with FLAG-tag. Although knockout of TgClpB1 or TgClpB2 did not affect growth under normal circumstances, TgClpB1 was required for T. gondii thermotolerance. The growth, replication, and invasion capabilities of TgClpB1-deficient mutants were significantly inhibited after extracellular parasites were pretreated at 45°C. Moreover, TgClpB1 were observed at the poles of the ΔTgClpB1 FLAG-tagged strain treated at 42°C.
Keywords: ClpB, caseinolytic proteases and chaperones, AAA+ ATPases, thermotolerance, toxoplasma gondii
INTRODUCTION
Caseinolytic proteases and chaperones (Clp) play an important role in protein homeostasis and cellular survival during both stress and optimal growth conditions [1]. The functions of Clp enzymes include the maintenance and repair of salvageable proteins in cells exposed to stress, as well as, disaggregation, translocation, unfolding, and degradation of abnormal or misfolded proteins. Clp chaperone-protease complexes, such as ClpAP, ClpPX, and ClpQY (HslVU), are multimeric ATP-dependent protease systems that are ubiquitous in both prokaryotes and eukaryotes. [2, 3]. These complexes comprise an ATPase and substrate-binding subunit (ClpA, ClpX, ClpY) joined with a proteolytic subunit (ClpP, ClpQ) [4, 5]. An adaptor protein may also be associated, such as ClpS, which interacts with the N-terminal domain of chaperone ClpA and influences the ClpAP complex [6].
T. gondii is a unicellular intracellular protozoan belonging to the parasitic phylum Apicomplexa and subclass Coccidium [7]. It is one of the most widespread parasites in the worldwide, potentially capable of infecting all birds and mammals including humans [8]. The parasite must adapt to a board range of temperatures during its complex life cycle, existing in hosts such as cattle (36.7-39.1°C), cats (38.6-40.1°C), dogs (37.9-39.9°C), chickens (39.6-43.6°C), and geese (40.0-44.0°C). Furthermore, the parasitic infection might induce a systemic inflammatory reaction that can elevate the temperature of the host. High temperature stress has a wide range of effects on cells, including changes in membrane fluidity and structure, cell cycle arrest, protein denaturation, and cell death. Such issues can occur when essential proteins are damaged by high temperatures, or when aggregates of damaged proteins become toxic to the cell [9, 10]. Clp superfamily proteins are crucial for protein quality control under these conditions, and yet little is known about this family of proteins in T. gondii.
ClpB and its orthologs (ClpA, ClpC, ClpD, ClpX, and ClpY), also known as heat shock protein 100 (HSP100)/Clp, are members of the ATPases Associated with diverse cellular Activities (AAA) superfamily, and is able to recover aggregated proteins in an ATP-dependent manner [11]. Notably, ClpB family members are specific to bacteria, cyanobacteria, yeast, plants, and parasitic protozoans and are not present in animals or humans [12]. Yeast ClpB and its bacterial homologs can disaggregate insoluble proteins caused by severe stresses [13]. Plasmodium falciparum ClpB1 (PfClpB1) is located at the parasite’s apicoplast and is involved in metabolic pathways necessary for the organism’s survival [14–16]. PfClpB2 is targeted to the parasitophorous vacuole membrane (PVM), where it acts as an essential subunit of the Plasmodium translocon of exported proteins (PTEX) complex [17]. In Leishmania major, the ClpB proved essential for the growth and survival of parasites infecting mouse macrophages in vitro [18, 19]. We sought to determine whether ClpB family members identified by searching the T. gondii genome, might also be involved in maintaining parasitic proteostasis. In this study, we present an identification and functional analysis of T. gondii’s Clp superfamily of proteins. Our results demonstrate that TgClpB1, TgClpB2 and TgClpB3 localize in the cytosol, mitochondria and apicoplast of T. gondii, respectively. Additionally, we found that TgClpB1 plays an essential role when cells are exposed to extreme heat.
RESULTS
The Clp superfamily proteins in T. gondii
Ten putative clp genes were identified in T. gondii (Figure 1). All sequences were found on the nuclear chromosomes. Two T. gondii sequences (TGGT1_318320, TgClpP and TGGT1_272230, TgClpQ) were found to have a high level of sequence homology with P. falciparum 3D7 ATP-dependent CLP protease (PfClpP, PF3D7_0307400) and ATP-dependent protease subunit ClpQ (PfClpQ, PF3D7_1230400), respectively. TgClpP was identified as caseinolytic protease by its S14_ClpP_2 domain (residues 193 to 477), containing conserved active site residues and an oligomer interface. We found that TgClpQ had a PRK05456 domain (residues 136 to 306), and therefore classified the protein as a homolog of the ATP-dependent protease subunit HslV.
Figure 1.
(A) Comparison of the conserved domains of the Toxoplasma gondii Clp proteases and ATPases. The ClpB1 protein of Class I can be identified by their two AAA domains, whereas Class II proteins contain a single AAA domain. The AAAs (AAA-1, orange; AAA-2, olive) are highly conserved, whereas the N domains (yellow), M domains (gray), and C domains (green) have low sequence homology. Other identified domains include ClpB_D2 depicted in purple, Clp_N in light blue, and the Clp protease in dark orange. (B) Table 1. Bioinformatics analysis of the Clp superfamily proteins in T. gondii.
T. gondii Clp Class I include TgClpB1, TgClpB2, TgClpB3, TgClpB4 and TgClpB5. All of these proteins are predicted to contain two ATP nucleotide-binding domains (AAA domains) that have characteristic Walker A and Walker B nucleotide-binding and -recognition motifs. As DNASTAR program protean prediction, the five TgClpBs present the long coiled-coil domains inserted between the two AAA domains, which is a characteristic of ClpB. The parasite contains two members of Clp ATPases Class II, which we named TgClpX and TgClpY. ClpY is the ATPase and the substrate-binding subunit. The ClpY molecule is distinguished by two domains: the AAA domain (residues 225 to 312). ClpS shares a very high degree of homology with P. falciparum ClpS (PF3D7_1320100). Of the family members we have identified, only TgClpS, TgClpB4, and TgClpB5 contain predicted apicoplast-targeting sequences [20]. SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) predicted the absence of signal peptide cleavage sites in amino acid sequences of these TgClp family proteins.
Generation of TgClpB1, TgClpB2 and TgClpB3 knockout and complementing strains
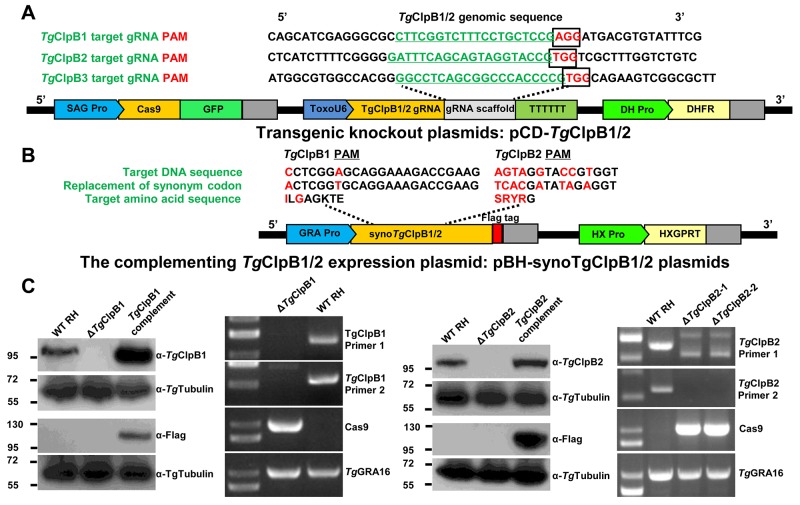
To investigate the function of TgClpB1, TgClpB2, and TgClpB3 in T. gondii, we proceeded to generate the knockout mutants (ΔTgClpB1, ΔTgClpB2 and ΔTgClpB3). The gene-targeting plasmids, designated pCD-TgClpB1, pCD-TgClpB2 and pCD-TgClpB3 (Figure 2A), were constructed using specific gRNAs with a DHFR selectable marker cassette. Plasmid pBluescript II containing codons synonymous to TgClpB1, TgClpB2 and TgClpB3 was then fused with FLAG-tag and a hypoxanthine xanthine guanine phosphoribosyl transferase (HXGPRT) expression cassette, designated pBS-syno-TgClpB1, pBS-syno-TgClpB2 and pBS-syno-TgClpB3 (Figure 2B). After electroporation and selection, ΔTgClpB1 and ΔTgClpB2, their complement, and the Cas9 control strains were all verified by Western blot analysis (Figure 2). Protein bands of approximately 100 kDa were observed in the lysate of the Cas9 control cell line, but not in the ΔTgClpB1 or ΔTgClpB2 strains. This size correspond to the predicted size of the proteins (104311.39 Da and 114596.39 Da) listed in ToxoDB. The 100 kDa band was also observed after probing the complementing strain lysates with anti-TgClpB1 or TgClpB2 serum, as appropriate. The presence of the proteins was additionally confirmed using anti-FLAG monoclonal antibody. To further verify TgClpB1 and TgClpB2 knockout, the mutant strains were also examined by PCR assay (Figure 2). Together, these experiments confirmed the respective absence of TgClpB1 and TgClpB2 in the ΔTgClpB1 and ΔTgClpB2 strains. We also attempted to generate the TgClpB3 knockout strain by introducing the pCD-TgClpB3 plasmid in three independent experiments. However, the transfected parasites failed to grow after addition of pyrimethamine. Our findings suggest that TgClpB3 may be crucial for the parasites survival.
Figure 2.
(A) The TgClpB1 and TgClpB2 disruption sites are shown in their genomic context. The target guide RNAs (gRNA) of TgClpB1 and TgClpB2 are underlined, and the protospacer adjacent motif (PAM) for each is framed. The pCD-TgClpB1 and pCD-TgClpB2 expression plasmids contain three important cassettes in addition to the TgClpB1, TgClpB2 or TgClpB3 gRNA: GFP, Cas9, and dihydrofolate reductase (DHFR). (B), (C) PCR and Western blot analysis confirmed the respective absence of TgClpB1 and TgClpB2 in the ΔTgClpB1 andΔTgClpB2 strains as well as the presence of the proteins in the parental HX-RH, ΔTgClpB1::TgClpB1-FLAG and ΔTgClpB2::TgClpB2-FLAG tachyzoites.
TgClpB1 and TgClpB2 are not required for trafficking of TgGRA16 and TgGRA24 to the host cell
TgClpB1 and TgClpB2 were found to have a high level of sequence homology with P. falciparum ClpB2/HSP101 (PF11_0175), which is an essential protein for export of diverse malaria effectors into host erythrocytes (Supplementary Figure 1). We suspected that TgClpB1 and TgClpB2 may have a similar function in assisting the transport of TgGRA16 and TgGRA24 from the PVM to the host nucleus. We therefore examined the location of TgGRA16 and TgGRA24 in the ΔTgClpB1/2 mutants by immunofluorescence in infected cells. To do this, we introduced a plasmid expressing either HA-tagged TgGRA16 or HA-tagged TgGRA24 into the RH-WT and ΔTgClpB1/2 strains. Infected cells were stained at 24 hpi, and this revealed that TgGRA16 and TgGRA24 from both knockout strains were still able to locatize to the host cell’s nucleus. This indicates that TgClpB1 and TgClpB2 are not required for exporting proteins from T. gondii to host cells (Supplementary Figure 2).
TgClpB1 is cytoplasmic and redundant under optimal conditions
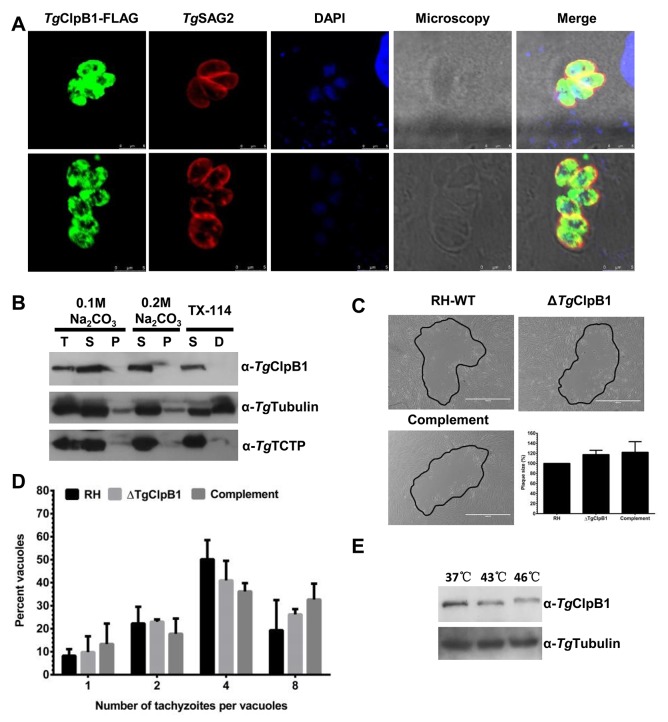
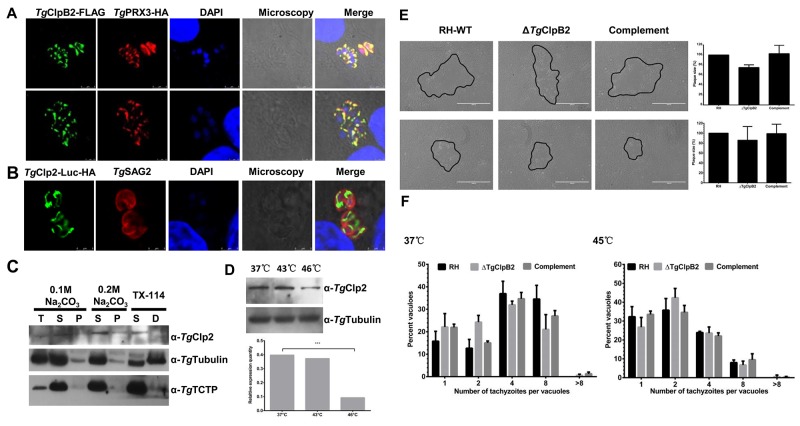
Unlike PfClpB2, TgClpB1 was not detected in the PVM of T. gondii. TgClpB1-FLAG instead presented as a soluble protein in the parasitic cytoplasm of T. gondii (Figure 3A and 3B). Growth of the ΔTgClpB1 mutant as measured by plaque formation and intracellular replication was normal when the extracellular parasites were maintained under the standard conditions of 37°C, 30 min (Figure 3C and 3D). The egress rate of ΔTgClpB1 was also not significantly different compared with RH-WT and TgClpB1 complement strain at optimal condition (data not shown). To explore the knockout parasite’s response to heat stress, we next characterized the expression levels of TgClpB1 after heat treatment at 43°C or 46°C for 4 h. We observed that the protein band corresponding to TgClpB1 shifted to a higher molecular weight at 46°C, but not at 43°C (Figure 3B).
Figure 3.
(A) TgClpB1-FLAG colocalizes with the cytosolic marker TgSAG2. (B) TgClpB1 is a soluble protein. Cytosolic proteins were extracted from RH-WT parasites and separated by either sodium carbonate treatment into soluble (S) and membrane pellet (P) fractions or by Triton X-114 phase partitioning into soluble (S) and detergent (D) fractions. The first lane displays total protein extract (T). (C) Wild-type, knockout, and complemented strains were grown in HFF cells for 7 days. A representative plaque is shown for each, outlined in black. (D) Intracellular replication was not changed at 37°C for the ΔTgClpB1 strain. (E) TgClpB1 was detected by Western blot in extracellular parasites treated at 37°C, 43°C, or 46°C for 4 h.
TgClpB1 is necessary for thermotolerant growth and replication of T. gondii tachyzoites
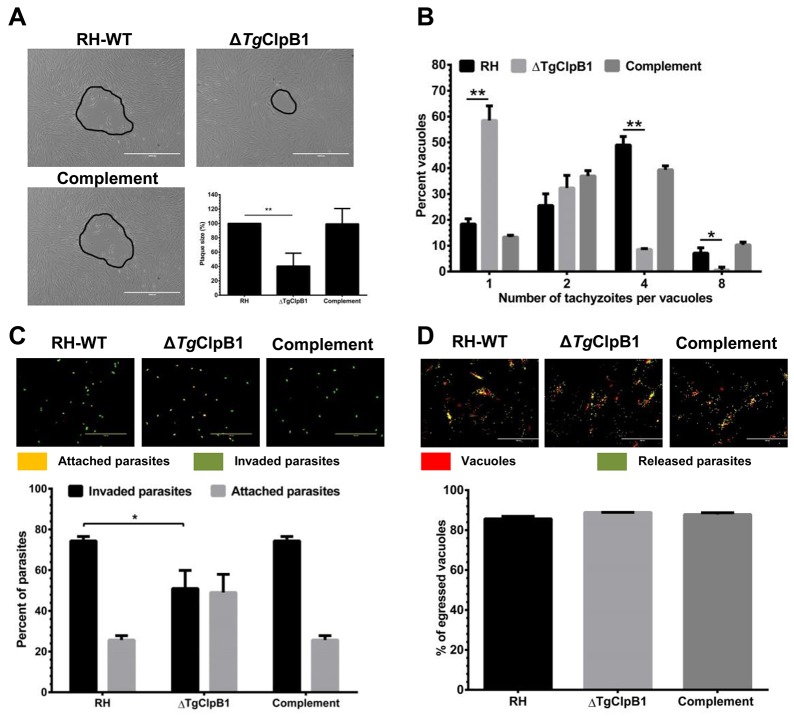
Aiming to further understand how TgClpB1 influences parasite viability, we examined the growth and replication capacities of the ΔTgClpB1 strain under heat shock conditions. The gradient temperature actions were performed to treat freely egressed RH-WT, ΔTgClpB1 and complement parasites at 37, 39, 42, 45, 47 or 50°C. The results showed that the growth of the parasites was universally inhibited at 47°C. However, apparent differences were able to be observed between RH-WT and ΔTgClpB1 strains at 45°C (Supplementary Figure 3). Thereafter, compared with its controls, growth of the ΔTgClpB1 strain was severely impaired after treatment at 45°C for 30 min. We observed a marked reduction in plaque formation consistent with the decreased intracellular replication of the ΔTgClpB1 mutant. This further supported the crucial role of ΔTgClpB1 in the growth and replication of T. gondii tachyzoites under heat stress conditions (Figure 4B).
Figure 4.
(A) When parasites were grown in HFF cells after the released parasites treated at 45°C, ΔTgClpB1 led to a reduction in plaque formation compared with RH-WT. Representative plaques are shown outlined in black. (B) The ΔTgClpB1 strain displays decreased intracellular replication at the same condiction. Parasites were allowed to invade Vero cells for 3 h. After 24 h, the parasites were fixed and stained with 4% paraformaldehyde. The percentage of vacuoles containing one, two, four, or eight parasites was determined. Results are shown as an average of three independent experiments. (C) The invasion efficiency of the ΔTgClpB1 strain was significantly reduced when the extracellular parasites were treated at 45°C. Below, mean values of three independent experiments are graphed with bars representing standard deviation. (D) ΔTgClpB1 mutant does not affect in ability to egress from host cells following stimulation of calcium signaling with calcium ionophore. A23187.
Loss of TgClpB1 reduced the invasion rate of the parasites after the heat stress
To investigate whether the TgClpB1 knockout strain had any defect in infectivity at 45°C, we performed an attachment/invasion assay. Evaluating the cellular invasion rate of the knockout parasites showed a significant difference in attachment/invasion behavior between RH-WT and ΔTgClpB1 strains (P < 0.05) (Figure 4C). These results indicate that the loss of ΔTgClpB1 reduces the parasite’s invasion ability under heat stress in vitro. Furthermore, the results reveal that ΔTgClpB1 did not affect the egress capacity of the parasites after the heat stress treatment (Figure 4D).
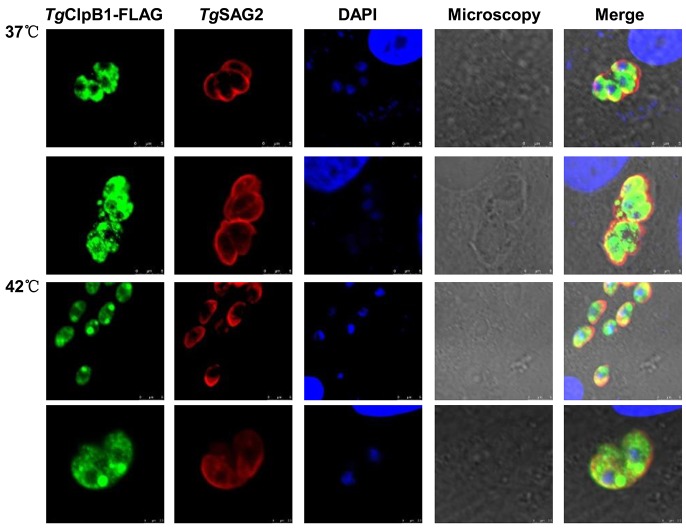
Heat shock induces polar aggregation within the parasites
To investigate whether TgClpB1 is activated upon heat shock, the TgClpB1-FLAG was detected by immunofluorescence in intracellular parasites. When incubated at 42°C, the TgClpB1-FLAG proteins display an increased localization at the poles of the parasites when compared with the diffuse cytosolic staining observed at 37°C. In individual parasites, one or two inclusion bodies were observed (Figure 5).
Figure 5. TgClpB1-FLAG aggregates at the parasitic poles under heat stress conditions.
Vero cells were infected with parasites expressing Flag-tagged TgClpB1. After treatment at 42°, the cultures were washed, fixed, and stained with antibodies to the Flag tag and TgSAG2.
TgClpB2 has mitochondrial localization and is not required for thermotolerance
The localization of TgClpB2-FLAG was observed at the mitochondria of parasites with TgPRX3-HA (Figure 6A). To characterize the expression profile of TgClpB2, the parasite lysates were detected by Western blot analysis using mouse anti-TgClpB2 antibody. TgClpB2 was detectable within both the soluble and insoluble fractions (Figure 6C). We also observed that TgClpB2 expression declined when the parasites were exposed to higher tempretures (Figure 6D). However, when the growth and replication rates of ΔTgClpB2 strains were examined under optimal and heat shock conditions, no significant change was observed as compared with the RH-WT and complementing strains.
Figure 6.
(A) Colocalization of TgClpB2-FLAG with the mitochondrial marker TgPRX3-HA suggests that TgClpB2 is targeted to the mitochondrion. (B) When fused to the reporter protein Luc-HA, the predicted transit peptide TgClpB2 (1-113) is sufficient to induce mitochondrial localization of the construct. (C) TgClpB2 is present in both the soluble and insoluble fractions of the parasitic extract. (D) TgClpB2 is detectable by Western blot in extracellular parasites treated at 37°C, 43°C, or 46°C for 4 h. (E) T. gondii thermotolerance is independent of TgClpB2. When grown in HFF cells for 7 days, no significant alteration of plaque formation is seen with the ΔTgClpB2 strain as compared with its controls. Representative plaques are outlined in black. (F) Loss of TgClpB2 does not significantly alter intracellular replication rate at either 37°C or 45°C.
N terminal signal of TgClpB2 targets the mitochondria of parasites
To test whether TgClpB2 contains a targeting sequence for the T. gondii mitochondria, Luc fusion protein constructs were made using the TgClpB2 pre-sequence (the first 1113 amino acids). This gene fusion construct successfully yielded a pattern of mitochondrial fluorescence when introduced to the parasite. Therefore, the first 113 amino acids of the TgClpB2 sequence appear to be sufficient to target proteins to the mitochondria of T. gondii (Figure 6B).
TgClpB3 is a membrane associated protein located at the apicoplast of the parasites
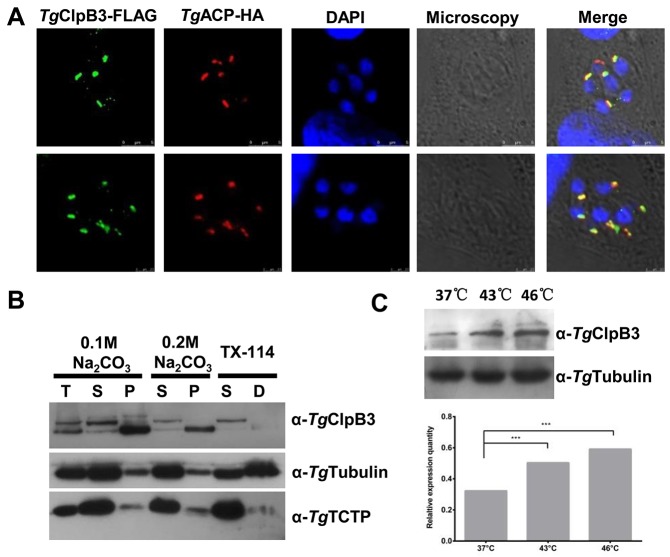
To examine the location of TgClpB3, HFF monolayers were infected with a parasitic TgACP-HA overexpression strain. The TgClpB3 was then detected by mouse anti-TgClpB3 polyclone antibody. The protein appeared to colocalize with the TgACP-HA marker at the apicoplast of the parasites (Figure 7A). To evaluate whether TgClpB3 is membrane associated, we next performed sodium carbonate extraction and Triton X-114 phase partitioning. Western blot analyses revealed that TgClpB3 was present in both unprocessed and mature forms in the total parasite lysate. The unprocessed version was mainly exhibited at the soluble fraction (Figure 7B), whereas the mature form was mainly detected at the insoluble and membrane fractions. Furthermore, TgClpB3 appeared to be strongly up-regulated after heat treatment at 43°C or 46°C for 4 h (Figure 7C).
Figure 7.
(A) Colocalization of TgClpB3-FLAG with the marker TgACP-HA suggests that TgClpB3 is targeted to the apicoplast of T. gondii. (B) TgClpB3 is a membrane-associated protein. Proteins were extracted from RH-WT and either separated into soluble (S) and membrane pellet (P) fractions by sodium carbonate treatment or into soluble (S) and detergent (D) phases by Triton X-114 phase partitioning. Total protein extract (T) is shown in the first lane. (C) Western blot detection of TgClpB3 in extracellular parasites treated at 37°C, 43°C, or 46°C for 4 h.
DISCUSSION
Here, we have provided the first integrated survey of the Clp chaperones and proteases in T. gondii. We found that the parasite contains two proteolytic subunits, which we term TgClpP and TgClpQ. Five Class I Clp chaperones (TgClpB1, TgClpB2, TgClpB3, TgClpB4 and TgClpB5), and two Class II Clp chaperones (TgClpX and TgClpY). TgClpS, which is responsible for the specific recognition of proteins bearing an N-terminal destabilizing residue, was also identified. In P. falciparum, PfClpQ directly interact with the C-terminus of PfClpY to form a protein complex in mitochondria, which is crucial for the growth and survival of asexual stage parasites [21, 22]. Generally, ClpXP is known to be an ATP-powered protease that can unfold and degrade aggregated proteins, performing essential functions of protein quality control as it participates in numerous regulatory circuits from bacteria to humans [23]. Although the T. gondii ClpP, ClpQ, ClpX and ClpY proteins were not characterized in this study, the parasite’s genome harbors two potential Clp protease systems: ClpXP and ClpQY. These prokaryotic-specific ATP-dependent protease complexes may represent potential drug targets in T. gondii.
ClpB and its orthologs involved in cellular quality control belong to the AAA superfamily. These enzymes are typified by an N-terminal domain and two ATP-binding domains (AAA domains) essential for hexamerization and chaperone function [12]. T. gondii contain five ClpB proteins. In Leishmania, ClpB is membrane-associated and crucial for the secretion of exosomes [19]. For P. falciparum, ClpB2 is a major component of PTEX and is required to mediate protein export to parasite’s host cell [17]. Although TgClpB1 and TgClpB2 both have a high level of sequence homology with PfClpB2 (PF11_0175), TgClpB1 and TgClpB2 were localized at the cytoplasm and mitochondria of parasite, respectively. Neither of the knockout strains limited the export of TgGRA16 and TgGRA24 into the nucleus of the host cells. The T. gondii homologues of PfEXP2 are TgGRA17 and TgGRA23, which located at the PVM and are responsible for the transfer of small molecules though the PVM [24]. In a recent report, MYR1 is suggested to be a component of machinery that media the transportation of GRA proteins into the cytosol of host cells [25]. Bath on our results, it is possible that T. gondii utilizes a complete different pathway for the transportation of the proteins from the PVM.
The formation of misfolded proteins is an inevitable process in living cells, which can cause cellular toxicity if left unchecked. Misfolding can be enhanced by internal and environmental stresses such as heat shock [26, 27]. ClpB acts to counter protein aggregation in the cytosol of bacteria, yeast, and plants, and thereby ensures the proper functioning of proteins in their cellular environment [28–31]. Through FLAG-tag constructs, we determined that the subcellular localization of TgClpB1 and TgClpB2 is, respectively, within the cytoplasm and mitochondria of the parasites. Moreover, the shifted molecular weight was also observed at the TgClpB1 after heat treatment. In Chlamydomonas, the molecular weight of HSF1 protein is observed to increase slightly between 15 and 30 min after the onset of heat shock, a change that has been demonstrated to be due to phosphorylation [32]. The higher molecular weight may be due to modifications of the activated protein after heat shock. As in the previous study, the higher molecular weight of TgClpB1 may be due to an activating modification that occurs as a result of heat shock. Furthermore, we discovered that disruption of TgClpB1 results in a severe defect in T. gondii invasion and intracellular growth after following heat shock. The specific action of ClpB1 in T. gondii may be similar to that of its homologous cytoplasmic protein in yeast, since both are directly involved in the organism’s thermotolerance.
We additionally determined that TgClpB1 is soluble, not a membrane-associated protein. Immunofluorescence analysis revealed that TgClpB1 migrates and displays polar localization after heat shock. Although the 42°C treatment used for our experiments has no growth effect on wild-type T. gondii, it does induce observable, polar-localized protein aggregations in the parasites. To our knowledge, this is the first observation of these inclusion bodies in T. gondii. In other organisms, previous reports in bacteria have detailed misfolded proteins observed to aggregate in inclusion bodies given various conditions. Moreover, yeast cells also accumulate aggregated, insoluble proteins at the perivacuolar insoluble protein deposit (IPOD) [31]. In mammals, an aggresome is assembled with misfolded and ubiquitylated proteins at an indentation of the nucleus surrounding the centriole [33]. Taken together, our results suggest that T. gondii may have a protein quality control mechanism similar to the yeast and bacterial response to protein aggregation.
TgClpB2 were detected at the mitochondria of the parasites by tagging the respective parasite genes with FLAG tag. To test the putative transit peptide of TgClpB2 present on nuclear-encoded mitochondrion proteins mediate targeting, Luc fusion proteins were constructed by using the putative N-terminal targeting transit peptides. The results demonstrated that the 113-aa N-terminal presequence of TgClpB2 is sufficient to target a protein into the mitochondrion. The declined expression of TgClpB2 after heat treatment was observed in this study. In previous study, the decreased expression of heat shock protein 70 mRNA and protein were also observed after heat treatment in cells of aged rats although the mechanism is still unclear [34]. On the other hand, the Mitochondrial ClpB Homolog Hsp78 of the Saccharomyces cerevisiae is indispensable for the resolubilization of protein aggregates generated by heat stress under in vivo conditions. However, our results reveal that the disruption of TgClpB2 does not affect the growth and replication capacities of T. gondii under optimal and heat stress conditions. The function of TgClpB2 needs to be characterized in the further study.
Finally, we have been able to determine that the TgClpB3 is located at the apicoplast of the parasites. A previous bioinformatic study implied that TgClpB4 and TgClpB5 are located in the apicoplast of parasites using a parametric model for ApicoTPs [20]. We found by Western blot analysis that the unprocessed and mature forms of TgClpB3 are separately in soluble and insoluble fraction, respectively. Although the TgClpB3 does not have transmembrane domain, in the present study the mature form appeared to be sequestered in the insoluble and membrane-associated fractions. This suggests that TgClpB3 may be a part of an insoluble complex on the membrane of the parasitic apicoplast. The apicoplasts of Apicomplexa parasites are organelles that originated from an alga through secondary endosymbiosis. They contain several essential metabolic pathways such as heme, isoprenoid, and fatty acid biosynthesis. A resent study demonstrated that TIC/TOC machinery is involved in the import mechanisms across the innermost membranes of primary and secondary plastids [35]. The ClpB/HSP100 is thought to associate with TIC machinery and function in protein folding [36]. It is therefore possible that TgClpB3/4/5 may be involved with the import of apicoplast proteins. Phenotype score of TgClpB3 is -4.75 based on a previous study that a genome-scale screen measures the contribution of each T. gondii’s gene [37]. Additionally, the failure to construct a viable TgClpB3-deficient parasite suggests an important function for this protein.
This study represents the first complete analysis of Clp chaperones and proteases in T. gondii. We showed that TgClpB1 and TgClpB2 are respectively present in parasitic cytoplasm and the mitochondria of T. gondii. Although the knockouts of TgClpB1/2 have no observed effect on the growth and replication of parasites at normal temperature, the loss of TgClpB1 resulted in a reduction of T. gondii replication and invasion rates after treatment at high temperature. TgClpB1 therefore plays a crucial role for the thermotolerant survival and growth of T. gondii tachyzoites. We also discovered a novel apicoplast-localized ClpB, TgClpB3, which is necessary for parasite viability.
MATERIALS AND METHODS
Ethics statement
Laboratory animal care and experimentation were performed in accordance with the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. All protocols used for animal studies were approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS) (approval number BRDW-XBS-12).
Cultivation of host cells and transfection of T. gondii tachyzoites
Human foreskin fibroblasts (HFFs) and Vero cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco-BRL), 2 mM glutamine, and 0.5% penicillin-streptomycin. All mammalian cells were cultured at 37°C and 5% CO2 in tissue culture plates. Tachyzoites of T. gondii RH ΔHXGPRT- or RH, as well as derived transgenic strains generated in this study, were propagated in Vero cells under standard procedures. Heat shock experiments for extracellular parasites were performed by incubating at various temperatures prior to harvest.
To generate TgClpB1/2/3 knockout parasites, pCD-TgClpB1/2/3 plasmids were transfected into 1×107 parasites by electroporation and selected by 1 μM pyrimethamine. To generate TgClpB1/2 complementing parasites, pBH-synoTgClpB1/2 plasmids were transfected into 1×107 ΔTgClpB1/2 and selected by mycophenolic acid (20 μg/ml) and xanthine (50 μg/ml). Stable clones were isolated by limiting dilution in 96-well plates. Single clones were amplified and screened by PCR and Western blotting. Transgenic parasites overexpressing TgClpB1/2 were performed by electroporation of pBH-synoTgClpBs into 107 HX tachyzoites. The pBS-DHFR-TgPRX3-HA and pBS-DHFR-TgACP-HA plasmids were transfected into TgClpB2 and TgClpB3 respectively transgenic parasites and selected by 1 μM pyrimethamine.
Bioinformatic procedures
Protein sequences from Plasmodium falciparum ClpP (accession no. PFC0310c), PfClpR (PF14_0348), PfClpB1 (PF08_0063), PfClpC (PF14_0063), PfClpQ (XP_001350699), PfClpY (PF3D7_0907400), Escherichia coli ClpP (P0A6G7), EcClpA (M31045), Synechococcus elongatus ClpX (PCC7942) and Arabidopsis thaliana ClpS1 (NP_564937) were retrieved from the GenBank and used in BLASTp queries of the ToxoDB (http://ToxoDB.org). Conserved regions in the catalytic domains of the Clp superfamily were analyzed by the NCBI conserved domain search program.
Cloning procedures and plasmids construction
All the PCR amplifications were performed with either Ex Taq (TaKaRa) or the KOD-Plus-Neo (TOYOBO) kit. Primers are listed in Supplementary Table 1. The partial TgClpB1, TgClpB2 and TgClpB3 were individually cloned into pET-28a. Construction of the target gRNA expression constructs (TgClpB1/2; Figure 2) was performed by hierarchical fusion PCR assembly of the ToxoU6 promoter. TgClpB1, TgClpB2 and TgClpB3 target sequences along with the gRNA scaffold were then cloned into the PmeI site of pCD-Cas9 using the ClonExpress II Kit [38, 39]. The plasmids containing TgClpB1, TgClpB2 and TgClpB3 targeting fragments, respectively designated pCD-TgClpB1/2/3, were then used to establish knockout strains stably expressing GFP. The full ORFs of TgClpB1-FLAG, TgClpB2-FLAG, TgClpB3-FLAG, TgGRA16-HA and TgGRA24-HA were amplified and inserted into the EcoRV site of the pBS-HXGPRT vector under control of the TgGRA1 promotor. TgPRX3 and TgACP open reading frames with a C-terminal HA tag were inserted downstream of the TgGRA1 promoter, and a dihydrofolate reductase (DHFR) cassette was inserted upstream, to generate pBS-DHFR-TgPRX3-HA and pBS-DHFR-TgACP-HA.
Production of TgClp1, TgClp2 and TgClp3 mouse antisera
RNA was isolated from a RH strain of T. gondii, and cDNAs were created using the SuperScript III First Strand Kit (Invitrogen). The truncated TgClpB1 (amino acids 82-260, TGGT1_268650), TgClpB2 (amino acids 65-312, TGGT1_268650) and TgClpB3 (amino acids 472-654, TGGT1_275690) were each amplified by PCR, cloned into the pET28 vector, and used to transform BL21 (DE3) competent E. coli. Recombinant TgClpB1, TgClpB2 and TgClpB3 were purified by a Ni-NTA affinity chromatography according to the manufacturer’s instructions (Amersham Pharmacia Biotech, USA). The purified rTgClpB1, rTgClpB2 and rTgClpB3 emulsified with Freund’s complete adjuvant (Sigma Chemicals, USA) and immunized into BALB/c mice intraperitoneally. Three months after the immunization, antisera were collected and probed using Western blotting and an indirect fluorescent-antibody test (IFAT). The anti-TgSAG2, anti-Tgtubulin, anti-TgGRA7 and anti-TgTCTP (TGGT1_251680) obtained by rabbit and mouse immunization.
Immunofluorescence assay and confocal microscopy
HFF cells infected with T. gondii were fixed with 4% paraformaldehyde for 30 min and then permeablized with 0.03% Triton X-100 in PBS at 4°C for 10 min. The cells were washed in PBS three times and blocked with 3% bovine serum albumin (BSA) at room temperature for 1 h. TgClpB1, TgClpB2 and TgClpB3 were detected with mouse polyclonal anti-FLAG M2 primary antibody and secondary antibodies conjugated to Alexa Fluor 488 fluorescent (life technology). TgSAG2 protein was detected by rabbit polyclonal anti-TgSAG2 and Alexa Fluor 647 conjugated goat anti-rabbit IgG. TgPRX3-HA as T. gondii mitochondria marker and TgACP-HA as an apicoplast marker. Both were detected with mouse anti-HA primary antibody and a secondary antibody conjugated to Alexa Fluor 647 fluorescent (life technology), respectively. Nuclei were visualized with 4’, 6-diamidino-2-phenylindole (DAPI) stain. Cells were mounted with ProLong Diamond Antifade Mountant (Thermo Fisher Scientific). The samples were imaged using confocal laser-scanning microscopy (Leica).
Improved mitochondrial targeting sequence
The fusion protein with HA tag was generated by linking the first 113 amino acids of to the full open reading frame of luciferase. The TgClpB2 transit sequence was amplified from tachyzoite TgClpB2 containing the full open reading frame by using sense primer TgLucb-DHFR-1F and antisense primer TgLucb-DHFR-1R for TgClpB2113-Luc-HA plasmid. T. gondii RH-WT were transfected and selected by pyrimethamine. Immunolabeling of TgClpB2113-Luc-HA fusion proteins was performed using the anti-HA monoclonal antibody and anti-TgSAG2 antibody. Fluoresce was observed by a confocal microscope.
Western blot analyses
Extracellular free parasites were harvested by centrifuging at 3000 g for 10 min and then lysed by RIPA (Sigma) for 10 min in ice. The samples were loaded on 10% SDS-PAGE and transferred at 60 mA for 150 min to the PVDF membrane Immobilon-PSQ (Millipore). The membrane was blocked for 60 min at room temperature with 3% skim milk in PBS. The primary antibodies were diluted as follows: mouse anti-TgClpB1 (1:500), mouse anti-TgClpB2, mouse anti-TgClpB3, rabbit anti-TgTubulin (1:2000), mouse anti-TgTCTP (1:500) and mouse anti-FLAG (1:2000) (Sigma). A horseradish peroxidase-conjugated goat anti-mouse antibody (1:4000) was used as the secondary antibody. The immunoblots were visualized on blue-sensitive x-ray film using the SuperSignal West Pico Chemiluminescent Substrate kit (Thermo Fisher Scientific).
Plaque assay
HFF monolayers grown in 6-well plates were infected with 100 parasites per well. After incubation for 3 h, wells were washed with PBS and fresh media was added. The parasites were grown for 1 week before being fixed with cold methanol for 30 min. Documentation was performed with EVOS FL Auto Cell Imaging System at 40-fold magnification. Images were further processed with Photoshop.
Intracellular replication assay
To determine the replication efficiency, the freshly egressed parasites were plated onto HFF cells in 6-well plates and allowed to invade for 3 h. After washing with PBS three times, parasites were allowed to grow for 24 h. IFA was performed using the anti-TgSAG2 antibody (1:1000). The number of parasites per vacuole was determined in 100 vacuoles for three independent experiments.
Red/green invasion assays
The 5×105 freshly egressed parasites were added to confluent HFF cells in 12-well plates. Parasites were allowed to invade for 2 h and were then fixed with 4% paraformaldehyde. After blocking with 3% BSA in PBS, The attached parasites were labeled with rabbit polyclonal anti-TgSAG2 antibody followed by Alexa Fluor 594 conjugated goat anti-rabbit IgG without permeabilization. Furthermore, the parasite infected cells were permeabilized with 0.03% Triton X-100 in PBS at 4°C for 10 min. Released parasites were labeled with rabbit polyclonal anti-TgSAG2 followed by Alexa Fluor 488 conjugated goat anti-rabbit IgG. The number of extracellular and intracellular parasites counted in 100 parasites for three independent experiments.
Induced egress assay
Freshly released parasites were allowed to invade the confluent HFF monolayers grown on 6 well plates for 36 h. The parasite infected HFF were artificially inducing egress with prewarmed egress media containing 0.06% DMSO or 1 μM A23187 (Sigma) calcium ionophore for 10min at 37°C in the dark. Parasites were fixed with cold methanol for 30min and were stained with anti-TgGRA7 and anti-TgSAG2 (1:1000) antibodies after blocking with 2% BSA in PBS. The average number of egressed vacuoles was determined by counting 100 vacuoles for three independent experiments for each condition.
Statistical analysis
The graphs and All statistical analyses used in the work were performed by GraphPad Prism version 5.00 (GraphPad Software, San Diego, CA). The various assay conditions used herein were evaluated with a Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test with a 95% confidence interval. Values of P < 0.05 were considered statistically significant.
SUPPLEMENTARY MATERIALS FIGURES AND TABLE
ACKNOWLEDGMENTS AND FUNDING
The plasmid pCD-TgClpB1, pCD-TgClpB2 and pCD-TgClpB3 are modified from pSAG1::CAS9-U6::sgUPRT (Addgene, #54467). We thank Dr. David Sibley for providing this plasmid. This work was supported by a grant from National Key Research and Development program of China (No. 2017YFD0500404).
Footnotes
CONFLICTS OF INTEREST
The authors have declared that no competing interest exists.
REFERENCES
- 1.Gur E, Ottofueling R, Dougan DA. Machines of destruction–AAA+ proteases and the adaptors that control them. Regulated proteolysis in microorganisms. Springer; 2013. pp. 3–33. [DOI] [PubMed] [Google Scholar]
- 2.Kirstein J, Molière N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat Rev Microbiol. 2009;7:589–99. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 3.Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 4.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–88. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 5.Liberek K, Lewandowska A, Ziętkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–35. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, Dougan DA, Bukau B. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–56. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- 7.Levine ND. Progress in taxonomy of the Apicomplexan protozoa. J Protozool. 1988;35:518–20. doi: 10.1111/j.1550-7408.1988.tb04141.x. [DOI] [PubMed] [Google Scholar]
- 8.Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30:1217–58. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riezman H. Why do cells require heat shock proteins to survive heat stress? Cell Cycle. 2004;3:61–63. [PubMed] [Google Scholar]
- 10.Imai J, Yashiroda H, Maruya M, Yahara I, Tanaka K. Proteasomes and molecular chaperones: cellular machinery responsible for folding and destruction of unfolded proteins. Cell Cycle. 2003;2:585–90. [PubMed] [Google Scholar]
- 11.Zolkiewski M, Zhang T, Nagy M. Aggregate reactivation mediated by the Hsp100 chaperones. Arch Biochem Biophys. 2012;520:1–6. doi: 10.1016/j.abb.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–96. [PubMed] [Google Scholar]
- 13.Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–15. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 14.AhYoung AP, Koehl A, Cascio D, Egea PF. Structural mapping of the ClpB ATPases of Plasmodium falciparum: targeting protein folding and secretion for antimalarial drug design. Protein Sci. 2015;24:1508–20. doi: 10.1002/pro.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Bakkouri M, Pow A, Mulichak A, Cheung KL, Artz JD, Amani M, Fell S, de Koning-Ward TF, Goodman CD, McFadden GI, Ortega J, Hui R, Houry WA. The Clp chaperones and proteases of the human malaria parasite Plasmodium falciparum. J Mol Biol. 2010;404:456–77. doi: 10.1016/j.jmb.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 16.Ngansop F, Li H, Zolkiewska A, Zolkiewski M. Biochemical characterization of the apicoplast-targeted AAA+ ATPase ClpB from Plasmodium falciparum. Biochem Biophys Res Commun. 2013;439:191–95. doi: 10.1016/j.bbrc.2013.08.064. [DOI] [PubMed] [Google Scholar]
- 17.Beck JR, Muralidharan V, Oksman A, Goldberg DE. PTEX component HSP101 mediates export of diverse malaria effectors into host erythrocytes. Nature. 2014;511:592–95. doi: 10.1038/nature13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krobitsch S, Brandau S, Hoyer C, Schmetz C, Hübel A, Clos J. Leishmania donovani heat shock protein 100. Characterization and function in amastigote stage differentiation. J Biol Chem. 1998;273:6488–94. doi: 10.1074/jbc.273.11.6488. [DOI] [PubMed] [Google Scholar]
- 19.Krobitsch S, Clos J. A novel role for 100 kD heat shock proteins in the parasite Leishmania donovani. Cell Stress Chaperones. 1999;4:191–98. doi: 10.1379/1466-1268(1999)004<0191:anrfkh>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cilingir G, Broschat SL, Lau AO. ApicoAP: the first computational model for identifying apicoplast-targeted proteins in multiple species of Apicomplexa. PLoS One. 2012;7:e36598. doi: 10.1371/journal.pone.0036598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain S, Rathore S, Asad M, Hossain ME, Sinha D, Datta G, Mohmmed A. The prokaryotic ClpQ protease plays a key role in growth and development of mitochondria in Plasmodium falciparum. Cell Microbiol. 2013;15:1660–73. doi: 10.1111/cmi.12142. [DOI] [PubMed] [Google Scholar]
- 22.Ramasamy G, Gupta D, Mohmmed A, Chauhan VS. Characterization and localization of Plasmodium falciparum homolog of prokaryotic ClpQ/HslV protease. Mol Biochem Parasitol. 2007;152:139–48. doi: 10.1016/j.molbiopara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman S, Roche E, Zhou Y, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–47. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold DA, Kaplan AD, Lis A, Bett GC, Rosowski EE, Cirelli KM, Bougdour A, Sidik SM, Beck JR, Lourido S, Egea PF, Bradley PJ, Hakimi MA, et al. The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell Host Microbe. 2015;17:642–52. doi: 10.1016/j.chom.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franco M, Panas MW, Marino ND, Lee MC, Buchholz KR, Kelly FD, Bednarski JJ, Sleckman BP, Pourmand N, Boothroyd JC. A novel secreted protein, MYR1, is central to Toxoplasma’s manipulation of host cells. MBio. 2016;7:e02231–15. doi: 10.1128/mBio.02231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hombach A, Ommen G, MacDonald A, Clos J. A small heat shock protein is essential for thermotolerance and intracellular survival of Leishmania donovani. J Cell Sci. 2014;127:4762–73. doi: 10.1242/jcs.157297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–64. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12:479–92. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eriksson MJ, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–46. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274:28083–86. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]
- 31.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz-Raffelt M, Lodha M, Schroda M. Heat shock factor 1 is a key regulator of the stress response in Chlamydomonas. Plant J. 2007;52:286–95. doi: 10.1111/j.1365-313X.2007.03228.x. [DOI] [PubMed] [Google Scholar]
- 33.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–95. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fargnoli J, Kunisada T, Fornace AJ, Jr, Schneider EL, Holbrook NJ. Decreased expression of heat shock protein 70 mRNA and protein after heat treatment in cells of aged rats. Proc Natl Acad Sci USA. 1990;87:846–50. doi: 10.1073/pnas.87.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fellows JD, Cipriano MJ, Agrawal S, Striepen B. A Plastid Protein That Evolved from Ubiquitin and Is Required for Apicoplast Protein Import in Toxoplasma gondii. MBio. 2017;8:e00950–17. doi: 10.1128/mBio.00950-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 37.Sidik SM, Huet D, Ganesan SM, Huynh MH, Wang T, Nasamu AS, Thiru P, Saeij JP, Carruthers VB, Niles JC. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell. 2016;166:1423–35.e12. doi: 10.1016/j.cell.2016.08.019. https://doi.org/10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen B, Brown KM, Lee TD, Sibley LD. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio. 2014;5:e01114–14. doi: 10.1128/mBio.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J, Jia H, Zheng Y. Knockout of leucine aminopeptidase in Toxoplasma gondii using CRISPR/Cas9. Int J Parasitol. 2015;45:141–48. doi: 10.1016/j.ijpara.2014.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.