Abstract
Exoribonucleases play an important role in all aspects of RNA metabolism. Biochemical and genetic analyses in recent years have identified many new RNases and it is now clear that a single cell can contain multiple enzymes of this class. Here, we analyze the structure and phylogenetic distribution of the known exoribonucleases. Based on extensive sequence analysis and on their catalytic properties, all of the exoribonucleases and their homologs have been grouped into six superfamilies and various subfamilies. We identify common motifs that can be used to characterize newly-discovered exoribonucleases, and based on these motifs we correct some previously misassigned proteins. This analysis may serve as a useful first step for developing a nomenclature for this group of enzymes.
INTRODUCTION
The metabolism of RNA molecules is now known to encompass a wide variety of reactions and to require a large number of distinct ribonucleases (RNases). Following the original identification and characterization of these RNases, primarily from Escherichia coli, work in this area has expanded to include many other prokaryotic and eukaryotic systems. In particular, the explosion of genome sequences in recent years has led to the identification of homologs of many RNases in a variety of diverse organisms. Such studies have served to highlight the frequent similarities, as well as some important differences, between the RNases of E.coli and those of other organisms. A detailed review dealing with the functional roles of exoribonucleases has appeared recently (1).
Eight 3′ to 5′ exoribonucleases have been characterized in E.coli. These are polynucleotide phosphorylase (PNPase), RNase II, RNase D, RNase BN, RNase T, RNase PH, RNase R and oligoribonuclease. This group of eight enzymes may account for all the exoribonuclease activities present in an E.coli cell (1). In addition, although studies of exoribonuclease activities in other prokaryotes have been relatively limited, many RNase genes homologous to those of the E.coli enzymes have been identified. Likewise, many of the eukaryotic RNase genes known at present are also related to those of the E.coli proteins. Thus, considering the wealth of information now available from genome sequencing, it seemed an appropriate time to catalog all the known exoribonucleases and their homologs into families based on sequence analysis and catalytic properties. Furthermore, the grouping of exoribonucleases into families could serve as a first step in developing a nomenclature for these enzymes.
The results presented here, based on exhaustive data mining, have led to the identification of six superfamilies, as well as various subfamilies, that encompass all the known exoribonucleases. These groupings, based on common structural features derived from sequence analysis, are consistent with the known catalytic properties of the various enzymes. Moreover, examination of the distribution of individual superfamilies and subfamilies among diverse organisms has provided a phylogenetic framework with interesting evolutionary implications.
While some of the sequence alignments and superfamilies reported here have been described previously (2), this work includes for the first time all the known exoribonucleases and their homologs. Every family member included here was firmly established by well-conserved sequence patterns; in earlier work, some false positives that are functionally unrelated and lack well-defined motifs appear to have been included (3). Thus, we define specific sequence motifs for each family and subfamily of RNases, which also should aid in cataloging newly-identified exoribonucleases. Detailed sequence alignments are shown only for those superfamilies that have not been described previously or for which significant new information has become available. It will be of interest to determine whether new RNases all fall into the groupings described here or whether introduction of additional enzyme superfamilies will be necessary.
An overview of the six exoribonuclease superfamilies is presented in Table 1. These groupings, which are discussed in detail below, consist of the RNR, DEDD, RBN, PDX, RRP4 and 5PX families, and a variety of subfamilies. The identities of the E.coli and Saccharomyces cerevisiae family members are detailed in Table 1, as are their distributions in other organisms and their catalytic features. As can be seen, one of the superfamilies, RBN, is restricted to eubacteria, whereas two other superfamilies, RRP4 and 5PX, are absent from eubacteria, but are found in eukaryotes and archaea or in eukaryotes, respectively. In addition, many of the subfamilies displayed limited phylogenetic distribution. Interestingly, this information indicates that eubacteria and archaea lack 5′ to 3′ exoribonucleases.
Table 1. Summary of exoribonuclease superfamilies.
 |
|
RNR FAMILY
Members of the RNR family, exemplified by E.coli RNase II and RNase R, are non-specific, processive, 3′ to 5′ exoribonucleases (4,5). The enzymes display maximal activity with both a monovalent cation and Mg2+, releasing nucleoside 5′ monophosphates. In addition to the E.coli enzymes, only a few other members of the family have been shown by direct assay to be exoribonucleases. These are the YvaJ (RNR_BACSU) gene product of Bacillus subtilis (6) and Rrp44p of S.cerevisiae (7). However, in view of the high degree of sequence conservation among the many members of this family (see below), it is likely that additional ones will also turn out to be RNases. In fact, a number of other members of the family are known to be involved in RNA metabolic events (8–11).
RNR family members are widely distributed among eubacteria and eukaryotes, but they have not been seen in any archaea except Halobacterium NRC-1. Among sequenced genomes of eubacteria, all lineages show the existence of RNR members except for the Actinobacteria and Borrelia. Interestingly, of the eight known bacterial exoribonucleases, Mycoplasma contain only RNase R. This may make RNase R a worthwhile target for anti-mycoplasma drugs. While γ division Proteobacteria often contain both RNase II and RNase R homologs, outside of this lineage, the single family member present more closely resembles RNase R than RNase II. In B.subtilis, for example, no RNase II activity is present (12), and the single RNR family member is a homolog of RNase R (37% identity) (6). In contrast, eukaryotic species normally contain three or more RNR family members (the S.cerevisiae members are shown in Table 1).
Enzymes of the RNR family are all large polypeptides with multiple domains. However, the members can be grouped into several subfamilies related either to eubacterial RNase II or RNase R, or to each of the S.cerevisiae proteins, Rrp44p, Msu1p and Ssd1p. Thus, while N-terminal sequences are conserved within subfamilies, they may be quite variable in both length and sequence from one subfamily to another. On the other hand, based on sequence alignments and secondary structure predictions, all RNR family members contain a C-terminal S1 RNA-binding domain (13). In the central region of the RNR proteins a highly conserved stretch of ∼400 amino acids is present which contains four prominent sequence motifs. These motifs, shown in Figure 1, have been noted previously (2,14), and it was suggested that motif IV could be considered an RNase II signature (2). In fact, motifs II and IV are highly conserved among RNR members, whereas motifs I and III are less conserved in a number of prokaryotic and eukaryotic members. At present, nothing is known about the relation of these motifs to exoribonuclease activity.
Figure 1.
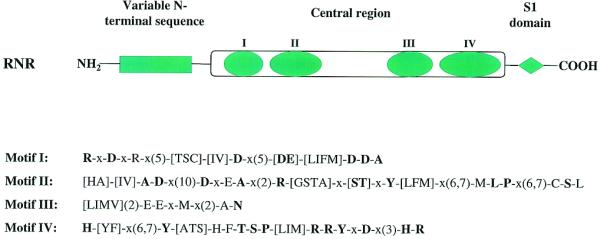
Schematic representation of the structure of RNR family proteins. RNR proteins normally contain a variable N-terminal sequence which is conserved within subfamilies, a conserved central region featuring four conserved sequence motifs, and a C-terminal S1 RNA-binding domain. The conserved sequence patterns of the four motifs in the central region are also shown. The syntax of the patterns follows that used in PHI-BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/pattern.html). Those residues conserved in >80% of the analyzed sequences are presented in bold lettering. Motif IV, which contains a long stretch of highly conserved or invariant residues, was designated as an RNase II signature in Prosite (http://expasy.cbr.nrc.ca/cgi-bin/nicedoc.pl?PDOC00904).
DEDD FAMILY
Exoribonucleases in this family actually are part of a much larger exonuclease superfamily, which includes the proofreading domains of many DNA polymerases as well as other DNA exonucleases (3,15). The proteins of this superfamily have a characteristic core comprised of four invariant acidic amino acids plus several other conserved residues distributed in three separate sequence motifs (Fig. 2) (15). We have named this the DEDD family after the four invariant acidic residues. The nucleases of this superfamily may share a common catalytic mechanism characterized by the involvement of two metal ions (16).
Figure 2.
Sequence conservation patterns in the DEDD family. This abbreviated sequence alignment was generated by ClustalX with some editing based on BLAST searches. The focus is on the DEDD RNases, but some DEDD DNases are also listed for comparison. Sequences are from the NCBI non-redundant protein sequence database, unless otherwise stated. Emphasis has been placed on presenting a wide variety of sequences, rather than on completeness due to space limitations. Included are the eight E.coli DEDD family members: EX1_ECOLI, DNA exonuclease I (accession no. G462029); EXOX_ECOLI, DNA exonuclease X (G9789746); DP3E_ECOLI, DNA polymerase III ɛ subunit (G118805); RNT_ECOLI, RNase T (G266952); ORN_ECOLI, oligoribonuclease (G1730261); RND_ECOLI, RNase D (G133152); DPO1_ECOLI, DNA polymerase I (G118825); DPO2_ECOLI, DNA polymerase II (G118829); six S.cerevisiae DEDD RNases: ORN_YEAST, yeast oligoribonuclease (G1730818); REX1_YEAST, yeast RNA exonuclease 1 (G2131716); REX3_YEAST, yeast RNA exonuclease 3 (G6323136); REX4_YEAST, yeast RNA exonuclease 4 (G6324493); PAN2_YEAST, Pan2p subunit of yeast poly(A)-binding protein-dependent poly(A) nuclease (G1709565); RRP6_YEAST, yeast exosome component Rrp6p (G6324574); and proteins from other model organisms: EX1_HAEIN, H.influenzae DNA exonuclease I (G1169569); DP3E_AQUAE, Aquifex aeolicus DNA polymerase III ɛ subunit (G6014995); DPO3_BACSU, B.subtilis DNA polymerase III α chain (G118793); DP3E_HAEIN, H.influenzae DNA polymerase III ɛ subunit (G1169396); RNT_VIBCH, Vibrio cholerae RNase T (G9655469); RNT_BUCSP, Buchnera sp. RNase T (G10038870); RNT_XYLFA, Xylella fastidiosa RNase T (G9107281); RNT_PSEAE, Pseudomonas aeruginosa RNase T (G9949677); RNT_HAEIN, H.influenzae RNase T (G1173107); ORN_HAEIN, H.influenzae oligoribonuclease (G1176352); ORN_MYCTU, Mycobacterium tuberculosis oligoribonuclease (G7227904); ORN_HUMAN, human oligoribonuclease (G7227908); YAA4_SCHPO, S.pombe Pan2p homolog (G1175466); YPO4_CAEEL, C.elegans Pan2p homolog (G1730942); PAN2_DROME, Drosophila melanogaster Pan2p homolog (G7303975); PAN2_HUMAN, human Pan2p homolog (translated from AC023500 with confirmation from ESTs, missing the very C-terminus in G7662258); PARN_HUMAN, human DAN nuclease, a poly(A)-specific 3′ to 5′ exoribonuclease (G4505611); PARN_SCHPO, S.pombe DAN nuclease homolog (G7491557); PARN_CAEEL, C.elegans DAN nuclease homolog (G7505706); RND_HAEIN, H.influenzae RNase D (G1173094); RND_RICPR, Rickettsia prowazekii RNase D (G7467941); RND_MYCTU, M.tuberculosis Rv2681 protein, a RNase D homolog (G7477317); PMC2_HUMAN, human polymyositis-scleroderma overlap syndrome-related nucleolar 100 kDa protein (PM/Scl autoantigen P100) (G8928564); PMC2_DROME, D.melanogaster homolog of PM/Scl autoantigen P100 (G7299933); DPOL_ARCFU, Archaeoglobus fulgidus DNA polymerase (G3122019); DPOD_HUMAN, human DNA polymerase δ catalytic chain (G118839); DPOE_HUMAN, human DNA polymerase ɛ catalytic subunit A (G1352309); EGL_DROME, D.melanogaster egl gene product (G7291631); WRN_HUMAN, human Werner syndrome helicase (G6136393); RND_SYNY3, Synechocystis hypothetical protein (G1001530); RP422_RICPR, R.prowazekii hypothetical protein RP422 (G7467752). The three conserved DEDD motifs are indicated at the top. Highly conserved residues among all family members are highlighted in red. The star at the top marks the fifth highly-conserved acidic residue (highlighted in red) between motifs II and III. Number of residues from the N-termini or residues between conserved blocks are indicated in parentheses. Residues that are highly conserved only within a subfamily are highlighted in blue. Yellow squares highlight some of the characteristic sequence motifs of subfamilies such as two of the three positively-charged, aromatic motifs specific to RNase T proteins, the sequence motif around the fifth conserved acidic residue in oligoribonucleases, and the characteristic motif III of RNase D.
As new groups of proteins have been added to the superfamily and as mutagenesis studies have confirmed their importance, the three characteristic sequence motifs and the four conserved acidic residues, first identified in DNA polymerases (15), have become better defined (Fig. 2). Several years ago, a detailed computer analysis identified additional family members, although some may be false positives (3). Besides the four invariant acidic residues present in the three motifs, a fifth acidic residue is also highly conserved between motifs II and III (17), and this has led to some confusion as to the identity of motif III (18,19). However, based on the structure of DNA polymerase (20), this fifth acidic residue is not at the active site and does not interact directly with the substrate, although it may affect exonuclease activity by interacting with residues in motif II. In fact, mutation of the corresponding residues in B.subtilis DNA polymerase III (17), in bacteriophage PRD1 DNA polymerase (21) and in E.coli RNase T (22; data not shown) completely eliminates the exonuclease activity. We have defined motif III as originally described (15).
Conservation patterns of these motifs may vary among different subfamilies. For example, in motif III these variations fall into two major groups. Instead of the conserved Y-x(3)-D sequence originally identified, many family members are now known to have a H-x(4)-D sequence. This variant of motif III was seen previously in some proteins, and was called the exo IIIɛ motif after the founding member, the E.coli DNA polymerase III ɛ subunit (17). Here, we use the names DEDDh and DEDDy to distinguish the many examples of these two motif III variants. The significance of the difference between a histidine and a tyrosine residue is still unclear. The conserved tyrosine residue has been shown in the Klenow fragment to help direct an activated H2O molecule for nucleophilic attack at the terminal phosphodiester bond (23). Thus, replacement of histidine with tyrosine would be expected to have a profound effect on catalytic activity. However, it cannot be responsible for the difference between DNases and RNases since both classes of enzymes are found in the DEDDh and DEDDy groups.
An abbreviated alignment of sample sequences from the DEDD superfamily is shown in Figure 2. Of the many subfamilies represented, we will focus on those known to be, or likely to be, exoribonucleases.
RNase D
RNase D is a 3′ to 5′ exoribonuclease that requires a divalent cation for activity (24). The E.coli enzyme participates in the maturation of small stable RNAs (25), and in vitro is active on denatured tRNA molecules and on tRNAs carrying residues following the CCA sequence (26). Members of the RNase D subfamily are DEDDy group exonucleases, but the proteins are not as highly conserved. For example, the Haemophilus influenzae protein shares only 41% sequence identity with its E.coli homolog. However, all RNase D subfamily members share a characteristic motif III with the sequence [DN]-W-x(2)-R-P-[LI]-x(6)-Y-A-x(2)-D (Fig. 2). Proteins of the RNase D subfamily are among the smaller members of the DEDDy group, containing ∼400 amino acids in bacteria and ∼800 in eukaryotes. Since the DEDD domain is only ∼150–200 amino acids in length, the role of a major portion of these proteins remains unknown.
Among sequenced bacterial genomes, RNase D homologs have been found only in Proteobacteria (α and γ divisions) and Mycobacteria. One protein in Synechocystis, RND_SYNY3, was annotated as an RNase D in GenBank (accession no. 1001530), but it is unlikely to be an RNase D ortholog. It is a much shorter protein with only the DEDD domain sequence, and it lacks the aforementioned RNase D motif III. RNase D-like proteins have not been found in any of the completed archaeal genomes. On the other hand, all eukaryotes seem to contain an RNase D subfamily member. Both the yeast exosome component RRP6p and the human PM-Scl 100 kDa autoantigen are close homologs of RNase D, and RRP6p has been shown to have exoribonuclease activity (27). Considering the phylogenetic distribution of RNase D (few in bacteria and universal in eukaryotes), its presence in bacteria that normally live in eukaryotic hosts, and the observation that the proteobacterial proteins are no more closely related to the mycobacterial ones than they are to those in eukaryotes, all suggest the possibility that RNase D has been horizontally transferred from eukaryotes to bacteria.
RNase T
RNase T, the second member of the DEDD family, is also a 3′ to 5′ exoribonuclease that requires a divalent cation for activity (28). The enzyme has many essential functions in E.coli including the 3′ maturation of small, stable RNAs and 23S rRNA, and it participates in the end turnover of tRNA (25,29,30). Yet, despite these many important roles, RNase T homologs have been found in only a small group of bacteria, the γ division of Proteobacteria. RNase T proteins are members of the DEDDh group of exonucleases. They are small polypeptides of ∼220 amino acids. The E.coli protein is known to function as an α2 dimer (31).
RNase T is closely related to the proofreading domains/subunits of bacterial DNA polymerase III (32) and, interestingly, E.coli RNase T also displays strong DNA exonuclease activity (33,34). RNase T subfamily members and DNA polymerase III proofreading domains/subunits share sequence patterns around the three DEDD exonuclease motifs (Fig. 2): motif I, [IVLF](3)-D-[TV]-E-T-[TA]-G; motif II, [IVLFA](3)-H-N-[AS]-x-F-D-x(2)-F; motif III, H-x-A-x(2)-D. Besides these common motifs, RNase T subfamily members share a number of other sequence features that distinguish them from DNA polymerase III. For example, R-F-R-G-[YF]-[YFL]-P immediately preceding DEDD motif I; [FY]-x(3)-[RF](2)-x(2)-K-x(3)-C-x-R-[GA] before DEDD motif II; and R-x(3)-K-R-x-P-F-H-P-F between motifs II and III. From this information it is apparent that RNase T homologs share a high degree of sequence identity. Modeling of RNase T on the known structure of the Klenow fragment (20), a DEDD family member, suggests that the three positively-charged aromatic motifs specific to RNase T proteins may cluster in the tertiary structure. It will be interesting to see if this prediction is confirmed by structural studies of RNase T currently underway. Also, considering that RNase T is confined to such a small group of bacteria, it is tempting to speculate that it may have originated from DNA polymerase III at the time the γ division Proteobacteria diverged from other bacteria.
Oligoribonuclease
Oligoribonuclease, the third member of the DEDD family, is a 3′ to 5′ exonuclease specific for small oligonucleotides that require a divalent cation for activity (35). It is the only one of the eight E.coli exoribonucleases to be required for growth, presumably because none of the other enzymes can act efficiently on its small substrates (36,37). In the absence of oligoribonuclease, small oligoribonucleotides 2–5 residues in length and derived from mRNA, accumulate (37). In Streptomyces oligoribonuclease is not essential, but its absence leads to slow growth (38). Interestingly, the human homolog is also able to hydrolyze small DNA oligomers (19). It appears that the ability to act on DNA may be a recurrent theme for DEDD exoribonucleases. The oligoribonucleases are members of the DEDDh family. They are small proteins of ∼200 amino acids, although slightly longer variants may be present in mitochondria (19). The E.coli protein, at least, is isolated as an α2 dimer (39).
Of the sequenced bacterial genomes, oligoribonuclease genes have been found only in Proteobacteria (β and γ divisions) and Actinomycetes. They have not been seen in archaea. In contrast, these genes are present in all the eukaryotic genomes examined. Oligoribonucleases are very highly conserved over their entire sequence. Any two of the proteins, regardless of their source, will share at least 40–50% identity. This observation strongly suggests that the bacterial enzymes may have arisen by horizontal transfer from their hosts. The high degree of sequence conservation is present throughout the DEDD domain and includes the following regions unique to oligoribonucleases (Fig. 2): motif I, [LIMV](2)-W-[IV]-D-[LC]-E-M-T-G-L; motif II, [IL]-A-G-N-S-[IV]-x(2)-D-x(2)-F-x(4)-M-P; motif III, K-x(3)-H-x-A-x(2)-D-I-x-E-S-[VI]-x-E-L-x(2)-Y-[RK]; and a sequence motif around the fifth conserved acidic residue between motifs II and III, Y-R-x-[LIV]-D-V-S-[ST]-[LIV]-K-x-L-x(2)-R-W-x-P. The functional significance of these motifs is unknown.
Other DEDD exoribonucleases
DAN nuclease, a poly(A)-specific 3′ to 5′ exoribonuclease was originally purified from calf thymus (40). The human version of this protein has also been cloned (PARN_HUMAN) (41). This protein is a member of the DEDDh family; however, it differs from other DEDDh proteins in that it contains a long insertion between motifs I and II (Fig. 2). DAN nuclease was originally claimed to be homologous to RNase D (41), but obviously this homology is very distant because RNase D is a member of the DEDDy group. DAN nuclease homologs have been found in many eukaryotes, e.g. Schizosaccharomyces pombe and Caenorhabditis elegans, but they are not present in S.cerevisiae.
Another DEDDh protein, the Pan2p subunit of the poly(A)-binding protein-dependent poly(A) nuclease from yeast was also suggested to be an exoribonuclease because of its homology with RNase T (42). However, Pan2p does not contain the RNase T specific sequence pattern, and it is a multidomain protein, in contrast to the single domain of RNase T (Fig. 2). Pan2p homologs may be present in every eukaryotic genome. While DAN nuclease and Pan2p are both large DEDDh proteins, they are not closely related to each other.
Many other DEDD domain-containing genes have been identified in eukaryotic cells, some of which may be exoribonucleases that do not fall into the RNase D, T or oligoribonuclease subfamilies. The most prominent of these are the Rex proteins (Fig. 2). Besides Rex2, which appears to be an oligoribonuclease ortholog, another three to four Rex genes (43) containing the DEDDh domain are present in each eukaryotic genome. They are closely related to each other, and also to Pan2 proteins. Various Rex proteins have been shown to be required for the processing of many RNA species (43), and in many respects they appear to function similarly to E.coli RNase T for stable RNA maturation. The Rex proteins have been placed in the RNase D family (43), but since they lack the DEDDy signature, such an assignment is not warranted.
RBN FAMILY
Of the six exoribonuclease superfamilies, the RNase BN family is the only one without identifiable homologs in eukaryotes. Moreover, except for E.coli RNase BN, nothing is known about the functions of these proteins, and even the E.coli enzyme is quite unusual (1). While it can act as a 3′ to 5′ exoribonuclease with a high degree of specificity for tRNAs with substitutions in the CCA sequence, it is most active with Co2+ as the divalent cation, it has a low pH optimum and it also requires a monovalent cation (44). RNase BN is essential for the 3′ maturation of certain phage-encoded tRNAs, but its role in the host is unknown (1). Although it is absent from eukaryotes and archaea, close homologs of RNase BN have been identified in many eubacterial species.
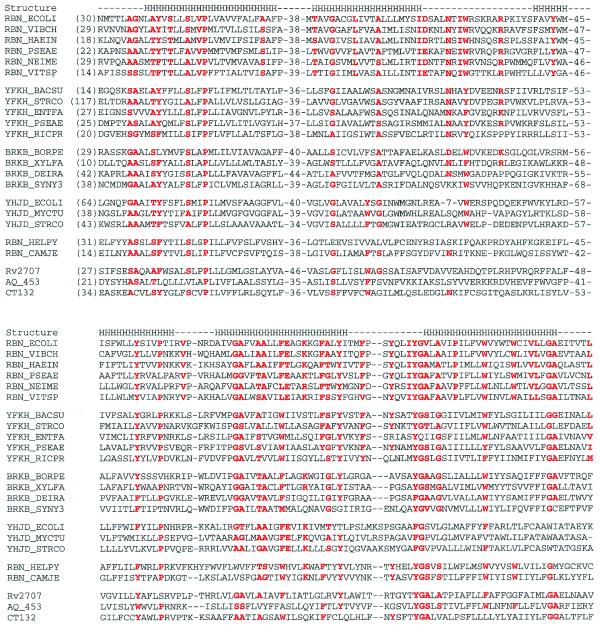
Unlike other exoribonucleases which normally contain multiple, invariant or highly conserved acidic residues needed for metal ion binding, such residues are not evident in the RBN family (Fig. 3). Instead, members of this family are hydrophobic proteins that are predicted to have similar secondary structures with a high helical content, and possibly containing six transmembrane helices (Fig. 3). In fact, one distant member of this family, BRKB_BORPE, was identified as a membrane protein (45). Among the closest members of the RBN family, a number of conserved sequence motifs can be identified. These are presented in Figure 3.
Figure 3.
Sequence comparisons of RBN family proteins. This abbreviated sequence alignment was generated by ClustalX with some editing based on BLAST searches. The sequences are from the NCBI non-redundant protein sequence database. The sequences included are: RBN_ECOLI, E.coli RNase BN (accession no. G418487); RBN_VIBCH, V.cholerae RNase BN (G9657340); RBN_HAEIN, H.influenzae RNase BN (G1176326); RBN_PSEAE, P.aeruginosa RNase BN (G9946857); RBN_NEIME, Neisseria meningitidis RNase BN (G7225749, G7379426); RBN_VITSP, Vitreoscilla sp. RNase BN (G3493604); YFKH_BACSU, B.subtilis transporter homolog yfkH (G7475937); YFKH_STRCO, Streptomyces coelicolor A3(2) yfkH homolog (G6425606); YFKH_ENTFA, Enterococcus faecalis yfkH homolog (G3608389); YFKH_PSEAE, P.aeruginosa yfkH homolog (G9948830); YFKH_RICPR, R.prowazekii hypothetical protein RP496 (G7467777); BRKB_BORPE, Bordetella pertussis brkB protein (G2120987); BRKB_XYLFA, X.fastidiosa brkB homolog (G9105274); BRKB_DEIRA, Deinococcus radiodurans brkB homolog (G7473422); BRKB_SYNY3, Synechocystis sp. (strain PCC 6803) brkB homolog (G7469712); YHJD_ECOLI, E.coli yhjD gene product (G586684); YHJD_MYCTU, M.tuberculosis hypothetical protein Rv3335c (G7477578); YHJD_STRCO, S.coelicolor A3(2) putative integral membrane protein (G7649576); RBN_HELPY, H.pylori hypothetical protein HP1407 (G7463952); RBN_CAMJE, Campylobacter jejuni putative RNase BN (G6968645); Rv2707, M.tuberculosis (strain H37Rv) hypothetical protein Rv2707 (G7477329); AQ_453, A.aeolicus hypothetical protein aq_453 (G7517605); CT132, Chlamidia trachomatis hypothetical protein CT132 (G7468636). The secondary structure noted by the letter H for helix is that predicted for E.coli RNase BN by TMHMM (http://www.cbs.dtu.dk/services/TMHMM-1.0/). All other superfamily members are predicted to have a similar secondary structure. Secondary structure predicted by PredictProtein (http://cubic.bioc.columbia.edu/predictprotein/) gave similar results, but with more helices at the N-terminus and the putative loop regions. Residues that are highly conserved in RNase BN and its homologs are highlighted in red. Corresponding residues in other subfamilies are also highlighted if they are conserved.
PDX FAMILY
Members of this family are distinguished by the fact that they are 3′ to 5′ phosphate-dependent exoribonucleases that release nucleoside diphosphates, rather than monophosphates, as products. The PDX family can be divided into two major groups, exemplified by PNPase and RNase PH. The former group consists of large, multidomain polypeptides, whereas the latter are generally smaller, single-domain proteins.
Close homologs of PNPase have been found in every sequenced bacterial genome except Mycoplasma. They are also present in plants as nuclear-encoded chloroplast proteins, but in these cases a signal peptide is present at the N-terminus (46–48). Interestingly, a PNPase homolog was recently found in the completed Drosophila genome (49), and it is also observed in the human sequence. It will be interesting to ascertain whether these gene products actually have PNPase activity. The PNPases are highly conserved proteins with even the most divergent sequences sharing 30–40% identical residues. In fact, the phylogenetic tree generated from PNPase sequences may well represent evolutionary distances among species. PNPase polypeptides contain at least four domains, each of them highly conserved (Fig. 4). The N-terminal domain of ∼300 amino acids is of unknown function. The central region contains the putative PDX catalytic domain shared by all PDX family members. Both KH and S1 RNA binding domains are found in the C-terminal region. The structure of each of these RNA binding domains has been determined (13,50).
Figure 4.
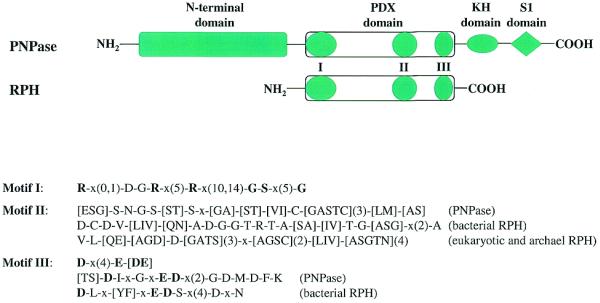
Schematic representation of the structure of PDX family proteins. Besides the PDX domain, PNPases contain extra domains at both termini: a highly conserved N-terminal domain of unknown function, and two C-terminal RNA-binding domains, KH and S1. Both KH and S1 RNA-binding domains are highly conserved among PNPases. Other PDX family members (RPH homologs) normally contain only a single highly-conserved PDX domain characterized by three conserved sequence motifs. The sequence patterns of these three motifs are shown including some specific patterns for subfamilies. The syntax of the patterns follows that used in PHI-BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/pattern.html). Those residues conserved in >80% of the analyzed sequences are shown in bold type.
RNase PH homologs are widely distributed in all kingdoms. In fact, of the eight known bacterial exoribonucleases, RNase PH is the only one with close homologs in the archaea. Despite this wide distribution, several eubacterial species within the Mycoplasma, Spirochetes, Chlamydia and also Helicobacter pylori lack the enzyme. Likewise, while most archaeal genomes contain two tandem RNase PH homologs, they are absent from Methanococcus jannashii and Halobacterium NRC-1. In eukaryotes, the number of RNase PH homologs expands to as many as six per genome. Small PDX members from various sources can be grouped into subfamilies, e.g. the bacterial ones, some eukaryotic and archaeal, and others. They all contain a single domain of 200–400 amino acids that in most cases spans the entire length of the polypeptide.
The presence of a common PDX domain among all family members was suggested previously (Fig. 4) (2); it contains three characteristic motifs. At the N-terminus is motif I: R-x(0,1)-D-G-R-x(5)-R-x(10,14)-G-S-x(5)-G. This motif is positively-charged and may bind the phosphate substrate and thereby define the exoribonuclease activity of this superfamily. At the C-terminal end of the PDX domain is acidic motif III which has the core sequence D-x(4)-E-[DE] with additional conserved residues in the PNPase and bacterial RPH subfamilies (Fig. 4). Between motifs I and III is a variable motif II, which is rich in small amino acids, but differs among subfamilies as shown in Figure 4.
RRP4 FAMILY
Mutations of the S.cerevisiae Rrp4 gene were found to lead to a defect in the 3′ maturation of 5.8S rRNA and to cessation of growth (51). Additional studies of this protein and its involvement in 5.8S RNA processing led to the discovery of the exosome, a eukaryotic complex of multiple exoribonucleases (7). Moreover, direct assay of a his-tagged form of Rrp4p showed that it has a distributive 3′ to 5′ exoribonuclease activity (6). Database searching revealed that Rrp4p is homologous to another exosome component, Rrp40p, and iterative searches with PSI-BLAST indicated that Rrp4p is also related to a third exosome component, Cs14p. These proteins and their homologs constitute an additional exoribonuclease superfamily.
Rrp4p orthologs probably exist in all eukaryotic species. In fact, the human counterpart can complement growth of yeast Rrp4 mutants (6), indicating their functional relationship. Rrp40p and Cs14p homologs are also seen in all the sequenced eukaryotic genomes. In contrast, while archaea have Rrp4p and Cs14p homologs, they appear to lack Rrp40 proteins. No close homologs of any RRP4 superfamily member can be identified in eubacteria.
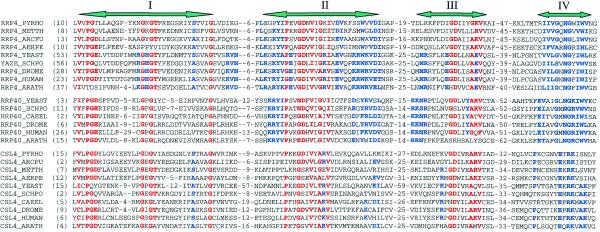
A multiple sequence alignment of the RRP4 superfamily is shown in Figure 5. Three subfamilies can be distinguished, exemplified by the yeast proteins, Rrp4p, Rrp40p and Csl4p. Proteins of this superfamily are distinguished by four sequence motifs which may vary among the subfamilies. Motif I, at the N-terminus of all three subfamilies, contains a P-G-[DE] sequence and several highly-conserved glycine residues. Motif II, which is highly conserved between the Rrp4 and Rrp40 families, is characterized by the sequence Y-x-[PG]-x(2)-[GN]-D-x-[LIV](2)-[GA]-x(9)-[WYF]-x-[LIV]-[DE]. This sequence is only partially conserved in the Csl4 subfamily. Motif III features a GD doublet shared by all the subfamilies. Motif IV, at the C-terminus, shows the sequence pattern [GAS]-N-G-x(2)-W in the Rrp4 and Rrp40 families, but E-x-R-K in the Csl4 family. PSI-BLAST searches suggest that motifs II and III may form an S1-like domain in the center of the proteins.
Figure 5.
Sequence comparisons of RRP4 family proteins. This abbreviated sequence alignment was generated by ClustalX with some editing based on BLAST searches. The sequences are from the NCBI non-redundant protein sequence database unless noted. Due to space limitations, only sequences from completed genomes and some model organisms are included here: RRP4_YEAST, S.cerevisiae exosome component Rrp4p (accession no. G6321860); YA2E_SCHPO, S.pombe rrp4p (G1175376); RRP4_DROME, D.melanogaster Rrp4p (G7291605); RRP4_HUMAN, human Rrp4p (alternative splicing product from U07561, missing 30 amino acids in G7657528); RRP4_ARATH, Arabidopsis thaliana Rrp4p homolog (G3850568); RRP4_PYRHO, Pyrococcus horikoshii Rrp4p homolog (G7519189); RRP4_METTH, Methanobacterium thermoautotrophicum Rrp4p homolog (G7482237); RRP4_ARCFU, A.fulgidus Rrp4p homolog (G7483018); RRP4_AERPE, Aeropyrum pernix Rrp4p homolog (G7516506); RRP40_YEAST, S.cerevisiae exosome component Rrp40p (G6324430); RRP40_SCHPO, S.pombe Rrp40p (G7490257); RRP40_CAEEL, C.elegans Rrp40p (G7504749); RRP40_DROME, D.melanogaster Rrp40p (translated from AE003585); RRP40_HUMAN, human Rrp40p (G8927588); RRP40_ARATH, A.thaliana Rrp40p homolog (alternative splicing product from AC006300); CSL4_YEAST, S.cerevisiae exosome component Cs14p (G1730832); CSL4_SCHPO, S.pombe Cs14p (G7491862); CSL4_CAEEL, C.elegans Cs14p (G7509960); CSL4_DROME, D.melanogaster Cs14p (G7297825); CSL4_HUMAN, human Cs14p (G7705612); CSL4_ARATH, A.thaliana Cs14p (translated from G2656024, missing C-term in G9758066); CSL4_PYRHO, P.horikoshii Cs14p homolog (G7429750); CSL4_ARCFU, A.fulgidus Cs14p homolog (G7429751); CSL4_METTH, M.thermoautotrophicum Cs14p homolog (G7429752); CSL4_AERPE, A.pernix Cs14p homolog (G7515775). The four sequence motifs are indicated at the top. Residues highly conserved among all RRP4 family members are highlighted in red, whereas those highlighted in blue indicate residues conserved within subfamilies. Numbers shown are the residues from the N-terminus or between blocks.
5PX FAMILY
Nucleases in this family are distinguished by their catalytic properties, degrading RNA to 5′ mononucleotides in a 5′ to 3′ direction. There are two known members of this family in a eukaryotic cell, termed Xrn1 and Rat1 in S.cerevisiae. Xrn1p and Rat1p are functionally interchangeable exoribonucleases, but >90% of Xrn1p is located in the cytoplasm, while Rat1p, at only 5–10% of the level of Xrn1p, is localized to the nucleus (52,53). At the sequence level, even though highly homologous, Xrn1p and Rat1p are distinguishable in that the former has long C-terminal sequences. At the very N-terminus there are also some subtle, but prominent, sequence differences between Xrn1p and Rat1p.
Both Xrn1p and Rat1p are conserved throughout eukaryotes, although Xrn1p may be absent from higher plants (54). However, close homologs of these 5′ exoribonucleases have not been found in any sequenced eubacterial or archaeal genomes. The proteins of this family are all large and sequence alignments and protease sensitivity suggest several domains (55): two highly conserved, acidic N-terminal domains and two basic C-terminal domains, which are weakly conserved and are much longer in the Xrn1 proteins (Fig. 6). The acidic N-terminal domains are believed to be responsible for the exonuclease activity because they are common to both families. Multiple sequence alignment of the N-terminal domains of Xrn1p and Rat1p and their homologs indicates that the N1 domain is more highly conserved than the N2 domain. For example, N1 domains from any two Xrn1 proteins or Rat1 proteins share >50% identity, and Xrn1 and Rat1 proteins share >40% sequence identity between their respective N1 domains. In contrast, the respective numbers for N2 domains are ∼40 and 30%. A phylogenetic tree based on the sequences in the N1 and N2 domains clearly shows that Xrn1p and Rat1p belong to two distinct subgroups. Interestingly, a virus protein EXO2_IRV6 (GenBank accession no. 5725643) does not fall into either group. EXO2_IRV6 contains only the N1 and N2 domains, and consequently is much shorter than either Xrn1 or Rat1.
Figure 6.
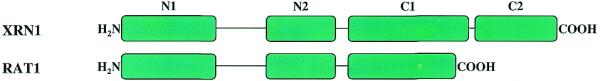
Schematic representation of the structure of 5PX family proteins. 5PX family members share two highly-conserved acidic N-terminal domains, N1 and N2, whereas the C-terminus is not conserved between Xrn1 and Rat1 subfamilies. The longer Xrn1 subfamily members have two basic C-terminal domains, C1 and C2. C1 is weakly conserved among Xrn1 proteins, whereas C2, which is proline-rich, is barely conserved.
Mutational analysis confirms that the N1 domain is important for the 5′ to 3′ RNase and DNase activities in 5PX proteins (55,56). Thus, a screen for non-complementing missense mutations of Xrn1 identified 14 single point mutations that reduced or abolished exonuclease activity. Each of these point mutations altered a highly conserved amino acid within the N1 domain (55). Also, these ‘important’ residues are among the limited number of invariant residues conserved in the virus protein EXO2_IRV6.
It is logical to think that there may be a relationship between these 5′ to 3′ exoribonucleases and exonucleases acting on DNA. However, it has not been possible to link Xrn1p and Rat1p to these other 5′ to 3′ exonucleases using the commonly available algorithms. In addition, a relationship between the 5PX exoribonucleases and other 5′ to 3′ exonucleases was not supported by secondary structure analysis.
CONCLUSIONS
Eight distinct exoribonucleases have been identified in E.coli, although many of them appear to overlap functionally. In contrast, many fewer exoribonucleases seem to exist in other eubacteria except for the γ division Proteobacteria, the close relatives of E.coli. In fact, some species seem to make do with only one exoribonuclease, e.g., only RNase R is found in Mycoplasma, and only PNPase is present in Borrelia burgdorferi. How the many RNA metabolic processes are handled in these organisms remains to be determined.
In archaea, the putative exoribonucleases present include two RNase PH homologs in tandem in each genome, and Rrp4p and Cs14p homologs. However, none of these four genes was found in the complete M.jannaschii or Halobacterium NRC-1 genomes. On the other hand, Halobacterium does contain a compact version of RNase R lacking an S1 domain. Like eubacteria, 5′ to 3′ exoribonucleases are not recognizable in archaea, raising many interesting questions regarding the evolution and functions of this group of nucleases.
Not surprisingly, eukaryotes have the largest arsenal of exoribonucleases. Of the functionally-characterized bacterial exoribonucleases, only RNase BN and RNase T do not have close homologs in eukaryotes. Some of the exoribonuclease superfamilies are greatly expanded in eukaryotes, e.g., at least five PDX members and three RNR members are present in a eukaryotic cell. The DEDD family is even more astonishing as there are more than 10 members in a eukaryotic cell. However, it is not clear how many of the eukaryotic DEDD members are exoribonucleases. We have raised the possibility that both the bacterial RNase D and oligoribonuclease may have been horizontally transferred into those organisms from eukaryotes.
There is very limited sequence homology among the exoribonuclease superfamilies except, perhaps, for some RNA-binding domains, such as the S1 RNA-binding domain. However, most exoribonucleases contain multiple invariant or highly-conserved acidic residues, which might be involved in metal ion-binding, consistent with the fact that all the known exoribonucleases require divalent cations for their activity. Two metal ion catalysis is probably a common feature of exonucleases (16).
The question also arises as to how exoribonucleases distinguish RNA molecules from DNA molecules. No easy answer to this question is evident from the sequence analysis. Theoretically, any exoribonuclease recognizing single-stranded RNA might also act on single-stranded DNA to some degree, and this idea is confirmed by the findings that RNase T and oligoribonuclease are also DNA exoribonucleases, and that yeast Xrn1p also has DNase activity. Moreover, it is known that the DEDD superfamily contains both RNases and DNases. It will be interesting to examine whether there are any additional examples of such cross-specificity among exoribonuclease superfamilies.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grant GM16317 from the National Institutes of Health.
References
- 1.Deutscher M.P. and Li,Z. (2000) Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acids Res. Mol. Biol., 66, 67–105. [DOI] [PubMed] [Google Scholar]
- 2.Mian I.S. (1997) Comparative sequence analysis of ribonucleases HII, III, II, PH and D. Nucleic Acids Res. 25, 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moser M.J., Holley,W.R., Chatterjee,A. and Mian,I.S. (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res., 25, 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen V. and Schlessinger,D. (1982) RNase I, II and IV of Escherichia coli. In Boyer,P.D. (ed.), The Enzymes, vol. XV part B. Academic Press, New York, pp. 501–515.
- 5.Kasai T., Gupta,R.S. and Schlessinger,D. (1977) Exoribonucleases in wild type Escherichia coli and RNase II-deficient mutants. J. Biol. Chem., 252, 8950–8956. [PubMed] [Google Scholar]
- 6.Oussenko I.A. and Bechhofer,D.H. (2000) The yvaJ gene of Bacillus subtilis encodes a 3′-to-5′ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J. Bacteriol., 182, 2639–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 8.Garriga G., Bertrand,H. and Lambowitz,A.M. (1984) RNA splicing in Neurospora mitochondria: nuclear mutants defective in both splicing and 3′ end synthesis of the large rRNA. Cell, 36, 623–634. [DOI] [PubMed] [Google Scholar]
- 9.Dobinson K.F., Henderson,M., Kelley,R.L. Collins,R.A. and Lambowitz,A.M. (1989) Mutations in the nuclear gene cyt-4 of Neurospora crassa result in pleiotropic defects in processing and splicing of mitochondrial RNAs. Genetics, 123, 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luukkonen B.G. and Seraphin,B. (1999) A conditional U5 snRNA mutation affecting pre-mRNA splicing and nuclear pre-mRNA retention identifies SSD1/SRK1 as a general splicing mutant suppressor. Nucleic Acids Res., 27, 3455–3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dziembowski A., Malewicz,M., Minczuk,M., Golik,P., Dmochowska,A. and Stepien,P.P. (1998) The yeast nuclear gene DSS1, which codes for a putative RNAse II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol. Gen. Genet., 260, 108–114. [DOI] [PubMed] [Google Scholar]
- 12.Deutscher M.P. and Reuven,N.B. (1991) Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl Acad. Sci. USA, 88, 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bycroft M., Hubbard,T.J., Proctor,M., Freund,S.M. and Murzin,A.G. (1997) The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell, 88, 235–242. [DOI] [PubMed] [Google Scholar]
- 14.Uesono Y., Toh-e,A. and Kikuchi,Y. (1997) Ssd1p of Saccharomyces cerevisiae associates with RNA. J. Biol. Chem., 272, 16103–16109. [DOI] [PubMed] [Google Scholar]
- 15.Bernad A., Blanco,L., Lazaro,J., Martin,G. and Salas,M. (1989) A conserved 3′–5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell, 59, 219–228. [DOI] [PubMed] [Google Scholar]
- 16.Steitz T.A. and Steitz,J.A. (1993) A general two metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA, 90, 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnes M.H., Spacciapoli,P., Li,D.H. and Brown,N.C. (1995) The 3′–5′ exonuclease site of DNA polymerase III from gram-positive bacteria: definition of a novel motif structure. Gene, 165, 45–60. [DOI] [PubMed] [Google Scholar]
- 18.Koonin E.V. (1997) A conserved ancient domain joins the growing superfamily of 3′–5′ exonucleases. Curr. Biol., 7, R604–R606. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen L.H., Erzbergr,J.P., Root,J. and Wilson,D.M.,III (2000) The human homolog of Escherichia coli orn degrades small single-stranded RNA and DNA oligomers. J. Biol. Chem., 275, 25900–25906. [DOI] [PubMed] [Google Scholar]
- 20.Brautigam C.A., Sun,S., Piccirilli,J.A. and Steitz,T.A. (1999) Structures of normal single-stranded DNA and deoxyribo-3′-S-phosphorothiolates bound to the 3′–5′ exonucleolytic active site of DNA polymerase I from Escherichia coli. Biochemistry, 38, 696–704. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W. and Ito,J. (1995) Family A and family B DNA polymerases are structurally related: evolutionary implications. Nucleic Acids Res., 22, 5177–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S. and Deutscher,M.P. (1992) Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J. Biol. Chem., 267, 25609–25613. [PubMed] [Google Scholar]
- 23.Beese L.S. and Steitz,T.A. (1991) Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J., 10, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cudny H., Zaniewski,R. and Deutscher,M.P. (1981) Escherichia coli RNase D. Purification and structural characterization of a putative processing nuclease. J. Biol. Chem., 256, 5627–5632. [PubMed] [Google Scholar]
- 25.Li Z., Pandit,S. and Deutscher,M.P. (1998) 3′ exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl Acad. Sci. USA, 95, 2856–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cudny H., Zaniewski,R. and Deutscher,M.P. (1981) Escherichia coli RNase D: catalytic properties and substrate specificity. J. Biol. Chem., 256, 5633–5637. [PubMed] [Google Scholar]
- 27.Burkard K.T. and Butler,J.S. (2000) A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with poly(A) polymerase and the hnRNA protein Np13p. Mol. Cell. Biol., 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutscher M.P. and Marlor,C.W. (1985) Purification and characterization of Escherichia coli RNase T. J. Biol. Chem., 260, 7067–7071. [PubMed] [Google Scholar]
- 29.Li Z., Pandit,S. and Deutscher,M.P. (1999) Maturation of 23S ribosomal RNA requires the exoribonuclease RNase T. RNA, 5, 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deutscher M.P., Marlor,C.W. and Zaniewski,R. (1985) RNase T is responsible for the end-turnover of tRNA in Escherichia coli. Proc. Natl Acad. Sci. USA, 82, 6427–6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang S. and Deutscher,M.P. (1992) Sequence and transcriptional analysis of the Escherichia coli rnt gene encoding RNase T. J. Biol. Chem., 267, 25609–25613. [PubMed] [Google Scholar]
- 32.Koonin E.V. and Deutscher,M.P. (1993) RNase T shares conserved sequence motifs with DNA proofreading exonucleases. Nucleic Acids Res., 21, 2521–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswanathan M., Dower,K.W. and Lovett,S.T. (1998) Identification of a potent DNase activity associated with RNase T of Escherichia coli. J. Biol. Chem., 273, 35126–35131. [DOI] [PubMed] [Google Scholar]
- 34.Zuo Y. and Deutscher,M.P. (1999) The DNase activity of RNase T and its application to DNA cloning. Nucleic Acids Res., 27, 4077–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niyogi S.K. and Datta,A.K. (1975) A novel oligoribonuclease of Escherichia coli. I. Isolation and properties. J. Biol. Chem., 250, 7307–7312. [PubMed] [Google Scholar]
- 36.Yu D. and Deutscher,M.P. (1995) Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J. Bacteriol ., 177, 4137–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh S. and Deutscher,M.P. (1999) Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl Acad. Sci. USA, 96, 4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohnishi Y., Nishiyama,Y., Sato,R., Kameyama,S. and Horinouchi,S. (2000) An oligoribonuclease gene in Streptomyces griseus. J. Bacteriol., 182, 4647–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Zhu,L. and Deutscher,M.P. (1998) Oligoribonuclease is encoded by a highly conserved gene in the 3′–5′ exonuclease superfamily. J. Bacteriol., 180, 2779–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korner C.G. and Wahle,E. (1997) Poly(A) tail shortening by a mammalian poly(A)-specific 3′ exoribonuclease. J. Biol. Chem., 272, 10448–10456. [DOI] [PubMed] [Google Scholar]
- 41.Korner C.G., Wormington,M., Muckenthaler,M., Schneider,S., Dehlin,E. and Whale,E. (1998) The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J., 17, 5427–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boeck,R., Tarun,S.,Jr, Rieger,M., Deardorff,J.A., Muller-Auer,S. and Sachs,A.B. (1996) The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J. Biol. Chem., 271, 432–438. [DOI] [PubMed] [Google Scholar]
- 43.van Hoof A., Lennertz,P. and Parker,R. (2000) Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J., 19, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Callahan C., Neri-Cortes,D. and Deutscher,M.P. (2000) Purification and characterization of the tRNA-processing enzyme RNase BN. J. Biol. Chem., 275, 1030–1034. [DOI] [PubMed] [Google Scholar]
- 45.Fernandez R.C. and Weiss,A.A. (1994) Cloning and sequencing of a Bordetella pertussis serum resistance locus. Infect. Immun., 62, 4727–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayes R., Kudla,J., Schuster,G., Gabay,L., Maliga,P. and Gruissem,W. (1996) Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J., 15, 132–141. [PMC free article] [PubMed] [Google Scholar]
- 47.Lisitsky I., Kotler,A. and Schuster,G. (1997) The mechanism of preferential degradation of polyadenylated RNA in the chloroplast. The exoribonuclease 100RNP/polynucleotide phosphorylase displays high binding affinity for poly(A) sequence. J. Biol. Chem., 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- 48.Li Q.S., Gupta,J.D. and Hunt,A.G. (1998) Polynucleotide phosphorylase is a component of a novel plant poly(A) polymerase. J. Biol. Chem., 273, 17539–17543. [DOI] [PubMed] [Google Scholar]
- 49.Adams M.D., Celniker,S.E., Holt,R.A., Evans,C.A., Gocayne,J.D., Amanatides,P.G., Scherer,S.E., Li,P.W., Hoskins,R.A., Galle,R.F., George,R.A., Lewis,S.E., Richards,S., Ashburner,M. et al. (2000) The genome sequence of Drosophila melanogaster. Science, 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 50.Musco G., Kharrat,A., Stier,G., Fraternali,F., Gibson,T.J., Nilges,M. and Pastore,A. (1997) The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome. Nature Struct. Biol., 4, 712–716. [DOI] [PubMed] [Google Scholar]
- 51.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 52.Heyer W.D., Johnson,A.W., Reinhart,U. and Kolodner,R.D. (1995) Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol. Cell. Biol., 15, 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poole T.L. and Stevens,A. (1995) Comparison of features of the RNase activity of 5′-exonuclease-1 and 5′-exonuclease-2 of Saccharomyces cerevisiae. Nucleic Acids Symp. Ser., 33, 79–81. [PubMed] [Google Scholar]
- 54.Kastenmayer J.P. and Green,P.J. (2000) Novel features of the XRN-family in Arabidopsis: evidence that ATXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc. Natl Acad. Sci. USA, 97, 13985–13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Page A.M., Davis,K., Molineus,C., Kolodner,R.D. and Johnson,A.W. (1998) Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res., 26, 3707–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solinger J.A., Pascolini,D. and Heyer,W.D. (1999) Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol. Cell. Biol., 19, 5930–5942. [DOI] [PMC free article] [PubMed] [Google Scholar]