ABSTRACT
Lactococcus lactis subsp. cremoris strains typically carry many dairy niche-specific adaptations. During adaptation to the milk environment these former plant strains have acquired various pseudogenes and insertion sequence elements indicative of ongoing genome decay and frequent transposition events in their genomes. Here we describe the reactivation of a silenced plant sugar utilization cluster in an L. lactis MG1363 derivative lacking the two main cellobiose transporters, PtcBA-CelB and PtcBAC, upon applying selection pressure to utilize cellobiose. A disruption of the transcriptional repressor gene llmg_1239 by an insertion sequence (IS) element allows expression of the otherwise silent novel cellobiose transporter Llmg_1244 and leads to growth of mutant strains on cellobiose. Llmg_1239 was labeled CclR, for cellobiose cluster repressor.
IMPORTANCE Insertion sequences (ISs) play an important role in the evolution of lactococci and other bacteria. They facilitate DNA rearrangements and are responsible for creation of new genetic variants with selective advantages under certain environmental conditions. L. lactis MG1363 possesses 71 copies in a total of 11 different types of IS elements. This study describes yet another example of an IS-mediated adaptive evolution. An integration of IS981 or IS905 into a gene coding for a transcriptional repressor led to activation of the repressed gene cluster coding for a plant sugar utilization pathway. The expression of the gene cluster allowed assembly of a novel cellobiose-specific transporter and led to cell growth on cellobiose.
KEYWORDS: Lactococcus lactis, cellobiose, plant, dairy, reverse evolution, IS element, sRNA, CclR, PTS, sugar metabolism
INTRODUCTION
Lactococcus lactis demonstrates an impressive genome plasticity and polymorphism. Various L. lactis strains can be found in many different environments, such as plants, milk, animals, and related niches (1, 2). Although a number of L. lactis strains have been “domesticated” and cultivated in milk for many years, thorough analyses of L. lactis genomes reveals the presence of multiple silent gene clusters and pseudogenes with predicted functions in plant sugar utilization as remnants of their plant niche-related history (2–7). Point mutations and genome rearrangements occasionally lead to activation of silent genes and gene clusters, sometimes even granting them new functions. Adaptive evolution in lactococcal strains occurs mostly by horizontal gene transfer and by intracellular activity of the mobilome (7, 8). Insertion sequences (IS) are the simplest type of mobile genetic elements. A typical IS element usually encodes one or two proteins necessary for its transposition and contains (near) perfect inverted repeats at its termini (8). The genome of Lactococcus lactis subsp. cremoris MG1363 contains 11 different types of IS elements and 71 copies in total. Recombination between identical IS elements and their ability to transpose from one location to another in a genome provide great genetic flexibility. IS elements have often been described to be involved in adaptation of L. lactis strains to unfavorable conditions during starvation or oxidative stress (8–10).
The metabolism of the plant disaccharide cellobiose by some lactococcal laboratory strains is a good example of an ongoing adaptive evolution process. After constant propagation under laboratory conditions, these strains lost the capability to efficiently take up and utilize cellobiose, and certain changes in their genomes are necessary to restore these deficiencies. L. lactis MG1363 can take up and utilize cellobiose when one of two dedicated phosphoenolpyruvate:carbohydrate phosphotransferase systems (PTSs) is functional. Activation of the PtcBA-CelB system in L. lactis MG1363 requires a celB promoter up-mutation (11), and PtcABC is assembled in NZ9000 (an isogenic derivative of MG1363) when a cre site in the promoter region of ptcC is disrupted and the gene becomes constitutively expressed (12; see Fig. 4). Besides cellobiose, PtcBA-CelB can transport lactose, while PtcABC usually imports glucose (13, 14).
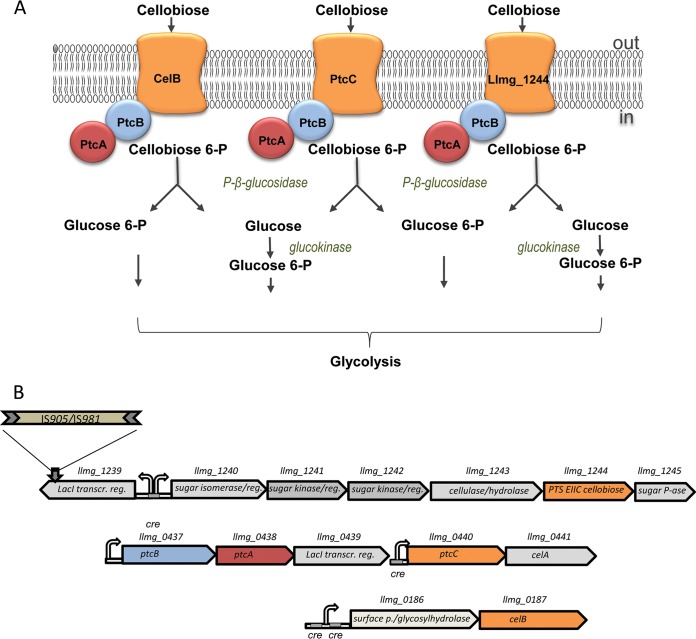
FIG 4.
Cellobiose-specific uptake and metabolism in L. lactis MG1363. (A) Three cellobiose-specific PTSs and subsequent metabolic routes for degradation of this sugar. P, phosphate. (B) Gene clusters containing cellobiose transporter genes. Cellobiose-specific EIIC component Llmg_1244 is expressed only when llmg_1239, coding for a transcriptional repressor, is disrupted.
We found that even after deletion of the membrane-integrated EII subunit C (EIIC) components of these two PTSs, namely, CelB and PtcC, L. lactis MG1363 was able to restore growth in a medium supplemented with cellobiose as the sole carbon source. Two types of IS elements were discovered that had integrated at different positions relative to a gene coding for a putative LacI-type transcriptional repressor in the Cel+ isolates. This gene is located upstream of a putative cellobiose-, beta-glucoside-, or lichenan-catabolic gene cluster. The aim of this study was to assess the effect of an IS integration in the putative repressor gene and to identify the silent cellobiose transporter responsible for the restored Cel+ phenotype of L. lactis MG1363 Cel− strains.
RESULTS
Cultivation of Cel− strains in cellobiose-containing medium leads to a Cel+ phenotype in L. lactis MG1363.
An L. lactis MG1363 derivative lacking the two main cellobiose transporters (PtcBA-CelB and PtcBAC), strain MGΔptcCΔcelB, does not grow in a chemically defined medium with cellobiose (CDM-Cel) as the sole carbon source for around 24 h. However, after a very long lag phase, an increase in culture optical density can be observed. Recently, van der Meulen et al. described Llmg_0963, which is annotated as a cellobiose-specific EIIC PTS component. A cellobiose-inducible small RNA (sRNA), LLMGnc_147, enhances expression of this protein (15). To find out whether expression of Llmg_0963 was responsible for the growth of MGΔptcCΔcelB in CDM-Cel, llmg_0963 was deleted in MGΔcelBΔptcC and growth of the triple mutant was monitored in CDM-Cel. After a lag phase of 24 h the strain started growing, demonstrating that Llmg_0963 does not play a role in the observed phenomenon (Fig. 1).
FIG 1.
(A to D) Growth of L. lactis MGΔcelBΔptcCΔllmg_0963 on M17 agar with or without cellobiose for 24 h. (A) Two types of colonies are formed by MGΔcelBΔptcCΔllmg_0963 on M17-Cel. (B) Growth of the same strain on solidified M17. (C) Only large colonies appear when a large colony from panel A is restreaked on solidified M17-Cel. (D) Two types of colonies appear when a small colony from panel A is restreaked on solidified M17-Cel. (E) Growth of L. lactis strains in CDM-Cel. Circles, MG1363 carrying a Pcel promoter up-mutation (growth rate [k] = 0.26 h−1); triangles, MGΔcelBΔptcC (k = 0.28 h−1); squares, MGΔcelBΔptcCΔllmg_0963 (k = 0.2 h−1). Growth curves show means for 3 replicates.
When MGΔcelBΔptcCΔllmg_0963 was streaked out on a rich M17 agar with cellobiose, two types of colonies appeared: small ones which were visible after overnight incubation and presumably grew on residual carbon sources in M17 and large ones (Fig. 1). The latter had (re)gained the ability to metabolize cellobiose and became distinguishable only after 24 h of incubation. When large colonies were restreaked on new M17-Cel agar plates, they retained the large colony Cel+ phenotype. When small colonies were restreaked, they again diversified into two phenotypes and formed small and large colonies. The inheritable Cel+ phenotype suggested that genetic changes were involved. In order to identify these changes, the whole genomes of colony-purified Cel+ strains with different genetic backgrounds (MGΔcelB and MGΔcelBΔptcCΔllmg_0963) were sequenced and compared to that of the parental strain, MG1363. For the purpose of the current study, a cellobiose-positive variant of MG1363 with a celB promoter up-mutation (Pcel+) was used (11). Cel+ isolates were picked from M17-Cel agar plates with L. lactis MGΔcelB and MGΔcelBΔptcCΔllmg_0963. The number of Cel+ colonies suggested that the genetic changes had occurred at very high frequencies, which pointed to possible involvement of a mobile genetic element(s).
Analyses of genome sequences of the Cel+ isolates revealed that, indeed, some genetic changes had occurred. Two types of IS elements, namely, IS981 and IS905N, had integrated in llmg_pseudo_39 of MGΔcelB and MGΔcelBΔptcCΔllmg_0963, respectively. In fact, the frameshift that was annotated in the sequence of llmg_pseudo_39 of L. lactis MG1363 (4) was not present in any of our sequenced strains, meaning that the gene is intact in these strains. The L. lactis MG1363 strain sequenced by Linares et al. did not contain the frameshift either, but the annotation of the gene was not changed (12). Since the gene encodes an intact protein of 348 amino acids in our strains, throughout this paper we label it llmg_1239, not pseudo_39. Sequence analyses of the PCR-amplified chromosomal regions of many other Cel− and Cel+ isolates revealed that the IS elements had integrated in llmg_1239 of Cel+ isolates only while all Cel− strains possessed an intact gene. The gene, which is identical to LLNZ_06380 of L. lactis NZ9000, codes for a transcriptional regulator of the LacI family and lies upstream of a putative plant sugar utilization gene cluster (Fig. 2). L. lactis NZ9000 is a derivative of MG1363 in which two genes (nisRK) were integrated into the chromosome as a part of the nisin-inducible gene expression system (16).
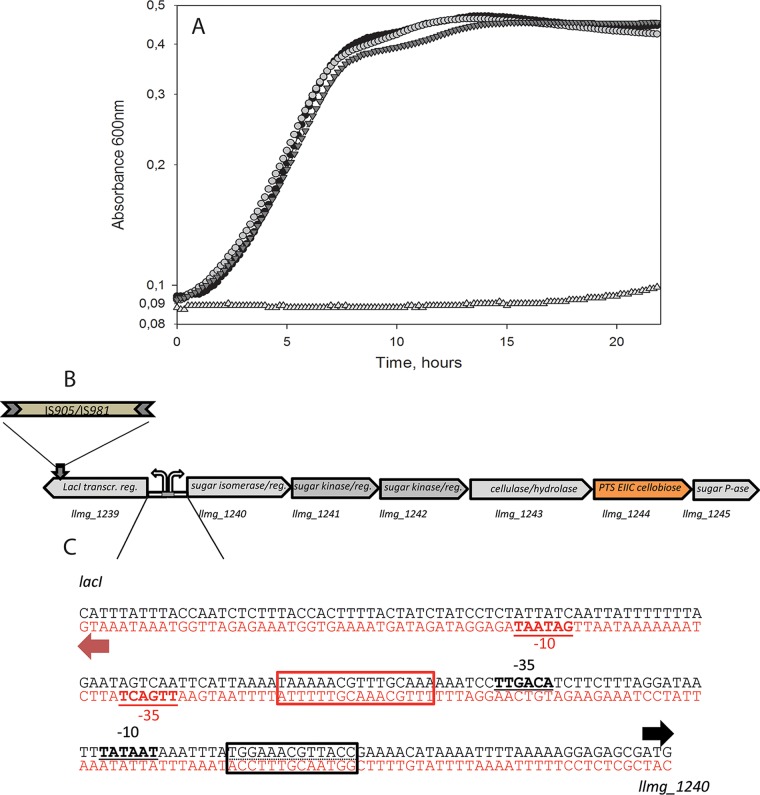
FIG 2.
Integration of IS/GFP into llmg_1239 and growth of L. lactis derivatives in CDM-Cel. (A) Black circles, MG1363 carrying a Pcel promoter up-mutation (k = 0.25 h−1); light gray circles, MGΔcelBΔptcCΔllmg_0963 llmg_1239::IS905 (M-IS) (k = 0.24 h−1); dark gray inverted triangles, MGΔcelBΔptcCΔllmg_0963 llmg_1239::gfp (M-GFP) (k = 0.23 h−1); light gray triangles, MGΔcelBΔptcCΔllmg_0963 (k = 0.0039 h−1). Growth curves show means for 3 replicates. (B) Genetic cluster llmg_1240-45 containing genes with predicted functions in sugar utilization and the predicted transcriptional repressor gene llmg_1239, located upstream. transcr. reg, transcription regulator; P-ase, phosphatase. (C) llmg_1239-llmg_1240 intergenic region. Translation start sites (ATG) are indicated by arrows, −10 and −35 sequences are underlined, and predicted operator sequences to which LacI family regulators can bind are shown in boxes. Black box, CcpA; red box, GalR of E. coli (note that L. lactis MG1363 does not possess GalR). The prediction was made using http://genome2d.molgenrug.nl.
Besides IS integration, one change in the promoter region of the catabolite control protein CcpA-encoding gene was discovered in the chromosomes of two Cel+ isolates. A transversion from an A to a T had occurred in the −10 region of the promoter, changing it from TATAAT into TATATT. The mutation might affect transcription of the ccpA gene, lowering the number of regulator molecules, since the changed −10 box is less similar to the consensus −10 sequence.
Disruption of llmg_1239 leads to a Cel+ phenotype of L. lactis MGCel−.
Closer analyses of the MGΔcelBΔptcCΔllmg_0963 Cel+ genome sequence showed that the integration of IS905N did not result in a premature translation stop but changed the last seven amino acids of the C terminus of the putative LacI family transcriptional regulator. These substitutions occurred in the putative ligand molecule binding domain. The changes in this domain, which most likely binds sugar, might affect the function of the regulator. MGΔcelB Cel+ colonies possessed a truncated version of the same regulator after the integration of IS981 into llmg_1239. The IS981 integration introduced a stop codon, leading to a truncated version of the putative regulator of 290 amino acids instead of the full-length 348-amino-acid protein. Again, the insertion only disrupted the ligand binding domain and left the putative DNA-binding helix-turn-helix motif at the N terminus of the protein intact. We hypothesized that integration of the IS element in llmg_1239 resulted in a relief of repression of an unknown sugar utilization cluster, which would allow transcription of a cellobiose-specific transporter and metabolic genes. Such Cel+ L. lactis mutants could be responsible for growth of the bacterial culture in cellobiose-containing medium and would form the larger colonies on M17-Cel plates.
To test the hypothesis that disruption of the putative LacI family regulator resulted in a Cel+ phenotype, a green fluorescent protein (GFP)-encoding gene was integrated in llmg_1239 of MGΔcelBΔptcCΔllmg_0963. To mimic the result of the IS integration, the first 179 amino acids and, thus, the DNA-binding domain were left intact and growth of the resulting strain in a medium with cellobiose was examined. Integration of gfp in llmg_1239 restored the ability of the strain to grow on cellobiose. MGΔcelBΔptcCΔllmg_0963llmg_1239::gfp (M-GFP) grew rapidly in both liquid and solid M17 supplemented with cellobiose, in contrast to the parental strain (Fig. 2). Integration of gfp into llmg_1239 resulted in a phenotype similar to that of an IS integration into the same region.
Transcriptome analyses hint at a novel PTS-Cel in L. lactis.
To assess the regulon of Llmg_1239 and to identify the unknown cellobiose transporter, the transcriptomes of L. lactis MG1363 (wild type [WT]), MGΔcelBΔptcCΔllmg_0963Cel+llmg_1239::IS905 (M-IS), and MGΔcelBΔptcCΔllmg_0963llmg_1239::gfp were compared to each other. Cells of all strains were cultured in rich M17 medium supplemented with 1% cellobiose. To ensure that cells were utilizing cellobiose, not the uncharacterized carbon sources in M17, they were harvested in the mid-exponential growth phase at an optical density at 600 nm (OD600) of 0.6 for total RNA isolation. One operon was clearly the most highly expressed in MGCel+ cells, harboring either an IS element or gfp in llmg_1239. The operon consists of six genes, the cluster from llmg_1240 to llmg_1245 (llmg_1240-45), and is located downstream of llmg_1239 (Fig. 3; Table 1).
FIG 3.
Comparison of the transcriptome of L. lactis M-IS (IS) and M-GFP (GFP) with that of MG1363 (WT) during growth in M17-Cel. Volcano plots show only the most highly upregulated genes.
TABLE 1.
Relevant genes that were differentially expressed in M-GFP and M-IS in comparison with the WTa
| Locus | Gene | Predicted function | Fold difference from WT |
|
|---|---|---|---|---|
| M-GFP | M-IS | |||
| llmg_1242 | llmg_1242 | Sugar kinase/transcriptional regulator | 52.5 | 20.7 |
| llmg_1240 | llmg_1240 | Sugar isomerase/transcriptional regulator | 22.0 | 12.8 |
| llmg_1244 | llmg_1244 | Cellobiose-specific PTS EIIC | 16.4 | 9.3 |
| llmg_1245 | llmg_1245 | Sugar phosphatase | 20.8 | 8.3 |
| llmg_1241 | llmg_1241 | Sugar kinase/transcriptional regulator | 14.3 | 7.3 |
| llmg_1243 | llmg_1243 | Cellulase/hydrolase | 8.1 | 2.9 |
| llmg_0437 | ptcB | Cellobiose-specific PTS EIIB | 2.2 | |
| llmg_0438 | ptcA | Cellobiose-specific PTS EIIA | 3.4 | |
| llmg_0439 | llmg_0439 | LacI family transcriptional regulator | 1.9 | |
| llmg_2311 | arcD1 | Arginine/ornithine antiporter | 9.1 | 2.2 |
| llmg_2310 | arcC1 | Carbamate kinase | 4.9 | 2.2 |
| llmg_2312 | arcB | Ornithine carbamoyltransferase | 4.3 | |
| llmg_2309 | arcC2 | Carbamate kinase | 2.7 | |
| llmg_2313 | arcA | Arginine deiminase | 2.4 | |
| llmg_1559 | flpA | Transcriptional regulator, Crp/Fnr family | 4.9 | |
| llmg_0456 | pgmB | Phosphoglucomutase | 3.0 | |
| llmg_0452 | treR | Trehalose operon transcriptional repressor | 2.1 | |
| LLMGnc_172 | sRNA | 13.3 | ||
| LLMGnc_008 | sRNA | 3.2 | ||
| LLMGnc_004 | 6S RNA | 2.4 | ||
| LLMGnc_131 | sRNA | 2.4 | ||
| LLMGnc_089 | sRNA | 2.3 | ||
| LLMGnc_133 | sRNA | 2.1 | ||
| LLMGnc_080 | sRNA | 2.1 | ||
| LLMGnc_139 | sRNA | 2.1 | ||
| LLMGnc_150 | sRNA | 2.0 | ||
| LLMGnc_002 | sRNA | 2.0 | ||
| LLMGnc_026 | sRNA | 2.0 | ||
| LLMGnc_032 | sRNA | 2.5 | ||
| LLMGnc_099 | sRNA | 2.2 | ||
| LLMGnc_044 | sRNA | 2.2 | ||
| LLMGnc_173 | sRNA | 2.2 | ||
| LLMGnc_150 | sRNA | 2.2 | ||
| LLMGnc_090 | sRNA | 2.1 | ||
| LLMGnc_134 | sRNA | 2.1 | ||
| LLMGnc_013 | sRNA | 2.0 | ||
| LLMGnc_056 | sRNA | 2.0 | ||
| LLMGnc_045 | sRNA | 2.0 | ||
| llmg_0190 | bglS | P-β-glucosidase BglS | −2.2 | |
| llmg_0188 | llmg_0188 | Function unknown | −5.1 | −5.1 |
| llmg_0189 | llmg_0189 | Function unknown | −6.6 | −9.6 |
| llmg_0186 | llmg_0186 | Surface protein/glycosylhydrolase | −22.7 | −12.4 |
| llmg_0187 | celB | Cellobiose-specific PTS EIIC | −14.4 | −20.8 |
| tnp904 | tnp904 | Transposase | −4.0 | −4.8 |
| tnp905 | tnp905 | Transposase | −4.2 | −4.1 |
| llmg_0550 | tnp891 | Transposase | −10.3 | −3.2 |
| llmg_0775 | ccpA | Catabolite control protein A | −1.4 | −2.8 |
| llmg_1165 | llmg_1165 | ABC-type sugar transport system | −17.0 | −7.3 |
| llmg_2539 | gapB | Glyceraldehyde 3-phosphate dehydrogenase | −9.5 | −10.9 |
| llmg_0099 | llmg_0099 | Ribosomal protein | −3.5 | −16.5 |
| llmg_1208 | llmg_1208 | Ribosomal protein | −15.0 | |
| llmg_2356 | llmg_2356 | Ribosomal protein | −6.8 | −9.1 |
| llmg_2357 | llmg_2357 | Ribosomal protein | −8.4 | −9.2 |
| llmg_2374 | llmg_2374 | Ribosomal protein | −6.4 | −6.6 |
| llmg_2366 | llmg_2366 | Ribosomal protein | −5.5 | |
| llmg_2365 | llmg_2365 | Ribosomal protein | −5.3 | −5.3 |
| llmg_1493 | llmg_1493 | Ribosomal protein | −6.0 | −9.5 |
| llmg_2354 | rpoA | RNA polymerase | −6.0 | −8.1 |
| llmg_2353 | llmg_2353 | Ribosomal protein | −4.1 | −10.1 |
Only significantly expressed genes are included (Bayesian P < 0.001).
A putative cellobiose-specific membrane-spanning PTS EIIC component is encoded by one of the genes in the operon, llmg_1244. Homologous proteins can be found only in L. lactis NZ9000, KW2, and IO-1, while similar transporters are encoded in Listeria monocytogenes genomes (60% nucleotide sequence identity). The latter are annotated as cellobiose-, beta-glucoside-, or lichenan-specific permeases. Less homology is observed with Enterococcus transporter genes (around 50% sequence similarity). Llmg_1240 contains a phosphosugar-binding domain found in sugar isomerases or regulatory proteins. Llmg_1241 is similar to a ROK (repressor, open reading frame, kinase) family transcriptional repressor; it possesses a sugar kinase motif and an ATPase domain. The short putative protein Llmg_1242, consisting of only 73 amino acids, resembles a sugar kinase or a transcriptional regulator found in Enterococcus and Listeria monocytogenes strains. The gene llmg_1243 encodes a protein with a putative cellulose/glycoside hydrolase domain. Llmg_1245 is a protein with putative haloacid dehalogenase-hydrolase activity, similar to trehalose/sucrose phosphatase. All these genes were upregulated around 3- to 20-fold in llmg_1239::IS905-carrying and 8- to 52-fold in llmg_1239::gfp-carrying Cel+ strains. Two cellobiose-specific EII cytoplasmic AB components containing ptcBA were upregulated in the M-GFP strain compared to their expression in the WT. The most highly upregulated gene cluster in the WT relative to its expression in M-IS and M-GFP was the celB operon (up to 20- to 22-fold), confirming its importance in cellobiose uptake in this strain.
Besides sugar utilization genes, the operon encoding the arginine deiminase pathway enzymes was overexpressed in M-GFP and M-IS cells. A small RNA molecule that is the product of the argR transcript, LLMGnc_172 (15), was also detected in the transcriptome of the M-GFP strain. sRNA LLMGnc_147, which was previously reported to be activated in the presence of cellobiose (15), was not differentially expressed in the strains compared.
Another regulatory RNA abundant in M-GFP cells was 6S RNA. This regulatory small RNA is known to be upregulated in L. lactis MG1363 during the exponential growth phase when galactose or cellobiose is the carbon source (15). It is interesting that although many genes show similar expression patterns in M-IS and M-GFP strains, different sets of sRNAs seem to be present in the two strains. Presumably, changes in sRNA expression are more dynamic and their expression differences are caused by minor variances in RNA sampling time.
Cellobiose utilization in L. lactis MG1363.
A cre site (TAAAAACGTTTGCAAAAAAT) is present in the llmg_1239-1240 intergenic region, suggesting global regulation of the operon by a LacI family protein, carbon catabolite repression (CCR) mediator CcpA (prediction performed with an online tool [http://genome2d.molgenrug.nl]) (Fig. 2). Regulation of catabolic genes and operons by CCR is very common in L. lactis and ensures dominant utilization of glucose when it is present in the growth medium. On the other hand, a weak basal expression of the repressor-encoding gene llmg_1239 can be observed in transcriptomics and RNA-seq data even in the presence of glucose (15, 17, 18). Another LacI family operator-like palindrome, TGGAAACGTTACCA, was identified downstream of the cre site in the llmg_1239-1240 intergenic region. The operator is similar to the consensus sequence recognized by GalR of Escherichia coli (18; predicted using the tool at http://genome2d.molgenrug.nl). Typical LacI family repressors bind their operator sites as dimers or tetramers in the absence of the effector molecules. The binding of the effector molecule (very often a carbohydrate) reduces the DNA binding affinity, releasing the regulator from the operator and allowing transcription (19).
Based on the data generated in this study, we propose that three cellobiose transporters can be activated in L. lactis MG1363 (Fig. 4).
DISCUSSION
The L. lactis subsp. cremoris MG1363 genome contains multiple silent gene clusters and pseudogenes with predicted functions in plant sugar utilization, as a heritage of its plant-related origin (4). Nowadays many of the L. lactis subsp. cremoris strains are adapted to a milk environment and are used for industrial fermentations. It has been suggested that the high number of pseudogenes could indicate ongoing specialization of the strains to their current niche, with constant degeneration of dispensable genes and their encoded functions (5, 7). Genome decay was reproduced in an experimental evolution study through cultivation of the plant isolate Lactococcus lactis subsp. lactis KF147 in milk for 1,000 generations (20). The outcome of the experiment was isolation of strains which had lost or had downregulated plant polymer utilization genes and gene clusters (20).
Here we describe a “reverse evolution” experiment: a reactivation of a silenced plant sugar utilization pathway when selection pressure to utilize cellobiose is applied. Disruption of the repressor gene llmg_1239 by an IS element allowed expression of the otherwise silent cellobiose transporter Llmg_1244 and led to survival of mutant strains on cellobiose. LacI family transcriptional regulators homologous to that encoded by llmg_1239 are present in the genomes of only a few L. lactis strains: NZ9000 and KW2 of L. lactis subsp. cremoris (gene sequence homology of 99%) and IO-1 of L. lactis subsp. lactis (gene sequence homology of 88%; KEGG database). While L. lactis NZ9000 is of dairy origin, KW2 is isolated from fermented corn and the xylose-utilizing strain IO-1 was first isolated from a drain pit of a kitchen sink (16, 21, 22). The transcriptional regulator is homologous to PurR and GalR from Cedecea, Klebsiella, and Listeria. It does not resemble any of the other cellobiose metabolism regulators, such as Llmg_0439 (LacI), CcpA, or ClaR (DexR) of L. lactis or CelR of Streptococcus. Due to the lack of clear homology to any other transcriptional regulator, we propose to designate Llmg_1239 CclR, for cellobiose cluster repressor.
It remains unclear to which effector molecule CclR/Llmg_1239 responds. Apparently, the absence of glucose and the presence of cellobiose in the medium were not the right cues for the relief of repression of the catabolic gene cluster llmg_1240-45. The reason for such tight repression could be the lack either of the proper effector molecule or of an additional coregulator protein that might have been lost during evolution. Alternatively, the ability of the regulator to bind the inducer may have been lost.
A functional PTS EII consists of three components. Once the sugar molecule is imported via the membrane-spanning EII component C, it needs to be phosphorylated by the cytosolic EIIB subunit. Phosphorylation of sugar lowers the affinity of EIIC for it, and the molecule is released to the cytosol (23). The EIIB component receives its phosphoryl group from an EIIA protein. Both of them are sugar specific and usually interact with only one EIIC. As the llmg_1240-45 cluster contains no genes potentially encoding EIIAB subunits, Llmg_1244 presumably works with EII components of another PTS. PtcAB are promiscuous EIIAB proteins and phosphorylate carbohydrates imported via PtcC, CelB, and, presumably, Llmg_0963. All of these EIIC components are annotated as cellobiose specific, although they can import other sugars (glucose and galactose) as well (14, 15). The ability of PtcBA to interact with more than one EIIC component is very unusual for bacterial PTSs. Genes coding for PtcBA form a separate cluster and can be transcribed independently from their membrane-integrated partners (Fig. 4). The transcriptome data showed an around 2- to 3-fold upregulation of ptcBA in the M-GFP strain compared to their expression in the WT strain. We propose that the full novel cellobiose PTS consists of Llmg_1244PtcBA.
Possibly because of the change from the consensus −10 box TATAAT into a less perfect TATATT in its promoter, ccpA expression is slightly lower in the M-GFP and M-IS strains than it is in the WT strain. Presumably, related to that, expression of the arc and the trehalose (pgm and treR) operons and that of the 6S RNA are higher in M-GFP and M-IS. Cellobiose has not been shown to cause CCR, but it consists of two glucose molecules and is metabolized by phospho (P)-β-glucosidases rather quickly in Cel+ strains (Fig. 2), making it plausible that some level of catabolite repression on cellobiose remains. In accordance with higher ccpA expression, glycolysis (gapB) and protein synthesis (RNA polymerase RpoA- and ribosomal protein-encoding genes) are more intense in the WT strain. To evaluate the role of the PccpA mutation in the strength of CCR, the level of induction of Parc and PackA in the presence of glucose was studied in L. lactis MG1363 (WT) and MGΔcelBΔptcC using promoter-gfp fusion constructs. Parc and PackA are strong promoters that contain cre sites and are repressed by CcpA in L. lactis (24). Fluorescence intensities driven from Parc and PackA differ between the WT and MGΔcelBΔptcC, especially in glucose-containing media (where CCR is active), although the difference is not very pronounced (data not shown). More studies are needed in order to elucidate the role of the change in the sequence of PccpA.
WT cells also strongly express a putative sugar ABC permease, Llmg_1165. The gene for this transporter is located in a putative sugar utilization cluster, llmg_1164-aguA. The cluster codes for two ABC transporters (Llmg_1164 and Llmg_1165), two putative endoglucanases, polysaccharide (chitin and xylan) deacetylase, and a putative α-glucuronidase, AguA. The role of this gene cluster in L. lactis MG1363 has not yet been studied. It is not clear whether the expression of llmg_1165 in WT cells is the result of the Llmg_1239 regulation or is an indirect effect of possible slight variations in growth rates or CcpA levels among the strains. Changes in expression were observed in a set of sRNAs in the strains studied. Unfortunately, little is known about the exact functions of the regulatory RNAs in L. lactis. The housekeeping 6S RNA, which was upregulated in M-GFP, was shown to be expressed during the exponential growth phase in cells utilizing cellobiose (15). The expression of LLMGnc_172 was associated with that of the arc cluster (15), which was also overexpressed in M-GFP. It is possible that the type of disruption of llmg_1239 (by an IS905 or via gfp integration) is the cause of differences in sRNA expression.
The catabolic cluster llmg_1240-45 and the repressor gene seem to have been acquired by horizontal gene transfer. The Llmg_1244 protein sequence shows closest similarity to a lichenan-specific EIIC component, LicC, or a cellobiose-specific CelB from various Listeria, Enterococcus, and Lactobacillus strains. Moreover, comparison of L. lactis MG1363 PTS components with Llmg_1244 demonstrates that it is not related to PtcC and CelB of this organism (32% and 29% similarity, respectively). Lactococci are in contact with other bacterial species in both industrial and natural environments. Bacterial genomes can be altered not only by point mutations or genome rearrangements but also by acquisition of foreign DNA (1). Although L. lactis strains possess elements of the competence machinery, it is not functional (25). Transduction by phages and transfer mediated by transposons or conjugative plasmids thus serve as the main sources of foreign DNA (3, 26). Integration of the acquired DNA can occur via homologous recombination of common IS elements.
IS elements play an important role in the evolution of lactococci and other bacteria. They facilitate DNA rearrangements and are responsible for creation of new genetic variants with selective advantages under certain environmental conditions. During adaptation of lactococci to the milk environment, IS elements most likely helped to acquire and rapidly disseminate such traits as lactose metabolism, casein protease activity, nisin production and immunity, and bacteriophage resistance (8). Horizontal gene transfer is obviously an essential mechanism for lactic acid bacteria (LAB) to adapt and therefore survive in their respective environments. L. lactis MG1363 possesses 71 copies in a total of 11 different types of IS elements, of which IS981 (27) and IS905 (28) are the most abundant (14 copies and 16 copies, respectively) (4). IS981 has been described to be involved in adaptation of L. lactis MG1363 and IL1403 to unfavorable conditions during starvation or oxidative- or bacteriocin-induced stress (9, 10, 29). A recombination event between two IS905 elements has been shown to be responsible for a large chromosomal inversion in L. lactis NCDO763, although no resulting phenotypic changes could be observed in comparison to MG1363. The two IS905 elements between which the inversion had occurred were fixed in these L. lactis strains, presumably because of their positive effects for the cell: one of the IS elements provided a −35 sequence for psp (pyrrolidone carboxyl peptidase gene), and the other one disrupted a gene for a phage terminase subunit and thus protected the cell from phage-caused lysis (30). The chromosomal region of L. lactis MG1363 harboring the cluster llmg_1240-45 is surrounded by transposases and helper proteins for both IS981 and IS905; e.g., llmg_1234 and llmg_1264 code for transposases for IS981, while llmg_1319 specifies a transposase for IS905. The abundance of mobile elements in the vicinity of the llmg_1240-45 cluster could have influenced the high frequency of their integration into llmg_1239. Since the integrants acquire a substantial growth advantage in cellobiose-containing medium, they are rapidly selected for.
The fact that llmg_1239 contains a frameshift and is annotated as a pseudogene in the first sequenced L. lactis MG1363 strain (4) may not be coincidental. Silencing of the repressor gene by such a mutation could be another mechanism to activate the llmg_1240-45 cluster. The frameshift was identified in a stretch of adenines (6 in a row) making the region prone to slipped-strand mispairing during replication. It is also known that this MG1363 derivative did not possess the PcelB activating mutation, meaning that it could not use PtcBA-CelB for the import of cellobiose. If this Cel− strain has been exposed to cellobiose before sequencing, it could have acquired the llmg_1239 frameshift mutation that helped it to survive on this disaccharide. None of the L. lactis MG1363 strain stocks sequenced in our laboratory harbored the frameshift in llmg_1239. The same was reported for the isolate sequenced by Linares et al. (12). Strains evolve with time, and many processes might have changed in what we call “model strains” because of long laboratory propagation. It is also clear that these model strains vary per laboratory. We show here that it is important to resequence llmg_1239 when studying the metabolism of cellobiose or other sugars in L. lactis MG1363 and its derivatives. While it was determined in this study that the silent sugar utilization cluster can be activated when cellobiose is present in the environment, the real inducer molecule that binds the repressor CclR remains to be elucidated.
MATERIALS AND METHODS
Microbial strains used and growth conditions.
L. lactis MG1363 (31) derivatives (Table 2) were grown as standing cultures at 30°C in M17 broth (Difco, Sparks, MD) or in chemically defined medium (CDM PC) (32) supplemented with 1% cellobiose or 0.5% glucose. M17 agar plates were prepared by adding 1.5% (wt/vol) agar to M17 supplemented with either glucose (0.5%) or cellobiose (1%). When appropriate, erythromycin (Sigma-Aldrich, St. Louis, MO) was used at 1 μg ml−1. E. coli DH5α (Invitrogen, Carlsbad, CA) was used as a cloning host and was grown in LB medium at 37°C or on LB medium solidified with 1.5% (wt/vol) agar. For plasmid selection, 150 μg ml−1 of erythromycin was added.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 rec1A end1A hsdR17 gyrA96 supE44 thi-1 relA1 | Invitrogen (Carlsbad, CA) |
| L. lactis strains | ||
| MG1363 (WT) | L. lactis subsp. cremoris plasmid-free derivative of NCDO712 carrying a Pcel up-mutation | 11 |
| MGΔcelB | MG1363 carrying a chromosomal deletion of celB | 11 |
| MGΔcelBΔptcC | MG1363 carrying chromosomal deletions of celB and ptcC | Gift from B. Martinez |
| MGΔcelBΔptcCΔllmg_0963 | MG1363ΔcelBΔptcC carrying a chromosomal deletion of llmg_0963 | This study |
| MGΔcelBllmg_1239::IS981 | MG1363ΔcelB with IS981 integrated in llmg_1239 | This study |
| MGΔcelBΔptcCΔllmg_0963 llmg_1239::IS905 (M-IS) | MGΔcelBΔptcCΔllmg_0963 with IS905 integrated in llmg_1239 | This study |
| MGΔcelBΔptcCΔllmg_0963 llmg_1239::gfp (M-GFP) | MGΔcelBΔptcCΔllmg_0963 with PackA-gfp integrated in llmg_1239 | This study |
| Plasmids | ||
| pCS1966 | Integration vector for L. lactis | 29 |
| pCS1966-0963′ | pCS1966 containing llmg_0963 deletion construct | This study |
| pSeudo39 | Integration vector for L. lactis carrying the flanking regions of llmg_1239 | Gift from J. Siebring |
| pSeudo39PackA-gfp | pSeudo39 containing PackA-gfp | 40 |
To study the growth of various L. lactis strains, the strains were grown in 0.2 ml of CDM supplemented with 1% cellobiose in 96-well microtiter plates at 30°C. Growth was monitored by measuring the OD600 with an Infinite 200 PRO 16 microplate spectrophotometer (Tecan Group Ltd., Mannedorf, Switzerland).
General DNA techniques.
DNA manipulations were executed essentially as described previously (33). Plasmid DNA and PCR products were isolated and purified using the High Pure plasmid isolation kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions.
Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were obtained from Thermo Fisher Scientific Baltics (Vilnius, Lithuania) and used according to the supplier's instructions. Phusion DNA polymerase was purchased from New England BioLabs (Ipswich, MA). PCR were performed in a Bio-Rad thermal cycler (Hercules, CA) using L. lactis MG1363 chromosomal DNA as the template.
Construction of the L. lactis deletion strain.
The flanking regions of llmg_0963 were amplified using Ko09631F (AAAATTCTAGAAATTATGGTATGCATTATAG)/Ko09632R (TGCATGGATCCTCTTTAGATTCACTCCTTTAAC) and Ko09633F (TGCATGGATCCGAATTAGAGGGTTCAGAAAC)/Ko09634R (TTTTAGGTACCAGACTTCTACTGACAGATTC). These fragments were ligated into pCS1966 (34, 35) via XbaI/BamHI and BamHI/KpnI restriction sites. The resulting vector was designated pCS1966-0963′. The pCS1966 derivative was obtained and maintained in E. coli DH5α.
Vector pCS1966-0963′ was introduced in L. lactis MG1363ΔcelBΔptcC (a kind gift from B. Martinez) via electroporation (36); cells in which the two-step homologous recombination event had occurred were selected by growing them on selective SA medium plates (37) supplemented with 20 μg ml−1 of 5-fluoroorotic acid hydrate (29, 32). The obtained strain was labeled MG1363ΔcelBΔptcCΔ0963. The chromosomal structure of the deletion strain was confirmed by PCR analysis and sequencing of the modified regions.
Construction of the gfp integration strain.
L. lactis plasmid pSeudo39PackA-gfp was constructed by introducing PackA and sfgfp(Bs) (38) into pSeudo39 vector carrying two flanking regions of llmg_1239 (a kind gift from J. Siebring) via SmaI/XhoI and XhoI/BamHI restriction sites, respectively. The PackA promoter fragment was obtained by PCR using primer pair 5′-GCATCCCGGGATCTTTATGGAAGAATTTAC-3′/5′-CGATCTCGAGTTTGGTCATGTTTAATAAAC-3′ (underlined sequences represent the SmaI and XhoI restriction sites, respectively) and the chromosomal DNA of L. lactis MG1363 as a template. The resulting vector was integrated into the chromosome of MG1363ΔcelBΔptcCΔ0963. The strain obtained after a two-step recombination event via the llmg_1239-homologous regions (as described above) was designated MG1363ΔcelBΔptcCΔ0963 llmg_1239::gfp (M-GFP).
Transcriptome analyses.
Transcriptome analysis was performed using full-genome L. lactis MG1363 DNA microarrays (Agilent Technologies, Santa Clara, CA) as described previously (39). Single colonies of the WT, M-GFP, and M-IS were used to inoculate 10 ml of M17 supplemented with 1% cellobiose. After overnight growth, the cultures were diluted with fresh medium 10× and harvested at the mid-exponential growth phase, at an OD600 of 0.6. RNA from M-GFP and M-IS was compared to RNA from MG1363 (WT). DNA Microarray slides were scanned with a Genepix 4200 laser scanner (Molecular Devices, Sunnyvale, CA). Slide images were analyzed using GenePix Pro v.6.0 software. Background subtraction and LOWESS (locally weighted scatterplot smoothing) normalization were done using the standard routines provided by GENOME2D software available at http://genome2d.molgenrug.nl/index.php/analysis-pipeline. A gene was considered differentially expressed when the Bayesian P value was <0.001, and a fold change cutoff of 2.5 was applied.
Accession number(s).
Gene expression data were deposited in the GEO database under accession number GSE103707.
ACKNOWLEDGMENTS
We thank A. Ravi and M. Tufano for their help with the transcriptome analyses. We thank B. Martinez (IPLA-CSIC, Spain) for kindly providing L. lactis MG1363ΔcelBΔptcC. We thank J. Siebring for kindly providing plasmid pSeudo39.
This work was supported by a grant from NWO-STW Technology Foundation (project number 10619).
REFERENCES
- 1.Quiberoni A, Rezaiki L, El Karoui M, Biswas I, Tailliez P, Gruss A. 2001. Distinctive features of homologous recombination in an ‘old’ microorganism, Lactococcus lactis. Res Microbiol 152:131–139. doi: 10.1016/S0923-2508(01)01183-4. [DOI] [PubMed] [Google Scholar]
- 2.Siezen RJ, Bayjanov JR, Felis GE, van der Sijde MR, Starrenburg M, Molenaar D, Wels M, van Hijum SAFT, Vlieg JETV. 2011. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb Biotechnol 4:383–402. doi: 10.1111/j.1751-7915.2011.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res 11:731–753. doi: 10.1101/gr.GR-1697R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Passerini D, Coddeville M, Le Bourgeois P, Loubiere P, Ritzenthaler P, Fontagne-Faucher C, Daveran-Mingot ML, Cocaign-Bousquet M. 2013. The carbohydrate metabolism signature of Lactococcus lactis strain A12 reveals its sourdough ecosystem origin. Appl Environ Microbiol 79:5844–5852. doi: 10.1128/AEM.01560-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siezen RJ, Renckens B, van Swam I, Peters S, van Kranenburg R, Kleerebezem M, de Vos WM. 2005. Complete sequences of four plasmids of Lactococcus lactis subsp. cremoris SK11 reveal extensive adaptation to the dairy environment. Appl Environ Microbiol 71:8371–8382. doi: 10.1128/AEM.71.12.8371-8382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelleher P, Bottacini F, Mahony J, Kilcawley KN, van Sinderen D. 2017. Comparative and functional genomics of the Lactococcus lactis taxon; insights into evolution and niche adaptation. BMC Genomics 18:267. doi: 10.1186/s12864-017-3650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romero DA, Klaenhammer TR. 1993. Transposable elements in lactococci: a review. J Dairy Sci 76:1–19. doi: 10.3168/jds.S0022-0302(93)77318-X. [DOI] [PubMed] [Google Scholar]
- 9.de Visser JA, Akkermans AD, Hoekstra RF, de Vos WM. 2004. Insertion-sequence-mediated mutations isolated during adaptation to growth and starvation in Lactococcus lactis. Genetics 168:1145–1157. doi: 10.1534/genetics.104.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bongers RS, Hoefnagel MH, Starrenburg MJ, Siemerink MA, Arends JG, Hugenholtz J, Kleerebezem M. 2003. IS981-mediated adaptive evolution recovers lactate production by ldhB transcription activation in a lactate dehydrogenase-deficient strain of Lactococcus lactis. J Bacteriol 185:4499–4507. doi: 10.1128/JB.185.15.4499-4507.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solopova A, Bachmann H, Teusink B, Kok J, Neves AR, Kuipers OP. 2012. A specific mutation in the promoter region of the silent cel cluster accounts for the appearance of lactose-utilizing Lactococcus lactis MG1363. Appl Environ Microbiol 78:5612–5621. doi: 10.1128/AEM.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linares DM, Kok J, Poolman B. 2010. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J Bacteriol 192:5806–5812. doi: 10.1128/JB.00533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aleksandrzak-Piekarczyk T, Polak J, Jezierska B, Renault P, Bardowski J. 2011. Genetic characterization of the CcpA-dependent, cellobiose-specific PTS system comprising CelB, PtcB and PtcA that transports lactose in Lactococcus lactis IL1403. Int J Food Microbiol 145:186–194. doi: 10.1016/j.ijfoodmicro.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 14.Pool WA, Neves AR, Kok J, Santos H, Kuipers OP. 2006. Natural sweetening of food products by engineering Lactococcus lactis for glucose production. Metab Eng 8:456–464. doi: 10.1016/j.ymben.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 15.van der Meulen SB, de Jong A, Kok J. 2016. Transcriptome landscape of Lactococcus lactis reveals many novel RNAs including a small regulatory RNA involved in carbon uptake and metabolism. RNA Biol 13:353–366. doi: 10.1080/15476286.2016.1146855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 17.Brouwer RWW. 2014. Computational methods for the analysis of bacterial gene regulation. PhD thesis. University of Groningen, Groningen, the Netherlands. [Google Scholar]
- 18.Pinto J. 2015. In principio erat Lactococcus lactis: towards a membrane protein overproducer host. PhD thesis. University of Groningen, Groningen, the Netherlands. [Google Scholar]
- 19.Weickert MJ, Adhya S. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem 267:15869–15874. [PubMed] [Google Scholar]
- 20.Bachmann H, Starrenburg MJC, Molenaar D, Kleerebezem M, Vlieg JETV. 2012. Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res 22:115–124. doi: 10.1101/gr.121285.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato H, Shiwa Y, Oshima K, Machii M, Araya-Kojima T, Zendo T, Shimizu-Kadota M, Hattori M, Sonomoto K, Yoshikawa H. 2012. Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of l-lactic acid. J Bacteriol 194:2102–2103. doi: 10.1128/JB.00074-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly WJ, Altermann E, Lambie SC, Leahy SC. 2013. Interaction between the genomes of Lactococcus lactis and phages of the P335 species. Front Microbiol 4:257. doi: 10.3389/fmicb.2013.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zomer AL, Buist G, Larsen R, Kok J, Kuipers OP. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:1366–1381. doi: 10.1128/JB.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wydau S, Dervyn R, Anba J, Dusko Ehrlich S, Maguin E. 2006. Conservation of key elements of natural competence in Lactococcus lactis ssp. FEMS Microbiol Lett 257:32–42. doi: 10.1111/j.1574-6968.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- 26.Machielsen R, Siezen RJ, van Hijum SAFT, Vlieg JETV. 2011. Molecular description and industrial potential of Tn6098 conjugative transfer conferring alpha-galactoside metabolism in Lactococcus lactis. Appl Environ Microbiol 77:555–563. doi: 10.1128/AEM.02283-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polzin KM, McKay LL. 1991. Identification, DNA sequence, and distribution of IS981, a new, high-copy-number insertion sequence in lactococci. Appl Environ Microbiol 57:734–743.1645511 [Google Scholar]
- 28.Dodd HM, Horn N, Gasson MJ. 1994. Characterization of IS905, a new multicopy insertion sequence identified in lactococci. J Bacteriol 176:3393–3396. doi: 10.1128/jb.176.11.3393-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roces C, Perez V, Campelo AB, Blanco D, Kok J, Kuipers OP, Rodriguez A, Martinez B. 2012. The putative lactococcal extracytoplasmic function anti-sigma factor Llmg2447 determines resistance to the cell wall-active bacteriocin Lcn972. Antimicrob Agents Chemother 56:5520–5527. doi: 10.1128/AAC.01206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daveran-Mingot ML, Campo N, Ritzenthaler P, Le Bourgeois P. 1998. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J Bacteriol 180:4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis Ncdo-712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goel A, Santos F, Vos WM, Teusink B, Molenaar D. 2012. Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl Environ Microbiol 78:134–143. doi: 10.1128/AEM.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 34.Solem C, Defoor E, Jensen PR, Martinussen J. 2008. Plasmid pCS1966, a new selection/counterselection tool for lactic acid bacterium strain construction based on the oroP gene, encoding an orotate transporter from Lactococcus lactis. Appl Environ Microbiol 74:4772–4775. doi: 10.1128/AEM.00134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Defoor E, Kryger MB, Martinussen J. 2007. The orotate transporter encoded by oroP from Lactococcus lactis is required for orotate utilization and has utility as a food-grade selectable marker. Microbiology 153:3645–3659. doi: 10.1099/mic.0.2007/005959-0. [DOI] [PubMed] [Google Scholar]
- 36.Holo H, Nes IF. 1995. Transformation of Lactococcus by electroporation. Methods Mol Biol 47:195–199. [DOI] [PubMed] [Google Scholar]
- 37.Jensen PR, Hammer K. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl Environ Microbiol 59:4363–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Overkamp W, Beilharz K, Detert Oude Weme R, Solopova A, Karsens H, Kovacs A, Kok J, Kuipers OP, Veening JW. 2013. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol 79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuipers OP, de Jong A, Baerends RJ, van Hijum SA, Zomer AL, Karsens HA, den Hengst CD, Kramer NE, Buist G, Kok J. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113–122. doi: 10.1023/A:1020691801251. [DOI] [PubMed] [Google Scholar]
- 40.Solopova A, van Gestel J, Weissing FJ, Bachmann H, Teusink B, Kok J, Kuipers OP. 2014. Bet-hedging during bacterial diauxic shift. Proc Natl Acad Sci U S A 111:7427–7432. doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]