ABSTRACT
To provide food security, innovative approaches to preventing plant disease are currently being explored. Here, we demonstrate that lytic bacteriophages and phage lysis proteins are effective at triggering lysis of the phytopathogen Agrobacterium tumefaciens. Phages Atu_ph02 and Atu_ph03 were isolated from wastewater and induced lysis of C58-derived strains of A. tumefaciens. The coinoculation of A. tumefaciens with phages on potato discs limited tumor formation. The genomes of Atu_ph02 and Atu_ph03 are nearly identical and are ∼42% identical to those of T7 supercluster phages. In silico attempts to find a canonical lysis cassette were unsuccessful; however, we found a putative phage peptidoglycan hydrolase (PPH), which contains a C-terminal transmembrane domain. Remarkably, the endogenous expression of pph in the absence of additional phage genes causes a block in cell division and subsequent lysis of A. tumefaciens cells. When the presumed active site of the N-acetylmuramidase domain carries an inactivating mutation, PPH expression causes extensive cell branching due to a block in cell division but does not trigger rapid cell lysis. In contrast, the mutation of positively charged residues at the extreme C terminus of PPH causes more rapid cell lysis. Together, these results suggest that PPH causes a block in cell division and triggers cell lysis through two distinct activities. Finally, the potent killing activity of this single lysis protein can be modulated, suggesting that it could be engineered to be an effective enzybiotic.
IMPORTANCE The characterization of bacteriophages such as Atu_ph02 and Atu_ph03, which infect plant pathogens such as Agrobacterium tumefaciens, may be the basis of new biocontrol strategies. First, cocktails of diverse bacteriophages could be used as a preventative measure to limit plant diseases caused by bacteria; a bacterial pathogen is unlikely to simultaneously develop resistances to multiple bacteriophage species. The specificity of bacteriophage treatment for the host is an asset in complex communities, such as in orchards where it would be detrimental to harm the symbiotic bacteria in the environment. Second, bacteriophages are potential sources of enzymes that efficiently lyse bacterial cells. These phage proteins may have a broad specificity, but since proteins do not replicate as phages do, their effect is highly localized, providing an alternative to traditional antibiotic treatments. Thus, studies of lytic bacteriophages that infect A. tumefaciens may provide insights for designing preventative strategies against bacterial pathogens.
KEYWORDS: Agrobacterium, bacteriophage, endolysin, lysis, phage genomics
INTRODUCTION
Crop damage caused by bacterial phytopathogens poses a threat to food security worldwide (1). Agrobacterium tumefaciens is one of the top three scientifically and economically most important bacterial plant pathogens (2) and is responsible for significant economic losses in stone fruit and nut production (3, 4). A. tumefaciens causes crown gall disease by transforming plant cells to constitutively express genes for the production of phytohormones and opines. Opines serve as a custom food source for the bacteria. Increased hormone production causes plant cells to locally overproliferate (5, 6), leading to tumor formation and the reduced transport of water and nutrients throughout the plant. Thus, infected plants often do not achieve maximal crop yields.
Current commercially available biocontrol involves the application of Agrobacterium radiobacter strain K84, which releases a bacteriocin called agrocin to outcompete A. tumefaciens. However, only a limited number of A. tumefaciens strains are sensitive to agrocin (7, 8), and sensitive strains of A. tumefaciens can become resistant (9). Therefore, alternative methods of biocontrol are emerging, including the selection and breeding of resistant crops (10–12), chemical treatments (13), and the isolation of additional antagonist organisms (14). In this work, we consider the possibility of lytic bacteriophages and phage-encoded lysis proteins as options for A. tumefaciens biocontrol.
Bacteriophages and phage-derived proteins have recently been employed against several plant pathogens (15, 16). Lytic phages contain a large reservoir of genes specifically involved in killing their host cells and are attractive as biocontrol agents. Lytic bacteriophages have shown promise in protecting tomato plants from wilting caused by Ralstonia solanacearum (17, 18), protecting oranges from citrus canker caused by Xanthomonas axonopodis (19), protecting leeks from bacterial blight caused by Pseudomonas syringae pv. porri (20), and protecting kiwi from canker caused by Pseudomonas syringae pv. actinidiae (21). While these studies indicate the potential of bacteriophages to serve as biocontrol agents, further optimization of phage replication and lysis will be necessary to effectively scale for use in crop fields and orchards. As of yet, bacteriophages have not been employed as a biocontrol against A. tumefaciens.
As research on bacteriophages as biocontrol agents has recently grown in popularity, so has the study of the phage proteins responsible for their antimicrobial activity (22). The term “enzybiotics,” first coined in 2001, refers to the direct application of phage endolysins, peptidoglycan hydrolase enzymes that target the bacterial cell wall, to susceptible hosts (23). The benefits of using endolysins include their direct mode of action, low incidence of resistance, and potential for protein optimization (24). The use of exogenously applied peptidoglycan hydrolases to directly kill bacteria is more established for Gram-positive bacteria, since they lack an outer membrane barrier; however, in at least some cases, the exogenous application of endolysins has been shown to effectively lyse Gram-negative bacteria (25). Additional strategies for targeting Gram-negative pathogens include the coapplication of an endolysin with an outer membrane permeabilizer such as EDTA (26) or engineering an endolysin to gain the ability to lyse bacteria from the outside by absorption through the outer membrane (27).
Lytic bacteriophages that infect A. tumefaciens have been isolated from soil and sewage samples (28–30), suggesting that there is untapped potential for using bacteriophages or endolysins as biocontrol agents against the pathogen; however, only Agrobacterium sp. strain H13-3 phage 7-7-1 has been subject to genomic characterization (31). In this work, we isolate and describe 2 lytic bacteriophages that specifically infect a subset of A. tumefaciens strains. These bacteriophages are closely related and contain a novel endolysin with potential antimicrobial activity.
RESULTS AND DISCUSSION
Isolation and characterization of bacteriophages that infect A. tumefaciens strain C58.
Wild-type strains of A. tumefaciens are well known for their ability to cause crown gall disease, and this ability is dependent on the presence of the tumor-inducing plasmid, pTi (3, 32, 33). In this work, A. tumefaciens strain C58 was selected as the host strain for the isolation of bacteriophage, since it was isolated from a cherry tree tumor (34), the complete genome sequence is available (35, 36), and it has been widely studied as a pathogen (37).
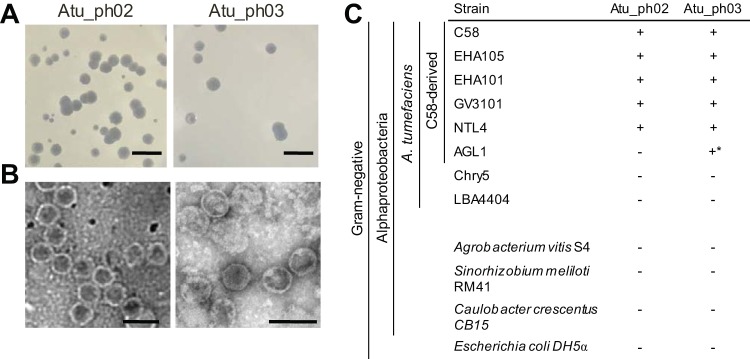
Using a modified phage enrichment protocol (38), we isolated two bacteriophages, called Atu_ph02 and Atu_ph03, from samples obtained from the Columbia, MO, regional wastewater treatment plant. The presence of bacteriophage in supernatants from cleared A. tumefaciens cultures was confirmed by spot and classic plaque assays. Virions were concentrated and partially purified by precipitation with polyethylene glycol (PEG) and differential centrifugation. Phages Atu_ph02 and Atu_ph03 form large and clear plaques on a lawn of A. tumefaciens (Fig. 1A). Transmission electron microscopy (TEM) of the virions revealed icosahedral heads with diameters of ∼58 nm and short tails (Fig. 1B). Subterminal tail fibers are not visible through TEM. This morphology suggests that these bacteriophages are podoviruses (39).
FIG 1.
Characterization of plaque and bacteriophage morphologies. (A) Plaques formed on a lawn of Agrobacterium tumefaciens strain C58 are shown for each bacteriophage. Bars, 10 mm. (B) Transmission electron micrographs reveal the morphology of each bacteriophage. Bars, 100 nm. (C) Specificities of bacteriophages were determined by spotting dilutions of phage on a lawn of the host bacterium. The + indicates that plaques were observed and − indicates that plaques were not observed. *, plaques were observed only at titers ∼1,000 times higher than required for plaque formation on other host strains. Strain AGL1 contains an insertion mutation in recA to stabilize recombination plasmids.
The host ranges of the phages were assessed by spotting phage stock dilutions on a range of bacteria (Fig. 1C). Each bacteriophage exhibits a narrow host range, only infecting a subset of A. tumefaciens strains. Both phages infected C58-derived strains (C58, EHA105, EHA101, GV3101, and NTL4) with the exception of AGL1, which carries a mutation in recA (40). This suggests that RecA, an enzyme responsible for homologous recombination and DNA repair, may be required for efficient bacteriophage infection. Atu_ph02 and Atu_ph03 did not infect A. tumefaciens strains that were not derived from C58 (i.e., LBA4404 and Chry5) or other tested species, including Agrobacterium vitis, Sinorhizobium meliloti, Caulobacter crescentus, and Escherichia coli. A narrow host range is considered to be an important asset when assessing the potential of bacteriophages as biocontrol agents against phytopathogens, as it minimizes harm to other beneficial microbes in the rhizosphere. Ideally, a cocktail of lytic bacteriophages requiring different host factors would be deployed to reduce the incidence of host resistance (21).
Phage treatment causes cell lysis and results in reduced pathogenicity of A. tumefaciens.
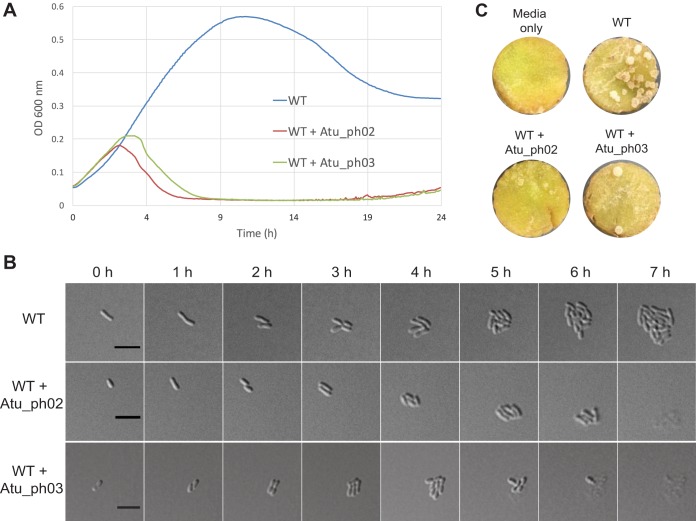
To further assess the potential of these bacteriophages as biocontrol agents, we measured the effect of phage infection on A. tumefaciens grown in liquid medium and on agarose pads, as well as A. tumefaciens-induced tumor formation on potato discs. Bacterial growth curves indicate the rate at which phages can inhibit the growth of their bacterial hosts. A. tumefaciens cells infected with Atu_ph02 or Atu_ph03 at a multiplicity of infection (MOI) of 0.001 grew for ∼3 h postinfection prior to the onset of cell lysis (Fig. 2A). Time-lapse microscopy showed uninfected wild-type (WT) cells form microcolonies within 7 h (Fig. 2B, top; see also Movie S1 in the supplemental material), whereas cells infected with Atu_ph02 (Fig. 2B, center; see also Movie S3) or Atu_ph03 (Fig. 2B, bottom; see also Movie S2) at an MOI of 0.01 initially grew and divided but began lysing 5 h after infection. Since phages are released after the first cell lyses, the remaining cells are subsequently infected, and all cells in the field are lysed within the next 2 h (Fig. 2B, center and bottom; Movie S2). In other representative fields (Movies S2 and S3), cells which were not initially infected formed relatively large microcolonies; however, cells on the periphery of these microcolonies later lysed. Since this lysis event was never observed in the absence of phage, we inferred that these cells were susceptible to the phage particles which had likely diffused through the agarose.
FIG 2.
Bacteriophages Atu_ph02 and Atu_ph03 lyse A. tumefaciens cells. (A) Growth curves of A. tumefaciens strain C58 (WT) infected with Atu_ph02 and Atu_ph03 at MOIs of 0.001. Results from representative growth curves are shown. Each line is the average from four replicate wells. (B) Time-lapse microscopy showing the growth of uninfected WT cells (top), WT cells infected with Atu_ph02 at an MOI of 0.01 (middle), and WT cells infected with Atu_ph03 at an MOI of 0.01 (bottom). Bars, 5 μm. (C) Representative potato discs treated with medium (top left), WT A. tumefaciens cells (top right), a mixture of WT A. tumefaciens cells and Atu_ph02 at an MOI of 1.0 (bottom left), and a mixture of WT A. tumefaciens cells and Atu_ph03 at an MOI of 1.0 (bottom right) after 14 days of incubation in a humid chamber. White spots on the potato discs are A. tumefaciens-induced tumors.
The qualitative potato tumor assay uses potato discs to mimic wound sites and evaluate the virulence of A. tumefaciens cells on plant hosts (41). Potato discs inoculated with A. tumefaciens formed tumors after 14 days of infection (Fig. 2C, top right). A coinoculation with Atu_ph02 or Atu_ph03 at an MOI of 1.0 reduced the number of tumors formed on the potato disc (Fig. 2C, bottom). Since bacteria readily evolve resistance to individual bacteriophages, phage Atu_ph02 or Atu_ph03 alone is unlikely to be an effective biocontrol agent. However, these phages may be valuable as components of a bacteriophage cocktail. Together, the results from the growth curve, microscopy, and potato tumor assay in Fig. 2 show that Atu_ph02 and Atu_ph03 are lytic phages capable of rapidly killing A. tumefaciens and potentially protecting plants from infection.
Phages Atu_ph02 and Atu_ph03 belong to the T7 supercluster.
Although Atu_ph02 and Atu_ph03 are similar in morphology (Fig. 1B), lysis rate (Fig. 2A), and genome size (see Fig. S1A), the genomes are not identical on the basis of a restriction fragment pattern analysis (Fig. S1B). Therefore, we sequenced both genomes to gain insights into the mechanism of phage-mediated host cell lysis.
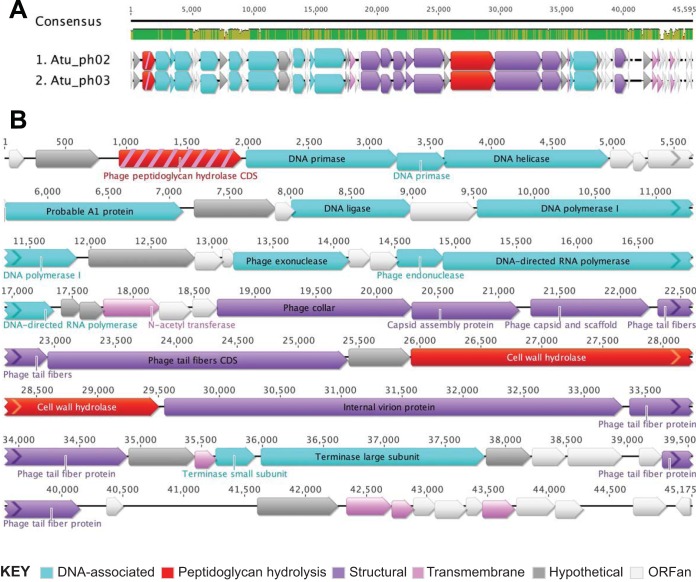
Phages Atu_ph02 and Atu_ph03 contain nearly identical small genomes of ∼45 kbp comprising 55 and 58 open reading frames (ORFs), respectively (Fig. 3, Table 1; see also Table S1). A dot plot analysis of the Atu_ph02 and Atu_ph03 sequences revealed that these genomes are almost entirely syntenic, with only a few regions indicative of small deletions or insertions (see Fig. S2A). A comparison of the 52 shared protein sequences revealed a high degree of similarity; 23 are 100% identical, another 23 are 90 to 99.88% identical, and the remaining 6 are 49% to 88% identical (Fig. S2B and Table S1).
FIG 3.
Genome organization of Atu_ph02 and Atu_ph03. (A) Genome alignments of Atu_ph02 and Atu_ph03 created using the MUSCLE plugin in Geneious. Consensus identities: green, 100% identical; gold, 30 to 100%; red, <30%; no color, 0%. (B) Gene annotations for the Atu_ph03 phage genome. Color coding indicates functional classifications of the open reading frames. Protein PPH, which is classified as both a peptidoglycan hydrolysis protein (red) and a transmembrane protein (pink), is colored red with pink stripes.
TABLE 1.
Summary of key genomic features
| Bacteriophage | Genome length (bp) | GC content (%) | No. of ORFs | Coding density (%) | No. of hypothetical proteins | No. of ORFansa |
|---|---|---|---|---|---|---|
| Atu_ph02 | 45,423 | 54.8 | 55 | 92.9 | 32 | 23 |
| Atu_ph03 | 45,175 | 54.7 | 58 | 93.8 | 36 | 26 |
ORFans are predicted proteins that do not have significant hits in the nonredundant (nr) database. Hypothetical proteins share homology with proteins in the nr database (42).
The genomes of Atu_ph02 and Atu_ph03 are organized in functional blocks (Fig. 3; see also Table S2) including genes encoding DNA-associated proteins (Fig. 3B, light blue arrows) and genes predicted to function in phage morphogenesis (Fig. 3B, purple arrows). Remarkably, 60% of the ORFs encode hypothetical proteins of unknown function (Fig. 3B, white and gray arrows), ∼70% of which (23 in Atu_ph02 and 26 in Atu_ph03) are ORFs with no significant homology to existing proteins in the nonredundant database (ORFans) (Fig. 3B, white arrows) (42).
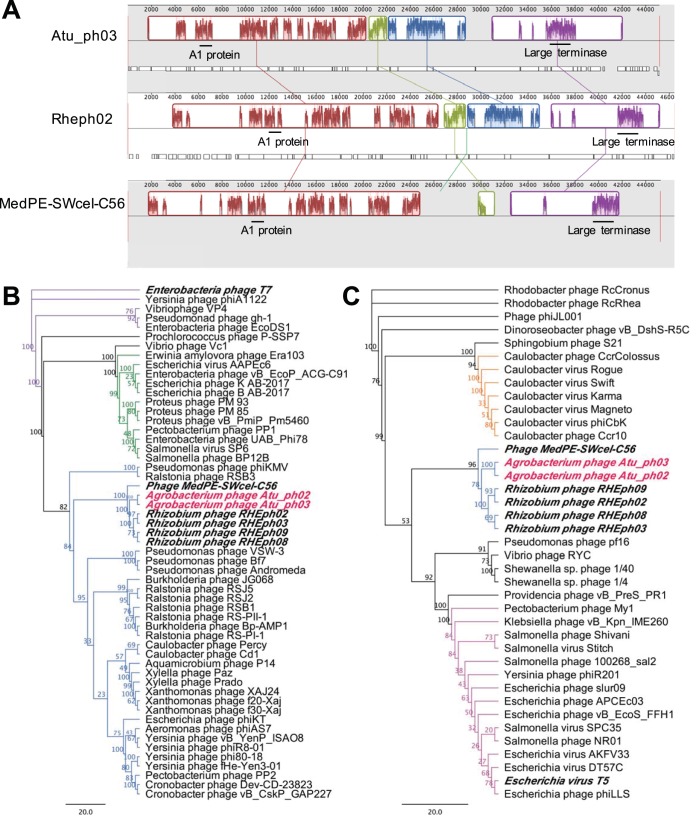
Whole-genome comparisons using the Atu_ph03 nucleic acid sequence revealed that this phage genome is ∼42% identical to both the T7-like Rhizobium etli phage RHEph02 and phage MedPE-SWcel-C56. Whole-genome alignments identified syntenic regions among these phage genomes (Fig. 4A). Similar to Rhizobium etli phages RHEph02, RHEph03, RHEph08, and RHEph09 (38), phages Atu_ph02 and Atu_ph03 can be classified as members of the T7 supercluster on the bases of the similarity of their genome organizations and the presence of core T7 genes. There are 4 conserved core T7 genes, which are predicted to encode the T7-like RNA polymerase, large terminase, and structural proteins (Table S2). These 4 core genes are also present in RHEph02, RHEph08, and phage MedPE-SWcel-C56 (Table S2). A phylogenetic analysis using the gene encoding the large terminase revealed that Agrobacterium phages Atu_ph02 and Atu_ph03, Rhizobium etli phages, and phage Med-SWcel-C56 form a distinct clade and share an ancestor with other T7-like bacteriophages that target nonenteric hosts, including Pseudomonas phage ϕKMV (Fig. 4B). Furthermore, this phylogeny supports the classification of these Agrobacterium phages within the ϕKMV-like cluster of phages, which includes characterized phages that infect Alphaproteobacteria such as Caulobacter phages ϕCd1 (43) and Percy (44), and Ralstonia phage ϕRSB1 (45). Furthermore, whole-genome comparisons using Atu_ph03 and Pseudomonas phage ϕKMV revealed that the genomes are 42% identical. A total of 13 of the predicted proteins in the Atu_ph03 genome have homologous proteins in ϕKMV that share at least 24% identity, including key proteins that function in DNA metabolism and virion structure and assembly (Table S2).
FIG 4.
Genome-wide syntenic mapping and key protein phylogenies. (A) Whole-genome alignments of Atu_ph03 with Rhizobium phage RHEph02 and phage MedPE-SWcel-C56. Positions of genes encoding the A1 protein and large terminase are shown with black bars. (B) Phylogenetic analysis of the large terminase subunits from Atu_ph02 and Atu_ph03 and other T7-like phages. Purple nodes indicate T7-like phages, green nodes indicate Sp-6-like phages, and blue nodes indicate ϕKMV-like phages. (C) Phylogenetic analysis of the probable A1 protein from Atu_ph02 and Atu_ph03, closely related Rhizobium phages, phage MedPE-SWcel-C56, and phages belonging to the T5 family. Blue nodes indicate ϕKMV-like phages, orange nodes indicate ϕCBK-like phages, and pink nodes indicate T5-like phages. Scale bars represent the number of amino acid substitutions per site. Numerical value on each node represents the bootstrap value of 100 replicates.
Phages Atu_ph02 and Atu_ph03 contain a putative A1 protein.
A surprising observation within the genomes of Atu_ph02 and Atu_ph03 is the presence of a homolog of the A1 protein from phage T5 (Fig. 3B; Table S2). The probable A1 protein is conserved within the clade containing the Rhizobium etli phages and phage MedPE-SWcel-C56, suggesting that the ancestor of these phages acquired the gene horizontally, since it is not prevalent among ϕKMV-like bacteriophages (Fig. 4C; Table S2). The putative A1 protein (Gp10) from Atu_ph03 is 486 amino acids and is 38% identical to the phage T5 A1 protein (Table S2). In phage T5, A1 mutants are defective in multiple processes, including degrading host DNA, downregulating pre-early gene expression (46), and completing T5 DNA transfer into the host cell (47). The probable A1 proteins have no readily identifiable functional domains, transmembrane domains, or signal peptides; thus, we cannot speculate on the function of the A1 protein in phages Atu_ph02 and Atu_ph03. Homologs to the A1 protein are found in other non-T5 phages, including Caulobacter phage ϕCBK (48, 49) (Fig. 4C). In phage ϕCBK and related phages, the A1 protein is located with the DNA replication module, suggesting that this protein may function to alter ϕCBK gene expression through an interaction with the host RNA polymerase (49). The putative A1 proteins in Agrobacterium phages Atu_ph02 and Atu_ph03 join a small family of A1-related proteins found in distinct clades of non-T5-like phages (Fig. 4C) and bacterial genomes, though the function remains uncharacterized in all cases (48).
Atu_ph02 and Atu_ph03 phage lysis proteins.
Many bacteriophages that infect Gram-negative hosts contain lysis cassettes consisting of endolysins and accessory proteins (50). Most of these endolysins have globular structures, each containing a single enzymatic active domain (EAD), and cannot reach the periplasm independently (50–52). In the canonical holin-endolysin system, endolysins accumulate in the cytoplasm until sufficient quantities of holins are inserted in the inner membrane to form homooligomeric pores that allow endolysins to enter the periplasm (50). An alternative strategy is used by signal-arrest-release (SAR) endolysins that contain an N-terminal type II signal anchor, which embeds the inactive enzyme in the inner membrane until pinholins cause membrane depolarization and release of the endolysin to the periplasm (50). Both the holin-endolysin and pinholin-SAR endolysin systems rely on spanins to fuse the inner and outer membranes to complete cell lysis. In addition to these strategies, in silico and experimental analyses suggest that some endolysins contain N-terminal signal sequences that may enable delivery to the periplasm via the Sec machinery (51, 53, 54).
Among the phage lysis proteins, endolysins are most readily identified in phage genomes due to the presence of peptidoglycan hydrolase domains. Thus, the genomes of Atu_ph02 and Atu_ph03 were searched for putative endolysin proteins. All of the predicted ORFs were translated and searched for the presence of domains encoding peptidoglycan hydrolases. Two candidate peptidoglycan hydrolases were identified (Fig. 3, red arrows; Table S2). Gp35 contains a putative cell wall hydrolase (pfam07486) and is located in close proximity to the putative tail proteins and internal virion proteins, suggesting that this hydrolase may function as a virion-associated lysin. Gp3 contains a putative N-acetylmuramidase domain (DUF3380), suggesting that it may hydrolyze peptidoglycan.
To determine if either Gp35 or Gp3 is part of a canonical lysis cassette, we searched for potential accessory proteins, such as holins or pinholins, by screening each putative protein sequence for the presence of transmembrane domains. Phage genome Atu_ph03 encodes a total of 6 predicted transmembrane proteins (Fig. 3, pink arrows); however, none of these proteins are in close proximity to the putative peptidoglycan hydrolases. This observation suggests that Atu_ph03 may not contain a canonical lysis cassette. Remarkably, one of the transmembrane-containing proteins also contains the putative phage peptidoglycan hydrolase domain DUF3380 (Fig. 3, red and pink-striped arrows). The sequences of this phage peptidoglycan hydrolase (PPH) in phages Atu_ph02 and Atu_ph03 are 100% identical (Fig. 3A; Table S1), and it is not present in the genomes of the closely related Rhizobium etli phages or phage MedPE-SWcel-C56 (Table S2). To determine if PPH contributes to cell lysis, pph (encoding Gp3) was subjected to bioinformatic and genetic characterizations.
Phage peptidoglycan hydrolase is sufficient to induce A. tumefaciens cell lysis.
PPH contains a domain (DUF3380) found in bacterial and viral proteins that binds peptidoglycan (Fig. 5A; see also Fig. S3). A recent characterization of an endolysin (Gp110) from a Salmonella phage 10 containing DUF3380 revealed that this domain functions as an N-acetylmuramidase and cleaves the β1 to 4 glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine in peptidoglycan (55). The predicted PPH transmembrane topology according to a hidden Markov model (56) oriented the transmembrane domain with the EAD in the periplasm and the short C-terminal tail in the cytosol (Fig. 5A). Remarkably, if PPH functions as an endolysin, this suggests that PPH may utilize an atypical mechanism of entering the periplasm and cause host cell lysis.
FIG 5.
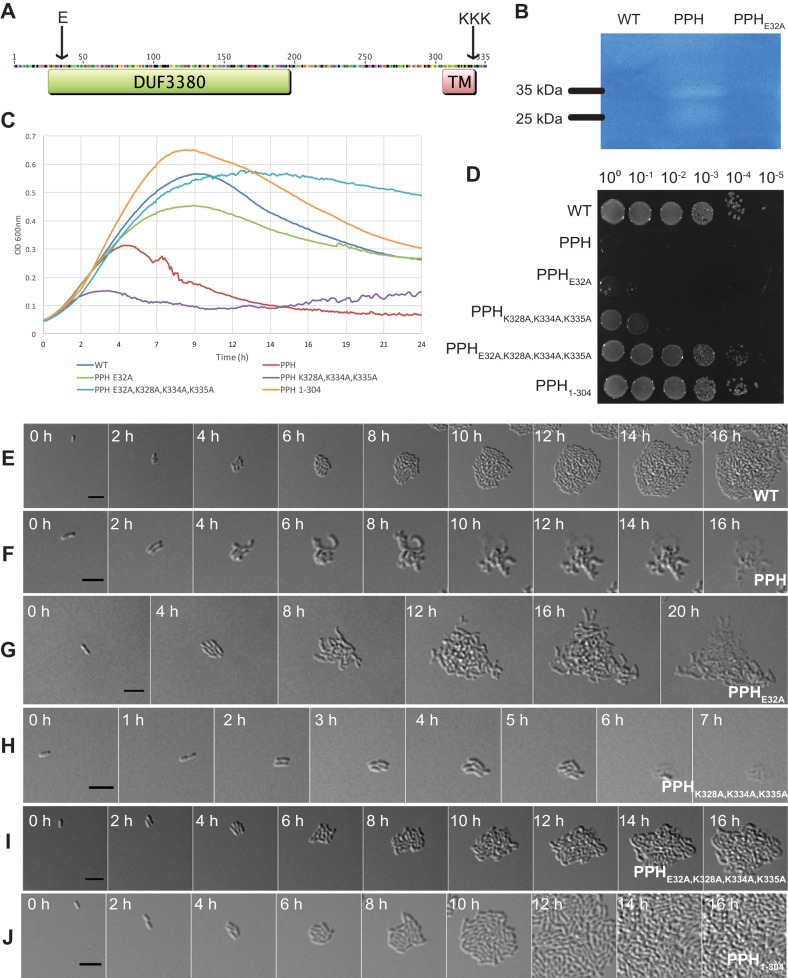
Characterization of phage peptidoglycan hydrolase (PPH) and its effect on A. tumefaciens. (A) Predicted PPH protein topology. Arrows indicate sites subject to point mutagenesis. (B) Zymogram of 30-μg whole-cell lysates from A. tumefaciens lacking PPH, expressing PPH, and expressing PPHE32A. (C) Growth curves of A. tumefaciens growth when expressing plasmid pSRKKm-Plac with variants of pph under induced conditions. (D) Cell viability of A. tumefaciens containing plasmids to express unmutated and variant PPH grown under induced conditions. Time-lapse microscopy of an A. tumefaciens cell containing an empty pSRKKm-Plac vector (E), pSRKKm-Plac-PPH (F), pSRKKm-Plac-PPHE32A (G), pSRKKm-Plac-PPHK328A,K334A,K335A (H), pSRKKm-Plac-PPHE32A,K328A,K334A,K335A (I), and pSRKKm-Plac-PPH1–304 (J). Cells were induced for PPH expression for 1 h prior to initiation of time-lapse microscopy. Bars, 5 μm.
In light of these observations, we sought to determine if the expression of PPH is sufficient for cell lysis. A plasmid containing pph under the control of a lac promoter (pSRKKm-Plac-PPH) was introduced into WT A. tumefaciens cells, enabling observations of cell viability, cell growth, cell morphology, and cell lysis under conditions where pph is induced by the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) (Fig. 5). A growth curve analysis showed that the induction of pph led to growth inhibition within 4 h (Fig. 5C), in contrast to the normal growth exhibited by the uninduced cells (see Fig. S4A). To measure viability, we grew A. tumefaciens cells containing pSRKKm-Plac-PPH in the absence of IPTG to mid-exponential phase. Next, we spotted dilutions of A. tumefaciens containing pSRKKm-Plac-PPH onto plates with and without IPTG. In the presence of the inducer, there was a 5-log loss in viability of A. tumefaciens cells (Fig. 5D) compared to that of the same strain in the absence of inducer (Fig. S4B). These results suggest that an accumulation of PPH is sufficient to inhibit A. tumefaciens growth. Next, time-lapse microscopy was used to determine if pph induction triggers the lysis of A. tumefaciens cells. A. tumefaciens cells with an empty pSRKKm-Plac plasmid grew and divided to form microcolonies (Fig. 5E; see also Movie S4). In contrast to cells infected with Atu_ph02 or Atu_ph03, which lyse rapidly with little change in cellular morphology (Fig. 2C; Movies S2 and S3), PPH induction causes cells to elongate and branch prior to cell lysis (Fig. 5F; see also Movie S5). The branching phenotype observed when pph was induced in A. tumefaciens cells is reminiscent of cells exhibiting a block in cell division (57–63). This observation suggests that PPH may have a dual function, namely, blocking cell division and triggering cell lysis.
We hypothesized that the peptidoglycan hydrolase activity is necessary for PPH induction to trigger cell lysis. Alignments of DUF3380 sequences from bacterial and viral proteins revealed the presence of a conserved glutamate (E), which is presumed to be a catalytic residue, followed by a conserved serine (S) (55) (Fig. S3). To determine if the predicted catalytic glutamate functions in cell lysis during PPH induction, we mutated this residue in PPH (PPHE32A) and characterized the cell growth (Fig. 5C), cell viability (Fig. 5D), and cell lysis of the PPHE32A strain (Fig. 5G; see also Movie S6). Cultures of A. tumefaciens cells producing PPHE32A became turbid and increased in optical density on the basis of the growth curve analysis (Fig. 5C), but were not viable when spotted on medium containing the inducer (Fig. 5D). Time-lapse microscopy of cell growth when PPHE32A was induced explains these seemingly contradictory observations (Fig. 5G; Movie S6). When PPHE32A is expressed, A. tumefaciens cell division is blocked and large extensively branched cells form. This unusual morphology causes both an increase in light scattering in the growth curve analysis and a marked decrease in cell viability. Cells lysed ∼4 h later postinduction when expressing PPHE32A than when they expressed PPH (compare Fig. 5F to G; Movies S5 and S6). Together, these observations suggest that the predicted N-acetylmuramidase domain contributes to PPH-mediated cell lysis. To further test this hypothesis, we assessed the ability of PPH to clear peptidoglycan embedded in an SDS-PAGE gel. A zymogram loaded with equal amounts of protein from whole-cell lysates of WT cells, WT cells expressing PPH, and WT cells expressing PPHE32A revealed bands of clearing at the expected size of 35 kDa when PPH was expressed (Fig. 5B; see also Fig. S5). A smaller band of clearing (∼25 kDa) was also observed when PPH was expressed and may indicate a degradation product of the PPH. In contrast, no clearing was observed when the PPHE32A variant was expressed (Fig. 5B; Fig. S5). The lack of clearing for the PPHE32A variant shows the important role this residue plays in peptidoglycan cleavage. These results suggest that PPH has peptidoglycan hydrolyzing activity, although it is possible that PPH stimulates other peptidoglycan hydrolases in A. tumefaciens. Future work using purified proteins will be necessary to confirm these results.
The observation that the expression of PPHE32A causes a dramatic cell morphology, including very large branched cells (Fig. 5G), suggests that PPH causes a block in cell division that is independent of the peptidoglycan hydrolase activity. Since divisome assembly is initiated in the cytoplasm (64) and the predicted topology of PPH suggests that only the C-terminal tail would extend into the cytoplasm, we examined this sequence for any remarkable features and observed that this region is lysine rich. To determine if the positively charged C terminus functions in the regulation of cell division and the timing of cell lysis, we constructed a plasmid encoding a PPH variant in which the lysines have been mutated to alanines (PPHK328A,K334A,K335A) and assessed cell growth (Fig. 5C), cell viability (Fig. 5D), and cell lysis (Fig. 5H; see also Movie S7). The growth curve revealed that expression of PPHK328A,K334A,K335A caused a growth defect that is significantly more pronounced than that of the wild-type PPH (Fig. 5C) and that spotting of cells expressing PPHK328A,K334A,K335A under inducing conditions resulted in a decrease in cell viability (Fig. 5D). Remarkably, time-lapse microscopy revealed that cells producing PPHK328A,K334A,K335A lysed rapidly (Fig. 5H; Movie S7). This phenotype is strikingly similar to that of cells infected with phages Atu_ph02 and Atu_ph03 (Fig. 2C; Movies S2 and S3). Unlike the cells expressing PPH, cells that expressed PPHK328A,K334A,K335A lysed rapidly without a block in cell division. This observation suggests that the positively charged lysines function in the regulation of PPH-mediated cell killing by contributing to the block in cell division but are not required for lysis.
To confirm the roles of the catalytic E32 and the positively charged lysine residues, the loss-of-function mutations were combined to create PPHE32A,K328A,K334A,K335A. The induction of PPHE32A,K328A,K334A,K335A restored normal growth (Fig. 5C) and viability (Fig. 5D). Time-lapse microscopy revealed that cells producing PPHE32A,K328A,K334A,K335A continued to elongate and divide, producing microcolonies (Fig. 5; see also Movie S8). Some cells expressing PPHE32A,K328A,K334A,K335A appeared to be overly curved, swollen, or bulging, suggesting that this variant of PPH causes relatively minor defects in the cell wall or cell growth. Together, these data suggest that we have identified important residues responsible for the PPH-mediated block in cell division and cell lysis.
Finally, we sought to determine the contribution of the C-terminal transmembrane domain in PPH-mediated cell lysis. We truncated PPH to remove the C-terminal TM domain and cytosolic tail and expressed PPH1–304 in A. tumefaciens. The expression of PPH1–304 did not impair growth (Fig. 5C), viability (Fig. 5D), or the ability to form microcolonies (Fig. 5J; see also Movie S9). Consistent with the predicted topology of PPH, these observations suggest that the cells do not lyse, since the peptidoglycan hydrolase does not reach the periplasm. We hypothesize that these cells do not exhibit a block in cell division, since the positively charged residues at the extreme C terminus of PPH are also absent.
While the possibility of a dual function of PPH in peptidoglycan hydrolysis and blocking cell division is intriguing, additional work is needed to determine if a block in cell division occurs during infection with phage Atu_ph02 or Atu_ph03 and if PPH contributes to a delay in cell division. Since we are artificially expressing PPH in A. tumefaciens, we cannot yet determine if this phenotype is an artifact of protein expression or representative of PPH induction during phage infection; however, the ability to abolish the cell division defect and induce rapid lysis by mutating the positively charged C terminus may suggest a biological role for this region of the peptide. Dual functions for PPH in peptidoglycan hydrolysis and blocking cell division are consistent with descriptions of single lysis proteins in phages with smaller genomes. For example, coliphages ϕX174, MS2, and Qβ encode lysis proteins E, L, and A2, respectively (65–68). Protein E causes host cell lysis by inhibiting the activity of a host protein involved in peptidoglycan biosynthesis (66). The C terminus of PPH may have a similar function leading to the inhibition of a cell division protein. Indeed, A. tumefaciens cells expressing PPH exhibited a branching phenotype similar to that in FtsZ-depleted cells (63) (Fig. 5F; Movie S5). The inhibition of cell division causes cells to increase in volume, which may benefit the phage by maximizing burst size (69). While it remains unclear if or how the phages Atu_ph02 and Atu_ph03 regulate the timing of lysis during infection, it appears that the C terminus of PPH may contribute to the regulation of cell lysis.
Overall, we find that PPH is a potent inhibitor of A. tumefaciens cell growth and viability, revealing the possibility that this protein may be engineered to be an even more potent antimicrobial. Future studies will be aimed at characterizing the enzymatic activity of PPH, identifying host factors required for PPH-mediated cell division blockade, and addressing the specificity of PPH as an antimicrobial.
Conclusions.
Our laboratory has isolated and characterized two closely related bacteriophages that specifically infect A. tumefaciens strains derived from C58. These phages, Atu_ph02 and Atu_ph03, are lytic and lead to host cell lysis. While the potato tumor assay shows that these phages offer partial protection from tumor formation, the use of these phages for biocontrol may be limited unless additional lytic phages are used in combination therapies. To investigate the mechanism of host cell lysis, the genomes Atu_ph02 and Atu_ph03 were sequenced, revealing the presence of a putative atypical endolysin, termed phage peptidoglycan hydrolase (PPH). PPH is sufficient for the lysis of A. tumefaciens cells and appears to have a dual function in disrupting the divisome assembly or function and triggering cell lysis. Mutational analyses suggest that a putative N-acetylmuramidase domain contributes to cell lysis, while a positively charged C terminus causes a block in cell division. The transmembrane domain is hypothesized to aid in delivering the peptidoglycan hydrolase domain to the periplasm and is necessary for rapid PPH-induced cell lysis. Understanding the mechanism by which PPH blocks cell division, the specific host factor it targets, and its enzymatic activity on the cell wall will elucidate the mode of action of PPH and determine if PPH shows promise as an enzybiotic. More detailed characterization will be necessary to confirm if PPH disrupts cell division during phage infection and to determine if PPH functions as an endolysin late during the phage infection cycle. This work demonstrates that bacteriophages have evolved additional mechanisms to kill their host cells and illustrates the value of exploring bacteriophage genomes as a source of candidate enzybiotics.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in the study are listed in Table 2. Agrobacterium tumefaciens strains and Sinorhizobium meliloti were cultured in Luria-Bertani (LB) broth, with the exception of A. tumefaciens LBA4404, which was cultured using yeast mannitol (YM) medium (70). Agrobacterium vitis was grown in potato dextrose media (Difco), and Caulobacter crescentus was cultured on peptone-yeast extract (PYE) medium (71). All of these strains were grown in liquid cultures at 28°C with shaking. Escherichia coli was cultured in LB broth at 37°C. When necessary, solid medium was prepared with 1.5% agar. Kanamycin was used at a working concentration of 300 μg/ml for A. tumefaciens and 50 μg/ml for E. coli. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used as an inducer at a concentration of 1 mM.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Growth medium | Reference or source |
|---|---|---|---|
| Plasmids | |||
| pSRKKm-Plac-sfgfp | Kmr, broad host range vector containing lacIq and lac promoter | 63 | |
| pSRKKm-Plac-PPH | PPHa inserted into pSRKKm-Plac-sfgfp | This study | |
| pSRKKm-Plac-PPHE32A | PPH predicted catalytic residue mutated | This study | |
| pSRKKm-Plac-PPHK328A,K334A,K335A | PPH regulatory residues mutated | This study | |
| pSRKKm-Plac-PPHE32A,K328A,K334A,K335A | PPH catalytic and regulatory residues mutated | This study | |
| pSRKKm-Plac-PPH1–304 | PPH truncation to remove TM domain | This study | |
| E. coli strains | |||
| DH5α | Cloning strain, gammaproteobacterium | LB | Life Technologies |
| S17-1 | Smr, RP4-2, Tc::Mu, Km-Tn7, for plasmid mobilization | LB | 83 |
| A. tumefaciens strains | |||
| C58 | Nopaline type strain; pTiC58; pAtC58 | LB | 32 |
| C58(pSRKKm-Plac-sfgfp) | C58 transformed with empty pSRKKm plasmid | LB | 63 |
| C58(pSRKKm-Plac-PPH) | C58 transformed with pSRKKM-Plac-PPH | LB | This study |
| C58(pSRKKm-Plac-PPHE32A) | C58 transformed with pSRKKm-Plac-PPHE32A | LB | This study |
| C58(pSRKKm-Plac-PPHK328A,K334A,K335A) | C58 transformed with pSRKKm-Plac-PPHK328A,K334A,K335A | LB | This study |
| C58(pSRKKm-Plac-PPHE32A,K328A,K334A,K335A) | C58 transformed with pSRKKm-Plac-PPHE32A,K328A,K334A,K335A | LB | This study |
| C58(pSRKKm-Plac-PPH1–304) | C58 transformed with pSRKKm-Plac-PPH1–304 | LB | This study |
| EHA105 | C58 derived, succinamopine strain, T-DNA deletion derivative of pTiBo542 | LB | MU plant transformation core facility |
| EHA101 | C58 derived, nopaline strain, T-DNA deletion derivative of pTiBo542 | LB | MU plant transformation core facility |
| GV3101 | C58 derived, nopaline strain | LB | MU plant transformation core facility |
| NTL4 | C58 derived, nopaline-agrocinopine strain, ΔtetRA | LB | 84 |
| AGL-1 | C58 derived, succinamopine strain, T-DNA deletion derivative of pTiBo542 ΔrecA | LB | MU plant transformation core facility |
| LBA4404 | Ach5 derived, octopine strain, T-DNA deletion derivative of pTiAch5 | YM | MU plant transformation core facility |
| Chry5 | Succinamopine strain, pTiChry5 | LB | 85 |
| Other bacterial strains | |||
| Agrobacterium vitis S4 | Vitopine strain, pTiS4, pSymA, pSymB | Potato dextrose | 86 |
| Sinorhizobium meliloti RM41 | Rhizopine strain, pSymA, pSymB, pRme41a | LB | 87 |
| Caulobacter crescentus CB15 | Alphaproteobacterium | PYE | 88 |
PPH, phage peptidoglycan hydrolase.
Clonal isolation of bacteriophage strains.
Bacteriophages capable of infecting A. tumefaciens strain C58 were isolated from wastewater samples using an enrichment protocol adapted from Santamaría et al. (38) and described in detail in the supplemental material.
Plaque assays.
Classic whole-plate plaque assays were performed using the soft agar overlay method (72). To complete whole-plate plaque assays, 100 μl cells (optical density at 600 nm [OD600] of ∼0.2) were incubated with 100 μl of diluted phage for 15 min at 28°C. Phage solutions were serially diluted in phage dilution buffer, Dulbecco's phosphate-buffered saline with calcium and magnesium (DPBS; Mediatech, Inc., Manassas, VA, USA) and 0.1% gelatin. The mixtures of cells and phage were each then added to 3 ml of soft agar prior to overlay. The soft agars containing bacteriophage and cells were poured onto room-temperature LB plates containing 1% agar and were swirled gently to spread the soft agar evenly across the plates. For host range testing, plaque assays were completed by spotting phage on lawns of bacterial cells. In the spot assays, 100 μl cells (OD600 of ∼0.2) was added to 0.3% soft agar and overlaid on solid medium. After solidification, 10 μl samples of phage serial dilutions in phage dilution buffer were spotted on the soft agar. Plates were incubated for 1 to 2 days and observed for plaque formation. Spot assays were used for host range testing with appropriate adjustments to the base medium and soft agar. The media used for each of the strains are listed in Table 2.
Partial purification of virions and preparation of virion DNA.
Virions from 1-liter cleared lysates were enriched and concentrated to 1.5 ml by 2 successive precipitations with 10% polyethylene glycol (73) and differential centrifugation (17,000 × g for 10 min and 288,000 × g for 2 h), as detailed in the supplemental material. Since some nonvirion lysate components copurify with virions during this procedure, we consider these virions to be only partially purified. Virion DNA was prepared from partially purified virions by 2 phenol extractions, chloroform extraction, and ethanol precipitation, as detailed in the supplemental material.
Transmission electron microscopy.
Virion morphology was observed by applying a small volume of concentrated partially purified virions onto a carbon-coated Formvar grid and negatively staining them with 2% uranyl acetate. Specimens were observed on a JEOL JEM-1400 transmission electron microscope at 120 kV. Capsid diameters of 10 virions from each phage strain were measured using ImageJ (74).
Growth curves.
Growth curves were performed in LB medium by infecting C58 cells at an optical density (OD600) of 0.05 with bacteriophage at an MOI of 0.001 in liquid culture. The turbidity of these cultures, represented by their OD600, was recorded every 5 min during a 24-h interval while the cells grew at 28°C. Cultures were shaken for 1 min prior to each reading. The OD600 was measured using a BioTek Synergy H1 Hybrid reader. For the growth curve with induction of PPH or PPH variants, cells were grown in LB medium without inducer for 16 h, and then diluted to an OD600 of 0.05. Where indicated, 1 mM IPTG was added to the cultures just prior to taking the initial reading.
Time-lapse microscopy.
A. tumefaciens strain C58 cells were grown to an OD600 of 0.2 and infected with Atu_ph02 and Atu_ph03 at MOIs of 0.01. Infected cells were incubated at room temperature for 15 min to allow phage attachment before 1-μl portions were added to a 1% agarose pad containing LB as described previously (57, 75). Cells were imaged using a 60× oil immersion objective (1.4 numerical aperture [NA]) by differential interference microscopy every 5 to 10 min for 24 h using a Nikon Eclipse TiE equipped with a QImaging Rolera EM-C2 1 K electron-multiplying charge-coupled-device (EMCCD) camera and Nikon Elements imaging software. Cells containing pSRKKm, pSRKKm-PPH, pSRKKm-PPHE32A, pSRKKm-PPHK328A,K334A,K335A, pSRKKm-PPHE32A,K328A,K334A,K335A, and pSRKKm-PPH1–304 were grown to OD600 values of 0.2 and induced with 1 mM IPTG. Cells were placed in the 28°C shaker for 1 h prior to imaging.
Potato tumor assay.
To test for phage protection from A. tumefaciens-mediated plant transformation, potato tumor assays adapted from Morton and Fuqua (41) were used. Briefly, red-skinned organically grown potatoes were rinsed, peeled, and sterilized prior to cutting discs. Sterilization consisted of soaking the potatoes in 1.05% sodium hypochlorite for 20 min and exposing each side to UV light for 20 min using a Cole-Parmer SK-97505-30 lamp emitting at 254 nm with an irradiance of 900 μW/cm2 at the work surface. Potatoes were then cut into cylinders and sliced into discs with diameters of 2 cm and thicknesses of 0.5 cm. Discs were overlaid with 100 μl cells (OD600 of 0.2) resuspended in DPBS-gelatin. When indicated, cells were premixed with Atu_ph02 or Atu_ph03 at MOIs of 1.0. Potatoes were incubated at room temperature for 10 to 20 days in a humid chamber and tumor formation was observed.
Cell viability assays.
Serial dilutions of A. tumefaciens cells containing plasmids for the expression of pph and pph variants were spotted on plates in the presence and absence of IPTG to test cell viability during pph induction. Colonies were inoculated overnight in LB with kanamycin and diluted to an OD600 of 0.05. Cells were then serially diluted, and 4 μl of each dilution was spotted onto LB plates containing kanamycin (Kan) and either IPTG (for induction) or 1% glucose (for maximal repression). Plates were grown for 2 days at 28°C and imaged.
Genome sequencing and assembly.
Libraries for genome sequencing were constructed from virion DNA following the manufacturer's protocol and reagents supplied in Illumina's TruSeq DNA PCR-free sample preparation kit (FC-121-3001). Briefly, 1 μg of DNA was sheared using standard Covaris methods to generate average fragmented sizes of 350 bp. The resulting 3′ and 5′ overhangs were converted to blunt ends by an end repair reaction using 3′-to-5′ exonuclease and polymerase activities, followed by size selection (350 bp) and purification with magnetic sample purification beads. A single adenosine nucleotide was added to the 3′ ends of the blunt fragments followed by the ligation of Illumina indexed paired-end adapters. The adaptor-ligated library was purified twice with magnetic sample purification beads. The purified library was quantified using a KAPA library quantification kit (KK4824), and library fragment sizes were confirmed by Fragment Analyzer (Advanced Analytical Technologies, Inc.). Libraries were diluted, pooled, and sequenced using a paired-end 75-base read length according to Illumina's standard sequencing protocol for the MiSeq. Library preparation and sequencing were conducted by the University of Missouri DNA core facility.
Genome annotation.
Protein-coding regions were annotated by RAST server (76). Proteins of interest were analyzed by TMHMM (56) and SignalP 4.1 (77). Whole-genome alignments were created using the Mauve (78) plugin in Geneious version 8.1. Phylogenetic trees were constructed using a ClustalW (79) protein alignment and PhyML version 3.0 (80) as a Geneious plugin using the Geneious Tree Builder with the settings set for a Le Gascuel substitution model with 100 bootstrap models. Nucleotide alignments were used to determine the percent identities between genomes using the MUSCLE (81) alignment in Geneious.
Construction of plasmids for characterization of PPH.
See Table 3 for a list of all primers used in plasmid construction and sequencing. All variants of pph were cloned into the vector pSRKKm-Plac-sfgfp, which allows for inducible expression of target genes under the control of the lac promoter using IPTG as the inducer (63). To construct pSRKKm-PPH, PCR using Phusion high-fidelity DNA polymerase (Thermo Scientific) was performed on the Atu_ph02 genomic DNA using primers PPH NdeI F and PPH BamHI R. PCR products were gel purified using the GeneJET gel extraction kit (Thermo Scientific). Amplified gene products and the pSRKKm-Plac-sfgfp plasmid (63) were digested with NdeI and BamHI overnight at 37°C and subsequently gel purified. The digested vector and insert were ligated using T4 DNA ligase (Invitrogen). The ligation reaction mixture was incubated at 4°C overnight. The ligation was transformed into DH5α chemically competent E. coli cells (Invitrogen) and selected for on LB agar plates containing kanamycin. Plasmid DNA was extracted using the GeneJET plasmid miniprep kit (Thermo Scientific) prior to electroporating competent A. tumefaciens cells as described previously (82). The pSRKKm-PPH vector was sequenced using the pSRK forward sequencing primer at the MU DNA core facility. To perform site-directed mutagenesis on PPH, the Q5 site-directed mutagenesis kit was used according to the manufacturer's protocol (New England BioLabs). To construct PPHE32A, primers PPH E32A F and PPH E32A R were used. For PPHK328A K334A K335A, the PPH K328A F and PPH K328A R primers were used first, followed by primers PPH K334A F and PPH K334A R, and lastly PPH K335A F and PPH K335A R. To construct PPH1–304, the PPH NdeI F and PPH1–304 BamHI R primers were used. Generated constructs were sequenced at the MU DNA core facility using the pSRKKm forward sequencing, PPH NdeI F, and PPH linker for NdeI primers.
TABLE 3.
Synthesized DNA primers used in this study
SDS-PAGE and zymography.
Whole-cell lysates were prepared using 100-ml cultures of exponential-phase cells (OD600 of 0.3 to 0.6) grown with inducer for 3 h for WT cells, WT cells expressing PPH, or WT cells expressing PPHE32A. Cells were centrifuged at 4,300 × g for 15 min at 4°C, and the cell pellets stored at −20°C overnight. The next day, pellets were resuspended in 8 ml B-PER bacterial protein extraction reagent (Thermo Scientific) with the addition of one-sixth of a crushed protease inhibitor tablet (Thermo Scientific). Cells were lysed by sonication (4 pulses comprising a 10-s burst, followed by a 20-s burst). Cell debris were pelleted at 17,000 × g for 15 min at 4°C. The soluble proteins in the supernatants were quantified using the Pierce BCA protein assay kit (Thermo Scientific). Thirty micrograms of total protein from each whole-cell lysate sample was boiled for 5 min and loaded onto two gels (one with embedded peptidoglycan), which ran at 30 V for 30 min, followed by 100 V for 90 min. For zymography, SDS-PAGE gels were embedded with peptidoglycan. A 1-liter culture of A. tumefaciens strain C58 was autoclaved, and peptidoglycan was harvested by centrifugation at 7,000 × g for 20 min at 4°C. The pellets containing peptidoglycan were resuspended in 10 ml 1× DPBS with calcium and magnesium (Corning Cellgro). Five hundred microliters of peptidoglycan was added to a 12% SDS polyacrylamide gel. After running the gel, the running gel was incubated in 25 mM Tris (pH 8) with 1% Triton X-100 at 28°C overnight to renature the proteins and enable peptidoglycan hydrolysis. The gels were stained in 1:50 0.1% methylene blue in 0.01% KOH for 3 h and destained in 0.01% KOH. For SDS-PAGE, the running gels were incubated in Coomassie blue dye (0.25% [wt/vol] brilliant blue R-250 [FisherBiotech], 10% acetic acid, 5% methanol) for 30 s, and then destained (7.5% acetic acid, 50% methanol) overnight with shaking.
Accession number(s).
Genome sequences of phages Atu_ph02 and Atu_ph03 have been deposited in the GenBank database with nucleotide accession numbers MF403005 and MF403006, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhanyuan Zhang and the MU plant transformation core facility for providing Agrobacterium strains, and Kenya Phillips and Courtney Buchanan for assisting in the characterization of Atu_ph03. We thank Tommi White, Martin Schauflinger, and DeAna Grant of the MU electron microscopy core for help with the transmission electron microscopy. We thank Nathan Bivens and the MU DNA core for assistance with sequencing the bacteriophages and William Spollen and the MU research informatics core for assistance with genome assembly and GenBank submission. Finally, we thank members of the Brown lab, especially Helen Blaine, and Chiqian Zhang for feedback during the preparation of the manuscript.
This research is supported by startup funds, a research council grant (URC 14-051), and a research board grant (3786-2) from the University of Missouri to P.J.B.B. H.A. has been supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number T32GM008396. J.R. was supported by a Monsanto undergraduate research fellowship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01498-17.
REFERENCES
- 1.Strange RN, Scott PR. 2005. Plant disease: a threat to global food security. Annu Rev Phytopathol 43:83–116. doi: 10.1146/annurev.phyto.43.113004.133839. [DOI] [PubMed] [Google Scholar]
- 2.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escobar MA, Dandekar AM. 2003. Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci 8:380–386. doi: 10.1016/S1360-1385(03)00162-6. [DOI] [PubMed] [Google Scholar]
- 4.Pulawska J. 2010. Crown gall of stone fruits and nuts, economic significance and diversity of its causal agents: tumorigenic Agrobacterium spp. J Plant Pathol 92:S87–S98. [Google Scholar]
- 5.Bourras S, Rouxel T, Meyer M. 2015. Agrobacterium tumefaciens gene transfer: how a plant pathogen hacks the nuclei of plant and nonplant organisms. Phytopathology 105:1288–1301. doi: 10.1094/PHYTO-12-14-0380-RVW. [DOI] [PubMed] [Google Scholar]
- 6.Kado CI. 1976. The tumor-inducing substance of Agrobacterium tumefaciens. Annu Rev Phytopathol 14:265–308. doi: 10.1146/annurev.py.14.090176.001405. [DOI] [Google Scholar]
- 7.Kerr A, Panagopoulos CG. 1977. Biotypes of Agrobacterium radiobacter var. tumefaciens and their biological control. J Phytopathol 90:172–179. doi: 10.1111/j.1439-0434.1977.tb03233.x. [DOI] [Google Scholar]
- 8.Kerr A, Roberts WP. 1976. Agrobacterium: correlations between and transfer of pathogenicity, octopine and nopaline metabolism and bacteriocin 84 sensitivity. Physiol Plant Pathol 9:205–211. doi: 10.1016/0048-4059(76)90041-2. [DOI] [Google Scholar]
- 9.Sule S, Kado CI. 1980. Agrocin resistance in virulent derivative of Agobacterium tumefaciens harboring the pTi plasmid. Physiol Plant Pathol 17:347–356. doi: 10.1016/S0048-4059(80)80028-2. [DOI] [Google Scholar]
- 10.Beneddra T, Picard C, Petit A, Nesme X. 1996. Correlation between susceptibility to crown gall and sensitivity to cytokinin in aspen cultivars. Phytopathology 86:225–231. doi: 10.1094/Phyto-86-225. [DOI] [Google Scholar]
- 11.Bliss FA, Almehdi AA, Dandekar AM, Schuerman PL, Bellaloui N. 1999. Crown gall resistance in accessions of 20 Prunus species. HortScience 34:326–330. [Google Scholar]
- 12.Reynders-Aloisi S, Pelloli G. 1998. Tolerance to crown gall differs among genotypes of rose rootstocks. HortScience 33:296–297. [Google Scholar]
- 13.Anand A, Uppalapati SR, Ryu CM, Allen SN, Kang L, Tang Y, Mysore KS. 2008. Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol 146:703–715. doi: 10.1104/pp.107.111302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dandurishvili N, Toklikishvili N, Ovadis M, Eliashvili P, Giorgobiani N, Keshelava R, Tediashvili M, Vainstein A, Khmel I, Szegedi E, Chernin L. 2011. Broad-range antagonistic rhizobacteria Pseudomonas fluorescens and Serratia plymuthica suppress Agrobacterium crown gall tumours on tomato plants. J Appl Microbiol 110:341–352. doi: 10.1111/j.1365-2672.2010.04891.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones JB, Vallad GE, Iriarte FB, Obradovic A, Wernsing MH, Jackson LE, Balogh B, Hong JC, Momol MT. 2012. Considerations for using bacteriophages for plant disease control. Bacteriophage 2:208–214. doi: 10.4161/bact.23857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buttimer C, McAuliffe O, Ross RP, Hill C, O'Mahony J, Coffey A. 2017. Bacteriophages and bacterial plant diseases. Front Microbiol 8:34. doi: 10.3389/fmicb.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iriarte FB, Obradovic A, Wernsing MH, Jackson LE, Balogh B, Hong JA, Momol MT, Jones JB, Vallad GE. 2012. Soil-based systemic delivery and phyllosphere in vivo propagation of bacteriophages: two possible strategies for improving bacteriophage persistence for plant disease control. Bacteriophage 2:215–224. doi: 10.4161/bact.23530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara A, Fujisawa M, Hamasaki R, Kawasaki T, Fujie M, Yamada T. 2011. Biocontrol of Ralstonia solanacearum by treatment with lytic bacteriophages. Appl Environ Microbiol 77:4155–4162. doi: 10.1128/AEM.02847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balogh B, Canteros BI, Stall KE, Jones JB. 2008. Control of citrus canker and citrus bacterial spot with bacteriophages. Plant Dis 92:1048–1052. doi: 10.1094/PDIS-92-7-1048. [DOI] [PubMed] [Google Scholar]
- 20.Rombouts S, Volckaert A, Venneman S, Devlercq B, Vandenheuvel D, Allonsius CN, Van Malderghem C, Jang HB, Briers Y, Noben JP, Klumpp J, Van Vaerenbergh J, Maes M, Lavigne R. 2016. Characterization of novel bacteriophages for biocontrol of bacterial blight in leek caused by Pseudomonas syringae pv. porri. Front Microbiol 7:279. doi: 10.3389/fmicb.2016.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frampton RA, Taylor C, Holguin Moreno AV, Visnovsky SB, Petty NK, Pitman AR, Fineran PC. 2014. Identification of bacteriophages for biocontrol of the kiwifruit canker phytopathogen Pseudomonas syringae pv. actinidiae. Appl Environ Microbiol 80:2216–2228. doi: 10.1128/AEM.00062-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roach DR, Donovan DM. 2015. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 5:e1062590. doi: 10.1080/21597081.2015.1062590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci U S A 98:4107–4112. doi: 10.1073/pnas.061038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keary R, McAuliffe O, Ross RP, Hill C, O'Mahony J, Coffey A. 2013. Bacteriophages and their endolysins for control of pathogenic bacteria, p 1028–1040. In Méndez-Vilas A. (ed), Microbial pathogens and strategies for combating them: science, technology and education. Formatex Research Center, Badajoz, Spain. [Google Scholar]
- 25.Lim JA, Shin H, Heu S, Ryu S. 2014. Exogenous lytic activity of SPN9CC endolysin against gram-negative bacteria. J Microbiol Biotechnol 24:803–811. doi: 10.4014/jmb.1403.03035. [DOI] [PubMed] [Google Scholar]
- 26.Guo M, Feng C, Ren J, Zhuang X, Zhang Y, Zhu Y, Dong K, He P, Guo X, Qin J. 2017. A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Front Microbiol 8:293. doi: 10.3389/fmicb.2017.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S, Gu J, Lv M, Guo Z, Yan G, Yu L, Du C, Feng X, Han W, Sun C, Lei L. 2017. The antibacterial activity of E. coli bacteriophage lysin lysep3 is enhanced by fusing the Bacillus amyloliquefaciens bacteriophage endolysin binding domain D8 to the C-terminal region. J Microbiol 55:403–408. doi: 10.1007/s12275-017-6431-6. [DOI] [PubMed] [Google Scholar]
- 28.Boyd RJ, Hildebrandt AC, Allen ON. 1970. Specificity patterns of Agrobacterium tumefaciens phages. Arch Mikrobiol 73:324–330. doi: 10.1007/BF00412299. [DOI] [PubMed] [Google Scholar]
- 29.Roslycky EB, Allen ON, McCoy E. 1965. Growth characteristics of phages of Agrobacterium radiobacter. Can J Microbiol 11:95–101. doi: 10.1139/m65-012. [DOI] [PubMed] [Google Scholar]
- 30.Lotz W, Mayer F. 1972. Electron microscopical characterization of newly isolated Rhizobium lupini bacteriophages. Can J Microbiol 18:1271–1274. doi: 10.1139/m72-196. [DOI] [PubMed] [Google Scholar]
- 31.Kropinski AM, Van den Bossche A, Lavigne R, Noben JP, Babinger P, Schmitt R. 2012. Genome and proteome analysis of 7-7-1, a flagellotropic phage infecting Agrobacterium sp H13-3. Virol J 9:102. doi: 10.1186/1743-422X-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson B, Currier TC, Gordon MP, Chilton MD, Nester EW. 1975. Plasmid required for virulence of Agrobacterium tumefaciens. J Bacteriol 123:255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christie PJ, Gordon JE. 2014. The Agrobacterium Ti plasmids. Microbiol Spectr 2. doi: 10.1128/microbiolspec.PLAS-0010-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton RH, Fall MZ. 1971. The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia 27:229–230. doi: 10.1007/BF02145913. [DOI] [PubMed] [Google Scholar]
- 35.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 36.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 37.Gohlke J, Deeken R. 2014. Plant responses to Agrobacterium tumefaciens and crown gall development. Front Plant Sci 5:155. doi: 10.3389/fpls.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santamaría RI, Bustos P, Sepulveda-Robles O, Lozano L, Rodriguez C, Fernandez JL, Juarez S, Kameyama L, Guarneros G, Davila G, Gonzalez V. 2014. Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl Environ Microbiol 80:446–454. doi: 10.1128/AEM.02256-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann HW. 2009. Phage classification and characterization. Methods Mol Biol 501:127–140. doi: 10.1007/978-1-60327-164-6_13. [DOI] [PubMed] [Google Scholar]
- 40.Lazo GR, Stein PA, Ludwig RA. 1991. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- 41.Morton ER, Fuqua C. 2012. Phenotypic analyses of Agrobacterium. Curr Protoc Microbiol Chapter 3:Unit 3D.3. doi: 10.1002/9780471729259.mc03d03s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin Y, Fischer D. 2008. Identification and investigation of ORFans in the viral world. BMC Genomics 9:24. doi: 10.1186/1471-2164-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West D, Lagenaur C, Agabian N. 1976. Isolation and characterization of Caulobacter crescentus bacteriophage phi Cd1. J Virol 17:568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lerma RA, Tidwell TJ, Cahill JL, Rasche ES, Kuty Everett GF. 2015. Complete genome sequence of Caulobacter crescentus podophage Percy. Genome Announc 3:e01373-15. doi: 10.1128/genomeA.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawasaki T, Shimizu M, Satsuma H, Fujiwara A, Fujie M, Usami S, Yamada T. 2009. Genomic characterization of Ralstonia solanacearum phage phiRSB1, a T7-like wide-host-range phage. J Bacteriol 191:422–427. doi: 10.1128/JB.01263-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCorquodale DJ, Lanni YT. 1970. Patterns of protein synthesis in Escherichia coli infected by amber mutants in the first-step-transfer DNA of T5. J Mol Biol 48:133–143. doi: 10.1016/0022-2836(70)90224-X. [DOI] [PubMed] [Google Scholar]
- 47.McCorquodale DJ, Warner HR. 1988. Bacteriophage T5 and related phages, p 439–475. In Calendar R. (ed), The bacteriophages. Plenum Press, New York, NY. [Google Scholar]
- 48.Davison J. 2015. Pre-early functions of bacteriophage T5 and its relatives. Bacteriophage 5:e1086500. doi: 10.1080/21597081.2015.1086500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gill JJ, Berry JD, Russell WK, Lessor L, Escobar-Garcia DA, Hernandez D, Kane A, Keene J, Maddox M, Martin R, Mohan S, Thorn AM, Russell DH, Young R. 2012. The Caulobacter crescentus phage phiCbK: genomics of a canonical phage. BMC Genomics 13:542. doi: 10.1186/1471-2164-13-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young R. 2014. Phage lysis: three steps, three choices, one outcome. J Microbiol 52:243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliveira H, Melo LD, Santos SB, Nobrega FL, Ferreira EC, Cerca N, Azeredo J, Kluskens LD. 2013. Molecular aspects and comparative genomics of bacteriophage endolysins. J Virol 87:4558–4570. doi: 10.1128/JVI.03277-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catalão MJ, Gil F, Moniz-Pereira J, Sao-Jose C, Pimentel M. 2013. Diversity in bacterial lysis systems: bacteriophages show the way. FEMS Microbiol Rev 37:554–571. doi: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 53.Kakikawa M, Yokoi KJ, Kimoto H, Nakano M, Kawasaki K, Taketo A, Kodaira K. 2002. Molecular analysis of the lysis protein Lys encoded by Lactobacillus plantarum phage phig1e. Gene 299:227–234. doi: 10.1016/S0378-1119(02)01076-4. [DOI] [PubMed] [Google Scholar]
- 54.São-José C, Parreira R, Vieira G, Santos MA. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J Bacteriol 182:5823–5831. doi: 10.1128/JB.182.20.5823-5831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodríguez-Rubio L, Gerstmans H, Thorpe S, Mesnage S, Lavigne R, Briers Y. 2016. DUF3380 domain from a Salmonella phage endolysin shows potent N-acetylmuramidase activity. Appl Environ Microbiol 82:4975–4981. doi: 10.1128/AEM.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 57.Brown PJ, de Pedro MA, Kysela DT, Van der Henst C, Kim J, De Bolle X, Fuqua C, Brun YV. 2012. Polar growth in the alphaproteobacterial order Rhizobiales. Proc Natl Acad Sci U S A 109:1697–1701. doi: 10.1073/pnas.1114476109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujiwara T, Fukui S. 1972. Isolation of morphological mutants of Agrobacterium tumefaciens. J Bacteriol 110:743–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujiwara T, Fukui S. 1974. Unidirectional growth and branch formation of a morphological mutant, Agrobacterium tumefaciens. J Bacteriol 120:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kahng LS, Shapiro L. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J Bacteriol 183:3065–3075. doi: 10.1128/JB.183.10.3065-3075.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Latch JN, Margolin W. 1997. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol 179:2373–2381. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su S, Stephens BB, Alexandre G, Farrand SK. 2006. Lon protease of the alpha-proteobacterium Agrobacterium tumefaciens is required for normal growth, cellular morphology and full virulence. Microbiology 152:1197–1207. doi: 10.1099/mic.0.28657-0. [DOI] [PubMed] [Google Scholar]
- 63.Figueroa-Cuilan W, Daniel JJ, Howell M, Sulaiman A, Brown PJ. 2016. Mini-Tn7 Insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the phytopathogen Agrobacterium tumefaciens. Appl Environ Microbiol 82:5015–5025. doi: 10.1128/AEM.01392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lutkenhaus J, Pichoff S, Du S. 2012. Bacterial cytokinesis: from Z ring to divisome. Cytoskeleton (Hoboken) 69:778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernhardt TG, Roof WD, Young R. 2000. Genetic evidence that the bacteriophage phi X174 lysis protein inhibits cell wall synthesis. Proc Natl Acad Sci U S A 97:4297–4302. doi: 10.1073/pnas.97.8.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernhardt TG, Struck DK, Young R. 2001. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J Biol Chem 276:6093–6097. doi: 10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- 67.Chamakura KR, Tran JS, Young R. 2017. MS2 lysis of Escherichia coli depends on host chaperone DnaJ. J Bacteriol 199:e00058–17. doi: 10.1128/JB.00058-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reed CA, Langlais C, Wang IN, Young R. 2013. A(2) expression and assembly regulates lysis in Qbeta infections. Microbiology 159:507–514. doi: 10.1099/mic.0.064790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tran TA, Struck DK, Young R. 2005. Periplasmic domains define holin-antiholin interactions in T4 lysis inhibition. J Bacteriol 187:6631–6640. doi: 10.1128/JB.187.19.6631-6640.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vincent J. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific, Oxford, United Kingdom. [Google Scholar]
- 71.Poindexter JS. 1964. Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams MH. 1959. Bacteriophages. Interscience Publishers, Inc., New York, NY. [Google Scholar]
- 73.Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G. 1970. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- 74.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Howell M, Daniel JJ, Brown PJB. Live cell fluorescence microscopy to observe essential processes during microbial cell growth. J Vis Exp, in press. [DOI] [PMC free article] [PubMed]
- 76.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 78.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 80.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 81.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morton ER, Fuqua C. 2012. Genetic manipulation of Agrobacterium. Curr Protoc Microbiol Chapter 3:Unit 3D.2. doi: 10.1002/9780471729259.mc03d02s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 84.Luo ZQ, Clemente TE, Farrand SK. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol Plant Microbe Interact 14:98–103. doi: 10.1094/MPMI.2001.14.1.98. [DOI] [PubMed] [Google Scholar]
- 85.Bush AL, Pueppke SG. 1991. Characterization of an unusual new Agrobacterium tumefaciens strain from Chrysanthemum morifolium Ram. Appl Environ Microbiol 57:2468–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Slater SC, Goldman BS, Goodner B, Setubal JC, Farrand SK, Nester EW, Burr TJ, Banta L, Dickerman AW, Paulsen I, Otten L, Suen G, Welch R, Almeida NF, Arnold F, Burton OT, Du Z, Ewing A, Godsy E, Heisel S, Houmiel KL, Jhaveri J, Lu J, Miller NM, Norton S, Chen Q, Phoolcharoen W, Ohlin V, Ondrusek D, Pride N, Stricklin SL, Sun J, Wheeler C, Wilson L, Zhu H, Wood DW. 2009. Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191:2501–2511. doi: 10.1128/JB.01779-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weidner S, Baumgarth B, Gottfert M, Jaenicke S, Puhler A, Schneiker-Bekel S, Serrania J, Szczepanowski R, Becker A. 2013. Genome sequence of Sinorhizobium meliloti Rm41. Genome Announc 1:e00013-12. doi: 10.1128/genomeA.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nierman WC, Feldblyum TV, Laub MT, Paulsen IT, Nelson KE, Eisen JA, Heidelberg JF, Alley MR, Ohta N, Maddock JR, Potocka I, Nelson WC, Newton A, Stephens C, Phadke ND, Ely B, DeBoy RT, Dodson RJ, Durkin AS, Gwinn ML, Haft DH, Kolonay JF, Smit J, Craven MB, Khouri H, Shetty J, Berry K, Utterback T, Tran K, Wolf A, Vamathevan J, Ermolaeva M, White O, Salzberg SL, Venter JC, Shapiro L, Fraser CM. 2001. Complete genome sequence of Caulobacter crescentus. Proc Natl Acad Sci U S A 98:4136–4141. doi: 10.1073/pnas.061029298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.