Significance
Alternative splicing is a regulatory mechanism by which multiple protein isoforms can be generated from one gene. Despite its biological importance, there has been no systematic approach that facilitates characterizing functional roles of protein isoforms in human metabolism. To this end, we present a systematic framework for the generation of gene-transcript-protein-reaction associations (GeTPRA) in human metabolism. The framework involves a generic human genome-scale metabolic model (GEM) that is an excellent framework to investigate genotype–phenotype associations. We show that a biochemically consistent and transcript-level data-compatible human GEM can be used to generate GeTPRA, which can be deployed to further upgrade the human GEM. Personal GEMs generated with GeTPRA information enabled more accurate simulation of cancer metabolism and prediction of anticancer targets.
Keywords: alternative splicing, gene-transcript-protein-reaction associations, human genome-scale metabolic model, protein isoform, Recon
Abstract
Alternative splicing plays important roles in generating different transcripts from one gene, and consequently various protein isoforms. However, there has been no systematic approach that facilitates characterizing functional roles of protein isoforms in the context of the entire human metabolism. Here, we present a systematic framework for the generation of gene-transcript-protein-reaction associations (GeTPRA) in the human metabolism. The framework in this study generated 11,415 GeTPRA corresponding to 1,106 metabolic genes for both principal and nonprincipal transcripts (PTs and NPTs) of metabolic genes. The framework further evaluates GeTPRA, using a human genome-scale metabolic model (GEM) that is biochemically consistent and transcript-level data compatible, and subsequently updates the human GEM. A generic human GEM, Recon 2M.1, was developed for this purpose, and subsequently updated to Recon 2M.2 through the framework. Both PTs and NPTs of metabolic genes were considered in the framework based on prior analyses of 446 personal RNA-Seq data and 1,784 personal GEMs reconstructed using Recon 2M.1. The framework and the GeTPRA will contribute to better understanding human metabolism at the systems level and enable further medical applications.
Alternative splicing of a gene generates multiple transcripts, which are translated to protein isoforms (1, 2). In human cells, 92–97% of the multiexon genes undergo alternative splicing events (3) and achieve functional diversity by providing alternative functional proteins and domains (4). Gene transcripts encoding protein isoforms can be categorized into principal and nonprincipal transcripts (PTs and NPTs). PTs are representative transcripts of a gene with a major biochemical function, whereas all the other alternative transcripts are currently NPTs requiring further study (5). The biological roles of protein isoforms need to be precisely understood, as they are highly associated with human cell metabolism and disease progression (4, 6, 7). Recent advances in next-generation sequencing including RNA-Seq and proteome/protein localization technologies have facilitated functional characterization of an increasing number of protein isoforms; these include computational prediction of biological functions of protein isoforms (4), analysis of recurrent switches of protein isoforms in tumor versus nontumor samples (8), annotation of PTs for each gene (9), and proteomic investigation of subcellular localization (SL) of protein isoforms (10). With increasing volumes of such transcript-level data, an upgraded systems biology framework is needed to facilitate characterizing functional roles of protein isoforms by linking transcript-level information (e.g., RNA-Seq data) to a higher biological phenotype, metabolism.
Human genome-scale metabolic models (GEMs) provide a systematic framework to investigate genotype–phenotype associations and can be considered to characterize protein isoforms (11–16). GEMs in general describe genome-wide metabolic pathways encoded by the target organism’s genome, using stoichiometric coefficients of associated metabolites (17, 18). To date, a series of comprehensive human GEMs has been released, including Recon 1 (19), Recon 2 (20), a revised Recon 2 (hereafter, Recon 2Q) (13), and Recon 2.2 (21), as well as human metabolic reaction (HMR) series (22, 23). These human GEMs have been employed to predict anticancer targets (24–26) and oncometabolites (27), characterize metabolism of abnormal human myocyte with type 2 diabetes (28), investigate roles of gut microbiota in host glutathione metabolism (29), predict biomarkers in response to drugs (30), predict essentiality of human genes having diverse numbers of transcript variants (31), identify poor prognosis in patients with breast cancer (32), and predict tumor sizes and overall survival rates of patients with breast cancer (33). Despite such a wide application scope, currently available human GEMs cannot be used to address transcript–phenotype associations beyond genotype–phenotype links because the human GEMs have incomplete gene-protein-reaction (GPR) associations that cannot be integrated with transcript-level data. Although previously released human GEMs Recon 1 and 2 claimed that transcript-level information was incorporated in their GPR associations (19, 20), the transcript identifiers (IDs) used in these models (e.g., Entrez gene ID x.1, x.2, …, etc.) do not match with those described in major genome annotation databases (34) (Results). Thus, gene-transcript-protein-reaction associations (GeTPRA) should be systematically defined to characterize functional roles of protein isoforms generated from different transcripts of metabolic genes.
Here, we introduce a systematic framework that evaluates metabolic functions of protein isoforms to generate GeTPRA, which is subsequently used to update a human GEM. In this study, the framework generated 11,415 GeTPRA corresponding to 1,106 metabolic genes. To establish the framework, a generic human GEM Recon 2M.1 was first developed that is biochemically consistent and transcript-level data compatible. Also, the importance of PTs and NPTs in human metabolism was analyzed using 446 personal RNA-Seq data (Dataset S1) and reconstruction of 1,784 personal GEMs based on the Recon 2M.1; consequently, both PTs and NPTs were considered in the framework. The framework for the GeTPRA was subsequently used to upgrade Recon 2M.1 toward Recon 2M.2. We discuss how the framework and its resulting GeTPRA can contribute to better understanding the biological roles of transcripts in human metabolism and enable further medical applications.
Results
Generation of a Generic Human GEM Recon 2M.1 That Is Biochemically Consistent and Transcript-Level Data Compatible.
Recon 2M.1, a biochemically consistent and transcript-level data-compatible generic human GEM, was first generated by systematically refining Recon 2Q (13). This model refinement is not just to enable model integration with transcript-level data but also to use Recon 2M.1 to evaluate whether reactions mediated by protein isoforms carry fluxes (Fig. 1 A and B). For the latter purpose, the blocked reactions in Recon 2Q were resolved by removing or gap-filling them. Although revised, Recon 2Q still appeared to have a large number of blocked reactions: 1,544 blocked reactions were found in Recon 2Q, corresponding to 21.1% of all of the metabolic reactions, according to flux variability analysis (see SI Appendix, SI Materials and Methods for details; Dataset S2). A large number of blocked reactions came from the previously released GEMs used to reconstruct Recon 2 (i.e., Ac-FAO, Edinburgh Human Metabolic Network, HepatoNet1, and hs_eIEC611; see SI Appendix, SI Materials and Methods). Systematic refinement of the Recon 2Q toward Recon 2M.1 and further incorporation of the updates made in Recon 2.2 (21) are detailed in SI Appendix, SI Materials and Methods and Dataset S2.
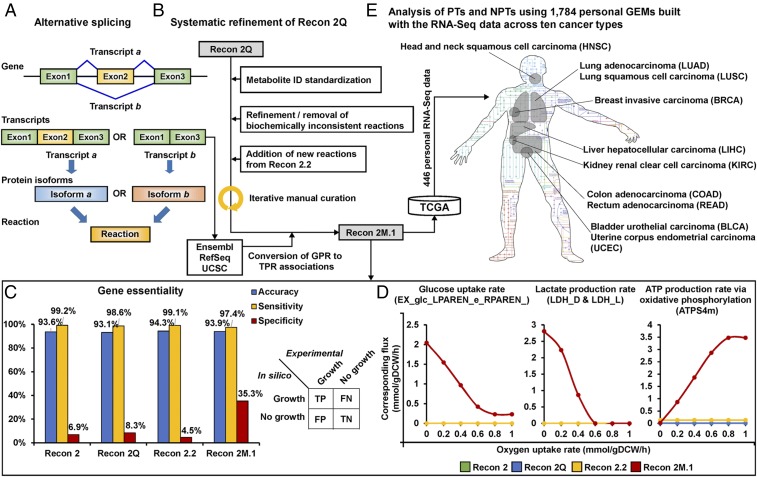
Fig. 1.
A scheme of Recon 2M.1 development and its use in reconstructing personal GEMs. (A) A concept of alternative splicing of human genes and its use in TPR associations of Recon 2M.1. Transcript IDs were obtained from Ensembl (37), RefSeq (38), and UCSC (39) databases. (B) A procedure of systematic refinement of the Recon 2Q. Biochemically inconsistent reactions include unbalanced, artificial, blocked, and/or redundant reactions. Iterative manual curation was conducted while validating the Recon 2M.1. (C) Results of the gene essentiality simulation using Recon 2 (20), Recon 2Q (13), Recon 2.2 (21), and Recon 2M.1. Experimental information on a total of 870 essential and 15,425 nonessential genes was obtained from Wang et al. (35). Accuracy, sensitivity, and specificity values were calculated using equations defined in SI Appendix, SI Materials and Methods, along with the prediction results. FN, false negatives; FP, false positives; TN, true negatives; TP, true positives. (D) Predicted profiles of glucose uptake rate and lactate and ATP production rates when maximizing the cell growth rate of the four human GEMs along the oxygen uptake rate. Relevant reaction IDs are shown in parentheses. (E) Reconstruction of 1,784 personal GEMs using Recon 2M.1 and 446 TCGA personal RNA-Seq data across the 10 cancer types, and their use to analyze principal and nonprincipal transcripts.
The resulting Recon 2M.1 appeared to be biochemically more consistent than the previous versions (Fig. 1 C and D and Table 1). In Recon 2M.1, the numbers of blocked reactions and dead-end metabolites were reduced, and the percentages of gene-mediated reactions and metabolites with annotations (i.e., chemical formula, compound IDs of public chemical databases, and structural descriptions) were all increased compared with previous Recon models (Table 1). This improved model statistic was accompanied with a reduction in the model size of Recon 2M.1. More important, Recon 2M.1 was found to give much improved simulation performance compared with the previous Recon versions (Fig. 1 C and D). Recon 2.2 and Recon 2M.1 showed biologically reasonable ATP production rates under aerobic or anaerobic conditions in a defined minimal medium containing one of 35 different carbon sources, while Recon 2 and Recon 2Q failed to predict biologically reasonable ATP production rates (Datasets S3 and S4). Furthermore, Recon 2M.1 showed more reliable predictions in gene essentiality than the previous human GEMs, according to experimental gene essentiality data recently released (35) (Fig. 1C and SI Appendix, SI Materials and Methods for the definition of essential and nonessential genes). Gene essentiality simulations of four models showed that all the four Recon models showed comparable accuracy of 93.1–94.3% and sensitivity values of 97.4–99.2%. However, Recon 2M.1 showed the greatest specificity value of 35.3%; the other three models had specificity values of 4.5–8.3% (Fig. 1C and SI Appendix, SI Materials and Methods for definitions of accuracy, sensitivity and specificity). Also, Recon 2M.1 was the only model that generated biologically reasonable profiles of glucose uptake rate and lactate and ATP production rates via oxidative phosphorylation in response to changes in oxygen uptake rate (Fig. 1D). These profiles altogether can be interpreted as anaerobic glycolysis in normal cells, for example, during extensive exercises, or as aerobic glycolysis (or Warburg effect) in cancer cells (36).
Table 1.
Comparison of model properties of Recon 2, Recon 2Q, Recon 2.2, Recon 2M.1, and Recon 2M.2
| Property | Recon 2 | Recon 2Q | Recon 2.2 | Recon 2M.1 | Recon 2M.2 |
| Total no. of reactions | 7,440 | 7,327 | 7,785 | 5,825 | 5,842 |
| Total no. of metabolites | 5,063 | 4,962 | 5,324 | 3,368 | 3,368 |
| No. of unique metabolites | 2,626 | 2,531 | 2,652 | 1,735 | 1,735 |
| No. of transcripts | 2,194 | 2,167 | N/A | 15,692 (Ensembl) | 15,597 (Ensembl) |
| 4,028 (RefSeq) | 4,005 (RefSeq) | ||||
| 14,094 (UCSC) | 14,004 (UCSC) | ||||
| No. of unique genes | 1,789 | 1,775 | 1,674 | 1,682 | 1,663 |
| No. of blocked reactions (% of all reactions) | 1,603 (21.5%) | 1,546 (21.1%) | 1,863 (23.9%) | 744 (12.8%) | 737 (12.6%) |
| No. of dead-end metabolites (% of all metabolites) | 1,176 (23.2%) | 1,066 (21.5%) | 1,088 (20.4%) | 239 (7.1%) | 240 (7.1%) |
| No. of balanced reactions (% of all reactions) | 6,340 (85.2%) | 6,242 (85.2%) | 7,035 (90.4%) | 4,568 (78.4%) | 4,574 (78.3%) |
| No. of gene-mediated reactions (% of all reactions) | 3,918 (52.7%) | 3,879 (52.9%) | 4,727 (60.7%) | 3,914 (67.2%) | 3,940 (67.4%) |
| No. of metabolites with chemical formula (% of all metabolites) | 4,877 (96.3%) | 4,807 (96.9%) | 5,318 (99.9%) | 3,310 (98.3%) | 3,310 (98.3%) |
| No. of metabolites with compound IDs of ChEBI, HMDB, HumanCyc and/or KEGG (% of all metabolites) | 3,081 (60.9%) | 3,341 (67.3%) | 3,274 (61.0%) | 2,321 (68.9%) | 2,321 (68.9%) |
| No. of metabolites with structural information using InChI and/or SMILES (% of all metabolites) | 2,914 (57.6%) | 3,023 (60.9%) | 3,360 (63.1%) | 2,333 (69.3%) | 2,333 (69.3%) |
On validation of the biochemical consistency of Recon 2M.1, its GPR associations were updated according to the latest GPR information from the Virtual Metabolic Human database (https://vmh.uni.lu/). Entrez gene IDs used in the GPR associations of Recon 2M.1 were subsequently converted to three different types of transcript IDs for three major genome annotation databases [i.e., Ensembl (37), RefSeq (38), and UCSC (39); Fig. 1A and SI Appendix, Fig. S1]. Therefore, the Recon 2M.1 now has TPR associations in which each reaction is associated with transcript IDs in place of gene IDs (Fig. 1A). When the Recon 2M.1 gets integrated with RNA-Seq data, transcript expression values can be mapped onto these transcript IDs in the TPR associations.
Relative Expression Levels of PTs in Human Metabolic Genes.
To generate the GeTPRA for human metabolic genes, we questioned whether, for each metabolic gene, it would be sufficient to consider only PTs, or whether entire transcripts (both PTs and NPTs) should be considered. This is because systems-level studies so far have focused only on PTs with respect to the functions and SLs of their corresponding protein products, while only a limited number of NPTs were characterized. Thus, we examined how important it is to consider NPTs at the systems level in generating accurate GeTPRA. To answer this question, relative expression levels of PTs compared with total transcript levels were first examined for each metabolic gene to define fractional expression (FE; Fig. 2A). Subsequently, the FEs of metabolic genes in the 446 TCGA personal RNA-Seq data were investigated (Fig. 1E and Dataset S1). PTs were chosen for each gene on the basis of the annotation available at the APPRIS database (9); it should be noted that a gene can have multiple PTs, and 12.4% of human genes have a single PT.
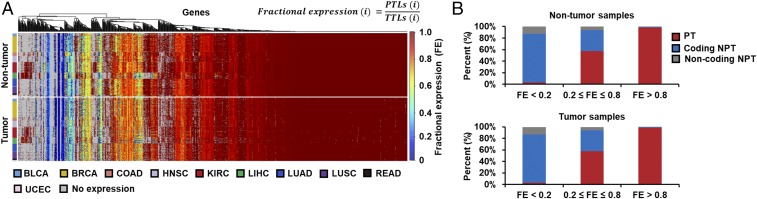
Fig. 2.
Relative expression levels of principal transcripts (PTs) in metabolic genes from 446 TCGA personal RNA-Seq data across the 10 cancer types. (A) Heat maps showing the distribution of FEs for all the metabolic genes from nontumor and tumor samples of the RNA-Seq data. FE is calculated by dividing principal transcript levels (PTLs) by total transcript levels (TTLs) for each metabolic gene, i. Metabolic genes in blue regions have high TTLs but low PTLs (i.e., low FE values), whereas metabolic genes in red regions have both consistent TTLs and PTLs (i.e., high FE values). Unexpressed genes are shown in gray. (B) Relative expression levels of PTs, coding NPTs, and noncoding NPTs for three different ranges of FEs in the nontumor and tumor samples.
Overall distribution of FEs of the metabolic genes in both nontumor and tumor samples suggests that PTs of each metabolic gene exert different levels of metabolic activities (Fig. 2A and SI Appendix, Fig. S2). Approximately 75% of metabolic genes appeared to have FEs >0.8 (Dataset S5). Among them, essential genes reported in Wang et al. (35) have an average FE value of 0.93 and suggest that essential genes operate reactions mainly through their PTs (SI Appendix, Fig. S3). Meanwhile, ∼6% of the metabolic genes were found to have FEs <0.2 (blue regions in Fig. 2A) in both nontumor and tumor samples. For these genes exhibiting such low FEs, NPTs rather than PTs are likely playing more important roles. This suggestion is also based on an observation that almost all the NPTs of genes showing FEs <0.2 actually encode protein products (Fig. 2B). Genes having FEs <0.2 are mainly involved in extracellular transport reactions and keratan sulfate synthesis reactions. Further evidence of changes in expression levels of PTs decoupled from total transcript levels in nontumor and tumor samples is available in SI Appendix, Fig. S4. These lines of evidence suggest NPTs should also be considered in addition to PTs to precisely describe GeTPRA of various types of cells (e.g., normal and cancer cells).
Reconstruction and Analysis of 1,784 Personal GEMs.
To date, personal GEMs have been reconstructed by including a particular metabolic reaction when the total transcript level of the corresponding gene is greater than a certain threshold value (11). The importance of PTs in personal GEMs is obvious, but the relative importance of NPTs is not known. On the basis of the above results showing different levels of importance exerted by NPTs, two different types of personal GEMs, one with total transcript-level data (T-GEM) and the other with PT-level data (P-GEM), were reconstructed to further gauge potential influence of NPTs in human metabolism. Recon 2M.1 was integrated with 446 TCGA personal RNA-Seq data to generate personal GEMs using the task-driven Integrative Network Inference for Tissues (tINIT) method (24, 40). A modified weight function was used for the implementation of the tINIT method to minimize the effects of outliers in the RNA-Seq data and sample variations (Fig. 3A and SI Appendix, SI Materials and Methods and Figs. S5–S7 and Dataset S6). Through this procedure, a total of 1,784 personal GEMs were reconstructed: 446 T-GEMs and 446 P-GEMs each for both nontumor and tumor samples (Fig. 3A). All these personal GEMs were found to satisfy essential metabolic tasks (i.e., generation of ATP, biomass, 8 nucleotides and 10 key intermediates; see SI Appendix, SI Materials and Methods for details; Dataset S7).
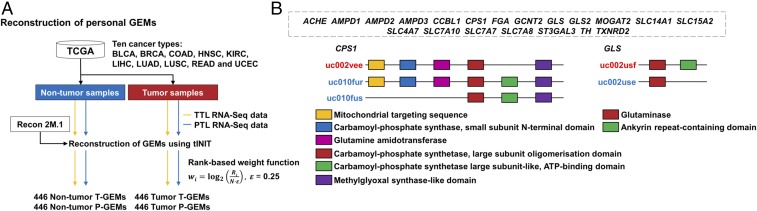
Fig. 3.
Consideration of PTs and NPTs in personal GEMs built with TCGA personal RNA-Seq data. (A) Reconstruction of a total of 1,784 personal GEMs through integration of Recon 2M.1 with 446 personal RNA-Seq data across the 10 cancer types. Information on total transcript and principal transcript levels (TTLs and PTLs) from the TCGA RNA-Seq data were used to build T-GEMs and P-GEMs, respectively, for personal nontumor and tumor samples. All personal GEMs were built using the tINIT algorithm (24, 40) with a rank-based weight function (SI Appendix, SI Materials and Methods and Fig. S5). Abbreviations of the cancer type names and the number of personal GEMs reconstructed for each cancer type are available in Fig. 1E and Dataset S1, respectively. (B) Twenty metabolic genes (dotted gray box) enriched in nontumor and/or tumor T-GEMs. Information on protein domains of these enriched metabolic genes was obtained from Ensembl (52). Protein isoforms of CPS1 and GLS have different sets of protein domains and subcellular localizations. Blue and red UCSC transcript IDs indicate PTs and NPTs, respectively.
To confirm the overall model validity, these 1,784 personal GEMs were examined by comparing T-GEMs and P-GEMs, as well as by analyzing them in the context of the FEs of metabolic genes for both nontumor and tumor samples (Fig. 2A). Obviously, T-GEMs had greater metabolic contents than P-GEMs in both nontumor and tumor samples of all cancer types (SI Appendix, Fig. S8). Pairwise comparison of T-GEMs and P-GEMs revealed that 475 and 462 reactions (associated with 34 and 35 metabolic pathways, respectively) were exclusively present in T-GEMs reconstructed for nontumor and tumor samples, respectively (SI Appendix, Fig. S9). The corresponding metabolic pathways of reactions unique to nontumor and tumor T-GEMs were overall redundant (i.e., 28 metabolic pathways being shared between the two types of T-GEMs); such a high degree of similarity between the nontumor and tumor T-GEMs was consistent with the overall similar distribution of FEs of all of the metabolic genes in both nontumor and tumor samples (Fig. 2A). Reactions unique to both T-GEMs were largely involved in extracellular transports, which were also consistent with the observations made for the FEs <0.2. This observation is attributed to the fact that extracellular transport contains the greatest number of reactions among pathways (SI Appendix, Fig. S10) and possesses the greatest number of reactions associated with genes having FEs <0.2 in Recon 2M.1 (SI Appendix, Fig. S11). However, there are also many metabolic genes exhibiting FEs <0.2 that play major roles in metabolic reactions (SI Appendix, Fig. S11). Thus, it could be concluded that the 1,784 personal GEMs reflected the observed characteristics of the FEs of metabolic genes in nontumor and tumor samples and were considered to be suitable for further examination of potential roles of NPTs in human GEMs.
Importance of Considering NPTs in Human GEMs.
Presence of a large number (tens to hundreds) of NPT-associated genes in the T-GEMs discussed here and comparative gene enrichment analysis of T-GEMs and P-GEMs revealed that NPT-associated genes/reactions should not be ignored in the reconstruction of personal GEMs. Although genes with FEs <0.2 make up only 6% of the entire metabolic genes considered (Fig. 2A), the number of such genes is not insignificant; overall, 446 nontumor T-GEMs had 44–214 genes with FEs <0.2 (Dataset S8). This trend also stands true for the tumor samples, as 446 tumor T-GEMs had 44–216 genes with FEs <0.2 (Dataset S8). This observation indicates that metabolic activities exerted by NPT-encoded proteins in both nontumor and tumor samples should not be ignored. These genes with FEs <0.2 included in T-GEMs were further selected and analyzed by comparing T-GEMs and P-GEMs for both nontumor and tumor samples [blue regions in SI Appendix, Fig. S12; comparative gene enrichment analysis using Fisher’s exact test with false discovery rate (FDR)-corrected P value < 0.05]. A total of 20 genes with FEs <0.2 appeared to be significantly enriched in T-GEMs for both nontumor and tumor samples. Among them, protein isoforms for sufficiently expressed PTs and NPTs of ACHE, AMPD1, AMPD2, AMPD3, CCBL1, FGA, GCNT2, GLS2, MOGAT2, SLC14A1, SLC15A2, SLC4A7, SLC7A10, SLC7A7, SLC7A8, ST3GAL3, TH, and TXNRD2 genes were found to share the same sets of protein domains (Fig. 3B and Dataset S9). In contrast, protein isoforms of CPS1 and GLS genes do not, which indicates that a single metabolic gene might play multiple biological roles, if functional, through different sets of protein domains and SL sequences in their protein isoforms (Fig. 3B). If the information on NPTs (i.e., 44–214 genes in 446 nontumor T-GEMs and 44–216 genes in 446 tumor T-GEMs) was not considered in reconstructing personal GEMs as in P-GEMs, the chance to identify GeTPRA would be highly limited. These results suggest NPTs should also be considered in reconstructing human GEMs when their expression levels are sufficiently high. This information is reflected in the reconstruction of human Recon 2M.2, as described here.
Systematic Generation of GeTPRA.
On the basis of the three studies presented here, we designed a framework that systematically generates GeTPRA as a resource for the study of human metabolic genes and network (Fig. 4A). Establishment of the framework for the GeTPRA was motivated by the fact that protein isoforms of a single metabolic gene can have multiple SL sequences and/or different domains (e.g., CPS1 and GLS in Fig. 3B). The framework uses software tools that extract catalytic and compartmental information of each protein isoform; EFICAz2.5 (41) and Wolf PSort (42) were used in this study, which predict EC numbers and SLs for each given peptide sequence, respectively. The results (Dataset S10) suggest the EFICAz and Wolf PSort are reliable for the prediction of GeTPRA, although they have room to be further improved. As a first step of the framework, the peptide sequences of 2,688 PT- and 4,594 NPT-encoded proteins resulting from 7,282 transcripts of 1,682 metabolic genes defined in Recon 2M.1 were retrieved from Ensembl and subjected to EFICAz2.5 and Wolf PSort analyses; genes were considered only if they have Ensembl and Entrez gene IDs consistently cross-referenced. As a result, 1,106 metabolic genes could be assigned for their EC numbers and SLs, and 576 genes could not (top pie chart in Fig. 4A). Among 1,106 metabolic genes, 556 (50%) genes were predicted to have a single consistent SL and an EC number for their 1,037 protein isoforms. A total of 468 (42%) metabolic genes generated 2,048 protein isoforms with a single EC number, but multiple SLs. Also, 21 (2%) metabolic genes generated protein isoforms with a single SL, but multiple EC numbers. Finally, 61 (6%) metabolic genes generated protein isoforms with multiple SLs and EC numbers (Fig. 4A). Thus, protein isoforms belonging to the last three categories can carry out multiple metabolic functions. Those unassigned 576 metabolic genes were not considered in GeTPRA (Dataset S11) because their protein isoforms could not be assigned with EC numbers and/or SLs.
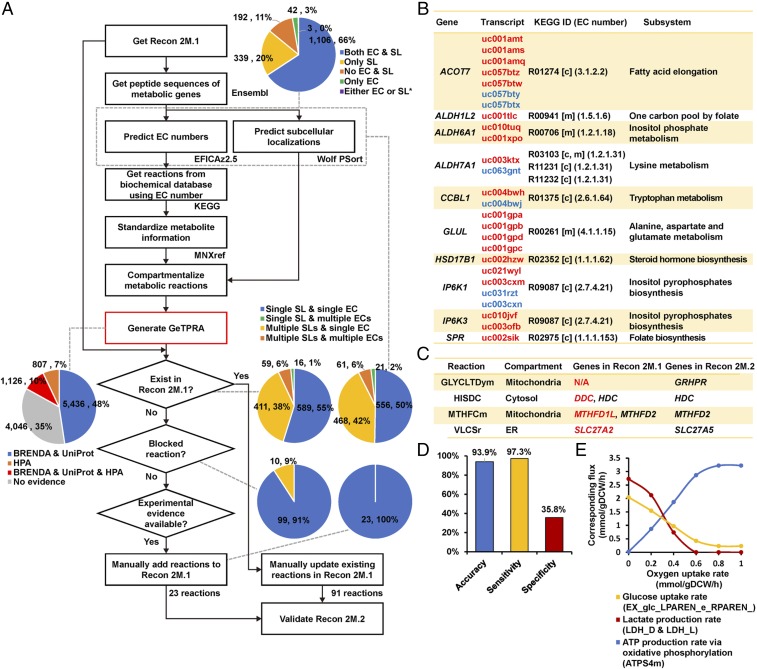
Fig. 4.
Systematic generation of GeTPRA. (A) A framework that generated the GeTPRA (red box; Dataset S11). Peptide sequences of metabolic genes defined in Recon 2M.1 were retrieved from Ensembl (52). EC numbers and SLs of all the protein isoforms of metabolic genes in Recon 2M.1 were predicted using EFICAz2.5 (41) and Wolf PSort (42), respectively. The top pie chart right to the flowchart shows the number and percentage of the genes having protein isoforms with availability of corresponding predictions (i.e., EC numbers and/or SLs). “Either EC or SL” with an asterisk indicates genes that have multiple protein isoforms, where some protein isoforms were assigned with only EC numbers, whereas others were assigned with only SLs. Reactions were retrieved from KEGG database (43) and standardized with MNXM IDs from MNXref namespace (53, 54). A pie chart left to the flowchart represents the number and percentage of the GeTPRA with availability of corresponding experimental evidence [i.e., HPA (10, 46) for SLs, and BRENDA (44) and UniProt (45) for EC numbers]. Four pie charts on the right side represent the number and percentage of the remaining metabolic genes after each process in the framework. The remaining metabolic genes may have a single and/or multiple EC number or numbers and SL or SLs through protein isoforms. (B) Ten representative metabolic reactions which were newly added to Recon 2M.1 according to the GeTPRA (see the full list of 23 newly added reactions in Dataset S12). Blue and red UCSC transcript IDs indicate PTs and NPTs, respectively. Metabolic reactions are represented with KEGG reaction IDs. (C) Four representative metabolic reactions whose GPR/TPR associations were updated based on the GeTPRA. For better readability, gene symbols are presented here instead of transcript IDs. Updated gene symbols in the Recon 2M.1 are shown in red. (D) Results of the gene essentiality simulation using the Recon 2M.2. Information on essential and nonessential genes used for the Recon 2M.1 validation was also used here (Fig. 1C). (E) Predicted profiles of glucose uptake rate and lactate and ATP production rates via oxidative phosphorylation when maximizing the cell growth rate of the Recon 2M.2 along the oxygen uptake rate. Relevant reaction IDs are shown in parentheses.
With EC numbers predicted for each protein isoform, their corresponding metabolic reactions were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (43), using KEGG API (www.kegg.jp/kegg/rest/) (Fig. 4A). The retrieved metabolic reactions were subsequently standardized using MNXM IDs from MNXref namespace and assigned with compartments according to the predicted SLs. As a result, a total of 11,415 GeTPRA including 2,976 candidate unique reactions were generated as an output of the framework (Fig. 4A). Overall, 65% of GeTPRA have experimental evidence available at BRENDA (44), UniProt (45), and/or Human Protein Atlas (HPA) (10, 46), while the remaining 35% GeTPRA are not experimentally supported (leftmost pie chart in Fig. 4A). It should be noted that contents and accuracies of GeTPRA appeared to be barely influenced by the version of Ensembl in the framework (Dataset S10).
On the basis of these results, GeTPRA was used to upgrade Recon 2M.1. The candidate reactions from GeTPRA, which were not available in Recon 2M.1, were added to the model one by one if they had experimental evidence, and examined with flux variability analysis to confirm whether they actually carry metabolic fluxes (Dataset S11). As a result, 25 candidate reactions were predicted to carry metabolic fluxes. They were manually curated, and 23 metabolic reactions were finally added to Recon 2M.1 (Fig. 4B and SI Appendix, Fig. S13 and Dataset S12). The 23 reactions are mediated by 43 PT- and 14 NPT-encoded proteins of 21 metabolic genes (i.e., ACOT7, ALDH1L2, ALDH6A1, ALDH7A1, BLVRA, CCBL1, DCTPP1, GLUL, GRHPR, HSD17B1, IP6K1, IP6K3, ISYNA1, ME2, MPST, PPOX, SPR, SULT2A1, TST, UCK1, and UCK2). The corresponding reactions added were involved in rather diverse metabolic pathways encompassing metabolisms of amino acids, carbohydrates, energy, lipids, nucleotides, xenobiotics, cofactors and vitamins. Despite the potential biochemical importance of these 23 reactions, they were not present in the previous Recon models.
Having used 23 metabolic reactions of GeTPRA, the remaining 2,953 reactions of GeTPRA were further employed to correct existing reactions in Recon 2M.1 that have obvious errors. Among the remaining 2,953 reactions, GPR/TPR associations of 272 metabolic reactions in Recon 2M.1 were found to be inconsistent with the remaining 2,953 reactions of the GeTPRA. Thus, GPR/TPR associations of these 272 reactions were manually curated. As a result, GPR/TPR associations of 85 reactions in Recon 2M.1 were modified, and six reactions were removed according to the GeTPRA, all having experimental evidence (Fig. 4C and Dataset S13). The resulting updated human GEM Recon 2M.2 was validated in the same way as Recon 2M.1 (Fig. 1 C and D). Gene essentiality analysis showed that the specificity obtained with Recon 2M.2 was 35.8% (Fig. 4D), which was slightly better than that (35.3%) obtained with Recon 2M.1 (Fig. 1C). Also, Recon 2M.2 was able to generate a biologically reasonable glucose uptake rate and lactate and ATP production rates in response to changes in oxygen uptake rate (Fig. 4E). Although we used Recon 2M.1 as an input for the GeTPRA framework, any Recon model can also be considered for an update if the model has consistent TPR associations and biologically reasonable simulation performance (Figs. 1 C and D and 4 D and E).
Simulation of Cancer Metabolism Using T-GEMs Built with Recon 2.2, 2M.1, and 2M.2.
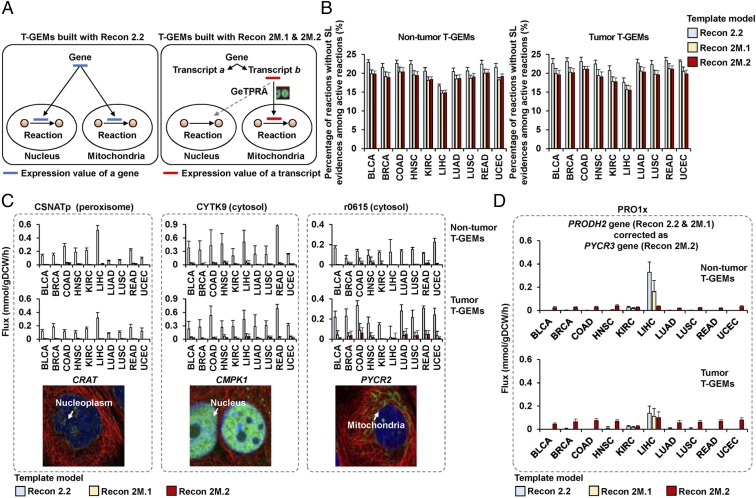
Next, we simulated cancer metabolism using T-GEMs built with Recon 2.2, 2M.1, and 2M.2 as template models to further validate GeTPRA Recon 2M.1 and 2M.2. For this, 446 nontumor and 446 tumor T-GEMs were first built using Recon 2.2, 2M.1 (already reconstructed earlier; Fig. 3A), and 2M.2 as template models and by using the tINIT method and 446 TCGA personal RNA-Seq data (SI Appendix, SI Materials and Methods). Upon generation of the personal nontumor/tumor T-GEMs, their metabolic fluxes were predicted by using the expression data from nontumor and tumor samples of the 446 TCGA personal RNA-Seq data as constraints and implementing the least absolute deviation method (47, 48) (SI Appendix, SI Materials and Methods). In this process of setting constraints for the Recon 2M.1- and 2M.2-based T-GEMs, the GeTPRA dataset (Dataset S11) was also used to specifically map transcripts to their corresponding reactions with correct compartments (Fig. 5A and SI Appendix, SI Materials and Methods). In case of T-GEMs built with Recon 2.2, gene information was mapped to all the relevant reactions (24). As a result, T-GEMs built with Recon 2.2, 2M.1 and 2M.2 all generated biologically reasonable flux profiles of lactate dehydrogenase (LDH_L), pyruvate kinase (PYK), and ATP synthase (ATPS4m) in tumors versus nontumors (SI Appendix, Fig. S14). However, for the entire metabolism, nontumor and tumor T-GEMs built with Recon 2.2 had the greatest percentage of reactions missing relevant SL evidence among active reactions (carrying fluxes) (Fig. 5B); SL evidence was obtained from HPA (www.proteinatlas.org/). T-GEMs built with Recon 2M.2 had the lowest percentage of reactions that are not experimentally supported. As a consequence, the use of GeTPRA more likely prevents reactions from carrying fluxes if relevant experimental evidence (e.g., SL data) is not available (Fig. 5 B and C and Dataset S14). In addition, modification of GPR/TPR associations in Recon 2M.2 through the GeTPRA (Fig. 4) enabled correct prediction of additional reaction flux values compared with those built with Recon 2.2 and 2M.1, as seen in the case of the reaction PRO1x (Fig. 5D). In conclusion, simulations with the T-GEMs built with Recon 2M.2 overall provided more reliable flux distributions. A series of these simulations demonstrates that GeTPRA, Recon 2M.1 and 2M.2 can be used to understand more accurately the effects of transcript-level changes on metabolic fluxes at the systems level, and consequently allow studying nondiseased and diseased states for further medical applications.
Fig. 5.
Simulation of cancer metabolism using T-GEMs built with Recon 2.2, 2M.1 and 2M.2. (A) Prediction of reaction fluxes using nontumor and tumor samples of the 446 TCGA personal RNA-Seq data. For T-GEMs built with Recon 2M.1 and 2M.2, the GeTPRA dataset serves to specifically map transcripts to their corresponding reactions with correct compartments. For T-GEMs built with Recon 2.2, expression values of genes are directly mapped to all of the relevant reactions. The black arrows from a gene or a transcript to reaction compartments indicate integration of expression values with a reaction. The gray dotted arrow indicates no integration for the transcript expression value, as the protein isoform does not have relevant SL evidence. (B) Average percentage of reactions without SL evidence among active reactions for all of the T-GEMs in each cancer type. SL evidence was collected from the HPA (v16.proteinatlas.org) (10, 46). (C) Three representative reactions having high flux values in the nontumor/tumor T-GEMs built with Recon 2.2. These flux values should be low or zero because these reactions do not have relevant SL evidence (see Dataset S14 for details). The reaction r0615 for pyrroline-5-carboxylate reductase 2 (PYCR2) is located in cytosol in Recon models, whereas its protein isoform was experimentally located to be in mitochondria (image). This inconsistency was also observed for the reactions CYTK9 for cytidine/uridine monophosphate kinase 1 (CMPK1) and CSNATp for carnitine O-acetyltransferase (CRAT). Legends for bars are same as in B. (Top and Bottom) Nontumor and tumor T-GEMs, respectively. All of the shown immunohistochemistry images were obtained from the HPA (10, 46). Antibody IDs of the shown immunohistochemistry images are HPA022815 for CRAT; HPA053730 for CMPK1; and HPA056873 for PYCR2. (D) Correct prediction of the PRO1x flux values by correcting its gene PRODH2 (encoding proline dehydrogenase 2) in Recon 2.2 and 2M.1 to PYCR3 (encoding pyrroline-5-carboxylate reductase 3) in Recon 2M.2 based on the GeTPRA. The high flux value of PRO1x in nontumor T-GEMs built with Recon 2.2 and 2M.1 for the LIHC was caused by the incorrect assignment of PRODH2 that is known to be highly expressed in liver according to the HPA. PYCR3 is moderately expressed in all cell types. Error bars mean ± SD.
Prediction of Anticancer Targets Using Tumor T-GEMs Built with Recon 2.2 and 2M.2.
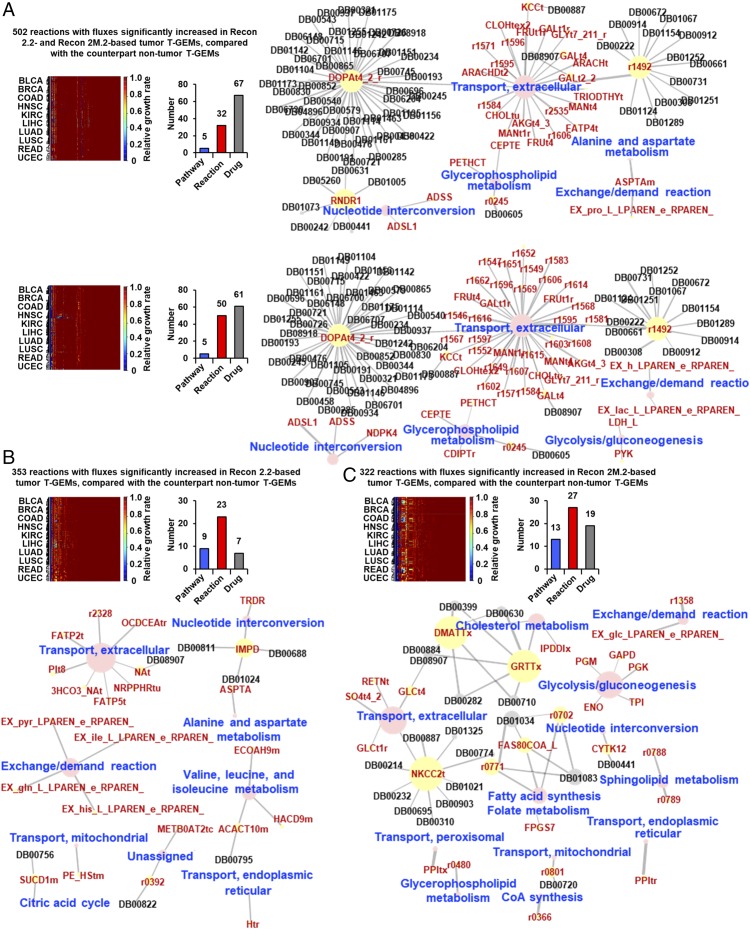
As an example of applications of T-GEMs developed by incorporating GeTPRA, the tumor T-GEMs built with Recon 2.2 and 2M.2 were compared in predicting anticancer targets. The potential anticancer targets were first selected by identifying those metabolic reactions that had fluxes predicted to be significantly increased in tumor T-GEMs in comparison with the counterpart nontumor T-GEMs across the 10 cancer types. These reactions were obtained from the previous section (Fig. 5) and subsequently subjected to single-knockout simulations (see SI Appendix, SI Materials and Methods for the knockout simulation method). Recon 2.2- and 2M.2-based T-GEMs had 502 reactions in common that showed increased fluxes in tumor T-GEMs (Fig. 6A), whereas Recon 2.2-based T-GEMs had a unique set of 353 such reactions (Fig. 6B). T-GEMs built with Recon 2M.2 had 322 unique reactions (Fig. 6C). These reactions were deemed final anticancer targets if their single knockouts reduced growth rates of tumor T-GEMs to less than 5% (25, 31) of their normal growth rates. As a result, Recon 2M.2-based T-GEMs generated greater numbers of both anticancer targets and approved drugs inhibiting the predicted targets: a total of 77 targets and 80 drugs from Recon 2M.2-based tumor T-GEMs versus a total of 55 targets and 74 drugs from Recon 2.2-based tumor T-GEMs. It should be noted that although these drugs are known to inhibit the predicted anticancer targets, they are not necessarily anticancer drugs. Therefore, the drugs known to inhibit the predicted targets could be considered as anticancer drugs (49, 50) if they were initially developed for diseases other than cancers. Recon 2.2- and 2M.2-based tumor T-GEMs generated 32 and 50 reactions as anticancer targets, respectively, on single knockouts of the 502 common reactions; 67 and 61 approved drugs were found to inhibit these anticancer targets from the Recon 2.2- and 2M.2-based tumor T-GEMs, respectively (Fig. 6A). Meanwhile, tumor T-GEMs built with Recon 2M.2 generated 27 anticancer targets across 13 metabolic pathways by knocking out the 322 reactions, nine of which appeared to be inhibited by 19 approved drugs (Fig. 6C). Tumor T-GEMs built with Recon 2.2 generated 23 anticancer targets across nine metabolic pathways, but only seven approved drugs were found to inhibit these targets (Fig. 6B). Interestingly, the anticancer targets that reduce the ratio of glycolytic to oxidative ATP flux (AFR) (25) were predicted only from the Recon 2M.2-based T-GEMs (SI Appendix, SI Materials and Methods): d-glucose exchange (EX_glc_LPAREN_e_RPAREN_), enolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, PYK, and triose-phosphate isomerase. A decrease in the AFR value on perturbation of a reaction is a strong indicator of an anticancer target, as the AFR value is positively correlated with cancer cell migration. Among the predicted targets reducing the AFR, glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase were previously validated by experiments (25). These targets were not predicted using Recon 2.2-based T-GEMs, suggesting that Recon 2M.2-based T-GEMs that incorporate GeTPRA allow more accurate prediction.
Fig. 6.
Prediction of anticancer targets using T-GEMs built with Recon 2.2 and 2M.2. (A) Heat maps representing relative growth rates (see SI Appendix, SI Materials and Methods for the definition) of tumor T-GEMs built with Recon 2.2 (Upper Left) and 2M.2 (Bottom Left) on single knockouts of 502 metabolic reactions. These 502 reactions were the ones having fluxes predicted to be commonly significantly increased in tumor T-GEMs built with Recon 2.2 and Recon 2M.2 compared with their counterpart nontumor T-GEMs across the 10 cancer types (Fig. 5; FDR-corrected P value < 0.01). Relationships among the reactions predicted as anticancer targets (yellow node with red letters), their corresponding pathways (pink node with blue letters), and approved drugs inhibiting the corresponding reactions (DrugBank IDs, “DB” followed by five digits in black) are shown in the form of network next to each heat map. (B) A heat map representing relative growth rates of tumor T-GEMs built with Recon 2.2 on single knockouts of 353 reactions with fluxes predicted to be significantly increased in comparison with the counterpart nontumor T-GEMs across the 10 cancer types (FDR-corrected P value < 0.01). The same type of network shown in A is also shown here below the heat map. (C) A heat map representing relative growth rates of tumor T-GEMs built with Recon 2M.2 on single knockouts of 322 reactions with fluxes predicted to be significantly increased in comparison with the counterpart nontumor T-GEMs across the 10 cancer types (FDR corrected P value < 0.01). The relative growth rates were obtained in the same manner as A. The same type of network shown in A is also shown here below the heat map. Information on the approved drugs and their targets was obtained from DrugBank 5.0 (55).
Discussion
In this work, we established a framework to generate GeTPRA and demonstrated its use in upgrading human GEMs and further application studies. The GeTPRA presented in this article are based on the transcript, EC number, KEGG reaction, and protein SL data available up to now, and thus can be continuously updated. Toward the development of more thorough and robust GeTPRA and reconstruction of a better human GEM, the following things need to be considered. First, the definitions of PT and NPT, although we used the most up-to-date information from APPRIS, are still being updated. Current definition of PTs for human metabolic genes can be ambiguous because their relative contribution to metabolic activities in comparison with total transcript levels (i.e., FEs presented in Fig. 2A) can vary significantly across environmental and biological conditions. It should be emphasized that functionally unknown NPTs should not be ignored, as they might play important roles in human metabolism, as we reported in this study (Fig. 3B). Second, GeTPRA need to be continuously updated, as mentioned earlier. Particular attention should be paid to the GeTPRA data that were removed from further consideration in our study. Such GeTPRA data representing blocked reactions in the Recon 2M.1 or not having experimental evidence do not necessarily mean they are biologically irrelevant in human metabolism. Rather, they should also be considered for future biochemical studies, including experimental validation, depending on a research purpose. Thus, GeTPRA can be used as a conceptual framework to further explore biological roles of transcripts generated from human metabolic genes. Of course, every time human genome annotation gets updated, the GeTPRA should be updated through reexecution of the framework. Third, quality control and quality assurance tests need to be established for Recon development. Despite continued efforts in updating the metabolic contents in the Recon models, simulation performance has been shown to be rather poor (Fig. 1 C and D). However, it is nice to see that more and more useful genetic and biochemical data, such as the gene essentiality data (35) used in this study, are becoming available to perform appropriate quality control and quality assurance tests. We might humbly suggest that Recon 2M.1 and 2M.2 serve as template models for reconstructing future Recon models.
It is hoped that the GeTPRA framework, the current version of GeTPRA, Recon 2M series (i.e., 2M.1 and 2M.2), and source codes to generate them will serve as community resources for fundamental studies on human metabolic genes and network. Beyond basic research, application studies such drug targeting, as showcased earlier, and identification of disease-related metabolic reactions can be performed using GeTPRA and Recon 2M.2 based on diseased cell-specific transcriptome data because many diseases and alternative splicing events are highly associated with each other. Such application studies might further contribute to exploring the effects of novel bioactive compounds on human metabolism to present novel ways of treating diseases (51). Through further community effort based on our study, it is believed that upgraded human GEMs will become available.
Materials and Methods
All the materials and methods conducted in this study are detailed in SI Appendix, SI Materials and Methods: standardization of metabolite IDs with MNXM IDs defined in the MNXref namespace, refinement or removal of biochemically inconsistent reactions (Datasets S2 and S15), validation of Recon 2M.1 (Datasets S2–S4), conversion of GPR to TPR associations in Recon 2M.1, acquisition of 446 TCGA personal RNA-Seq data across 10 cancer types and statistical comparative expression analyses (SI Appendix, Fig. S4), reconstruction of 1,784 personal GEMs across 10 cancer types (SI Appendix, Figs. S5–S7, Datasets S3 and S6), simulation of cancer metabolism using T-GEMs built with Recon 2.2, 2M.1 and 2M.2, prediction of anticancer targets using tumor T-GEMs built with Recon 2.2 and 2M.2, and metabolic simulations in General (Dataset S3).
Eight versions of COBRA-compliant SBML files are available for Recon 2M.1 and Recon 2M.2 at https://zenodo.org/record/583326, depending on the use of MNXref versus BiGG IDs and of Entrez gene IDs (GPR associations) versus Ensembl transcript IDs versus RefSeq transcript IDs versus UCSC transcript IDs (TPR associations for the last three database IDs). COBRA-compliant SBML files of the 1,784 personal GEMs (both P-GEMs and T-GEMs) built with Recon 2M.1, 892 personal GEMs (only T-GEMs) built with Recon 2M.2, and 892 personal GEMs (only T-GEMs) built with Recon 2.2 are also available as zip files at https://zenodo.org/record/583326. Source codes used in this study are available (https://bitbucket.org/kaistmbel/recon-manager) for the collection of scripts used to generate and simulate Recon 2M.1 and 2M.2 and for the implementation of the GeTPRA framework (https://bitbucket.org/kaistmbel/getpra).
Supplementary Material
Acknowledgments
We acknowledge contributions from the TCGA Research Network. The results published here are in whole or part based on data generated by the TCGA Research Network: https://cancergenome.nih.gov/. This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea.
Footnotes
Conflict of interest statement: S.Y.L. and H.U.K. have coauthored publications with J.N. and N.D.P. These were Commentary articles and did not involve any research collaboration.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713050114/-/DCSupplemental.
References
- 1.Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 2.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Li HD, Menon R, Omenn GS, Guan Y. The emerging era of genomic data integration for analyzing splice isoform function. Trends Genet. 2014;30:340–347. doi: 10.1016/j.tig.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez JM, Carro A, Valencia A, Tress ML. APPRIS webserver and webservices. Nucleic Acids Res. 2015;43:W455-9. doi: 10.1093/nar/gkv512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medina MW, Krauss RM. Alternative splicing in the regulation of cholesterol homeostasis. Curr Opin Lipidol. 2013;24:147–152. doi: 10.1097/MOL.0b013e32835cf284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calabretta S, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031–2039. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebestyén E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res. 2015;43:1345–1356. doi: 10.1093/nar/gku1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez JM, et al. APPRIS: Annotation of principal and alternative splice isoforms. Nucleic Acids Res. 2013;41:D110–D117. doi: 10.1093/nar/gks1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 11.Ryu JY, Kim HU, Lee SY. Reconstruction of genome-scale human metabolic models using omics data. Integr Biol. 2015;7:859–868. doi: 10.1039/c5ib00002e. [DOI] [PubMed] [Google Scholar]
- 12.Mardinoglu A, Gatto F, Nielsen J. Genome-scale modeling of human metabolism–A systems biology approach. Biotechnol J. 2013;8:985–996. doi: 10.1002/biot.201200275. [DOI] [PubMed] [Google Scholar]
- 13.Quek LE, et al. Reducing Recon 2 for steady-state flux analysis of HEK cell culture. J Biotechnol. 2014;184:172–178. doi: 10.1016/j.jbiotec.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Mardinoglu A, Nielsen J. New paradigms for metabolic modeling of human cells. Curr Opin Biotechnol. 2015;34:91–97. doi: 10.1016/j.copbio.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Hyötyläinen T, et al. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat Commun. 2016;7:8994. doi: 10.1038/ncomms9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma H, et al. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol Syst Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HU, Sohn SB, Lee SY. Metabolic network modeling and simulation for drug targeting and discovery. Biotechnol J. 2012;7:330–342. doi: 10.1002/biot.201100159. [DOI] [PubMed] [Google Scholar]
- 18.Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15:107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- 19.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiele I, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swainston N, et al. Recon 2.2: From reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardinoglu A, et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol Syst Biol. 2013;9:649. doi: 10.1038/msb.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mardinoglu A, et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 24.Agren R, et al. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10:721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yizhak K, et al. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol Syst Biol. 2014;10:744. doi: 10.15252/msb.20134993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam H, et al. A systems approach to predict oncometabolites via context-specific genome-scale metabolic networks. PLoS Comput Biol. 2014;10:e1003837. doi: 10.1371/journal.pcbi.1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Väremo L, et al. Proteome- and transcriptome-driven reconstruction of the human myocyte metabolic network and its use for identification of markers for diabetes. Cell Rep. 2015;11:921–933. doi: 10.1016/j.celrep.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 29.Mardinoglu A, et al. The gut microbiota modulates host amino acid and glutathione metabolism in mice. Mol Syst Biol. 2015;11:834. doi: 10.15252/msb.20156487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blais EM, et al. Reconciled rat and human metabolic networks for comparative toxicogenomics and biomarker predictions. Nat Commun. 2017;8:14250. doi: 10.1038/ncomms14250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu JY, Kim HU, Lee SY. Human genes with a greater number of transcript variants tend to show biological features of housekeeping and essential genes. Mol Biosyst. 2015;11:2798–2807. doi: 10.1039/c5mb00322a. [DOI] [PubMed] [Google Scholar]
- 32.Leoncikas V, Wu H, Ward LT, Kierzek AM, Plant NJ. Generation of 2,000 breast cancer metabolic landscapes reveals a poor prognosis group with active serotonin production. Sci Rep. 2016;6:19771. doi: 10.1038/srep19771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Megchelenbrink W, Katzir R, Lu X, Ruppin E, Notebaart RA. Synthetic dosage lethality in the human metabolic network is highly predictive of tumor growth and cancer patient survival. Proc Natl Acad Sci USA. 2015;112:12217–12222. doi: 10.1073/pnas.1508573112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfau T, Pacheco MP, Sauter T. Towards improved genome-scale metabolic network reconstructions: Unification, transcript specificity and beyond. Brief Bioinform. 2016;17:1060–1069. doi: 10.1093/bib/bbv100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–D669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruitt KD, et al. RefSeq: An update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karolchik D, et al. University of California Santa Cruz The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agren R, et al. Reconstruction of genome-scale active metabolic networks for 69 human cell types and 16 cancer types using INIT. PLoS Comput Biol. 2012;8:e1002518. doi: 10.1371/journal.pcbi.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar N, Skolnick J. EFICAz2.5: Application of a high-precision enzyme function predictor to 396 proteomes. Bioinformatics. 2012;28:2687–2688. doi: 10.1093/bioinformatics/bts510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton P, et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheer M, et al. BRENDA, the enzyme information system in 2011. Nucleic Acids Res. 2011;39:D670–D676. doi: 10.1093/nar/gkq1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 47.Lee D, et al. Improving metabolic flux predictions using absolute gene expression data. BMC Syst Biol. 2012;6:73. doi: 10.1186/1752-0509-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HU, Kim TY, Lee SY. Framework for network modularization and Bayesian network analysis to investigate the perturbed metabolic network. BMC Syst Biol. 2011;5(Suppl 2):S14. doi: 10.1186/1752-0509-5-S2-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ozsvári B, Lamb R, Lisanti MP. Repurposing of FDA-approved drugs against cancer–Focus on metastasis. Aging (Albany NY) 2016;8:567–568. doi: 10.18632/aging.100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanhaelen Q, et al. Design of efficient computational workflows for in silico drug repurposing. Drug Discov Today. 2017;22:210–222. doi: 10.1016/j.drudis.2016.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Kim HU, Ryu JY, Lee JO, Lee SY. A systems approach to traditional oriental medicine. Nat Biotechnol. 2015;33:264–268. doi: 10.1038/nbt.3167. [DOI] [PubMed] [Google Scholar]
- 52.Kinsella RJ, et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bernard T, et al. Reconciliation of metabolites and biochemical reactions for metabolic networks. Brief Bioinform. 2014;15:123–135. doi: 10.1093/bib/bbs058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moretti S, et al. MetaNetX/MNXref–Reconciliation of metabolites and biochemical reactions to bring together genome-scale metabolic networks. Nucleic Acids Res. 2016;44:D523–D526. doi: 10.1093/nar/gkv1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Law V, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.