Abstract
Sensory rhodopsins (SRs) belong to a subfamily of heptahelical transmembrane proteins containing a retinal chromophore. These photoreceptors mediate the cascade of vision in animal eyes and phototaxis in archaebacteria and unicellular flagellated algae. Signal transduction by these photoreceptors occurs by means of transducer proteins. The two archaebacterial sensory rhodopsins SRI and SRII are coupled to the membrane-bound HtrI and HtrII transducer proteins. Activation of these proteins initiates phosphorylation cascades that modulate the flagellar motors, resulting in either attractant (SRI) or repellent (SRII) phototaxis. In addition, transducer-free SRI and SRII were shown to operate as proton pumps, analogous to bacteriorhodopsin. Here, we present the x-ray structure of SRII from Natronobacterium pharaonis (pSRII) at 2.1-Å resolution, revealing a unique molecular architecture of the retinal-binding pocket. In particular, the structure of pSRII exhibits a largely unbent conformation of the retinal (as compared with bacteriorhodopsin and halorhodopsin), a hydroxyl group of Thr-204 in the vicinity of the Schiff base, and an outward orientation of the guanidinium group of Arg-72. Furthermore, the structure reveals a putative chloride ion that is coupled to the Schiff base by means of a hydrogen-bond network and a unique, positively charged surface patch for a probable interaction with HtrII. The high-resolution structure of pSRII provides a structural basis to elucidate the mechanisms of phototransduction and color tuning.
Archaeal rhodopsins are photoactive heptahelical transmembrane proteins containing a retinal chromophore that is bound to a Lys in helix G. Despite their common structural fold, their functions are distinctly different: bacteriorhodopsin (BR) and halorhodopsin (HR) are light-driven ion pumps transporting protons and chloride ions, respectively. The two archaebacterial sensory rhodopsins (SRs) SRI and SRII are blue- and green-sensitive photosensors that transduce light signals into attractant and repellent migratory responses, respectively (1, 2). Signal transduction by these photoreceptors occurs by means of the membrane-bound HtrI and HtrII transducer proteins, whose activation initiates phosphorylation cascades that modulate the flagellar motors (3). In addition, analogous to BR, both SRI and SRII can operate as proton pumps when not bound to their respective transducers (2), suggesting that key elements of the mechanism of energy transduction in BR have been conserved in SRs. In particular, activation of these photoreceptors and also of visual rhodopsins from animal eyes induces tilting of helices on the cytoplasmic side (4). The mechanism by which this structural change in SRII is translated into proton pumping or signaling is not understood yet. Moreover, the physical mechanism of the spectral shift of SRII, which absorbs maximally near 500 nm (as compared with other archaeal rhodopsins whose absorption maxima are >560 nm) has not been defined. A comprehensive understanding of the mechanism of action of SR requires a knowledge of its high-resolution structure. Here, we present the x-ray structure of SRII from Natronobacterium pharaonis (pSRII) at 2.1-Å resolution, revealing a unique molecular architecture of the retinal-binding pocket that is essential for elucidating the mechanism of phototransduction and color tuning as well as a putative positively charged binding site for the transducer HtrII.
Methods
Protein Expression, Purification, and Crystallization.
The pSRII gene was amplified by PCR with genomic DNA extracted from N. pharaonis. The PCR was carried out with primers that introduced NcoI and XhoI sites. The PCR product was digested with NcoI and XhoI and ligated into pET-28b(+) (Novagen), which contains a sequence coding for a terminal His tag. The fidelity of the PCR product was confirmed by sequence analysis. For overexpression, transformed Escherichia coli BL21 were grown at 37°C in medium supplemented with 50 μg/ml kanamycin. Expression was induced by the addition of 1 mM isopropyl β-d-thiogalactoside (IPTG) and 10 μM all-trans retinal. After 2.5 hr, cells were harvested and broken up in a French press. Purification of pSRII was carried out as described (5). In brief, membranes were isolated by centrifugation, and solubilized with 1.5% n-dodecyl β-d-maltoside. pSRII was purified by applying the detergent-solubilized membranes to a Ni-nitrilotriacetic acid agarose column. Purified pSRII was mixed with polar lipids isolated from Halobacterium salinarum at a lipid:protein molar ratio of 15:1. Crystallization trials of pSRII from a 60% (wt/wt) lipidic–cubic phase at 20°C were set up in 72-microwell plates or in glass vials with cubic phase volume of about 0.2 μl and 5.0 μl, respectively. Final concentrations were 4.5 mg/ml pSRII, 0.005% n-dodecyl β-d-maltoside, 75 mM NaOAc at pH 4.6, and 1.5 M NaCl. Crystals formed after a few weeks and grew as needles with lengths up to 300 μm.
X-Ray Data Collection.
X-ray diffraction data were collected at beam lines ID14-EH2 and ID13 of the European Synchrotron Radiation Facility (ESRF), Grenoble, France, using ADSC (Quantum, Durham, NC) and MAR CCD (MAR-Research, Hamburg, Germany) x-ray detectors, respectively. Data set 1 was collected to 2.1-Å resolution on ID14-EH2 and consisted of a continuous data collection of 120 frames with 1° oscillations. Data set 2 was collected to 2.3-Å resolution on the ID13 microfocus beam line in several passes of 10 frames of 1° oscillation. Crystals were aligned along the spindle axis. During data collection, crystals were translated so as to be sequentially exposed to the x-ray beam at various spots along the needle. The data obtained exhibited low mosaicity (0.5°).
Data Processing.
Data were processed with the software program hkl package (6) and the ccp4 suite (7). Crystals belong to space group C2221 (a = 84.8 Å, b = 128.7 Å, c = 50.7 Å) with one monomer per asymmetric unit.
Crystallographic Refinement.
The structure of pSRII was solved by molecular replacement with the program amore (8). BR model 1QHJ was used as the search model (9) and was restricted to the seven transmembrane helices in which all residues were set to Ala. Loops and side chains were progressively built during subsequent positional and individual B-factor refinement rounds with programs cns (10) and o (11). Simulated annealing rounds were regularly performed. The final model to 2.1-Å resolution consists of residues 2–219 (of 239 residues), the retinal, one chloride ion, and 27 water molecules.
Results and Discussion
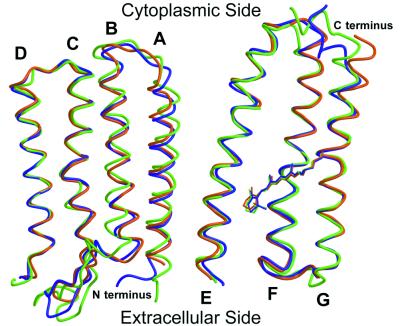
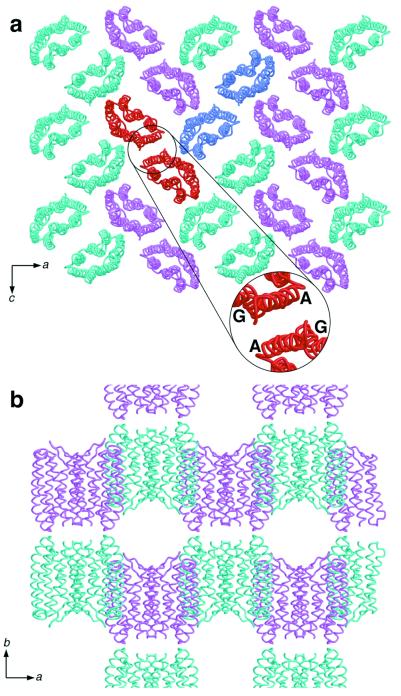
Crystals of pSRII were grown in a lipidic–cubic phase (12) and diffracted to 2.1-Å resolution (Table 1). The x-ray structure of pSRII was solved by molecular replacement using the ground-state BR structure (9) as the initial search model. Despite their different functions, the overall structures of pSRII, BR (13), and HR (14) are conserved: they are composed of seven transmembrane helices, and the chromophore, an all-trans retinal, is bound by a Schiff base to a Lys on helix G (Fig. 1). Helices C–G display the highest similarity, with an rms deviation (rmsd) on the main chain atoms of 0.77 Å (pSRII-BR) and 0.89 Å (pSRII-HR), respectively. Residual electron density in 2Fobs−Fcalc and Fobs−Fcalc maps revealed the presence of alkyl chains next to helices A and G on one side of the protein, and next to helices B and C on the other side. Aromatic residues corresponding to Tyr-64 and Trp-80 in BR, which interact strongly with lipids and contribute to trimer stabilization (9), are absent from pSRII. Like BR and HR, pSRII forms a layered packing arrangement in lipidic–cubic phase-grown crystals. In contrast to BR and HR, pSRII packs pairwise within the crystals, with helices A and G at the interface (Fig. 2).
Table 1.
Diffraction data and refinement statistics
| Beam line measurement | Data set 1, ID14-EH2 | Data set 2, ID13 |
|---|---|---|
| Wavelength, Å | 0.933 | 0.964 |
| Resolution, Å | 2.1 | 2.3 |
| Unit cell, Å* | 84.8, 128.7, 50.7 | 85.9, 129.8, 51.2 |
| Mosaicity, ° | 1.7 | 0.5 |
| No. of observations | 99,617 | 43,109 |
| No. of unique reflections | 15,962 | 12,594 |
| Rsym, %† (outer shell)‡ | 7.2 (42.5) | 8.5 (38.4) |
| I/σ (I) (outer shell)‡ | 8.4 (1.8) | 7.2 (1.8) |
| Completeness (outer shell)‡ | 96.1 (80.1) | 96.7 (96.4) |
| Wilson B factor, Å2 | 23.7 | 19.9 |
| Number of atoms | ||
| Total (protein, retinal, ion, water) | 1,682 (1,634, 20, 1, 27) | 1670 (1623, 20, 1, 26) |
| Average B factors, Å2 | ||
| Total (protein, retinal, ion, water) | 37.3 (37.3, 28.2, 38.1, 40.3) | 31.1 (31.1, 20.9, 34.2, 33.2) |
| Rcryst, %§ (outer shell)¶ | 23.7 (27.3) | 22.7 (23.2) |
| Rfree, %‖ (outer shell)¶ | 25.6 (29.5) | 24.2 (24.7) |
| rmsd of bond lengths, Å, and angles, ° | 0.006, 1.0 | 0.007, 1.0 |
Data set 1 was taken on the ID-EH2 beam line; data set 2 was taken on the ID13 beam line.
The space group is C2221.
Rsym = ΣjΣh|Ih,j − 〈Ih〉|/ΣjΣh Ih,j.
Outer shells were 2.21/2.10 Å and 2.42/2.30 Å for data sets 1 and 2, respectively.
Rcryst = Σh∥Fobs(h)| − |Fcalc(h)∥/Σh|Fobs(h)|.
Outer shells for refinement were 2.23/2.10 Å and 2.44/2.30 Å for data sets 1 and 2, respectively.
Calculated from a set of 5% randomly selected reflections that were excluded from refinement.
Figure 1.
Superposition of the Cα traces and the retinals of pSRII (orange), BR (9) (purple), and HR (14) (green). Helices C–G, which delineate the retinal-binding pocket, are structurally conserved among the three archaerhodopsins. Marked differences are visible in the BC loop and in the N and C termini. Figures were drawn with a modified version of molscript (27) and were rendered with RASTER 3D (28).
Figure 2.
Packing arrangement of pSRII. (a) Arrangement parallel to the ac plane, viewed along the b axis. Monomers are shown as cyan and pink ribbon models. The protein forms crystallographic dimers, and two dimers are highlighted in red and blue. (Inset) Dimer interface, consisting of helices A and G from both monomers. (b) Arrangement parallel to the ab plane viewed along the c axis showing the layer-like packing of pSRII molecules in the crystal.
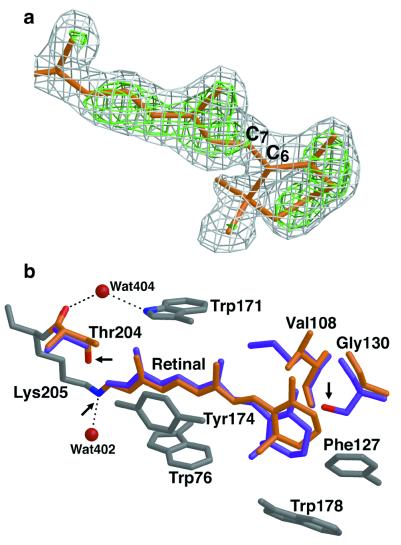
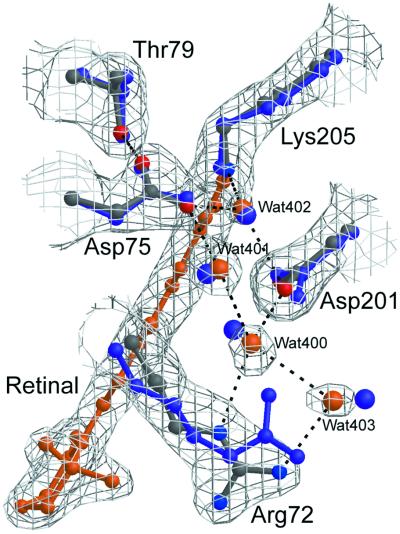
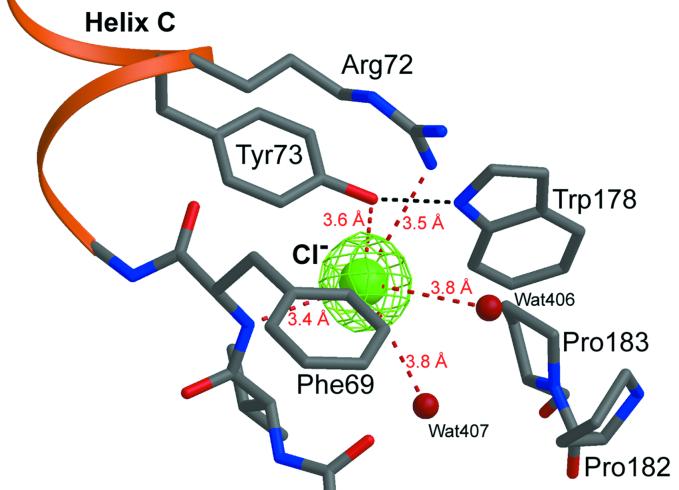
The structure at 2.1-Å resolution highlights similarities and differences in the retinal-binding pocket and along the putative proton pathway among archaerhodopsins of known structures. Most importantly, the retinal-binding pocket in pSRII reveals unique structural features that provide the structural basis for color tuning. Analogous to BR and HR, the β-ionone ring in pSRII is in the 6-s-trans configuration, as shown by the 2Fobs−Fcalc and Fobs−Fcalc omit electron-density maps, which were calculated before the inclusion of the retinal atoms into the model (Fig. 3a). In contradistinction to BR, the retinal chromophore in pSRII is largely unbent (Fig. 3b), which could contribute to the blue-shifted spectrum of pSRII. The hydroxyl group of Thr-204 alters the polarity of the retinal-binding pocket, contributing to the blue-shifted spectrum of pSRII, as suggested from mutagenesis experiments (15). Interestingly, Thr-204 does not perturb the π-bulge in helix G, a trademark in BR (16) and HR (14). Unlike its orientation in BR, the guanidinium group of Arg-72 in pSRII points toward the extracellular side of the membrane (Fig. 4), resulting in a larger distance to Asp-75. This larger distance may account for the elevated pKa of Asp-75 (5.6), as compared with a pKa of 2.2 of the corresponding Asp-85 in BR (17). Maps of 2Fobs−Fcalc and Fobs−Fcalc reveal an unusually high electron density near the extracellular side of helix C (Fig. 5). We suggest that this density corresponds to a chloride ion, because crystals of pSRII were grown in the presence of 1.5 M NaCl. This interpretation is in agreement with recent studies indicating large chloride-induced spectral shifts of pSRII (17, 18). The putative chloride ion is coordinated to Arg-72, Tyr-73, waters 406 and 407, and the N—H of the backbone amide of Phe-69 and is coupled to the Schiff base by means of a hydrogen-bond network (Fig. 5). However, analogously to BR, the protonated Schiff base in pSRII is also H-bonded to a water molecule (Wat-402), which in turn is H-bonded to two Asp residues, Asp-75 and Asp-201 (Fig. 4).
Figure 3.
The retinal in pSRII. (a) Configuration of the β-ionone ring of the retinal. The 2Fobs−Fcalc (gray) and Fobs−Fcalc (green) electron-density maps contoured at 1.0 and 3.0 σ, respectively, were calculated before the inclusion of the retinal atoms into the model. They are superimposed on the refined model (orange). This view shows unambiguously that the retinal in pSRII is in the 6-s-trans configuration. (b) Comparison of the retinal-binding pockets in pSRII and BR. The retinal in pSRII (orange) is less bent than that in BR (purple). Conserved residues among archaerhodopsins are shown in gray. The nonconserved pSRII residues (Val-108, Gly-130, and Thr-204) are shown in orange, and the respective BR residues (Met-118, Ser-141, and Ala-215) are depicted in purple. Arrows indicate the Schiff-base nitrogen and the oxygens of Thr-204 in pSRII and of Ser-141 in BR. Modifications of residues at both ends of the retinal induce change of polarity in its binding pocket. Figures were drawn with a modified version of molscript (27) and were rendered with RASTER 3D (28).
Figure 4.
Detailed view of the active sites of pSRII and BR. The 2Fobs−Fcalc electron-density map of pSRII (gray) contoured at 1.2σ is superimposed on its model (protein: gray, blue, and red; retinal and water: orange). The model for BR (9), including waters, is depicted in purple. The hydrogen-bond networks in the two proteins are similar from the Schiff base to Wat-403. Arg-72 in pSRII is oriented toward the extracellular medium as opposed to the orientation of the equivalent residue Arg-82 in BR.
Figure 5.
Putative chloride-binding site. The chloride ion (green) is coordinated to five polar atoms and is surrounded by hydrophobic residues. Fobs−Fcalc electron-density map calculated without the ion and contoured at 3σ is shown in green.
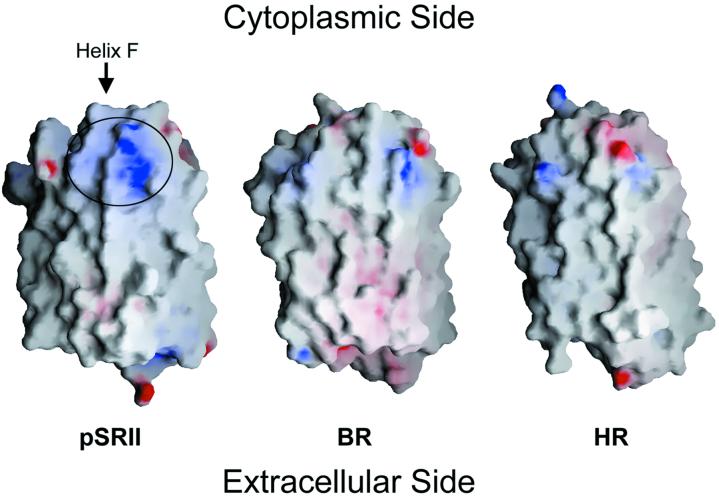
The surface of pSRII reveals a unique patch of charged and polar residues at the cytoplasmic ends of helices F (Lys-157, Ser-158, Arg-162, Arg-164, and Asn-165) and G (Asp-214) (Fig. 6). This patch is not present in BR or HR. It is, therefore, probable that these positively charged residues interact electrostatically with the negatively charged cytoplasmic domain Gly-Asp-Gly-Asp-Leu-Asp of transmembrane helix 2 of HtrII, which is conserved among various species (19). The extraordinary stability of the pSRII–HtrII complex (4) may be explained by this electrostatic interaction, because it is further enhanced by the low-dielectric environment of the lipid bilayer. Interestingly, Ser-158 and Phe-210 in helices F and G, respectively, point outwards. We propose that transmission of the light signal from the receptor to HtrII is mediated by means of the interaction of Ser-158 and Phe-210 with the transmembrane helices of HtrII. This model is in agreement with recent results from EPR experiments (4), indicating that light-induced mobility of spin-labeled Ser-158 is restrained in the presence of HtrII, and also in agreement with the general concept that pSRII transmits the light signal to HtrII by means of helix–helix interactions (20). A movement of helices F and/or G during the photocycle, as in BR (21), would trigger the signal transduction.
Figure 6.
Comparison of the electrostatic surface potentials of pSRII, BR, and HR. Electrostatic surface potentials were generated with grasp (29). Negative and positive electrostatic surface potentials are shown in red and blue, respectively. pSRII displays a patch of residues on the cytoplasmic side, shown in the circle, which is significantly more positively charged than in BR and HR. This patch is a putative binding site for HtrII.
A fundamental goal in photobiology is the elucidation of factors that determine color tuning in rhodopsins. The absorption maximum of pSRII at 497 nm is considerably blue-shifted relative to that of BR, HR, and SRI (λ > 560 nm). Chromophore–opsin interactions that produce spectral shifts might include the polarity near the β-ionone ring, interactions between the Schiff base and its counter ion, and retinal bending (22). Structural comparison of the retinal-binding pockets in archaerhodopsins reveals that whereas most residues are conserved (Tyr-73, Trp-76, Thr-80, Gly-112, Trp-171, Tyr-174, Pro-175, and Trp-178), three residues vary—Val-108, Gly-130, and Thr-204 in pSRII (Met, Ser, and Ala in BR, HR, and SRI, respectively). These three residues play a key role in color tuning as they modify the polarity of the retinal-binding pocket. Thus, the Schiff-base nitrogen is located 4.5 Å from the hydroxyl group of Thr-204, and the β-ionone ring is flanked by Gly-130 and Ser-141 in pSRII and BR, respectively. Indeed, replacement of pSRII residues Val-108, Gly-130, and Thr-204 by the respective BR residues (Met-118, Ser-141, and Ala-215) resulted in mutants exhibiting red-shifted absorption maxima up to 515 nm (15). Moreover, introduction of the pSRII residue Val-108 into the corresponding site in BR (Met-118) resulted in a dramatic blue-shifted absorption maximum down to 480 nm (23). Furthermore, in contrast to BR and HR, the structure of pSRII reveals a largely unbent retinal, which might also contribute to the blue-shifted spectrum of pSRII (24). The polyene chains in pSRII, BR (Fig. 3b), and HR are firmly constrained at the Schiff-base end by conserved aromatic residues (Trp-76, Trp-171, and Tyr-174). Close to the β-ionone ring, the substitution of Met-118 and Ser-141 in BR by Val-108 and Gly-130 in pSRII modifies the steric environment so as to release constraints imposed on the polyene chain, resulting in a less bent conformation of the retinal (Fig. 3b).
As pSRII and BR display a high degree of structural similarity, it was suggested that pSRII also may act as a light-driven proton pump. Indeed, wild-type transducer-free pSRII and its F86D mutant were shown recently to pump protons (25), although at a markedly lower rate than BR. Whereas the structures of the proton pathway in pSRII and BR are nearly identical from the Schiff base to Wat-403 (Fig. 4), significant differences can be identified in the extracellular and cytoplasmic regions. In contrast to Arg-82 in BR's ground-state structure, the side chain of the corresponding Arg-72 in pSRII is oriented toward the extracellular side. In addition, the proton release dyad in BR (Glu-204 and Glu-194) is replaced by a single negatively charged residue (Asp-193) in pSRII. On the cytoplasmic side, pSRII is markedly more hydrophobic than BR, with Phe-86 and Leu-40 replacing Asp-96 and Thr-46 in BR. Because the Schiff base in BR is reprotonated from Asp-96, this step is less efficient in pSRII and in the D96N BR mutant, as indicated by a slower photocycle. Conversely, the pSRII double mutant F86D/L40T exhibits a photocycle rate comparable to that of BR (26). The extended duration of the photocycle of pSRII reflects the need for a long-lived signaling intermediate.
Conclusions
The high-resolution structure of pSRII reveals distinct features in the retinal-binding pocket. Specifically, the unbent conformation of the retinal in pSRII relative to that in BR and HR, the hydroxyl group of Thr-204 in the vicinity of the Schiff base, and the outward orientation of the side chain of Arg-72 are unique. These features are likely to be important for elucidating the mechanisms of color tuning, light-driven proton pumping, and photo-signaling mediated by the same photopigment. A positively charged surface patch on the cytoplasmic side of pSRII has been identified, and is proposed to play a role in the interaction with the membrane-bound transducer-protein HtrII. Furthermore, a putative chloride ion is found in the extracellular part of the protein, and its role remains to be determined. The high degree of structural similarity between BR, HR, and pSRII highlights the remarkable fact that minor changes in structure suffice to achieve the functionally divergent tasks of vectorial and selective ion transport and photo-signaling. This principle of minor variation achieving diversity is shared by the superfamily of heptahelical transmembrane receptors, for which subtle structural changes result in a plethora of biological responses. In particular, from slight structural variation within the family of visual rhodopsins emerges the phenomenon of color vision.
Acknowledgments
We thank G. Altenberg, S. Arzt, H. Belrhali, D. Bourgeois, M. Burghammer, S. Hayashi, C. Maxwell, E. Mitchell, C. Riekel, K. Schulten, E. Tajkhorshid, and T. Ursby for experimental contributions and insightful discussions. We are indebted to M. Engelhard and A. Wegener for providing pSRII for the initial screening. Support from the European Union-BIOTECH, the Swedish Natural Science Research Council (NFR), the French Ministry of Education and Research (MENR), the Swiss National Science Foundation's Swiss Priority Programmme BioTechnology, the Welch Foundation, and the Howard Hughes Medical Institute is greatly acknowledged. J.N. was the recipient of a National Institutes of Health Fogarty International Fellowship. P.N. is a fellow of the Human Frontiers Research Science Organization.
Abbreviations
- BR
bacteriorhodopsin
- HR
halorhodopsin
- SR
sensory rhodopsin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1h68).
References
- 1.Spudich J L, Yang C-S, Jung K-H, Spudich E N. Annu Rev Cell Dev Biol. 2000;16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki J, Spudich J L. Biochim Biophys Acta. 2000;1460:230–239. doi: 10.1016/s0005-2728(00)00142-0. [DOI] [PubMed] [Google Scholar]
- 3.Spudich J L. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 4.Wegener A-A, Chizhov I, Engelhard M, Steinhoff H-J. J Mol Biol. 2000;301:881–891. doi: 10.1006/jmbi.2000.4008. [DOI] [PubMed] [Google Scholar]
- 5.Hohenfeld I P, Wegener A A, Engelhard M. FEBS Lett. 1999;442:198–202. doi: 10.1016/s0014-5793(98)01659-7. [DOI] [PubMed] [Google Scholar]
- 6.Otwinowski Z. In: Data Collection and Processing. Sawyer L, Isaacs N W, Bailey S, editors. Cheshire, U.K.: Daresbury Laboratory; 1993. pp. 55–62. [Google Scholar]
- 7.Collaborative Computational Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 8.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 9.Belrhali H, Nollert P, Royant A, Menzel C, Rosenbusch J P, Landau E M, Pebay-Peyroula E. Structure (London) 1999;7:909–917. doi: 10.1016/s0969-2126(99)80118-x. [DOI] [PubMed] [Google Scholar]
- 10.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 11.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 12.Landau E M, Rosenbusch J P. Proc Natl Acad Sci USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pebay-Peyroula E, Rummel G, Rosenbusch J P, Landau E M. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 14.Kolbe M, Besir H, Essen L-O, Oesterhelt D. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 15.Shimono K, Iwamoto M, Sumi M, Kamo N. Photochem Photobiol. 2000;72:141–145. doi: 10.1562/0031-8655(2000)072<0141:eotcaa>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 16.Luecke H, Schobert B, Richter H T, Cartailler J-P, Lanyi J. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 17.Chizhov I, Schmies G, Seidel R, Sydor J R, Luttenberg B, Engelhard M. Biophys J. 1998;75:999–1009. doi: 10.1016/S0006-3495(98)77588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimono K, Kitami M, Iwamoto M, Kamo N. Biophys Chem. 2000;87:225–230. doi: 10.1016/s0301-4622(00)00195-2. [DOI] [PubMed] [Google Scholar]
- 19.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X N, Zhu J, Spudich J L. Proc Natl Acad Sci USA. 1999;96:857–862. doi: 10.1073/pnas.96.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kochendoerfer G G, Lin S W, Sakmar T P, Mathies R A. Trends Biochem Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 23.Greenhalgh D A, Farrens D L, Subramaniam S, Khorana H G. J Biol Chem. 1993;268:20305–20311. [PubMed] [Google Scholar]
- 24.Subramaniam S, Henderson R. Nature (London) 2000;406:653–657. doi: 10.1038/35020614. [DOI] [PubMed] [Google Scholar]
- 25.Schmies G, Luttenberg B, Chizhov I, Engelhard M, Bamberg E. Biophys J. 2000;78:967–976. doi: 10.1016/S0006-3495(00)76654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwamoto M, Shimono K, Sumi M, Kamo N. Biophys Chem. 1999;79:187–192. doi: 10.1016/s0301-4622(99)00054-x. [DOI] [PubMed] [Google Scholar]
- 27.Esnouf R M. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 28.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 29.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]