Abstract
We propose a multiscale weighted principal component regression (MWPCR) framework for the use of high dimensional features with strong spatial features (e.g., smoothness and correlation) to predict an outcome variable, such as disease status. This development is motivated by identifying imaging biomarkers that could potentially aid detection, diagnosis, assessment of prognosis, prediction of response to treatment, and monitoring of disease status, among many others. The MWPCR can be regarded as a novel integration of principal components analysis (PCA), kernel methods, and regression models. In MWPCR, we introduce various weight matrices to prewhitten high dimensional feature vectors, perform matrix decomposition for both dimension reduction and feature extraction, and build a prediction model by using the extracted features. Examples of such weight matrices include an importance score weight matrix for the selection of individual features at each location and a spatial weight matrix for the incorporation of the spatial pattern of feature vectors. We integrate the importance score weights with the spatial weights in order to recover the low dimensional structure of high dimensional features. We demonstrate the utility of our methods through extensive simulations and real data analyses of the Alzheimer’s disease neuroimaging initiative (ADNI) data set.
Keywords: Alzheimer, Feature, Principal component analysis, Regression, Spatial, Supervised
1 Introduction
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) study began in 2004 and is the first “Big Data” project for Alzheimer’s disease (AD), which has been a groundbreaking project. It has collected imaging, genetic, clinical, and cognitive data from thousands of subjects in order to delineate the complex relationships among the clinical, cognitive, imaging, genetic and biochemical biomarker characteristics of the entire spectrum of AD as the pathology evolves from normal aging (NC), to mild cognitive impairment (MCI), to dementia or AD. This paper is motivated by the joint analysis of fluorodeoxyglucose positron emission tomography (FDG-PET) data and clinical and behavioral variables from n = 196 subjects in the ADNI study. After applying a standard preprocessing pipeline, the dimension of the processed FDG-PET images is 79 × 95 × 69. We are particularly interested in addressing two questions:
(Q1) the first one is to identify FDG-PET imaging biomarkers for classifying subjects to either AD or NC group;
(Q2) the second one is to identify FDG-PET imaging biomarkers observed at baseline to accurately predict the change in the Alzheimer’s Disease Assessment Scale-Cognitive (ADAS-Cog) test score at least two years later after initial assessment.
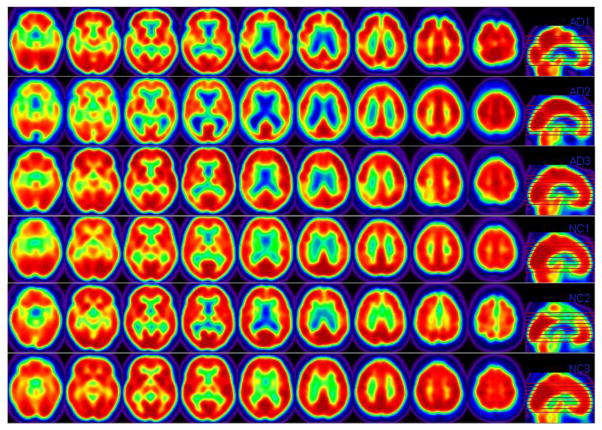
Statistically, these questions of interest can be formulated as the use of a high-dimensional vector of features (or FDG-PET), denoted as x = (xg: g ∈ 𝒢), to predict an outcome variable, denoted as y, where 𝒢 = {g1, …, gp} is a set of locations, in which p is the total number of locations in 𝒢: In this case, x is a vector of FDG-PET imaging measures on a 3-dimensional (3D) lattice and y is either disease status in (i) or the change in the ADAS-cog score in (ii). Figure 1 shows some selected slices of the processed PET images from 3 randomly selected Alzheimer’s Disease (AD) subjects and 3 randomly selected normal control (NC) subjects.
Figure 1.
ADNI PET Data. Each row consists of pre-selected 2-dimensional (2D) slides obtained from a randomly selected subject. The first three rows come from 3 randomly selected AD subjects and the last three rows come from 3 randomly selected NC subjects.
To answer questions (Q1) and (Q2), we develop a multiscale weighted principal component regression (MWPCR) framework to deal with three challenges arising from the use of high-dimensional x with strong spatial features (e.g., FDG-PET) to predict y. Such challenges include (i) noisy functional data, (ii) complex spatial information, and (iii) the remarkable variability of brain structure and function across subjects. For instance, in most neuroimaging studies, the dimension of neuroimaging data (or x) can be much larger than the number of subjects, which varies from several dozens to a few thousands. Moreover, different components of x may be highly correlated with each other and share some specific spatial structures (Friston, 2009; Vincent et al., 2011; Hinrichs et al., 2009; Cuingnet et al., 2012).
Many existing supervised learning and variable selection methods (Hastie et al., 2009; Clarke et al., 2009; Fan and Fan, 2008; Bickel and Levina, 2004; Buhlmann et al., 2012; Tibshirani, 1996), however, can be sub-optimal for high-dimensional prediction problem considered here, since the effect of high dimensional data x (e.g., image biomarker) on y is often non-sparse (Li et al., 2015; Zhou et al., 2013; Friston, 2009; Hinrichs et al., 2009). First, the existing unstructured regularization methods can suffer from diverging spectra and noise accumulation in high dimensional feature space (Reiss and Ogden, 2010; Bickel and Levina, 2004; Buhlmann et al., 2012; Fan and Fan, 2008), whereas the structured ones (e.g., fused Lasso or Ising prior) can be computationally challenging for high-dimensional imaging predictor (Vincent et al., 2011; Cuingnet et al., 2012; Fan et al., 2012; Goldsmith et al., 2014). Alternatively, it is imperative to use some dimension reduction methods, such as principal component analysis and/or screening methods, to extract and select important ‘low-dimensional’ features, while eliminating redundant features (Skocaj et al., 2007; Bair et al., 2006; Fan and Fan, 2008; Krishnan et al., 2011; Zhao et al., 2012). Moreover, most supervised learning methods coupled with dimension reduction methods do not account for the strong spatial features of high-dimensional imaging data as discussed above (Allen et al., 2014; Guo et al., 2015).
A general framework of MWPCR is developed to address some of the challenges discussed above. The MWPCR provides a simple solution to the problem of interest by hierarchically and spatially extracting low-dimensional ‘transformed’ variables from x in order to dramatically improve prediction accuracy. Compared with the existing literature (Allen et al., 2014; Guo et al., 2015; Shen and Zhu, 2015), we make several major contributions as follows:
(i) MWPCR provides a comprehensive and powerful dimension reduction framework for integrating feature selection, smoothing, and feature extraction for continuous and discrete response variables (e.g., binary response for classification).
(ii) We evaluate the finite sample properties of MWPCR by using both simulation studies and the analysis of ADNI data. Our numerical results reveal that MWPCR significantly outperforms many competing methods under some scenarios.
(iii) We systematically investigate the theoretical properties of MWPCR under the high-dimensional binary classification setting. Specifically, we are able to reveal the importance of incorporating different types of weights for improving classification accuracy.
(iv) The code for MWPCR was written in Matlab, which along with its documentation will be freely accessible from the public website http://www.nitrc.org and our lab website http://odin.mdacc.tmc.edu/bigs2/.
The paper is organized as follows. In Section 2, we introduce the model setup of MWPCR. We discuss various strategies of determining global and local weights that account for an association between y and each individual feature xg across g ∈ 𝒢 and the spatial patterns of x. In Section 3, simulation studies are conducted to examine the finite sample performance of MWPCR. We conduct real data analysis in Section 4 based on ADNI data to address the two questions (Q1) and (Q2) discussed above. We give some concluding remarks in Section 5. We also investigate some theoretical properties of MWPCR under the high-dimensional binary classification setting and put them in the supplementary document.
2 Multiscale Weighted Principal Component Regression
In this section, we describe data structure and then introduce the model setup and estimation method of MWPCR.
2.1 Data Structure
Consider data from n independent subjects. For each subject, we observe a qy × 1 vector of discrete or continuous responses, denoted by yi = (yi,1, …, yi,qy)T, a qz × 1 vector of discrete and/or continuous clinical covariates, denoted by zi = (zi,1, …, zi,qz)T, and a p × 1 vector of data xi = {xi,g: g ∈ 𝒢} measured on 𝒢 for i = 1, …, n. Let XT = (x1| …|xn) be a p × n matrix. In many cases, both qy and qz are relatively small compared with n, whereas p is much larger than n. For instance, in many imaging studies, it is common to use high dimensional imaging data to classify a class variable, such as disease status. In this case, qy is as small as one, whereas p can be several millions. Moreover, 𝒢 = {g1, …, gp} is a set of prefixed locations, such as voxels in 3D lattices, so it is possible to define an edge set 𝒮 = {(gk, gj): gk, gj ∈ 𝒢} associated with 𝒢. For instance, in spatial statistics and imaging analysis, one often uses pixels and their first-order (or high-order) neighboring pixels to construct edges in 𝒮.
2.2 Model Setup
The proposed MWPCR consists of two components: a low-rank model for multi-scale weighted PCA (MWPCA) and a prediction model. Let Q(ℓ) be a p × p weight matrix at the ℓ–th scale for ℓ = 1, …, L. The low-rank model for MWPCA can be written as
| (1) |
for ℓ = 1, …, L, where E(xi) = μ, K ≤ min(n, p), and is an n × p matrix of measurement errors that follows a matrix-variate distribution with mean 0n,p and an arbitrary covariance matrix. Moreover, , and are, respectively, n × K, K × K, and p × K matrices such that diag(D(ℓ)) ≥ 0 and U(ℓ)TU(ℓ) = V(ℓ)TV(ℓ) = IK, where IK is a K × K identity matrix.
We combine all {U(ℓ)}ℓ≥1 from different scales into an n × (KL) matrix given by UC = (uC,1 ··· uC,n)T = (U(1), …, U(L)). We then build a prediction model R(yi; uC,i, zi, θ) with yi as response and uC,i and zi as covariates, where θ is a vector of unknown (finite-dimensional or non-parametric) parameters. For instance, when qy = 1, a popular prediction model is the generalized linear model given by
| (2) |
where ϕ is a dispersion parameter and b(·) and s(·, ·) are known functions. Moreover, it is assumed that ḃ(ηi) = db(ηi)/dηi = E(yi|uC,i, zi) satisfies , where βz and βu are coefficient vectors associated with zi and uC,i, respectively, and h(·) is a link function. In this case, we have θ = (ϕ, βz, βu). Our prediction model can be various parametric and nonparametric regression models for continuous and discrete responses and multivariate and univariate responses, such as survival data and classification problems (Hastie et al., 2009; Clarke et al., 2009).
The key novelty of MWPCR is the use of MWPCA to extract important low-dimensional features of x that are predictive of y. Our MWPCA can be regarded as a novel extension of various supervised and unsupervised dimension reduction models for matrix decomposition (Allen et al., 2014; Skocaj et al., 2007; Huang et al., 2009). Specifically, the three key features of MWPCA include the integration of importance score weights and spatial weights, a multiscale strategy for feature extraction, and its computational efficiency. In contrast, although a general duality diagram method (Dray and Jombart, 2011; Skocaj et al., 2007) explicitly incorporates two weight matrices, it only accounts for structural dependencies (e.g., smoothness) in x.
2.3 Estimation Procedure
We introduce a three-stage algorithm for MWPCR as follows.
Stage 1. Build an importance score vector (or function) WI = (wI,g): 𝒢 → R+ and a spatial weight matrix WE = (wE,gg′): 𝒢 × 𝒢 → R.
Stage 2. At the ℓ–th scale, use WE and WI to build a spatial weight matrix Q(ℓ) and then compute the first K principal components in U(ℓ) according to model (1). Repeat it for ℓ = 1, …, L.
Stage 3. Build the prediction model R(y; uC, z, θ).
We slightly elaborate on these stages. In Stage 1, the importance scores wI,g play an important feature screening role in MWPCR and they can be learnt directly either from {x, y} or other sources. Examples of wI,g in the literature are primarily based on some statistics (e.g., Pearson correlation or distance correlation) between xg and y at each location g used in the sure independence screening (Bair et al., 2006; Li et al., 2012). However, most importance scores wI,g are independently calculated at each location, so they largely ignore complex spatial structures at different locations.
In Stage 1, WE = (wE,gkgj) ∈ Rp×p can be either symmetric or asymmetric. The elements wE,gkgj are usually calculated by using various similarity criteria, such as Gaussian similarity from Euclidean distance, local neighborhood relationship, correlation, and prior information obtained from other data (Yan et al., 2007). Then, we can threshold WE to create an adjacency matrix with elements of either 1 or 0, which leads to 𝒮, depending on whether the corresponding correlation value exceeds a prefixed threshold or not. By choosing different thresholds, we can obtain different edge sets 𝒮. In Section 2.4, we will discuss how to determine WE and WI, while explicitly accounting for the complex spatial structure among different locations.
In Stage 2, we construct the weight matrix Q(ℓ) at the ℓ–th scale as follows. To extract important features from x, we construct a matrix , where 1{·} is an indicator function and sI,1 ≤ … ≤ sI,L are pre-specified thresholds. The use of is similar to various marginal screening methods (Fan and Lv, 2008; Fan and Fan, 2008; Bair et al., 2006). By tuning the value of sI,ℓ, we can screen out ‘uninformative’ features at different scales.
To capture the spatial features of x, we may construct a spatial similarity matrix , where sE,ℓ = (sE,ℓ;1, sE,ℓ;2)T and D(gk, gj) is a specific distance (e.g., Euclidean) between gk and gj. The value of sE,ℓ;2 controls the number of locations in {gj ∈ 𝒢: D(gk, gj) ≤ sE,ℓ;2}, which is a patch set at gk (Taylor and Meyer, 2012), whereas sE,ℓ;1 is used to shrink small |wE,gkgj | to zero.
Given and , we may set Q(ℓ) as either or . Specifically, corresponds to selecting important features from x first and then smoothing those selected features. In contrast, corresponds to smoothing x first and then extracting important features from the smoothed x. According to our experiences, outperforms in terms of prediction accuracy in many scenarios, even though the use of can be computationally demanding when p is extremely large.
Given Q(ℓ), we can ‘prewhiten’ (X − 1nμT) and calculate X̃(ℓ) and its singular value decomposition (SVD) (U(ℓ), D(ℓ), V(ℓ)) in (1). In practice, a simple criterion for determining K is to include all components up to a prefixed proportion of the total variance, say 85%. For high dimensional data, we consider a regularized PCA by iteratively solving a single-factor two-way regularized matrix factorization. Specifically, for a given K, we minimize with respect to (U(ℓ), D(ℓ), V(ℓ)) the following objective function given by
| (3) |
subject to and for all k, where λv and λu are two tuning parameters and P1(·) and P2(·) are two penalty functions. We use adaptive Lasso penalties for P1(·) and P2(·) and then iteratively solve (3) (Aharon et al., 2006). For each k0, we use the sparse method in Lee et al. (2010) to estimate ( ). In this way, we can sequentially compute ( ) for k = 1, …, K.
In Stage 3, based on {(yi, uC,i, zi)}i≥1, we use an estimation method to estimate θ as follows:
| (4) |
where ρ(…) is a loss function, λ is a tuning parameter and P3(·) is a penalty function. Given test vectors x* and z*, we can do prediction as follows:
Calculate by setting u(ℓ)* = (x* − μ)TQ(ℓ)V(ℓ){D(ℓ)}−1, in which μ, Q(ℓ), V(ℓ), and D(ℓ) are learnt from the training data.
Optimize an objective function based on to calculate an estimate of y.
2.4 Importance Score Weights and Spatial Weights
There are two sets of weights in MWPCR, including (i) importance score weights enabling a selective treatment for individual features and (ii) spatial weights accommodating the underlying spatial dependence among features across neighboring locations. As shown in simulation studies, the use of the two sets of weights can dramatically improve prediction accuracy. Below, we propose several specific strategies to determine them.
2.4.1 Importance Score Weights
As discussed in Section 2.3, at each location g, wI,g is calculated based on a statistical model between (xg, z) and y in order to perform feature selection according to each feature’s discriminative importance. Statistically, most existing methods (Bair et al., 2006; Li et al., 2012) use a marginal model by assuming
| (5) |
where β = (β(g): g ∈ 𝒢) and β(g) is introduced to quantify the association between yi and xi,g at each location g ∈ 𝒢. At the g–th location, wI,g is a statistic based on the marginal model . A simple example is to use the Pearson correlation between each feature and class label as the importance score weight. Noninformative features (e.g., correlation less than a given threshold) can be simply discarded by setting wI,g = 0. However, those wI,g’s largely ignore complex spatial structure, such as homogenous patches defined below, across all locations (Bair et al., 2006; Li et al., 2012).
It is common to assume that β(g) across all locations are naturally clustered into G homogeneous patches, denoted by {𝒢j: j = 1, …, G}, such that
| (6) |
Note that a patch 𝒢j consists of a set of locations that are spatially connected through edges in 𝒮. It has been shown that algorithms based on patch information have led to state-of-the art techniques for classification and denoising (Taylor and Meyer, 2012; Li et al., 2011; Polzehl and Spokoiny, 2006; Arias-Castro et al., 2012).
We propose two strategies to learn the homogenous patches 𝒢j in (6) by jointly modelling (xi, zi) and yi. The first strategy is to model the conditional distribution of xi given yi and zi, denoted by f(xi|yi, zi, β). The second strategy is to model the conditional distribution of yi given xi and zi, denoted by f (yi|xi, zi, β). Finally, we can learn patches 𝒢j from the estimated β and then construct importance score weights.
The first strategy is to model f(xi|yi, zi, β). Let 𝒮g(h) be an edge set at scale h at each location g. We consider a sequence of nested edge sets across multiscales hs such that h0 = 0 ≤ h1 ≤ … ≤ hS and 𝒮g(h0) = {g} ⊂ … ⊂ 𝒮g(hS). To learn the homogeneous patches, a general framework of Multiscale Adaptive Regression Model (MARM) developed in Li et al. (2011) is to maximize a sequence of weighted functions as follows:
| (7) |
where ω(g, g′; h) characterizes the similarity between the observations at g′ and those at g with ω(g, g; h) = 1. If ω(g, g′; h) ≈ 0, then the observations at g′ do not provide information on β(g). Therefore, ω(g, g′; h) can prevent incorporation of locations, whose observations do not contain information on β(g) and preserve the edges of homogeneous regions.
Let D1(g, g′) and D2(β̂(g; hs−1), β̂(g′; hs−1)) be, respectively, the spatial distance between locations g and g′ and a similarity measure between β̂ (g; hs−1) and β̂(g′; hs−1). The ω(g, g′; hs) can be defined as
| (8) |
where K1(·) and K2(·) are two nonnegative kernel functions and γn is a bandwidth parameter that may depend on n. The weights K1 (D1(g, g′)/hS) give less weight to location g′ ∈ 𝒮g(hS), which is far from the location g. The weights K2(u) downweight location g′ with large D2(β̂(g; hS), β̂ (g′; hS)), which indicates a large difference between β̂(g′; hS) and β̂(g; hS). Moreover, by following Li et al. (2011) and Polzehl and Spokoiny (2006), we set K1(x) = (1−x)+ and K2(x) = exp(−x). See the detailed algorithm of MARM in Li et al. (2011).
The second strategy is to model f(yi|xi, zi, β) and the prior distribution of β, given by f (β). Since xi is often high dimensional, it is much difficult to carry out statistical inference based on f(yi|xi, zi, β) compared with f (xi|yi, zi, β). Moreover, our primary goal is to perform feature selection in order to eventually use a small subset of xi to predict yi, while correcting for zi. Similar to the first strategy, we also take the marginal method and then incorporate a specific structure to estimate β as follows:
| (9) |
where 𝒩g is a set of the neighboring locations of location g.
Similar to the first strategy, we propose an adaptive smoothing algorithm to estimate β as follows. Consider a sequence of nested edge sets 𝒮g(h0) = {g} ⊂ … ⊂ 𝒮g(hS) for h0 = 0 ≤ h1 ≤ … ≤ hS.
[Step (i)] Calculate β̂(g; h0) and Cov(β̂(g; h0)) according to across all locations g.
[Step (ii)] Smooth {β̂(g; h0): g ∈ 𝒢} to sequentially estimate β̂(g; hs) for s = 1, …, S across all g ∈ 𝒢. Candidate methods include local polynomial, nonlocal mean, and propagation-separation, among others (Polzehl and Spokoiny, 2006; Arias-Castro et al., 2012).
For both strategies, after the iteration hS, we can obtain β̂(g; hS) and its covariance matrix, denoted by Cov(β̂(g; hS)), across all g ∈ 𝒢. Finally, we calculate wI,g as a function of β̂(g; hS) and Cov(β̂(g; hS)), such as the Wald test and its p-value. Then, we use a clustering algorithm, such as the K-mean algorithm, to group {β̂(g; hS): g ∈ 𝒢} into several homogeneous clusters (Hastie et al., 2009), in which β̂(g; hS) varies very smoothly in each cluster.
2.4.2 Spatial Weights
As discussed in Section 2.3, wE,gg′ often characterizes the degree of certain ‘similarity’ between locations g and g′. We consider three spatial weight matrices, including (i) the precision matrix, (ii) a locally spatial weight matrix, and (iii) a cluster-based spatial weight matrix as follows.
For the precision matrix, let Σ be the covariance matrix of xi, we can set ; thus, is the precision matrix of xi. When Σ−1 has certain sparsity structures (e.g., factor model), various estimation methods have been developed even for extremely large p.
The locally spatial weight matrix consists of non-negative weights assigned to the spatial neighboring locations of each location. Specifically, we set wE,gg′ as
| (10) |
in which ω(g, g′; hS) is defined in (8). Thus, we have wE,gg′ = 0 for all g′ ∉ 𝒮g(hS) and Σg′∈𝒢 wE,gg′ = 1.
The cluster-based spatial weight matrix consists of non-negative weights assigned to locations in the same homogeneous cluster. Specifically, we use the Laplace-Beltrami operator to construct WE (Luxburg, 2007). It is assumed that each edge between two locations g and g′ carries a non-negative weight wgg′. Thus, matrix W = (wgg′) is a weighted adjacency matrix of 𝒢. The degree of a location g ∈ 𝒢 is defined as dg = Σg′∈𝒢 wgg′ and the degree matrix WD is given by WD = diag(dg1, …, dgp).The unnormalized Laplacian matrix L of the graph 𝒢 is defined as WL = WD − W, which can be regarded as a discrete representation of the Laplace-Beltrami operator. Finally, we set WE = exp(−0.5WL/γ), where exp(·) denotes the matrix exponential. In practice, when p is extremely large, it is computationally infeasible to directly use the huge p × p matrix WE. In this case, based on the clustering results in (6), we only consider locations in each cluster and each cluster forms a connected subgraph, which leads to dramatically computational savings (Cuingnet et al., 2012).
2.4.3 Weights Selection
A critical question is how to select spatial weights and/or importance score weights for constructing Q(ℓ) in different applications. Ideally, we may either use one of them or combine some of them together to construct Q(ℓ). Theoretically, we have investigated the effects of applying importance score weights and different spatial weights in MWPCR on classification accuracy for high dimensional binary classification and put them in the supplementary document. We have three key theoretical results as follows.
The use of feature selection can substantially improve classification accuracy for high dimensional binary classification.
The use of spatial kernel weights and importance score weights in MWPCR can substantially improve classification accuracy even when signals are weak.
The use of the true Σ−1/2 can improve classification accuracy, where Σ is the covariance matrix of x.
Based on these results, we suggest to first apply the locally spatial weight matrix (or the cluster-based spatial weight matrix) and then use the importance score weights based on β̂(g; hS). Although the use of Σ−1/2 can improve classification accuracy, estimating Σ−1/2 can be very challenging when p is even moderate. Thus, we avoid estimating Σ−1/2 in all simulations and real data analysis.
3 Simulation Studies: Binary Outcome
We use two sets of simulation studies, including binary and continuous outcomes, to examine the finite sample performance of MWPCR under different scenarios. We demonstrate that MWPCR outperforms or at least is compatible with many state-of-the-art methods. For the sake of space, we include all simulation results for continuous outcome in the supplementary document.
We applied MWPCR to a high-dimensional binary classification problem as follows. We simulated 20 × 20 × 10 3D-images from a linear model given by
| (11) |
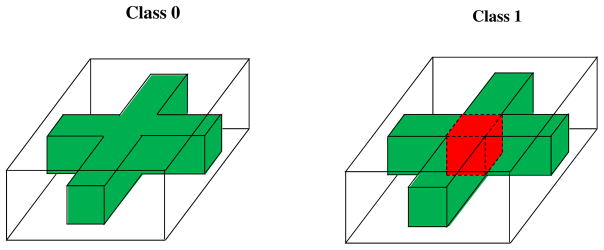
where li is the class label coded as either 0 or 1 and εi(g) are random variables with zero mean. Figure 2 presents the true mean images of class li = 0 and class li = 1, in which a red cuboid 3 × 3 × 4 region characterizes the maximum difference 1 between classes 0 and 1. In this case, we have p = 4, 000. Then, we set n = 100 with 60 images from Class 0 and the rest from Class 1.
Figure 2.
True mean images for the first set of simulations: Class 0 in the left panel and Class 1 in the right panel. The white, green, and red colors, respectively, correspond to 0, 1, and 2.
We consider three types of noise εi(g) in (11). First, were independently generated from a N(0, 22) generator across all voxels. Second, were generated from by introducing the short range spatial correlation, where || · ||1 is the L1 norm of a vector and mg is the number of locations in the set {|| g′−g ||1≤ 1}. Third, to introduce the long range spatial correlation, were generated according to , where g = (g1, g2, g3)T and ξi,k for k = 1, 2, 3 were independently generated from a N(0, 1) generator. Moreover, the noise variances in all voxels of the red cuboid region equal 4, 4/6, and 4{sin(πg1/10)2 + cos(πg2/10)2 + sin(πg3/5)2} + 4 for Type I, II, and III noises, respectively. Therefore, among the three types of noise, Type III noise has the smallest signal-to-noise ratio and Type II noise has the largest one.
We ran the three stages of MWPCR as follows. In Stage 1, let {hs = 1.2s, s = 0, 1, …, S = 5}, and for each g ∈ 𝒢, we set wI,g = −p log(p(g))/{−Σg∈𝒢 log(p(g))}, where p(g) is the p-value of Wald test β1(g) = 0 in (11) at voxel g. The spatial weight WE is given by (10). We set the spatial weight WE according to (10) and (8). Specifically, we considered three types of spatial weights WE, including MWPCR1: only the location kernel function K1(.) in (8); MWPCR2: only the similarity kernel function K2(.) in (8); and MWPCR3: the combination of kernel functions K1(.) and K2(.) in (8). Then, we selected the bandwidth {hs = 1.2s, s = 0, …, S = 5} in these kernel functions in order to determine WI and WE. In Stage 2, we used different numbers of principal components in MWPCA to reconstruct the low dimensional representation of simulated images. In Stage 3, we tried different classification methods, including linear regression, k-nearest neighbor (k-NN) and support vector machine (SVM), on these low dimensional representations. Since their performances are similar to each other, we only report the results based on the linear regression throughout the paper. The linear regression uses class label li as dependent variable and principal components as explanatory variables. An image is classified as Class 0, if its predictive value is less than 0, and as Class 1, otherwise.
We first used the leave-one-out cross validation to calculate the misclassification rates for MWPCR1, MWPCR2, MWPCR3, and a standard principal component analysis (PCA). Table 1 presents the classification results based on 5, 7 and 10 principal components. The misclassification errors for all MWPCR methods are quite stable for different numbers of principal components under different types of noise. All MWPCR methods perform relatively well for Type II noise compared with Type I and III noises, since Type II noise has the largest signal-to-noise ratio. Moreover, MWPCR3 is slightly better than MWPCR1 and MWPCR2, which may be due to the fact that MWPCR3 combines both the local smooth and similarity kernels. Moreover, it seems that MWPCR3 is very robust to the long-range correlation structure of Type III noise. Compared with all MWPCR methods, PCA performs very poor, since it does not incorporate the class label information.
Table 1.
Classification results for the first set of simulations: misclassification rates for MWPCR1, MWPCR2, MWPCR3, and PCA based on three numbers of principal components under three types of noise.
| Noise | Number of PCs | PCA | MWPCR1 | MWPCR2 | MWPCR3 |
|---|---|---|---|---|---|
|
| |||||
| Type I | 5 | 0.47 | 0.11 | 0.09 | 0.10 |
| 7 | 0.48 | 0.13 | 0.11 | 0.10 | |
| 10 | 0.49 | 0.13 | 0.11 | 0.10 | |
|
| |||||
| Type II | 5 | 0.41 | 0.04 | 0.08 | 0.03 |
| 7 | 0.39 | 0.03 | 0.09 | 0.04 | |
| 10 | 0.42 | 0.03 | 0.07 | 0.04 | |
|
| |||||
| Type III | 5 | 0.27 | 0.13 | 0.10 | 0.09 |
| 7 | 0.26 | 0.13 | 0.10 | 0.10 | |
| 10 | 0.28 | 0.13 | 0.10 | 0.10 | |
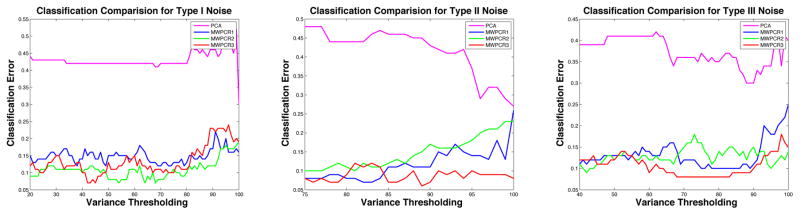
Second, we used the same variance thresholding to compare the three MWPCR methods with PCA. Figure 3 shows that the classification error (magenta curve) for PCA is much larger than that for all other methods. For each fixed variance threshold, the number of extracted principal components from PCA is less than that of MWPCR1, MWPCR2 and MWPCR3. Overall, MWPCR3 outperforms all other methods for all three types of noises. The variance threshold in the middle panel of Figure 3 starts from 70%, since the first principal component of PCA almost accounts for 70% of the total variance for Type II noise.
Figure 3.
Classification results for the first set of simulations: classification rate curves for MW- PCR1, MWPCR2, MWPCR3, and PCA based on the variance thresholding method for the three types of noise. Overall classification errors for MWPCR3 (red curve) are smaller than those of others, confirming the good performance of MWPCR3. Also MWPCR3 is quite robust to different variance thresholds. The performance of PCA is very poor and its classification error (magenta curve) is much larger than all MWPCR methods for the three types of noises.
Third, we compared MWPCR3, in which 5 principal components were used, with eight other state-of-the-art classification methods. These eight classification methods include sparse discriminant analysis (sLDA) (Clemmensen et al., 2011), sparse partial least squares (SPLS) analysis (Chun and Keles, 2010), sparse logistic regression (SLR) (Yamashita, 2011), support vector machine (SVM) (Chang and Lin, 2011), regularized optimal affine discriminant (ROAD) (Fan et al., 2012), wavelet-based multicscale PCA (WMSPCA)(Bakshi, 1998), the combination of sure independence screening (SIS) (Fan et al., 2010) and principal component analysis (PCA) (SIS+PCA), and graph-constrained elastic-net (GraphNet) (Grosenick et al., 2013). We chose these classification methods due to their excellent performance in various simulated and real data sets.
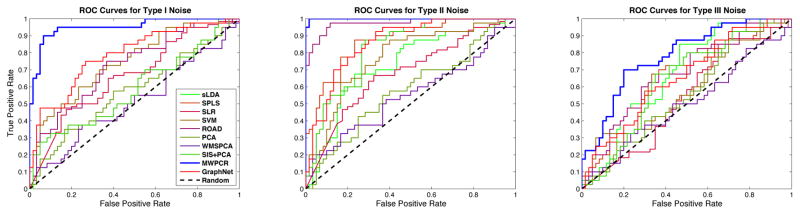
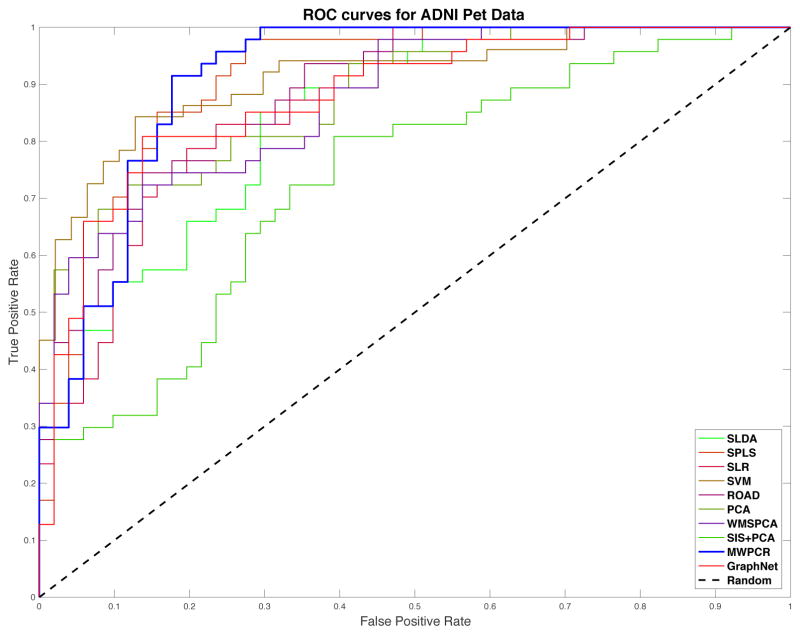
Fourth, for all classification methods, we first calculated their misclassification rates by using the leave-one-out cross validation and then generated the receiver operating characteristics (ROC) curves of all nine methods. For ROC, we used model (11) to independently generate a testing set with the same sample size and the same proportion of Class 0 to Class 1 as the training set. For each method, we applied 10-fold cross validation to the training set in order to select the tuning parameter(s) and build the model based on the training set. Then, we applied the fitted model to the testing set in order to generate the ROC curves of all nine classification methods in Figure 4. Based on these ROC curves, we calculated their area under curve (AUC) values (Fawcett, 2006).
Figure 4.
ROC curves of different classification methods for the three types of noise in the first set of simulations. The blue curves correspond to MWPCR and have the highest AUC value.
Table 2 presents the classification results, including both misclassification rates and AUC values. Table 2 reveals that MWPCR outperforms all other classification methods, especially when the signal-to-noise ratio is low for Type I and II noises. Except WMSPCA, SIS+PCA, and MWPCR, all other classification methods are also sensitive to the presence of the long-range correlation structure in Type III noise. However, if high dimensional features do not have strong spatial structures, then it is expected that MWPCR may perform worse than other competing classification methods.
Table 2.
Misclassification Rates (MRs) and AUC values for the first set of simulations: comparison between MWPCR and eight other state-of-the-art classification Methods. sLDA denotes sparse discriminant analysis; SPLS denotes sparse partial least squares; SLR denotes sparse logistic regression; SVM denotes support vector machine; ROAD denotes regularized optimal affine discriminant; WMSPCA denotes wavelet based multiscale principal component analysis; SIS+PCA combines sure independence screening (SIS) and principal component analysis; and GraphNet denotes graph-constrained elastic-net.
| Type/Measure | sLDA | SPLS | SLR | SVM | ROAD | WMSPCA | SIS+PCA | GraphNet | MWPCR |
|---|---|---|---|---|---|---|---|---|---|
| I/MR | 0.28 | 0.43 | 0.45 | 0.38 | 0.36 | 0.20 | 0.33 | 0.32 | 0.10 |
| II/MR | 0.27 | 0.08 | 0.18 | 0.26 | 0.08 | 0.13 | 0.46 | 0.22 | 0.03 |
| III/MR | 0.52 | 0.30 | 0.61 | 0.60 | 0.50 | 0.21 | 0.10 | 0.35 | 0.09 |
| I/AUC | 0.59 | 0.59 | 0.66 | 0.75 | 0.72 | 0.52 | 0.59 | 0.79 | 0.95 |
| II/AUC | 0.73 | 0.87 | 0.68 | 0.77 | 0.97 | 0.55 | 0.83 | 0.86 | 0.99 |
| III/AUC | 0.68 | 0.65 | 0.54 | 0.59 | 0.68 | 0.50 | 0.64 | 0.66 | 0.78 |
4 Real Data Analysis
4.1 ADNI PET Data
Alzheimer’s disease (AD) is the most common form of dementia and results in the loss of memory, thinking and language skills. AD is an escalating national epidemic and a genetically complex, progressive, and fatal neurodegenetive disease. The incidence of AD doubles every five years after the age of 65 and the number of AD patients has dramatically increased recently, which has caused a heavy socioeconomic burden. AD is the sixth-leading cause of death in the United States, while there is no means to prevent, cure or even slow its progression.
The development of MWPCR is motivated by using the baseline FDG-PET data set to address questions (Q1) and (Q2). The ADNI PET data set downloaded from the ADNI web site (www.loni.usc.edu/ADNI) consists of 196 subjects with 102 NCs and 94 AD subjects. There are three subjects, missing the gender and age information. Among all the rest of the subjects, there are 117 males whose mean age is 76.20 years with standard deviation 6.06 years and 76 females whose mean age is 75.29 years with standard deviation 6.29 years. FDG-PET images acquired 30–60 minutes post-injection were processed by using a standard image processing pipeline. A detailed description of PET protocols and acquisition can be found at www.adni-info.org. Such pipeline consists of average, spatially alignment, interpolation to a standard voxel size, intensity normalization, and smoothing to a common resolution of 8-mm full width at half maximum.
4.2 Binary Classification
The first goal is to use MWPCR to classify subjects from ADNI to either AD or NC group based on their FDG-PET images. It is associated with the second primary objective of ADNI aiming at developing new diagnostic methods for AD intervention, prevention, and treatment. We first applied MWPCR3 to ADNI and used the same setting as simulations in Section 3 except that we considered a linear model for f(xi,g|yi, zi, β(g)), in which zi includes both age and gender and yi is diagnosis status (AD versus NC). We also compare MWPCR3 with nine other classification methods, including PCA and the eight state-of-the-art classification methods discussed in Section 3. For the PCA method, we applied PCA with five principal components, which account for around 90% of the total variance, and then used the same linear regression as MWPCR3 to perform classification analysis. Figure 5 presents three selected slices of the weight matrix WI. The red regions, such as supramarginal gyrus right, correspond to the voxels with large importance score weights and contain the most important information for classification.
Figure 5.
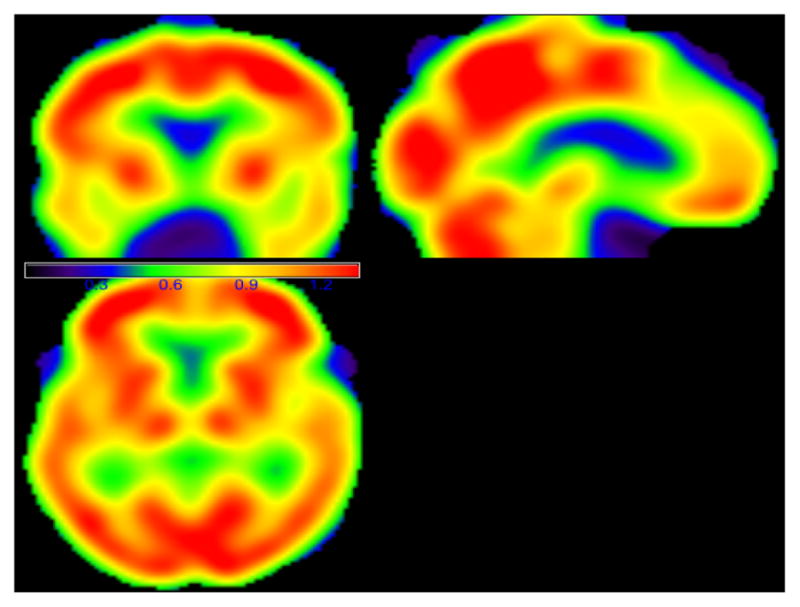
The images of the importance score weight matrix for the ADNI binary classification analysis. The red regions have large weight score values and contain the important classification information.
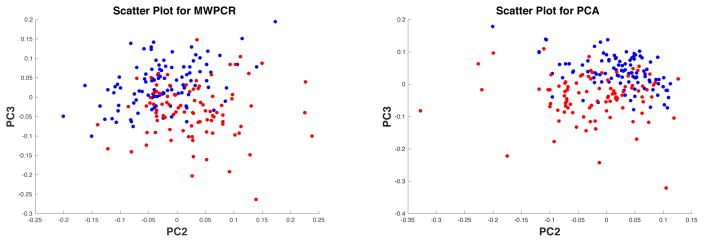
Second, for both PCA and MWPCA, we extracted their corresponding first five principal component scores and directions. Figure 6 shows the scatter plot of PC2 and PC3 scores for PCA and that for MWPCA, in which blue and red points correspond to NC and AD subjects, respectively, where PC2 and PC3 represent the second and third principal components, respectively. It seems that compared with PCA, the blue and red points are more separable for MWPCA. Furthermore, Figure 7 presents some selected slides of the principal directions corresponding to PC2 and PC3 for MWPCA. We are able to identify several key regions of interest, such as “ supramarginal gyrus”, “ superior temporal gyrus”, and “inferior frontal gyrus”. For instance, the superior temporal gyrus is in the temporal lobe of the human brain and contains several important structures of the brain, including Brodmann areas 41, 42, and 22p. It is probably involved with language perception and processing (Marcus et al., 2014). Moreover, within the brain, the anatomical regions that show the greatest decrease in FDG uptake with aging are the bilateral superior medial frontal, motor, anterior, and middle cingulate and bilateral parietal cortices. Among them, the superior temporal pole was found to be particularly affected.
Figure 6.
ADNI binary classification results: scatter plots of PC2 and PC3 scores for MWPCR (left panel) and PCA (right panel). Blue and red points in both panels correspond to NC and AD subjects, respectively.
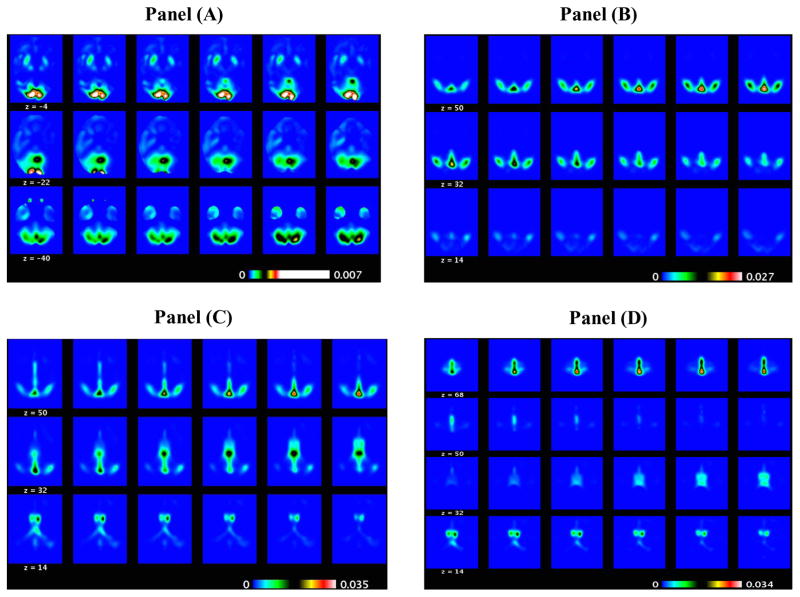
Figure 7.
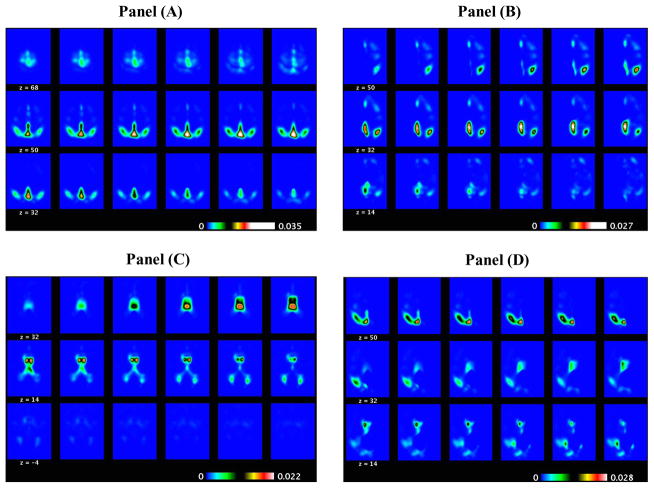
ADNI PET binary classification results: the selected slides of the PC2 direction image (positive elements in Panel (A) and negative elements in Panel (B)) and those of the PC3 direction image (positive elements in Panel (C) and negative elements in Panel (D)) obtained from MWPCR.
Third, similar to Section 3, we calculated the misclassification rates of all classification methods by using the leave-one-out cross validation and then generated their receiver operating characteristics (ROC) curves. For the ROC analysis, we randomly and proportionally split the data set into 2 parts, a training set and a testing set. For each part, the sample sizes are same (98/98). Within each part, the proportion of AD to NC remains the same. For each classification method, we used 10-fold cross validation on the training set to select the tuning parameter(s) and build the model, and then we applied the fitted model to the testing set in order to calculate the relative scores. Subsequently, we generated all ROC curves and their AUC values.
Table 3 presents the classification results based on classification error and AUC, while Figure 8 presents the ROC curves of all ten classification methods. sLDA and SIS+PCA perform much worse than all other methods. In general, SPLS, SVM and WMSPCA are comparable with each other, but they outperform SLR and ROAD. In terms of misclassification rate, MWPCR outperforms all nine other classification methods. In contrast, in terms of AUC, MWPCR, SPLS, and SVM are compatible with each other. It may indicate that the classification accuracy can be significantly improved by incorporating spatial smoothness and correlation.
Table 3.
Misclassification Rates (MRs) and AUC values of different classification methods for ADNI PET data
| sLDA | SPLS | SLR | SVM | ROAD | PCA | WMSPCA | SIS+PCA | GraphNet | MWPCR | |
|---|---|---|---|---|---|---|---|---|---|---|
| MR | 0.255 | 0.163 | 0.179 | 0.168 | 0.189 | 0.194 | 0.168 | 0.255 | 0.128 | 0.117 |
| AUC | 0.845 | 0.912 | 0.863 | 0.912 | 0.878 | 0.877 | 0.873 | 0.730 | 0.879 | 0.913 |
Figure 8.
ADNI PET binary classification results: ROC curves of the ten different classification methods. The blue line corresponds to MWPCR.
4.3 ADAS-Cog Score Prediction
The second goal is to use MWPCR to identify FDG-PET imaging biomarkers observed at baseline to accurately predict the change in the ADAS-Cog test score (or TOTAL11) at least two years later after initial assessment. The TOTAL11, which measures the cognitive performance of each subject, was calculated from the 11-item ADAS-Cog, such as Word Recall, whose details can be found in http://adni.loni.usc.edu/data-samples/data-faq/. Since three subjects are missing gender and age information and ten other subjects only have the baseline TOTAL11, we only use 183 subjects in this analysis.
We ran MWPCR as follows. We first fitted a linear model with the TOTAL11 score at the latest time point as response and the baseline TOTAL11 score, age, gender, time since baseline, and years of education, and then we used the residual obtained from the linear model as the response y and the FDG-PET image as x. In Stage 1, we fitted a linear model for f(xi,g|yi, β(g)), in which we dropped off zi. Then, WI is calculated based on the p-value of Wald test associated with the correlation between xi,g and yi at each voxel g. In Stage 2, following the simulations in Section 3, we chose MWPCR3 with different numbers of principal components for MWPCA in order to construct the low-dimensional latent variables {uk,i}. In Stage 3, we fitted a linear latent variable regression given by to do prediction.
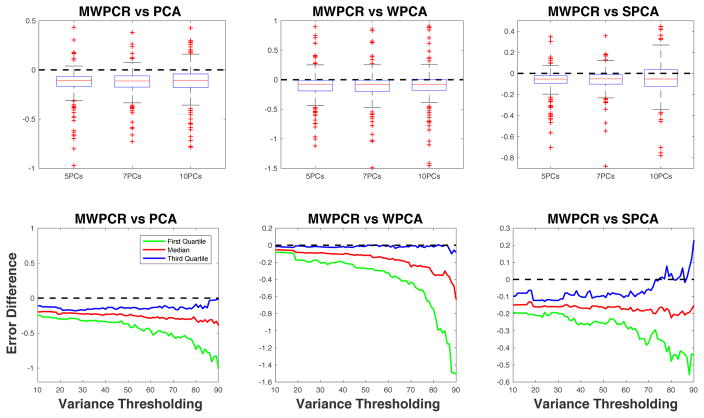
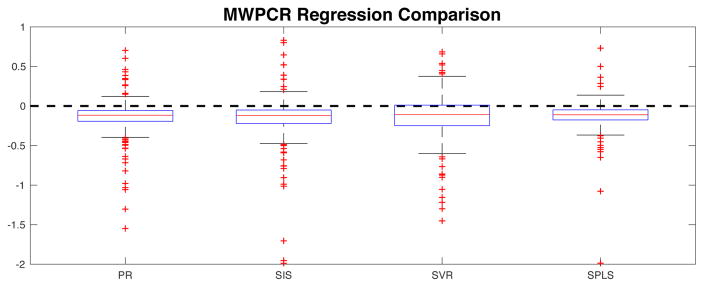
Second, we compared MWPCR and three other dimensional reduction methods including PCA, weighted PCA (WPCA) (Skocaj et al., 2007), and supervised PCA (SPCA) (Bair et al., 2006). We used the leave-one-out cross validation method to compute the prediction errors of all methods. Let ŷi be the fitted response value based on the linear latent variable regression, we define the prediction error as |ŷi − yi|/|yi|. Subsequently, we calculated the prediction error differences between MWPCR and all other three methods and their quantile curves across different numbers of principal components and variance thresholds. Figure 9 presents the comparison results based on the prediction error differences and their quantile curves. Both the error differences and the quantile curves are less than 0 (below the dashed line), confirming the better performance of MWPCR in predicting changes in ADAS-Cog score.
Figure 9.
ADAS-Cog Score Prediction for ADNI PET Data: comparison between MWPCR with PCA, WPCA, and SPCA. The panels in the first row show the boxplots of error differences between MWPCR and PCA (WPCA and SPCA) for different numbers of principal components. The panels in the second row show the first, second and third quantile curves of error differences between MWPCR and PCA (WPCA and SPCA) for different variance thresholds.
Third, for MWPCA, we extracted their corresponding first five principal component scores and directions. Figure 10 presents some selected slides of the principal directions corresponding to PC1 and PC5 for MWPCA, where PC1 and PC5 represent the first and fifth principal components, respectively. We are able to identify several key regions of interest, such as “right lateral ventricle”, “right middle temporal gyrus”, “right fornix”, and “ right middle frontal gyrus”. For instance, the fornix is on the medial aspects of the cerebral hemispheres connecting the medial temporal lobes to the hypothalamus. Since the fornix serves a vital role in memory functions, it has become the subject of recent research emphasis in Alzheimer’s disease (AD) and mild cognitive impairment (MCI) (Nowrangi and Rosenberg, 2015).
Figure 10.
ADAS-Cog Score Prediction for ADNI PET Data: the selected slides of the PC1 direction image (positive elements in Panel (A) and negative elements in Panel (B)) and those of the PC5 direction image (positive elements in Panel (C) and negative elements in Panel (D)) obtained from MWPCR.
Finally, we compare MWPCR with four other high-dimensional regression methods including penalized regression (PR) (Tibshirani, 1996), sure independence screening (SIS) regression (Fan and Lv, 2008), support vector regression (SVR) (Basak et al., 2007), and SPLS (Chun and Keles, 2010). Figure 11 shows the boxplots of the prediction error differences between MWPCR and all the other regression methods, indicating that MWPCR outperforms all other regression methods.
Figure 11.
ADAS-Cog score prediction for ADNI PET Data: comparison of MWPCR with the four other regression methods, including PR, SIS, SVR and SPLS.
5 Discussion
We have developed a general MWPCR framework for the use of high-dimensional data on graph to predict a low-dimensional response. MWPCR enables an efficient and selective treatment of individual features, accommodates the complex dependence among features, and has the ability of utilizing the underlying spatial pattern possessed by image data. MWPCR integrates feature selection, smoothing, and feature extraction in a single framework. In the simulation studies and real data analyses, MWPCR shows substantial improvement over many state-of-the-art methods for high-dimensional problems. Moreover, both theoretically and numerically, we have demonstrated the importance of using both importance score weights and spatial weights in prediction problems.
Supplementary Material
Acknowledgments
Dr. Zhu’s work was partially supported by NIH grants MH086633, NSF grants SES-1357666 and DMS-1407655, and a grant from Cancer Prevention Research Institute of Texas. This material was based upon work partially supported by the NSF grant DMS-1127914 to the Statistical and Applied Mathematical Science Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We are grateful for the many valuable suggestions from referees, associated editor, and editor.
We would like to thank the Editor, the anonymous Associate Editor, and the referees for their suggestions that have led to a much improved paper.
“Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp - content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
“Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year publicprivate partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), positron emission tomography (PET), other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of mild cognitive impairment (MCI) and early Alzheimer’s disease (AD). Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California, San Francisco. ADNI is the result of efforts of many coinvestigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 subjects but ADNI has been followed by ADNI-GO and ADNI-2. To date these three protocols have recruited over 1500 adults, ages 55 to 90, to participate in the research, consisting of cognitively normal older individuals, people with early or late MCI, and people with early AD. The follow up duration of each group is specified in the protocols for ADNI-1, ADNI-2 and ADNI-GO. Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see www.adni-info.org.”
Contributor Information
Hongtu Zhu, Professor of Biostatistics, Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, TX, 77230, and University of North Carolina, Chapel Hill, NC, 27599.
Dan Shen, Assistant Professor in Interdisciplinary Data Sciences Consortium and Department of Mathematics and Statistics, University of South Florida, Tampa, FL 33620.
Leo Yufeng Liu, Doctoral student under the supervision of Dr. Hongtu Zhu.
References
- Aharon M, Elad M, Bruckstein A. K-SVD: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans on Signal Processing. 2006;54:4311–4322. [Google Scholar]
- Allen GI, Grosenick L, Taylor J. A generalized least squares matrix decomposition. Journal of the American Statistical Association. 2014;109:145–159. [Google Scholar]
- Arias-Castro E, Salmon J, Willett R. Oracle Inequalities and Minimax Rates for Nonlocal Means and Related Adaptive Kernel-Based Methods. SIAM J Imaging Sci. 2012;5:944–992. [Google Scholar]
- Bair E, Hastie T, Paul D, Tibshirani R. Prediction by supervised principal components. J Amer Statist Assoc. 2006;101:119–137. [Google Scholar]
- Bakshi BR. Multiscale PCA with application to multivariate statistical process monitoring. American Institute of Chemical Engineers AIChE Journal. 1998;44:1596. [Google Scholar]
- Basak D, Pal S, Patranabis DC. Support vector regression. Neural Information Processing-Letters and Reviews. 2007;11:203–224. [Google Scholar]
- Bickel P, Levina E. Some theory for Fisher’s linear discriminant function, ‘naive Bayes’, and some alternatives when there are many more variables than observations. Bernoulli. 2004;10:989–1010. [Google Scholar]
- Buhlmann P, Rutimann P, Van de Geer S, Zhang CH. Correlated variables in regression: clustering and sparse estimation. Tech rep, ETH Zurich 2012 [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2:27. [Google Scholar]
- Chun H, Keles S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J Roy Statist Soc Ser B. 2010;72:3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke B, Fokoue E, Zhang HH. Principles and Theory for Data Mining and Machine Learning. New York: Springer Verlag; 2009. [Google Scholar]
- Clemmensen L, Hastie T, Witten D, Ersbøll B. Sparse discriminant analysis. Technometrics. 2011;53:406–413. [Google Scholar]
- Cuingnet R, Glaunes JA, Chupin M, Benali H, Colliot O ADNI. Spatial and anatomical regularization of SVM: a general framework for neuroimaging data. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2012 doi: 10.1109/TPAMI.2012.142. [DOI] [PubMed] [Google Scholar]
- Dray S, Jombart T. Revisiting Guerry’s data: introducing spatial constraints in multivariate analysis. Annals of Applied Statistics. 2011;5:2278–2299. [Google Scholar]
- Fan J, Fan Y. High-dimensional classification using features annealed independence rules. Ann Statist. 2008;36:2605–2637. doi: 10.1214/07-AOS504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Feng Y, Samworth R, Wu Y. R package version 0.6. 2010. SIS: Sure Independence Screening. [Google Scholar]
- Fan J, Feng Y, Tong X. A road to classification in high dimensional space: the regularized optimal affine discriminant. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2012;74:745–771. doi: 10.1111/j.1467-9868.2012.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2008;70:849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern recognition letters. 2006;27:861–874. [Google Scholar]
- Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- Goldsmith J, Huang L, Crainiceanu CM. Smooth scalar-on-image regression via spatial Bayesian variable selection. Journal of Computational and Graphical Statistics. 2014;23:46–64. doi: 10.1080/10618600.2012.743437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenick L, Klingenberg B, Katovich K, Knutson B, Taylor JE. Interpretable whole-brain prediction analysis with GraphNet. NeuroImage. 2013;72:304–321. doi: 10.1016/j.neuroimage.2012.12.062. [DOI] [PubMed] [Google Scholar]
- Guo R, Ahn M, Zhu H. Spatially weighted principal component analysis for imaging classification. Journal of Computational and Graphical Statistics. 2015;24:274–296. doi: 10.1080/10618600.2014.912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2. Hoboken, New Jersey: Springer; 2009. [Google Scholar]
- Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC ADNI. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. NeuroImage. 2009;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JZ, Shen H, Buja A. The analysis of two-way functional data using two-way regularized singular value decompositions. Journal of the American Statistical Association. 2009;104:1609–1620. [Google Scholar]
- Krishnan A, Williams L, McIntosh A, Abdi H. Partial least squares (PLS) methods for neuroimaging: a tutorial and review. Neuroimage. 2011;56:455–475. doi: 10.1016/j.neuroimage.2010.07.034. [DOI] [PubMed] [Google Scholar]
- Lee M, Shen H, Huang JZ, Marron JS. Biclustering via Sparse Singular Value Decomposition. Biometrics. 2010;66:1087–1095. doi: 10.1111/j.1541-0420.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang T, Wang Q, Gonzalez MZ, Maresh EL, Coan JA, et al. Spatial Bayesian variable selection and grouping for high-dimensional scalar-on-image regression. The Annals of Applied Statistics. 2015;9:687–713. [Google Scholar]
- Li RZ, Zhong W, Zhu L. Feature screening via distance correlation learning. Journal of American Statistical Association. 2012;107:1129–1139. doi: 10.1080/01621459.2012.695654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu H, Shen D, Lin W, Gilmore JH, Ibrahim JG. Multiscale adaptive regression models for neuroimaging data. Journal of the Royal Statistical Society: Series B. 2011;73:559–578. doi: 10.1111/j.1467-9868.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxburg UV. A tutorial on spectral clustering. Statistics and Computing. 2007;17:395–416. [Google Scholar]
- Marcus C, Mena E, Subramaniam RM. Brain PET in the diagnosis of Alzheimer’s disease. Clinical nuclear medicine. 2014;39:e413. doi: 10.1097/RLU.0000000000000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrangi MA, Rosenberg PB. The fornix in mild cognitive impairment and Alzheimer’s disease. Frontiers in aging neuroscience. 2015;7:1. doi: 10.3389/fnagi.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polzehl J, Spokoiny VG. Propagation-separation approach for local likelihood estimation. Probab Theory Relat Fields. 2006;135:335–362. [Google Scholar]
- Reiss P, Ogden R. Functional generalized linear models with images as predictors. Biometrics. 2010;66:61–69. doi: 10.1111/j.1541-0420.2009.01233.x. [DOI] [PubMed] [Google Scholar]
- Shen D, Zhu H. International Conference on Information Processing in Medical Imaging. Springer; 2015. Spatially Weighted Principal Component Regression for High-Dimensional Prediction; pp. 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skocaj D, Leonardis A, Bischof H. Weighted and robust learning of subspace representations. Pattern Recogn. 2007;40:1556–1569. [Google Scholar]
- Taylor KM, Meyer FG. A random walk on image patches. SIAM J Imaging Sciences. 2012;5:688–725. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J Roy Statist Soc Ser B. 1996;58:267–288. [Google Scholar]
- Vincent M, Gramfort A, Varoquaux G, Eger E, Thirion B. Total variation regularization for fMRI-based prediction of behavior. IEEE Transactions on Medical Imaging. 2011;30:1328–1340. doi: 10.1109/TMI.2011.2113378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita O. Quick manual for sparse logistic regression toolbox ver1.2.1. 2011 software at http://www.cns.atr.jp/~oyamashi/SLR_WEB/
- Yan S, Xu D, Zhang B, Zhang HJ, Yang Q, Lin S. Graph embedding and extensions: a general framework for dimensionality reduction. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2007;29:40–51. doi: 10.1109/TPAMI.2007.12. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ogden RT, Reiss PT. Wavelet-based LASSO in functional linear regression. Journal of Computational and Graphical Statistics. 2012;21:600–617. doi: 10.1080/10618600.2012.679241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Li L, Zhu HT. Tensor regression with applications in neuroimaging data analysis. Journal of Americal Statistical Association. 2013;108:540–552. doi: 10.1080/01621459.2013.776499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.