Abstract
Ser-Arg-rich (SR) proteins play numerous roles in spliceosome assembly and the regulation of splice-site selection. Whereas considerable attention has focused on the mechanistic details of SR protein activities, little is known concerning how these splicing regulators are controlled by the cell. Here we examined the subcellular localization of precursor mRNA splicing factors during early development of the nematode Ascaris lumbricoides. In the early embryo, before major zygotic gene activation, most SR proteins, along with RNA polymerase II, are localized in the cytoplasm. As development proceeds, we observe a significant decrease in the cytoplasmic levels of these factors and a concomitant increase in nuclear localization. In contrast, trimethylguanosine-capped small nuclear ribonucleoproteins are predominantly localized in the nucleus throughout this period. We previously showed that the phosphorylation state and activity of SR proteins are regulated during A. lumbricoides embryogenesis. These changes correlate with the onset of precursor mRNA splicing and zygotic transcription. Thus, a coordinate change in the subcellular localization of SR proteins and RNA polymerase II occurs at the transition from reliance on maternally deposited factors to embryonic expression. We propose that before zygotic gene activation, SR proteins and RNA polymerase II are stockpiled in the cytoplasm of early embryos, awaiting signals that lead to their activation.
Early embryogenesis can be divided into two distinct phases. Initially, maternally deposited factors such as mRNAs and proteins program development. The transition to zygotic control of embryogenesis occurs after a species-specific number of reductive cell divisions. Both degradation of maternally deposited mRNAs and a robust increase in transcription by RNA polymerase II (Pol II) are hallmarks of zygotic gene activation (ZGA) (1, 2). In early mammalian embryos, Pol II is localized in the cytoplasm with nuclear translocation occurring concomitant with ZGA (3). Thus, spatial and temporal regulation of Pol II contributes to the activation of zygotic genes during early embryogenesis. We previously demonstrated that precursor (pre)-mRNA splicing activity, like transcription, is also tightly regulated during embryogenesis in the parasitic nematode Ascaris lumbricoides (4). Our data suggest that the onset of pre-mRNA splicing is coupled with ZGA.
The nuclear organization of the gene-expression machinery, and splicing factors in particular, has been an area of intense research in recent years. Most splicing factors are localized throughout the nucleus in both a speckled pattern as well as in a diffuse nucleoplasmic pool (for review, see ref. 5). One class of spliceosomal components exhibiting this distribution is the Ser/Arg-rich (SR) non-small nuclear ribonucleoprotein (non-snRNP)-associated splicing factors (SR proteins). SR proteins function to initiate spliceosome assembly through proper splice-site recognition and pairing (for review, see ref. 6) and are recruited from speckles to nascent transcripts in the nucleoplasm in a manner dependent upon the carboxyl-terminal domain (CTD) of Pol II (7). Thus, speckles can be thought of as storage sites for pre-mRNA splicing factors in the nucleus of interphase cells. Although SR proteins accumulate in the nucleus, a subset appear to shuttle continuously and rapidly between the nucleus and the cytoplasm, possibly escorting the spliced mRNA to its final destination in the cytoplasm (8, 9). The nucleocytoplasmic shuttling activity of SR proteins appears to be linked to their state of phosphorylation and may prove to be a key regulatory target for the cell. Because splice-site selection is sensitive to the concentration of SR proteins both in vitro and in vivo (10, 11), modulation of SR protein phosphorylation and localization may be an effective way for the cell to control patterns of pre-mRNA splicing.
Previously, we demonstrated that SR protein phosphorylation and activity are regulated during A. lumbricoides embryogenesis (4). Before ZGA, SR proteins are hyperphosphorylated and inactive in in vitro splicing assays. After ZGA, SR proteins become partially dephosphorylated and active in vitro. Thus, changes in the phosphorylation state and activity of SR proteins correlate with the onset of pre-mRNA splicing during A. lumbricoides embryogenesis. In this study, we have taken advantage of the A. lumbricoides embryo system to address questions relating to the subcellular localization of SR proteins during development. We show by two distinct methods, immunofluorescence microscopy and subcellular fractionation, that a large cytoplasmic pool of SR proteins are present in early embryos. Concomitant with ZGA the cytosolic levels decrease, presumably because of their import into newly formed nuclei created from the reductive cell divisions of embryogenesis. When examined after subcellular fractionation, a subset of SR proteins that accumulate in the nucleus are partially dephosphorylated. We propose that SR proteins are stockpiled in the cytoplasm of embryos before ZGA in a hyperphosphorylated and inactive state with nuclear translocation occurring after ZGA. This process, coupled to SR protein partial dephosphorylation, may play a role in the activation of the pre-mRNA splicing machinery during early development.
Materials and Methods
Embryo Culture and Fixation.
Fertilized eggs were collected from the uteri of live mature female A. lumbricoides (Spear Products, Quakertown, PA). The eggs were then allowed to develop upon incubation at 30°C with agitation. Embryos were collected at the indicated times and the outer layers of the egg shell were removed as previously described (12). Stripped embryos were then treated with methanol/chloroform/acetic acid/Earl's balanced salts solution (Sigma), 6:1:1:1, for 10 s, then postfixed in 4% formaldehyde (made fresh from paraformaldehyde) for 5 min. After formaldehyde fixation, the embryos were incubated for 5 min in ice-cold 100% methanol, then for 5 min in 100% acetone. The cells were then serially rehydrated in 90%, 75%, 50%, and 25% methanol/Tris-buffered saline (25 mM Tris⋅HCl, pH 7.4/137 mM NaCl/2.68 mM KCl) containing 0.1% Tween 20 (TBST) for 10 min each and finally washed two times in TBST for 5 min. Cells were often stored in 90% methanol/TBST at −20°C for several weeks.
Antibodies and Reagents.
The pan-SR protein antibodies mAb104 (hybridoma) and mAb1H4 (Zymed) have been described previously (13, 14). The anti-trimethylguanosine (anti-TMG) cap antibody was described previously and was obtained from Oncogene Research Products (La Jolla, CA) (15). The anti-Pol II antibody, mAbH14, was obtained from Babco (Richmond, CA) and described in detail elsewhere (16). Goat anti-mouse IgG and goat anti-mouse IgM conjugated to FITC were obtained from Vector Laboratories.
Immunofluorescence Microscopy.
Fixed, permeabilized embryos were adhered to microscope slides coated in 1% polylysine (Sigma). The embryos were then blocked with 10% FBS for 30 min at room temperature before addition of the primary antibody. Primary antibodies were added as either undiluted cell-culture supernatant (mAb104) or diluted in 10% FBS as follows: mAb1H4 (1:200), anti-TMG (1:10), and mAbH14 (1:100). Staining with primary antibodies was performed at room temperature for 1 h, then the embryos were washed two times in TBST. For negative controls, the primary antibodies were omitted. FITC-conjugated secondary antibodies were used at 1:200 dilution. Samples were incubated with secondary antibodies at room temperature for 1 h, then washed four times in TBST. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) before the samples were mounted in Slow Fade mounting medium (Molecular Probes). Immunostained embryos were examined by using a 100× differential interference contrast (DIC) objective on a Zeiss Axiophot microscope. Images were captured by using a Hamamatsu cooled charge-coupled device camera controlled by the qed image acquisition software package.
Fractionation of A. lumbricoides Embryos.
Nuclear and cytosolic fractions were prepared as described (17) with the following exceptions. MgCl2 was omitted from all buffers and the phosphatase inhibitors β-glycerophosphate and KF were included in all buffers at a final concentration of 20 mM. Additionally, the volume of the nuclear fractions was adjusted to be made equal to the volume of the cytosolic fractions.
Localization of SR Proteins in Isolated Nuclei.
To immunostain SR proteins present in nuclei isolated from 1-cell-stage embryos, nuclei were fixed for 5 min in 3.5% formaldehyde at room temperature. Nuclei were then permeabilized in 0.2% Triton X-100 for 5 min on ice. Fixed, permeabilized nuclei were processed for immunofluorescence with the anti-SR protein antibody 1H4 as described above.
Western Blot Analysis of Fractionated A. lumbricoides Embryos.
Western blot analysis of SR proteins was performed as described previously (12) with the exception that SR proteins were detected with mAb1H4. Because the volumes of each fraction had been equalized, gels were loaded to reflect changes in the nuclear and cytosolic volumes occurring during the reductive cell divisions of A. lumbricoides embryogenesis. Thus, 1-cell nuclear and cytosolic fractions were loaded in a 1:4 ratio (vol/vol), and the 32- to 64-cell nuclear and cytosolic fractions were loaded at a 1.15:1 ratio (vol/vol) (17).
In Vitro Kinase Reactions.
Phosphorylation of SR proteins present in nuclear and cytosolic fractions was performed as described previously (4). Approximately 1.2 μg of total protein from each fraction was incubated with 2 units of SR protein kinase 1 (SRPK1) for 40 min at room temperature. Radiolabled proteins were resolved by 12% SDS/PAGE and visualized by PhosphorImager.
Results
Immunofluorescence Analysis of SR Protein Localization in Developing Embryos.
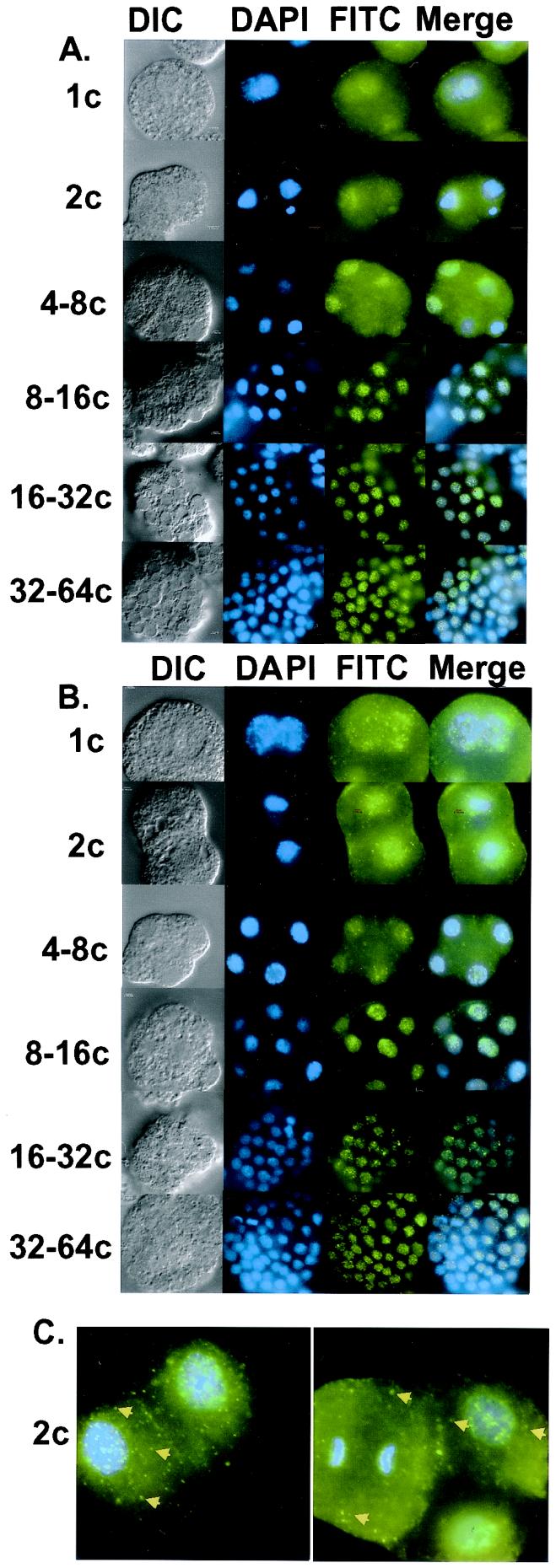
In cultured cells, SR protein kinases such as SRPK1 and Clk/Sty can induce the redistribution of shuttling SR proteins from their steady-state nuclear, speckled pattern to a predominantly cytoplasmic localization (8, 18–21). We therefore investigated whether changes in the phosphorylation state of SR proteins that occur during A. lumbricoides embryogenesis influence their subcellular localization. Synchronous embryo preparations were grown to the 1-cell, 2-cell, 4- to 8-cell, 8- to 16-cell, 16- to 32-cell and 32- to 64-cell stages, then fixed and permeabilized as described in Materials and Methods. The subcellular localization of SR proteins before and after the onset of zygotic gene expression (4- to 8-cell stage) (22, 23) was determined by indirect immunofluorescence with mAb104, which recognizes a highly conserved phosphoepitope (13). We observed that SR proteins were present in both the nuclei and cytosol of the 1-cell, 2-cell, and 4- to 8-cell embryos (Fig. 1A). As development proceeds, the cytosolic pool of SR proteins decreases and staining by mAb104 becomes increasingly localized to the nuclei (see Fig. 1A; 8–16c, 16–32c, and 32–64c). We obtained analogous results with a related, isotypically distinct pan-SR protein monoclonal antibody, 1H4 (Fig. 1B). Again we observed that SR proteins were present in both the nuclei and cytoplasm of pre-ZGA embryos; however, as development proceeds the vast majority of the signal accumulates in the nuclei. Control experiments using the FITC-conjugated secondary antibodies alone demonstrated very low levels of nonspecific fluorescence detectable under our immunolabeling conditions (data not shown). Additionally, because mAb104 and mAb1H4 are two distinct Ig classes (IgM and IgG, respectively), it is unlikely that the changes in SR protein localization observed during embryogenesis are due to differences in antibody accessibility.
Figure 1.
Immunolocalization of SR proteins during A. lumbricoides embryogenesis. (A) SR proteins were visualized by immunolabeling fixed, permeabilized embryos with mAb104 and detected with anti-mouse IgM conjugated to FITC. (B) SR proteins were visualized by immunolabeling fixed, permeabilized embryos with mAb1H4 and detected with anti-mouse IgG conjugated to FITC. (C) Pre-ZGA embryos contain mitotic interchromatin granule (MIG)-like structures. Embryos were stained with mAb1H4 as above. Arrows designate MIG-like structures. DNA was counterstained with DAPI in all images. Lowercase “c” after numerals stands for “cell.”
Closer examination of embryos stained for SR proteins with either mAb104 or mAb1H4 revealed an interesting morphology. In early embryos before ZGA, cytosolic SR proteins were distributed both diffusely as well as in a speckled pattern (2-cell stage; Fig. 1C, Left, arrows). These cytoplasmic speckles appear to be similar to mitotic interchromatin granule (MIG) clusters that are distributed throughout mitotic cells and contain at least the SR protein SC-35, snRNP antigens, U2AF65 and Pol II (24–27). Near the end of mitosis, after reformation of the nuclear membrane, snRNPs relocate to the nucleus whereas SR proteins continue to reside in MIGs. By late telophase, SC-35 returns to the nucleus. Here, we observe MIG-like structures before ZGA even in interphase cells. Fig. 1C (Right) shows a striking example of this pattern. Here, in an early embryo, we observe two interphase cells (to the right, only the upper one is in focus) and one mitotic cell that is in anaphase (to the left). As expected, intense cytoplasmic staining is observed in the mitotic cell; additionally, numerous foci, presumed MIG-like structures, are observed throughout the cytosol. Similarly, strong cytoplasmic staining and foci are also observed in the interphase cell. These data suggest that a subset of SR proteins may be stored in MIG-like structures before ZGA.
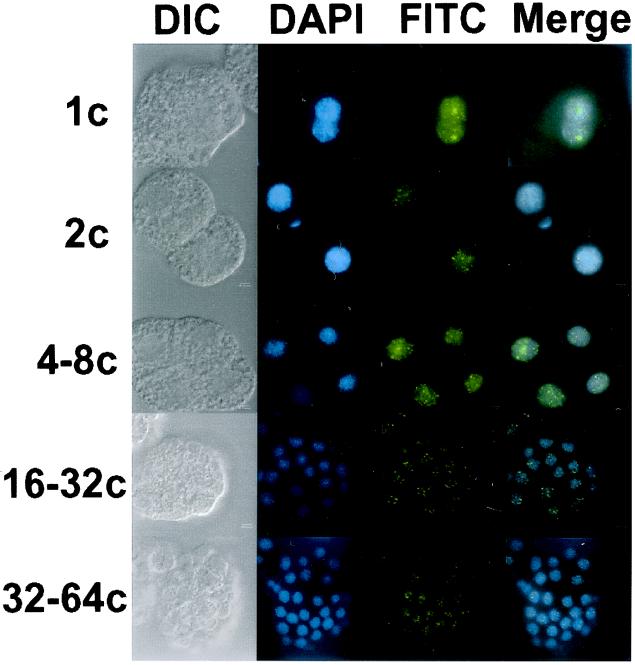
To further examine the fate of the pre-mRNA splicing machinery during nematode embryogenesis, we localized the TMG-capped snRNPs. Indirect immunofluorescence experiments with the anti-TMG cap antibody, K121, demonstrated that snRNPs appear to be localized predominantly to the nuclear compartment at all stages of embryogenesis (Fig. 2). These data are consistent with a number of studies examining the distribution of snRNPs during embryogenesis in other systems and provide a control for our anti-SR protein localization experiments (28–31).
Figure 2.
Immunolocalization of TMG-capped snRNP particles in A. lumbricoides embryos. snRNP particles were visualized by indirect immunofluorescence in fixed, permeabilized embryos with the anti-TMG antibody K121 followed by anti-mouse IgG conjugated to FITC. Lowercase “c” after numerals stands for “cell.”
Localization of Pol II During Embryogenesis.
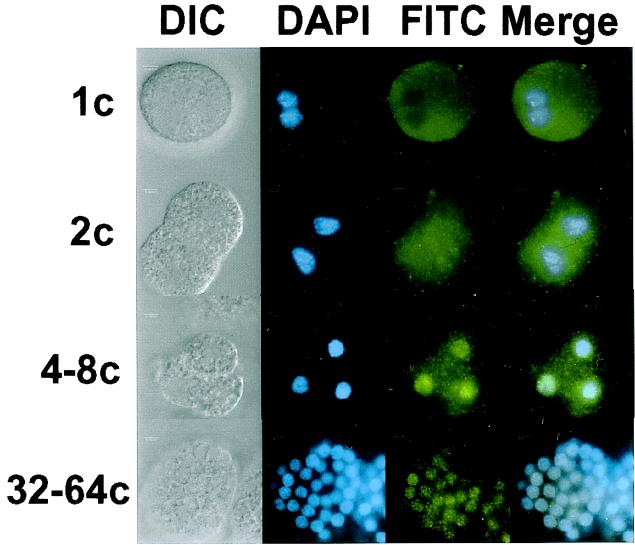
ZGA is characterized by a burst of transcription from Pol II. Repression of zygotic gene expression during mammalian embryogenesis appears to involve the sequestration of Pol II in the cytoplasm before ZGA. Accumulation of Pol II in the nucleus occurs concomitant with ZGA. Like SR proteins, there is evidence that Pol II is hyperphosphorylated before activation of zygotic transcription (3, 4). Dephosphorylation of the CTD of Pol II can be correlated with both nuclear translocation and the timing of ZGA. Previous studies demonstrated that major ZGA in A. lumbricoides occurs at the 4- to 8-cell stage (22, 23). We examined the subcellular localization of Pol II during this phase of embryogenesis by indirect immunofluorescence with the monoclonal antibody H14. H14 recognizes a specific phosphoepitope present in the phylogenetically conserved CTD (16). Fig. 3 shows that, as in mammalian embryos, Pol II is distributed throughout the cytoplasm before ZGA in nematodes. Nuclear accumulation of the H14 antigen initiates near the 4- to 8-cell stage (only three cells of the embryo are in focus in the image depicted in Fig. 3, the 4- to 8-cell stage), suggesting that the subcellular localization of Pol II is developmentally regulated.
Figure 3.
Immunolocalization of Pol II in A. lumbricoides embryos. Pol II was detected with the mouse monoclonal antibody H14 followed by anti-mouse IgM conjugated to FITC. Lowercase “c” after numerals stands for “cell.”
Analysis of Individual SR Protein Localization by Subcellular Fractionation.
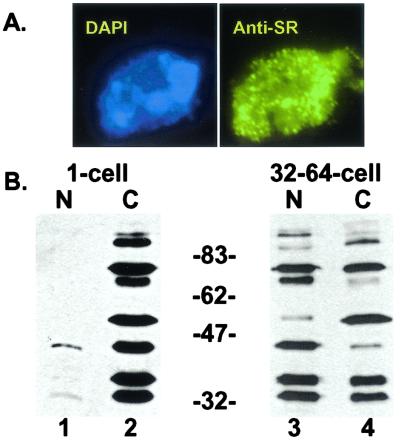
To extend our analysis of SR protein localization to a biochemical level, we used an A. lumbricoides embryo fractionation procedure (17). Briefly, 1-cell or 32- to 64-cell embryos were separated into nuclear and cytosolic fractions. Microscopic analysis of the nuclear and cytosolic fractions after DAPI staining verified that the nuclei were both intact and absent from the cytosolic fraction (data not shown). Furthermore, we were able to isolate stoichiometric amounts of nuclei from 1-cell-stage embryos, demonstrating that the early nuclei were not damaged during the procedure (data not shown). To control for the possibility of SR protein leakage from nuclei prepared from 1-cell-stage embryos, purified nuclei were fixed, permeabilized, and stained for SR proteins with mAb1H4. We observed that isolated nuclei from 1-cell embryos exhibit identical morphology to nuclei in intact embryos at the same developmental stage, suggesting that leakage of SR proteins from the nuclei does not occur (Fig. 4A).
Figure 4.
Subcellular fractionation of 1-cell and 32- to 64-cell A. lumbricoides embryos. (A) Isolated 1-cell stage nuclei are intact (representative nucleus, 100×). Nuclei were fixed, permeabilized, and stained with the anti-SR protein antibody mAb1H4 (Right) and counterstained with DAPI (Left). (B) Western blot analysis (mAb1H4) of nuclear (lanes headed by “N”) and cytosolic (lanes headed by “C”) fractions prepared from 1-cell and 32- to 64-cell embryos. Because the volumes of each fraction were equalized (see Materials and Methods), gels were loaded to reflect changes in the nuclear and cytosolic volumes occurring during the reductive cell divisions of A. lumbricoides embryogenesis.
We then determined the distribution of nuclear and cytosolic SR proteins from 1-cell and 32- to 64-cell embryos by Western blot analysis with mAb1H4 (Fig. 4B). At the 1-cell stage, the majority of SR proteins can be found in the cytosolic fraction, whereas considerably fewer are found in the nuclear fraction (Fig. 4B, compare lanes 1 and 2). Importantly, we detect a complete array of SR proteins in the nuclear fraction when blots are overexposed (data not shown). By the 32- to 64-cell stage however, equivalent total levels of SR proteins can be found in both compartments (Fig. 4B, compare lanes 3 and 4). These data indicate that whereas the nuclei at any given cell stage contain SR proteins, significantly higher relative levels of SR proteins are found in the cytoplasm of early, pre-ZGA embryos.
A closer inspection of the SR proteins present in each fraction reveals several interesting results. First, it is evident that both the 70-kDa and 90-kDa 1H4-reactive proteins migrate as doublets in the 1-cell cytosolic fraction. It is unclear whether the differences in the mobility of these proteins are due to posttranslational modifications, such as their state of phosphorylation, or whether the bands represent distinct SR proteins. Second, it appears that specific SR proteins may accumulate in the nucleus differentially. For example the 45-kDa species is predominantly cytosolic in the 1-cell embryo and nuclear in the 32–64-cell-stage embryo. By contrast, the 55-kDa species shows relatively moderate nuclear accumulation by the 32- to 64-cell stage. The same observation is made when examining the 70-kDa doublet. At the 32- to 64-cell stage the lower band accumulates predominantly in the nuclear fraction, whereas the upper band still has a large cytosolic component. These results, from subcellular fractionation, correlate with our immunofluorescence data in that the majority of SR proteins are cytoplasmic before ZGA and the amount of nuclear SR proteins increases relative to the cytoplasmic pool as development proceeds. Although the pan-SR protein antibodies used in this study recognize phosphoepitopes, changes in the extent of SR-protein phosphorylation previously observed during this phase of A. lumbricoides embryogenesis (4) do not alter antibody reactivity (data not shown). Thus, we can directly compare the levels of SR proteins present in each subcellular fraction by Western blot analysis.
Nuclear Accumulation of Specific SR Proteins Coincides with Dephosphorylation.
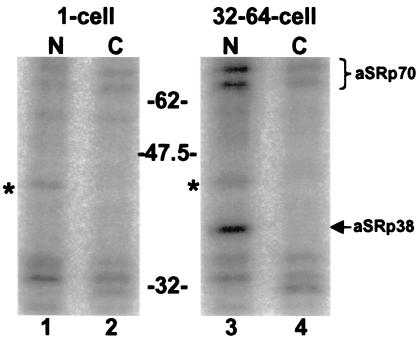
Previously, we demonstrated that SR proteins purified from A. lumbricoides embryos before ZGA are more highly phosphorylated than those isolated from later, post-ZGA embryos (4). We took advantage of subcellular fractionation to investigate whether nuclear accumulation of specific SR proteins correlated with changes in their level of phosphorylation. SRPK1 was used to phosphorylate endogenous SR proteins in the nuclear and cytosolic fractions prepared from 1- and 32–64-cell embryos with [γ-32P]ATP (Fig. 5). In this assay, 32P incorporation is inversely proportional to the native phosphorylation state of the SR proteins (4). Interestingly, aSRp38 and aSRp70 showed the highest levels of label incorporation in the 32- to 64-cell stage nuclear fractions, whereas the 1-cell and 32- to 64-cell stage cytosolic fractions showed considerably less 32P incorporation (Fig. 5, compare lane 3 to lanes 2 and 4). Although Western blot analysis demonstrates that the cytoplasmic fractions are enriched in both aSRp38 and aSRp70 (see Fig. 4B), they remain poor substrates for SRPK1. Importantly, SRPK1 can phosphorylate exogenous, purified 32-cell stage SR proteins that have been added to the cytosolic fractions, demonstrating that an inhibitor of SRPK1 is not present (data not shown). These data suggest that cytosolic aSRp38 and aSRp70 are more highly phosphorylated then their nuclear counterparts. Thus, nuclear translocation of aSRp38 and aSRp70 may require partial dephosphorylation.
Figure 5.
Partially dephosphorylated forms of aSRp38 and aSRp70 accumulate in the nucleus after ZGA. Endogenous SR proteins present in each fraction were phosphorylated with [γ-32P]ATP by SRPK1. The bands marked with asterisks (*) are phosphorylated independent of SRPK1 (data not shown).
Discussion
We have demonstrated that SR proteins are stockpiled in the cytoplasm of early A. lumbricoides embryos. Indirect immunofluorescence microscopy with two related but distinct anti-SR-protein antibodies labeled both the cytosolic and nuclear compartments in embryos prepared before ZGA. As development proceeds, the cytosolic pool of SR proteins appears to be distributed to newly formed daughter nuclei that result from reductive cell divisions that are characteristic of early embryogenesis. In contrast, TMG-capped snRNPs are localized to the nucleus throughout embryogenesis. Nuclear localization of Pol II correlates with ZGA and closely parallels that of SR proteins, suggesting that the onset of these two aspects of gene expression may be coordinately regulated. Finally, subcellular fractionation of A. lumbricoides embryos demonstrates that specific SR proteins differentially accumulate in the nucleus after ZGA and that, for a subset of these splicing factors, partial dephosphorylation may be required for nuclear translocation.
Cytoplasmic Stockpiling of SR Proteins and Pol II.
We have observed that the majority of both Pol II and SR proteins are present in the cytoplasm of embryos before ZGA. In effect, this may be a way for the embryo to maintain a store of SR proteins until they are required to process the burst of pre-mRNAs synthesized after ZGA (2, 3). The presence of SR proteins in the nucleus of early embryos may reflect a requirement for the splicing of low levels of transcripts that are synthesized before the major activation of zygotic genes at the 4- to 8-cell stage. Our data suggest that activation of zygotic gene expression and splicing may be coordinated. This idea is supported by the discovery that Pol II is intimately involved in numerous aspects of pre-mRNA processing (32). For example, the CTD of Pol II is thought to integrate pre-mRNA splicing, capping, and polyadenylation of nascent transcripts by functioning as a molecular adapter for pre-mRNA processing factors. Interaction between the splicing apparatus and the CTD of Pol II is well documented and contributes to the hypothesis that pre-mRNA splicing occurs cotranscriptionally (for review, see ref. 33). Previous studies have shown that interactions between Pol II and SR proteins can occur independent of ongoing transcription, as Pol II coimmunoprecipitates with SR proteins from mitotic extracts and colocalizes with SR proteins in MIGs (26). Thus before ZGA, it is possible that SR proteins and Pol II are present in a cytoplasmic complex; however, we have as yet been unable to detect interactions between these factors in cytosolic fractions prepared from pre-ZGA embryos. Nonetheless, the coordinated import of SR proteins and Pol II into the nucleus is coincident with the onset of ZGA.
Whereas immunofluorescence microscopy yielded a wealth of data concerning the cellular localization of SR proteins during A. lumbricoides embryogenesis, subcellular fractionation allowed us to obtain information concerning specific SR proteins. Western blot analysis of fractionated embryos strongly supports the hypothesis that SR proteins are stockpiled in the cytoplasm of early, pre-ZGA embryos. Specifically, both the nuclear and cytosolic fractions contain the same array of SR proteins; however, their concentration is much higher in the cytoplasm of early embryos. After ZGA, there appear to be roughly equivalent levels of SR proteins in each compartment, suggesting that the cytoplasmic pool of SR proteins has become distributed to the newly formed nuclei arising from the reductive cell divisions of embryogenesis. Of particular interest is the observation that nuclear translocation of SR proteins appears to occur with specificity. These data suggest that nuclear translocation of specific SR proteins may be differentially regulated during embryogenesis. Perhaps regulation of the nuclear import of discrete SR proteins may have important implications for developmental control of alternative splicing, thus providing an additional regulatory mechanism to control posttranscriptional gene expression.
Regulation of SR Proteins by Phosphorylation.
Phosphorylation plays a critical role in regulating both the biochemical activities of SR proteins as well as their subcellular localization (for review, see ref. 5). In fact, the current view of spliceosome assembly in vivo takes both of these factors into consideration. At the biochemical level, phosphorylation of RS-domain-containing splicing factors is important to initiate spliceosome assembly and to enhance sequence-specific RNA binding (34–36). In addition, protein–protein interactions between RS-domain-containing splicing factors as well as their overall activity in pre-mRNA splicing can be regulated through reversible phosphorylation (4, 12, 37–42). In the context of the living nucleus, the use of the green fluorescent protein as a fusion partner for SR proteins led to the discovery that SR proteins are released from speckles into the nucleoplasm in a phosphorylation-dependent manner (43). Thus, phosphorylation can both release SR proteins from speckles and increase their affinity for the pre-mRNA and for components of the splicing machinery. Likewise, dephosphorylation of SR proteins is believed to facilitate disassembly of splicing complexes and to recycle SR proteins back to speckles (35, 43, 44). This current study suggests that SR proteins localized to the cytoplasm before ZGA are hyperphosphorylated and that nuclear translocation of a subset of SR proteins may require partial dephosphorylation. These results are consistent with several studies showing that SR protein kinases can induce redistribution of SR proteins to the cytoplasm (8, 20, 21).
Although the phosphorylation state of SR proteins clearly influences their subcellular localization, the mechanism through which this is accomplished is unclear. Recently two SR protein specific transporters, TRN-SR and TRN-SR2, have been identified by yeast two-hybrid screens (21, 45). Both are members of the importin-β family of nuclear transport proteins and interact with the RS domain of SF2/ASF. Additionally, phosphorylation of the RS domain is required for interactions between SR proteins and TRN-SR2. Therefore, phosphorylation must be required for import of SR proteins to the nucleus. This appears to be true for nuclear targeting of SR proteins when expressed in the yeast Saccharomyces cerevisiae (46). Although budding yeast does not have classical SR-protein homologues, it does express a kinase, SKY1, with significant homology to the mammalian kinase SRPK1 (47). Nuclear targeting of heterologous SR proteins in yeast requires functional SKY1p or mammalian SRPK1. Thus, it is likely that phosphorylation of a subset of SR proteins is important for nuclear targeting.
If RS domain phosphorylation is necessary for nuclear import of SR proteins, how does the expression of an SR protein kinase induce cytoplasmic accumulation of shuttling SR proteins? Several distinct hypotheses have been proposed that would support a role for RS domain phosphorylation in nuclear import and the induction of cytoplasmic accumulation of shuttling SR proteins by increased kinase activity. First, there is evidence that shuttling SR proteins might be retained in the cytoplasm by means of direct interactions with SR protein kinases (20). Second, the net charge of SR proteins may influence their cellular localization. When SR proteins are basally phosphorylated, they are imported to the nucleus and targeted to nuclear speckles. When SR proteins are hyperphosphorylated, they might be exported from the nucleus or retained in the cytoplasm by a sequestration factor (8, 46). Another possibility is that specific sites of phosphorylation may influence the cellular localization and activity of SR proteins. In yeast for example, the cellular localization and activity of the transcription factor Pho4 are regulated by distinct combinations of specific phosphorylation sites, thus allowing for multiple levels of regulation (48). It is conceivable that SR proteins are regulated in a similar manner, as there are numerous potential phosphorylation sites that are recognized by several distinct kinase activities. Finally, it has recently been reported that the subcellular localization of the yeast protein Npl3, a distant relative of SR proteins, is regulated by SKY1 (the SRPK1 homologue) (49). Additionally, phosphorylation of Npl3 by SKY1 appears to be antagonized by arginine methylation (50). Thus, antagonistic types of posttranslational modifications may play a role in regulating the localization and activity of shuttling proteins, although it is unclear whether a similar mode of regulation occurs in mammalian cells. Our results demonstrate a correlation between cytoplasmic localization and hyperphosphorylation of SR proteins. It is clear that a more detailed analysis of SR-protein phosphorylation sites is required to begin to elucidate the mechanism through which phosphorylation regulates the activity and subcellular localization of these critical splicing factors.
Acknowledgments
We thank Xiang-Dong Fu, Greg Matera, Tim Nilsen, Charles Romfo, JoAnn Wise, and members of the Bruzik lab for critical reading of the manuscript. Also, we thank Piet deBoer, Sue Burden-Gulley, Susann Brady-Kalnay, and Mark Frey for assistance with immunofluorescence microscopy. J.R.S. is partially supported by National Research Service Award Institutional Training Grant T32 GM 08056. J.P.B. is funded by National Institutes of Health Grant R01–54204.
Abbreviations
- Pol II
RNA polymerase II
- ZGA
zygotic gene activation
- pre-mRNA
precursor mRNA
- SR
Ser-Arg-rich
- snRNP
small nuclear ribonucleoprotein
- CTD
carboxyl-terminal domain
- TMG
trimethylguanosine
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- SRPK1
SR protein kinase 1
- MIG
mitotic interchromatin granule
References
- 1.Duval C, Bouvet P, Omilli F, Roghi C, Dorel C, LeGuellec R, Paris J, Osborne H B. Mol Cell Biol. 1990;10:4123–4129. doi: 10.1128/mcb.10.8.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nothias J Y, Majumder S, Kaneko K J, DePamphilis M L. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- 3.Bellier S, Chastant S, Adenot P, Vincent M, Renard J P, Bensaude O. EMBO J. 1997;16:6250–6262. doi: 10.1093/emboj/16.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanford J R, Bruzik J P. Genes Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Misteli T, Spector D L. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 6.Graveley B R. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misteli T, Spector D L. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 8.Cáceres J F, Screaton G R, Krainer A R. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y, Steitz J A. Mol Cell. 2001;7:899–905. doi: 10.1016/s1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 10.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 11.Cáceres J F, Stamm S, Helfman D M, Krainer A R. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 12.Sanford J R, Bruzik J P. RNA. 1999;5:918–928. doi: 10.1017/s1355838299990234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth M B, Zahler A M, Stolk J A. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neugebauer K M, Roth M B. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 15.Krainer A R, Maniatis T. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 16.Bregman D B, Du L, van der Zee S, Warren S L. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidl C, Moritz K B. Nucleic Acids Res. 1998;26:768–777. doi: 10.1093/nar/26.3.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gui J-F, Tronchére H, Chandler S D, Fu X-D. Proc Natl Acad Sci USA. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colwill K, Pawson T, Andrews B, Prasad J, Manley J L, Bell J C, Duncan P I. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- 20.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer A R, Hagiwara M. J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- 21.Lai M C, Lin R I, Huang S Y, Tsai C W, Tarn W Y. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 22.Cleavinger P J, McDowell J W, Bennett K L. Dev Biol. 1989;133:600–604. doi: 10.1016/0012-1606(89)90062-6. [DOI] [PubMed] [Google Scholar]
- 23.Spicher A, Etter A, Bernard V, Tobler H, Müller F. Dev Biol. 1994;164:72–86. doi: 10.1006/dbio.1994.1181. [DOI] [PubMed] [Google Scholar]
- 24.Leser G P, Fakan S, Martin T E. Eur J Cell Biol. 1989;50:376–389. [PubMed] [Google Scholar]
- 25.Spector D L, Fu X-D, Maniatis T. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E, Du L, Bregman D B, Warren S L. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gama-Carvalho M, Krauss R D, Chiang L, Valcarcel J, Green M R, Carmo-Fonseca M. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lobo S M, Marzluff W F, Seufert A C, Dean W L, Schultz G A, Simerly C, Schatten G. Dev Biol. 1988;127:349–361. doi: 10.1016/0012-1606(88)90321-1. [DOI] [PubMed] [Google Scholar]
- 29.Dean W L, Seufert A C, Schultz G A, Prather R S, Simerly C, Schatten G, Pilch D R, Marzluff W F. Development (Cambridge, UK) 1989;106:325–334. doi: 10.1242/dev.106.2.325. [DOI] [PubMed] [Google Scholar]
- 30.Dean W L, Schultz G A. Cell Differ Dev. 1990;31:43–51. doi: 10.1016/0922-3371(90)90089-f. [DOI] [PubMed] [Google Scholar]
- 31.Watson A J, Wiemer K E, Arcellana-Panlilio M, Schultz G A. Mol Reprod Dev. 1992;31:231–240. doi: 10.1002/mrd.1080310402. [DOI] [PubMed] [Google Scholar]
- 32.Hirose Y, Tacke R, Manley J L. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirose Y, Manley J L. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 34.Tacke R, Chen Y, Manley J L. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao W, Jamison S F, Garcia-Blanco M A. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao S-H, Manley J L. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 37.Wu J Y, Maniatis T. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 38.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Nature (London) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 39.Xiao S H, Manley J L. EMBO J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roscigno R F, Garcia-Blanco M A. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 41.Hertel K J, Maniatis T. Proc Natl Acad Sci USA. 1999;96:2651–2655. doi: 10.1073/pnas.96.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prasad J, Colwill K, Pawson T, Manley J L. Mol Cell Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misteli T, Cáceres J F, Clement J Q, Krainer A R, Wilkinson M F, Spector D L. J Cell Biol. 1998;143:297–307. doi: 10.1083/jcb.143.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mermoud J E, Cohen P T W, Lamond A I. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kataoka N, Bachorik J L, Dreyfuss G. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeakley J M, Tronchere H, Olesen J, Dyck J A, Wang H Y, Fu X D. J Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siebel C W, Feng L, Guthrie C, Fu X D. Proc Natl Acad Sci USA. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komeili A, O'Shea E K. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 49.Gilbert W, Siebel C W, Guthrie C. RNA. 2001;7:302–313. doi: 10.1017/s1355838201002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun C Y, Fu X D. J Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]