Abstract
Claudins, the major transmembrane proteins of tight junctions, are members of the tetraspanin superfamily of proteins that mediate cellular adhesion and migration. Their functional importance is demonstrated by mutations in claudin genes that eliminate tight junctions in myelin and the testis, abolish Mg2+ resorption in the kidney, and cause autosomal recessive deafness. Here we report that two paralogs among 15 claudin genes in the zebrafish, Danio rerio, are expressed in the otic and lateral-line placodes at their earliest stages of development. Related claudins in amphibians and mammals are expressed in a similar manner in vertebrate primordia such as sensory placodes, branchial arches, and limb buds. We also show that the claudin gene family may have expanded along the chordate stem lineage from urochordates to gnathostomes, in parallel with the elaboration of vertebrate characters. We propose that tight junctions not only form barriers in mature epithelia, but also participate in vertebrate morphogenesis.
Compartments bounded by epithelia are a fundamental feature of any metazoan. Compartmentalization overcomes the limitations imposed by diffusion and facilitates the control of cellular environments, the division of labor among differentiated cell types, and the efficient transport of nutrients and information over long distances. To form effective diffusional barriers, epithelia use two types of intercellular seals, tight junctions and septate junctions (1, 2). Both contain matching strands of intramembrane particles in the apposed cell membranes. Both are selective and dynamic rather than impermeable and static barriers. Both are linked by intracellular membrane-associated proteins to signaling pathways and the cytoskeleton. The two types of junctions are distinguished by their ultrastructure and their distribution among metazoan phyla. At tight junctions the intercellular space is wholly obliterated, whereas at septate junctions the apposed cell membranes are separated by distinct septa and remain 12–18 nm apart. Tight junctions are in general characteristic of chordates, whereas septate junctions are found in nonchordates (1). However, tight junctions also have been found in the blood–brain and blood–testis barriers of adult arthropods (3) and septate junctions occur at the nodes of Ranvier in the vertebrate nervous system (4).
The intercellular barrier in mammalian tight junctions is formed by claudin proteins (2), members of the tetraspanin superfamily of integral membrane proteins that mediate cellular adhesion and migration (5). The mammalian claudin family comprises at least 20 members (6). Mutations in claudin genes eliminate tight junctions in myelin and the testis (7), abolish Mg2+ resorption in the kidney (8), and cause autosomal recessive deafness (9). Claudins by themselves can form the strands of membrane particles that are characteristic of the vertebrate tight junction (10). For septate junctions and arthropod tight junctions, in contrast, the molecular nature of the barrier remains unknown (4).
Compartmentalization is not only a hallmark of metazoans, but also may have facilitated the increase in diversity and complexity during their evolution (11, 12). Compartments in a broad sense—of body plans, genomes, and developmental programs—allowed variation of individual modules while protecting others from the potentially deleterious effects of mutations. During the evolution of vertebrates, for example, the chordate body plan was retained while novel structures such as the cranium, paired sensory organs, and paired appendages were added (13, 14). This increase in anatomical complexity likely reflected an increase in genetic complexity as many gene families expanded by retrotransposition, tandem duplication, and duplication of whole genomes (15, 16). The redundancy of the duplicated genes presumably allowed them to evolve independently and to acquire novel functions or perform old functions in novel combinations (14, 17). During the embryonic development of chordates, members of all subphyla have been postulated to pass after gastrulation through a common phylotypic period (11, 18). Only then do the primordia of characteristic vertebrate structures become apparent and only thereafter do features specific to the various orders, classes, and families develop.
Vertebrate tight junctions had been until recently investigated in detail only in adults or during early cleavage of the embryo (19). During a cDNA subtraction screen in the zebrafish, Danio rerio, we identified two claudin genes that are expressed after gastrulation with exquisite specificity in sensory placodes and the pronephros. This expression during the phylotypic period in characteristic vertebrate structures and the importance of tight junctions for compartmentalization suggested a connection between claudins and vertebrate evolution and ontogeny. We therefore investigated the phylogeny of claudins and their expression in vertebrate primordia.
Materials and Methods
Subtractive cDNA Hybridization.
Zebrafish embryos at about prim-6 stage were cut transversely with ultra-fine scissors (Fine Science Tools, Foster City, CA) anterior and posterior to the otic vesicle and at the seventh somite (Fig. 1a). The hindbrain region containing the otic vesicles was removed (“ear” sample), as were the anterior part of the head, the trunk, and the tail (“head-and-tail” sample). From 150 embryos, 2 μg of total ear RNA and 70 μg of total head-and-tail RNA were obtained by purification with Trizol (Life Technologies, Rockville, MD), digestion with DNaseI, and centrifugation through a cesium trifluoracetate cushion. One microgram from each sample was used for cDNA synthesis and limited amplification with a Smart PCR cDNA synthesis kit followed by subtractions with a PCR-Select cDNA subtraction kit (CLONTECH).
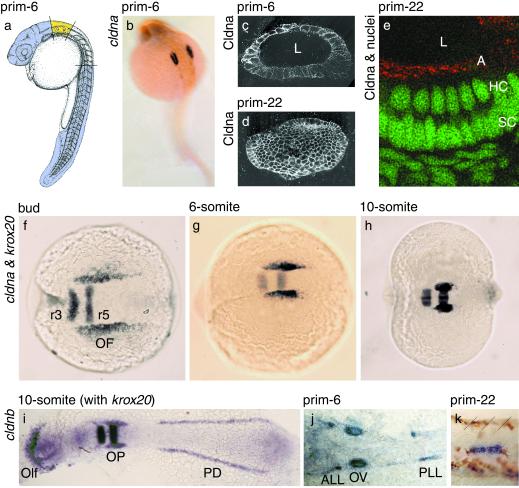
Figure 1.
Zebrafish cldna and cldnb mark the earliest developmental stages of the acoustico-lateralis system. (a) Material dissected for the subtractive cDNA hybridization: ear region in yellow; head and tail regions in mauve. [Reprinted with permission from ref. 48 (Copyright 1995, Wiley-Liss, a subsidiary of John Wiley & Sons, Inc.)]. (b and f–h) Whole-mount in situ hybridizations with antisense probes for cldna, which labels the developing ear, and krox20, which marks hindbrain rhombomeres 3 and 5. (c–e) Immunofluorescence detection of Cldna protein throughout the cell membranes of the otic vesicle after acetone permeabilization (c and d), but only at the apical surface of the sensory epithelium, the location of tight junctions, after detergent extraction (e; Cldna in red, nuclei in green). Each panel shows the combination of several adjacent confocal sections; in d, the vesicle was sectioned tangentially. (i–k) Whole-mount in situ hybridizations with antisense probes for cldnb, which labels placodal structures, including those of the ear and lateral-line organ, and for krox20. (k) Migrating placode of the posterior lateral line, with brown pigment cells and boundaries between somites 19 and 22 marked by slanted lines. Views are lateral (a, c–e, and k), dorsolateral (b), or dorsal (f–j), all with anterior to the left, and of whole embryos (a, b, and f–i) or details (c–e, j, and k); in i and j, the specimens have been flattened. L, lumen of the otic vesicle; A, apical surface of the otic vesicle's epithelium; HC, hair-cell nuclei; SC, supporting-cell nuclei; r3 and r5, hindbrain rhombomeres 3 and 5; OF, otic field; Olf, olfactory placode; OP, otic placode; PD, pronephric duct; ALL, anterior lateral-line placode; OV, otic vesicle; PLL, posterior lateral-line placode.
Differential Filter Hybridization.
A plasmid library was prepared with the cDNAs that remained after the forward subtraction with ear cDNA as tester. The PCR-amplified inserts of 376 clones were spotted onto duplicate filter sets and hybridized with radioactively labeled probes from either the forward or the reverse subtraction. For 32 clones, the hybridization signal was more than one standard deviation above average with the forward-subtracted probe, but within one standard deviation of the average with the reverse-subtracted probe.
In Situ Hybridization.
Whole-mount in situ hybridizations were conducted as described (20–22). For double labeling, a digoxigenin-labeled probe for zebrafish cldna or cldnb mRNA and a fluorescein-labeled probe for krox20 mRNA were detected sequentially with the same blue chromogen. No labeling was observed in control hybridizations with sense probes.
Cloning and Sequencing.
The partial cldna cDNA from the subtractive hybridization was used to screen a bacteriophage library from 24-h-old zebrafish embryos (Stratagene) and to isolate a full-length cDNA, as confirmed by Northern blotting. A partial cDNA for Xenopus laevis cldna was amplified with the degenerate primers 5′-TGGGARGGNHTNTGGATG-3′ and 5′-RYNADNGGRTTRAARTCNYG-3′ and used to isolate a full-length cDNA from an embryonic stage 30 library (Stratagene). Zebrafish and mouse cDNAs identified by blast searches (23) of GenBank were purchased from Incyte Genomics (St. Louis). The zebrafish and frog cDNAs were sequenced completely on both strands.
Nomenclature.
We numbered each nonmammalian claudin gene like the human ortholog with the lowest suffix. If an ortholog could not be identified unambiguously, a character suffix was assigned provisionally. If a subsequent study identifies the human ortholog, a character suffix may be changed to the appropriate number.
Immunofluorescence.
The coding sequence of zebrafish cldna was amplified with the PCR primers 5′-CGGGATCCAATTGTTCCACCATGGTATCA-3′ and 5′-GCAAGCTTGCTCTAGATCAGACATACCCCTTGGTTCC-3′ and inserted between the BamHI and HindIII sites of the vector pET-28a(+) (Novagen). The resulting fusion protein was expressed in Escherichia coli BL21(DE3), purified by affinity chromatography under denaturing conditions on Ni2+-Sepharose (Novagen), and used to immunize rabbits. Antibodies specific for the carboxyl terminus of Cldna were isolated by affinity chromatography (24) with the peptide NH2-CANSPPQDQYKATYTARSGGTKGYV-COOH linked to Sulfolink gel (Pierce). On a Western blot, the affinity-purified antiserum recognized the recombinant full-length Cldna protein, but not a deletion mutant without the carboxyl terminus. After fixation, zebrafish embryos were permeabilized by extraction with acetone at −20°C for 7 min or with 1% (vol/vol) Triton X-100 in PBS at room temperature for 1 h, and labeled as described (ref. 25, protocol 6.12) with a 1:10,000 dilution of the anti-Cldna serum, 3 μg/ml Alexa-488-conjugated goat anti-rabbit antiserum, and 1 μg/ml of the nuclear stain To-Pro-3 (Molecular Probes). Fluorescence was detected by confocal microscopy.
Phylogenetic Analysis.
Protein sequences were aligned with CLUSTALX 1.81 (26), and transmembrane segments were predicted with TMHMM 2.0 (27). The aligned claudin sequences in fasta format are available as supplemental text, which is published on the PNAS web site, www.pnas.org. Pseudoreplicate data sets were generated with seqboot from the PHYLIP 3.573c package (28); columns of the alignment that contain an initial methionine or a gap were disregarded. Maximum-likelihood distances and tree probabilities were calculated with TREE PUZZLE 5.0 (29) by using the wag substitution matrix and a discrete Γ distribution with four bins. PAUP 4.0β4a (30) was used in heuristic searches for optimal tree topologies with minimum evolution as the objective function and to draw a consensus tree.
Locating Introns.
For the human claudin genes, we deduced exon boundaries in the coding regions from the literature (31) or published mRNA and genomic sequences. For mouse Cldn13 and the zebrafish claudins, we used PRIMER3 (www-genome.wi.mit.edu/genome_software/other/primer3.html) to design primer pairs that flanked the intron positions observed in the human genes (Table 2, which is published as supplemental material). To amplify products shorter than 1 kb, we conducted PCRs containing 0.035 units/μl Amplitaq Gold enzyme (Applied Biosystems), 1× Amplitaq Gold buffer, 2.5 mM MgCl2, 0.2 mM each dNTP, 0.8 μM each primer, and 2.5 ng/μl genomic DNA (Research Genetics, Huntsville, AL) with the following protocol: 95°C for 10 min; 30 cycles of 94°C for 30 s, 68°C for 30 s, 72°C for 2 min; and 72°C for 3.5 min. To amplify products longer than 1 kb, we conducted PCRs containing 0.15 μl Expand long-template enzyme mixture (Roche Diagnostics) per 10 μl, 1× Expand buffer 2, 0.5 mM each dNTP, 0.3 μM each primer, and 5 ng/μl genomic DNA with the following protocol: 92°C for 2 min; 10 cycles of 92°C for 10 s, 68°C for 30 s, 68°C for 15 min; 20 cycles of 92°C for 10 s, 68°C for 30 s, 68°C for 15 min plus a 20-s extension per cycle; and 68°C for 7 min. The PCR products were sized by gel electrophoresis and sequenced across the exon-intron boundaries. In all cases, splice donor (GT) and acceptor (AG) sequences bounded the introns.
Physical Mapping.
The chromosomal locations of the human claudin genes were obtained from the locuslink database (32) and blast searches of the human genome sequence. Zebrafish genes were mapped with the T51 radiation-hybrid panel (Research Genetics), the primers shown in Table 2, and Amplitaq Gold under the conditions described above, except for an annealing temperature of 60°C. The scoring data were analyzed by use of the mapping server at the Max-Planck-Institut für Entwicklungsbiologie in Tübingen, Germany (33). Zebrafish orthologs of genes within 2 Mb of human CLDN3 and CLDN4 were found by iterative searches of GenBank. Orthology was assumed if the predicted zebrafish protein in a reciprocal blastp search matched the human protein most closely, with at least 60% pairwise sequence identity, and clustered with the human protein on a phylogenetic tree of similar sequences from any organism.
Results and Discussion
To study the formation of the vertebrate ear from its primordium, the otic placode (34), we conducted a screen for molecular markers in the zebrafish that comprised a cDNA subtraction (35), a differential filter hybridization, and in situ hybridizations at prim-6 stage (Fig. 1a). At this stage, when the first sensory hair cells have formed (36), one of the genes we identified was expressed exclusively in the otic vesicle (Fig. 1b). Because the protein encoded by a full-length cDNA from this gene is most similar to mammalian claudins, we refer to the zebrafish gene as claudin a (cldna; see Materials and Methods for nomenclature). Consistent with the presence of the protein Cldna in tight junctions, an antiserum against its unique carboxyl terminus detected the protein after detergent extraction solely at the apical surface of the otic vesicle's epithelium (Fig. 1 c–e). Expression in the ear has previously been described only for mouse Cldn14 and only after birth (9). Immature tight junctions, however, have been observed in the chicken's ear as early as embryonic stage 15, before the otic cup closes (37).
Because the mammalian claudin family comprises at least 20 members (6), we speculated that zebrafish also might harbor multiple claudins and that more than one gene might be expressed in the developing ear. blast searches of GenBank with all known human claudin sequences revealed 14 additional zebrafish genes that encode claudin-like proteins; reciprocal database searches with the zebrafish protein sequences always identified a mammalian claudin as the closest match, with at least 43% sequence identity (Fig. 5, which is published as supplemental material). Although our set of 15 zebrafish claudins may be incomplete, it is comparable in size to the mammalian gene family. In whole-mount in situ hybridizations with probes for the 14 additional zebrafish claudins, we found that one of them, cldnb, also is expressed in the ear at prim-6 stage (Fig. 1j).
During the first 36 h of development, the expression patterns of cldna and cldnb are similar, but not identical. We first detected the mRNAs of both genes at the end of gastrulation (Fig. 1f and data not shown), shortly after the time of otic induction (38). Their initial expression domains, diffuse stripes along both sides of the hindbrain primordium, coincide with the presumptive otic fields. At the 10-somite stage, concomitant with the invagination of the otic placode, these domains have shrunk and sharpened (Fig. 1 g and h). Unlike cldna, which is expressed exclusively in the ear at least until prim-22 stage, cldnb is expressed also in the olfactory placode and the pronephric duct (Fig. 1i). In addition, we detected cldnb mRNA after prim-6 stage in patches rostral and caudal to the otic vesicles (Fig. 1j) that represent placodes of the anterior and posterior lateral lines (39). At later stages, the caudal boundary of the posterior patch coincides with that of the migrating primordium of the posterior lateral line (Fig. 1k). Zebrafish cldna and cldnb thus mark the initial stages of the acoustico-lateralis system; cldna is one of the earliest molecular markers exclusive to the ear (34).
Sensory placodes and the pronephros, the domains of cldna and cldnb expression, are characters of vertebrate embryos that arose along the chordate stem lineage (13). Tight junctions, the cellular structures in which claudins occur, are likewise characteristic of chordates (1). Suspecting a link between the evolution of vertebrates and claudins, we therefore investigated the phylogeny of the claudin gene family, with an emphasis on zebrafish cldna and cldnb.
First, we asked whether any nonvertebrate claudins were known. Outside the Phylum Chordata we and others (40) found no sequences in GenBank with significant similarity to claudins, even in the sequenced genomes of Drosophila melanogaster and Caenorhabditis elegans. The closest match we found, with a blastp expectation value of 0.0002, was the hypothetical protein F53B3.5 from C. elegans, which lacks several of the characteristic residues of claudin proteins (Fig. 5) and is equally similar to other members of the tetraspanin superfamily (5). Among nonvertebrate chordates, we found a single claudin gene from the urochordate Halocynthia roretzi (Fig. 5). These findings are consistent with the presence of claudins only in chordates.
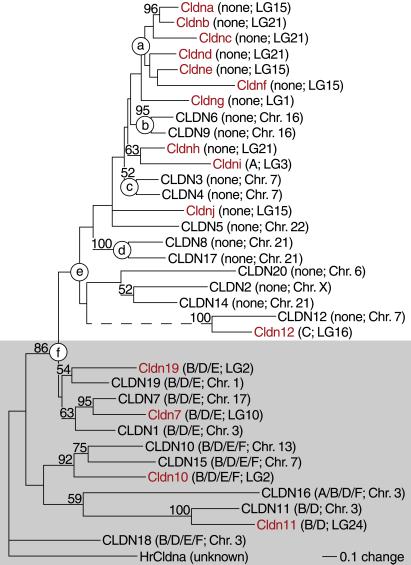
We next asked when the claudin gene family had diversified. For a representative set of 36 claudins, we analyzed the aligned protein sequences, intron positions, and chromosomal locations. From the sequence alignment (Fig. 5), we reconstructed a 50%-majority consensus tree with compatible groupings by using maximum-likelihood distances and a heuristic tree-search algorithm (41). The resulting phylogram (Fig. 2) is supported by three independent lines of evidence. First, we obtained essentially the same topology after we replaced the human sequences with their mouse orthologs (data not shown). Next, all claudin genes with multiple introns in the coding region cluster together (Table 1 and Fig. 2, gray shading). The remaining genes lack introns in the coding region save for zebrafish cldni and cldn12, each of which contains a single intron in an unusual position (Table 1). The bipartite configuration of the phylogram strongly suggests a concerted loss of introns by retrotransposition between nodes e and f. Finally, sets of genes whose chromosomal locations coincide, presumably because of recent tandem gene duplications, also cluster on the tree (Fig. 2, nodes a to d). Because the zebrafish and human claudins are mostly interspersed in this phylogram, we conclude that by the time ray-finned fishes diverged from the tetrapod lineage, vertebrates already possessed an abundance of claudin genes.
Figure 2.
Zebrafish paralogs cldna and cldnb are orthologs of mammalian claudins 3–6 and 9. This 50% majority consensus tree with compatible groupings was reconstructed from the alignment in Fig. 5 by using maximum-likelihood distances and a heuristic tree-search algorithm (41) and rooted for display purposes with HrCldna from Halocynthia roretzi as the outgroup. Numbers indicate the percentage of 1,000 bootstrap replicates that support the adjacent node. Zebrafish proteins are shown in red, human and urochordate in black; introns in the coding region (Table 1) and the linkage group or chromosome of the corresponding gene (Table 2) are listed in parentheses. Claudins whose genes contain multiple introns in their coding regions are shaded in gray. CLDN12 and Cldn12 appear to be tetraspanins, but only distant relatives of the other claudins; their common branch (dashed line) was shortened to two-fifths to conserve space. Mouse Cldn13 was omitted from this tree because its placement, either with CLDN3 and CLDN4 or with CLDN12 and Cldn12, was the least consistent among bootstrap replicates and its omission markedly increased the tree's likelihood.
Table 1.
Conservation of intron positions in the coding regions of human and zebrafish claudins
| Intron* | Gene† | Exon boundaries‡ |
|---|---|---|
| A | CLDN16 | Leu-Glu108 —Val109-Ser |
| cldni | Ala-Gln44 —Val45-Ser | |
| B | CLDN1 | Leu-Ser-S —er75-Thr-Leu |
| CLDN7 | Leu-Ser-A —la75-Ala-Leu | |
| cldn7 | Leu-Asp-S —er75-Ala-Leu | |
| CLDN10 | Leu-Asp-G —ly74-Tyr-Ile | |
| cldn10 | Leu-Pro-V —al73-His-Ile | |
| CLDN11 | Leu-Pro-G —ly76-Tyr-Val | |
| cldn11 | Ile-Pro-A —la76-Tyr-Ile | |
| CLDN15 | Leu-Ser-G —ly74-Tyr-Ile | |
| CLDN16 | His-Pro-L —eu143-Lys-Leu | |
| CLDN18 | Leu-Pro-A —la74-Met-Leu | |
| CLDN19 | Leu-Asp-G —ly75-His-Ile | |
| cldn19 | Leu-Pro-G —ly75-Glu-Ile | |
| C | cldn12 | Pro-Thr-G —ly93-Leu-Leu |
| D | CLDN1 | Leu-Ala-G —ly130-Leu-Ala |
| CLDN7 | Val-Ala-G —ly130-Leu-Ala | |
| cldn7 | Val-Gly-A —la130-Leu-Cys | |
| CLDN10 | Leu-Ser-G —ly128-Leu-Cys | |
| cldn10 | Ile-Gly-G —ly128-Leu-Cys | |
| CLDN11 | Leu-Leu-A —la131-Leu-Cys | |
| cldn11 | Val-Ile-S —er131-Leu-Cys | |
| CLDN15 | Leu-Ala-G —ly128-Ile-Cys | |
| CLDN16 | Ile-Ala-G —ly198-Thr-Pro | |
| CLDN18 | Val-Ser-G —ly129-Leu-Cys | |
| CLDN19 | Leu-Ala-G —ly130-Leu-Cys | |
| cldn19 | Thr-Gly-G —ly130-Leu-Phe | |
| E | CLDN1 | Asn-Ala-Ar —g158-Tyr-Glu |
| CLDN7 | Asn-Ile-Ly —s158-Tyr-Glu | |
| cldn7 | Asn-Thr-Ly —s158-Tyr-Glu | |
| CLDN10 | Glu-Gln-Ly —s155-Tyr-Glu | |
| cldn10 | Gly-Val-Ar —g155-Phe-Glu | |
| CLDN15 | Gly-Thr-Ly —s155-Tyr-Glu | |
| CLDN18 | Gln-Thr-Ar —g168-Tyr-Thr | |
| CLDN19 | Asn-Ala-Ar —g158-Tyr-Glu | |
| cldn19 | Asn-Ala-Ar —g158-Tyr-Glu | |
| F | CLDN10 | Thr-Pro-Ar —g191-Tyr-Thr |
| cldn10 | Glu-Lys-Gl —y193-Ala-Tyr | |
| CLDN15 | Ala-Ala-Se —r194-Ala-Arg | |
| CLDN16 | Phe-Lys-A —sp262-Val-Gly | |
| CLDN18 | Glu-Thr-As —n205-Tyr-Lys |
See Fig. 5 for the location of the introns in the sequence alignment.
Symbols capitalized for human and lowercase for zebrafish.
Numbers denote the position of the amino acids whose codons border or straddle the intron (—).
We then examined the homologs of cldna and cldnb in the zebrafish and in mammals. The phylogram provides strong bootstrap support for cldna and cldnb being closest paralogs (Fig. 2) that arose from a recent duplication of a gene cluster now located on both zebrafish linkage groups LG15 and LG21 (Fig. 3a). The overlapping but distinct expression patterns of these two genes may reflect degenerative mutations of tissue-specific regulatory elements (17). The phylogram also suggests that mammalian claudins 6 and 9, 3 and 4, and 5 are orthologs of zebrafish cldna and cldnb. In support of this proposition, we detected conserved syntenies between zebrafish LG15 and LG21, where cldna and cldnb occur, and human chromosome 7, where CLDN3 and CLDN4 reside (Fig. 3a). These findings indicate that the claudin family continued to grow through gene or genome duplications after the divergence of ray-finned fishes and tetrapods.
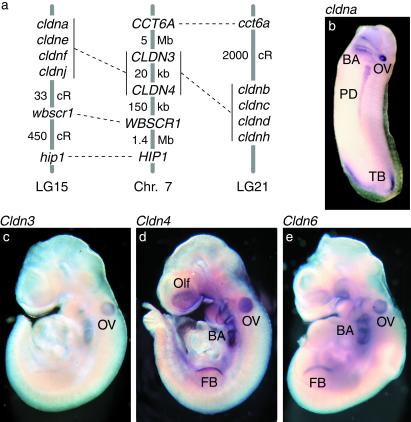
Figure 3.
Orthologs of zebrafish cldna and cldnb are expressed in vertebrate primordia. (a) Conserved syntenies between zebrafish linkage groups 15 and 21, which harbor paralogous clusters of claudin genes that include cldna and cldnb, and human chromosome 7, which contains CLDN3 and CLDN4. We mapped putative zebrafish orthologs of six genes from the vicinity of CLDN3 and CLDN4 (45); two of them, hip1 and wbscr1, occur in LG15 near cldna (Table 2). In addition, zebrafish LG21 harbors the ortholog of the human CCT6A gene on 7p11 (46). (b) In situ hybridization of a stage 30 X. laevis embryo with an antisense probe for frog cldna. (c–e) In situ hybridizations of mouse embryos between embryonic days 9.0 and 9.5 with antisense probes for Cldn3, Cldn4, and Cldn6. All views are lateral. BA, branchial arches; OV, otic vesicle; PD, pronephric duct; TB, tail bud; Olf, olfactory placode; FB, forelimb bud.
Finally, we asked whether the orthologs of zebrafish cldna and cldnb exhibit similar expression patterns early in organogenesis. In whole-mount in situ hybridizations with mice between embryonic days 9.0 and 9.5, we detected Cldn3, Cldn4, and Cldn6 mRNAs in the otic vesicle as well as in the olfactory placode, branchial arches, and forelimb bud (Fig. 3 c–e). At a comparable stage of development in Xenopus we detected expression of its cldna gene, which is most similar to mammalian claudins 4 and 6, in the otic vesicle, branchial arches, pronephric duct, and tailbud (Fig. 3b; see also ref. 42). These results demonstrate that claudin genes closely related to zebrafish cldna and cldnb are expressed specifically in primordia characteristic of the vertebrate embryo.
We conclude that claudins emerged around the same time as chordates (Fig. 4), when tight junctions replaced septate junctions (1). Like many other gene families (16), the claudins then expanded as the vertebrate body plan evolved from the chordate stem configuration through the addition of novel structures such as the cranium, paired sensory organs, and paired appendages (13, 14). A more precise specification of this model will require exhaustive screens for claudin genes in the nongnathostome clades shown in Fig. 4. What were the claudins' precursors in nonchordates and were they associated with septate junctions? How many claudin genes did the common chordate ancestor harbor? Did the expansion of the claudin gene family result from whole-genome duplications in the vertebrate lineage? The strong and specific expression of claudins in vertebrate primordia reported here and the randomization of the embryo's left-right axis upon overexpression of frog cldna (42) suggest that tight junctions play an active role in morphogenesis. Such a role may explain some of the morphological and neurological abnormalities of individuals with Williams–Beuren syndrome, in which the CLDN3 and CLDN4 genes are excised as part of a large deletion (43). The diversification of the claudin gene family may have facilitated the formation of tight junctions during organogenesis at different times and at different sites. Alternatively, the various claudins may mediate selective cell adhesion (44) in vertebrate primordia.
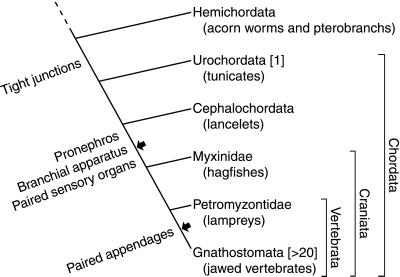
Figure 4.
Concurrent evolution of the claudin family and the vertebrate body plan. The number of distinct claudin genes found to date in each clade is given in brackets. The appearance of those vertebrate characters that we found to be associated with claudin expression is indicated beside the stem lineage (13). Arrows indicate two presumptive genome duplications (16); a third genome duplication may have taken place in ray-finned fishes alone (15, 47).
Supplementary Material
Acknowledgments
We thank Dr. Robert Geisler for the analysis of radiation-hybrid mapping data, Dr. Ali Hemmati-Brivanlou for access to his Xenopus facility, and Dr. Kathleen Millen for suggestions about mouse in situ hybridizations. Ms. Jennifer Nuñez, Ms. Carrie Rosenberger, and Mr. Jak Fak contributed at the beginning of this project. Mr. Adam Kuhl, Ms. Andrea Seymour, Mr. David Chosid, and Ms. Jennifer Voisine maintained the zebrafish colony. Dr. Hemmati-Brivanlou, Dr. Axel Meyer and members of our laboratory group provided comments on the manuscript. This research was supported by a grant from the American Hearing Research Foundation to R.K. and by National Institutes of Health Grant DC00317 to A.J.H., who is an Investigator of the Howard Hughes Medical Institute.
Note Added in Proof.
An additional zebrafish claudin, Cldn8, is encoded by expressed sequence tags fl13b04 and fl24f07. The cldn8 gene contains no introns within the coding region and resides in LG15.
Footnotes
References
- 1.Green C R, Bergquist P R. J Cell Sci. 1982;53:279–305. [Google Scholar]
- 2.Tsukita S, Furuse M, Itoh M. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 3.Lane N J. In: Tight Junctions. Cereijido M, editor. Boca Raton, FL: CRC; 1992. pp. 23–48. [Google Scholar]
- 4.Bellen H J, Lu Y, Beckstead R, Bhat M A. Trends Neurosci. 1998;21:444–449. doi: 10.1016/s0166-2236(98)01267-3. [DOI] [PubMed] [Google Scholar]
- 5.Bronstein J M. Curr Opin Neurobiol. 2000;10:552–557. doi: 10.1016/s0959-4388(00)00125-2. [DOI] [PubMed] [Google Scholar]
- 6.Tsukita S, Furuse M. Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 7.Gow A, Southwood C M, Li J S, Pariali M, Riordan G P, Brodie S E, Danias J, Bronstein J M, Kachar B, Lazzarini R A. Cell. 1999;99:649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- 8.Simon D B, Lu Y, Choate K A, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, et al. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 9.Wilcox E R, Burton Q L, Naz S, Riazuddin S, Smith T N, Ploplis B, Belyantseva I, Ben-Yosef T, Liburd N A, Morell R J, et al. Cell. 2001;104:165–172. doi: 10.1016/s0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 10.Furuse M, Sasaki H, Fujimoto K, Tsukita S. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirschner M, Gerhart J. Proc Natl Acad Sci USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll S B. Nature (London) 2001;409:1102–1109. doi: 10.1038/35059227. [DOI] [PubMed] [Google Scholar]
- 13.Maisey J G. Cladistics. 1986;2:201–256. doi: 10.1111/j.1096-0031.1986.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 14.Shimeld S M, Holland P W. Proc Natl Acad Sci USA. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer A, Schartl M. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 16.Holland P W H. Semin Cell Dev Biol. 1999;10:541–547. doi: 10.1006/scdb.1999.0335. [DOI] [PubMed] [Google Scholar]
- 17.Force A, Lynch M, Pickett F B, Amores A, Yan Y L, Postlethwait J. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richardson M K. Dev Biol. 1995;172:412–421. doi: 10.1006/dbio.1995.8041. [DOI] [PubMed] [Google Scholar]
- 19.Fleming T P, Papenbrock T, Fesenko I, Hausen P, Sheth B. Semin Cell Dev Biol. 2000;11:291–299. doi: 10.1006/scdb.2000.0179. [DOI] [PubMed] [Google Scholar]
- 20.Harland R M. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 21.Parr B A, Shea M J, Vassileva G, McMahon A P. Development (Cambridge, UK) 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- 22.Thisse C, Thisse B. In: The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Westerfield M, editor. Eugene: Univ. of Oregon Press; 1998. p. 9.82. [Google Scholar]
- 23.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. [Google Scholar]
- 25.Jowett T. Methods Cell Biol. 1999;59:63–85. doi: 10.1016/s0091-679x(08)61821-x. [DOI] [PubMed] [Google Scholar]
- 26.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer E L L. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 29.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 30.Swofford D L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 31.Paperna T, Peoples R, Wang Y-K, Kaplan P, Francke U. Genomics. 1998;54:453–459. doi: 10.1006/geno.1998.5619. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler D L, Church D M, Lash A E, Leipe D D, Madden T L, Pontius J U, Schuler G D, Schriml L M, Tatusova T A, Wagner L, et al. Nucleic Acids Res. 2001;29:11–16. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geisler R, Rauch G J, Baier H, van Bebber F, Brobeta L, Dekens M P, Finger K, Fricke C, Gates M A, Geiger H, et al. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- 34.Torres M, Giraldez F. Mech Dev. 1998;71:5–21. doi: 10.1016/s0925-4773(97)00155-x. [DOI] [PubMed] [Google Scholar]
- 35.Diatchenko L, Lau Y-F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, et al. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haddon C, Lewis J. J Comp Neurol. 1996;365:113–128. doi: 10.1002/(SICI)1096-9861(19960129)365:1<113::AID-CNE9>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Ginzberg R D, Gilula N B. Dev Biol. 1979;68:110–129. doi: 10.1016/0012-1606(79)90247-1. [DOI] [PubMed] [Google Scholar]
- 38.Mendonsa E S, Riley B B. Dev Biol. 1999;206:100–112. doi: 10.1006/dbio.1998.9134. [DOI] [PubMed] [Google Scholar]
- 39.Raible D W, Kruse G J. J Comp Neurol. 2000;421:189–198. [PubMed] [Google Scholar]
- 40.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 41.Swofford D L, Olsen G J, Waddell P J, Hillis D M. In: Molecular Systematics. Hillis D M, Moritz C, Mable B K, editors. Sunderland, MA: Sinauer; 1996. pp. 407–514. [Google Scholar]
- 42.Brizuela B J, Wessely O, De Robertis E M. Dev Biol. 2001;230:217–229. doi: 10.1006/dbio.2000.0116. [DOI] [PubMed] [Google Scholar]
- 43.Morris C A, Mervis C B. Annu Rev Genomics Hum Genet. 2000;1:461–484. doi: 10.1146/annurev.genom.1.1.461. [DOI] [PubMed] [Google Scholar]
- 44.Furuse M, Sasaki H, Tsukita S. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valero M C, de Luis O, Cruces J, Perez Júrado L A. Genomics. 2000;69:1–13. doi: 10.1006/geno.2000.6312. [DOI] [PubMed] [Google Scholar]
- 46.Barbazuk W B, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell J A, McPherson J D, Johnson S L. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wittbrodt J, Meyer A, Schartl M. BioEssays. 1998;20:511–515. [Google Scholar]
- 48.Kimmel C B, Ballard W W, Kimmel S R, Ullmann B, Schilling T F. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.