Abstract
This review assessed evidence of disparities in benefits of pharmacogenomics related to ‘model performance’ in subgroups of patients and studies which reported impact on health inequalities. ‘Model performance’ refers to the ability of algorithms including clinical, environmental and genetic information to guide treatment. A total of 4978 abstracts were screened by one reviewer and 30% (1494) were double screened by a second independent reviewer, after which data extraction was performed. Additional forward and backward citation searching of reference lists was conducted. Investigators independently double rated study quality and applicability of included studies. Only five individual studies were identified which met our inclusion criteria, but were contradictory in their conclusions. While three studies of genotype-guided dosing of warfarin reported that ethnic disparities in healthcare may widen, two other studies (one reporting on warfarin and reporting on clopidogrel) suggested that disparities in healthcare may reduce. There is a paucity of studies which evaluates the impact of pharmacogenomics on health disparities. Further work is required not only to evaluate health disparities between ethnic groups and countries but also within ethnic groups in the same country and solutions need to be identified to overcome these disparities.
Keywords: : disparities, efficacy, equity, genetics, pharmacogenomics
Minimizing modifiable health disparities is deemed fundamental for equitable and progressive achievement of universal health coverage [1]. Equity refers to social justice or fairness; it is an ethical concept which is based on the principles of distributive justice [2]. Equity in health can largely be defined as the absence of systematic disparities in health between social groups who have different levels of underlying social advantage, or different positions in social hierarchy [2]. Equity in healthcare relates service provision to need and therefore equity which focuses resources on those in greatest need is inherently just. Inequities in health systematically put people who may already be socially disadvantaged at further disadvantage in terms of health and thus the term ‘inequity’ has a moral and ethical dimension [2]. Inequity refers to differences which are unnecessary and avoidable but, in addition these differences may also be considered unfair and unjust [1,2].
Precision medicine (PM) has the potential to revolutionize the health sector through improving the effectiveness of treatments while simultaneously reducing adverse effects and avoiding inappropriate treatment [3]. PM tailors care for each individual patient based on clinical, environmental and genetic information [3]. In PM, the use of biomarkers (which to date have been mainly molecular) can be used for the purpose of risk assessment, diagnosis, prognosis, monitoring and guiding therapeutic decisions [3]. Perhaps the most advanced area of PM is pharmacogenomics, which aims to relate genetic differences in the way drugs are handled (pharmacokinetic determinants) and how they act (pharmacodynamics determinants) to the differences in drug efficacy and safety observed between different individuals [4].
Concerns have been raised by the European Personalized Medicine Association (EPEMED) and National Institute on Minority Health and Disparities (NIMHD) among others that the rate at which PM is implemented may vary. In particular new therapies or tests may be taken up less quickly and in lower numbers in more deprived populations due to challenges in access, availability and ability to pay privately and understanding of healthcare information compared with more educated and wealthier populations [3,5]. In order to address this, as part of the US Precision Medicine Initiative, the ‘All of Us’ research program aims to reflect the diversity of the US population from varied social, racial/ethnic and ancestral populations living in a variety of geographies, social environments, economic circumstances, age groups and health statuses [5].
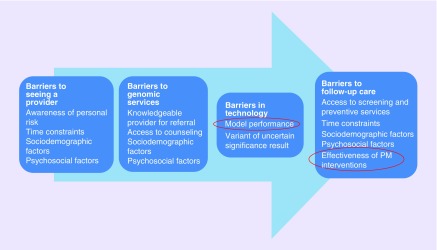
Barriers to seeing a provider may occur due to differential awareness of personal risk, inability to attend appointments due to time constraints, sociodemographic factors and psychosocial factors [6]. Sociodemographic factors include education level, socioeconomic status, marital status, age, sex, ethnicity and parenthood. Psychosocial factors include interest in PM, disease-specific stress, perceived risk, knowledge, perceived benefits, perceived limitations, anxiety, depression and general distress. Additional barriers include accessibility to providers with appropriate expertise and available resources [6].
The remit of this review was to systematically review published evidence which considers whether PM interventions either widen or reduce health disparities [7]. The review followed PRISMA-E guidelines and the format of the review is described in Figure 1. Given that PM as an overall area is relatively broad, we have focused the scope of the review on pharmacogenomics. Specifically, our research question examines barriers in effectiveness of pharmacogenomics related to ‘model performance’ in subgroups of patients. ‘Model performance’ refers to the ability of algorithms including clinical, environmental and genetic information to guide treatment. Our review also aims to identify studies which assessed the ‘effectiveness of pharmacogenomic interventions’ in subgroups of patients and reported impact on disparities in healthcare. These two factors are highlighted (circled) in Figure 2.
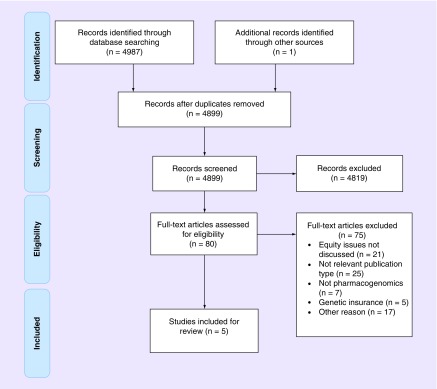
Figure 1. . PRISMA flow diagram displaying articles included and excluded in this review.
Figure 2. . Barriers to the delivery of pharmacogenomic interventions.
Methods
The systematic review protocol was registered with PROSPERO, the international database of prospectively registered systematic reviews (identification number CRD42016032517), conducted according to the Centre for Reviews and Dissemination's guidance for undertaking reviews in healthcare and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis with a focus on health equity (PRISMA-E) guidelines [8,9].
The scope of the research question was informed by the Campbell and Cochrane Equity Methods Group and the Cochrane Public Health Group who recommend the PROGRESS-Plus approach to analyses of health inequality information. PROGRESS-Plus is an acronym for place of residence, race/ethnicity, culture/language, occupation, gender/sex, religion, socioeconomic status and social capital and ‘Plus’ captures other characteristics which may indicate a disadvantage, such as age and disability [7]. This approach summarizes a number of social stratification factors understood to influence health opportunities, including the chance to participate and benefit from PM (see Supplementary Table 1 for definition of PROGRESS-Plus factors).
Data sources & searches
The website of PharmGKB was searched to identify a total of 201 drugs that had labels containing pharmacogenetic information with differing approvals between European Medicines Agency (EMA); US FDA; Pharmaceuticals and Medical Device Agency, Japan (PMDA); and Health Canada, Santé Canada (HCSC). The types of regulatory recommendations provided were split into informative pharmacogenetic test, actionable pharmacogenetic test, genetic testing recommended and genetic testing required.
We searched the following databases (via Ovid) between January 2006 and January 2016: Embase, MEDLINE, PubMed, The Cochrane Library and Cochrane Central Register of Controlled Trials (CENTRAL) and Web of Science. The search terms consisted of two clauses combined with the Boolean ‘AND’ operator. These pertained to a list of drugs identified from the PharmGKB website and health disparity terms. The search was restricted to studies of human subjects and written in the English language. Further articles were identified from searching the grey literature and backward citation searching of the reference lists of included studies and from forward citation searching using google scholar. The full search strategy is detailed in Supplementary Table 2.
Study selection
Studies published over the past 10 years were reviewed to assess the most relevant evidence. Randomized controlled trials (RCTs), prospective and retrospective cohort studies, case–control studies and cross-sectional studies were included if they considered the impact of pharmacogenomic interventions on health disparities. It is recommended that a wider array of evidence is included (such as non-randomized studies) in systematic reviews which have a focus on health equity compared with other types of systematic reviews due to varied reporting of equity evidence [7]. However, we excluded editorials, letters, historical articles, reviews and abstracts.
We included English language studies only because no evidence of a systematic bias exists from the use of English language restrictions in systematic review-based meta-analyses in conventional medicine [11]. We included studies on persons aged 16 years and above that measured outcomes directly and indirectly (e.g., self-reported). Studies that focused on genotyping to guide efficacy and discussed health equity issues were included. Studies were included if terms relating to impact on health disparities were stated in the title, abstract or keywords of the publication. Studies which discussed equity issues related to uptake rates associated with sociodemographic and psychosocial factors were outside the scope of this review (further information about our inclusion criteria is provided in Supplementary Table 3).
Screening, data extraction & quality assessment
Titles and abstracts were screened by one reviewer (AM) and 30% were double screened by a second independent reviewer (JD). Full texts were retrieved where reviewers were in agreement that the article met the inclusion criteria (99.4% agreement between reviewers of the 30% studies double screened) and consensus was reached by discussion between the two reviewers. After determining article inclusion, one reviewer entered study data into evidence tables (AM); a second senior reviewer checked the information for accuracy and completeness (JD).
Data were extracted on the following study characteristics: year of publication, setting, study type, sample population, genetic test and health equity comment. We classified the included studies based on PROGRESS-Plus items (see Supplementary Table 2 online for PROGRESS-Plus definitions of measures) [21].
AM graded the strength of the evidence using the Grading of Recommendations, Assessment, Development and Evaluations (GRADE), quality assessment method (see Supplementary Table 4). There were four categories of quality ratings in GRADE – ‘high’, ‘moderate’, ‘low’ and ‘very low’. Briefly, the default quality rating was ‘high’ for evidence from randomized controlled (including cluster) trials and ‘low’ for evidence from observational studies. Evidence for each study was examined for risks of bias, inconsistency, indirectness, imprecision and publication bias. Quality may be rated down if there is evidence of any of these five factors; for example, an observational study where risk of bias is judged to be serious would be rated down by one point from ‘low’ to ‘very low’. Alternatively, quality may be modified upward if there is a large magnitude of effect or if any plausible confounders were likely to minimize the observed effect. The quality of reporting of individual studies was expanded further in the narrative and additional information on how ratings were determined for each study was provided [12].
Data synthesis & analysis
Results are presented as summaries of individual studies and reported in the context of the impact of pharmacogenomics on health disparities. An overview of study quality using the GRADE method and equity considerations is provided. Evidence of equity impact was classified as:
Pro-equity (symbol: +) which refers to reduction in health disparities compared with the general population;
Anti-equity (symbol: -) which refers to an increase in health disparities, and;
Mixed equity (symbol: ?) which refers to some improvement in an outcome for a vulnerable group but health disparities still persist and increase in other areas.
Results
Study selection & characteristics
A total of 4978 papers were identified by the search of electronic databases. We retrieved 80 full-text articles, of which four met the inclusion criteria for the review and one additional article was identified from a hand search of reference lists and therefore five studies were included in the review [13–17]. Reasons for exclusion are presented in Figure 1. The heterogeneity in outcomes, methodologies and settings precluded meta-analysis.
The characteristics of the included studies are presented in Table 1. Of studies that were identified: four studies were based on evaluating genotype-guided dosing of warfarin to provide predictive information on anticoagulation response [13–16], while the fifth focused on the use of genotyping to guide treatment of the antiplatelet drug clopidogrel [17]. No other drug-gene studies met our inclusion criteria.
Table 1. . Summary of included studies .
| n | Test type | Average age | Age range or SD | Women (%) | Description of sample | Country | Equity comment | GRADE | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|
| T = 362; C = 194; AA = 168 | CYP2C9 (*2 and *3) and VKORC (11173 C/T) genotype | 58.7 | 45–72 | 30.9 | A prospective cohort of patients (of various indications) with a target international normalized ratio of between 2.0 and 3.0 were genotyped | USA | Uncertain usefulness of variants in African–Americans to provide predictive information in anticoagulation response | Ethnicity - | 3 | [13] |
| T = 575; EA = 302; AA = 273 | CYP2C9 (*2, *3, *5, *6 and *11) and VKORC1 (1173C/T, 3730G/A, 2255C/T, 1542G/C) genotype | 61.1 | SD +14.7–16.0 | 48.7 | A prospective cohort of patients (of various indications) with a target international normalized ratio of between 2.0 and 3.0 were genotyped | USA | Inconsistent ability to provide predictive information in anticoagulation response | Ethnicity - | 3 | [14] |
| T = 366; M = 49; PI = 21; CH = 9 | CYP2C9 (*2 and *3) and VKORC1 (1639G/A) genotype | 68.6 | SD +9–12 | – | A prospective cross-sectional simulation study of patients with severe coronary disease | New Zealand | Study guides warfarin maintenance dose and therefore may reduce ethnic disparities in treatment outcomes | Ethnicity + | 2 | [15] |
| T = 3672; W = 2543; B = 639; J = 227; CH = 263 | CYP2C9 (*2 and *3) and VKORC1 (1639AA) genotype | – | – | – | Patients included in IWPC dataset from 22 study sites with a target international normalized ratio of between 2.0 and 3.0 | Singapore | Warfarin PGx testing reduces inaccurate dosing in white patients but black, Japanese and Chinese do not benefit | Ethnicity - | 3 | [16] |
| T = 13608; E=-; M=-; ETA=-; PI=- | CYP2C19 (*2) genotype | – | 45–80 | – | Cost–effectiveness analysis using international multicenter RCT data of genetic testing for CYP2C19 variants to guide thienopyridine treatment patients with ACS | New Zealand | Treatment strategy has potential to reduce ethnic health disparities | Ethnicity + | 2 | [17] |
AA: African–American; ACS: Acute coronary syndrome; B: Black; C: Caucasian; CH: Chinese; E: European; EA: European–American; ETA: East Asian; IWPC: International warfarin pharmacogenetics consortium; J: Japanese; M: Mãori; PGx: Pharmacogenetic test; PI: Pacific Islander; SD: Standard deviation; T: Total; W: White.
Our final included studies comprised of two from the USA [13,14], two studies from New Zealand [15,17] and one study from Singapore [16]. Of the included studies, four were prospective cohort studies [13–16] and one study by Panattoni et al. [17] was a cost–effectiveness analysis. The quality of reporting is shown in Table 1 according to the GRADE criteria. Initially, four of the studies; 13–16 were rated up due to consistent identification of association between CYP2C9 enzyme and VKORC1 gene and warfarin dose requirements to achieve anticoagulation control. While, the fifth study [17] was rated up due to consistent identification of association between CYP2C19 enzyme and metabolism rates of clopidogrel. There was limited risk of bias due to study design, importance of recruitment setting, presence of self-reported information and response rate. However, of the five studies, two studies [15,17] were subsequently rated down due to use of prediction modelling simulation and possible indirectness issues. Both indirectness (e.g., comparators used) and imprecision (e.g., due to inaccuracies in measurement) were not substantial concerns for other included studies. Overall, three studies were deemed to be of ‘moderate quality’ [13,14,16] and the remaining two studies were deemed of ‘low quality’ [15,17].
Of five studies identified, three studies of genotype-guided dosing of warfarin reported that ethnic disparities in healthcare may widen [13,14,16]. In contrast, one study of genotype-guided dosing of warfarin and one study of genotype-guided antiplatelet therapy reported that ethnic disparities in healthcare may reduce [15,17]. In all five studies, the impact on disparities in healthcare was discussed but none of the studies quantified the resulting predicted increase or reduction in disparities in healthcare.
Predicted impact of genotype-guided dosing of warfarin & health disparities
A study by Gladding et al. [15] reported that genotype-guided dosing of warfarin could theoretically reduce ethnic health disparities as personalizing treatment using pharmacogenetics enables improved treatment by addressing underlying genetic differences between individuals. The study tested the frequency of known important variants within the VKORC1 (1639G/A) and CYP2C9 (*2 and *3) genes in a population of cardiac patients and then performed a simulation, based on genetic and personal factors, to estimate the mean dose of warfarin for different South Pacific ethnic groups [15]. The authors report that while genetic variability within an ethnic group can be greater than between ethnic groups, no data existed for dose requirements in populations of Maori or Pacific Islander in New Zealand, nor have these groups been studied extensively in terms of pharmacogenetics. Gladding and colleagues highlight the importance of trial participation and long-term data collection for potentially underserved groups [15]. Moreover, lack of trial participation and data collection can lead to mistreatment for understudied groups and therefore the potential treatment benefits may not be fully generalizable [18].
In studies by Kealey et al. [13], Limdi et al. [14] and Chan et al. [16] the authors highlighted inconsistencies in the ability to provide predictive information for anticoagulation response with genotype-guided dosing of warfarin for certain ethnic groups compared with Caucasians. Moreover, these studies reported that health disparities widen due to poor characterization of genetic variants for certain group of patients. Kealey et al. reported that at the time, no studies had been completed to evaluate whether genetic variation in CYP2C9 was useful in predicting warfarin response in African–Americans [13]. A later study by Limdi et al. [14] found that CYP2C9 and VKORC1 accounted for up to 30% of the variability in warfarin dose among European–Americans and 10% among African–Americans. Limdi and colleagues concluded that although CYP2C9 and VKORC1 genotyping has the potential to facilitate the development of individually tailored warfarin dose in both African–Americans and European–Americans, the ability to predict overanticoagulation risk was limited to European–Americans [14]. A further study by Chan et al. [16] also found that there are different dose requirements between different races and there was considerable overlap in dose distributions depending on genotype combinations. Chan and colleagues revealed that genotype-guided dosing of warfarin can reduce inaccurate dosing by 18–24% in white individuals, whereas black, Japanese and Chinese individuals were not found to benefit from genotype-guided dosing of warfarin over standard dosing algorithms [16].
Genotype-guided treatment compared with treatment of clopidogrel or prasugrel alone & health disparities
A clinical trial found that patients with acute coronary syndromes and reduced function allele CYP2C19*2 (*2 allele) who are treated with thienopyridines (antiplatelet medications), have an increased risk of adverse cardiac events with clopidogrel, but not prasugrel because prasugrel activation is not predominantly dependent on oxidation by the enzyme CYP2C19 [17]. A study by Panattoni et al. [17] found that genotype-guided treatment compared with standard treatment of clopidogrel is a potentially cost-effective strategy for the entire New Zealand population and in particular for Maoris and Pacific Islander patients. It was reported that Maori and Pacific Islander ethnicities have a relatively high incidence of CYP2C19*2 allele and therefore poor metabolizers of clopidogrel were more commonly identified. Increased cost–effectiveness was found in Maori and Pacific Islander ethnicities due to enhanced efficacy [19]. Therefore, the authors concluded that the introduction of genotype-guided clopidogrel dosing has the potential to reduce ethnic disparities in healthcare as a result of enhanced treatment efficacy within a disadvantaged population group [17].
Discussion
Our review to explore the impact of differing treatment responses on disparities in healthcare has highlighted a paucity of evidence with only five studies identified. The data that were available centered on differing pharmacogenomic treatment responses in different ethnic groups and the authors discussed how this may lead to disparities in healthcare. The case studies of genotype-guided dosing of warfarin and genotype guided treatment of clopidogrel revealed several barriers which need to be overcome in order to fully realize potential treatment benefits [18]. Most striking from our analysis is that no papers were identified which determined whether there were health disparities in the same ethnic group within the same country. Inequalities in health are determined by many different factors including socioeconomic and more work will be needed in this area as PM approaches become implemented into practice.
Research on the impact of pharmacogenomics on disparities in healthcare may have been hindered by the relative lack of implementation into clinical practice due to associated lack of cost–effectiveness evidence. A systematic review found robust evidence of cost–effectiveness for pharmacogenomic testing for prevention of adverse drug reactions only for a limited number of drugs (abacavir, allopurinol, carbamazepine, clopidogrel and irinotecan) even though over 200 drugs were identified with labels containing pharmacogenetic information [20].
Warfarin has a narrow therapeutic index and thus getting the dose correct is crucial to prevent either excessive or insufficient anticoagulation [21,22]. It was estimated in the USA that hospital admissions related to warfarin complications were estimated to cost on average US$10,819 per patient and that the cost of drug-related morbidity and mortality exceeded US$177 billion [23]. Recently the results of two large RCTs which evaluated genotype-guided dosing of warfarin were published, one which was conducted in Europe (EU-PACT) and the other in the USA (COAG) [22]. EU-PACT demonstrated that genotype-guided dosing compared with fixed loading dose regimen in newly diagnosed patients with thromboembolic disorder in the UK and Sweden found an improved achievement of the primary outcome of percentage time within International Normalized Ratio (INR) target time within Therapeutic Range (TTR) evaluated over 3 months [21]. COAG failed to show an improvement in TTR compared with a standard clinical dosing algorithm [22]. African–American patients in COAG were less likely to achieve TTR in the genotyped arm compared with the control arm [22]. While EU-PACT consisted of an ethnically homogenous cohort (97% white), COAG was an ethnically heterogeneous cohort (67% white, 27% black, 6% Hispanic). CYP2C9 *2 and *3 allele frequencies are lower in African–Americans than European–Americans (1 and 2%, respectively and 6 and 10%, respectively), while other SNPs are present in African–Americans but are rare in Caucasians (CYP2C9*8 and *11). The latter SNPs however were not assessed to inform dosing African–American patients in COAG [21,22]. A cost–effectiveness analysis of the EU-PACT trial showed that genotype-guided dosing in the UK and Swedish populations was cost-effective when compared with current standard clinical care [24]. It is now clear that ethnicity-stratified analysis can improve dose prediction across ethnic groups when compared with ethnicity-combined analysis [25]. Such population-specific warfarin pharmacogenomic dosing algorithms are likely to address, at least in part, a source of health disparities in pharmacogenetically underserved groups [25–27].
Clopidogrel is a commonly prescribed antiplatelet drug. Clopidogrel is a prodrug which is metabolized by CYP2C19 to become active. While patients with reduced-function variants (*2, *3, *4 and *8) require higher doses of the drug, patients with a gain-of-function variant (*17) require lower dose [28]. Individuals may have a combination of variants and the Clinical Pharmacogenetics Implementation Consortium (CPIC) has developed guidelines for differing metabolism rates to inform treatment strategies [29]. The prevalence of variants differ by ethnicity; the most common CYP2C19 loss-of-function allele is *2 with allele frequencies of 29–35% in Asians and only approximately 15% in Caucasians and Africans [29]. The study by Panattoni et al. [19] highlights how variation in the frequency of the variant allele in certain ethnic populations can confer benefits in that population and potentially reduce health inequalities.
It is important to note that clopidogrel treatment failure is multifactorial as noncompliance, drug–drug interactions and comorbidities may also have a clinically significant impact on health outcomes [30]. Therefore to date, due to a lack of prospective data from RCTs which would adjust for confounding factors, genotyping to identify CYP2C19 loss-of-function alleles is not yet widely recommended as part of routine clinical care [30]. However, two large prospective RCTs (TAILOR-PCI and POPular Genetics study) are underway to address this gap in clinical evidence [30].
From both pharmacogenomic examples, it is apparent that identifying genetic variants in genes across an ethnically diverse population can improve treatment algorithms to optimize care. A report by the Precision Medicine Initiative Working Group further explains that knowledge of genetic variability in six different ethnic groups is necessary to identify variations in disease etiology and course [5]. The results of this review identified a small number of studies which have focused on the impact of pharmacogenomics on health disparities. However, from our findings it is difficult to state with any degree of certainty as to whether health disparities have been widened or reduced by pharmacogenomics. Indeed, the results from our review should be treated with caution as ‘absence of evidence is not evidence of absence’.
Access to knowledge-based disparities in implementation of pharmacogenomics in clinical practice may be compounded by a paucity of evidence of clinical effectiveness in underserved groups. Furthermore, provider and patient relations are paramount for the potential realization of PM. There is known substantial disparity in uptake of genetic tests, which has been found to be associated with a range of psychosocial, sociodemographic factors and clinical factors [6]. Therefore, proactive initiatives to minimize and prevent disparities caused by these factors should be encouraged. This should include conscious decisions to ensure that there is participation across sociodemographic groups during the development phase to ensure that the potential benefits from pharmacogenomic research are fully realized [18,31].
A limitation of the scope of this review is that impact on health disparities is rarely reported in primary clinical studies. Moreover, the electronic search may have failed to identify potential sources of evidence if terms relating to impact on health disparities were not stated in the title, abstract or keywords of the publication. Furthermore, none of the included studies attempted to quantify the impact of pharmacogenomic-guided treatment on health disparities [13–16,19]. Despite these limitations it is important to emphasize again that we did not identify any studies that specifically evaluated health disparities caused by pharmacogenomics within the same ethnic group in the same country. A report by the WHO Commission on Social Determinants of Health (2008) highlighted the overarching importance of improving daily living conditions and combating the inequitable distribution of money, power and resources in reducing health disparities [32]. While pharmacogenomics is not a main determinant for population health, since translation of research is at such an early stage, a proactive approach provides an opportunity to ensure future advancements benefit disadvantaged populations.
Conclusion & future perspective
In summary, our review has highlighted that there is limited analysis and reporting of impact on health disparities in pharmacogenomics studies. In the literature, it is widely acknowledged that concerted efforts are required to ensure that the underserved and vulnerable populations also have future access to clinical innovations. However, whether this is happening is unclear at present and thus future pharmacogenomics studies should incorporate equity assessments to address the existing gap in evidence.
Summary points.
Minimizing modifiable health disparities is fundamental for an equitable & progressive achievement of healthcare coverage
There is concern that innovative medicines may be taken up less quickly and in lower numbers in different populations due to challenges in access, availability and understanding of health information.
This systematic literature review considers evidence of the impact of pharmacogenomic interventions on widening or reducing health disparities.
Systematic literature review methodology
The review was informed by the Centre for Reviews and Dissemination's guidance for undertaking reviews in healthcare and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses with a focus on health equity guidelines.
Over 200 drugs which include pharmacogenomic information in the drug label were identified from the PharmGKB website. A search of major databases was conducted using two clauses combined which included pharmacogenomic drugs and health disparity terms.
Results of the systematic literature review
A total of 4978 abstracts were screened and 80 articles were full text screened.
Four met the inclusion criteria and one additional article was identified from a hand search of references.
Predicted impact of genotype-guided dosing of warfarin & health disparities
Genotype-guided dosing of warfarin could theoretically reduce ethnic health disparities as personalizing treatment improves health outcomes by addressing underlying genetic differences between individuals.
Inconsistencies in the ability to provide predictive information for anticoagulation response with genotype-guided dosing of warfarin for certain ethnic groups.
Genotype-guided treatment compared with treatment of clopidogrel or prasugrel alone & health disparities
Maori and Pacific Islander ethnicities have a relatively high incidence of CYP2C19*2 allele and therefore poor metabolizers of clopidogrel were more commonly identified.
Genotype-guided clopidogrel treatment has the potential to reduce ethnic disparities in healthcare as a result of enhanced treatment efficacy within a disadvantaged population group.
Conclusion
The review highlights a paucity of research which investigates the impact of pharmacogenomics on health disparities.
Lack of trial participation and data collection can lead to mistreatment for understudied groups and therefore the potential treatment benefits may not be fully generalizable.
Knowledge of how pharmacogenomic innovations translate within different subgroups is paramount to ensure beneficial aggregate population effects do not conceal widening disparities.
Footnotes
Financial & competing interests disclosure
This study was supported by the NIH Research Collaboration for Leadership in Applied Health Research and Care North West Coast (NIHR CLAHRC NWC). The investigators were solely responsible for the content and the decision to submit the manuscript for publication. The funding source had no role in the selection, critical appraisal, or synthesis of evidence. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. The authors have no other relevant affiliations or financialinvolvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/pgs-2017-0076
References
- 1.Hosseinpoor AR, Bergen N, Koller T, et al. Equity-oriented monitoring in the context of universal health coverage. PLoS Med. 2014;11:e1001727. doi: 10.1371/journal.pmed.1001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitehead M. The concepts and principles of equity and health. Health Promot. Int. 1991;6:217–228. [Google Scholar]
- 3.EPEMED. Personalized Medicine in Europe – Enhancing Patient Access to Pharmaceutical Companion Products. European Personalized Medicine Association; 2014. www.epemed.org/online/www/content2/104/107/910/pagecontent2/4339/791/ENG/EpemedWhitePaperNOV14.pdf [Google Scholar]
- 4.Shargel L, Wu-Pong S, ABC Y. Applied Biopharmaceutics and Pharmacokinetics, 5th ed. McGraw-Hill; NY, USA: 2005. pp. 258–267. [Google Scholar]
- 5.NIH. Precision Medicine Initiative Cohort Program. 2016. www.nih.gov/precision-medicine-initiative-cohort-program
- 6.Sweeny K, Ghane A, Legg AM, Huynh HP, Andrews SE. Predictors of genetic testing decisions: a systematic review and critique of the literature. J. Genet. Couns. 2014;23:263–288. doi: 10.1007/s10897-014-9712-9. [DOI] [PubMed] [Google Scholar]
- 7.Welch VA, Petticrew M, O'Neill J, et al. Health equity: evidence synthesis and knowledge translation methods. Syst. Rev. 2013;2:43. doi: 10.1186/2046-4053-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.CRD. Systematic reviews: CRD's guidance for undertaking reviews in healthcare. www.york.ac.uk/media/crd/Systematic_Reviews.pdf [Google Scholar]
- 9.Welch V, Petticrew M, Tugwell P, et al. PRISMA-Equity 2012 Extension: reporting guidelines for systematic reviews with a focus on health equity. PLoS Med. 2012;9:1–2. doi: 10.1371/journal.pmed.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.PharmGKB. 2015. www.pharmgkb.org/view/drug-labels.do Drug Labels. 1.
- 11.Morrison A, Polisena J, Husereau D, et al. The effect of English-language restriction on systematic review-based meta-analyses: a systematic review of empirical studies. Int. J. Technol. Assess. Healthcare. 2012;28:138–144. doi: 10.1017/S0266462312000086. [DOI] [PubMed] [Google Scholar]
- 12.Guyatt G, Oxman AD, Akl EA, et al. GRADE guidelines: 1. introduction - GRADE evidence profiles and summary of findings. J. Clin. Epidemiol. 2011;64:383–394. doi: 10.1016/j.jclinepi.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 13.Kealey C, Chen Z, Christie J, et al. Warafin and cytochrome P450 2C9 genotype: possible ethic variation in warafin sensitivity. Pharmacogenomics. 2007;8:217–225. doi: 10.2217/14622416.8.3.217. [DOI] [PubMed] [Google Scholar]
- 14.Limdi NA, Arnett DK, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European–Americans and African–Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gladding P, Stewart R, Webster M, White H. A simulation of warfarin maintenance dose requirement using a pharmacogenomic algorithm in an ethnically diverse cohort. Hear. Lung Circ. 2009;18:S11. doi: 10.2217/pme.10.24. [DOI] [PubMed] [Google Scholar]
- 16.Chan SL, Suo C, Chia KS, Teo YY. The population attribu fraction as a measure of the impact of warfarin pharmacogenetic testing. Pharmacogenomics. 2012;13:1247–1256. doi: 10.2217/pgs.12.104. [DOI] [PubMed] [Google Scholar]
- 17.Panattoni L, Brown PM, Ao B, Te Webster M, Gladding P. The cost effectiveness of genetic testing for CYP2C19 variants to guide thienopyridine treatment in patients with acute coronary syndromes: a New Zealand evaluation. Pharmacoeconomics. 2012;30:1067–1084. doi: 10.2165/11595080-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 18.Manrai AK, et al. Genetic misdiagnoses and the potential for health disparities. N. Engl. J. Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panattoni L, Brown PM, Ao B, Te Webster M, Gladding P. The cost effectiveness of genetic testing for CYP2C19 variants to guide thienopyridine treatment in patients with acute coronary syndromes: a New Zealand evaluation. Pharmacoeconomics. 2012;30:1067–1084. doi: 10.2165/11595080-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Plumpton CO, Roberts D, Pirmohamed M, Hughes DA. A systematic review of economic evaluations of pharmacogenetic testing for prevention of adverse drug reactions. Pharmacoeconomics. 2016;34(8):771–793. doi: 10.1007/s40273-016-0397-9. [DOI] [PubMed] [Google Scholar]
- 21.Anderson JL, Gage BF, Rosenberg YD, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmel SE, French BF, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N. Engl. J. Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finlayson AE, Godman B, Paterson K, et al. Personalizing healthcare: from genetics through payment to improving care? J. R. Soc. Med. 2013;106:41–44. doi: 10.1258/jrsm.2012.120193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhoef TI, Redekop WK, Langenskiold S, et al. Cost–effectiveness of pharmacogenetic-guided dosing of warfarin in the United Kingdom and Sweden. Pharmacogenomics J. 2016;16:478–484. doi: 10.1038/tpj.2016.41. [DOI] [PubMed] [Google Scholar]
- 25.Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African–American individuals: a genome-wide association study. Lancet. 2013;382:790–796. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubo K, Ohara M, Tachikawa M, et al. Population differences in S-warfarin pharmacokinetics among African–Americans, Asians and whites: their influence on pharmacogenetic dosing algorithms. Pharmacogenomics J. 2016 doi: 10.1038/tpj.2016.57. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 27.Daneshjou R, Gamazon ER, Burkley B, et al. Genetic variant in folate homeostasis is associated with lower warfarin dose in African–Americans. Blood. 2014;124:2298–2306. doi: 10.1182/blood-2014-04-568436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N. Engl. J. Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 29.Scott SA, Sangkuhl K, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 2013;94:317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira NL, Geske JB, Mayr M, Shah SH, Rihal CS. Pharmacogenetics of clopidogrel. Circ. Cardiovasc. Genet. 2016;9:185–188. doi: 10.1161/CIRCGENETICS.115.001318. [DOI] [PubMed] [Google Scholar]
- 31.Tan DSW, Mok TSK, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J. Clin. Oncol. 2016;34:91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 32.WHO. Closing the gap in a generation. Health Equity Through Action on the Social Determinants of Health; 2008. www.who.int/social_determinants/final_report/csdh_finalreport_2008.pdf [DOI] [PubMed] [Google Scholar]