Abstract
C4 photosynthesis is a carbon-concentrating mechanism that evolved independently more than 60 times in a wide range of angiosperm lineages. Among other alterations, the evolution of C4 from ancestral C3 photosynthesis requires changes in the expression of a vast number of genes. Differential gene expression analyses between closely related C3 and C4 species have significantly increased our understanding of C4 functioning and evolution. In Chenopodiaceae, a family that is rich in C4 origins and photosynthetic types, the anatomy, physiology and phylogeny of C4, C2, and C3 species of Salsoleae has been studied in great detail, which facilitated the choice of six samples of five representative species with different photosynthetic types for transcriptome comparisons. mRNA from assimilating organs of each species was sequenced in triplicates, and sequence reads were de novo assembled. These novel genetic resources were then analyzed to provide a better understanding of differential gene expression between C3, C2 and C4 species. All three analyzed C4 species belong to the NADP-ME type as most genes encoding core enzymes of this C4 cycle are highly expressed. The abundance of photorespiratory transcripts is decreased compared to the C3 and C2 species. Like in other C4 lineages of Caryophyllales, our results suggest that PEPC1 is the C4-specific isoform in Salsoleae. Two recently identified transporters from the PHT4 protein family may not only be related to the C4 syndrome, but also active in C2 photosynthesis in Salsoleae. In the two populations of the C2 species S. divaricata transcript abundance of several C4 genes are slightly increased, however, a C4 cycle is not detectable in the carbon isotope values. Most of the core enzymes of photorespiration are highly increased in the C2 species compared to both C3 and C4 species, confirming a successful establishment of the C2 photosynthetic pathway. Furthermore, a function of PEP-CK in C2 photosynthesis appears likely, since PEP-CK gene expression is not only increased in S. divaricata but also in C2 species of other groups.
Keywords: Caryophyllales, evolution, leaf, photorespiration, photosynthesis, RNA-Seq, Salsola
Introduction
The convergent evolution of complex traits challenges evolutionary biologists since evolutionary stable intermediate steps seem to be required to accomplish the transition to complex phenotypes (Washburn et al., 2016). In plants, one prime example of such a complex trait is C4 photosynthesis. C4 photosynthesis evolved more than 60 times in various angiosperm lineages including monocots and eudicots (Sage, 2016). These multiple independent origins of C4 photosynthesis from the ancestral C3 pathway allow investigating the acquisition of the C4 syndrome in individual plant groups and, subsequently, to integrate all acquired components from the different plant groups for understanding the whole complexity and variability of C4 evolution. Specifically, C4 is a carbon-concentrating mechanism that evolved to cope with decreasing atmospheric CO2 concentration (Ehleringer et al., 1991), a condition which would otherwise favor photorespiration particularly in subtropical regions (Bauwe et al., 2010). In mesophyll tissue, atmospheric CO2 is initially fixed by phosphoenolpyruvate carboxylase (PEPC), yielding a compound consisting of four carbon atoms, the key and name-giving step of C4 (Hatch, 1987). This C4 compound is then modified, transported into the bundle sheath tissue and eventually decarboxylated to increase CO2 concentration, allowing a high carboxylation:oxygenation ratio of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) in the Calvin-Benson cycle, which results in drastically reduced photorespiration (Hatch, 1987).
The current model of C4 evolution predicts a gradual establishment of the C4 cycle from C3 through a limited number of evolutionary steps (Sage et al., 2012; Bräutigam and Gowik, 2016). Here, the formation of a photorespiratory CO2 pump—operating via glycine shuttling by restricting the combined glycine decarboxylase complex (GDC) and serine hydroxymethyltransferase (SHMT) reactions to the bundle sheath—is a major landmark (Sage et al., 2012; Bräutigam and Gowik, 2016). Species exhibiting this glycine shuttle are characterized by lower CO2 compensation points than C3 plants, a leaf anatomy that is intermediate between C3 and C4 species, and no or low C4 cycle activity (Edwards and Ku, 1987; Sage et al., 2012). The C3-C4 intermediate species of Flaveria that mostly rely on the photorespiratory CO2 pump and where C4 cycle activity is low are called C2 species (Sage et al., 2012). Whether C3-C4 intermediate phenotypes with or without a photorespiratory CO2 pump represent true evolutionary intermediate states for the complex C4 syndrome is still under debate and subject of recent investigations (Monson et al., 1984; Monson and Moore, 1989; Sage, 2004; Heckmann et al., 2013; Williams et al., 2013; Bräutigam and Gowik, 2016; Schlüter and Weber, 2016; Kadereit et al., 2017). Although rare in comparison to C3 and C4 species, the extant C2 species clearly represent an established photosynthetic pathway by their mere existence, and also by their phylogenetic age (i.e., several million years) in some lineages (Christin et al., 2011; reviewed in Sage et al., 2012).
Evolution of C4 or C2 from C3 photosynthesis requires many changes, including alterations in leaf anatomy, physiology and gene regulation (Gowik and Westhoff, 2011; Langdale, 2011). Analyses of leaf transcriptomes provided broad knowledge of gene expression in C4 and C2 photosynthesis by comparing closely related C3, C4 and/or C2 species (Bräutigam et al., 2011; Külahoglu et al., 2014; Mallmann et al., 2014; van den Bergh et al., 2014; Ding et al., 2015; Aubry et al., 2016; Schlüter et al., 2016a,b), leading to the identification of many genes and proteins which function in C4 or C2 photosynthesis. One general trend, for example, seems to be that most of the key genes of photorespiration, while transcriptionally downregulated in C4, are highly expressed in C2 when compared to C3 plants (reviewed in Bräutigam and Gowik, 2016). Besides genes encoding proteins of the C4 cycle and photorespiration, genes related to photosynthesis strongly differ between C3 and C4 species, at least in the genera Cleome sensu lato and Flaveria (Bräutigam et al., 2011; Gowik et al., 2011; Aubry et al., 2014; Külahoglu et al., 2014; Kümpers et al., 2017). Gene expression analyses also revealed that C4 genes were mostly recruited from expression domains with housekeeping functions (Külahoglu et al., 2014). Additionally, known and novel transporters could be identified, and transport processes in general seem to be very important in C4 photosynthesis (Schlüter et al., 2016b).
The goosefoot family (Chenopodiaceae) is an outstanding system to study C4 photosynthesis, because it comprises the largest number of both C4 species and independent C4 origins in the eudicots, with an outstanding diversity of the C4 phenotype (Kadereit et al., 2003, 2010, 2014; Kadereit and Freitag, 2011; Sage, 2016). Additionally, Chenopodiaceae contain a number of unique study systems, e.g., single cell C4 plants, like the genus Bienertia (Freitag and Stichler, 2002) or Suaeda aralocaspica (Freitag and Stichler, 2000), the stem succulent C4 hygro-halophytes Tecticornia indica and T. bibenda (Shepherd and van Leeuwen, 2007; Voznesenskaya et al., 2008) and the species of tribe Salsoleae that conduct C3 in cotyledons before they switch to C4 in leaves or assimilating shoots (Voznesenskaya et al., 2013; Li et al., 2015; Lauterbach et al., 2016). Furthermore, Salsoleae seem particularly suitable to study the evolution of C2 and C4 photosynthesis because the tribe contains a comparatively large number of C2 species (summarized in Voznesenskaya et al., 2013 and Schüssler et al., 2016). Salsoleae are widespread in semi-deserts, deserts and coastal regions of Eurasia and well-adapted to dry and saline conditions (Akhani et al., 2007). Their leaves and/or shoots show a central water storage tissue. Often the leaves are reduced and photosynthesis is taken over by the shoots (Schüssler et al., 2016). Leaves or assimilating shoots often have a multi-layered epidermis and a hypodermis. The chlorenchyma forms a continuous layer surrounding the entire leaf and consists of 2-3 mesophyll layers in C3 species and one outer mesophyll layer and a Kranz layer in C4 species (Salsoloid leaf anatomy), respectively (Carolin et al., 1975). In the C3-C4 intermediate species in general, the Kranz layer is either continuous or interrupted by water storage cells, and the mesophyll can consist of two layers (Voznesenskaya et al., 2013; Schüssler et al., 2016). Recently, a molecular phylogeny including the major lineages of Salsoleae has been published (Schüssler et al., 2016) which is the basis for the phylogenetically informed sampling of this study. Despite the high diversity in this group, large-scale genomic analyses of coding sequence information are currently limited to two taxa, Haloxylon ammodendron (Li et al., 2015) and Salsola soda (Lauterbach et al., 2016).
Here we present transcriptome de novo assemblies and the analyses of differential gene expression of representative species of Salsoleae with different photosynthetic types, including C3, C4, and C2 photosynthesis. In particular, we sequenced the transcriptomes of the main assimilating organs (i.e., leaf or assimilating shoot) of the C3 species Salsola webbii, two distinct populations of the C2 species S. divaricata, the C4 species S. oppositifolia, and Hammada scoparia, which conducts C3 in cotyledons, but C4 in assimilating shoots. Additionally, we included publicly available leaf transcriptome data from Salsola soda, which also exhibits C3 in cotyledons and C4 in leaves (Lauterbach et al., 2016). Transcriptomes of all five species were assembled de novo and gene expression patterns between the species/populations were compared with a focus on genes involved in C4 photosynthesis and photorespiration to address the question whether gene expression profiles of C3, C2, and C4 species of Salsoleae are comparable to other study systems like Cleome or Flaveria.
Materials and methods
Plant material
Seeds were taken from plants collected in the field, and vouchers of these collections are deposited at the herbarium of Johannes Gutenberg-University Mainz (MJG; see Table 1 for further information and a comment on taxonomic and nomenclatural issues in Salsoleae). Plants of H. scoparia, Salsola divaricata (from two different populations: population 184 (Pop-184) located in Lanzarote and population 198 (Pop-198) located in Gran Canaria), S. oppositifolia, and S. webbii were grown from seeds in potting soil (custom mixed soil from the Botanic Garden, Johannes Gutenberg-University Mainz) in a glasshouse with an additional light intensity of ca. 300 μmol m−2 s−1. Samples were harvested between 16th April and 16th May 2014 between 10:30 and 13:00, immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. A highly reduced phylogenetic tree based on the results of Schüssler et al. (2016) including only species of the current study is shown in Figure 1.
Table 1.
Species of Salsoleae sensu stricto included in the study.
| Species* | PS type | δ13C of leaf (n = 3) | Herbarium and voucher ID |
|---|---|---|---|
| Hammada scoparia (Pomel) Iljin** | C4 | −18.759 | MJG living collection no. 87 (source MSB serial no. 89920; Morocco: Taroudannt) |
| Salsola divaricata Moq.; Population184 | C2 | −32.208 | MJG Herbarium no. 014225, living collection no. 184 (source: Canary Islands: Lanzarote, Orzóla) |
| Salsola divaricata Moq.; Population198 | C2 | −31.759 | MJG living collection no. 198 (source: Canary Islands: Gran Canaria, Cuesta Ramón, Jinamar) |
| Salsola oppositifolia Desf. | C4 | −17.514 | MJG Herbarium no. 013564, living coll. No. 173 (source: SW Morocco, 18 km N Agadir, near Tamrhakt) |
| Salsola soda L. | C4 | −15.434*** | MJG no. 014562 (see Lauterbach et al., 2016) |
| Salsola webbii Moq. | C3 | −31.712 | MJG living collection no. 67 (source: G. Edwards lab, Pullman, Washington, originally collected in S. Spain) |
The highly polyphyletic genus Salsola and also the Salsoleae are currently experiencing dramatic taxonomic and nomenclatural rearrangements (compare Akhani et al., 2007, 2016; Hernández-Ledesma et al., 2015; Mosyakin et al., 2017). For the sake of easy comparability for non-Salsoleae experts we therefore prefer to use the established names in this paper.
sometimes treated as synonym of Hammada articulata O. Bolòs and Vigo.
measured in Lauterbach et al. (2016).
Figure 1.
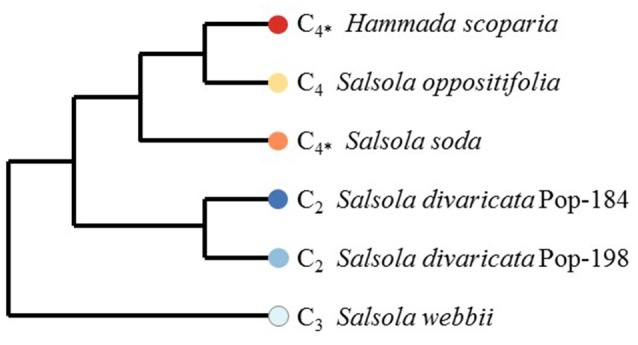
Highly reduced phylogenetic tree including only species of the current study [adapted and modified from Open Access article by Schüssler et al. (2016), which permits unrestricted reuse; C4*, species with C4 photosynthesis in leaves/assimilation shoots but C3 in cotyledons].
Carbon isotope discrimination measurements
Leaves or, in case of H. scoparia, assimilation shoots of all five species were harvested and dried for several days in silica gel. Dry leaf samples were pulverized using the mixer mill MM 301 (Retsch). Approximately 200 mg of each sample were used to determine stable carbon isotope ratios (i.e., 13C/12C) relative to the Pee Dee belemnite standard (Craig, 1957) by the Institute for Geosciences at Johannes Gutenberg-University Mainz. For each sample, technical triplicates were measured.
RNA sequencing
RNA extraction, library preparation and mRNA sequencing were performed as described in Lauterbach et al. (2016). In brief, total RNA was extracted from 9 to 90 mg cotyledon or leaf tissue using the RNeasy Plant Mini Kit (Qiagen), including DNase I digestion with RNase-Free DNase Set (Qiagen). After quality control of RNA using the 2100 Bioanalyzer (Agilent Technologies), NanoDrop (Thermo Fisher Scientific) and Qubit (Life Technologies), 500 ng of total RNA were used for cDNA library preparation with the TruSeq RNA Sample Preparation Kit (Illumina Inc.), following the Low Sample Protocol (TruSeq RNA Sample Preparation v2, May 2012). Sequencing of 101 bp single-end reads was performed on an Illumina HiSeq2000 platform.
For RNA-Seq, different tissues were sampled: (1) assimilation shoots of H. scoparia, (2) cotyledons and leaves of S. divaricata Pop-184, (3) cotyledons and leaves of S. divaricata Pop-198, (4) cotyledons and leaves of S. oppositifolia, (5) cotyledons and leaves of S. soda (RNA-Seq data taken from Lauterbach et al., 2016), (6) leaves of S. webbii (see Table 1 for further details). Three biological replicates (i.e., triplicates) per species/population and organ were sequenced, except for S. webbii where only two biological replicates and one technical replicate were sequenced due to the lack of a third individual in the living collection at Botanic Garden, Johannes Gutenberg-University Mainz. Additionally, for H. scoparia and S. webbii no seeds were available, thus only leaves could be sampled. Both cotyledons were sampled for cotyledon samples while a single leaf was sampled for leaf samples.
Processing of raw reads and de novo transcriptome assembly
Sequence reads were quality controlled with the FASTQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and filtered and trimmed using the tools “fastx_clipper” (option -M 15 -l 20) to remove adapter sequences, “fastx_trimmer” (option -f 15) to remove the first 14 bases of the 5' end of all reads, “fastq_quality_trimmer” (option -t 20 -l 20) to remove low quality bases (below PHRED score of 20) from the 3' end, and “fastq_quality_filter” (option -q 20 -p 80) to remove all reads with an overall PHRED score below 20. All tools are from the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) and were used in the same order as listed.
Initially, we conducted de novo transcriptome assemblies for each species with two different tools, SOAPdenovo-Trans v1.04 (Xie et al., 2014) and Trinity v2.1.1 (Grabherr et al., 2011), since both were reported to perform well in de novo assembling transcriptome data (Honaas et al., 2016; Wang and Gribskov, 2016). Default parameters were used, except for kmer size = 73 in SOAPdenovo-Trans-127mer. The qualities of de novo transcriptome assemblies were assessed using three different approaches. Firstly, BUSCO v.2.0 (Benchmarking Universal Single-Copy Orthologs; Simão et al., 2015) with the plant early release data set was used as a proxy for the completeness of the assembled transcriptomes. The BUSCO tool evaluates the completeness of the expected gene content with hidden Markov models (using HMMER v3.1; hmmer.org) and lineage-specific BUSCO profiles (Simão et al., 2015). Secondly, we mapped all quality controlled reads back to the corresponding de novo transcriptome assembly using bowtie2 v.2.1.0 (Langmead and Salzberg, 2012) for inferring the percentage of back-mapped reads as a measure of quality. Finally, we performed a sequential BLASTx search (cut-off e-value of 1e-10, and keeping only best hit) against coding sequences of the minimal Arabidopsis thaliana transcriptome (Bräutigam et al., 2011), the Beta vulgaris transcriptome (version “BeetSet-2”, Dohm et al., 2014) and the UniProtKB database (http://www.uniprot.org/) to see how many contigs were most similar to genes with a known function/annotation (i.e., putative genes). Here, all contigs firstly were mapped again Arabidopsis, the remaining unmapped contigs were mapped against B. vulgaris, and again remaining unmapped contigs were mapped against UniProtKB. A. thaliana was chosen because this species has the best-annotated plant genome, and B. vulgaris was chosen because this is the phylogenetically closest species to tribe Salsoleae with a sequenced genome.
After deciding to use the Trinity assemblies (see next paragraph), functional annotation of transcripts was performed via BLAST-based comparison (see above) against the Arabidopsis and B. vulgaris transcriptomes and the UniProtKB database. Sets of annotated transcripts served as reference transcriptomes for inferring differential gene expression (see below). Contigs from different species producing the same annotation match in either Arabidopsis, B. vulgaris or UniProtKB databases were defined as putative orthologs. As additional assessment for defined putative orthologs, pairwise reciprocal BLAST (tBASTx, cut-off e-value of 1e-10) between all six de novo assemblies were performed, and read counts especially of genes involved in C4 photosynthesis and photorespiration were manually double-checked. To exclude the presence of multiple putative loci per real locus and due to that erroneous read counts, unique presence of genes encoding proteins involved in C4 photosynthesis and photorespiration were manually checked. Venn diagrams showing the overlap of the putative orthologous contigs between all assemblies were calculated and drawn using the “venn” package (by Adrian Dusa, University of Bucharest, Romania) in R (R Core Team, 2016).
Quality assessment of de novo transcript assemblies
De novo assemblies were conducted with at least 97.8 million (S. webbii) and up to 205 million (S. oppositifolia) quality-controlled reads. While we only used data from leaf samples for differential gene expression analysis, we included additional reads from cotyledon samples (Lauterbach et al., in preparation) for the de novo assemblies of S. divaricata Pop-184 and Pop-198, S. oppositifolia, and S. soda transcriptomes (see above).
Results of both assemblers, SOAPdenovo-Trans and Trinity, yielded partly different results (Table 2, Supplementary Table S1, Supplementary Figure S1). We decided to continue downstream analyses with the Trinity assemblies for several reasons. First, the number of assembled bases was at least twice as high in the Trinity assemblies compared to SOAPdenovo-Trans assemblies. Second, assessment with BUSCO showed that the completeness in terms of expected gene content was considerably higher in the Trinity assemblies (Supplementary Figure S2). Third, the amount of mappable reads was higher in the Trinity assemblies, which is an important metric to identify leading assemblers. Finally, contigs of the Trinity assemblies matched more proteins with known annotation of the three references (Supplementary Figure S3).
Table 2.
Summary of statistics of de novo transcriptome assemblies using SOAPdenovo-Trans and Trinity (BUSCO, Benchmarking Universal Single-Copy Orthologs).
| No. of assembled contigs | Mean length (in bp) | No. of assembled bases (in million) | Complete BUSCOs (in %) | Fragmented BUSCOs (in %) | Missing BUSCOs (in %) | Back mapping rate (in %) | |
|---|---|---|---|---|---|---|---|
| SOAPdenovo-Trans | 56,756–186,466 | 333.87–682.05 | 37–84.4 | 47.9–87.4 | 6.3–37.8 | 6.3–17.5 | 84.9–96.3 |
| Trinity | 127,382–465,856 | 659.49–951.37 | 119.1–302 | 89.6–93.9 | 2.9–7.0 | 2.8–4.4 | 94.1–98.4 |
Statistics, data analysis, and differential gene expression
For studying differential gene expression, processed reads were mapped against the respective de novo transcriptome reference assemblies using bwa-mem v.0.7.13 (Li and Durbin, 2009). To infer quantitative information about transcript abundance, reads that uniquely mapped to one contig (i.e., excluding unmapped reads and multi-mapped reads) were extracted by Samtools v.1.3 (Li and Durbin, 2009). These counts were further used for comparison between replicates and species. RNA-Seq data of the six leaf samples were comparatively analyzed to detect differential gene expression. Pairwise comparisons between all six samples were statistically evaluated using “edgeR” (Robinson et al., 2010) in R. After normalization using the trimmed mean of M-values (TMM) method in edgeR, only contigs that had at least five reads per million in at least three samples (including replicates) were considered. Log2 transcript ratios were calculated and a log2 fold change (log2FC) of ≥1 was applied as threshold to call differentially expressed genes. A significance threshold of 0.01 was defined after Benjamini-Hochberg correction to account for multiple testing (Benjamini and Hochberg, 1995) and applied to call differentially expressed genes. To further analyze and visualize the data, log2 transformed read counts (transcripts per million) were used for hierarchical clustering using Pearson correlation and average linking method (MultiExperiment Viewer, MeV v.4.9, http://mev.tm4.org/) and principal component analysis (PCA, MeV). Additionally, hierarchical clusterings of transcript abundance of selected genes encoding known C4 cycle proteins (Lauterbach et al., 2016) and the eight core enzymes of photorespiration (Bauwe et al., 2010) were conducted (Pearson correlation, average linkage method; MeV).
Results
Carbon isotope discrimination
Carbon isotope values varied between −15.434 and −32.208 (Table 1). Leaves of S. divaricata Pop-184, S. divaricata Pop-198, and S. webbii had values of −31.712, −32.208, and −31.759, respectively, indicating the absence of a high-activity C4 cycle, while leaves of H. scoparia, S. oppositifolia and S. soda had values of −18.759, −17.514, and −15.434, respectively, indicating a predominating C4 metabolism (Table 1). The values for S. oppositifolia and H. scoparia appear a bit too negative, maybe this is due to the fact that these were relatively young plants raised in the greenhouse. According to values in the literature (Schüssler et al., 2016, and references therein) both species are clearly C4 plants.
RNA sequencing
In total, RNA sequencing yielded between 29.6 and 39.4 million single-end raw reads per replicate, with at least 97.8% reads remaining after quality filtering (Supplementary Table S1, Supplementary Figure S4). Raw data is available in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-5778.
Differences in gene expression between the species
After deciding to use the annotated Trinity assemblies as reference transcriptomes, quality-filtered reads of the different RNA-Seq samples were mapped and counted using bwa-mem. 48,818 contigs had a reliable annotation in one of the six samples; of these, 15,064 contigs have an Arabidopsis annotation, 19,108 contigs have a B. vulgaris annotation, and 14,646 contigs have a UniProtKB annotation. Of the 48,818 contigs with a reliable annotation in one of the six samples (i.e., putative genes), 34,022 had at least five reads per million in at least three samples, and downstream analyses were limited to this subset.
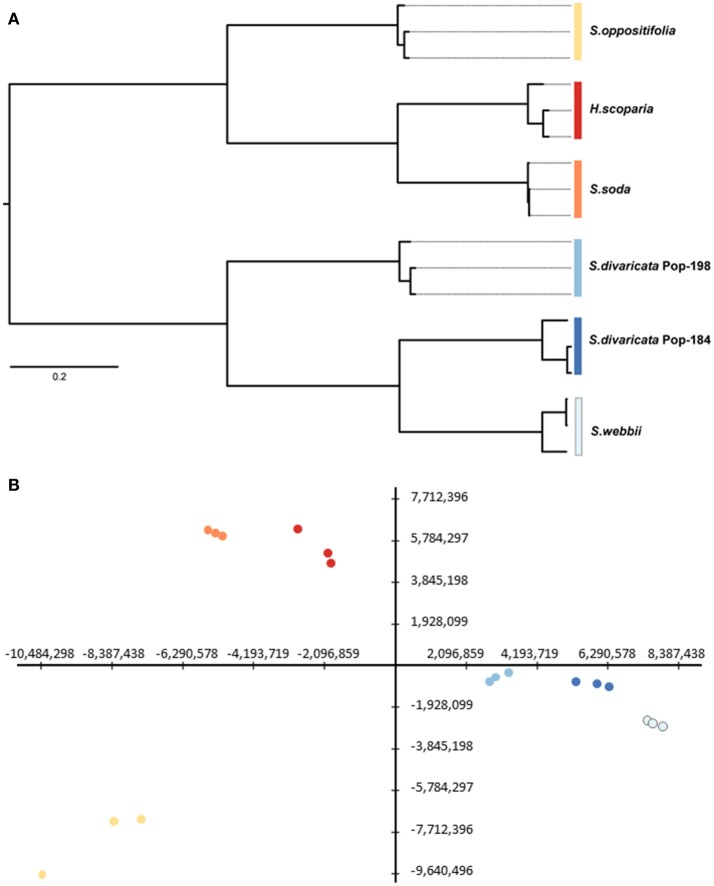
According to their transcript profiles, replicates of each sample clearly grouped together in hierarchical clustering (Figure 2A). The trend in the hierarchical clustering also mostly reflected the phylogenetic relatedness of samples (Figure 1). Thus, the two different populations of the C2 species S. divaricata grouped together, a result that reflects both phylogenetic relationship and photosynthetic type. The same was observed for the three C4 species H. scoparia, S. oppositifolia, and S. soda, which grouped together (Figures 1, 2A). H. scoparia and S. soda share a similar photosynthetic phenotype (i.e., C4 in leaves/assimilation shoots but C3 in cotyledons), and based on their transcript expression profiles they group together (Figure 2A).
Figure 2.
(A) Hierarchical clustering using Pearson correlation and (B) principal component analysis showing the first (X-axis) and second (Y-axis) components, which explain 57.41% of the total variation (see Supplementary Figure S5 for the first three components), of gene expression data. (The colors in B) are the same as in (A) and corresponds to replicates of the respective species).
Results of the PCA in general supported the results of hierarchical clustering (Figure 2B). All replicates of the same species grouped closely together. The first two components explained 60.58% (Figure 2B), and the first three components explained 76.32% (Supplementary Figure S5) of the total variance (principal component 1: 36.56%; principal component 2: 24.01%; principal component 3: 15.74%; Figure 2B, Supplementary Figure S5). The samples of H. scoparia and S. soda showed close similarity. The two populations of S. divaricata grouped together in hierarchical clustering, results of PCA showed S. divaricata Pop-184 as somewhat closer to the C3 species S. webbii than the Pop-198 sample to S. webbii. As in the hierarchical clustering, S. oppositifolia was clearly distinct from the other two C4 species and also from both populations of S. divaricata and from S. webbii. The first component of the PCA separated the species based on the photosynthetic type (the three C4 species on the left side, C3 species on the right, and the two samples of the C3-C4 intermediate species in between) (Figure 2).
Transcript abundance among photosynthetic types
Differentially expressed transcripts, defined by adjusted P-value ≤ 0.01 and log2FC ≥1, were identified by pairwise comparisons between all samples. By comparing differentially expressed transcripts between S. webbii (C3) and all three C4 species H. scoparia, S. oppositifolia and S. soda, 4,436 genes were found significantly up-regulated in S. webbii while 1,907 genes were up-regulated in all three C4 species (Supplementary Table S2). Comparing significantly differentially expressed transcripts between C3 (S. webbii) and C2 photosynthesis (S. divaricata populations), 6,304 genes were up- and 5,320 genes down-regulated in S. webbii (Supplementary Table S2). 1,714 genes were significantly up- and 856 down-regulated in C2 S. divaricata when compared with all three C4 species (Supplementary Table S2).
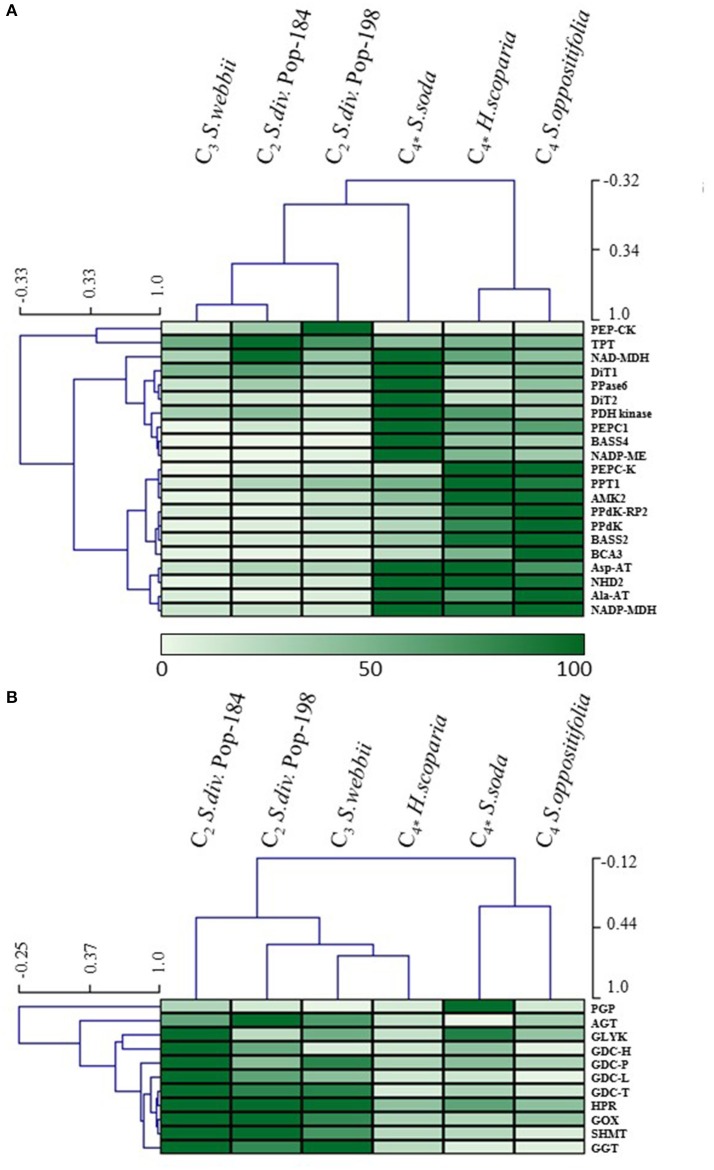
Transcript abundance of C4-related genes
Transcript abundances of 21 genes encoding known C4 cycle proteins were analyzed between the species with focus on the different photosynthetic types (Figure 3A). As expected, transcripts encoding the C4 cycle proteins alanine aminotransferase 1 (Ala-AT1), adenosine monophosphate kinase 2 (AMK2), aspartate aminotransferase 5 (Asp-AT5), bile acid:sodium symporter family protein (BASS) 2, BASS4, beta carbonic anhydrase 3 (BCA3), NADP-dependent malate dehydrogenase (NADP-MDH), NADP-malic enzyme (NADP-ME), PEPC1, PEPC kinase (PEPC-K), pyruvate orthophosphate dikinase (PPdK), PEP/phosphate translocator (PPT) 1, and sodium:hydrogen antiporter 2 (NHD2) were significantly (FDR q–values ≤ 0.01) more abundant in all three C4 species compared to C3 S. webbii (log2FC between 1.26 and 7.33; Supplementary Table S2). Increased abundance of transcripts encoding AMK2, BASS4, NHD2, PEPC1, PEPC-K, and PPT1 was found in the three C4 species and both populations of S. divaricata (C2) as compared to S. webbii (C3). No known C4 cycle gene was significantly up-regulated in the C3 species S. webbii when compared to all other samples.
Figure 3.
Hierarchical clustering and heatmap of relative transcript abundance of genes encoding (A) known C4 cycle proteins and (B) the eight core enzymes of photorespiration (C4*, species with C4 photosynthesis in leaves/assimilation shoots but C3 in cotyledons; S. div, S. divaricata; for description of abbreviated genes see text).
In addition to the genes that were increased in all three C4 species when compared to C3 S. webbii (see above), several C4 related genes were increased in one or two of the three C4 species only: Transcripts of dicarboxylate transporter (DiT) 2 and pyrophosphatase 6 (PPase6) were more abundant in S. oppositifolia and S. soda (log2FC between 1.08 and 2.35), and transcripts of PPdK regulatory protein (PPdK-RP) were more abundant in H. scoparia and S. oppositifolia (log2FC ≥2.41; Supplementary Table S2). Further, transcripts encoding NAD-dependent malate dehydrogenase (NAD-MDH) were more abundant in S. soda (log2FC ≥1.41). As expected in C4 species, many of the genes encoding C4 cycle proteins were among the most highly expressed transcripts in the three C4 species. For example, PPdK was the most highly expressed gene in S. oppositifolia and the second most highly expressed gene in H. scoparia. Other highly expressed genes in the three C4 species were Ala-AT, Asp-AT, NADP-ME, NADP-MDH, and PEPC. The NADP-ME subtype of C4 photosynthesis is known from C4 species of the Salsoleae (Akhani et al., 2007). In accordance, we found high abundance of transcripts encoding NADP-ME in the three C4 species, while those of NAD-malic enzyme (NAD-ME) and PEP-carboxykinase (PEP-CK) were low.
In the C2 species S. dicaricata, AMK2, BASS4, NHD2, PEPC1, PEPC-K, and PPT1 were significantly more abundant compared to S. webbii (log2FC between 1.12 and 4.43). Interestingly, PEP-CK was more abundant in S. divaricata compared to all other species (log2FC between 2.72 and 7.26). Additionally, in S. divaricata Pop-184, transcripts of NAD-MDH, PPase6, and triose phosphate transclocator (TPT) were significantly up-regulated when compared to S. webbii (log2FC ≥1.1), while NADP-ME and PPdK were up-regulated in S. divaricata Pop-198 in comparison to S. webbii (log2FC ≥1.5) (Supplementary Table S2). The gene encoding the transporter protein TPT, which is known to function in the C4 cycle, was highly abundant in all six samples (between 51st and 82nd highest expressed gene).
In addition to the 21 genes known to be involved in C4 photosynthesis (see above), transcripts of genes currently described to have an alleged function related to C4 metabolism (Lauterbach et al., 2016) were analyzed. Transcripts encoding pyruvate dehydrogenase (PDH) kinase, probably controlling metabolite exit from the C4 pathway via pyruvate decarboxylation (Bräutigam et al., 2014), were significantly more abundant in S. soda (log2FC 1.29) when compared to S. webbii, but still abundant in all other species, including C3 S. webbii. Phosphate transporter4.1 (PHT4.1) was significantly more abundant in all species (in S. soda with log2FC of 0.8) compared to S. webbii. The gene encoding PHT4.4 was highly upregulated in all three C4 species compared to S. webbii (log2FC ≥1.85). Levels of mRNA coding for asparagine synthetase (ASN) were increased in S. oppositifolia and S. soda when compared to S. webbii (log2FC 1.45 and 5.89, respectively). Also, ASN was the 14th highest expressed gene in S. soda (Supplementary Table S2).
Transcript abundance of genes encoding key enzymes of photorespiration
Since photorespiration has been reported as strongly modified between closely related C3 and C4 species (e.g., Cleome, Bräutigam et al., 2011) and also between C3, C2, and C4 species (e.g., Flaveria; Gowik et al., 2011; Mallmann et al., 2014), the transcript abundance of the eight core enzymes of photorespiration was analyzed and compared between species (Figure 3B). Transcripts encoding phosphoglycolate phosphatase (PGP) were significantly more abundant in all three C4 species compared to S. webbii (log2FC between 1.16 and 4.73), and gene expression of seven genes were increased in S. webbii in comparison to all three C4 species: serine:glyoxylate aminotransferase (AGT/SGT), GDC P-protein (GDC-P), GDC L-protein (GDC-L), GDC T-protein (GDC-T), glutamate:glyoxylate aminotransferase (GGT), NADP-dependent hydroxypyruvate reductase (HPR), and SHMT (Supplementary Table S2). The two genes encoding GDC H-protein (GDC-H) and PGP each were significantly up-regulated in S. divaricata compared to S. webbii (log2FC ≥1.96), the former also significantly up-regulated when compared to the three C4 species (log2FC ≥1.01). Further, S. divaricata Pop-184 showed an increase of transcripts encoding GDC-H, glycerate 3-kinase (GLYK) and PGP when compared to S. divaricata Pop-198 and S. webbii (Supplementary Table S2). In general, all eight genes encoding key enzymes of photorespiration were highly expressed in S. divaricata (Pop-184 and 198) and S. webbii. Interestingly, transcripts of GDC-P were not only highly abundant in the C2 and C3 species, but in all six samples.
Discussion
Both C2 and C4 photosynthesis are complex and require many changes in gene expression patterns compared to the ancestral C3 photosynthesis (Sage, 2004). RNA-Seq strongly promoted the understanding of C2 and C4 evolution in the last few years. In eudicots, the flagship group to decipher differential gene expression leading to C2 or C4 photosynthesis is Flaveria (Asteraceae), which includes, besides C3 species, many C2 and C4 species (Edwards and Ku, 1987). Likewise, tribe Salsoleae (Chenopodiaceae) comprises C3, some C2 and an immense diversity of C4 lineages (Schüssler et al., 2016). However, it differs significantly in leaf anatomy and ecology from the model genus Flaveria, making Salsoleae a promising candidate for increasing our knowledge of convergent acquisition of C2 and C4 photosynthesis.
A novel transcriptome data resource for salsoleae
Until now, no transcriptome assembly of tribe Salsoleae (Chenopodiaceae) has been available and large-scale gene expression studies based on RNA sequencing were for the most part absent in this group (Li et al., 2015; Lauterbach et al., 2016). Thus, we used a de novo assembly approach to produce reference transcriptomes of six samples representing five species of Salsoleae, thereby providing a novel and large mRNA data resource for this group. Initially applying two different tools, the Trinity assembler was selected because of superior results (see Results; Table 2). De novo assemblies of the two different populations of the same species, S. divaricata, yielded about the same amount of contigs, implying a consistent performance of Trinity. The number of cDNA contigs obtained did not correlate with the diversity of tissues included in the sequencing: While only stem or leaf tissue of adult plants was available for H. scoparia and S. webbii, respectively, the other four assemblies, which included cotyledon and adult leaf tissues, yielded fewer (S. soda) or more contigs (S. divaricata Pop-184, S. divaricata Pop-198, S. oppositifolia). Interestingly, we obtained more than twice as many contigs from S. oppositifolia compared to S. soda. This huge difference could potentially have been caused by different ploidy levels in these species: Salsola oppositifolia is octoploid (Lago and Castroviejo, 1992; Peruzzi and Cesca, 2004), while S. divaricata and S. webbii are tetraploid (G. Kadereit and D. Tefarikis; pers. observation; Padrón, 2012) and S. soda and H. scoparia are diploid (Löve, 1970, 1986; Tarnavschi and Lungeanu, 1982; Zakharyeva, 1985).
Physiological studies (Voznesenskaya et al., 2013) identified S. webbii as a typical C3 plant, which is why this species was selected as a C3 reference and gene expression in leaves of S. webbii was taken as exemplary for C3 in all downstream analyses. The transcriptome data of S. webbii differed strongly from all other species included (Figure 2), which reflects both phylogenetic position and photosynthetic type (Figure 1). Additionally, stable carbon isotope values of leaves (or in case of H. scoparia assimilation shoots) were measured since stable carbon isotopes can be used as an indicator of C4 metabolism (O'Leary, 1981; Cernusak et al., 2013). All measurements of the species of the current study were consistent with published data (Akhani et al., 1997; Pyankov et al., 2001; Voznesenskaya et al., 2013; Schüssler et al., 2016): H. scoparia, S. oppositifolia, and S. soda showed C4 values, while S. webbii and both populations of S. divaricata had C3-like stable carbon isotope values. Based on anatomical findings and gas exchange measurements, S. divaricata was previously shown to conduct C2 photosynthesis (Voznesenskaya et al., 2013), which is hardly detectable in stable carbon isotope measurements.
Gene expression broadly reflects photosynthesis type in salsoleae
Intriguingly, the first principal component of a PCA based on the gene expression data separated the species based on photosynthesis types, with both C2 S. divaricata samples lying in between the three C4 species and C3 S. webbii. Thus, other variables, for instance different life cycles (S. soda: annual vs. all other included species: perennial) or sampled organ (H. scoparia: stem vs. leaf in all others), seemed to be of minor importance for differences in gene expression, whereas photosynthesis type was the dominant factor. By comparison, in a PCA of gene expression data of several closely related species of Flaveria with different photosynthesis types, the first three components explained only 27% of the total variance (Mallmann et al., 2014). In contrast, the first three components in our own PCA explained about 73% of the total variance. On the one hand this difference could imply a lower gene regulatory diversity among Salsoleae species than among species of Flaveria. However, the older evolutionary age of Salsoleae would argue against this assumption. Another explanation might be that other confounding factors influenced gene expression in the Flaveria study, while in Salsoleae the different photosynthesis types largely shaped the major gene expression pattern. This is even true for the C4 taxa H. scoparia, S. oppositifolia, and S. soda, which differ from each other morphologically and physiologically: e.g., H. scoparia has completely reduced leaves and photosynthesis is taken over by the shoots, and S. soda has an annual life cycle while the other two are perennial. Also, H. scoparia and S. soda convert from C3 during cotyledon stage to C4 in mature stage (Lauterbach et al., 2016). In terms of C4 photosynthesis, however, the three species are alike in most characters. All have Salsoloid leaf (or stem) anatomy, which is the typical C4 anatomy in this group (Voznesenskaya et al., 2013; Lauterbach et al., 2016; Schüssler et al., 2016). Also, all belong to the NADP-ME subtype (Akhani et al., 2007; current study). Gene expression levels of most C4 genes were thus highly abundant, and at the same time expression of most of the major photorespiratory enzymes was significantly decreased in all three C4 species compared to C3 S. webbii. This important result is in full agreement with studies comparing closely related C3 and C4 species in spiderflowers (Cleome sensu lato, Bräutigam et al., 2011; Aubry et al., 2014; Külahoglu et al., 2014) and Flaveria (Gowik et al., 2011).
Transcriptome analysis reveals specific features of the photosynthesis phenotypes in salsoleae
One key enzyme of C4, PEPC, is known to be present in the plant genome in multiple copies, and while different isoforms function in different tissues, only one is recruited into the C4 cycle (Westhoff and Gowik, 2004). Here, we provide evidence that PEPC1 is the C4-specific isoform in Salsoleae. PEPC1 is one of the top-expressed genes in H. scoparia, S. oppositifolia, and S. soda and about 25-times less expressed in S. webbii, which is in accordance with high PEPC expression in other phylogenetically distant C4 species like Flaveria bidentis (Gowik et al., 2011). In agreement with these findings, leaves of S. oppositifolia were recently shown to have high levels of PEPC protein (Schüssler et al., 2016). Only few amino acid changes in exon 9 of PEPC1 seem to play a major role in altering the efficiency of the enzyme toward its function in the C4 pathway (Paulus et al., 2013). Inspection of the PEPC1 exon 9 amino acid sequence indicates that C4-typical changes, e.g., alanine to serine at position 780 (Zea mays; Bläsing et al., 2000) or 774 (Flaveria) and phenylalanine to valine at position 794 (Z. mays; Christin et al., 2007; Rosnow et al., 2014) are expectedly not present in the C3 and C2 Salsoleae species studied here, present in the two C4 species H. scoparia and S. oppositifolia, but are surprisingly absent in the C4 species S. soda. The entire amino acid sequence of PEPC1 exon 9 of S. soda is more similar to those of the C3 and C2 species than to the other studied Salsoleae C4 taxa. This indicates that either the PEPC efficiency in C4 function is improved by alternative amino acid changes (as described for Suaedoideae; Rosnow et al., 2015) or that the high expression of PEPC1 in the mesophyll cells is already sufficient for C4 function. PEPC1 has also been shown to be the C4-specific isoform in other C4 lineages of Caryophyllales, while e.g., in Flaveria (Asterales) a different PEPC isoform was recruited into the C4 cycle (Sunil et al., 2014).
Unexpectedly, the gene encoding the P-protein of the glycine cleavage system, GDC-P, which normally is involved in CO2 release in photorespiration, was expressed at a considerable amount not only in the C3 and C2 species but in all five Salsoleae species irrespective of the photosynthetic type. We therefore hypothesize that while most enzymes of the photorespiratory cycle should be absent from the mesophyll of C4 species because RuBisCO and the toxic products of its oxygenation reaction are lacking, others like GDC could still be present given their dual role also in C1 metabolism (Parys and Jastrzebski, 2008; Schulze et al., 2016).
Based on gene expression data, Lauterbach et al. (2016) speculated that the proteins encoded by PHT4.1 and PHT4.4 might be involved in the C4 syndrome, at least in S. soda. PHT4.1 and PHT4.4, which may function as ascorbate transporters, could play a role in protection against reactive oxygen species stress resulting from photostress in the mesophyll cells of NADP-ME C4 species by providing cells with the antioxidant ascorbate (Miyaji et al., 2015; Lauterbach et al., 2016). Consistent with the high expression level of PHT4.1 and PHT4.4 in the C4 species S. soda (Lauterbach et al., 2016), a significant increase of transcript abundance of these two genes was also observed in the two other C4 species, H. scoparia and S. oppositifolia (current study). Intriguingly, PHT4.1 transcripts were highly and PHT4.4 transcripts slightly more abundant in C2 S. divaricata, too. Thus, the results of the current study support a proposed function of the two PHT4 family proteins related to both C4 and C2 photosynthesis.
Asparagine synthetase, ASN, was found to be highly expressed in C4 leaves of S. soda when compared to C3 cotyledons of the same species (Lauterbach et al., 2016). ASN functions in ammonium metabolism, and asparagine is a key compound for nitrogen transport (Lam et al., 1996). Therefore, a possible functional connection between nitrogen metabolism and the switch from C3 to C4 was postulated (Lauterbach et al., 2016). In the current study, all C4 species showed high levels of transcripts encoding ASN, nevertheless transcript abundance in S. soda was much higher compared to the other species. In Flaveria, however, no significant differences of ASN expression between C3 F. pringlei and F. robusta, C2 F. ramosissima, and C4 F. bidentis and F. trinervia could be detected (see Supplementary Dataset 1, Gowik et al., 2011). In Cleomaceae on the other hand, ASN was also significantly upregulated in C4 Gynandropsis gynandra from leaf stage 3 onwards (i.e., leaves of an age 6 days onwards) compared to closely related C3 Tarenaya hassleriana (see Supplementary Datasets, Külahoglu et al., 2014). Thus, further investigation of up-regulation of ASN transcription in C4 species of Salsoleae might be worthwhile.
The PCA showed S. divaricata Pop-198 somewhat closer to the C4 species than S. divaricata Pop-184. Assuming that C2 species are true evolutionary intermediates with a gradual increase of C4-ness from C3 to C4 (Sage et al., 2012; Bräutigam and Gowik, 2016), gene expression profiles of the two populations of S. divaricata could imply that S. divaricata Pop-198 is closer to C4 than Pop-184. However, only two populations were studied and the variation of transcript abundance might as well reflect a generally higher plasticity of photosynthetic gene expression in C2 species. In a Flaveria study, a higher expression of NADP-ME was already present in more C3-like intermediate species, which linearly increased in the more advanced intermediates and peaked in the C4 species (Mallmann et al., 2014). This may be taken to indicate a more advanced C4 cycle in Pop-198 than in Pop-184, because NADP-ME is significantly more abundant in Pop-198. Nevertheless, a wide range of C4 genes, for example the first enzyme of the C4 cycle, PEPC, were highly expressed in both populations, implying a low-level but detectable C4 cycle in S. divaricata in general. However, a C4 cycle should result in less negative carbon isotope values; a result that was not observed in the current study. Transcript abundance of most of the core enzymes of photorespiration were highly increased in S. divaricata compared to both C3 and C4 species, which is consistent with expression patterns in C2 species of Flaveria (Gowik et al., 2011; Mallmann et al., 2014) and confirms the successful establishment of the C2 cycle in S. divaricata. These gene expression results thus complement anatomical findings, results of in situ immuno-localization with an antibody against GDC-P, and results of gas exchange measurements of S. divaricata, that all together identified S. divaricata as C2 species (Voznesenskaya et al., 2013).
C4 photosynthesis has been subdivided into three subtypes based on the main acting decarboxylases: NAD-ME, NADP-ME and PEP-CK. Recently, it was suggested to consider the PEP-CK type as a supplementary pathway to either NAD-ME or NADP-ME (Wang et al., 2014) because PEP-CK-only decarboxylation could not be observed. In subfamily Salsoloideae, the NAD-ME subtype is known from C4 species of the Caroxyloneae, and the NADP-ME subtype is known from C4 species of the Salsoleae (Akhani et al., 2007), whereas PEP-CK activity related to C4 photosynthesis has so far not been reported in this group. Interestingly, while according to their transcript patterns all of the three studied Salsoleae C4 species exclusively use NADP-ME as decarboxylase, we found PEP-CK abundant in both populations of S. divaricata. In Flaveria, PEP-CK was lowly expressed in C2 species and no differences in gene expression between the different photosynthetic types could be found (Gowik et al., 2011; Mallmann et al., 2014). In two C2 species of Moricandia, however, transcripts encoding PEP-CK showed enhanced abundance compared to C3 Moricandia moricandioides (Schlüter et al., 2016a). Also, Ala-AT transcripts were not increased in C2 Moricandia and NADP-ME transcripts only slightly enhanced (Schlüter et al., 2016a). Likewise, we observed that Ala-AT transcript abundance was not higher in S. divaricata and augmented transcript abundance of the decarboxylase NADP-ME was only observed in one population (Pop-198) when compared to S. webbii. It is tempting to speculate on a possible function of high PEP-CK abundance in a low level C4 cycle in the C2 species Moricandia arvensis, M. suffruticosa, and S. divaricata. One would expect, however, that genes encoding other proteins involved in the PEP-CK type C4 cycle, such as Ala-AT and Asp-AT, were also highly abundant, which at least in case of S. divaricata was not observed. High abundance of PEP-CK is inconsistent with a study investigating the decarboxylation enzymes of several plant groups, including few species of Salsoleae (Koteyeva et al., 2015). In this study, only low PEP-CK activity and PEP-CK protein abundance (using immunostaining) were found in S. divaricata (Koteyeva et al., 2015). These contrasting results could have different reasons. First, neither of the three anti-PEP-CK antibodies (anti-Megathyrsus maximum, anti-Oryza sativa, anti-Ananas comosus; Koteyeva et al., 2015) was able to bind to the PEP-CK gene that was upregulated in the current study. Second, post-transcriptional or post-translational regulation of PEP-CK could result in low abundance of PEP-CK protein. Third, variation in PEP-CK transcript abundance among populations of S. divaricata could occur. The latter explanation is at least partly supported because in the two studied populations transcript abundance of PEP-CK differed three-fold. Also, plasticity in the decarboxylation biochemistry of a supplemental PEP-CK pathway has been hypothesized (Furbank, 2011) and predicted in a modeling analysis (Wang et al., 2014) as well as observed in the C4 species maize (Pick et al., 2011) and C4 G. gynandra (Sommer et al., 2012). Information about the localization of PEP-CK within the leaf could clarify a possible function of PEP-CK in S. divaricata, and also in C3-C4 Moricandia species.
Conclusion
The overall gene expression pattern, as visualized by PCA, showed photosynthetic type as the main factor separating the different Salsoleae species. C2 type S. divaricata was intermediate between C3 S. webbii and the three C4 species. Despite major differences in life cycle, habit and photosynthesis during seedling stage, the C4 species showed similar gene expression profiles of C4 genes and photorespiratory genes. Most C4 genes were highly abundant in all three C4 species. Further, our results suggested that PEPC1 was the C4-specific isoform in Salsoleae, as found for other C4 lineages of Caryophyllales. The protein, however, lacks C4 typical amino acid changes in S. soda. Two recently proposed transporters from the PHT4 protein family might not only be involved in C4 photosynthesis, but also be active in C2 photosynthesis in Salsoleae. Here, they might be involved in protection against reactive oxygen species resulting from the absence of most photorespiratory reactions in the mesophyll. The transcript profile of the C2 species S. divaricata was mostly comparable with that observed in C2 species of Flaveria or Moricandia. Moreover, in one population of S. divaricata, Pop-198, the transcript pattern of C4 genes implied a slightly more advanced C4 cycle than in Pop-184. However, a C4 cycle is not detectable in the carbon isotope values, and this species is not functioning as a C4 plant physiologically (Voznesenskaya et al., 2013). Also, a function of PEP-CK in C2 photosynthesis in S. divaricata is likely, because PEP-CK was highly increased compared to C3 and C4 species of Salsoleae, a result that was also observed in C2 Moricandia, but needs further investigations.
Author contributions
ML designed the research, performed experiments, analyzed and interpreted the data, and wrote the manuscript. HS analyzed and interpreted the data, and critically revised the manuscript. KB performed experiments. TH designed the research and critically revised the manuscript. PW designed the research and critically revised the manuscript. UG designed the research, analyzed and interpreted the data, and critically revised the manuscript. GK conceived and designed the research, interpreted the data, and critically revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Millenium Seed Bank (MSB Kew), Jaime Gil González (Lanzarote), Magui Olangua Corral (Gran Canaria), Helmut Freitag (Kassel) and Elena Voznesenskaya (St. Petersburg) for the contribution of seed samples. We thank the “Genomics and Transcriptomics laboratory” of the “Biologisch-Medizinisches Forschungszentrum” (BMFZ) at the Heinrich-Heine-University Duesseldorf (Germany) for technical support and conducting the Illumina sequencing. Parts of this research were performed using the supercomputer Mogon and/or advisory services offered by Johannes Gutenberg University Mainz (hpc.uni-mainz.de), which is a member of the AHRP and the Gauss Alliance e.V. We gratefully acknowledge the support of Dr. Christian Meesters and Benjamin Rieger (Johannes Gutenberg University Mainz, Germany) in providing tools and guidance through high parallel computing of de novo transcriptome assembly and data processing. We thank C. Wild (Botanical Garden Univ. Mainz) for cultivating the plants. We also thank three reviewers for their constructive comments which greatly helped to improve the manuscript.
Footnotes
Funding. This work was funded by the Deutsche Forschungsgemeinschaft with grants to GK (KA1816/7-1, KA1816/9-1) and the Cluster of Excellence on Plant Sciences (CEPLAS) to KB and PW. TH acknowledges support by the University Center for Computational Sciences in Mainz (CSM).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2017.01939/full#supplementary-material
Details of de novo transcriptome assemblies using SOAPdenovo-Trans and Trinity.
Mapping of reads, transcript abundance and statistical tests of differentially gene expression using edgeR. Key C4 genes and core enzymes of photorespiration are annotated in column B and C. Hs, Hammada scoparia; Sd184, S. divaricata Pop-184; Sd198, S. divaricata Pop-198; So, S. oppositifolia; Ss, S. soda; Sw, S. webbii.
References
- Akhani H., Edwards G. E., Roalson E. H. (2007). Diversification of the Old World Salsoleae s.l. (Chenopodiaceae): molecular phylogenetic analysis of nuclear and chloroplast data sets and a revised classification. Int. J. Plant Sci. 168, 931–956. 10.1086/518263 [DOI] [Google Scholar]
- Akhani H., Khoshravesh R., Malekmohammadi M. (2016). Taxonomic novelties from Irano-Turanian region and NE Iran: Oreosalsola, a new segregate from Salsola s.l., two new species in Anabasis and Salvia, and two new combinations in Caroxylon and Sesel. Phytotaxa 249, 159 10.11646/phytotaxa.249.1.7 [DOI] [Google Scholar]
- Akhani H., Trimborn P., Ziegler H. (1997). Photosynthetic pathways in Chenopodiaceae from Africa, Asia and Europe with their ecological, phytogeographical and taxonomical importance. Plant Syst. Evol. 206, 187–221. 10.1007/BF00987948 [DOI] [Google Scholar]
- Aubry S., Aresheva O., Reyna-Llorens I., Smith-Unna R. D., Hibberd J. M., Genty B. (2016). A specific transcriptome signature for guard cells from the C4 plant Gynandropsis gynandra. Plant Physiol. 170, 1345–1357. 10.1104/pp.15.01203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S., Kelly S., Kümpers B. M. C., Smith-Unna R. D., Hibberd J. M. (2014). Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10:e1004365. 10.1371/journal.pgen.1004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H., Hagemann M., Fernie A. R. (2010). Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336. 10.1016/j.tplants.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300. [Google Scholar]
- Bläsing O. E., Weston P. H., Svensson P. (2000). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria-a conserved serine residue in the carboxyterminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275, 27917–27923. 10.1074/jbc.M909832199 [DOI] [PubMed] [Google Scholar]
- Bräutigam A., Gowik U. (2016). Photorespiration connects C3 and C4 photosynthesis. J. Exp. Bot. 67, 2953–2962. 10.1093/jxb/erw056 [DOI] [PubMed] [Google Scholar]
- Bräutigam A., Kajala K., Wullenweber J., Sommer M., Gagneul D., Weber K. L., et al. (2011). An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiol. 155, 142–156. 10.1104/pp.110.159442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A., Schliesky S., Külahoglu C., Osborne C. P., Weber A. P. M. (2014). Towards an integrative model of C4 photosynthetic subtypes: insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 65, 3579–3593. 10.1093/jxb/eru100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carolin R. C., Jacobs S. W. L., Vesk M. (1975). Leaf structure in Chenopodiaceae. Botanisches Jahrbuch für Systematische Pflanzengeschichte und Pflanzengeographie 95, 226–255. [Google Scholar]
- Cernusak L. A., Ubierna N., Winter K., Holtum J., A M., Marshall J. D., Farquhar G. D. (2013). Environmental and physiological determinants of carbon isotope discrimination in terrestrial plants. New Phytol. 200, 950–965. 10.1111/nph.12423 [DOI] [PubMed] [Google Scholar]
- Christin P.-A., Osborne C. P., Sage R. F., Arakaki M., Edwards E. J. (2011). C4 eudicots are not younger than C4 monocots. J. Exp. Bot. 62, 3171–3181. 10.1093/jxb/err041 [DOI] [PubMed] [Google Scholar]
- Christin P.-A., Salamin N., Savolainen V., Duvall M. R., Besnard G. (2007). C4 Photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. 17, 1241–1247. 10.1016/j.cub.2007.06.036 [DOI] [PubMed] [Google Scholar]
- Craig H. (1957). Isotopic standards for carbon and oxygen and correction factors for mass-spectrometric analysis of carbon dioxide. Geochim. Cosmochim. Acta 12, 133–149. 10.1016/0016-7037(57)90024-8 [DOI] [Google Scholar]
- Ding Z., Weissmann S., Wang M., Du B., Huang L., Wang L., et al. (2015). Identification of photosynthesis-associated C4 candidate genes through comparative leaf gradient transcriptome in multiple lineages of C3 and C4 species. PLoS ONE 10:e0140629. 10.1371/journal.pone.0140629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm J. C., Minoche A. E., Holtgräwe D., Capella-Gutiérrez S., Zakrzewski F., Tafer H., et al. (2014). The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505, 546–549. 10.1038/nature12817 [DOI] [PubMed] [Google Scholar]
- Edwards G. E., Ku M. S. (1987). Biochemistry of C3-C4 intermediates. Biochem. Plants 10, 275–325. [Google Scholar]
- Ehleringer J. R., Sage R. F., Flanagan L. B., Pearcy R. W. (1991). Climate change and the evolution of C4 photosynthesis. Trends Ecol. Evol. 6, 95–99. 10.1016/0169-5347(91)90183-X [DOI] [PubMed] [Google Scholar]
- Freitag H., Stichler W. (2000). A remarkable new leaf type with unusual photosynthetic tissue in a central asiatic genus of chenopodiaceae. Plant Biol. 2, 154–160. 10.1055/s-2000-9462 [DOI] [Google Scholar]
- Freitag H., Stichler W. (2002). Bienertia cycloptera Bunge ex Boiss., Chenopodiaceae, another C4 Plant without Kranz Tissues. Plant Biol. 4, 121–132. 10.1055/s-2002-20444 [DOI] [Google Scholar]
- Furbank R. T. (2011). Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J. Exp. Bot. 62, 3103–3108. 10.1093/jxb/err080 [DOI] [PubMed] [Google Scholar]
- Gowik U., Bräutigam A., Weber K. L., Weber A. P. M., Westhoff P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell 23, 2087–2105. 10.1105/tpc.111.086264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U., Westhoff P. (2011). The path from C3 to C4 photosynthesis. Plant Physiol. 155, 56–63. 10.1104/pp.110.165308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch M. D. (1987). C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Arch. Biochem. Biophys. 895, 81–106. 10.1016/0003-9861(88)90351-7 [DOI] [Google Scholar]
- Heckmann D., Schulze S., Denton A. K., Gowik U., Westhoff P., Weber A. P. M., et al. (2013). Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a mount fuji fitness landscape. Cell 153, 1579–1588. 10.1016/j.cell.2013.04.058 [DOI] [PubMed] [Google Scholar]
- Hernández-Ledesma P., Berendsohn W. G., Borsch T., Mering S., Von Akhani H., Arias S., et al. (2015). A taxonomic backbone for the global synthesis of species diversity in the angiosperm order Caryophyllales. Willdenowia 45:281 10.3372/wi.45.45301 [DOI] [Google Scholar]
- Honaas L. A., Wafula E. K., Wickett N. J., Der J. P., Zhang Y., Edger P. P., et al. (2016). Selecting superior de novo transcriptome assemblies: lessons learned by leveraging the best plant genome. PLoS ONE 11:e0146062. 10.1371/journal.pone.0146062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadereit G., Bohley K., Lauterbach M., Tefarikis D. T., Kadereit J. W. (2017). C3-C4 intermediates may be of hybrid origin - a reminder. New Phytol. 215, 70–76. 10.1111/nph.14567 [DOI] [PubMed] [Google Scholar]
- Kadereit G., Borsch T., Weising K., Freitag H. (2003). Phylogeny of Amaranthaceae and Chenopodiaceae and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164, 959–986. 10.1086/378649 [DOI] [Google Scholar]
- Kadereit G., Freitag H. (2011). Molecular phylogeny of Camphorosmeae implications for biogeography, evolution of C4 photosynthesis and taxonomy. Taxon 60, 51–78. Available online at: http://www.jstor.org/stable/41059822 [Google Scholar]
- Kadereit G., Lauterbach M., Pirie M. D., Arafeh R., Freitag H. (2014). When do different C4 leaf anatomies indicate independent C4 origins? Parallel evolution of C4 leaf types in Camphorosmeae (Chenopodiaceae). J. Exp. Bot. 65, 3499–3511. 10.1093/jxb/eru169 [DOI] [PubMed] [Google Scholar]
- Kadereit G., Mavrodiev E. V., Zacharias E. H., Sukhorukov A. P. (2010). Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): Implications for systematics, biogeography, flower and fruit evolution, and the origin of C4 photosynthesis. Am. J. Bot. 97, 1664–1687. 10.3732/ajb.1000169 [DOI] [PubMed] [Google Scholar]
- Koteyeva N. K., Voznesenskaya E. V., Edwards G. E. (2015). An assessment of the capacity for phosphoenolpyruvate carboxykinase to contribute to C4 photosynthesis. Plant Sci. 235, 70–80. 10.1016/j.plantsci.2015.03.004 [DOI] [PubMed] [Google Scholar]
- Külahoglu C., Denton A. K., Sommer M., Mass J., Schliesky S., Wrobel T. J., et al. (2014). Comparative transcriptome atlases reveal altered gene expression modules between two cleomaceae C3 and C4 plant species. Plant Cell 26, 3243–3260. 10.1105/tpc.114.123752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kümpers B. M. C., Burgess S. J., Reyna-Llorens I., Smith-Unna R., Boursnell C., Hibberd J. M. (2017). Shared characteristics underpinning C4 leaf maturation derived from analysis of multiple C3 and C4 species of Flaveria. J. Exp. Bot. 68, 177–189. 10.1093/jxb/erw488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago E., Castroviejo S. (1992). Nuevos datos cariológicos de Chenopodiaceae ibéricas. Nova Acta Científica Compostelana (Bioloxía) 3, 201–203. [Google Scholar]
- Lam H.-M., Coschigano K. T., Oliveira I. C., Melo-Oliveira R., Coruzzi G. M. (1996). The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 569–593. 10.1146/annurev.arplant.47.1.569 [DOI] [PubMed] [Google Scholar]
- Langdale J. A. (2011). C4 Cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. 10.1105/tpc.111.092098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbach M., Billakurthi K., Kadereit G., Ludwig M., Westhoff P. (2016). C3 cotyledons are followed by C4 leaves : intra-individual transcriptome analysis of Salsola soda (Chenopodiaceae). J. Exp. Bot. 68, 161–176. 10.1093/jxb/erw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Ma X., Zhao J., Xu J., Shi J., Zhu X.-G., et al. (2015). Developmental genetic mechanisms of C4 syndrome based on transcriptome analysis of C3 Cotyledons and C4 Assimilating Shoots in Haloxylon ammodendron. PLoS ONE 10:e0117175. 10.1371/journal.pone.0117175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löve A. (1970). IOPB chromosome number reports XXVI. Taxon 19, 264–269. Available online at: http://www.jstor.org/stable/1217966 [Google Scholar]
- Löve A. (1986). Chromosome number reports XCI. Taxon 35, 404–410. [Google Scholar]
- Mallmann J., Heckmann D., Bräutigam A., Lercher M. J., Weber A. P. M., Westhoff P., et al. (2014). The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. Elife 2014, 1–23. 10.7554/eLife.02478.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji T., Kuromori T., Takeuchi Y., Yamaji N., Yokosho K., Shimazawa A., et al. (2015). AtPHT4;4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nat. Commun. 6:5928. 10.1038/ncomms6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson R. K., Edwards G. E., Ku M. S. B. (1984). C3-C4 Intermediate Photosynthesis in Plants. Bioscience 34, 563–574. [Google Scholar]
- Monson R. K., Moore B. (1989). On the significance of C3-C4 intermediate photosynthesis to the evolution of C4 photosynthesis. Plant. Cell Environ. 12, 689–699. [Google Scholar]
- Mosyakin S. L., Freitag H., Rilke S. (2017). Kali versus Salsola: the instructive story of a questionable nomenclatural resurrection. Isr. J. Plant Sci. 9978, 1–13. 10.1080/07929978.2016.1256135 [DOI] [Google Scholar]
- O'Leary M. H. (1981). Carbon Isotope Fractionation in Plants. Phytochemistry 20, 553–567. [Google Scholar]
- Padrón M. A. (2012). Caracterización Morfológica y Genética de Especies Arbustivas de los Géneros SALSOLA L. y Suaeda Forssk. ex J. F. Gmel. en las Islas Canarias. [dissertation]. Santa Cruz de Tenerife, Spain: Universidad de La Laguna. [Google Scholar]
- Parys E., Jastrzebski H. (2008). Mitochondria from leaf mesophyll cells of C4 plants are deficient in the H protein of glycine decarboxylase complex. J. Plant Physiol. 165, 1061–1069. 10.1016/j.jplph.2007.07.015 [DOI] [PubMed] [Google Scholar]
- Paulus J. K., Schlieper D., Groth G. (2013). Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4:1518. 10.1038/ncomms2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peruzzi L., Cesca G. (2004). Chromosome numbers of flowering plants from Calabria, S Italy, II. Willdenowia 34, 353–360. 10.3372/wi.34.34203 [DOI] [Google Scholar]
- Pick T. R., Bräutigam A., Schlüter U., Denton A. K., Colmsee C., Scholz U., et al. (2011). Systems analysis of a maize leaf developmental gradient redefines the current C4 model and provides candidates for regulation. Plant Cell 23, 4208–4220. 10.1105/tpc.111.090324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyankov V. I., Ziegler H., Kuz'min A. N., Edwards G. E. (2001). Origin and evolution of C4 photosynthesis in the tribe Salsoleae (Chenopodiaceae) based on anatomical and biochemical types in leaves and cotyledons. Plant Syst. Evol. 230, 43–74. 10.1007/s006060170004 [DOI] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria: Available online at: https://www.R-project.org [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow J. J., Edwards G. E., Roalson E. H. (2014). Positive selection of Kranz and non-Kranz C4 phosphoenolpyruvate carboxylase amino acids in Suaedoideae (Chenopodiaceae). J. Exp. Bot. 65, 3595–3607. 10.1093/jxb/eru053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow J. J., Evans M. A., Kapralov M. V., Cousins A. B., Edwards G. E., Roalson E. H. (2015). Kranz and single-cell forms of C 4 plants in the subfamily Suaedoideae show kinetic C 4 convergence for PEPC and Rubisco with divergent amino acid substitutions. J. Exp. Bot. 66, 7347–7358. 10.1093/jxb/erv431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R. F. (2004). The evolution of C4 photosynthesis. New Phytol. 161, 341–370. 10.1046/j.1469-8137.2004.00974.x [DOI] [PubMed] [Google Scholar]
- Sage R. F. (2016). A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: Species number, evolutionary lineages, and Hall of Fame. J. Exp. Bot. 67, 4039–4056. 10.1093/jxb/erw156 [DOI] [PubMed] [Google Scholar]
- Sage R. F., Sage T. L., Kocacinar F. (2012). Photorespiration and the Evolution of C4 Photosynthesis. Annu. Rev. Plant Biol. 63, 19–47. 10.1146/annurev-arplant-042811-105511 [DOI] [PubMed] [Google Scholar]
- Schlüter U., Bräutigam A., Gowik U., Melzer M., Christin P.-A., Kurz S., et al. (2016a). Photosynthesis in C3-C4 intermediate Moricandia species. J. Exp. Bot. 68, 191–206. 10.1093/jxb/erw391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter U., Denton A. K., Bräutigam A. (2016b). Understanding metabolite transport and metabolism in C4 plants through RNA-seq. Curr. Opin. Plant Biol. 31, 83–90. 10.1016/j.pbi.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Schlüter U., Weber A. P. M. (2016). The Road to C4 Photosynthesis: evolution of a Complex Trait via Intermediary States. Plant Cell Physiol. 57, 881–889. 10.1093/pcp/pcw009 [DOI] [PubMed] [Google Scholar]
- Schulze S., Westhoff P., Gowik U. (2016). Glycine decarboxylase in C3, C4 and C3–C4 intermediate species. Curr. Opin. Plant Biol. 31, 29–35. 10.1016/j.pbi.2016.03.011 [DOI] [PubMed] [Google Scholar]
- Schüssler C., Freitag H., Koteyeva N., Schmidt D., Edwards G., Voznesenskaya E. V., et al. (2016). Molecular phylogeny and forms of photosynthesis in tribe Salsoleae (Chenopodiaceae). J. Exp. Bot. 68, 207–223. 10.1093/jxb/erw432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd K. A., van Leeuwen S. J. (2007). Tecticornia bibenda (Chenopodiaceae: Salicornioideae), a new C4 samphire from the Little Sandy Desert, Western Australia. Nuytsia 16, 383–391. Available online at: https://florabase.dpaw.wa.gov.au/nuytsia/article/471 [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Sommer M., Bräutigam A., Weber A. P. M. (2012). The dicotyledonous NAD malic enzyme C 4 plant Cleome gynandra displays age-dependent plasticity of C4 decarboxylation biochemistry. Plant Biol. 14, 621–629. 10.1111/j.1438-8677.2011.00539.x [DOI] [PubMed] [Google Scholar]
- Sunil M., Hariharan A. K., Nayak S., Gupta S., Nambisan S. R., Gupta R. P., et al. (2014). The draft genome and transcriptome of Amaranthus hypochondriacus: A C4 dicot producing high-lysine edible pseudo-cereal. DNA Res. 21, 585–602. 10.1093/dnares/dsu021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnavschi I. T., Lungeanu I. (1982). Bemerkungen über einige Chromosomenzahlen wildwachsender Arten aus Rumänien. Lucr. Grad. Bot. Bucuresti 1981, 17–20. [Google Scholar]
- van den Bergh E., Külahoglu C., Bräutigam A., Hibberd J. M., Weber A. P. M., Zhu X.-G., et al. (2014). Gene and genome duplications and the origin of C4 photosynthesis: Birth of a trait in the Cleomaceae. Curr. Plant Biol. 1, 2–9. 10.1016/j.cpb.2014.08.001 [DOI] [Google Scholar]
- Voznesenskaya E. V., Akhani H., Koteyeva N. K., Chuong S. D. X., Roalson E. H., Kiirats O., et al. (2008). Structural, biochemical, and physiological characterization of photosynthesis in two C4 subspecies of Tecticornia indica and the C3 species Tecticornia pergranulata (Chenopodiaceae). J. Exp. Bot. 59, 1715–1734. 10.1093/jxb/ern028 [DOI] [PubMed] [Google Scholar]
- Voznesenskaya E. V., Koteyeva N. K., Akhani H., Roalson E. H., Edwards G. E. (2013). Structural and physiological analyses in Salsoleae (Chenopodiaceae) indicate multiple transitions among C3, intermediate, and C4 photosynthesis. J. Exp. Bot. 64, 3583–3604. 10.1093/jxb/ert191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Gribskov M. (2016). Comprehensive evaluation of de novo transcriptome assembly programs and their effects on differential gene expression analysis. Bioinformatics 33, 327–333. 10.1093/bioinformatics/btw625 [DOI] [PubMed] [Google Scholar]
- Wang Y., Bräutigam A., Weber A. P. M., Zhu X.-G. (2014). Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65, 3567–3578. 10.1093/jxb/eru058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn J. D., Bird K. A., Conant G. C., Pires J. C. (2016). Convergent evolution and the origin of complex phenotypes in the age of systems biology. Int. J. Plant Sci. 177, 305–318. 10.1086/686009 [DOI] [Google Scholar]
- Westhoff P., Gowik U. (2004). Evolution of C4 phosphoenolpyruvate carboxylase. Genes and proteins: A case study with the genus Flaveria. Ann. Bot. 93, 13–23. 10.1093/aob/mch003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. P., Johnston I. G., Covshoff S., Hibberd J. M. (2013). Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. Elife 2013, 1–19. 10.7554/eLife.00961.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Wu G., Tang J., Luo R., Patterson J., Liu S., et al. (2014). SOAPdenovo-Trans: De novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30, 1660–1666. 10.1093/bioinformatics/btu077 [DOI] [PubMed] [Google Scholar]
- Zakharyeva O. I. (1985). Chromosome numbers of some flowering plants from the Caucasus and Middle Asia. Bot. Zhurn. SSSR 70, 1699–1701. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of de novo transcriptome assemblies using SOAPdenovo-Trans and Trinity.
Mapping of reads, transcript abundance and statistical tests of differentially gene expression using edgeR. Key C4 genes and core enzymes of photorespiration are annotated in column B and C. Hs, Hammada scoparia; Sd184, S. divaricata Pop-184; Sd198, S. divaricata Pop-198; So, S. oppositifolia; Ss, S. soda; Sw, S. webbii.