Abstract
Pseudouridine (Ψ) is the most abundant posttranscriptional modification in noncoding RNAs. Pseudouridines are often clustered in important regions of rRNAs (ribosomal RNAs), snRNAs (small nuclear RNAs), and tRNAs (transfer RNAs), contributing to RNA function. Pseudouridylation is governed by two independent mechanisms. The first involves single protein enzymes called pseudouridine synthases (PUSs) that alone recognize the substrate and catalyze the isomerization of uridine to pseudouridine (RNA-independent pseudouridylation). The second is an RNA-guided pseudouridylation by a family of box H/ACA RNPs (ribonucleoproteins), each of which consists of a unique RNA (box H/ACA RNA) and four common core proteins (Cbf5/NAP57/Dyskerin, Nhp2/L7Ae, Nop10, and Gar1). The RNA component serves as a guide that base pairs with the substrate RNA and directs the enzyme (Cbf5) to carry out the pseudouridylation reaction at a specific site. The crystal structures of many PUSs have been solved in numerous organisms including E. coli and human. Several partial and complete crystal structures of archaea and yeast box H/ACA RNPs are available, providing a rich source of information regarding the molecular interactions between protein components and box H/ACA RNA. Over the years, several experimental systems have been developed to study the mechanism and function of pseudouridylation. Apart from noncoding RNA pseudouridylation, recent experiments have provided evidence of mRNA pseudouridylation as well. Despite remarkable progress, there is a need to accelerate efforts in order to understand the detailed mechanisms and functions of RNA pseudouridylation.
1. INTRODUCTION
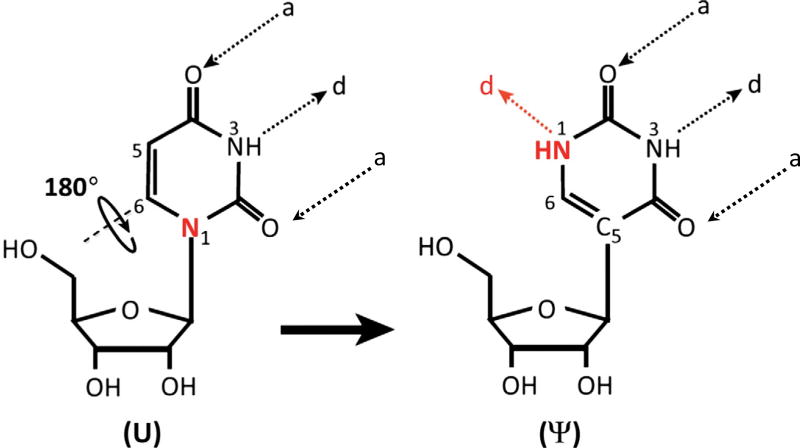
Ribonucleic acids (RNAs) undergo over 140 distinct types of posttranscriptional modifications [1]. Among them, pseudouridylation is the most abundant modification. Pseudouridine was initially found in total RNA hydrolysates of calf liver and considered as the fifth ribonucleoside due to its high abundance. Later it was renamed as pseudouridine, which is derived from the Greek letter psi (Ψ) [2,3]. In the formation of pseudouridine from uridine (pseudouridylation), the nitrogen–carbon (N1–C1′) bond, which links the uracil base to the ribose sugar, is first broken and the liberated uracil base is rotated 180 degree along the N3–C6 axis. The rotated base then establishes a new carbon–carbon (C5–C1′) bond between the base and the sugar (Fig. 1). Pseudouridine gains two new features that differentiate it from uridine. First, the canonical C–N glycosidic bond is changed to a more inert C–C bond [3]. Second, there is an extra hydrogen bond donor at the N1 of the pseudouridine base. These distinctions cause efficient base stacking and water coordination of pseudouridine, thereby increasing the rigidity of the phosohodiester backbone and thermodynamic stability of the Ψ-A base pair compared to U-A base pair [4].
Fig. 1.
The isomerization reaction of uridine to pseudouridine. The bond between the N1 of uridine base (marked with red) and the C1 of ribose sugar, is first broken. The liberated uridine base rotates 180 degree around the N3–C6 axis and attaches to the ribose sugar via a new bond between the C5 of the base and C1 of the ribose sugar. “a” and “d” stand for H-bonds acceptors and donors, respectively.
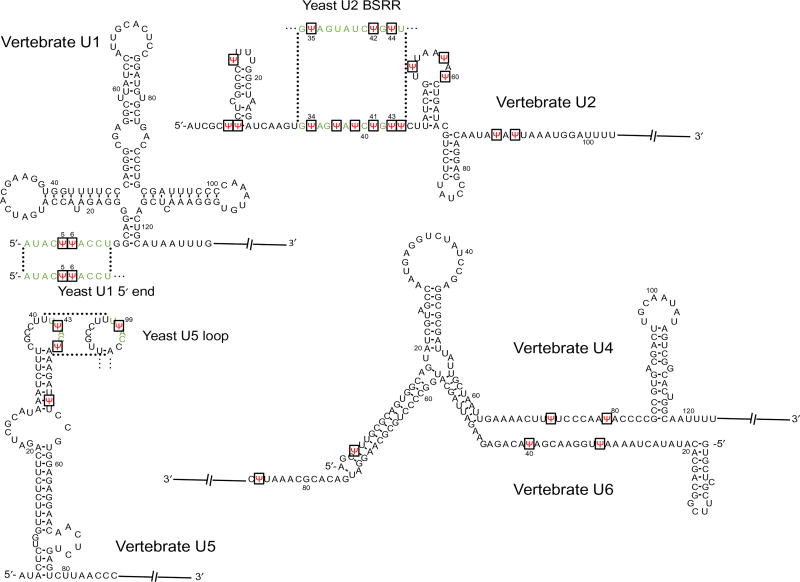
Pseudouridylation mainly occurs in noncoding RNAs, such as snRNAs (small nuclear RNAs), rRNAs (ribosomal RNAs), and tRNAs (transfer RNAs). For example, there are a total of 24 pseudouridines in the 5 vertebrate spliceosomal snRNAs (U1, U2, U4, U5, and U6), and 13 of them are concentrated in U2 snRNA [5]. In Saccharomyces cerevisiae (yeast), a total of six pseudouridines have been identified in snRNAs, including three in U2, two in U1, and one in U5 [6] (Fig. 2). Significantly, pseudouridine residues are concentrated in evolutionarily conserved and functionally important regions of snRNAs. For instance, the two pseudouridines in both vertebrate and yeast U1 snRNA are in the 5′ end region (Ψ5 and Ψ6), which forms base-pairing interactions with the 5′ splice site during spliceosome assembly. There are multiple pseudouridines in the branch site recognition region (BSRR) of yeast and vertebrate U2 snRNA that are involved in base pairing with the pre-mRNA branch site during splicing. There is a Ψ(Ψ43) in the conserved loop of vertebrateU5 snRNA that interacts with the 5′ and 3′ exon sequences. Ψ is also present in yeast U5 snRNA at the equivalent position (Ψ99) within the conserved loop (Fig. 2) [7]. Furthermore, there is experimental evidence available to show that pseudouridines contribute significantly to RNA function. For instance, the loss of pseudouridines at the BSRR of U2 impacts pre-mRNA splicing [8]. In a more detailed study, it has been demonstrated that yeast U2 lacking Ψ42 and Ψ44 shows reduced binding affinity with Prp5, an ATPase required for spliceosome assembly, thus resulting in inefficient spliceosome assembly and splicing [9]. In an exciting observation, pseudouridylation of U6 snRNA at U28 (catalyzed by Pus1p) is important to initiate the filamentous growth in yeast, demonstrating the impact of Ψ on cell growth program and development [10].
Fig. 2.
snRNA sequences of vertebrate and yeast counterparts. The vertebrate U1, U2, U4, U5, and U6 partial sequences with sites of pseudouridylation (red) shown. The important regions of vertebrate snRNAs and their yeast snRNAs counterparts, including the 5′ end region of U1, the BSRR of U2, and the conserved loop sequence of U5, are shown in green, and Ψs in these regions are also indicated in red along with the vertebrate sequences.
In rRNA, there are a large number of pseudouridylation sites as well. For instance, there are approximately 97 pseudouridines in mammalian rRNAs and 46 in yeast rRNAs [11]. As in snRNAs, the pseudouridine sites are clustered in functionally important regions of rRNAs. For instance, multiple Ψs are present in the ribosome peptidyl transferase center and blocking pseudouridylation in this region results in defects in protein translation and cell growth [12]. Furthermore, pseudouridines are abundant in the ribosome decoding center of 18S rRNA and A-site finger region of 25S rRNA; importantly, elimination of Ψs in these regions leads to functional defects [13]. Hypopseudouridylated rRNAs cause impaired ribosome–ligand interactions, resulting in decreased affinity for tRNAs and poor translational fidelity in yeast and mammalian cells [14]. Therefore, the presence of pseudouridine at strategic locations of snRNAs and rRNAs together with the relative high degree of conservation clearly suggests that pseudouridylation plays an important role in pre-mRNA splicing and protein synthesis.
2. MECHANISM OF PSEUDOURIDYLATION
The pseudouridylation of RNA substrates occurs via two main mechanisms. One is an RNA-independent mechanism that involves only a single protein enzyme called pseudouridine synthase (PUS). The other mechanism is an RNA-dependent mechanism involving an RNA–protein complex known as box H/ACA RNP. In prokaryotes, the reaction of pseudouridylation is catalyzed solely by the protein-only mechanism, while both mechanisms appear to coexist in eukaryotic organisms.
2.1 RNA-Independent Mechanism
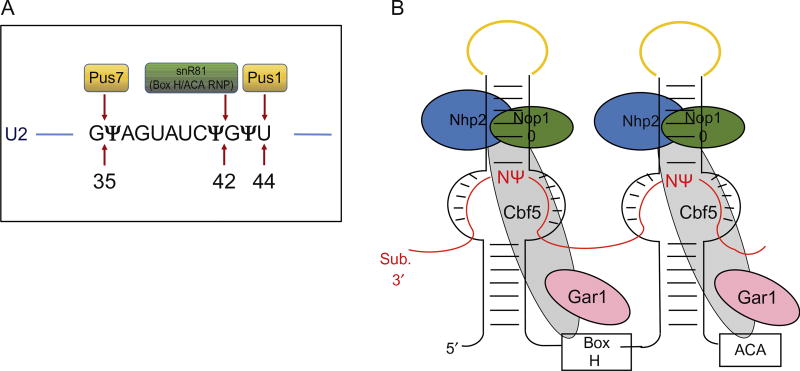
PUSs carry out both substrate recognition and catalysis of the isomerization reaction of uridine to pseudouridine without using any cofactors (Fig. 3A). Depending on the sequence and structure conservation, PUS enzymes are divided into six distinct families, TruA, RluA, RsuA, PUS10, TruB, and TruD, that are named after the founding protein member [15]. Despite the low conservation of structure and sequence, all of these PUS families catalyze the same reaction by using aspartate (Asp) as the nucleophile. The (crystal) structures of some PUS enzymes have been solved (see “The evolution of multi-substrate specificity by RNA modification enzymes” by Alfonzo). However, the exact mechanism of isomerization is poorly understood. It is known that pseudouridylation of tRNA in both eukaryotes and prokaryotes is catalyzed by PUS enzymes. Interestingly, pseudouridylation of some of the yeast snRNAs is catalyzed by both RNA-independent and RNA-dependent mechanisms [16,17]. For example, the Branlant group found that a pseudouridylase called Pus1p, which was earlier identified as a tRNA pseudouridylase, can also catalyze the pseudouridylation of yeast U2 snRNA at position 44 [18]. Later, using a GST-ORF fusion protein library, Ma et al. discovered that the position 35 (U35) of yeast U2 snRNA is also pseudouridylated by a PUS enzyme, Pus7p [19].
Fig. 3.
Schematic of uridine-to-pseudouridine conversion catalyzed by two mechanisms in Saccharomyces cerevisiae. (A). The pseudouridylation catalyzed by a standalone protein pseudouridine synthase 1 (Pus1p) and pseudouridine synthase 7 (Pus7p) in conversion of U44 to Ψ44 and U35 to Ψ35, respectively, in U2 snRNA. Pseudouridylation at position 42 by snR81 box H/ACA RNP is also indicated. (For the structure of a pseudouridine synthase, refer to chapter “The evolution of multi-substrate specificity by RNA modification enzymes” by Alfonzo) (B). The box H/ACA RNP, consisting of a small noncoding box H/ACA guide RNA that folds into a typical hairpin–hinge–hairpin–tail secondary structure forming two hairpins and four common core proteins. The core proteins are centromere-binding factor 5 (Cbf5; known as dyskerin/NAP57 in mammals), glycine–arginine-rich protein 1 (Gar1), nonhistone protein 2 (Nhp2), and nucleolar protein 10 (Nop10). The substrate RNA (indicated in red lines) drapes around the box H/ACA RNP complex via complementary base-pairing interactions with the guiding pockets of box H/ACA RNA, positioning the target uridine, and its 3′ adjacent nucleotide (N) at the base of the upper stem and leaving them unpaired. The apical loops of box H/ACA RNA that hold the CAB boxes are indicated in yellow.
2.2 RNA-Dependent Mechanism
The RNA-dependent pseudouridylation is catalyzed by box H/ACA RNPs, each of which contains an RNA component called a box H/ACA RNA and a set of common core proteins (Fig. 3B). Box H/ACA RNAs are noncoding RNAs that fold into a hairpin–hinge–hairpin–tail secondary structure. The hinge region and the tail region contain evolutionary conserved box H with the consensus sequence “ANANNA” and the trinucleotide Box “ACA,” respectively. The two hairpins each contain an internal loop called pseudouridylation guide pocket, which has a short specific sequence complementary to the substrate RNA. The guiding pockets recognize the sites of modifications through Watson–Crick base-pairing interactions with substrate RNAs, thereby positioning the uridine to be modified at the base of the upper stem and leaving it unpaired. This brings the target uridine 13–16 nucleotides upstream of either box H or box ACA [20]. Although the typical box H/ACA guide RNA consists of two hairpins, it can vary in some organisms. For example, in archaea, the number of hairpins can vary from one to three [21] while in humans there is a box H/ACA RNA that contains a structure of four hairpins [22].
All box H/ACA RNPs examined to date each also contain an evolutionarily conserved set of four core proteins [23]. These proteins are essential for the stability of each other and the stability of box H/ACA RNA. They are also essential for catalysis of pseudouridylation. In humans, the four core proteins are dyskerin (Cbf5p in S. cerevisiae, NAP57 in rats, and Nop60B in Drosophila), Gar1p, Nhp2p (L7Ae in archaea), and Nop10p. All of these core proteins are essential for cell viability and have been characterized in the context of box H/ACA RNA-guided pseudouridylation [24–28]. Among the four proteins, dyskerin (Cbf5p, NAP57, or Nop60B) is the pseudouridylation enzyme that catalyzes the reaction. The pseudouridylase activity of S. cerevisiae Cbf5p was initially identified by mutational analysis [29], and it was later realized that the rat NAP57 and the yeast Cbf5p were homologs with 71% sequence identity [30,31]. Interestingly, Cbf5p is also homologous to the bacterial PUS TruB [29]. Detailed analysis of Cbf5p revealed that it contained a catalytic domain, which is common to all known PUSs, as well as a carboxyl-terminal PUA domain (PUS and archaeosine transglycosylase). Both domains show a high degree of structural conservation among bacterial, archaeal, and yeast pseudouridylases [32,33].
While Cbf5p is the most well-studied protein component, the other three proteins are also important for RNA-guided pseudouridylation. Gar1p is a small protein consisting of two glycine–arginine-rich (GAR) domains that flank either side of the central core domain [34]. Structural analysis shows that Gar1p does not directly interact with box H/ACA RNA but is essential for substrate turnover in the reaction of pseudouridylation [32]. Nhp2p (nonhistone protein) is a small basic protein [23]. Recent fluorescent data on the archaeal homologue of Nhp2p, L7Ae, suggests that it might be involved in the correct positioning of the substrate uridine at the active site of the box H/ACA RNA-guiding pocket [35]. The smallest protein component, Nop10p consists of only 64 amino acids. To date, no recognizable or known motifs have been reported in Nop10p, but it has been shown that only Nop10p and Cbf5p proteins alone display a very low level of pseudouridylation activity when incubated with a box H/ACA guide RNA and a complementary substrate RNA. This low level of pseudouridylation was significantly increased when L7Ae protein was present [36].
Box H/ACA RNPs that are involved in the pseudouridylation of rRNAs are localized to the nucleolus (snoRNPs; small nucleolar RNP), whereas the box H/ACA RNPs that guide the pseudouridylation of spliceosomal snRNAs are localized to the Cajal bodies (scaRNPs; small Cajal body-specific RNP) [37]. The nucleolar targeting of box H/ACA snoRNPs requires an intact box H and box ACA together called the box H/ACA motif [38]. The localization of scaRNPs into Cajal bodies involves an additional element, called the CAB box, in the 5′ and 3′ apical loops of box H/ACA RNA, and a CAB-binding protein [39,40].
In catalyzing pseudouridylation, all pseudouridylation enzymes, including RNA dependent (box H/ACA RNPs) and independent (PUS), use a universally conserved aspartic acid at the active site to nucleophilically attack the target uridine. The site of attack, whether at the base (C6) or at the ribose (C1′), was under debate [41]. However, the structural analysis of archaeal box H/ACA RNP revealed a closer proximity of attacking aspartate to the C6 of the uridine base [42,43]. On the other hand, how the separation happens between the base and the sugar and how the rotation of the base occurs remain known. A thumb-loop element in the catalytic domain appears to play an important role in both substrate RNA binding and turnover [44,45].
3. STRUCTURE OF BOX H/ACA RNP
The crystal structure of many PUSs, derived from bacteria, archaea, and yeast, has been solved. The bacterial TruB, complexed with a part of tRNA, was the first to be crystallized [46]. Then, several partial and complete structures of archaea and yeast box H/ACA RNP with or without substrate RNA were solved [32,35,44,47–51]. For instance, the crystal structure of Pyrococcus furiosus (Pf) H/ACA RNP, assembled with a single-hairpin H/ACA RNA, was solved. The upper stem of guide RNA binds to Cbf5p, Nop10p, and L7Ae (Nhp2p) while the lower stem and ACA motif are bound to the PUA domain of Cbf5p. Gar1p directly contacts Cbf5p and is important for substrate turnover (loading and release) through controlling the confirmation of thumb-loop of Cbf5p. These interactions between the protein components and the lower stem, upper stem, or the H/ACA motif, specifically position the catalytic domain of Cbf5p over the pseudouridylation guide pocket, forming a molecular bracket [32,44,50,51].
The structure of eukaryotic box H/ACA RNP was poorly understood. However, recently the crystal structure of yeast box H/ACA RNPs with two-hairpin H/ACA RNA and recombinant proteins Cbf5p, Nop10p, Gar1p, and Nhp2p has been reconstituted, displaying a general similarity to that of archaea [33]. However, there are some major differences between the two structures, specifically in the region around the upper stem of the box H/ACA RNA. In archaea, there is a K-turn motif bound by L7Ae, which contacts Nop10p, but does not stably associate with Cbf5p–Nop10p in the absence ofRNA[21,32,36,52]. In contrast, no K-turn is present in the upper stem of H/ACA RNA in eukaryotic complex and Nhp2p stably interacts with Cbf5p–Nop10p protein complex [53]. Removal of archaeal L7Ae affects the basic catalytic activity and the substrate turnover, but Nhp2p in yeast seems less important compared to L7Ae because yeast box H/ACA RNP can assemble without Nhp2p and the complex lacking Nhp2 displayed noticeable but reduced activity in multiple-turnover reactions [33].
Even though the mammalian box H/ACA RNP structure is not yet available, a lot of information can be deduced from the available structures of other species, due to high degree of evolutional similarity. Based on the information, the structure of mammalian box H/ACA RNP has been modeled. Accordingly, NAP57 (or Dyskerin) forms the core of the RNP particle with NOP10 and GAR1 binding independently to the catalytic domain of NAP57. NHP2 appears to dock on NOP10 without contacting NAP57. The guide RNA is draped over the core trimer, which consists of NAP57, NOP10, and NHP2 and does not seem to contact GAR1 [6].
4. THREE ELEMENTS REQUIRED FOR BOX H/ACA RNP ACTIVITY
In order to understand the mechanism of uridine-to-pseudouridine conversion, a great deal of effort has been made to dissect the box H/ACA RNP system in detail. Based on the initial work, the conserved box H (in the hinge region) and box ACA (at the tail) appear to be important for proper nucleolar localization as well as for pseudouridylation [38,54]. Later, several reconstitution systems were developed to dissect the minute details in the function of box H/ACA RNP [36,52,55,56]. All these studies identified three important sequence and structural elements that are critical for RNA-guided pseudouridylation by box H/ACA RNA (Fig. 3B). The first required element is the stability of the hairpin structure harboring the pseudouridylation pocket. This includes the upper and lower stems in between the guide pocket (pseudouridylation pocket). The stable base-pairing interactions of these stems are important to maintain a proper guide pocket. The second element is the stability of base-pairing between the guide sequence and the substrate RNA. Stable interactions are necessary to position the target uridine at the catalytic center of pseudouridylase Cbf5p (Dyskerin/NAP57). The third element is the distance between the target uridine and box H or box ACA. Usually this distance is approximately 13–16 nucleotides [57]. This distance not only allows the upper stem of box H/ACA RNA to bind with Cbf5p, Nop10p and Nhp2, and the lower stem and box H or ACA to bind with the PUA domain of Cbf5p, but also establishes direct contact between the catalytic center of Cbf5p and the target uridine [44,55,58].
5. EXPERIMENTAL SYSTEMS TO STUDY PSEUDOURIDYLATION
Owing to the importance and abundance of the spliceosomal snRNA pseudouridylation, a great effort has also been made to elucidate the function of snRNA pseudouridylation. As a result, several reconstitution systems have been developed, including the mammalian in vitro system, the Xenopus oocyte microinjection system, and the yeast system.
5.1 The Mammalian in vitro System
In the early 1990s, Jeffery Patton carried out the first functional analysis of U2 snRNA modification using nuclear extracts prepared from HeLa cells. In these experiments, he showed that in vitro synthesized spliceosomal snRNA, when incubated with nuclear extracts under appropriate conditions, could be efficiently pseudouridylated. Interestingly, he demonstrated that 5-fluorouridine can block pseudouridylation when it is incorporated into U2 snRNA at the sites of pseudouridylation. Using this knowledge, he carried out a series of in vitro experiments and showed that the inhibition of pseudouridylation of spliceosomal snRNA impaired snRNP assembly, suggesting an important role of pseudouridylation in snRNP biogenesis and pre-mRNA splicing [59,60]. Later, in 2004, using functional reconstitution, the Luhrmann group provided experimental evidence suggesting that U2 snRNA pseudouridylation might indeed be important for pre-mRNA splicing. In this study, the endogenous U2 snRNP was depleted from HeLa cell nuclear extracts using antisense oligonucleotides and reconstituted using in vitro synthesized or cellularly derived U2 snRNA [61]. Using this strategy, they later identified Ψ6, Ψ7, and Ψ15 as important pseudouridines for pre-mRNA splicing [62].
5.2 The Xenopus Oocyte Microinjection System
The Xenopus oocyte microinjection system specifically relies on the fact that an endogenous spliceosomal snRNA can be depleted from oocytes upon injection of an antisense oligonucleotide that is complementary to the target snRNA, thereby directing an endogenous RNase H to degrade the RNA strand (snRNA) of the RNA–DNA hybrids. The antisense oligonucleotide itself is later degraded by an endogenous DNase activity. The depleted snRNA can be restored by injecting the respective in vitro synthesized snRNA. After a short reconstitution period pre-mRNA splicing can be assayed. Using this system, Yu et al. demonstrated that in vitro transcribed (and thus unmodified) U2 snRNA did not rescue pre-mRNA splicing in U2-depleted Xenopus oocytes, while the cellularly derived (and therefore modified) U2 effectively restored the splicing activity in the same oocytes [63]. Furthermore, using a series of chimeric U2 snRNAs that have both modified and unmodified parts, they identified many pseudouridines in the 5′ end region and in the BSRR of U2 snRNA to be important for splicing [63,64].
5.3 The Yeast System
In S. cerevisiae U2 snRNA, there are three pseudouridines at the BSRR. Ψ35 and Ψ44 are modified by the standalone protein enzymes Pus7p and Pus1p, respectively. The third site, Ψ42, is modified by snR81, a box H/ACA RNP; Ψ42 is the only RNA-guided modification site in the yeast U2 snRNA [18,19,56]. The identification of three enzymes responsible for pseudouridylation at three sites in U2 snRNA made it possible to carry out a genetic analysis to dissect the function of U2 pseudouridylation in yeast pre-mRNA splicing. The experiments showed that removal of pseudouridines, either individually or in combination, resulted in splicing defects (to various extents), which, in turn, led to growth defects [9,65].
6. INDUCIBLE PSEUDOURIDYLATION
Apart from the constitutive pseudouridylation discussed earlier, it has been shown that pseudouridylation can be induced as well. The first line of evidence on inducible pseudouridylation came from Wu et al., who showed that U2 snRNA in yeast can be pseudouridylated at novel sites upon changes in growth conditions [66]. In this study, they subjected yeast cells to nutrient deprivation stress by growing cells to saturation or using nutrient-depleted media. Total RNA was isolated from stressed cells and U2 pseudouridylation was assayed, and they detected novel modification sites, positions 56 and 93, which were previously identified as unmodified uridines. Further analysis showed that position 56 was pseudouridylated by the standalone protein enzyme Pus7p, which was previously known to constitutively pseudouridylate position 35 (Ψ35) in U2 snRNA. Position 93 was found to be pseudouridylated by the 3′ guide pocket of box H/ACA RNA snR81, which is normally responsible for the constitutive pseudouridylation of U1051 in 25S rRNA [Note: The 5′ guide pocket of snR81 is responsible for the constitutive pseudouridylation of U2 at position 42 (Ψ42)]. Interestingly, the sequences surrounding positions 56 and 93 were not identical but similar to the sequences surrounding the constitutively pseudouridylated sites, positions 35 of U2 and 1051 of 25S rRNA, respectively. Wu et al. further showed that the imperfect base pairing between the guide pocket of snR81 and substrate was necessary for induction of pseudouridylation at position 93 in U2 snRNA under stress conditions [66]. Interestingly, the Pus1p-catalyzed U6 snRNA pseudouridylation at position 28 (U28-to-Ψ28 conversion), which plays an important role in regulating the yeast filamentous growth program, appears to be developmentally induced as well [10].
7. mRNA PSEUDOURIDYLATION
Even though pseudouridylation was previously considered as a posttranscriptional modification that occurs exclusively in noncoding RNAs, Karijolich et al. demonstrated that mRNA can also be pseudouridylated by designer box H/ACA RNPs [67]. They inserted a premature termination codon (PTC) into CUP1 gene in the ACT1–CUP1 reporter mRNA [68] and engineered the yeast snR81 box H/ACA RNA (by changing its guide sequence) to base pair with the ACT1–CUP1 mRNA, thereby specifically targeting the uridine in the PTC for pseudouridylation. They showed that the PTC within the ACT1–CUP1 mRNA (and other mRNAs) can indeed be pseudouridylated [67]. Remarkably, pseudouridylation of PTC resulted in stop codon read through or nonsense suppression.
It had been reported that there are numerous putative box H/ACA RNAs with typical hairpin–hinge–hairpin–tail structure that fail to match any known pseudouridine sites in stable noncoding RNAs. These RNAs have thus been dubbed as “orphan” box H/ACA guide RNAs. However, the fact that designer box H/ACA RNAs can guide mRNA pseudouridylation at specific sites [67] raises a possibility that some of the naturally occurring “orphan” box H/ACA might target mRNA for pseudouridylation [69–71].
Excitingly, several groups recently presented evidence of naturally occurring mRNA pseudouridylation in S. cerevisiae and mammalian cells [72–75]. They developed pseudouridine-seq methods by coupling the conventional CMCT (N-cyclohexyl-N′-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate) modification–primer-extension, which is often used to detect pseudouridines in noncoding RNA, with deep sequencing and bioinformatics analysis. Using these methods, a large number of pseudouridines, ranging from 50–100 in yeast to 100–400 in human mRNAs, were identified. There is no positional bias of mRNA pseudouridylation and the pseudouridine residues were detected within the 5′ and 3′ untranslated regions (UTRs) and coding sequences. Interestingly, however, all the studies were in agreement with the fact that most of the mRNA pseudouridylation is catalyzed by the standalone PUS enzymes and only a few were catalyzed by box H/ACA RNPs. In addition, mRNA pseudouridylation was found to be highly inducible, as evidenced by the fact that the number of pseudouridines in yeast and human mRNAs increases upon nutritional stress and serum starvation, respectively.
8. CONCLUDING REMARKS
In this chapter, we have discussed one of the most abundant posttranscriptional RNA modifications, pseudouridylation. Since the discovery of the box H/ACA RNA family, RNA pseudouridylation has attracted much attention and has been studied extensively. However, even though we have come a long way in the field of RNA pseudouridylation, much still needs to be done to fully understand the mechanism and function of this modification. For instance, it is completely unclear as to whether mRNA pseudouridylation plays a role in mRNA processing and/or protein coding. At the same time, it has been reported that pseudouridylation (or pseudouridylation enzymes) can be linked to a range of diseases such as mitochondrial myopathy and sideroblastic anemia [76], and yet, the detailed roles of pseudouridylation in human diseases remain elusive. However, with recent rapid advances in the field of RNA modification, we are hopeful that some of the major questions related to RNA pseudouridylation will soon be addressed.
Acknowledgments
We thank the members of the Yu laboratory for critical reading of and helpful comments on the manuscript. The work carried out in the Yu laboratory was supported by grant GM104077 from NIH (to Y.-T.Y.).
References
- 1.Machnicka MA, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957;227(2):907–915. [PubMed] [Google Scholar]
- 3.Cohn WE. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim. Biophys. Acta. 1959;32:569–571. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 4.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341051. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 5.Reddy R, Busch H. Small nuclear RNAs: RNA sequences, structure, and modifications. In: Birnstiel ML, editor. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. Springer Berlin Heidelberg; Berlin, Heidelberg: 1988. pp. 1–37. [Google Scholar]
- 6.Yu Y-T, Meier UT. RNA-guided isomerization of uridine to pseudouridine—pseudouridylation. RNA Biol. 2014;11(12):1483–1494. doi: 10.4161/15476286.2014.972855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adachi H, Yu Y-T. Insight into the mechanisms and functions of spliceosomal snRNA pseudouridylation. World J. Biol. Chem. 2014;5(4):398–408. doi: 10.4331/wjbc.v5.i4.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, et al. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA. 2002;8(12):1515–1525. [PMC free article] [PubMed] [Google Scholar]
- 9.Wu G, et al. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J. 2016;35(6):654–667. doi: 10.15252/embj.201593113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep. 2014;8(4):966–973. doi: 10.1016/j.celrep.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA. 2006;12(1):15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.King TH, et al. Ribosome structure and activity Are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell. 2003;11(2):425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 13.Liang X-H, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28(6):965–977. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Jack K, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell. 2011;44(4):660–666. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 2014;11(12):1540–1554. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosjean H. Modification and editing of RNA: historical overview and important facts to remember. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. pp. 1–22. [Google Scholar]
- 17.Decatur WA, Schnare MN. Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Mol. Cell. Biol. 2008;28(10):3089–3100. doi: 10.1128/MCB.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massenet S, et al. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol. Cell. Biol. 1999;19(3):2142–2154. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma X, Zhao X, Yu Y-T. Pseudouridylation (Ψ) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 2003;22(8):1889–1897. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y-T, Terns RM, Terns MP. Mechanisms and functions of RNA-guided RNA modification. In: Grosjean H, editor. Fine-Tuning of RNA Functions by Modification and Editing. Springer Berlin Heidelberg; Berlin, Heidelberg: 2005. pp. 223–262. [Google Scholar]
- 21.Rozhdestvensky TS, et al. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31(3):869–877. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss AM, et al. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30(21):4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terns M, Terns R. Noncoding RNAs of the H/ACA family. Cold Spring Harb. Symp. Quant. Biol. 2006;71:395–405. doi: 10.1101/sqb.2006.71.034. [DOI] [PubMed] [Google Scholar]
- 24.Henras A, et al. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J. 1998;17(23):7078–7090. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watkins NJ, et al. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA. 1998;4(12):1549–1568. doi: 10.1017/s1355838298980761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dragon F, Pogačić V, Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000;20(9):3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20(23):9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe Y-I, Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: Cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res. 2000;28(12):2342–2352. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zebarjadian Y, et al. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell. Biol. 1999;19:7461–7472. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang W, et al. An essential yeast protein, CBF5p, binds in vitro to centromeres and microtubules. Mol. Cell. Biol. 1993;13(8):4884–4893. doi: 10.1128/mcb.13.8.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J. Cell Biol. 1994;127(6):1505–1514. doi: 10.1083/jcb.127.6.1505. (published erratum appears in J. Cell Biol. 140 (2) (1998) 447). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443(7109):302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 33.Li S, et al. Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev. 2011;25(22):2409–2421. doi: 10.1101/gad.175299.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Girard JP, et al. GAR1 is an essential small nucleolar RNP protein required for prerRNA processing in yeast. EMBO J. 1992;11(2):673–682. doi: 10.1002/j.1460-2075.1992.tb05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang B, et al. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nat. Struct. Mol. Biol. 2007;14(12):1189–1195. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- 36.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33(10):3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109(2):145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 38.Narayanan A, et al. Nucleolar localization signals of Box H/ACA small nucleolar RNAs. EMBO J. 1999;18(18):5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richard P, et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22(16):4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tycowski KT, et al. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol. Cell. 2009;34(1):47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spedaliere CJ, et al. The pseudouridine synthases: revisiting a mechanism that seemed settled. J. Am. Chem. Soc. 2004;126(40):12758–12759. doi: 10.1021/ja046375s. [DOI] [PubMed] [Google Scholar]
- 42.Gu X, Liu Y, Santi DV. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. U.S.A. 1999;96(25):14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J, Liang B, Li H. Functional and structural consequences of uridine substitutions on H/ACA ribonucleoprotein particle pseudouridine synthase. Biochemistry. 2010;49(29):6276–6281. doi: 10.1021/bi1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duan J, et al. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol. Cell. 2009;34(4):427–439. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Liang B, et al. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat. Struct. Mol. Biol. 2009;16(7):740–746. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoang C, Ferré-D’Amaré AR. Cocrystal structure of a tRNA Ψ55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107(7):929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 47.Rashid R, et al. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell. 2006;21(2):249–260. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 48.Hamma T, et al. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nat. Struct. Mol. Biol. 2005;12(12):1101–1107. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- 49.Manival X, et al. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5–aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic Acids Res. 2006;34(3):826–839. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye K. H/ACA guide RNAs, proteins and complexes. Curr. Opin. Struct. Biol. 2007;17(3):287–292. doi: 10.1016/j.sbi.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 51.Reichow SL, et al. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35(5):1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baker DL, et al. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes Dev. 2005;19(10):1238–1248. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C, Meier UT. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23(8):1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bortolin ML, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. EMBO J. 1999;18(2):457–469. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao M, et al. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA. 2009;15(1):176–186. doi: 10.1261/rna.1361509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma X. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J. 2005;24:2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 58.Ge J, Yu Y-T. RNA pseudouridylation: new insights into an old modification. Trends Biochem. Sci. 2013;38(4):210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patton JR. Ribonucleoprotein particle assembly and modification of U2 small nuclear RNA containing 5-fluorouridine. Biochemistry. 1993;32(34):8939–8944. doi: 10.1021/bi00085a027. [DOI] [PubMed] [Google Scholar]
- 60.Patton JR. Multiple pseudouridine synthase activities for small nuclear RNAs. Biochem. J. 1993;290(Pt. 2):595–600. doi: 10.1042/bj2900595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ségault V, et al. In vitro reconstitution of mammalian U2 and U5 snRNPs active in splicing: Sm proteins are functionally interchangeable and are essential for the formation of functional U2 and U5 snRNPs. EMBO J. 1995;14(16):4010–4021. doi: 10.1002/j.1460-2075.1995.tb00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DÖNmez G, Hartmuth K, LÜHrmann R. Modified nucleotides at the 5 0 end of human U2 snRNA are required for spliceosomal E-complex formation. RNA. 2004;10(12):1925–1933. doi: 10.1261/rna.7186504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998;17(19):5783–5795. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA. 2004;10:681–690. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang C, McPheeters DS, Yu Y-T. ψ35 in the branch site recognition region of U2 small nuclear RNA Is important for Pre-mRNA splicing in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280(8):6655–6662. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 66.Wu G, et al. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J. 2011;30(1):79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karijolich J, Yu Y-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–398. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133:851–863. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hüttenhofer A, et al. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20(11):2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vitali P, et al. Identification of 13 novel human modification guide RNAs. Nucleic Acids Res. 2003;31(22):6543–6551. doi: 10.1093/nar/gkg849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kiss AM, et al. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24(13):5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carlile TM, et al. Pseudouridine profiling reveals regulatedmRNApseudouridylation in yeast and human cells. Nature. 2014;515(7525):143–146. doi: 10.1038/nature13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwartz S, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159(1):148–162. doi: 10.1016/j.cell.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li X, et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015;11(8):592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- 75.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One. 2014;9(10):e110799. doi: 10.1371/journal.pone.0110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bykhovskaya Y, et al. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA) Am. J. Hum. Genet. 2004;74(6):1303–1308. doi: 10.1086/421530. [DOI] [PMC free article] [PubMed] [Google Scholar]