Summary
Mononuclear phagocytes are a heterogeneous family that occupy all tissues and assume numerous roles to support tissue function and systemic homeostasis. Our ability to dissect the roles of individual subsets is limited by a lack of technologies that ablate gene function within specific mononuclear phagocyte sub-populations. Using Nr4a1-dependent Ly6Clow monocytes we present a proof-of-principle approach that addresses these limitations. Combining ChIP-Seq and molecular approaches we identified a single, conserved, sub-domain within the Nr4a1 enhancer that was essential for Ly6Clow monocyte development. Mice lacking this enhancer lacked Ly6Clow monocytes but retained Nr4a1 gene expression in macrophages during steady state and in response to LPS. As Nr4a1 regulates inflammatory gene expression and differentiation of Ly6Clow monocytes, decoupling these processes allows Ly6Clow monocytes to be studied independently.
Graphical abstract
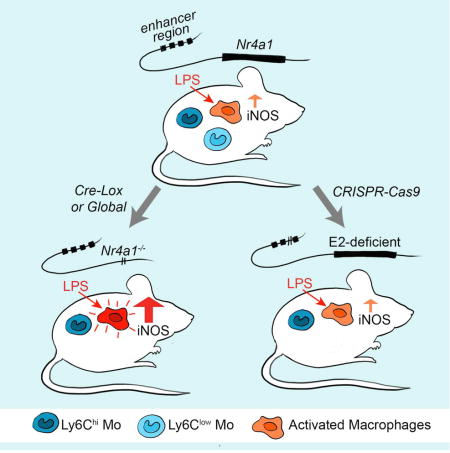
Introduction
Mononuclear phagocytes (MP) are an ontologically diverse family of cells comprised of macrophages and monocytes (Perdiguero and Geissmann, 2016). From a traditional standpoint the primary functions of MP involve recognition and clearance of invading pathogens, maintaining tissue integrity and resolving inflammation (Sica and Mantovani, 2012). To perform these diverse functions MP occupy all bodily tissues and display a tremendous degree of phenotypic plasticity. In recent years transcriptomic profiling efforts have revealed the full extent of this heterogeneity, and functional studies have unveiled diverse, specialized and often unexpected roles for individual MP subsets (Amit et al., 2016). In this context, tissue macrophages and blood monocytes may be considered accessory cells that allow optimal performance of the host tissue (Okabe and Medzhitov, 2016). In complement to their roles in immunity and host defense, tissue MP are now also recognized as major regulators of higher-order physiological processes including systemic energy balance (Odegaard and Chawla, 2011), intestinal peristalsis (Muller et al., 2014) and cognitive function (Parkhurst et al., 2013). Delineating the full extent of MP functions in health and disease represents a largely unexplored frontier in both immunology and physiology.
Monocytes constitute the blood-borne phase of the MP system and are composed of at least two subsets in both mouse and human that are believed to be conserved (Cros et al., 2010). Mouse monocyte subsets can be discriminated using the Ly6C surface antigen (Geissmann et al., 2010), Ly6Chi monocytes are progenitors of inflammatory, and some tissue-resident macrophages. Ly6Clow monocytes exhibit a patrolling behavior on the vascular endothelium and contribute to vascular homeostasis by maintaining the endothelial layer (Carlin et al., 2013). In addition, Ly6Clow monocytes have recently been shown to play a crucial role protecting against the seeding of tumor metastases in the lung (Hanna et al., 2015).
An indispensable approach to understand cellular behavior is through loss-of-function. The identification of lineage defining transcription factors (LDTFs) for MP subsets has led to new insights into the functions of these cells. For example, the transcription factor Nr4a1 is the master regulator of the Ly6Clow monocyte subset (Hanna et al., 2011). Nr4a1 is uniquely highly expressed in Ly6Clow monocytes, and is required in a cell-intrinsic fashion. Accordingly Nr4a1−/− mice were instrumental in revealing a role for Ly6Clow monocytes in tumor metastasis (Hanna et al., 2015). In another example, loss of the transcription factor Spic selectively ablates splenic red pulp macrophages revealing a role for this population in iron homeostasis (Kohyama et al., 2009). Similarly macrophage-specific deletion of Gata6 impairs the maturation of peritoneal macrophages leading to the discovery that these cells modulate B-1 cell IgA production (Okabe and Medzhitov, 2014).
Pleiotropy, exemplified by the differential action of a single gene in multiple cell types, is a major obstacle to understanding cell-specific gene function. To overcome this problem the Cre recombinase-loxP (Cre-Lox) system is routinely used to ablate genes containing loxP-flanked exons by controlling Cre enzyme expression with cell-specific promoters. The ability of the Cre-Lox system to excise gene expression in a cell-specific manner is limited by the cell-specificity of Cre transgenes and the time taken for recombination to occur (Yona et al., 2012). These considerations present problems when using the Cre-Lox system to study gene function within populations of closely related cells. A prime example of this problem is Nr4a1. Nr4a1−/− mice lack Ly6Clow monocytes, macrophage Nr4a1 is also induced by LPS and represses inflammatory gene expression (Hanna et al., 2012). These confounding influences limit the utility of Nr4a1−/− to study Ly6Clow monocytes. Furthermore, the current MP Cre transgenes (Lyz2-cre, Csf1r-cre, Cx3cr1-cre) cannot delete Ly6Clow monocyte Nr4a1 without also disrupting Nr4a1 across MP subsets. These pan-myeloid effects of the current myeloid Cre transgenes limit the ability of conditional deletion approaches to determine gene- and cell-type function within the diverse MP compartment.
Enhancers are genomic sequences that positively influence promoter activity and are critical determinants of signal-dependent and cell-specific gene expression (Andersson et al., 2014). While such elements are rigorously defined by deletion or mutation, they are associated with a distinct chromatin signature that includes enrichment of histone H3 lysine 4 monomethylation (H3K4me1) and (H3K4me2) (Heintzman et al., 2007; Heinz et al., 2010). MP enhancers are enriched for the LDTFs PU.1 and C/EBPβ, which instruct enhancer selection (Heinz et al., 2010). Enhancers are subject to additional regulation leading to their further classification as ‘poised’ or ‘active’. Enhancers are activated upon binding of signal dependent transcription factors (SDTFs), leading to acetylation at H3 lysine 27 (H3K27ac) and the increased expression of associated genes (Creyghton et al., 2010; Heinz et al., 2013). These chromatin features can be used to map enhancer-like regions on a genome-wide scale through the use of ChIP-sequencing. The enhancer landscapes between MP subsets show considerable diversity (Gosselin et al., 2014; Lavin et al., 2014) and result from the myriad environmental niches these populations are exposed to. As Nr4a1 shows unique expression characteristics in Ly6Clow monocytes, we hypothesized a SDTF acting at a cell-specific enhancer regulates transcription of the Ly6Clow monocyte LDTF Nr4a1.
Here we explore this hypothesis by mapping the Nr4a1 enhancer locus in Ly6Clow monocytes. We identified a single sub-domain 4kb upstream of the Nr4a1 transcription start site that was essential for Ly6Clow monocyte development. Dissection of this element provided insight into the transcriptional processes driving Ly6Clow monocyte development. Furthermore, using mice that lack this enhancer we have shown that macrophage Nr4a1 gene expression was unaffected both in response to inflammatory signaling and during steady state.
Results
Ly6Clow monocytes are ontological neighbors to Ly6Chi monocytes
Lineage tracing and BrdU pulse-chase studies have established a consensus that Ly6Chi monocytes give rise to Ly6Clow monocytes (Yona et al., 2012). Yet it remains possible that Ly6Clow monocytes arise independent of the Ly6Chi population. The monocyte dendritic cell precursor (MDP) that expresses Flt3 (CD135), Cx3cr1 and Kit (CD117) gives rise to all mouse monocytes (Hettinger et al., 2013). MDPs undergo further restriction towards the monocyte lineage upon their differentiation into the common monocyte progenitor (cMoP), at which point the expression of FLT3 is lost and Ly6C is gained (Hettinger et al., 2013).
We reasoned that if Ly6Clow monocytes arise independently of the Ly6Chi population that we would observe a signature that was common to the Ly6Clow monocyte and their progenitor, but not the Ly6Chi monocyte. To test this hypothesis we performed ChIP-Seq on MDP, cMoP, Ly6Chi and Ly6Clow monocytes (Figure 1a) using H3K4me2 to define enhancers, and activity with H3K27ac. To assess data quality we visually inspected loci associated with prototypical marker genes for the profiled cell types (Figure 1b). As expected MDPs showed enrichment for H3K27ac at the Flt3 locus whereas both MDPs and cMoPs showed H3K27ac at the Kit locus, both used to sort these populations. In contrast to Ly6Clow monocytes, Ly6Chi monocytes and cMoPs show enhancer activity at the Ly6c2 locus encoding for the Ly6C surface antigen. MDPs also showed activity at the Ly6c2 locus in spite of low mRNA and cell-surface protein expression (Figure 1a,b), perhaps indicating a priming of this locus prior to transcription. Finally, in Ly6Clow monocytes Cx3cr1 shows marked H3K27ac reflecting the robust Cx3cr1 expression in this population (Carlin et al., 2013).
Figure 1. Epigenomic profiling of Mo subsets and progenitors supports the model of Ly6Chi to Ly6Clow Mo conversion.
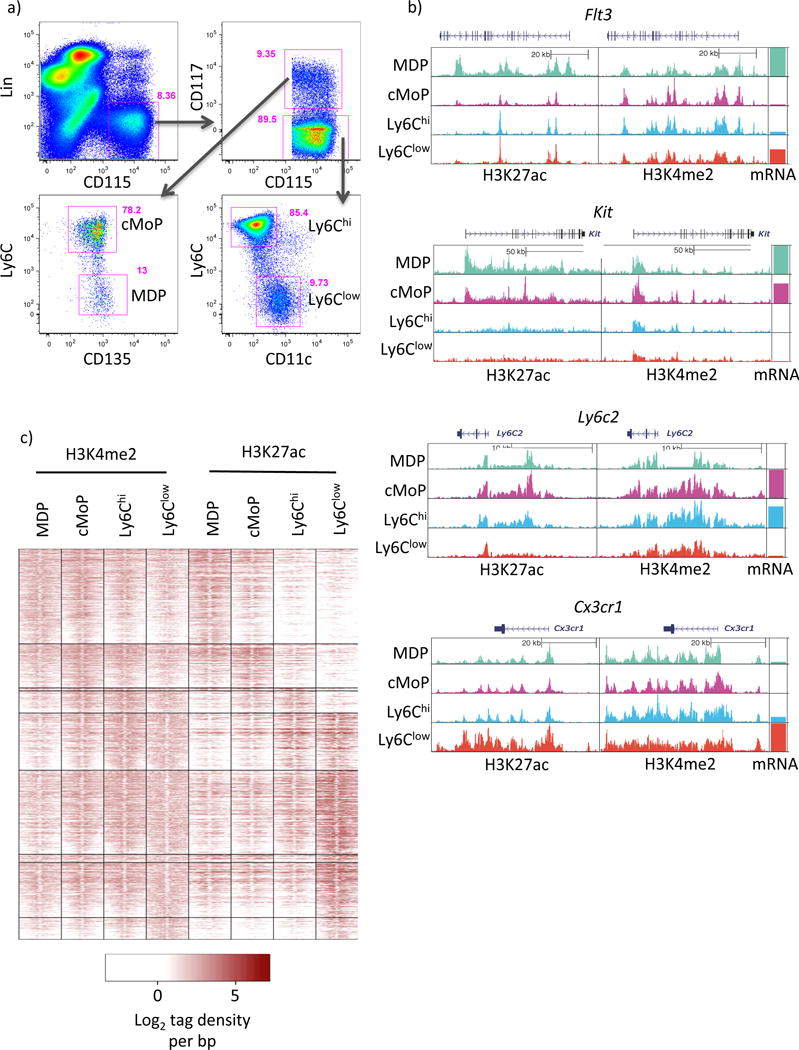
a) Gating strategy used to sort monocytes and upstream progenitors. Cells were previously gated on live singlets (using a FSC-W versus FSC-A gate). b) UCSC genome browser screenshots showing H3K27ac and H3K4me2 tag distributions at key lineage genes. c) Distribution of H3K4me2 and H3K27ac tags ±1kb from PU.1 peak centers in DE enhancers. Please see supplementary figures S1 and S2.
To gain a global overview of enhancer dynamics during monocyte development H3K4me2 and H3K27ac were quantitated at all 99,462 enhancers defined as H3K4me2 enriched regions greater than 2.5kb from the nearest transcription start site. Differential enrichment (DE) of histone marks was then computed between all pairwise comparisons identifying a total of 620 DE H3K4me2 regions (0.62% of total, Figure S1a) and 9,879 H3K27ac regions (9.93% of total, Figure S1b). Unbiased hierarchical clustering of DE enhancers (Figure 1c, S1c) showed that these changes in H3K27ac are mirrored by more subtle shifts in H3K4me2. De novo motif analysis within these defined clusters identified transcriptional processes driving monocyte differentiation (Figure S1d). All enhancer clusters were enriched for ETS (PU.1) motifs. Progenitor-associated enhancers showed restricted over-representation of Myb motifs, which were lost upon Ly6Chi monocyte differentiation, concomitant with the acquisition of CEBP, and KLF motifs. At the global acetylation level Ly6Clow monocytes enhancers were uniquely characterized by over-represented NR4A and Mef2a motifs. Of note we did not observe a signature of enhancer or motif usage that was shared between Ly6Clow monocytes and either progenitor but not Ly6Chi monocytes, nor did such a pattern emerge at the transcriptomic level (Figure S2). Instead, the patterns of differential enhancer usage were consistent with a continuous transition between the MDP and Ly6Clow monocytes with both the cMoP and Ly6Chi monocytes falling as intermediaries in this cascade. Collectively, these data strongly support the consensus model of monocyte development in which Ly6Clow monocytes derive directly from the Ly6Chi monocyte population.
Ly6Clow monocytes possess a cell-specific super-enhancer at the Nr4a1 locus
Super-enhancers (SE) are a recently defined class of highly cell-type specific enhancer (Whyte et al., 2013) that selectively mark genes involved in lineage specification and function. The defining features of SEs include their extended size, adornment with LDTFs and genomic correlates of high gene expression (Hnisz et al., 2013). As Ly6Clow monocytes highly express Nr4a1 and require it for their development (Hanna et al., 2011) we postulated that it is a SE regulated gene. We used H3K27ac to define SEs in Ly6Clow monocytes, confirming that Nr4a1 does overlap a SE (Nr4a1se; Figure 2a and S2). Validating the SE characteristics of Nr4a1se, we also observed a high density of the MP LDTFs PU.1 and CEBPβ (Figure S3). Enumeration of H3K4me2 and H3K27ac tag counts at Nr4a1se in MDP, cMoP, Ly6Chi and Ly6Clow monocytes revealed a selective and robust (5-fold) acquisition of H3K27ac in Ly6Clow monocytes, demonstrating that Nr4a1se is a Ly6Clow monocyte-specific SE region (Figure 2b, c).
Figure 2. Ly6Clow Mo possess a cell-specific SE at the Nr4a1 locus.
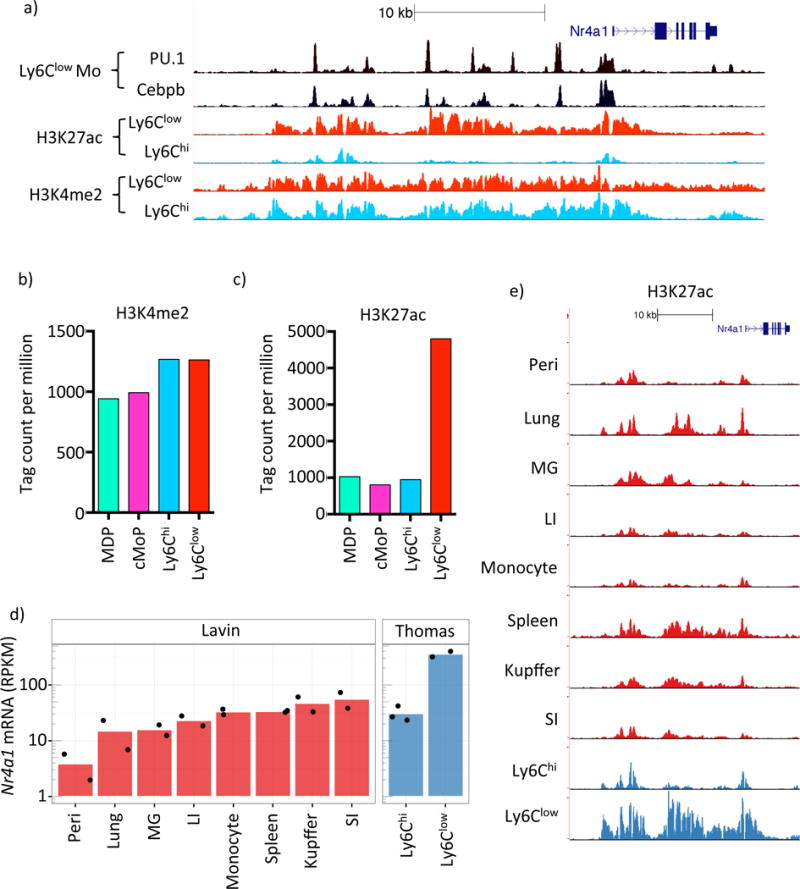
a) UCSC genome browser screenshot of the Nr4a1 locus depicting Ly6Clow Mo PU.1 and C/EBPβ binding and H3K4me2 and H3K27ac in both Ly6Chi and Ly6Clow Mo. b) and c) H3K4me2 and H3K27ac tag counts at Nr4a1se in MDP, cMoP, Ly6Chi and Ly6Clow Mo. d) mRNA-Seq expression of Nr4a1 in various tissue macrophage subsets taken from Lavin et al (2014), labeled ‘Lavin’, and from our data, labeled ‘Thomas’ (Peri= peritoneal macrophage, MG= microglia, LI= large intestine, SI= small intestine). e) H3K27ac at Nr4a1se for the same MP populations as in d). Please refer to supplementary figure S3.
The formation of SEs represents a confluence of developmental and environmental cues (Hnisz et al., 2015). We leveraged the phenotypic heterogeneity of the diverse MP family to address whether steady-state Nr4a1 expression may be regulated by distinct mechanisms. First we assessed Nr4a1 expression in MP populations taken from Lavin et al (2014) and our own monocyte subset data. We observed steady-state expression of Nr4a1 spanning three orders of magnitude (Figure 2d). Importantly Nr4a1 expression in Ly6Chi monocytes from both datasets (labeled Ly6Chi and Mononocytes) were consistent, facilitating the comparison between datasets. Consistent with previous observations we also observed by far the highest Nr4a1 expression in Ly6Clow monocytes (Hanna et al., 2011).
Next we questioned whether the range of Nr4a1 mRNA expression was associated with the continuous acquisition of a fixed enhancer signature at Nr4a1se, or if Ly6Clow monocytes possessed a unique H3K27ac profile. We visualized H3K27ac at Nr4a1se in MP populations corresponding to those in Figure 2d (Figure 2e). We observed comparable H3K27ac tag distributions between Ly6Chi monocyte populations, facilitating a qualitative comparison between datasets. Each population was found to differ qualitatively in both the enrichment of histone acetylation and, to varying degrees, also in the histone acetylation ‘fingerprint’ at Nr4a1se (Figure 2e). Notably, the most intense H3K27ac signature at Nr4a1se was observed in Ly6Clow monocytes (Figure 2d,e). Taken together, these findings imply that distinct mechanisms may regulate steady-state Nr4a1 expression between MP populations.
Nr4a1se domain E2 is a conserved SE sub-domain essential for Ly6Clow monocyte development
We sought to identify regions of Nr4a1se that regulate Nr4a1 gene expression in Ly6Clow monocytes. Based on PU.1 and C/EBPβ binding in Ly6Clow monocytes (Figure S4a) we cloned 12 candidate sequences into luciferase reporters containing a minimal Nr4a1 promoter (Figure S4b). Nr4a1se sub-domains E2, E6, E8 and E9 induced modest but consistent luciferase activity in the myeloid RAW264.7 cell line indicating that these regions may regulate Nr4a1 in Ly6Clow monocytes (Figure 3a). It has been suggested that human CD14dimCD16hi monocytes are homologous to mouse Ly6Clow monocytes. This notion is based on global gene expression profiling and observation of the characteristic patrolling behavior in CD14dimCD16hi monocytes (Cros et al., 2010). We reasoned that if CD14dimCD16hi and Ly6Clow monocytes are truly orthologous that Nr4a1 should be regulated by a conserved mechanism. To test this hypothesis we assessed the genetic regulatory state of human DNA sequences orthologous to mouse E2, E6, E8 and E9 in human monocytes using publicly available datasets (Boyle et al., 2008; Schmidl et al., 2014). Human monocyte DNase-Seq clearly shows open chromatin at orthologous regions to mouse E2, E6 and E9 (Figure 3b), however no ortholog for E8 was identified by nucleotide sequence alignment using BLAT. Human E2, E6 and E9 also possessed H3K27ac, and H3K27ac enrichment was greater at E2 and E6 in CD14dimCD16hi monocytes than CD14hiCD16neg monocytes, consistent with the pattern observed between mouse monocyte subsets (Figure 3b). This provides evidence that E2, E6 and E9 are functionally conserved enhancer elements between species, supporting the notion that these regions are important regulators of MP Nr4a1 gene expression.
Figure 3. Identification of a conserved SE sub-domain essential for Ly6Clow Mo development.
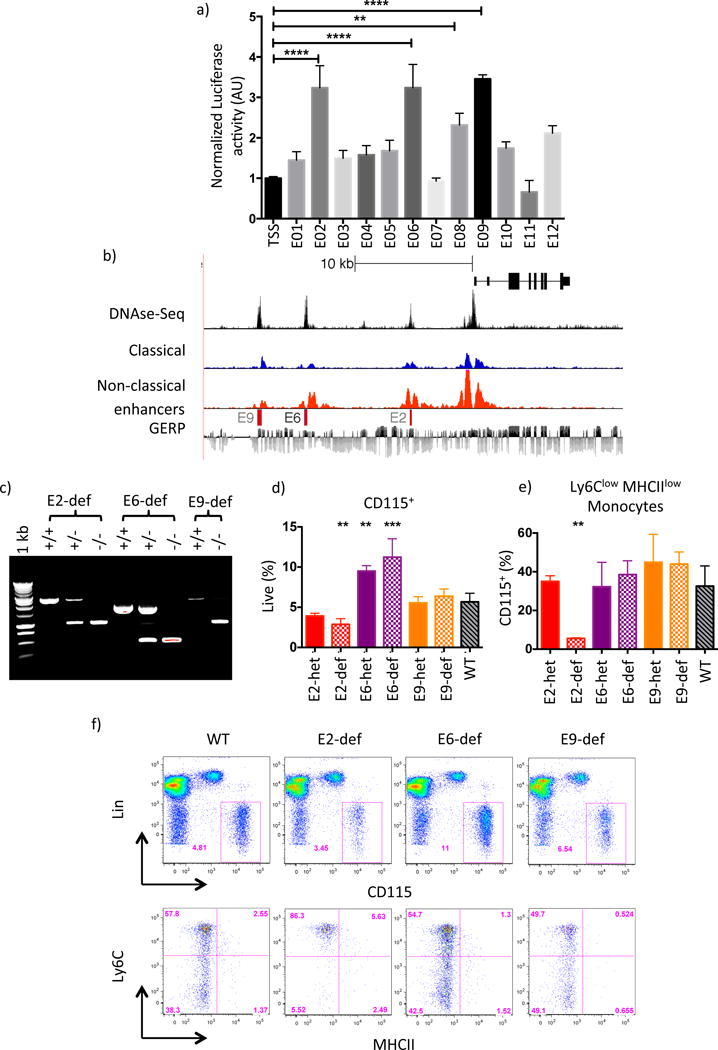
a) Relative luciferase activity in RAW264.7 cells of candidate enhancer regions cloned into pGL4.Nr4a1 vector. Results shown are averaged over 2 independent experiments. Error bars represent SD, P-values calculated by 2-way anova * P<0.05, **P<0.01, ****P<0.0001. b) UCSC genome browser screenshot of the human Nr4a1 locus. H3K27ac tracks for CD14+CD16dim (classical) and CD14dimCD16+ (non-classical) Mo are shown alongside BLAT alignments of nucleosome-free DNA sequence obtained from mouse E2, E6 and E9. Mo DNase-Seq data were taken from (Boyle et al, 2008); nucleotide sequence conservation is indicated using the GERP track. c) PCR Genotyping of E2, E6 and E9 heterozygous (het) and deficient (def) mice. d) and e) Enumeration of peripheral blood Mo in WT, E2, E6 and E9 domain-heterozygous (het) and -deficient (def) mice. Parametric t-tests performed relative to WT, ** p< 0.01 *** p<0.001. f) FACS gating and representative flow plot for each genotype in d) and e). Please refer to supplementary figure S4.
To test the in vivo functions of E2, E6 and E9 we used the CRISPR-Cas9 system to generate three mouse strains, each containing a deletion of enhancer sequence to give E2 domain-deficient, E6 domain-deficient and E9 domain-deficient mice respectively (Figure 3c, S4c). Flow cytometry analysis of peripheral monocytes revealed a reduction in monocyte frequencies in E2 domain-deficient, no difference in E9 domain-deficient, and a modest increase in E6 domain-deficient mice (Figure 3d). WT-like monocyte subset ratios were preserved in both E6 domain-deficient and E9 domain-deficient mice, however a striking deficit in Ly6Clow monocytes phenocopying the Nr4a1−/− strain was present in E2 domain-deficient mice (Figure 3e,f) (Hanna et al., 2011). Therefore the conserved super-enhancer sub-domain E2 is indispensible for Ly6Clow monocyte development.
Deletion of enhancer domain E2 decouples inflammation-associated Nr4a1 gene expression from Ly6Clow monocyte-associated Nr4a1 expression
Current genetic models to study MP Nr4a1 are based on global Nr4a1−/− mice or Cre recombinase mediated deletion of Nr4a1flox/flox alleles using myeloid specific Cre transgenes, which include, but are not restricted to Csf1r, Lyz2 (LysM) and Cx3cr1. While these tools ablate MP Nr4a1 they are non-specific with respect to individual MP subsets (Chow et al., 2011; Yona et al., 2012). Consequently, in models that lack Ly6Clow monocytes, Nr4a1 is also disrupted in macrophages (Figure 4a). As Ly6Clow monocytes are absent in E2 domain-deficient mice and Nr4a1 is a key suppressor of macrophage inflammatory gene expression we questioned whether E2 domain-deficient macrophages possess wild type-like responses to inflammatory stimuli.
Figure 4. E2 does not regulate Nr4a1 mRNA expression in response to inflammatory stimuli.
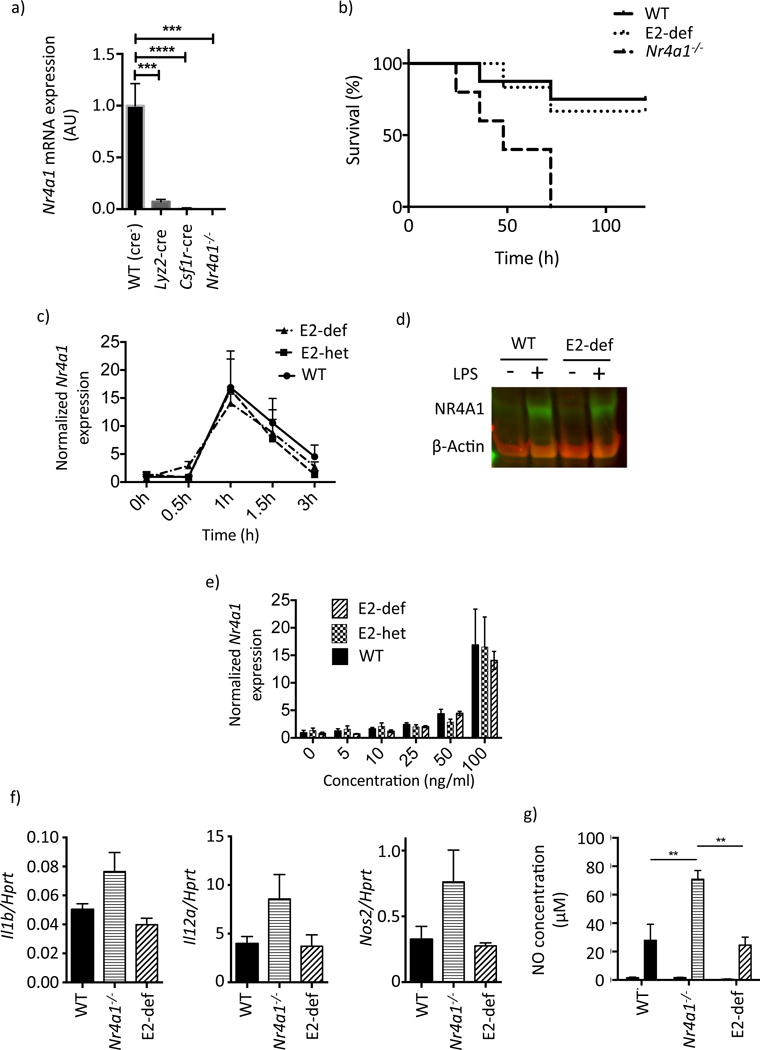
a) Nr4a1 mRNA expression in thioglycollate-elicited macrophages obtained from Nr4a1flox/flox mice crossed to Lyz2-cre and Csf1r-cre, WT (Cre− littermates) are shown as control. b) Kaplan-Mayer curve showing survival of WT, E2 domain-deficient and Nr4a1−/− mice in response to a single dose of 2.5mg/kg LPS I.P. Mantel-Cox (Log rank) test results: WT vs. Nr4a1−/− p<0.01, E2 domain-deficient vs. Nr4a1−/− p<0.05, E2 domain-deficient vs. WT p>0.05. c) RT-PCR time course of Nr4a1 mRNA in primary thioglycollate-elicited macrophages following LPS stimulation (100ng/ml). d) Immuno blot showing Nr4a1 expression 1h post LPS stimulation (100ng/ml). e) Nr4a1 mRNA expression in dose escalation at 1h post LPS stimulation. f) Inflammatory cytokine mRNA expression in primary peritoneal macrophages 1h post injection of 1ug LPS I.P. g) Nitric oxide in culture supernatant 96h after LPS stimulation.
During endotoxic shock macrophage mediated production of inflammatory cytokines ultimately precipitates organ failure and mortality (Jacob et al., 2007). As Nr4a1−/− mice are more sensitive to LPS-induced organ failure (Li et al., 2015) we decided to test the role of E2 in this setting. We administered a single high dose of LPS (2.5 mg/kg) to E2 domain-deficient, Nr4a1−/−, and WT mice by IP injection. All three groups displayed disheveled fur and shivering within the first 72 hours however Nr4a1−/− mice showed a significantly higher mortality than both WT and E2 domain-deficient groups, whose symptoms resolved in the 72–120-hour time window (Figure 4b). To understand the role of E2 in the regulation of Nr4a1 gene expression we measured Nr4a1 responses to LPS stimulation in thioglycollate-elicited macrophages. WT, E2 domain-heterozygous and E2 domain-deficient macrophages all showed the well-characterized peak of Nr4a1 mRNA at 1h followed by a return to baseline by 3h (Figure 4c, Pei et al., 2005). Immuno blot analysis also confirmed protein induction in WT and E2 domain-deficient macrophages at 1h following stimulation (Figure 4d). Furthermore, in a dose-response setting (Figure 4e) we observed no differences in Nr4a1 mRNA expression between WT, E2 domain-heterozygous and E2 domain-deficient macrophages. Finally, following LPS challenge we detected higher expression of Il12, Il1b and Nos2 mRNA and iNOS activity in Nr4a1−/− mice relative to E2 domain-deficient mice, which were comparable to wild type controls (Figure 4f,g). These studies confirm that the E2 region does not regulate Nr4a1 expression in response to TLR4 stimulation.
Differential PU.1 binding reveals candidate regulators of Ly6Clow monocyte gene expression
We sought to determine the molecular mechanisms regulating Nr4a1 expression in Ly6Clow monocytes. Cooperative interactions between PU.1 and secondary co-factors establish and maintain macrophage enhancer repertoires (Heinz et al., 2010, 2013). Furthermore, differences in SDTF activity elicited within tissue microenvironments are responsible for the diverse range of MP enhancers observed in vivo (Gosselin et al., 2014; Lavin et al., 2014). One mechanism for enhancer acquisition involves SDTF driving the formation of latent enhancers associated with de novo H3K4me2 and PU.1 binding (Kaikkonen et al., 2013; Ostuni et al., 2013). Thus the subset of de novo PU.1 binding events provides valuable information concerning secondary SDTF identity by virtue of motif enrichment (Gosselin et al., 2014).
To define motifs associated with Ly6Clow monocytes we determined PU.1 binding profiles in Ly6Chi and Ly6Clow monocytes, identifying a total of 65,070 PU.1 peaks. Strict criteria for differential binding led to the identification of 345 Ly6Clow monocyte specific PU.1 peaks (Figure 5a) that possess the hallmarks of a latent enhancer repertoire. These include increased H3K4me2 and nucleosome phasing at regions immediately surrounding the PU.1 peaks (Figure S5a); elevated H3K27ac (Figure 5b); and, increased expression of proximal mRNA transcripts (Figure 5c) in Ly6Clow monocytes relative to upstream progenitors. Thus, the Ly6Clow monocyte PU.1-specific peak profile was associated with enhancers that regulate the Ly6Clow monocyte gene expression program. Motifs implicated in Ly6Clow monocyte gene expression were identified using de novo motif enrichment analysis on this PU.1 peak set. In strong agreement with our earlier analysis (Figure S1c) we recovered ETS, C/EBP and NR4A motifs (Figure 5d)as expected, alongside IRF, KLF, RUNX and MEF2 motifs. Using our RNA-Seq dataset we identified transcription factors that were expressed in Ly6Clow monocytes and could bind to these motifs (Figure 5e), considering the most abundant as our primary candidate. This approach defined Cebpb, Irf5, Klf2, Mef2a, Nr4a1, Sfpi1 (PU.1) and Runx2 as principal regulators of Ly6Clow monocyte gene expression.
Figure 5. Identification of motifs associated with Ly6Clow Mo development.
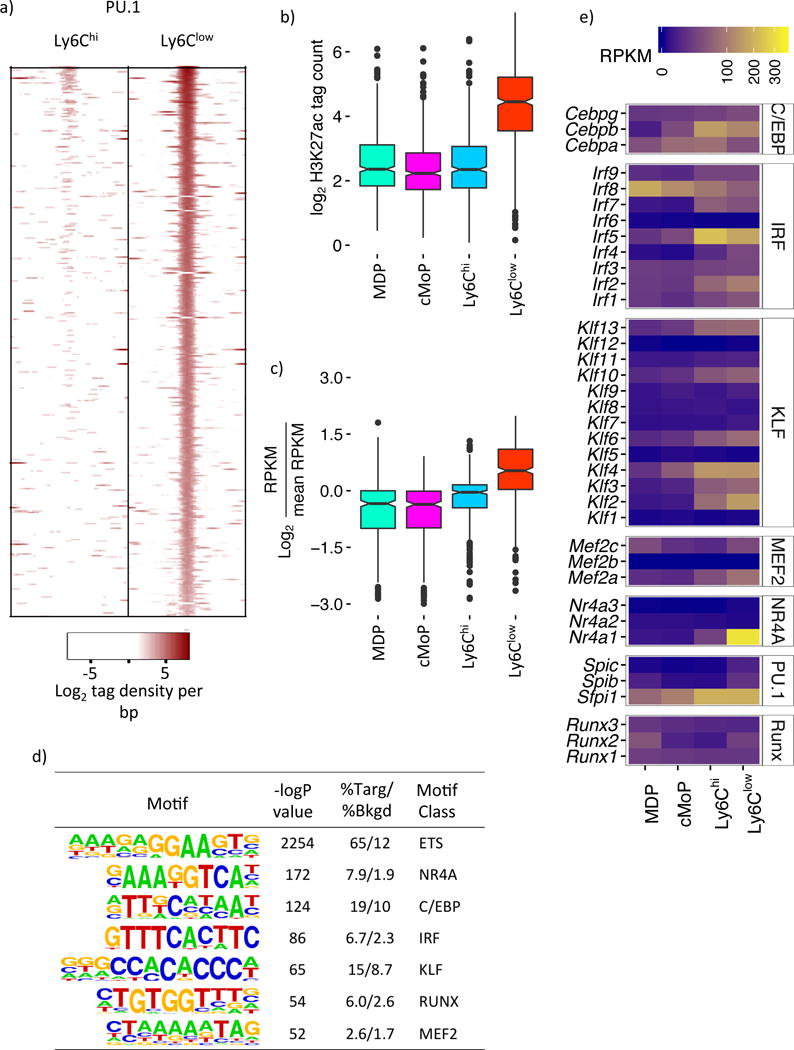
a) Normalized PU.1 ChIP-Seq signal in Ly6Chi and Ly6Clow Mo of Ly6Clow Mo-specific PU.1 sites ±400 bases from PU.1 peak center. b) H3K27ac ±500 bases of Ly6Clow Mo-specific PU.1 peaks. c) Relative expression of genes proximal to Ly6Clow Mo-specific PU.1 peaks. d) Overrepresented motifs in Ly6Clow Mo specific PU.1 peaks identified by de novo enrichment analysis. e) RNA-Seq expression amounts of TFs predicted to bind motifs in d). Please refer to supplementary figure S5a.
Klf2 regulates Ly6Clow monocyte conversion via E2
To identify transcription factors that regulate Nr4a1 expression via E2 we overexpressed each candidate with the E2 reporter. Cebpb, Irf5, Klf2, Mef2a, Nr4a1 and Runx2 were co-transfected into RAW264.7 macrophages alongside either the Nr4a1-TSS or E2-Nr4a1-TSS luciferase reporter plasmids. To account for promoter dependent effects of the cDNA and the intrinsic activity of the E2 sequence, an enhancer index was calculated that denotes the difference in the ratio of induced luciferase activity between E2-Nr4a1-TSS and Nr4a1-TSS. We found that only Klf2 drove E2-dependent luciferase expression (Figure 6a). Supporting this finding a motif search for the enriched IRF, KLF, RUNX and MEF2 motifs within the E2 region identified a cluster of 3 KLF motifs but failed to find any instances of the other motif classes (Figure S5b).
Figure 6. Klf2 drives Ly6Chi to Ly6Clow Mo conversion via E2.
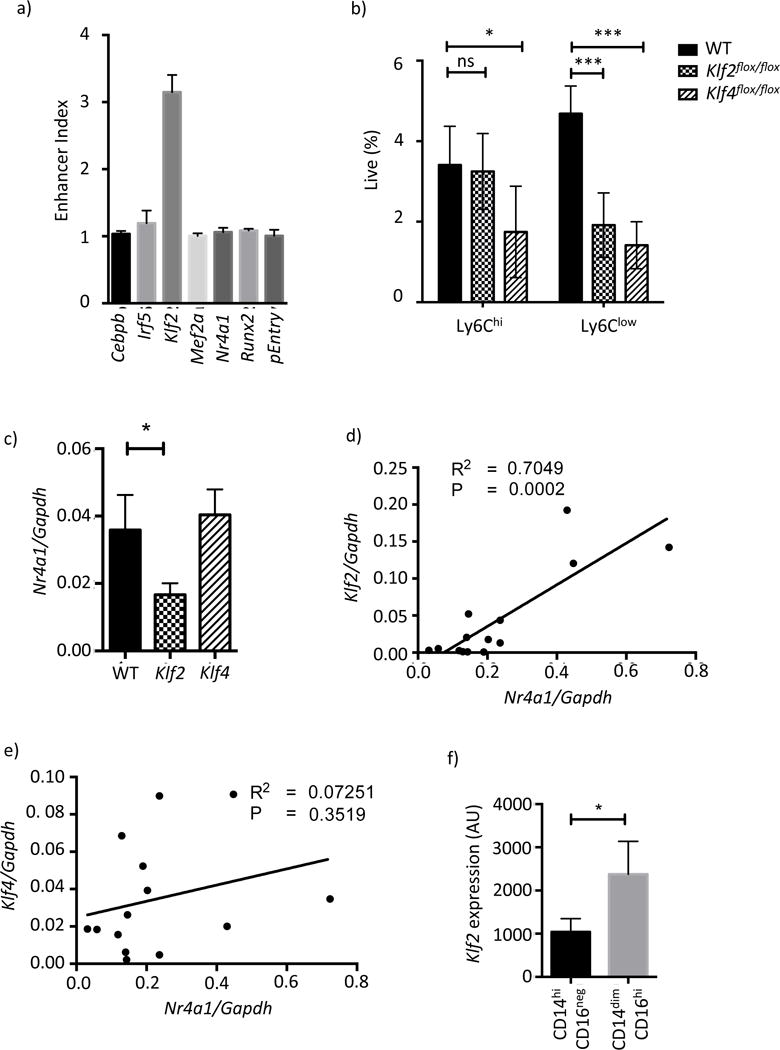
a) Overexpression of candidate TFs in the presence of pGL4.Nr4a1_E2 and pGL4.Nr4a1 reporter vectors in RAW264.7 cells. The enhancer index is calculated as described in the methods. b) Blood Mo frequencies in Lyz2-cre Klf2flox/flox and Lyz2-cre Klf4flox/flox mice. c) Nr4a1 mRNA expression amounts in primary Ly6Chi Mo d) Klf2 mRNA correlated against Nr4a1 in Ly6Clow Mo e) the same data as d) for Klf4 mRNA. f) Klf2 mRNA expression amounts in primary human Mo subsets as measured by microarray. Please refer to supplementary figures S5b and S6.
We chose to investigate the role of KLF factors in Ly6Clow monocyte development. KLFs are broadly expressed in Ly6Clow monocytes with Klf2 and Klf4 being the most abundant (Figure 5e). Owing to the perinatal and embryonic lethality of Klf2−/− and Klf4−/− mice we assessed monocyte frequencies in myeloid-specific Lyz2-cre Klf2flox/flox and Lyz2-cre Klf4flox/flox mice (Liao et al., 2011; Mahabeleshwar et al., 2011). Flow cytometric analysis of Lyz2-cre Klf4flox/flox blood monocytes showed a decrease in both populations (Figure 6b), consistent with previous reports (Alder et al., 2008). In Lyz2-cre Klf2flox/flox mice Ly6Chi monocytes were unaffected however Ly6Clow monocytes were partially reduced (Figure 6b). Similar results were also observed in the bone marrow of Lyz2-cre Klf2flox/flox and Lyz2-cre Klf4flox/flox mice and in bone marrow chimeric mice retrovirally transduced with shRNA targeting Klf2 and Klf4 (Figure S6a,b).
We have previously observed incomplete deletion of loxP-flanked genes in blood monocytes using the Lyz2-cre system (Hanna et al., 2015). Therefore the differences in monocyte frequencies between Lyz2-cre Klf2flox/flox and Lyz2-cre Klf4flox/flox mice may either reflect intrinsic differences in the ability of each gene to regulate monocyte development, or simply the abundance of each TF in each subset. To address this we measured Klf2 and Klf4 mRNA in Ly6Chi and Ly6Clow monocytes sorted from Lyz2-cre Klf2flox/flox and Lyz2-cre Klf4flox/flox mice using primers that span the loxP-flanked exon to determine recombination efficiency. In Lyz2-cre Klf2flox/flox mice 72% and 91% loss of Klf2 mRNA was observed for Ly6Chi and Ly6Clow monocytes respectively (Figure S6c). Thus the selective loss of Ly6Clow monocytes in Lyz2-cre Klf2flox/flox mice is not consistent with the incomplete deletion of Klf2 in Ly6Chi monocytes. A role for Klf2 in Ly6Chi to Ly6Clow monocyte conversion is also consistent with the induction of Klf2 gene expression in the transition between subsets (Figure 5e). In Lyz2-cre Klf4flox/flox mice Klf4 mRNA was reduced by 63% and 69% in Ly6Chi and Ly6Clow monocytes respectively (Figure S6d). Although Lyz2-cre Klf4flox/flox mice possessed fewer Ly6Clow monocytes it is not clear whether this resulted from fewer precursor Ly6Chi monocytes, a decrease in Ly6Chi to Ly6Clow monocyte conversion, or both. We reasoned that if Klf2 or Klf4 regulated Ly6Chi to Ly6Clow monocyte conversion via Nr4a1 that Klf2 or Klf4 gene expression would predict Nr4a1 expression. Indeed, Nr4a1 mRNA expression was lower in Ly6Chi monocytes derived from Lyz2-cre Klf2flox/flox mice, but not Lyz2-cre Klf4flox/flox mice (Figure 6c). Subsequent investigation revealed a significant positive correlation between Klf2 and Nr4a1 transcript expression in both monocyte subsets, however no such relationship was found between Klf4 and Nr4a1 gene expression (Figure 6d,e and S6e,f).
While of a preliminary nature, these correlative findings suggest that Klf2 regulates Ly6Clow monocyte development via E2. Given our evidence for a conserved mechanism of Nr4a1 gene expression between species (Figure 3b) we predict that KLF2 expression follows a similar pattern in human monocyte subsets.
Interrogation of microarray data for KLF2 in human monocytes confirmed this hypothesis, showing significantly higher KLF2 expression in human CD14dimCD16hi monocytes (Figure 6f).
E2 is a monocyte-specific enhancer
E2 regulates Nr4a1 expression in Ly6Clow monocytes but not in response to LPS stimulation. Furthermore, at steady state, macrophage Nr4a1 expression spanned three orders of magnitude. To determine the extent to which E2 was Ly6Clow monocyte specific we measured Nr4a1 expression in various MP populations from E2 domain-deficient mice. Blood monocytes, F4/80hi large, F4/80int small peritoneal macrophages (LPM and SPM respectively), CD11c+ lung alveolar macrophages, and F4/80+ splenic red pulp macrophages were selected (Figure 7a). In line with previous observations (Tacke et al., 2015) macrophage frequencies were largely unchanged in Nr4a1−/− mice, except for moderately fewer F4/80hi LPM (Figure S7a). RT-PCR analysis captured the expected range of Nr4a1 expression between macrophage populations (Figure 7b). Strikingly however no difference in Nr4a1 mRNA was detected between tissue macrophages, except for moderately lower expression in E2 domain-deficient splenic macrophages. In contrast to this a substantial reduction in steady state Nr4a1 mRNA expression was observed in both Ly6Chi and Ly6Clow monocytes (Figure 7c) showing that E2 is a monocyte-specific enhancer.
Figure 7. E2 is a monocyte-specific enhancer.
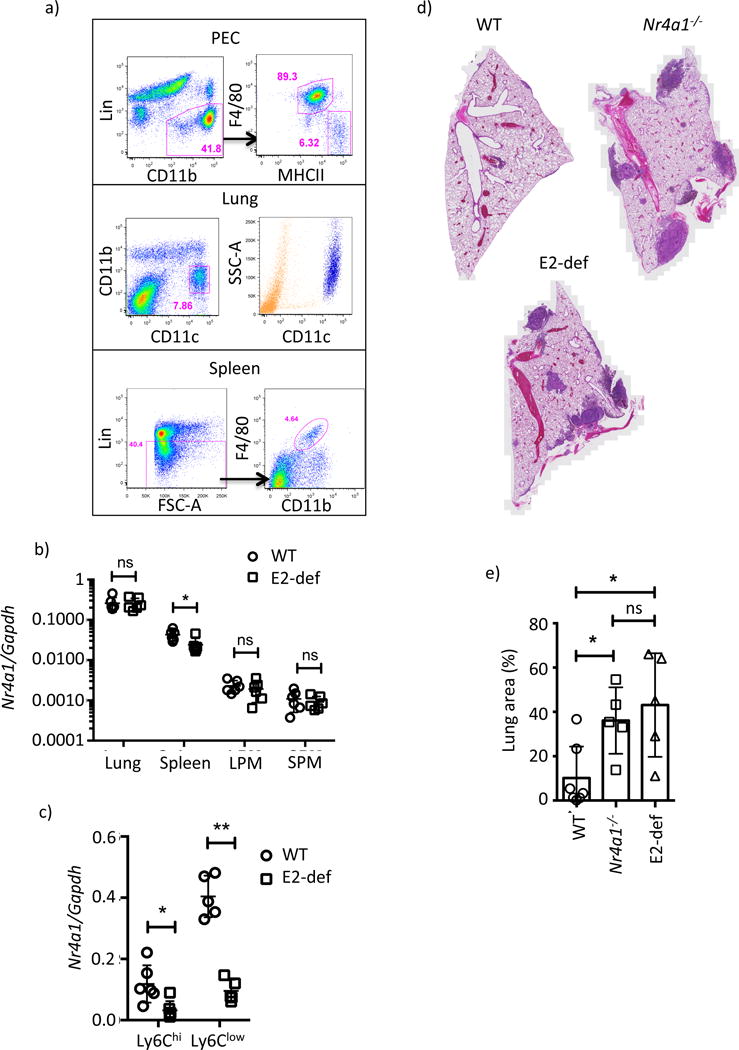
a) Gating scheme for tissue macrophage sorting. All cells were previously gated on live singlets. The side scatter high profile of lung CD11c+ macrophages is shown in the right panel (blue). b and c) Relative Nr4a1 mRNA expression in tissue macrophages and blood Ly6Chi and Ly6Clow Mo. Statistics for b) and c) measured by students t-test * p<0.05, ** p<0.01. d) Representative sections of B16F10 tumors in WT, Nr4a1−/− and E2 domain-deficient mice. e) and f) Quantification of cancer metastasis area, error bars represent SD statistics were analyzed by ANOVA * p<0.05. Please refer to supplementary figure S7.
We recently reported that Ly6Clow monocytes prevented cancer metastasis to the lung (Hanna et al., 2015). To demonstrate the utility of the E2 domain-deficient model to study Ly6Clow monocytes we assessed tumor burden in mice using a well-established model of metastasis. B16F10 melanoma cells were intravenously injected into WT, Nr4a1−/− and E2 domain-deficient mice. 18 days after challenge mice were sacrificed and tumor burden was measured by histology. Both Nr4a1−/− and E2 domain-deficient mice showed a loss of Ly6Clow monocytes (Figure S7b) and significantly higher tumor burden than WT (Figure 7d,e) however no difference was observed between Nr4a1−/− and E2 domain-deficient mice. Consistent with a critical role for Ly6Clow monocytes in controlling metastasis, the differences are explained by fewer tumor regions rather than tumor size (Figure S7c,d). Therefore the E2 domain-deficient model effectively decouples Nr4a1-dependent inflammatory phenotypes from Ly6Clow monocyte function, providing an improved tool to study the function of these cells. In contrast to conditional deletion strategies using the Cre-Lox system, enhancer targeting confers an unprecedented degree of specificity, enabling the causal inference of individual MP subsets in disease.
Discussion
During our investigation into the transcriptional regulatory pathways of Ly6Clow monocyte development we identified E2 as a single domain that is absolutely required for Ly6Clow monocyte development. We also observed distinct, cell-subset specific, patterns of H3K27ac at the Nr4a1se locus leading us to investigate whether Nr4a1 gene expression is controlled by distinct mechanisms between MP subsets. E2 domain-deficient mice lack Ly6Clow monocytes, yet macrophage Nr4a1 expression was largely unaffected during steady state and during inflammation. This implies a modular structure for the Nr4a1 locus control region Nr4a1se and is consistent with a recent analysis of several SEs showing that these regions consist of multiple sub-domains (Hnisz et al., 2015). Disrupting these sub-domains differentially affected target gene expression depending upon the locus. For example at Sik1, additive effects between sub-domains contribute towards the stable induction of gene expression, whereas at Prdm14, mRNA expression was regulated almost entirely by one of five sub-domains (Hnisz et al., 2015). Our study has identified three active and conserved MP enhancers upstream of Nr4a1; E2, E6 and E9, yet only E2 regulates Ly6Clow monocyte development. This highlights the importance of detailed molecular analyses to establish the relevance of individual enhancer elements, and TF binding motifs within these elements. Whether E6 and E9 regulate MP Nr4a1 gene expression in response to other stimuli is a line of future enquiry.
Our studies suggest that KLF transcription factors regulate Nr4a1 gene expression via E2. Ly6Clow monocyte enhancers were enriched for KLF motifs, and these motifs were present in E2. Multiple KLF family members are expressed in Ly6Clow monocytes, which may act redundantly at E2. In Ly6Clow monocytes Klf2 and Klf4 were the most abundant family members, thus we investigated the roles of these genes. Both Klf2 and Klf4 regulate macrophage inflammatory gene expression by inhibiting p300 and PCAF recruitment to NF-kB (Das et al., 2006; Liao et al., 2011). The different roles ascribed to each factor may arise from the different contexts in which they are expressed. Klf4 was induced during Ly6Chi monocyte differentiation, and was further up-regulated in macrophages by IL-4. In this context Klf4 regulates Ly6Chi monocyte development and facilitates the M2 program of macrophage activation (Feinberg et al., 2007; Liao et al., 2011). Klf2 however is expressed in circulating monocytes, and strongly reduced by hypoxia leading to increased NF-kB and Hif-1 activity (Mahabeleshwar et al., 2011).
Our findings suggest non-redundant roles for Klf2 and Klf4 in in monocyte development. Klf4 expression is high in both monocyte subsets and Lyz2-cre Klf4flox/flox mice have fewer monocytes. However, the ratio of Ly6Chi to Ly6Clow monocytes, and Ly6Clow monocyte Nr4a1 expression were not affected in Lyz2-cre Klf4flox/flox mice However, depletion of Klf2 facilitated a selective loss of Ly6Clow monocytes and Klf2 mRNA expression was predictive of Nr4a1 expression, implying a causal relationship between Klf2 and Nr4a1. Despite repeated attempts we were not able to obtain successful immunoprecipitation of KLF2 at the Nr4a1se locus due to an absence of high-quality commercial antibodies; neither was overexpression of Klf2 sufficient to up-regulate Nr4a1 mRNA in vitro (data not shown). These preliminary findings suggest that Klf2 acts via E2 locus and is necessary, but not sufficient, for Ly6Clow monocyte development. The identification of additional critical co-factors acting via E2, the Nr4a1 promoter or elsewhere will pave the way for a more detailed mechanistic understanding of how E2 regulates Nr4a1 gene expression in Ly6Clow monocyte development.
MP occupy all tissues of the body where these cells perform specialized functions to facilitate homeostasis. For example, Ly6Clow monocytes maintain vascular integrity by facilitating removal of damaged endothelium (Carlin et al., 2013), alveolar macrophages maintain lung function by clearing surfactants (Nakamura et al., 2013), red pulp macrophages contribute to iron homeostasis by recycling erythrocytes (Kohyama et al., 2009), whereas the microglia’s dedicated functions include supporting brain function by synaptic pruning (Paolicelli et al., 2011). These specialized functions are imparted by tissue-specific LTDFs; Ly6Clow monocytes require Nr4a1 (Hanna et al., 2011); alveolar macrophages rely upon PPARγ (Schneider et al., 2014); splenic red pulp-macrophages macrophages need Spic (Kohyama et al., 2009); while resident peritoneal macrophages depend upon the transcription factor GATA6 (Okabe and Medzhitov, 2014).
In some cases, such as for Gata6 and Spic, LDTF expression within the MP system is cell-specific such that macrophage-specific deletion of the factor is sufficient to study the function of the associated subset. However, for the most part both the expression and requirement of key MP TFs are not cell-specific. Epigenetic analyses of MP subsets reveals shared motif usage and co-expression of cognate TFs over multiple subsets (Lavin et al., 2014). For example PPAR motifs are enriched in alveolar and renal macrophage enhancers and PPARγ regulates both alveolar macrophage and osteoclast differentiation (Schneider et al., 2014; Wan et al., 2007). Disease processes complicate matters further by invoking dynamic transcription factor expression and altering requirements, in this context PPARγ is up-regulated by IL-4 and controls alternative macrophage activation (Odegaard et al., 2007). Similarly, Nr4a1 is required for Ly6Clow monocyte development and is also induced by LPS (Hanna et al., 2011; Pei et al., 2005). These pleiotropic effects limit our ability to study individual MP subsets and the lack of specificity provided by current myeloid Cre transgenes presents a problem when attempting to target individual subsets.
We have shown that enhancer targeting can overcome these limitations. Our approach leverages the unique enhancer repertoires resulting from the exclusive environment each MP subset occupies. We have shown that targeting cell-specific SE sub-domains within key LDTFs functionally decouples cell-specific aspects of gene expression while retaining physiological characteristics of gene function relevant to neighbouring cell subsets. In our case, targeting the E2 sub-domain provides a loss-of-function tool to study Ly6Clow monocytes that preserves both Nr4a1 expression in other MP populations and the rapid kinetics of Nr4a1 expression in response to LPS signaling. Conceptually this strategy can be applied to any scenario in which gene expression is regulated by distinct enhancer sequences as an approach to study gene function amongst closely related cell types.
Experimental procedures
Mice
C57BL/6J mice were purchased from the Jackson laboratory. Congenic Nr4a1−/− mice previously described (Hanna et al., 2011) and enhancer deficient mice were maintained in-house. All experiments followed guidelines of the La Jolla Institute for Allergy and Immunology (LIAI) Animal Care and Use Committee, and approval for use of rodents was obtained from LIAI according to criteria outlined in the Guide for the Care and Use of Laboratory Animals from the National Institutes of Health. Mice were euthanized by CO2 inhalation.
Cell sorting
Mice were sacrificed by CO2 inhalation and bone marrow or blood extracted into DPBS + 2mM ETDA. Following RBC lysis cells were blocked and stained for surface antigens and purified by flow cytometry as described in the figures as described in the extended experimental procedures.
Thioglycollate elicited macrophages
Thioglycollate elicited macrophages were elicited by intraperitoneal injection of 1ml 4% Brewer’s Thioglycollate Medium. 5 days after injection macrophages were harvested and cultured as described in the extended experimental procedures.
RNA isolation and RNA-Seq
Sorted cells were immediately spun down and stored in Trizol. RNA was extracted using DirectZol columns (Zymo Research). RNA was prepared for sequencing using the TruSeq v2 kit (Illumina).
ChIP-Seq
Histone ChIP was performed using the protocol described in (Gilfillan et al., 2012) and transcription factor ChIP performed essentially as described in (Gosselin et al., 2014), with modifications described in the extended methods. ChIP-Seq libraries were prepared using the ThruPlex-FD kit (Rubicon Genomics).
Molecular cloning and overexpression studies
Enhancers were amplified by PCR and cloned into the SalI and Bamh1 sites of a pGL4.10 series vector described in (Heinz et al., 2013) modified to contain an Nr4a1 minimal promoter (300 bp) and 5′UTR, please see table S1. Transfection assays were performed in murine RAW264.7 macrophages using Lipofectamine LTX. cDNA expression vectors were obtained from Origene. Full details are in the extended experimental procedures.
CRISPR mouse generation
Mouse genome editing was performed essentially as described with minor modifications (Concepcion et al., 2015; Wang et al., 2013). Four sgRNA (two pairs targeting each enhancer flanking region) were injected into embryos of supervoulated C57BL/6 mice along with Cas9 mRNA (Life Technologies) at the UCSD transgenic core facility. Please refer to table S2. Founder mice containing the desired deletions identified by PCR (table S3) were mated with C57BL/6 and offspring thereafter maintained via sibling mating.
Bone marrow chimeras
Bone marrow was shipped on ice as described in the extended experimental methods. Recipient mice were lethally irradiated and bone marrow transplanted by retroorbital injection. Mice were allowed to recover for at least 6 weeks before performing experiments.
Cancer study
300,000 B16F10 melanoma cells were injected via tail vein into recipient mice. Lungs were harvested in zinc buffered formalin and mounted into paraffin blocks. Sections were stained with H&E and slides were scanned with an AxioScan Z1 (Zeiss), see extended experimental procedures.
Endotoxin sensitivity assay
Ultrapure LPS-EB (Invivogen) was made up in PBS and injected IP. Mice were monitored three times daily for the first 72 hours and twice daily thereafter.
Bioinformatics data analysis
FASTQ files were mapped to the mouse mm10 reference genome using RNA-STAR for RNA-Seq experiments, or Bowtie for ChIP-Seq studies. RNA-Seq expression was quantitated using featureCounts and differential expression analyzed using edgeR. ChIP-Seq analysis was performed using HOMER. For full details see extended experimental procedures.
Supplementary Material
Acknowledgments
We would like to acknowledge the flow cytometry core and sequencing core facilities as well as the UCSD transgenic core facility. Funding sources: R01HLx118765, R01CA202987 and R01HL112039 to CCH; R01DK091183, R01CA17390 and R01HL088093 to CKG; R00HL123485 to CER; R01GM086912 to BAH; R01HL086548 and R01075427 to MKJ. AHA Fellowships 16POST27630002 to GDT and 12DG12070005 to RNH. KDR was supported in part by a Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Predoctoral Training Grant, T32 GM008666, from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Illumina sequencing for this project have been deposited at NCBI’s Gene Expression Omnibus (GEO) as a SuperSeries under the accession number GSE80040.
Author contributions
Conceptualization CCH, CKG and GDT; Methodology GDT, RNH, CER, KDR, BAH; Software, formal analysis and visualization GDT; Investigation GDT, AB and DY; Resources MKJ, VNT, ZM and SM; Writing - original draft GDT and CCH; Writing - Reviewing and Editing - all authors; Funding Acquisition CCH and GDT.
References
- Alder JK, Georgantas RW, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-Like Factor 4 Is Essential for Inflammatory Monocyte Differentiation In Vivo. J Immunol. 2008;180:5645–5652. doi: 10.4049/jimmunol.180.8.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, Hedrick CC, Cook HT, Diebold S, Geissmann F. Nr4a1-Dependent Ly6Clow Monocytes Monitor Endothelial Cells and Orchestrate Their Disposal. Cell. 2013;153:362–375. doi: 10.1016/j.cell.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Brown BD, Merad M. Studying the mononuclear phagocyte system in the molecular age. Nat Rev Immunol. 2011;11:788–798. doi: 10.1038/nri3087. [DOI] [PubMed] [Google Scholar]
- Concepcion D, Ross KD, Hutt KR, Yeo GW, Hamilton BA. Nxf1 natural variant E610G is a semi-dominant suppressor of IAP-induced RNA processing defects. PLoS Genet. 2015;11:e1005123. doi: 10.1371/journal.pgen.1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci. 2010 doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cros J, Cagnard N, Woollard K, Patey N, Zhang SY, Senechal B, Puel A, Biswas SK, Moshous D, Picard C, et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity. 2010;33:375–386. doi: 10.1016/j.immuni.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A. 2006;103:6653–6658. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, et al. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Hughes T, Sheng Y, Hjorthaug HS, Straub T, Gervin K, Harris JR, Undlien DE, Lyle R. Limitations and possibilities of low cell number ChIP-seq. BMC Genomics. 2012;13:645. doi: 10.1186/1471-2164-13-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, et al. Environment Drives Selection and Function of Enhancers Controlling Tissue-Specific Macrophage Identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, Hedrick CC. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RN, Shaked I, Hubbeling HG, Punt JA, Wu R, Herrley E, Zaugg C, Pei H, Geissmann F, Ley K, et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ Res. 2012;110:416–427. doi: 10.1161/CIRCRESAHA.111.253377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RN, Cekic C, Sag D, Tacke R, Thomas GD, Nowyhed H, Herrley E, Rasquinha N, McArdle S, Wu R, et al. Patrolling monocytes control tumor metastasis to the lung. Science. 2015 doi: 10.1126/science.aac9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. Effect of natural genetic variation on enhancer selection and function. Nature. 2013 doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, Feuerer M. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Schuijers J, Lin CY, Weintraub AS, Abraham BJ, Lee TI, Bradner JE, Young RA. Convergence of Developmental and Oncogenic Signaling Pathways at Transcriptional Super-Enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A, Zhou M, Wu R, Halpern VJ, Ravikumar TS, Wang P. PRO-INFLAMMATORY CYTOKINES FROM KUPFFER CELLS DOWNREGULATE HEPATOCYTE EXPRESSION OF ADRENOMEDULLIN BINDING PROTEIN-1. Biochim Biophys Acta. 2007;1772:766–772. doi: 10.1016/j.bbadis.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, Frazier WA, Murphy TL, Murphy KM. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu Y, Chen H, Li F, Wu J, Zhang H, He J, Xing Y, Chen Y, Wang W, et al. Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation. Nat Chem Biol. 2015;11:339–346. doi: 10.1038/nchembio.1788. [DOI] [PubMed] [Google Scholar]
- Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121:2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahabeleshwar GH, Kawanami D, Sharma N, Takami Y, Zhou G, Shi H, Nayak L, Jeyaraj D, Grealy R, White M, et al. The myeloid transcription factor KLF2 regulates the host response to polymicrobial infection and endotoxic shock. Immunity. 2011;34:715–728. doi: 10.1016/j.immuni.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Koscsó B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, Mortha A, Leboeuf M, Li XM, Mucida D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Ebina-Shibuya R, Itoh-Nakadai A, Muto A, Shima H, Saigusa D, Aoki J, Ebina M, Nukiwa T, Igarashi K. Transcription repressor Bach2 is required for pulmonary surfactant homeostasis and alveolar macrophage function. J Exp Med. 2013;210:2191–2204. doi: 10.1084/jem.20130028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue-Specific Signals Control Reversible Program of Localization and Functional Polarization of Macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. Tissue biology perspective on macrophages. Nat Immunol. 2016;17:9–17. doi: 10.1038/ni.3320. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent Enhancers Activated by Stimulation in Differentiated Cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, III, Lafaille JJ, Hempstead BL, Littman DR, Gan WB. Microglia Promote Learning-Dependent Synapse Formation through Brain-Derived Neurotrophic Factor. Cell. 2013;155:1596–1609. doi: 10.1016/j.cell.2013.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- Perdiguero EG, Geissmann F. The development and maintenance of resident macrophages. Nat Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Renner K, Peter K, Eder R, Lassmann T, Balwierz PJ, Itoh M, Nagao-Sato S, Kawaji H, Carninci P, et al. Transcription and enhancer profiling in human monocyte subsets. Blood blood. 2014 doi: 10.1182/blood-2013-02-484188. 2013–02 – 484188. [DOI] [PubMed] [Google Scholar]
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. doi: 10.1038/ni.3005. [DOI] [PubMed] [Google Scholar]
- Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Hilgendorf I, Garner H, Waterborg C, Park K, Nowyhed H, Hanna RN, Wu R, Swirski FK, Geissmann F, et al. The transcription factor NR4A1 is essential for the development of a novel macrophage subset in the thymus. Sci Rep. 2015;5:10055. doi: 10.1038/srep10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity. 2012 doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


