Abstract
There is an interest to develop sugar kelp (Saccharina latissima) cultivation in the rural, eastern Maine region of the USA. Future farming efforts would benefit from an understanding of the genetic diversity and population structure of kelp, to inform management and conservation, and to identify genetic resources. The purpose of the present study was to characterize the fine-scale population genetic structure of kelp in eastern Maine, using twelve microsatellite loci. A total of 188 samples were genotyped from five sampling locations. Overall, kelp exhibited relatively low genetic diversity and small but significant differentiation among populations (FST = 0.0157). The greatest genetic difference was detected between two geographically close populations in Penobscot and Frenchman Bays, which is likely due to patterns in the Eastern Maine Coastal Current that may limit meiospore recruitment. The population structure could not be fully explained by an isolation-by-distance model. Fine-scale structuring was also detected among populations along the more continuous, eastern Maine coastline. These differences highlight that sugar kelp populations are finely structured across small spatial scales, and that future management and farming efforts should aim to maintain genetic diversity and assess the culture potential of local populations.
Keywords: Gene flow, Genetic diversity, Kelp, Microsatellite, Polymorphism, Population structure
INTRODUCTION
The sugar kelp Saccharina latissima (Linnaeus) C.E.Lane, C.Mayes, Druehl & G.W.Saunders, previously known as Laminaria saccharina (Linnaeus) J.V.Lamouroux, is a brown macroalga broadly distributed in both temperate and polar rocky coastal sites in the northern hemisphere, and like many kelp species, plays important roles as nursery habitat, shelter, and substrate for many organisms (Bolton 2010, Devit & Saunders 2010). S. latissima can grow to immense sizes (>4 m in length) and occurs attached to rocks, mussels, and man-made substrates from the intertidal zone to 26 m depths (Borum et al. 2002; Bartsch et al. 2008; Mathieson & Dawes 2017). As a biennial or perennial species, this kelp is characterized by rapid, early seasonal growth rates and sporangium formation in the autumn and winter, when daylight and temperature decrease (Lüning 1979; Bartsch et al. 2008; Mathieson & Dawes 2017). Meiospore dispersal is limited, and largely driven by water currents (Billot et al. 2003; Bartsch et al. 2008). Haploid gametophytes mostly settle on hard substrates, or in some rarer cases can develop as endophytes inside other algal hosts, where fertilization takes place and new sporophytes form on the surface (Garbary et al. 1999; Lane & Saunders 2005; Bartsch et al. 2008).
Saccharina latissima has received considerable economic attention in recent years, due to a variety of uses, including in biofuels, bioremediation, feed supplements, and pharmaceutical products (Forbord et al. 2012; Marinho et al. 2015; Jahan et al. 2017). For example, S. latissima and other brown macroalgae contain biologically active phenolic compounds, such as phlorotannins, which are known to exhibit anti-diabetic, anticarcinogenic, and anti-human immunodeficiency virus (HIV) activities (Jormalainen & Honkanen 2004; Thomas & Kim 2011; Cornish et al. 2015; Vilg et al. 2015). In addition, S. latissima contains relatively high quantities of alginates, which function as storage polysaccharides and are used commercially as thickeners and emulsifiers in food industries (MacArtain et al. 2007). Consumption of kelp is also associated with several therapeutic and medicinal benefits, due in part to high quantities of alginates and mannitol (MacArtain et al. 2007; Kim & Bhatnagar 2011). Another abundant polysaccharide, laminarin, has been investigated in S. latissima as an alternative fermentation substrate for efficient bioethanol production (Adams et al. 2009). Due to these important uses, the demand for kelp is likely to increase in the future, and may exceed limited supplies from wild harvests and require new production from aquaculture (Peteiro & Freire 2013).
Recent cultivation efforts of S. latissima have led to many advances, including successful year-round meiospore induction, identification of optimal growing conditions at the nursery stage, and implementation in integrated multi-trophic aquaculture (IMTA) systems (Forbord et al. 2012; Sanderson et al. 2012; Rößner et al. 2014; Freitas et al. 2016). While many of these studies were conducted in Europe, sugar kelp aquaculture has also been investigated in both Canada and the USA (Brinkhuis et al. 1984; Troell et al. 2009; Redmond et al. 2014; Kim et al. 2015). Overall, however, seaweed industries in the northeast USA remains largely focused on wild harvests, with some recent efforts in aquaculture (Kim et al. 2014 2017; Johnson et al. 2014; Redmond et al. 2014; Augyte et al. 2017). In particular, there is an interest to develop S. latissima culture in the rural, economically impoverished region of eastern Maine (Hall-Arber et al. 2005), to both diversify local industries and preserve the traditional fishing lifestyle.
The development of a new regional industry in cultivation of S. latissima would benefit from a more thorough understanding of local population genetics, for proper management and conservation, as well as the identification of genetic resources and population dynamics, before farming techniques are established. To this end, the purpose of the present study was to characterize the fine-scale population genetic structure of sugar kelp in eastern Maine. Recently, twelve polymorphic microsatellite loci were identified in S. latissima and used to characterize the genetic population structure along a salinity gradient in the North Sea-Baltic transition zone (Nielsen et al. 2016). To our knowledge, however, these loci have not been assessed in boreal, northwest Atlantic populations, which also represents an opportunity to further evaluate the genetic diversity of this species.
MATERIALS AND METHODS
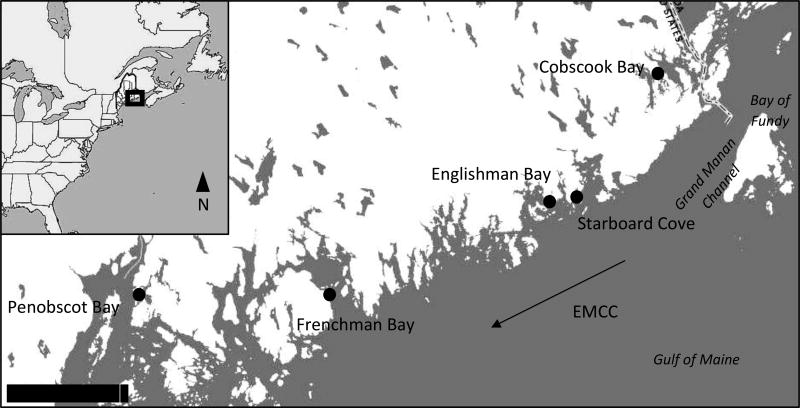
Adult sporophyte kelp samples were collected (approximately 40 individuals/site) from five intertidal sites in eastern Maine, spanning approximately 225 km, during the summer and early autumn of 2016 (Fig. 1). The sampling scheme included major bays in the region (Penobscot Bay, Frenchman Bay, and Cobscook Bay), as well as two coastal sites adjacent to the Gulf of Maine (Englishman Bay and Starboard Cove). Sampling was conducted at low tide and in a haphazard manner, to avoid collection of adjacent individuals that could originate from the same gametophyte. Blade fragments (~1 cm2) were removed from each individual and added to 1.5 ml microcentrifuge tubes with silica gel beads, to preserve and desiccate the samples, until DNA extractions were performed.
Fig. 1.
Map of sampling locations in eastern Maine. Scale bar refers to 30 km, and the arrow refers to the direction of the Eastern Maine Coastal Current (EMCC) in the region.
Since many brown algae contain abundant, viscous polysaccharides and polyphenols that may interfere with DNA extractions or downstream PCR applications (Snirc et al. 2010, Greco et al. 2014), genomic DNA was extracted from all kelp samples using a modified, combined cetyl trimethylammonium bromide (CTAB) and sodium dodecyl sulfate (SDS) protocol from Maeda et al. (2013). The protocol modifications included a different homogenization method, for ease of use, and several changes in reagent volumes and composition (β-mercaptoethanol and RNase A were not used). Briefly, a dried kelp sample fragment (~5 mm2) was added to a microcentrifuge tube with autoclaved sand (Sigma-Aldrich, St. Louis, MO, USA), 400 µl of extraction buffer (100 mM Tris-HCl pH 9.5, 50 mM EDTA pH 8.0, 500 mM NaCl), and 29 µl of 20% SDS. Each sample was fully homogenized by hand using sterile pestles (Nettleton et al. 2013) and incubated at 65°C for 30–45 min. Potassium acetate (5 M) was added (129 µl) to each sample, incubated at −20°C for 15 min, centrifuged (20,000 × g, 10 min), and the supernatant was transferred to a new tube. Cold isopropanol was added (280 µl), incubated at −20°C for 15 min, centrifuged again, and the DNA pellet was washed twice with 600 µl of cold 75% ethanol. Samples were centrifuged (20,000 × g, 5 min) between wash steps. The DNA pellets were vacuum-aspirated, resuspended in 100 µl of sterile water, and either stored at 4°C overnight or used immediately in successive steps. CTAB buffer (3% CTAB (w/v), 1% polyvinylpyrrolidone (w/v), 1.4 M NaCl, 20 mM EDTA pH 8.0, 100 mM Tris-HCl pH 9.5) was added (500 µl) to each sample, incubated at 65°C for 30 min, combined with 200 µl of 5 M potassium acetate, and incubated on ice for 15 min. Each sample was then centrifuged (20,000 × g, 10 min), the supernatant was transferred, and 650 µl of chloroform:isoamyl alcohol (24:1) was added. Samples were mixed vigorously by hand for 15 sec, centrifuged again, and the top aqueous layer was transferred to a new tube. The chloroform:isoamyl alcohol extractions were repeated twice more, 480 µl of cold isopropanol was then added, and samples were incubated at −20°C for 20 min. Samples were centrifuged again, DNA pellets were washed twice with 75% ethanol, as outlined above, and vacuum-aspirated prior to resuspension in sterile water. Extracted DNA was stored at −20°C.
Microsatellite analysis was performed using twelve loci for S. latissima previously characterized by Paulino et al. 2016. PCR was conducted in 15 µl volumes using 2 µl of DNA template, 1 × Promega GoTaq Flexi PCR buffer (Promega, Madison, WI, USA), 1.5–2.5 mM MgCl2, 0.2 µg/µl bovine serum albumin, 0.28 mM dNTPs, 0.1–0.3 µM of each primer, and 0.2 U GoTaq Flexi DNA polymerase. Multiplex PCRs were conducted following guidelines in Nielsen et al. 2016, except different fluorescent dyes were used to label either the forward or reverse primer: 1) SLN319 (FAM-labeled), SLN320 (FAM), SLN34 (HEX), and SLN35 (HEX), 2) SLN58 (FAM) and SLN62 (HEX), and 3) SLN32 (NED) and SLN36 (HEX). Other loci were used individually in PCR reactions, including SLN54 (HEX), SLN314 (FAM), SLN510 (FAM), and SLN511 (HEX). All PCR cycling conditions were conducted using a C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and followed established protocols by Paulino et al. 2016. PCR products were transported to the MDI Biological Laboratory (Salisbury Cove, ME, USA) for fragment analysis in an Applied Biosystems 3130XL capillary sequencer (Applied Biosystems, Foster City, CA, USA). Peaks were scored using PeakScanner 2.0 (Applied Biosystems), and raw scores were sorted manually into allelic bins. The same positive control sample was run on every PCR plate to ensure consistent scoring patterns and normalize interassay variation prior to binning.
Microsatellite data were evaluated for stuttering, large allele drop out, and the presence of null alleles using MICRO-CHECKER (Van Oosterhout et al. 2004). Tests for deviations from Hardy-Weinberg equilibrium and linkage disequilibrium were conducted in GENEPOP v3.4 (Raymond & Rousset 1995), using a Markov chain Monte Carlo (MCMC) method of 1,000 or 100 batches, respectively, and 10,000 iterations per test. Significance for multiple tests was adjusted following a standard Bonferroni correction (Rice 1989). Per locus FST values, a global FST, and tests of allelic differentiation using Fisher’s exact tests (1,000 batches and 10,000 iterations) were also conducted in GENEPOP. Observed and expected heterozygosities (Ho and He) and inbreeding coefficients (FIS values) for each locus and population were calculated using GDA (Lewis & Zaykin 2001). Significance of population FIS values were evaluated in GENETIX (Belkhir et al. 1996–2004). Number of alleles per locus and allelic richness were calculated using FSTAT 2.9.3 (Goudet 1995). To identify if the sampled kelp populations experienced a recent bottleneck effect, the program BOTTLENECK was used, with 1,000 iterations, all mutation models (88% stepwise mutations and 12% infinite allele mutations), and variance among multiple steps set at 12 (Piry et al. 1999). Wilcoxon sign-rank tests for significant heterozygosity excess and allele frequency distributions were used to detect evidence of a recent bottleneck (Luikart et al. 1998). The file conversion program CREATE was used to change input data file formats among all statistical tests (Coombs et al. 2008).
To identify genetic differences among the five populations, pairwise population FST values and their significance were evaluated in FSTAT. FST values were also used in principal component analysis (PCA) in GENALEX 6.503 (Peakall & Smouse 2012), to better visualize the population structure. An isolation-by-distance model for the genetic structure was evaluated using a Mantel test in GENALEX. The correlation between genetic and geographic distance was tested by comparing pairwise linearized FST values (FST(1−FST)) to the shortest coastal distances between sites (approximately within 5 km of coast). Geographic distances were calculated using the Measure feature in the ArcGIS Maine Basemap General viewer available from the State of Maine Office of GIS (http://maine.maps.arcgis.com/home/index.html).
To further characterize the population structure, without a priori clustering assumptions, a Bayesian approach in STRUCTURE 2.3.4 was used (Pritchard et al. 2000; Hubisz et al. 2009). The no admixture model with correlated allele frequencies and the LocPrior algorithm was used to infer population structure, since this is better suited to detect overall weak structuring (Hubisz et al. 2009; Pritchard et al. 2010) likely evident across the fine spatial scale in the present study. Five runs were conducted at each number of assumed populations (K), ranging one through five, with a 300,000 burn-in followed by 200,000 iterations. Data were uploaded to STRUCTURE HARVESTER (Earl & vonHoldt 2012) to determine the optimal number of clusters, using the ΔK method of Evanno et al. (2005). Following the identification of the optimal number of clusters, 25 additional runs were conducted in STRUCTURE using this number (K = 3), and the results were averaged using the greedy algorithm in CLUMPP 1.1.2 (Jakobsson & Rosenberg 2007). CLUMPP results were used to generate an averaged bar plot in DISTRUCT (Rosenberg 2004).
To further evaluate differences in population structure evident between FST- and STRUCTURE-based analyses, analyses of molecular variance (AMOVA) were conducted in ARLEQUIN 3.5.2.2 (Excoffier & Lischer 2010). Populations were grouped into either three (1-Penobscot Bay, 2-Cobscook Bay, and 3-Frenchman Bay-Englishman Bay-Starboard Cove) or four (1-Penobscot Bay, 2-Frenchman Bay, 3-Cobscook Bay, and 4-Englishman Bay-Starboard Cove) clusters, and global AMOVA results were averaged over all loci. Assignment tests in GENECLASS2 (Piry et al. 2004) were also performed, to further assess connectivity among populations and identify the ability of each individual to be assigned back to its correct population of origin. The Bayesian approach of Rannala & Mountain (1997), with a 0.05 assignment threshold, was used to calculate the log likelihood of an individual genotype originating from each population. Individuals were assigned to populations based on the greatest log likelihood value. To assess if the observed number of assignments to each population was significantly greater than by chance, chi-square goodness-of-fit tests (χ2) were conducted. The expected numbers of assignments were calculated by assuming equal proportions to each of the five populations (n/5).
RESULTS
Multilocus genotypes were obtained for 188 total samples, with at least 30 individuals from each site. Two microsatellite markers (SLN319 and SLN511) were not polymorphic in the sampled populations (fixed for 419 and 368 bp alleles, respectively) and were removed prior to analyses. In addition, SLN511 exhibited inconsistent scoring patterns among individuals, which likely indicated the presence of null alleles. At all other loci, only six total individuals exhibited missing data at one locus each, with three individuals at SLN35, one at SLN36, and two at SLN54. No evidence of stuttering, large allele dropout, or null alleles was detected at any locus in any population, except for SLN34 in Starboard Cove, where there was a significant homozygote excess (P < 0.05). Since an excess was not detected in other populations, however, the possibility of null alleles at SLN34 was not considered in later analyses. The final data set consisted of 10 polymorphic microsatellite markers and is available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.sp640.
Genetic diversity
No deviations from Hardy-Weinberg equilibrium were detected at any locus in any population (P > 0.05), and there was no evidence of linkage disequilibrium between pairs of loci following Bonferroni correction (P < 0.0011). The ten microsatellite loci exhibited variable levels of polymorphism (3–17 alleles) and allelic richness varied from 1.4 to 8.7 (Table 1). Observed and expected heterozygosities were also variable and ranged from 0.016 to 0.665 and 0.016 to 0.669, respectively (Table 1). FIS values for each locus were overall small but largest in SLN34 and SLN58 (Table 1). Locus FST values were also small and ranged from −0.0050 to 0.0170, with a global FST value of 0.0157. Most loci exhibited significant differentiation across all populations (at the α = 0.05 level), except for three loci with extremely low polymorphism (3–4 total alleles) (Table 1).
Table 1.
Number of alleles, allelic richness (AR), observed (Ho) and expected (He) heterozygosities, FIS, FST, and allelic differentiation P-values for 10 microsatellite markers.
| Locus | Alleles | AR | Ho | He | FIS | FST | P |
|---|---|---|---|---|---|---|---|
| SLN32 | 17 | 8.7 | 0.665 | 0.699 | 0.049 | 0.0170 | <0.0001 |
| SLN34 | 4 | 3.3 | 0.223 | 0.253 | 0.118 | 0.0032 | 0.2033 |
| SLN35 | 8 | 4.6 | 0.195 | 0.202 | 0.039 | 0.0300 | 0.0001 |
| SLN36 | 8 | 5.1 | 0.326 | 0.338 | 0.036 | 0.0035 | 0.0353 |
| SLN54 | 4 | 2.5 | 0.075 | 0.079 | 0.041 | 0.0049 | 0.1488 |
| SLN58 | 6 | 3.4 | 0.096 | 0.123 | 0.220 | 0.0203 | <0.0001 |
| SLN62 | 10 | 6.3 | 0.293 | 0.320 | 0.086 | 0.0092 | 0.0005 |
| SLN314 | 6 | 4.9 | 0.484 | 0.494 | 0.020 | 0.0388 | 0.0001 |
| SLN320 | 3 | 1.4 | 0.016 | 0.016 | −0.004 | −0.0050 | 0.7900 |
| SLN510 | 7 | 4.7 | 0.612 | 0.571 | −0.072 | 0.0062 | 0.0165 |
| All | 7.3 | 4.5 | 0.298 | 0.309 | 0.035 | 0.0157 | <0.0001 |
Bold indicates significance at the α = 0.05 level.
Genetic diversity was largely similar among the five sampled populations. Mean number of alleles and mean allelic richness ranged from 3.9 to 4.8 and 3.6 to 4.2, respectively (Table 2). Population observed and expected heterozygosities were overall small but largely similar to each other (0.283 to 0.339 and 0.273 to 0.340, respectively) and FIS values were not significantly different from zero (P > 0.05) (Table 2). Tests of bottleneck effects exhibited no evidence of heterozygosity excess in any population, under any mutation model (P > 0.90), and all allele frequencies exhibited normal L-shaped distributions (data not shown).
Table 2.
Geographic locations, sample sizes (n), mean number of alleles, mean allelic richness (AR), observed (Ho) and expected (He) heterozygosities, and FIS values for all populations.
| Site name | Latitude | Longitude | n | Mean alleles |
Mean AR |
Ho | He | FIS1 |
|---|---|---|---|---|---|---|---|---|
| Penobscot Bay | 44.387673 | −68.795717 | 39 | 3.9 | 3.6 | 0.283 | 0.273 | −0.037 |
| Frenchman Bay | 44.389029 | −68.199764 | 38 | 4.3 | 4.2 | 0.339 | 0.340 | 0.002 |
| Englishman Bay | 44.605977 | −67.467015 | 30 | 4.0 | 4.0 | 0.294 | 0.288 | −0.024 |
| Starboard Cove | 44.606107 | −67.396495 | 49 | 4.8 | 4.1 | 0.288 | 0.302 | 0.048 |
| Cobscook Bay | 44.883288 | −67.129206 | 32 | 4.4 | 4.2 | 0.288 | 0.326 | 0.116 |
FIS values were not significant at the α = 0.05 level.
Population structure
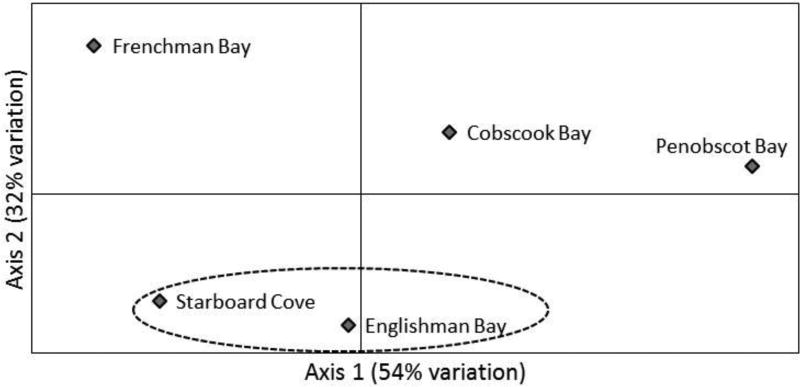
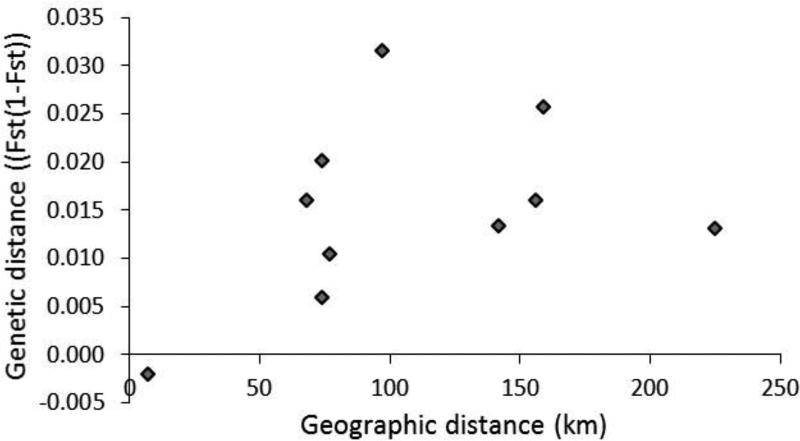
Significant population structuring was detected by FST analysis, with 8 out of 10 pairwise comparisons exhibiting significant differentiation following Bonferroni correction (P < 0.005) (Table 3). Significant pairwise FST values ranged from 0.0104 to 0.0316. In addition, the comparison between kelp from Englishman Bay and Cobscook Bay was significant at the α = 0.05 level (FST = 0.0061). Only Englishman Bay and Starboard Cove kelp exhibited a nonsignificant pairwise FST value (FST =−0.0020). The PCA exhibited clear separation between most populations, with no apparent clustering except for Englishman Bay and Starboard Cove. The greatest differentiation was evident between the Penobscot and Frenchman Bay populations (Fig. 2). The population structure could not be fully explained by an isolation-by-distance model, as the Mantel test identified no significant correlation between genetic and geographic distance among the five populations (P = 0.1290) (Fig. 3).
Table 3.
Pairwise FST values across 10 microsatellite markers for all kelp populations studied.
| Penobscot Bay | Frenchman Bay | Englishman Bay | Starboard Cove | |
|---|---|---|---|---|
| Frenchman Bay | 0.0316 | |||
| Englishman Bay | 0.0164 | 0.0202 | ||
| Starboard Cove | 0.0260 | 0.0104 | −0.0020 | |
| Cobscook Bay | 0.0133 | 0.0134 | 0.0061* | 0.0161 |
Bold indicates significance following standard Bonferroni correction (P < 0.005), while the asterisk indicates significance at the α = 0.05 level.
Fig. 2.
Principal component analysis of pairwise FST values for all sampled populations. The dotted circle encloses populations with a negative, non-significant (P > 0.05) pairwise value.
Fig. 3.
Mantel test of isolation-by-distance across all sampled kelp populations (P = 0.1290).
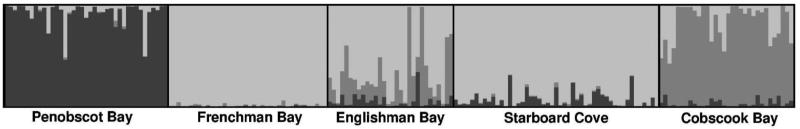
STRUCTURE analysis and the ΔK method identified three likely genetic clusters (K = 3). Sampling location information used in the LocPrior algorithm was informative for the population structure, as mean r = 1.14 across the 25 runs. The r parameter refers to how sampling locations deviate from the overall population. Large r values (>> 1) indicate that different locations likely have the same ancestry, while small values (near or below 1) indicate that location information was informative to the model and sampling locations differ in their ancestry (Hubisz et al. 2009). The three genetic clusters consisted of: 1) Penobscot Bay, 2) Cobscook Bay, and 3) Frenchman Bay, Englishman Bay, and Starboard Cove (Fig. 4). Kelp from Englishman Bay, however, also exhibited some evidence of mixed membership. Five individuals showed an elevated proportional membership (>59%) to the Cobscook Bay cluster, with two exhibiting >98% membership. In addition, two Cobscook Bay individuals and two Penobscot Bay individuals showed elevated proportional membership to the Frenchman Bay cluster (>52%, and 50%, respectively).
Fig. 4.
Genetic clustering identified through STRUCTURE analysis. Each vertical bar represents an individual kelp sample and its proportional membership to three clusters (light gray, gray, and dark gray). Black lines separate different populations.
Hierarchical AMOVA, using the three genetic clusters identified by STRUCTURE analysis, showed no evidence of significant genetic variation among the population clusters (P = 0.1020), while significant heterogeneity was present among populations within the Frenchman Bay cluster (P = 0.0112) (Table 4). In contrast, when the four genetic groupings evident by FST were used instead, significant variation existed among the clusters (P = 0.0064), with the Englishman Bay and Starboard Cove cluster being genetically homogenous (P = 0.6016) (Table 4). Both AMOVAs detected highly significant genetic variation within populations (>98% of total variation, P < 0.0001).
Table 4.
Analysis of molecular variance (AMOVA) averaged over 10 loci, using the five populations grouped into either three or four clusters. Df refers to degrees of freedom, while SS refers to sum of squares.
| Source of variation | df | SS | % variation | F-statistic | P-value |
|---|---|---|---|---|---|
| Three clusters1 | |||||
| Among clusters | 2 | 8.15 | 0.93 | 0.0093 | 0.1020 |
| Among populations within clusters | 2 | 5.34 | 0.96 | 0.0097 | 0.0112 |
| Within populations | 371 | 565.56 | 98.11 | 0.0189 | <0.0001 |
| Four clusters2 | |||||
| Among clusters | 3 | 12.20 | 2.01 | 0.0180 | 0.0064 |
| Among populations within clusters | 1 | 1.30 | −0.20 | −0.0021 | 0.6016 |
| Within populations | 371 | 565.56 | 98.20 | 0.0201 | <0.0001 |
Clusters: 1) Penobscot Bay, 2) Frenchman Bay, Englishman Bay, and Starboard Cove, and 3) Cobscook Bay
Clusters: 1) Penobscot Bay, 2) Frenchman Bay, 3) Englishman Bay and Starboard Cove, and 4) Cobscook Bay
Bold indicates significance at the α = 0.05 level.
Assignment tests also reflected evidence of small genetic divergence among the populations. The overall quality index for the assignment test was 40.66%, with only 45.2% of all individuals correctly assigned to their population of origin. Penobscot Bay kelp exhibited a relatively large correct assignment (71.8%), and its largest incorrect assignment was to Englishman Bay (12.8%) (Table 5). No Penobscot Bay individuals were assigned to Cobscook Bay. Frenchman Bay kelp exhibited a moderate correct assignment (50.0%), with a relatively large proportion assigned to another population in its STRUCTURE cluster (Starboard Cove: 26.7%). No Frenchman Bay individuals were assigned to Penobscot Bay. Englishman Bay individuals were equally assigned back to their original population or to Cobscook Bay (20% each), and a larger proportion was assigned to Starboard Cove (34.7%). Cobscook Bay kelp also exhibited only moderately correct assignments (46.9%), and the largest incorrect assignment was to Englishman Bay (18.8%). Proportional assignments for all populations were significantly greater than expected by chance, except for Englishman Bay individuals (P > 0.10), which indicated overall equal assignments across the five populations.
Table 5.
Percent assignment of all individuals in each population either: 1) back to their correct population of origin, or 2) incorrect assignment to a different population. P refers to probability thresholds associated with chi-square (χ2) values of each population test.
| % assignment | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Population | Penobscot Bay |
Frenchman Bay |
Englishman Bay |
Starboard Cove |
Cobscook Bay |
P |
| Penobscot Bay | 71.8 | 5.1 | 12.8 | 10.3 | 0.0 | < 0.0001 |
| Frenchman Bay | 0.0 | 50.0 | 10.5 | 26.3 | 13.2 | < 0.0001 |
| Englishman Bay | 16.7 | 6.7 | 20.0 | 36.7 | 20.0 | > 0.10 |
| Starboard Cove | 18.4 | 16.3 | 24.5 | 34.7 | 6.1 | < 0.05 |
| Cobscook Bay | 12.5 | 9.4 | 18.8 | 12.5 | 46.9 | < 0.01 |
Bold indicates significance at the α = 0.05 level.
DISCUSSION
Sugar kelp populations in eastern Maine were characterized by significant differentiation among sites but overall low levels of genetic diversity. Although no significant inbreeding was detected, low to moderate levels of polymorphism were evident at most microsatellite loci. In addition, two loci used in the present study were monomorphic across populations. Using the same markers, most individual kelp populations in European waters exhibited larger allelic richness and heterozygosity, irrespective of geographic location (Nielsen et al. 2016; Paulino et al. 2016). The only previously sampled European population, at these markers, to exhibit somewhat similar heterozygosities to those in the present study was found in the more isolated Vejle Fjord in Denmark (Ho = 0.318, He = 0.358), which was within the North Sea-Baltic salinity transition zone and characterized by a small population size and limited connectivity (Nielsen et al. 2016). Less genetic diversity was also detected in some populations of other algal species, including the kelp Laminaria digitata (Hudson) J.V.Lamouroux and the red alga Gigartina skottsbergii Setchell & N.L.Gardner, which were either spatially isolated by habitat discontinuities or in marginal zones of their geographic distribution, and exhibited significant, recent bottleneck effects (Billot et al. 2003; Faugeron et al. 2004; Robuchon et al. 2014). Kelp populations in eastern Maine, however, did not exhibit a recent bottleneck, and although they were located in boreal waters, were not at the southern edge of their distribution (Mathieson et al. 2008; Guiry & Guiry 2017; Mathieson & Dawes 2017). Instead, less genetic diversity in the present study may reflect a relatively recent colonization event in the northwest Atlantic after the last glacial maximum, as evidenced in another brown alga, Fucus vesiculosis Linnaeus (Coyer et al. 2011). F. vesiculosis populations along the Maine coast also exhibited relatively low genetic diversity at microsatellite loci, and were characterized by a high abundance of a single mitochondrial haplotype that implicated a European origin in glacial refugia (Muhlin & Brawley 2009; Coyer et al. 2011). Similarly, S. latissima populations in western Greenland waters exhibited fewer alleles and heterozygosities than European populations (Paulino et al. 2016). Reduced diversity of sugar kelp in eastern Maine, therefore, may reflect some ancestral genetic signature of a colonization and expansion event across the northwest Atlantic, rather than evidence of a marginal or bottlenecked population.
Kelp populations within the region exhibited significant differentiation, but at overall smaller levels than those previously detected in Saccharina spp. across broad spatial scales. For example, kelp population comparisons across greater geographic distances in the North and Baltic Seas, as well as in Chinese and Japanese waters, exhibited genetic differentiation an order of magnitude larger (FST > 0.1) than those observed in the present study (Liu et al. 2012; Nielsen et al. 2016). Small but significant population structuring, however, was still evident across the small spatial scale in eastern Maine waters, including among individuals in the major embayments of Penobscot, Frenchman, and Cobscook Bays. In particular, the greatest level of differentiation was detected between the Penobscot and Frenchman Bay populations, which were geographically closer to each other than several other comparisons, and contributed to an overall lack of evidence for a significant isolation-by-distance model to the population structure. In contrast, isolation-by-distance is relatively common in many kelp species, since meiospore dispersal is both spatially and temporally limited, and female gametophytes are more likely to be fertilized by nearby individuals (Bartsch et al. 2008; Alberto et al. 2010, 2011; Robuchon et al. 2014; Nielsen et al. 2016). Other geographical features, however, such as habitat continuity and local ocean currents, also greatly impact population structuring in macroalgae (Alberto et al. 2011). The relatively larger differentiation between the Penobscot and Frenchman Bay populations is likely driven by local patterns in the Eastern Maine Coastal Current (EMCC), which flows in a southwest direction along the Maine coast and is deflected offshore, by high outflow, in the vicinity of Penobscot Bay (Pettigrew et al. 2005). Meiospore recruitment from eastern locations into Penobscot Bay, therefore, is probably restricted, compared to other sites along the more continuous, eastern Maine coastline. In agreement, STRUCTURE-based analyses identified some evidence of genetic mixing among the Frenchman Bay, Englishman Bay, and Starboard Cove populations. Analyses of molecular variance, though, suggested that they did not represent a single cluster, since significant genetic variation was evident within the grouping. Rather, geographic distance likely contributed to their fine-scale structuring, as there was a larger separation between Frenchman Bay and the other two populations, which were close geographically and more genetically homogenous to each other.
The Cobscook Bay population exhibited significant differentiation from most other sites, likely due to reduced connectivity and partial geographic isolation, since the mouth of Cobscook Bay is connected to the Bay of Fundy and partially separated from the EMCC by the Grand Manan Channel. Some evidence of genetic mixing, however, was detected between the Cobscook and Englishman Bay populations. For example, a few individuals from Englishman Bay exhibited greater proportional membership to Cobscook Bay. In addition, assignment tests identified the Englishman Bay collection as a mixture of individuals from multiple populations, with assignments no greater than expected by chance, and equal individual assignments among itself and Cobscook Bay. This connectivity, however, did not include the centrally located Starboard Cove population, which in contrast, exhibited very small assignment scores to Cobscook Bay and a highly significant pairwise FST value. In many marine systems, population structure is characterized by asymmetrical transport, due in part to different habitat features and discontinuities among sites (Alberto et al. 2011). In the present study, the Englishman Bay site consists of predominately sandy substrate, with kelp sporadically attached to mussels and boulders, while all other locations exhibit more rocky substrates. In addition, Starboard Cove is a relatively sheltered site, compared to Englishman Bay, and receives a greater proportion of freshwater input from the local terrestrial environment. These local features may restrict meiospore recruitment, or perhaps, reduce gametophyte survival from a locally acclimated Cobscook Bay population that is likely exposed to stronger tidal currents and high nutrient levels (Mathieson et al. 2009). Kelp populations in other species, for instance, exhibit local acclimation with respect to latitude or habitat (Henkel & Hofmann 2008). More research, however, will be needed to elucidate the possible factors that drive these small, fine-scale differences in populations along the eastern Maine coastline.
The fine-scale genetic structure of kelp in this region is likely strongly influenced by the EMCC southwest current, as well as geographic isolation associated with major bays. Even along the fairly continuous coastline east of Penobscot Bay, some evidence of local differentiation was detected at small spatial scales, which is likely dependent on both habitat features and geographic distances that impact meiospore dispersal. Wild harvesting practices and future kelp cultivation should avoid fragmentation or extirpation of these local populations, especially considering the overall reduced genetic diversity when compared to European S. latissima populations. If future kelp aquaculture goals in this region are to be realized, management may be needed to maintain adequate diversity, and further research comparing local populations in culture is possibly warranted.
Acknowledgments
Funding was provided by the Maine Economic Improvement Fund – Small Campus Initiative. We thank Chris Smith from MDI Biological Laboratory for assistance with fragment analyses. Research reported in this project was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423.
References
- Adams JM, Gallagher JA, Donnison IS. Fermentation study on Saccharina latissima for bioethanol production considering variable pre-treatments. Journal of Applied Phycology. 2009;21:569–574. [Google Scholar]
- Alberto F, Raimondi PT, Reed DC, Coelho NC, Leblois R, Whitmer A, Serrão EA. Habitat continuity and geographic distance predict population genetic differentiation in giant kelp. Ecology. 2010;91:49–56. doi: 10.1890/09-0050.1. [DOI] [PubMed] [Google Scholar]
- Alberto F, Raimondi PT, Reed DC, Watson JR, Siegel DA, Mitarai S, Coelho N, Serrão EA. Isolation by oceanographic distance explains genetic structure for Macrocystis pyrifera in the Santa Barbara Channel. Molecular Ecology. 2011;20:2543–2554. doi: 10.1111/j.1365-294X.2011.05117.x. [DOI] [PubMed] [Google Scholar]
- Augyte S, Yarish C, Redmond S, Kim JK. Cultivation of a morphologically distinct strain of the sugar kelp, Saccharina latissima forma angustissima, from coastal Maine, USA, with implications for ecosystem services. Journal of Applied Phycology. 2017 doi: 10.1007/s10811-017-1102-x. [DOI] [Google Scholar]
- Bartsch I, Wiencke C, Bischof K, Buchholz CM, Buck BH, Eggert A, Feuerpfeil P, Hanelt D, Jacobsen S, Karez R, Karsten U, Molis M, Roleda MY, Schubert H, Schumann R, Valentin K, Weinberger F, Wiese J. The genus Laminaria sensu lato: recent insights and developments. European Journal of Phycology. 2008;43:1–86. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. Laboratorie Génome, Populations, Interactions, CNRS UMR 5171. Université de Montpellier II; Montpellier, France: 1996–2004. GENETIX 4.05.2 logiciel sous Windows TM pour la génétique des populations. [Google Scholar]
- Billot C, Engel CR, Rousvoal S, Kloareg B, Valero M. Current patterns, habitat discontinuities and population genetic structure: the case of kelp Laminaria digitata in the English Channel. Marine Ecology Progress Series. 2003;253:111–121. [Google Scholar]
- Bolton JJ. The biogeography of kelps (Laminariales, Phaeophyceae): a global analysis with new insights from recent advances in molecular phylogenetics. Helgoland Marine Research. 2010;64:263–279. [Google Scholar]
- Borum J, Pederson MF, Krause-Jensen D, Christensen PB, Nielsen K. Biomass, photosynthesis and growth of Laminaria saccharina in a high-arctic fjord, NE Greenland. Marine Biology. 2002;141:11–19. [Google Scholar]
- Brinkhuis BH, Mariani EC, Breda VA, Brady-Campbell MM. Cultivation of Laminaria saccharina in New York Marine Biomass Program. Hydrobiologia. 1984;116:226–282. [Google Scholar]
- Coombs JA, Letcher BH, Nislow KH. CREATE: a software to create input files from diploid genotypic data for 52 genetic software programs. Molecular Ecology Resources. 2008;8:578–580. doi: 10.1111/j.1471-8286.2007.02036.x. [DOI] [PubMed] [Google Scholar]
- Cornish ML, Critchley AT, Mouritsen OG. A role for dietary macroalgae in the amelioration of certain risk factors associated with cardiovascular disease. Phycologia. 2015;54:649–666. [Google Scholar]
- Coyer JA, Haorau G, Costa JF, Hogerdijk B, Serrão EA, Billard E, Valero M, Pearson GA, Olsen JL. Evolution and diversification within the intertidal brown macroalgae Fucus spiralis/F. vesiculosis species complex in the North Atlantic. Molecular Phylogenetics and Evolution. 2011;58:283–296. doi: 10.1016/j.ympev.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Devit DC, Saunders GW. A DNA barcode examination of the Laminariaceae (Phaeophyceae) in Canada reveals novel biogeographical and evolutionary insights. Phycologia. 2010;49:235–248. [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Faugeron S, Martínez EA, Correa JA, Cardenas L, Destombe C, Valero M. Reduced genetic diversity and increased population differentiation in peripheral and overharvested populations of Gigartina skottsbergii (Rhodophyta, Gigartinales) in southern Chile. Journal of Phycology. 2004;40:454–462. [Google Scholar]
- Forbord S, Skjermo J, Arff J, Handå A, Reitan KI, Bjerregaard R, Lüning K. Development of Saccharina latissima (Phaeophyceae) hatcheries with year-round production of zoospores and juvenile sporophytes on culture ropes for kelp aquaculture. Journal of Applied Phycology. 2012;24:393–399. [Google Scholar]
- Freitas JRC, Salinas Morrondo JM, Cremades Ugarte J. Saccharina latissima (Laminariales, Ochrophyta) farming in an industrial IMTA system in Galicia (Spain) Journal of Applied Phycology. 2016;28:377–385. [Google Scholar]
- Garbary DJ, Kim KY, Klinger T, Duggins D. Red algae as hosts for endophytic kelp gametophytes. Marine Biology. 1999;135:35–40. [Google Scholar]
- Goudet J. FSTAT (version 1.2): a computer program to calculate F-statistics. Journal of Heredity. 1995;86:485–486. [Google Scholar]
- Greco M, Sáez CA, Brown MT, Bitonti MB. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model brown alga. PLoS One. 2014;9:e96470. doi: 10.1371/journal.pone.0096470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiry MD, Guiry GM. World-wide electronic publication. National University of Ireland; Galway: 2017. AlgaeBase. http://www.algaebase.org; searched on 29 May 2017. [Google Scholar]
- Hall-Arber M, Dyer C, Poggie J, McNally J, Gagne R. New England’s fishing communities. MIT Sea Grant College Program; Massachusetts: 2005. p. 426. [Google Scholar]
- Henkel SK, Hofmann GE. Differing patterns of hsp70 gene expression in invasive and native kelp species: evidence for acclimation-induced variation. Journal of Applied Phycology. 2008;20:915–924. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan A, Ahmad IZ, Fatima N, Ansari VA, Akhtar J. Algal bioactive compounds in the cosmeceutical industry: a review. Phycologia. 2017;56:410–422. [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Johnson RB, Kim J-K, Armbruster LC, Yarish C. Nitrogen allocation of Gracilaria tikvahiae grown in urbanized estuaries of Long Island Sound and New York City, USA: a preliminary evaluation of ocean farmed Gracilaria for alternative fish feeds. Algae. 2014;29:227–235. [Google Scholar]
- Jormalainen V, Honkanen T. Variation in natural selection for growth and phlorotannins in the brown alga, Fucus vesiculosis. Journal of Evolutionary Biology. 2004;17:807–820. doi: 10.1111/j.1420-9101.2004.00715.x. [DOI] [PubMed] [Google Scholar]
- Kim S-K, Bhatnagar I. Physical, chemical, and biological properties of wonder kelp – Laminaria. In: Kim S-K, editor. Advances in food and nutrition research. Vol. 64. Academic Press; Massachusetts: 2011. pp. 85–95. [DOI] [PubMed] [Google Scholar]
- Kim J-K, Kraemer GP, Yarish C. Field scale evaluation of seaweed aquaculture as a nutrient bioextraction strategy in Long Island Sound and the Bronx River Estuary. Aquaculture. 2014;433:148–156. [Google Scholar]
- Kim J-K, Kraemer GP, Yarish C. Use of sugar kelp aquaculture in Long Island Sound and the Bronx River Estuary for nutrient extraction. Marine Ecology Progress Series. 2015;531:155–166. [Google Scholar]
- Kim J-K, Yarish C, Hwang EK, Park M, Kim Y. Seaweed aquaculture: cultivation technologies, challenges, and its ecosystem services. Algae. 2017;32:1–13. [Google Scholar]
- Lane CE, Saunders GW. Molecular investigation reveals epi/endophytic extrageneric kelp (Laminariales, Phaeophyceae) gametophytes colonizing Lessoniopsis littoralis thalli. Botanica Marina. 2005;48:426–436. [Google Scholar]
- Lewis PO, Zaykin D. Genetic data analysis: computer program for analysis of allelic data. [18 May 2016];Version 1.1. 2001 Available at: http://phylogeny.uconn.edu/software/
- Liu F, Yao J, Wang X, Repnikova A, Galanin DA, Duan D. Genetic diversity and structure within and between wild and cultivated Saccharina japonica (Laminariales, Phaeophyta) revealed by SSR markers. Aquaculture. 2012;358–359:139–145. [Google Scholar]
- Luikart G, Allendorf FW, Cornuet J-M, Sherwin WB. Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity. 1998;89:238–247. doi: 10.1093/jhered/89.3.238. [DOI] [PubMed] [Google Scholar]
- Lüning K. Growth strategies of three Laminaria species (Phaeophyceae) inhabiting different depth zones in the sublittoral region of Helgoland (North Sea) Marine Ecology Progress Series. 1979;1:195–207. [Google Scholar]
- MacArtain P, Gill CIR, Brooks M, Campbell R, Rowland IR. Nutritional value of edible seaweeds. Nutrition Reviews. 2007;65:535–543. doi: 10.1301/nr.2007.dec.535-543. [DOI] [PubMed] [Google Scholar]
- Maeda T, Kawai T, Nakaoka M, Yotsukora N. Effective DNA extraction method for fragment analysis using capillary sequencer of the kelp, Saccharina. Journal of Applied Phycology. 2013;25:337–347. [Google Scholar]
- Marinho GS, Holdt SL, Birkeland MJ, Angelidaki I. Commercial cultivation and bioremediation potential of sugar kelp, Saccharina latissima, in Danish waters. Journal of Applied Phycology. 2015;27:1963–1973. [Google Scholar]
- Mathieson AC, Hehre EJ, Dawes CJ, Neefus CD. An historical comparison of seaweed populations from Casco Bay, Maine. Rhodora. 2008;110:1–102. [Google Scholar]
- Mathieson AC, Dawes CJ, Hehre EJ, Harris LG. Floristic studies of seaweeds from Cobscook Bay, Maine. Northeastern Naturalist. 2009;16(Monograph 5):1–48. [Google Scholar]
- Mathieson AC, Dawes CJ. Seaweeds of the northwest Atlantic. University of Massachusetts Press; Massachusetts: 2017. p. 816. [Google Scholar]
- Muhlin JF, Brawley S. Recent vs relic: discerning the genetic signature of Fucus vesiculosis (Heterokontophyta: Phaeophyceae) in the Northwestern Atlantic. Journal of Phycology. 2009;45:828–837. doi: 10.1111/j.1529-8817.2009.00715.x. [DOI] [PubMed] [Google Scholar]
- Nettleton JC, Mathieson AC, Thornber C, Neefus CD, Yarish C. Introduction of Gracilaria vermiculophylla (Rhodophyta, Gracilariales) to New England, USA: estimated arrival times and current distribution. Rhodora. 2013;115:28–41. [Google Scholar]
- Nielsen MM, Paulino C, Neiva J, Krause-Jensen D, Bruhn A, Serrão EA. Genetic diversity of Saccharina latissima (Phaeophyceae) along a salinity gradient in the North Sea-Baltic Sea transition zone. Journal of Phycology. 2016;52:523–531. doi: 10.1111/jpy.12428. [DOI] [PubMed] [Google Scholar]
- Paulino C, Neiva J, Coelho NC, Aires T, Marbà N, Krause-Jensen D, Serrão EA. Characterization of 12 polymorphic microsatellite markers in the sugar kelp Saccharina latissima. Journal of Applied Phycology. 2016;28:3071–3074. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peteiro C, Freire O. Biomass yield and morphological features of the seaweed Saccharina latissima cultivated at two different sites in a coastal bay in the Atlantic coast of Spain. Journal of Applied Phycology. 2013;25:205–2013. [Google Scholar]
- Pettigrew NR, Churchill JH, Janzen CD, Mangum LJ, Signell RP, Thomas AC, Townsend DW, Wallinga JP, Xue H. The kinematic and hydrographic structure of the Gulf of Maine Coastal Current. Deep-Sea Research II. 2005;52:2369–2391. [Google Scholar]
- Piry S, Luikart G, Cornuet J-M. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. Journal of Heredity. 1999;90:502–503. [Google Scholar]
- Piry S, Alapetite A, Cornuet J-M, Paetkau D, Baudouin L, Estoup A. GeneClass 2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D. [18 May 2016];Documentation for structure software: version 2.3. 2010 Available at: http://pritch.bsd.uchicago.edu/structure/html.
- Rannala B, Mountain JL. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences USA. 1997;94:9197–9201. doi: 10.1073/pnas.94.17.9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2) population genetic software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Redmond S, Green L, Yarish C, Kim J, Neefus C. New England seaweed culture handbook. [26 May 2017];2014 Available at: http://digitalcommons.uconn.edu/seagrant_weedcult/1.
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Rößner Y, Krost P, Schulz C. Increasing seaweed crop yields through organic fertilisation at the nursery stage. Journal of Applied Phycology. 2014;26:753–762. [Google Scholar]
- Robuchon M, Le Gall L, Mauger S, Valero M. Contrasting genetic diversity patterns in two sister kelp species co-distributed along the coast of Brittany, France. Molecular Ecology. 2014;23:2669–2685. doi: 10.1111/mec.12774. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Sanderson JC, Dring MJ, Davidson K, Kelly MS. Culture, yield and bioremediation potential of Palmaria palmata (Linnaeus) Weber & Mohr and Saccharina latissima (Linnaeus) Aquaculture. 2012;354–355:128–135. [Google Scholar]
- Snirc A, Silberfeld T, Bonnet J, Tillier A, Tuffet S, Sun J-S. Optimization of DNA extraction from brown algae (Phaeophyceae) based on a commercial kit. Journal of Phycology. 2010;46:616–621. [Google Scholar]
- Thomas NV, Kim S-K. Potential pharmacological applications of polyphenolic derivatives from marine brown algae. Environmental Toxicology and Pharmacology. 2011;32:325–335. doi: 10.1016/j.etap.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Troell M, Joyce A, Chopin T, Neori A, Buschmann AH, Fang J-G. Ecological engineering in aquaculture – potential for integrated multi-trophic aquaculture (IMTA) in marine offshore systems. Aquaculture. 2009;297:1–9. [Google Scholar]
- Van Oosterhoult C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Vilg JV, Nylund GM, Werner T, Qvirist L, Mayers JJ, Pavia H, Undeland I, Albers E. Seasonal and spatial variation in biochemical composition of Saccharina latissima during a potential harvesting season for western Sweden. Botanica Marina. 2015;58:435–447. [Google Scholar]