Abstract
Purpose
Detecting signals of micrometastatic disease in patients with early breast cancer (EBC) could improve risk stratification and allow better tailoring of adjuvant therapies. We previously showed that postoperative serum metabolomic profiles were predictive of relapse in a single-center cohort of estrogen receptor (ER)–negative EBC patients. Here, we investigated this further using preoperative serum samples from ER-positive, pre-menopausal women with EBC who were enrolled in an international phase III trial.
Experimental Design
Proton nuclear magnetic resonance (NMR) spectroscopy of 590 EBC samples (319 with relapse or ≥6 years clinical follow-up) and 109 metastatic breast cancer (MBC) samples was performed. A Random Forest (RF) classification model was built using a training set of 85 EBC and all MBC samples. The model was then applied to a test set of 234 EBC samples, and a risk of recurrence score was generated on the basis of the likelihood of the sample being misclassified as metastatic.
Results
In the training set, the RF model separated EBC from MBC with a discrimination accuracy of 84.9%. In the test set, the RF recurrence risk score correlated with relapse, with an AUC of 0.747 in ROC analysis. Accuracy was maximized at 71.3% (sensitivity, 70.8%; specificity, 71.4%). The model performed independently of age, tumor size, grade, HER2 status and nodal status, and also of Adjuvant! Online risk of relapse score.
Conclusions
In a multicenter group of EBC patients, we developed a model based on preoperative serum metabolomic profiles that was prognostic for disease recurrence, independent of traditional clinicopathologic risk factors.
Introduction
In the treatment of early breast cancer (EBC), risk stratification based on prognostic features is critical to decisions about the appropriate adjuvant strategy, in particular whether or not chemotherapy is warranted. Molecular profiling of the primary tumor has improved on traditional clinicopathologic risk stratification, yet still a significant proportion of “high risk” patients do not relapse and may receive chemotherapy unnecessarily (1–3). In addition to focusing on the characteristics of the primary cancer, an improved method to detect the actual presence of micrometastatic disease would help to identify those who might benefit from adjuvant therapies and those who may not.
Metabolomics is the study of metabolites (small molecules) in blood, tissue, or other biological samples, where the presence and relative concentrations of these molecules can be used as evidence of cellular processes and functions. Given that cancer cells can have significantly altered metabolism, the pattern of metabolites produced can yield a “signature” that may indicate the cancer’s presence or behavior (4). Importantly, and in contrast to gene expression profiling as a risk stratifier, this is a signal that originates directly or indirectly from micrometastatic disease, rather than one derived from features of the primary tumor. Furthermore, the surrounding stroma and immune response may also contribute to an altered metabolomic profile, thus offering combined information on residual tumor and host response. A major challenge in metabolomics is detecting this signature against the dynamic sea of metabolic data from normal cellular function.
Several groups including our own have identified a metastatic “signature” in patients with advanced breast cancer, using nuclear magnetic resonance (NMR) spectra or mass spectrometry to analyze the metabolites in biological samples, primarily serum (5–7). We compared the NMR spectra of serum from a group of EBC patients and a group of metastatic breast cancer (MBC) patients and identified a metastatic signature that could differentiate the two groups (5).
From there, we hypothesized that EBC patients with micrometastatic disease may also have features of the metastatic signature in their metabolomic profile, whereas those with no micrometastatic disease would not, and that this signature would predict for relapse. This hypothesis was tested in a follow-up study using serum from a biobank of estrogen receptor–negative (ER−) patients from the Memorial Sloan Kettering Cancer Center (New York, NY), for whom clinical outcome (relapse at 5 years) was known (8). A model was built in which EBC patients were assigned a metabolomic risk score [Random Forest (RF) risk score], which was a function of the likelihood that they would be misclassified as metastatic based on their serum NMR spectra. Again, we were able to demonstrate that EBC and MBC profiles differed, but importantly, we also demonstrated that the RF risk score could predict relapse, independent of traditional clinicopathologic risk factors, in this single-center group of ER− EBC women.
In this current study, we aimed to test the RF risk score again as a predictor of relapse in a large group of premenopausal EBC patients with ER-positive (ER+) disease taking part in a multi-center adjuvant trial.
Patients and Methods
This retrospective study was a collaborative project among the International Breast Cancer Research Foundation, the University of Florence Magnetic Resonance Centre (Florence, Italy), and the Sandro Pitigliani Medical Oncology Department, Hospital of Prato (Prato, Italy). The study protocol received ethics approval from the ethics committee of the Hospital of Prato.
Patient selection
Serum samples for analysis were obtained from a bank of blood samples that had been collected during a phase III adjuvant breast cancer clinical trial (NCT00201851; ref. 9) and a parallel phase III MBC clinical trial (NCT00293540; ref. 10) conducted at centers across South East Asia. Both the trials were run by the International Breast Cancer Research Foundation.
In the adjuvant trial, 740 premenopausal women with stage II–IIIB hormone receptor (HR)–positive breast cancer received surgical oophorectomy at the time of breast cancer surgery (mastectomy), followed by tamoxifen for 5 years, to investigate the hypothesis that surgery performed during the luteal phase of the menstrual cycle would be associated with better outcomes. At the time of enrollment, 231 patients were estimated to be in the luteal phase and were scheduled for immediate surgery; 509 patients were estimated not to be in the luteal phase and were randomized to receive either immediate surgery or surgery scheduled to occur in the predicted mid luteal phase (9). Blood samples were collected preoperatively in fasted patients on the day of surgery. Frozen sera were initially stored at local sites and then shipped frozen to the United States. Subsequently specimens were shipped still frozen to Italy. No patients were recorded as diabetic. The trial was designed to follow patients for recurrence for at least 6 years, and deidentified clinical outcome data were made available for the purposes of this study. The study was approved at individual participating institutions in the Philippines, Vietnam, and Morocco and/or by supervising Institutional Review Boards for these institutions and at lead investigator’s American institutions. The consent processes addressed the use of samples for future research studies.
In the metastatic trial, premenopausal patients with ER+ MBC were randomized to undergo oophorectomy surgery as palliative endocrine therapy in either the follicular or the luteal phase of the menstrual cycle, followed by tamoxifen (10). Blood samples were collected preoperatively from fasted patients on the day of surgery. Frozen sera were initially stored at local sites and then shipped frozen to the United States. Subsequently specimens were shipped still frozen to Italy. Diabetic status of patients was not recorded.
NMR sample preparation
Frozen serum samples were thawed at room temperature and shaken before use and then were prepared according to standard operating procedures (11).
A total of 300 μL of sodium phosphate buffer (70 mmol/L Na2HPO4; 20% (v/v) 2H2O; 0.025% (v/v) NaN3; 0.8% (w/v) sodium trimethylsilyl [2,2,3,3-2H4]propionate pH 7.4) was added to 300 μL of each serum sample, and the mixture was homogenized by vortexing for 30 seconds. A total of 450 μL of this mixture was transferred into a 4.25-mm NMR tube (Bruker BioSpin srl) for the analysis.
NMR analysis
Monodimensional 1H NMR spectra for all samples were acquired using a Bruker 600 MHz spectrometer (Bruker BioSpin) operating at 600.13 MHz proton Larmor frequency and equipped with a 5-mm CPTCI 1H-13C-31P and 2H-decoupling cryoprobe, including a z-axis gradient coil, an automatic tuning-matching, and an automatic sample changer. A BTO 2000 thermocouple served for temperature stabilization at the level of approximately 0.1 K at the sample. Before measurement, samples were kept for at least 3 minutes inside the NMR probehead for temperature equilibration (310 K for serum samples).
According to standard practice (12, 13), three monodimensional 1H NMR spectra with different pulse sequences were acquired for each serum sample, allowing the selective detection of different molecular components:
a standard nuclear Overhauser effect spectroscopy pulse sequence NOESY 1Dpresat (noesygppr1d.comp; Bruker BioSpin) using 64 scans, 98,304 data points, a spectral width of 18,028 Hz, an acquisition time of 2.7 seconds, a relaxation delay of 4 seconds, and a mixing time of 0.1 second was applied to obtain a spectrum in which both signals of metabolites and high molecular weight macromolecules (lipids and lipoproteins) are visible.
a standard spin echo Carr–Purcell–Meiboom–Gill (CPMG; ref. 14; cpmgpr1d.comp; Bruker BioSpin) pulse sequence with 64 scans, 73,728 data points, a spectral width of 12,019 Hz, and a relaxation delay of 4 seconds was used for the selective observation of low molecular weight metabolites, suppressing signals arising from macromolecules.
a standard diffusion-edited (ledbgppr2s1d.comp; Bruker BioSpin; ref. 15) pulse sequence, using 64 scans, 98,304 data points, a spectral width of 18,028 Hz, and a relaxation delay of 4 seconds was applied to suppress metabolite signals.
Spectral processing
Free induction decays were multiplied by an exponential function equivalent to a 1.0-Hz line-broadening factor before applying Fourier transformation. Transformed spectra were automatically corrected for phase and baseline distortions and calibrated (anomeric glucose doublet at 5.24 ppm) using TopSpin 3.2 (Bruker Biospin srl). Each 1D spectrum in the range 0.2 to 10.00 ppm was segmented into 0.02-ppm chemical shift bins, and the corresponding spectral areas were integrated using AMIX software (version 3.8.4, Bruker BioSpin). Binning is a means to reduce the number of total variables and to compensate for small shifts in the signals, making the analysis more robust and reproducible (16, 17). Regions between 4.5 and 6.5 ppm containing residual water signal were removed, and the dimension of the system was reduced to 391 bins. The total spectral area was calculated on the remaining bins, and total area normalization was carried out on the data prior to pattern recognition.
Statistical analysis
Statistical analyses were planned prior to specimen retrieval, based on those performed in the previous study, including minimum number of samples required (8). All data analyses were performed using R (18). Principal component analysis (PCA) was used first as an unsupervised exploratory analysis to assess the presence of any clusters or outliers.
To confirm that serum metabolomic profiles can be used to distinguish patients with MBC from those with early disease, an RF classifier (19) was built to separate early and metastatic patients. For the initial model, the group of EBC patients who had relapsed or had minimum 5 years clinical follow-up was randomly split into two groups, to form a training set and a validation set, as in the previous study (8). Briefly, the RF classifier uses data from the metastatic and training set to build an ensemble of decision trees, where each tree contains a random sample of the original data, with only a small number of variables (bins) at each decision node, used to predict whether a sample is early or metastatic. For early patients, a score was created that expresses the extent to which the serum metabolomic profile appears to be metastatic, designated as the “RF risk score.” For each patient, three “RF risk scores” were derived using the three types of spectra (NOESY1D, CPMG, and diffusion-edited spectra). For all calculations, the R package “Random Forest” (20) was used to grow a forest of 1,000 trees, using the default settings.
The next step was to test the hypothesis that a metastatic metabolomic signature in early disease would be predictive of relapse and that higher RF relapse scores would correlate with higher risk of developing a relapse. Using ROC analysis, the performance of the RF risk score was compared with actual breast cancer outcome. A prognostic model was created using the CPMG RF risk score, which had the best performance in the training set. To delineate high risk of relapse, a cutoff for the RF risk score was calculated in the training set that optimized accuracy, sensitivity, and specificity, and the performance of the model was subsequently tested in the validation set.
Multivariate analysis of the impact of provenance of the sample was achieved using unsupervised PCA of the spectra. When this impact was found to be significant, the model for relapse prediction was redesigned:
We hypothesized that samples from different clinical sites had been collected or stored following different operating procedures (e.g., longer periods from collection to sera separation and freezing, or different freezing temperatures), and that this may be reflected in the metabolomic spectra. As reported in the literature (11), lactate (coupled with pyruvate and glucose) is the most sensitive marker for sample degradation. To overcome this influence, we removed the bins related to lactate from the data matrix.
The nonrelapsing patients included in the analysis were restricted to those with a minimum follow-up of 6 years, as HR+ breast cancer has a relatively steady relapse rate for at least 10 years.
Finally, we chose to include in the training set only women who had not developed a recurrence, to reduce the likelihood of confounding factors due to the presence of patients with micrometastases in the model. Thus, ROC analysis could only be carried out on the subsequent test set of relapsed and nonrelapsed patients.
Assessment of confounding factors (e.g., age, tumor size, nodal status, etc.) within the spectra was performed by using the multivariate RF classifier analysis to determine whether spectra could be predictive of each factor. The independent prognostic capacity of the redesigned RF risk score model was evaluated in a multivariate analysis controlling for standard prognostic features, which also included an Adjuvant! Online (AoL) risk of relapse score. The AoL score was calculated for 10-year risk of relapse assuming no adjuvant therapy and was used as a surrogate combined clinicopathologic risk.
For the analysis of individual metabolites, the spectral regions related to 22 metabolites were assigned in the 1H CPMG NMR profiles by using matching routines of AMIX 3.8.4 (Bruker BioSpin) in combination with the BBIOREFCODE (Bruker BioS-pin) and the Human Metabolome Database (21). The spectral regions were fitted and integrated to obtain the concentration in arbitrary units, and these data were used to compare metabolite concentrations between EBC and MBC patients. Wilcoxon signed-rank test (22) was chosen to perform the analysis on the biological asymptotic assumption that the metabolite concentrations are not normally distributed, and FDR correction was applied using the Benjamini–Hochberg method (23). P < 0.05 was deemed significant. Because of the method used to generate spectra, NMR profiles could not be used to measure individual lipid concentrations, nor metabolites in very small concentrations, such as acylcarnitines.
Results
Patients
Serum samples from 675 women with EBC and 125 with MBC were received. Of these, 101 samples were deemed nonevaluable for technical reasons (plasma instead of serum, inadequate amount of serum, hemolysis, and insufficient clinical information), leaving 590 EBC and 109 MBC samples suitable for NMR spectroscopy to build metabolomic profiles. Baseline characteristics are reported in Table 1.
Table 1.
Patients and tumor characteristics for EBC and MBC cohorts, including populations restricted to include only relapsed patients or those with clinical follow-up greater than 5 or 6 years
| Characteristic | EBC all | EBC – relapsed or follow-up ≥5 years | EBC – relapsed or follow-up ≥6 years | MBC |
|---|---|---|---|---|
| Number | 590 | 443 | 319 | 109 |
| Age, mean (range) | 42 (29–50) | 42 (29–50) | 42 (29–50) | 39 (22–53) |
| Tumor size, n (%) | ||||
| <2 cm | 35 (5.9) | 23 (5.2) | 11 (3.5%) | — |
| 2–5 cm | 396 (67.1) | 285 (64.3) | 203 (63.6%) | |
| >5 cm | 159 (27) | 135 (30.5) | 105 (32.9%) | |
| Grade, n (%) | ||||
| I | 74 (13) | 63 (14) | 46 (14) | — |
| II | 300 (51) | 224 (51) | 162 (51) | |
| III | 115 (19) | 89 (20) | 73 (23) | |
| Unknown | 101 (17) | 67 (15) | 38 (12) | |
| Lymph node status, n (%) | ||||
| 0 | 248 (42) | 166 (37.5) | 106 (33) | — |
| 1–3 | 157 (27) | 121 (27.5) | 83 (26) | |
| >3 | 185 (31) | 156 (35) | 130 (41) | |
| HER2, n (%) | ||||
| Positive | 108 (18) | 90 (20.5) | 76 (24) | — |
| Negative | 388 (66) | 298 (67) | 210 (66) | |
| Unknown | 94 (16) | 55 (12.5) | 33 (10) | |
| ER, n (%) | ||||
| Positive | 552 (93.6) | 410 (92.6) | 297 (93) | — |
| Negative | 37 (6.3) | 32 (7.2) | 22 (7) | |
| Unknown | 1 (0.2) | 1 (0.2) | 0 (0) | |
| PR, n (%) | ||||
| Positive | 545 (92.4) | 405 (91.4) | 291 (91) | — |
| Negative | 44 (7.4) | 37 (8.4) | 28 (9) | |
| Unknown | 1 (0.2) | 1 (0.2) | 0 (0) | |
| Treatment arm, n (%) | ||||
| A | 186 (31.5) | 142 (32.0) | 106 (33.2) | — |
| B | 216 (36.6) | 158 (35.7) | 111 (34.8) | |
| C | 188 (31.9) | 143 (32.3) | 102 (32.0) | |
| Dominant metastatic site, n (%) | ||||
| Soft tissue | — | — | — | 79 (72.5) |
| Bone | 17 (15.6) | |||
| Viscera | 13 (11.9) | |||
| Prior systemic treatment, n (%) | ||||
| No | — | — | — | 69 (63.3) |
| Yes | 40 (36.7) | |||
NOTE: Treatment arm A: not in luteal phase at the time of trial entry, randomized to luteal phase surgery; treatment arm B: not in luteal phase at the time of trial entry, randomized to immediate, non-luteal phase surgery; and treatment arm C: in luteal phase at the time of trial entry, immediate surgery in luteal phase.
Abbreviation: PR, progesterone receptor.
Provenance of samples
EBC samples came from 5 centers in the Philippines and 2 centers in Vietnam; MBC samples came from 5 centers in the Philippines, 3 centers in Bangladesh, and one in Nigeria (Table 2). Notably, no MBC samples came from Vietnam, and only 24 came from Philippine General Hospital in Manila, yet these centers contributed the majority of EBC samples, representing significant imbalance.
Table 2.
Distribution of EBC and MBC samples by treatment center
| Country | Samples, n | EBC samples, n | MBC samples, n |
|---|---|---|---|
| Vietnam, Hanoi - Hospital K | 228 | 228 | — |
| Vietnam, Danang - Danang General | 14 | 14 | — |
| Philippines, Manila - PGH | 302 | 278 | 24 |
| Philippines, Cebu - Vicente Sotto Hospital | 39 | 26 | 13 |
| Philippines, Manila - Santo Tomas Hospital | 9 | 3 | 6 |
| Philippines, Manila - Rizal | 20 | 15 | 5 |
| Philippines, Manila - East Avenue | 29 | 26 | 3 |
| Nigeria, Ibadan - University College Hospital | 8 | — | 8 |
| Bangladesh, Dhaka - Dhaka Medical College | 15 | — | 15 |
| Bangladesh, Khulna - Khulna Medical College | 28 | — | 28 |
| Bangladesh, Dhaka - BSMMU | 7 | — | 7 |
| Total | 699 | 590 | 109 |
Abbreviations: BSMMU, Bangabandhu Sheikh Mujib Medical University; PGH, Philippine General Hospital.
Discrimination between EBC and MBC patients
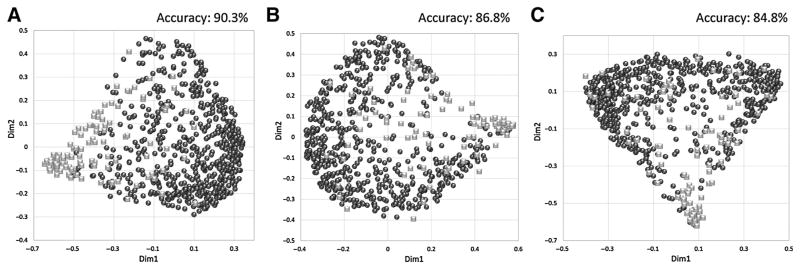
Using the RF classifier for supervised analysis, the metabolomic profiles of 590 EBC and 109 MBC patients were classified, and show significant differential clustering, with near-complete separation of the two groups (Fig. 1). Clustering was achieved by the CPMG, NOESY1D, and diffusion spectra.
Figure 1.
Clustering of serum metabolomic profiles. Discrimination between EBC (black circles, n = 590) and MBC (gray squares, n = 109) patients using the RF classifier. A–C, CPMG (A), NOESY1D (B), and diffusion (C).
As in the previous studies (5, 8), the clustering provided by the CPMG spectra shows the highest accuracy for predicting early or metastatic status, with accuracy of 90.3% [95% confidence interval (CI), 90.2%–90.4%], compared with 86.8% (95% CI, 86.7%–86.8%) for NOESY1D, and 84.4% (95% CI, 84.3%–84.5%) for diffusion edited. Only results for CPMG spectra will be reported from here on.
Relapse prediction by RF score
A metabolomic RF risk score for each EBC sample was generated on the basis of the probability that the NMR spectrum would be classified as metastatic. The initial model was built using the same parameters as in the previous study, using CPMG spectra and only including EBC samples from patients who either relapsed or were relapse free with a minimum of 5 years clinical follow-up data (total 443). The training set consisted of 68 relapsed and 41 nonrelapsed EBC patients chosen at random and all 109 metastatic patients. The validation set consisted of the remaining 124 relapsed and 210 nonrelapsed EBC patients. The AUC obtained for the training set was 0.644, and the accuracy of the RF risk score was maximized using a threshold of 0.18, which yielded sensitivity of 61.3% (95% CI, 60.3%–62.2%), specificity of 61.0% (95% CI, 60.6%–61.3%), and overall accuracy for predicting likelihood of relapse of 61.1% (95% CI, 60.6%–61.6%; Supplementary Fig. S1A). The model was then applied to the validation set, using the RF risk score threshold of 0.18, achieving a sensitivity, specificity, and predictive accuracy of 71.7%, 46.7%, and 62.4%, respectively, and an AUC of 0.631 (Supplementary Fig. S1B).
In view of the low AUC results, investigation of the effect of provenance (collection center) and length of follow-up was carried out.
Exploratory unsupervised PCA of the CMPG spectra showed marked differentiation among the different centers of collection (Supplementary Fig. S2A), with the spectral region of lactate resulting in the most relevant discrimination in the first two principal components. Lactate concentrations, calculated in arbitrary units from the spectra, differed significantly between EBC and MBC patients (Table 3), demonstrating the key role of lactate in both discrimination of EBC and MBC and in the identification of treatment centers. This finding was consistent with our hypothesis regarding differences in storage and handling between treatment centers in our samples.
Table 3.
Comparison of individual metabolite concentrations in arbitrary units in the serum NMR spectra of EBC and MBC patients, for all patients included in the study
| All hospitals | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| EBC | MBC | P | ||||
|
|
|
|||||
| Median | MAD | Median | MAD | |||
| Choline | Higher levels in MBC | 329.82 | 533.53 | 1,607.25 | 1,953.73 | 2.88E–16 |
| Acetate | 272.09 | 177.43 | 512.38 | 220.30 | 6.11E–14 | |
| Formate | 10.37 | 3.00 | 18.83 | 8.21 | 3.63E–20 | |
| Lactate | 1,240.10 | 334.19 | 2,117.70 | 659.71 | 1.57E–24 | |
| Glutamate | 269.60 | 121.22 | 431.99 | 154.55 | 1.59E–13 | |
| 3-Hydroxybutyrate | 88.30 | 58.74 | 134.81 | 81.05 | 8.53E–03 | |
| Phenylalanine | 253.18 | 71.80 | 374.63 | 106.38 | 1.17E–16 | |
| Glycine | 817.54 | 224.84 | 1,148.41 | 314.98 | 2.69E–14 | |
| Leucine | 585.41 | 178.33 | 770.75 | 288.17 | 4.59E–06 | |
| Alanine | 1,920.95 | 376.40 | 2,384.07 | 511.61 | 1.19E–07 | |
| Proline | 97.38 | 30.84 | 114.99 | 43.44 | 5.59E–03 | |
| Tyrosine | 156.48 | 36.38 | 182.26 | 42.73 | 4.59E–06 | |
| Isoleucine | 168.84 | 35.37 | 193.65 | 45.08 | 9.24E–04 | |
| Histidine | 162.96 | 39.65 | 186.24 | 47.26 | 1.60E–03 | |
| Creatine | 147.90 | 46.12 | 167.65 | 51.09 | 2.12E–02 | |
| Creatinine | 194.25 | 38.19 | 216.27 | 48.55 | 5.59E–03 | |
| Methionine | 118.72 | 27.45 | 127.43 | 32.47 | 3.65E–02 | |
| Citrate | 111.84 | 22.22 | 118.77 | 42.60 | >0.05 | |
| Valine | 1,138.91 | 171.58 | 1,169.77 | 211.08 | >0.05 | |
|
| ||||||
| Mannose | Higher levels in EBC | 68.19 | 13.57 | 64.38 | 14.09 | >0.05 |
| Glucose | 3,275.44 | 292.23 | 2,843.51 | 584.25 | 3.93E–08 | |
| Glutamine | 105.32 | 40.11 | 80.67 | 45.68 | 3.59E–02 | |
NOTE: Adjusted P values are for the significance of any difference between EBC and MBC. Abbreviation: MAD, median absolute deviation.
Relapse prediction by RF score—optimized model
To overcome the influence of lactate, we removed the bins related to this metabolite from the data matrix. The PCA score plot (Supplementary Fig. S2B) calculated using this reduced data matrix shows greatly reduced dispersion of the data points. This observation is confirmed by calculating the generalized variance (24) of the first three PCA components. This value (calculated as the determinant of the covariance matrix) represents the volume of the ellipsoid containing the data. Using the complete data matrix, we obtain a generalized variance of 16.8, whereas for the reduced data matrix, the generalized variance is 11.8, illustrating that removal of the bins corresponding to lactate indeed reduced spreading of the data, thus reducing the location effect.
The EBC cohort was restricted to those with relapse or minimum 6 years follow-up, which reduced the sample size to 319. In this new model, the training set consisted of 85 early patients without relapse (randomly selected) and all 109 metastatic patients. The test set contained 192 early patients that suffered relapse and the remaining 42 relapse-free early patients.
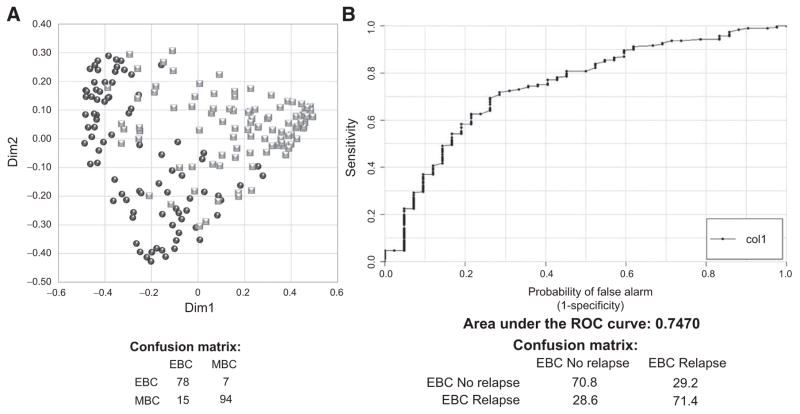
Using the CPMG NMR spectra, the RF classifier discriminated EBC from MBC patients in the training set with sensitivity, specificity, and predictive accuracy of 90.0% (95% CI 89.7%–90.3%), 84.9% (95% CI 84.7%–85.1%), and 87.1% (95% CI 86.9%–87.3%), respectively (Fig. 2A). This new model was then applied to the test set to assess ability to predict relapse, attaining an AUC of 0.747. The accuracy of the RF risk score was maximized using a threshold of 0.235, which yielded sensitivity of 70.8%, specificity of 71.4%, and overall accuracy for predicting likelihood of relapse of 71.3% (Fig. 2B). AUC scores for NOESY1D and diffusion-editing spectra were inferior, at AUC 0.706 and 0.617, respectively.
Figure 2.
A, Discrimination between EBC patients without relapse for at least 6 years (black circles, n =85) and MBC patients (gray squares, n =109) using the RF classifier on CPMG spectra. The confusion matrix is also reported. B, Prediction of relapse in the test set containing 192 EBC with relapse and 42 EBC without relapse up to 6 years. The ROC and AUC scores are presented for the RF risk score on CPMG spectra. The confusion matrix is also reported.
The AUC score calculated on the RF score was assessed for significance against the null hypothesis of no prediction accuracy in the data, by means of 10,000 randomized class permutation tests. The estimate AUC score obtained after randomization is 0.531 (95% CI, 0.53–0.531), demonstrating the significance of our result (AUC, 0.747; P = 1.63 × 10−20) despite the problems encountered.
Comparison with known prognostic factors
The known prognostic factors age, tumor size (0–2 cm, 2.1–5 cm, >5 cm), nodal status (0, 1–3, >3), histologic grade, and HER2 overexpression were compared with the CPMG RF risk score, calculated on the optimized set, in univariate and multivariate regression analyses (Table 4). We also compared the RF risk score with the 10-year risk of recurrence as calculated by AoL in a separate multivariate analysis. In all cases, the RF risk score maintained independent prognostic value.
Table 4.
ORs for prognostic features and RF risk score in the optimized set of EBC patients, using univariate and multivariate analysis
| Characteristics | OR (univariate) | P | OR (multivariate) | P | OR (multivariate) | P |
|---|---|---|---|---|---|---|
| Age | ||||||
| <43 | 1.0 | 1.0 | — | — | ||
| ≥43 | 0.5205 | 5.06E–03 | 0.5429 | 0.0536 | ||
| Tumor size | ||||||
| <2 cm | 1.0 | 1.0 | — | — | ||
| 2–5 cm | 2.1115 | 0.2447 | 3.5484 | 0.1458 | ||
| >5 cm | 4.8124 | 0.0181 | 6.1417 | 0.0450 | ||
| Grade | ||||||
| I | 1.0 | 1.0 | — | — | ||
| II | 2.2533 | 0.0165 | 2.4557 | 0.0472 | ||
| III | 2.2858 | 0.0319 | 2.0559 | 0.1512 | ||
| Lymph node status | ||||||
| 0 | 1.0 | 1.0 | — | — | ||
| 1–3 | 4.2742 | 3.40E–06 | 3.0661 | 0.0048 | ||
| >3 | 11.8770 | 5.57E–15 | 7.6775 | 1.38E–07 | ||
| HER2 | ||||||
| Negative | 1.0 | 1.0 | — | — | ||
| Positive | 2.3243 | 5.52E–03 | 2.4567 | 0.0170 | ||
| ER | ||||||
| Positive | 1.0 | 1.0 | — | — | ||
| Negative | 0.9522 | 0.9130 | 1.3370 | 0.6822 | ||
| PR | ||||||
| Positive | 1.0 | 1.0 | — | — | ||
| Negative | 2.6099 | 0.0437 | 4.3288 | 0.0320 | ||
| Treatment arm | ||||||
| A | 1.0 | 1.0 | — | — | ||
| B | 0.8449 | 0.5522 | 0.7493 | 0.4606 | ||
| C | 0.5563 | 0.0397 | 0.5666 | 0.1558 | ||
| AoL score | ||||||
| ≤52.7 | 1.0 | — | — | 1.0 | ||
| 52.7–73.7 | 3.9519 | 2.04E–06 | 3.8740 | 7.13E–06 | ||
| >73.7 | 12.9138 | 1.84E–13 | 15.308 | 1.18E–13 | ||
| RF risk score | ||||||
| <0.235 | 1.0 | 1.0 | 1.0 | |||
| >0.235 | 2.7991 | 1.59E–05 | 3.1224 | 0.0004 | 3.4768 | 7.84E–06 |
NOTE: AoL score split into tertiles. A multivariate analysis using only RF risk score and AoL score is also reported in the last two columns. Abbreviation: PR, progesterone receptor.
Similarly, using RF classification to predict individual prognostic features based on the CPMG NMR spectra, none of these features could be meaningfully discriminated (Supplementary Fig. S3). Only the tumor size showed a weak concordance with the CPMG RF risk score (coefficient of correlation = 0.18; P value corrected with Bonferroni = 0.02).
Metabolite analysis
NMR spectra were analyzed to identify which metabolites were contributing to discrimination of MBC and EBC profiles. In the combined multicenter populations (Table 3), compared with EBC patients, patients with MBC are characterized by higher serum levels (adjusted P < 0.05) of citrate, choline, acetate, formate, lactate, glutamate, 3-hydroxybutyrate, phenylalanine, glycine, leucine, alanine, proline, tyrosine, isoleucine, creatine, creatinine, and methionine and lower serum levels (adjusted P < 0.05) of glucose and glutamine. In single-center analysis (Supplementary Table S1), citrate, formate, methionine, and phenylalanine were significantly higher in MBC patients. Others were numerically higher, consistent with the multicenter populations, but low patient numbers limit statistical significance.
In the cohort of EBC patients with relapse or follow-up of at least of 6 years (those included in the RF models), the patients who developed a recurrence were characterized by higher serum levels (adjusted P < 0.05) of choline, phenylalanine, leucine, histidine, glutamate, glycine, tyrosine, valine, lactate, and isoleucine but lower levels of glutamate (Supplementary Table S2). In the RF risk score algorithm, bins corresponding to phenylalanine, histidine, a lipid fraction (undifferentiated), methionine, glutamate, acetone, and formate carried the most weight in descending order of rank.
Discussion
Risk stratification in EBC for the purpose of deciding whether to recommend adjuvant therapies is of great importance, not least because of the significant toxicity associated with such treatment. For those at low risk of relapse, the risk of harm may outweigh the absolute risk of benefit.
The purpose of adjuvant therapy is to treat suspected residual micrometastatic disease. Yet, current prognostic factors and algorithms, including modern genomic signatures, are extrapolated from features of the primary tumor, surrogate markers for the likelihood of micrometastatic disease being present and progressing to an incurable state. Importantly, however, even among high-risk populations, a substantial proportion of patients is cured by surgery alone. In the seminal trial of adjuvant CMF versus no further treatment in women with node-positive EBC after primary cancer resection, 22% of untreated patients remained alive and disease free after a median of 28.5 years (1). In another study of ER+, node positive, EBC patients stratified by Oncotype DX, 60% of those with a high Oncotype recurrence score remained disease free after 6 years, with only tamoxifen as adjuvant therapy (3). These nonrelapsing high-risk patients either had no micro-metastatic disease to begin with or were able to control it without the use of chemotherapy. The ability to detect the presence of micrometastatic disease could greatly refine the selection of high-risk patients.
A theoretical advantage of metabolomics as a residual disease detector is its potential to capture not only signals from the micrometastatic disease, but also the surrounding stroma and any inflammatory/immune response. Other liquid biopsies, such as circulating tumor cells or plasma tumor DNA, will miss these host factors, potentially reducing sensitivity.
In this study, we were once again able to identify a metabolomic signal in the sera of EBC patients associated with increased risk of disease recurrence that is independent of standard risk factors, this time in a large, multicenter population. Our previous study was limited to a set of ER− patients from the MSKCC biobank, with blood draws taken after resection of the primary cancer, but before commencement of adjuvant therapy (8). In the current study, all patients were HR+, blood samples had been taken preoperatively, and patients came from multiple clinical sites in several countries, making this a new exploratory study rather than a confirmatory one. These differences introduced new challenges.
The effect of serum sample provenance was found to be the most discriminating feature of spectra. Causes for this may be multiple and include differences in the populations, such as diet or ethnicity, but may also reflect non-patient–related factors, such as specimen handling. Delays in centrifugation, insufficient or variable cooling, and unintentional thaw refreeze can affect the metabolic composition dramatically.
As noted in Table 2, only 5 centers collected samples both from EBC and MBC patients, and these samples are very few with respect to the total population of this study, while Vietnam provided a large proportion of EBC samples and no MBC samples. This large discrepancy, and resultant effect on spectra, would therefore be expected to induce an error in the building of the models (i.e., we are discriminating collection centers rather than EBC and MBC). Limiting the model to samples from a single site to control for location gave too few samples for meaningful analysis.
Controlling for lactate, a metabolite associated with suboptimal handling, removed much of the bias associated with location and thus may explain the cause. Lactate may also be affected by other metabolic factors. In this study, all patients were fasted as per preoperative protocol, reducing a dietary effect. Similarly, diabetes can have an effect if uncontrolled; to the best of our knowledge, there were no patients with diabetes in the EBC trial, but these data were lacking from the MBC trial. However, lactate is known to be altered in metastatic disease (7, 8), and indeed in this study, it differed between EBC and MBC patients and relapsing and nonrelapsing patients.
Examination of individual metabolites was performed to compare with results from other studies, but data must be interpreted with caution in light of the strong observed effect of provenance. A comparison of metabolites between EBC and MBC patients from a single center was also included, but small numbers of MBC patients affect statistical significance. Large numbers of metabolites were at significantly higher concentrations in MBC (Table 3), of which nine also correlated with relapse in the refined EBC cohort (Supplementary Table S2). Higher levels of phenylalanine were seen in MBC, in line with other studies (5, 6), and correlated with relapse in EBC. Similarly, higher glutamate in MBC is consistent with Jobard and colleagues’ work (6) and correlated positively with relapse in EBC here. Histidine was higher in MBC compared with EBC, but in this case, it is at odds with three other studies (6–8) in which it was lower.
The fact that geographical differences impacted significantly on the construction of a discriminating model is reflective of a broader issue for this approach, related to the need to establish a specific metastatic profile for each new study population. For example, applying the RF risk model created on the MSKCC dataset (8) to the current study population yielded very poor accuracy, due to the fact that differences between samples were far greater than the differences between the respective EBC and MBC cohorts (Supplementary Fig. S4). This greatly limits the transferability of the current approach between populations, until common standard procedures for metabolomics are adopted and a more universal metastatic profile can be established. In this regard, further studies are warranted.
A limitation of the dataset used is the fact that a follow-up time of 5 or 6 years is insufficient to capture all relapses in an HR+ EBC cohort, where relapse rates remain fairly constant for at least 10 years. Thus, there will be a proportion of EBC patients labeled as nonrelapsed who are in fact destined to relapse. Limiting the EBC cohort to patients with longer follow-up would be expected to improve on this but comes at the expense of a reduced sample size and was not possible here.
All patients in this study received systemic adjuvant endocrine therapy, making it impossible to know whether the therapy was directly responsible for the lack of relapse. An ideal series would have an arm with no intervention, although this is unlikely to be feasible for high-risk patients.
In this study, we were able to detect a signal correlated with recurrence despite the fact that the primary tumor was in situ at the time of blood draw. The presence and stage of the primary tumor might be expected to alter the metabolomic profile, and yet, the model predicted poorly for tumor size and nodal status, with little or no correlation. Whether the small correlation with size was as a direct result of the primary tumor or related to the fact that larger tumors are associated with higher risk of relapse, and thus may be a surrogate for presence of micrometastatic disease, is unknown. Part of our hypothesis is that the metabolomic signal correlating with residual disease, and thus relapse, is as much a reflection of the host state as it is of the presence of the tumor cells themselves, and our results support this.
Given the challenges of this study, in particular, the diversity of the populations and the fact that the bulk of the MBC and EBC sera came from different clinical sites, respectively, it is all the more compelling that a signal could be identified that discriminated between early and late breast cancer patients and that the RF risk score, essentially a measure of the “metastatic-ness” of the sera, correlated with relapse. Indeed, we see this study as complementary to the previous one because it suggests that the metabolomic score is relevant in predicting relapse in both ER− and ER+ patients, in both a single-center and in a multicenter setting.
What remains to be determined is whether the signal that correlates with relapse is truly a marker of micrometastatic disease or in fact reflective of the biological state of the primary tumor. Tumor-based genomic profiling assays, such as the 21-gene assay or MammaPrint, assess the expression of a number of genes to arrive at a risk of recurrence score, with those genes relating to proliferation playing a significant role. It is possible that differences in tumor gene expression state are also reflected in the metabolome, contributing to the metabolomic profile, in which case the metabolomic signals correlating with relapse may be dependent on the gene expression profile. It is vital then that future studies address this question, to examine whether metabolomics may provide independent, relevant prognostic information in the setting of genomic risk stratification, and we are investigating this currently. Given that a substantial proportion of patients designated as high-risk by genomic assays will not relapse, it would be invaluable to have a biomarker that might identify and restratify these patients. Ideally, this would be performed in prospective trials in which there is prespecified stratification by genomic risk score, where the metabolomic risk score could be tested for prognostic power within each risk group.
Supplementary Material
Translational Relevance.
Adjuvant chemotherapy in early breast cancer improves survival by targeting micrometastatic disease. Because of difficulties in detecting such a disease in patients, there is a tendency to overtreat, meaning that many patients receive chemotherapy unnecessarily, with substantial morbidity. We hypothesize that the combined altered cellular behavior of micrometastatic disease, supporting stroma and host response, results in a unique, detectable pattern of metabolites (metabolomic profile) similar to that seen in advanced disease and that it correlates with relapse. Here, using serum taken from premenopausal women enrolled in two phase III trials, and using nuclear magnetic resonance spectroscopy, we show that patients with metabolomic profiles more resembling the metastatic profile have a higher rate of relapse. Metabolomics thus has the potential to identify patients with micrometastatic disease, improve risk stratification, and reduce overprescription of chemotherapy.
Acknowledgments
We acknowledge and thank the Sandro Pitigliani Foundation and the Breast Cancer Research Foundation for their generous support.
Grant Support
This study was supported by the Breast Cancer Research Foundation (to A. Di Leo). This work has also been partially supported by Fondazione Veronesi through the Post-Doctoral Fellowship-2015 (to L. Tenori).
Footnotes
Disclosure of Potential Conflicts of Interest
A. Di Leo is a consultant/advisory board member for AstraZeneca, Bayer, Eisai, Genomic Health, Ipsen, Lilly, Novartis, Pfizer, and Pierre Fabre. No potential conflicts of interest were disclosed by the other authors.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Contributions
Conception and design: T.V. To, L. Biganzoli, R.R. Love, C. Luchinat, A. Di Leo
Development of methodology: L. Tenori, T.V. To, R.R. Love, A. Di Leo
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A. Vignoli, G.L. Uy, T.V. To, C. Adebamowo, S.M. Hossain, R.R. Love
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C.D. Hart, A. Vignoli, L. Tenori, T.V. To, L. Biganzoli, R.R. Love, C. Luchinat, A. Di Leo
Writing, review, and/or revision of the manuscript: C.D. Hart, A. Vignoli, L. Tenori, T.V. To, C. Adebamowo, S.M. Hossain, L. Biganzoli, E. Risi, R.R. Love, C. Luchinat, A. Di Leo
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): T.V. To, S.M. Hossain, R.R. Love
Study supervision: T.V. To, C. Adebamowo, R.R. Love, C. Luchinat, A. Di Leo
References
- 1.Bonadonna G, Moliterni A, Zambetti M, Daidone MG, Pilotti S, Gianni L, et al. 30 years’ follow up of randomised studies of adjuvant CMF in operable breast cancer: cohort study. BMJ. 2005;330:217. doi: 10.1136/bmj.38314.622095.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 3.Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 2010;11:55–65. doi: 10.1016/S1470-2045(09)70314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claudino WM, Quattrone A, Biganzoli L, Pestrin M, Bertini I, Di Leo A. Metabolomics: available results, current research projects in breast cancer, and future applications. J Clin Oncol. 2007;25:2840–6. doi: 10.1200/JCO.2006.09.7550. [DOI] [PubMed] [Google Scholar]
- 5.Oakman C, Tenori L, Claudino WM, Cappadona S, Nepi S, Battaglia A, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann Oncol. 2011;22:1295–301. doi: 10.1093/annonc/mdq606. [DOI] [PubMed] [Google Scholar]
- 6.Jobard E, Pontoizeau C, Blaise BJ, Bachelot T, Elena-Herrmann B, Trédan O. A serum nuclear magnetic resonance-based metabolomic signature of advanced metastatic human breast cancer. Cancer Lett. 2014;343:33–41. doi: 10.1016/j.canlet.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Asiago VM, Alvarado LZ, Shanaiah N, Gowda GA, Owusu-Sarfo K, Ballas RA, et al. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70:8309–18. doi: 10.1158/0008-5472.CAN-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenori L, Oakman C, Morris PG, Gralka E, Turner N, Cappadona S, et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptornegativeearly breast cancer atincreased riskofdisease recurrence. Results from a retrospective study. Mol Oncol. 2015;9:128–39. doi: 10.1016/j.molonc.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Love RR, Laudico AV, Van Dinh N, Allred DC, Uy GB, Quang le H, et al. Timing of adjuvant surgical oophorectomy in the menstrual cycle and disease-free and overall survival in premenopausal women with operable breast cancer. J Natl Cancer Inst. 2015;107:djv064. doi: 10.1093/jnci/djv064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Love RR, Hossain SM, Hussain MM, Mostafa MG, Laudico AV, Siguan SS, et al. Luteal versus follicular phase surgical oophorectomy plus tamoxifen in premenopausal women with metastatic hormone receptor-positive breast cancer. Eur J Cancer. 2016;60:107–16. doi: 10.1016/j.ejca.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, Turano P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J Biomol NMR. 2011;49:231–43. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 12.Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 13.Gebregiworgis T, Powers R. Application of NMR metabolomics to search for human disease biomarkers. Comb Chem High Throughput Screen. 2012;15:595–610. doi: 10.2174/138620712802650522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meiboom S, Gill D. Modified spinecho method for measuring nuclear relaxation times. Rev Sci Instrum. 1958;29:688–91. [Google Scholar]
- 15.Wu DH, Chen A. Three-dimensional diffusion-ordered NMR spectroscopy: the homonuclear COSY-DOSY experiment. J Magn Reson Ser A. 1996;123:215–8. [Google Scholar]
- 16.Spraul M, Neidig P, Klauck U, Kessler P, Holmes E, Nicholson JK, et al. Automatic reduction of NMR spectroscopic data for statistical and pattern recognition classification of samples. J Pharm Biomed Anal. 1994;12:1215–25. doi: 10.1016/0731-7085(94)00073-5. [DOI] [PubMed] [Google Scholar]
- 17.Holmes E, Foxall PJ, Nicholson JK, Neild GH, Brown SM, Beddell CR, et al. Automatic data reduction and pattern recognition methods for analysis of 1H nuclear magnetic resonance spectra of human urine from normal and pathological states. Anal Biochem. 1994;220:284–96. doi: 10.1006/abio.1994.1339. [DOI] [PubMed] [Google Scholar]
- 18.Ihaka R, Gentleman RR. A language for data analysis and graphics. J Comput Stat Graph. 1996;5:299–314. [Google Scholar]
- 19.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 20.Liaw A, Wiener M. Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 21.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3. 0–The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilcoxon F. Individual comparisons by ranking methods. Biom Bull. 1945;1:80. [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 24.Bagai OP. The distribution of the generalized variance. Ann Math Stat. 1965;36:120–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.