Abstract
ATP-dependent degradation plays a critical role in the quality control and recycling of proteins in cells. However, complete degradation of membrane proteins by ATP-dependent proteases in bacteria is not well-studied. We discovered that the degradation of a multidomain and multispan integral membrane protein AcrB could be facilitated by the introduction of a ssrA-tag at the C-terminus of the protein sequence and demonstrated that the cytoplasmic unfoldase-protease complex ClpXP was involved in the degradation. This is the first report to our knowledge to reveal that the ClpXP complex is capable of degrading integral membrane proteins. The chaperone SspB also played a role in the degradation. Using purified proteins, we demonstrated that the addition of the ssrA-tag did not drastically affect the structure of AcrB, and the degradation of detergent solubilized AcrB by purified ClpXP could be observed in vitro.
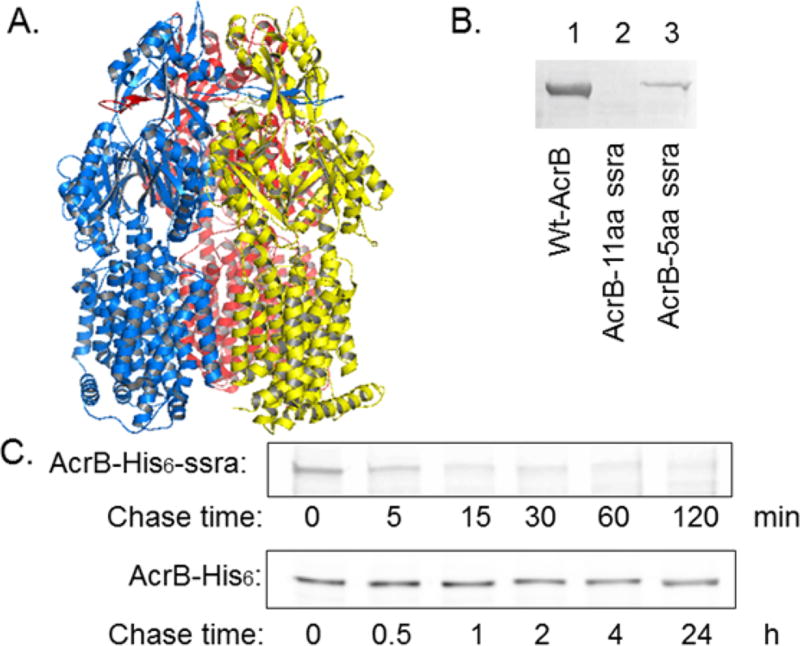
In bacteria, the major players in protein degradation are ATP-dependent proteases, including HslUV, ClpAP, ClpXP, Lon, FtsH, and their homologues.1–4 These proteases utilize their ATPase functions to facilitate the unfolding and translocation of substrate proteins. Among them, ClpAP and ClpXP are complexes of two proteins, an unfoldase ClpX/ClpA, and a peptidase ClpP.5–7 By itself ClpP can degrade small peptide substrates, but to degrade larger proteins it needs to form a complex with an ATPase, such as ClpA or ClpX, which dissociates stable protein complexes and unfolds proteins at the expense of ATP hydrolysis. One role of the ClpXP and ClpAP complexes is to degrade proteins bearing the ssrA-tag. In bacteria, the ssrA-tagging system has evolved to get rid of incompletely synthesized proteins which result from stalled synthesis of the ribosome.8–10 They are tagged for destruction by the cotranslational addition of an 11-residue peptide (AANDENYALAA) by an amazing molecule tm-RNA.11,12 This ssrA mechanism effectively degrades nonsense proteins and releases/recycles ribosomes.13,14 While the degradation of ssrA-tagged soluble proteins has been well studied, a similar process of membrane proteins has not been investigated. Here we report that the introduction of the ssrA-tag leads to the complete degradation of a large integral membrane protein AcrB. AcrB is a multidrug efflux pump conserved in all Gramnegative bacteria and is a major contributor to confer antibiotic resistance to bacteria.15–17 The structure and function unit of AcrB is a trimer, which associates with the peripheral protein AcrA and outer membrane protein TolC to form a complex that spans both the inner and outer membranes.18–22 Each AcrB protomer contains 1049 residues, with 12 transmembrane helices (TMH) and a large periplasmic domain, formed by two long periplasmic loops in between TMH1/2, and TMH 7/8 (Figure 1A).
Figure 1.
An ssrA-tag at the C-terminus facilitated the degradation of AcrB. (A) AcrB is a trimeric integral membrane protein with 12 transmembrane helices and a large periplasmic domain in each protomer (color-coded) (Created using The PyMOL Molecular Graphics System (Version 1.8 Schrödinger, LLC.) from 2GIF.pdb.32 (B) Anti-AcrB Western blot analyses of membrane extracts obtained from DL41ΔacrB strain containing plasmid- encoded wt-AcrB, AcrB- 11aa ssrA, or AcrB-5aa ssrA. (C) Degradation of S35-Met-AcrB-His6-ssra and S35-Met-AcrB-His6 determined by the pulse-chase experiment. Note the difference in the unit of the time.
AcrB can be purified and readily crystallizes, suggesting that its structure is intrinsically stable. However, the introduction of the oligonucleotide encoding the ssrA-tag into the acrB gene right before its stop codon rendered the protein no longer detectable in the cell lysate. We transformed plasmid pQE-AcrB or pQE-AcrB-ssrA into BW25113ΔacrB and examined protein expression under the basal condition. AcrB was detected using a Western blot with an anti-AcrB antibody raised against a peptide sequence corresponding to residues 1032–1045 of AcrB (AcrB-CT antibody).23 As shown in Figure 1B, wild type AcrB expressed well and yielded a clear band on the blot (lane 1). However, after we added the ssrA-tag to the protein, its expression was completely abolished (lanes 2). Earlier studies show that the last five residues YALAA of the ssrA tag is the critical factor, which also promotes degradation although at a lower efficiency.12 To confirm that the degradation was actually caused by the tag, we also constructed AcrB with the truncated ssrA tag (AcrB-5aa ssrA). As expected, the expression level of the protein was much lower than that of the wild type AcrB, but is still clearly observable (Figure 1B, lane 3). For the rest of the study, by AcrB-ssrA we indicate AcrB bearing the 11-residue ssrA tag at the C-terminus. Since the AcrB-CT antibody recognizes a specific peptide sequence close to the C-terminus of the protein, we have also repeated the experiment using an anti-AcrB antibody raised against the full-length AcrB (AcrB-FL antibody). We did not observe large AcrB fragment in the lysate, indicating that the degradation was complete (data not shown).
To confirm that the lack of AcrB-ssrA is a result of degradation, not a defect in translation, we conducted the S35 Met pulse chase experiment (Figure 1C). A His6 tag was inserted at the C-terminus of AcrB, right before the ssrA-tag. DL41ΔacrB transformed with plasmids pQE-AcrB-His6 or pQE-AcrB-His6-ssrA was cultured in the presence of S35 Met for 2 min and then chased using a large excess of cold Met. Samples were collected at different time points and analyzed. As discussed above, AcrB-His6 was very stable, and the intensity did not significantly decrease over the 24 h of this experiment. In the case of AcrB-His6-ssrA, a clear band could be seen at time 0, indicating normal expression, but the band intensity of AcrB-His6-ssrA dropped to the background level within ~15 min, indicating fast degradation.
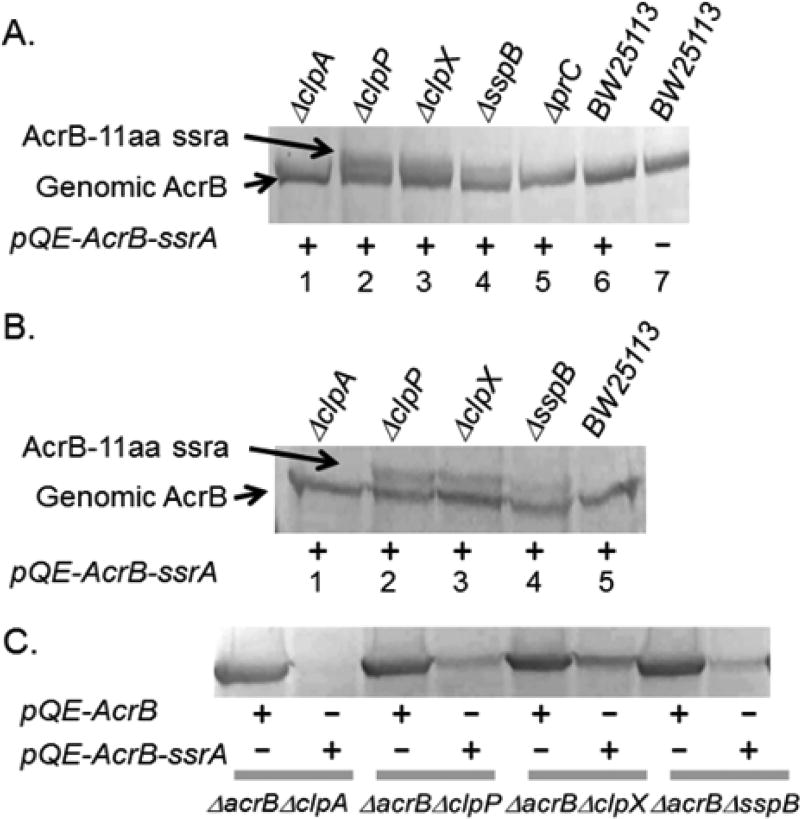
To investigate if the degradation is mediated by the ClpAP and/or ClpXP, we obtained single gene knockout strains lacking each gene from the Yale Coli Genetic Stock Center and transformed plasmid encoding AcrB-ssrA to examine the level of degradation (Figure 2A,B). We examined the effect of knocking out five single genes, including clpA, clpX, clpP, sspB, prc. Among them, ClpA is similar to ClpX and also functions with ClpP.8 SspB is a known chaperone that enhances the degradation of ssrA-tagged substrate by increasing the binding affinity and lowering the KM.24–27 Of the 11 residues in the ssrA-tag, only the terminal AA-COOH is directly involved in ClpX binding. Several residues upper stream in the tag (AANEDNY) mediate binding to SspB, which aids in the delivery of ssrA-tagged protein to ClpXP.27 Prc is a periplasmic protease (also known as Tsp) that degrades protein substrates in a carboxy-terminal-specific manner and is known to degrade ssrA tagged proteins that are exported to the periplasm.9 Plasmid pQE-AcrB-ssrA was transformed into the indicated strains, and the cellular AcrB-ssrA level was determined using anti-AcrB Western blot with the AcrB-CT-antibody. We found that residual AcrB-ssrA could be detected in three strains: ΔclpX, ΔclpP, and ΔsspB. Among them, ΔclpX contains the highest level of residual AcrB-ssrA. And because the strains still contain the genomic AcrB, a protein band at a slightly smaller molecular weight could be observed in all samples (the extra amino acids in the tag slowed down the migration of the protein in the gel). The presence of these two bands further confirmed that the production (or more likely, degradation) of the wild type AcrB was not affected in the knockout strains so the degradation is ssrA-tag specific. In the parent strain (lanes 6 and 7), the transformation with the plasmid did not lead to a detectable AcrB-ssrA band. The same was observed in the ΔclpA or Δprc strains, suggesting that ClpA and Prc are not involved.
Figure 2.
Anti-AcrB Western blot analyses of membrane vesicles. (A) Single gene knockout strains of BW25113 missing the indicated gene expressing AcrB-ssrA from a plasmid. Result was analyzed using 8% SDS-PAGE. The last two lanes are the parent strain with or without the plasmid, respectively. (B) Same as in A, but analyzed on a 6% gel to better separate the genomic AcrB and AcrB-ssrA. (C) Expression of AcrB or AcrB-ssrA in the indicated double knockout strain.
To determine the level of degradation, we first created double knockout strains to further eliminate the genomic acrB gene and then transformed plasmids pQE-AcrB or pQE-AcrB-ssrA into each strain for comparison (Figure 2C). Assuming the transcription and translation was not affected by the last few nucleotides/amino acids making up the ssrA-tag, the difference in the detected protein level should reveal information about the level of degradation. We found that the degradation level in ΔclpX is approximately 60–70%. In ΔclpP and ΔsspB, the degradation level reached 80–90%. These results indicate that (1) ClpXP and SspB are involved in the degradation of AcrBssrA; (2) there are likely other proteins involved in the degradation as well.
To demonstrate that the addition of the ssrA tag did not affect the membrane integration and folding of AcrB, we compared the CD spectra of purified AcrB-His6 and AcrB-His6-ssrA (Supplementary Figure 1). Since AcrB-ssrA is completely degraded in wild type Escherichia coli, we expressed and purified it in DL41ΔacrBΔclpX. A His6 tag was inserted between the last amino acid of AcrB and the ssrA tag to facilitate purification. As described above, the addition of his-tag did not affect the degradation of the protein. The expression and purification of AcrB-his6-ssrA were performed as described for AcrB-His6.28,29 AcrB-His6-ssrA was purified from membrane vesicles, indicating that the membrane integration was not compromised by the C-terminal tag. This is consistent with our expectation as the membrane integration of inner membrane proteins are cotranslational. For a large protein such as AcrB, the membrane insertion should be close to complete before the C-terminal ssrA-tag emerges from the exit tunnel of the ribosome. The CD spectra of detergent-solubilized AcrB-His6 and AcrB-His6-ssrA superimposed well onto each other, indicating the presence of the tag did not affect the overall folding of the protein.
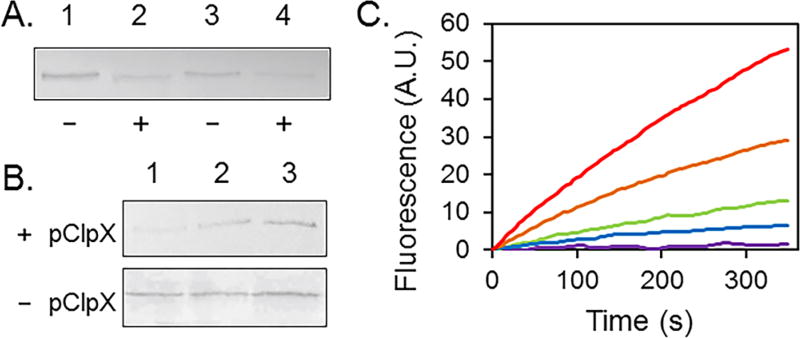
The ultimate test of the degradation of AcrB-ssrA by ClpXP is to purify each component and conduct the degradation assay in a test tube. Plasmids were also constructed to express ClpP and ClpX with N-terminal His6 tag. Expression and purification of His6-ClpX and His6-ClpP were conducted following published protocols.13 Two reactions were set up: one contains AcrB-His6-ssrA, ClpX, and ClpP at a molar ratio of (AcrB-His6-ssrA)3: (ClpX)6: (ClpP)14 = 1:5:15. The control sample was identical except that ClpX and ClpP were not present. After incubation overnight, the level of AcrB-His6-ssrA in the presence of ClpX/ClpP was determined using an anti-AcrB Western blot to be approximately 30–40% of the control sample (Figure 3A). The level of degradation could not be improved by the addition of more ClpXP or the increase of digestion time, indicating that under the current experimental condition the efficiency of the protease is quite low. Another possible explanation for the incomplete degradation is the damage of the C-terminal ssrA-tag during and after protein purification. Since the last few amino acids are critical for ClpXP-facilitated degradation, even the truncation of one Ala at the very end of the sequence would lead to a protein that is no longer degradable by the system. To better quantify the rate of digestion, a better in vitro assay is clearly necessary.
Figure 3.
Degradation of AcrB-His6-ssrA. (A) Anti-AcrB Western blot analysis of degradation of detergent solubilized AcrB-His6-ssrA in vitro. Lanes 1 and 2 contain 0.2 µg of AcrB-His6-ssrA, and lanes 3 and 4 contain 0.1 µg of AcrB-His6-ssrA. Lanes 2 and 4 also contain ClpX and ClpP. (B) Anti-AcrB Western blot analysis of whole cell lysate prepared from DL41ΔacrBΔclpX transformed with pQE-AcrB-His6-ssrA and pBAD-ClpX (pClpX) (top) or pQE-AcrB-His6-ssrA alone (bottom). AcrB-His6-ssrA was expressed at the basal level without induction, and then the cell culture was divided equally into three samples: arabinose was added into the first sample to induce the expression of ClpX (lane 1). Sample 1 and sample 2 (lane 2) were incubated for an additional 2 h at 28 °C, while sample 3 (lane 3) was left on ice. (C) EtBr accumulation assay of DL41ΔacrBΔclpX transformed with the indicated plasmids and cultured with or without ClpX induction. pQE70 (red, negative control), pQE-AcrB-His6-ssrA/pClpX with arabinose (orange), pQE-AcrB-His6-ssrA/pClpX without induction (green), pQE-AcrB-His6-ssrA (blue), and pQE-AcrB-His6 (purple, positive control).
As discussed above, we expect that membrane insertion should have occurred before the translation of the ssrA tag and the degradation should involve membrane-inserted (or partially inserted) substrate. However, direct capture of the nascent polypeptide from the ribosome for degradation is also possible. To determine if AcrB-His6-ssrA inserted into the cell membrane could be degraded by the ClpXP system, we took advantage of the fact that in the strain lacking ClpX, expression of AcrB-His6-ssrA could be detected (Figure 2). These AcrB-His6-ssrA molecules should be functional in conducting substrate efflux if and only if they are properly inserted into the cell membrane. The activity of AcrB could be measured using a convenient drug susceptibility test, in which the minimum inhibitory concentration (MIC) of known AcrB substrates could be determined for E. coli strains. A strain containing active AcrB displays significantly higher MIC than a strain deficient in AcrB. We determined the MIC of two well established AcrB substrates, novobiocin and erythromycin, for three strains: the positive control (DL41ΔacrBΔclpX transformed with plasmid pQE-AcrB-His6), the negative control (DL41ΔacrBΔclpX transformed with the empty vector pQE70), and DL41ΔacrBΔclpX transformed with plasmid pQE-AcrB-His6-ssrA. The MIC for the three strains are listed in Table 1. The residual amount of AcrB-His6-ssrA is clearly functional as they conferred elevated level of resistance to a strain previously lacking AcrB (DL41ΔacrBΔclpX) against both erythromycin and novobiocin.
Table 1.
MIC of Two AcrB Substrates of Strain DL41ΔacrBΔclpX Transformed with the Indicated Plasmid
| plasmid | erythromycin (µg/mL) |
novobiocin (µg/mL) |
|---|---|---|
| pQE-AcrB-His6 (positive cont.) | 80 | 160 |
| pQE70 (negative cont.) | 10 | 20 |
| pQE-AcrB-His6-ssrA | 80 | 40 |
Once we confirmed that AcrB-His6-ssrA was membrane inserted in DL41ΔacrBΔclpX strain, the next step is to examine if it could be degraded. To enable controlled degradation, we cotransformed DL41ΔacrBΔclpX with two plasmids: pQE-AcrB-His6-ssrA and pBAD-ClpX. We routinely express AcrB at the basal level without induction.30 The expression of ClpX from pBAD-ClpX could be induced by the addition of arabinose.31 The strain was cultured until its OD600 reached 0.6, and then the cell culture was divided into three samples and arabinose (0.1%, w/v) was added into the first sample. The first and second samples were incubated at 28 °C with shaking for an additional 2 h, while the third sample was left on ice and used as a “no further growth” control. Next, the AcrB levels in each sample were examined using anti-AcrB Western blot (Figure 3B). The DL41ΔacrBΔclpX strain containing only one plasmid pQE-AcrB-His6-ssrA was also treated similarly and used as a control. When pBAD-ClpX was cotransformed with pQE-AcrB-His6-ssrA, the addition of arabinose led to a significant drop of the AcrB level, which was not observed in the control strain lacking pBAD-ClpX. Samples 2 and 3 have similar AcrB levels. This result indicates that the induced expression of ClpX accelerated the degradation of AcrB-His6-ssrA, including the portion that was synthesized before the time of the induction.
To further confirm that the observed AcrB-His6-ssrA in the DL41ΔacrBΔclpX strain was actually membrane inserted under this experimental condition (with two plasmids cotransformed), we used a well-established ethidium bromide (EtBr) accumulation assay to examine the activity of AcrB. EtBr is a substrate of AcrB, and E. coli strains deficient in AcrB accumulate EtBr at a much faster rate than a wild type strain that contains AcrB, which leads to a quick increase of fluorescence signal when EtBr enters the cell and intercalates into nucleic acid. As shown in Figure 3C, EtBr accumulation was measured for DL41ΔacrBΔclpX transformed with different plasmids. pQE70 (red) and pQE-AcrB-His6 (purple) are negative and positive controls, respectively. The slope of fluorescence increase, which reflects the rate of EtBr accumulation, was much faster in the negative control. When the strain was transformed with pQE-AcrB-His6-ssrA (blue), the residual level of AcrB-His6-ssrA conferred significant resistance against EtBr accumulation, consistent with the result of the MIC measurement (Table 1). The strain containing both pQE-AcrB-His6-ssrA and pBAD-ClpX was first cultured to the log phase and then split into two samples, one cultured without induction (green) and the other with induction with arabinose (orange). The activities correlated well with the detected AcrB level in Figure 3A. EtBr accumulation was lower in the strain without ClpX induction, indicating that the induction of ClpX led to the degradation of functional (and membrane inserted) AcrB-His6-ssrA.
In summary, we discovered that the introduction of an extra 11 amino acid residues, the ssrA-tag, at the C-terminus of this large 1049-residue and highly stable integral membrane protein leads to its complete degradation. We have both established that ClpX, ClpP, and SspB are involved in the degradation of ssrA-tagged AcrB in cells and showed that ClpXP could degrade detergent-solubilized AcrB-ssrA using purified proteins. The addition of the tag did not have a detectable effect of the overall secondary structure composition of the protein. These results lead to an exciting new question: how does an integral membrane protein get efficiently degraded by a soluble protease complex in the cell? The required dislodging of the transmembrane helices of the protein substrate and even more strikingly, the long hydrophilic loop, through the cell membrane is an extra challenge for the system to handle. Many more studies are clearly required to elucidate this interesting and fundamentally important question.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Bob Sauer for the kind gift of plasmid for GFP-ssrA expression as well as his valuable suggestions. We sincerely thank Dr. Karl R. Schmitz for his helpful suggestions on the GFP-ssrA digestion assay condition.
Funding
This work was supported by the National Science Foundation (MCB 1158036, Y.W.) and National Institute of Allergy and Infectious Diseases (1R21AI103717, Y.W.; R01 AI051517, R.E.D.; and F31 fellowship AI120653-01, S.R.W.).
Footnotes
ASSOCIATED CONTENT
- Materials and methods (PDF)
The authors declare no competing financial interest.
References
- 1.Gottesman S. Annu. Rev. Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 2.Dougan DA, Mogk A, Bukau B. Cell. Mol. Life Sci. 2002;59:1607–1616. doi: 10.1007/PL00012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson PI, Whiteheart SW. Nat. Rev. Mol. Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 4.Sauer RT, Baker TA. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- 5.Baker TA, Sauer RT. Biochim. Biophys. Acta, Mol. Cell Res. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levchenko I, Luo L, Baker TA. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 7.Kruklitis R, Welty DJ, Nakai H. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman S, Roche E, Zhou YN, Sauer RT. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keiler KC, Waller PRH, Sauer RT. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 10.Moore SE, Sauer RT. Annu. Rev. Biochem. 2007;76:101–124. doi: 10.1146/annurev.biochem.75.103004.142733. [DOI] [PubMed] [Google Scholar]
- 11.Moore SD, Sauer RT. Mol. Microbiol. 2005;58:456–466. doi: 10.1111/j.1365-2958.2005.04832.x. [DOI] [PubMed] [Google Scholar]
- 12.Karzai AW, Roche ED, Sauer RT. Nat. Struct. Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 13.Farrell CM, Grossman AD, Sauer RT. Mol. Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- 14.Lies M, Maurizi MR. J. Biol. Chem. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikaido H. J. Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikaido H, Takatsuka Y. Biochim. Biophys. Acta, Proteins Proteomics. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikaido H, Pages JM. FEMS Microbiol. Rev. 2012;36:340–363. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okusu H, Ma D, Nikaido H. J. Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pos KM. Biochim. Biophys. Acta, Proteins Proteomics. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Tikhonova EB, Zgurskaya HI. J. Biol. Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- 21.Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. Proc. Natl. Acad. Sci. U. S. A. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zgurskaya HI. Future Microbiol. 2009;4:919–932. doi: 10.2217/fmb.09.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zgurskaya HI, Nikaido H. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7190–7195. doi: 10.1073/pnas.96.13.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levchenko I, Seidel M, Sauer RT, Baker TA. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- 25.Flynn JF, Levchenko I, Sauer RT, Baker TA. Genes Dev. 2004;18:2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinness KE, Baker TA, Sauer RT. Mol. Cell. 2006;22:701–707. doi: 10.1016/j.molcel.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 27.Bolon DN, Grant RA, Baker TA, Sauer RT. Mol. Cell. 2004;16:343–350. doi: 10.1016/j.molcel.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Lu W, Zhong M, Wei Y. J. Mol. Biol. 2011;411:264–274. doi: 10.1016/j.jmb.2011.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Lu W, Zhong M, Wei Y. Protein Pept. Lett. 2011;18:863–871. doi: 10.2174/092986611796011446. [DOI] [PubMed] [Google Scholar]
- 30.Ye C, Wang Z, Lu W, Zhong M, Chai Q, Wei Y. Biochemistry. 2014;53:3738–3746. doi: 10.1021/bi5000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guzman LM, Belin D, Carson MJ, Beckwith J. J. Bacteriol. 1995;177(14):4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.