Summary
Family B DNA polymerases (PolBs) play a central role during replication of viral and cellular chromosomes. Here, we report the discovery of a third major group of PolBs, which we denote primer-independent PolB (piPolB), that might be a link between the previously known protein-primed and RNA/DNA-primed PolBs. PiPolBs are encoded by highly diverse mobile genetic elements, pipolins, integrated in the genomes of diverse bacteria and also present as circular plasmids in mitochondria. Biochemical characterization showed that piPolB displays efficient DNA polymerization activity that can use undamaged and damaged templates and is endowed with proofreading and strand displacement capacities. Remarkably, the protein is also capable of template-dependent de novo DNA synthesis, i.e., DNA-priming activity, thereby breaking the long-standing dogma that replicative DNA polymerases require a pre-existing primer for DNA synthesis. We suggest that piPolBs are involved in self-replication of pipolins and may also contribute to bacterial DNA damage tolerance.
Keywords: DNA replication, translesion synthesis, primer-independent DNA synthesis, de novo DNA synthesis, family B DNA polymerase, self-replicating mobile element, DNA damage
Graphical Abstract
Highlights
-
•
Primer-independent PolBs (piPolBs) display templated de novo DNA synthesis capacity
-
•
piPolBs denote a third major group of family B DNA polymerases
-
•
piPolBs are the hallmark of pipolins, self-replicating mobile genetic elements
-
•
Pipolins are widespread among diverse bacterial phyla and mitochondria
Redrejo-Rodríguez et al. report and characterize a DNA polymerase group (piPolB) from the B family that can perform primer-independent DNA replication. PiPolBs are encoded by Pipolins, diverse self-replicating genetic elements that are widespread among bacterial phyla and in mitochondria.
Introduction
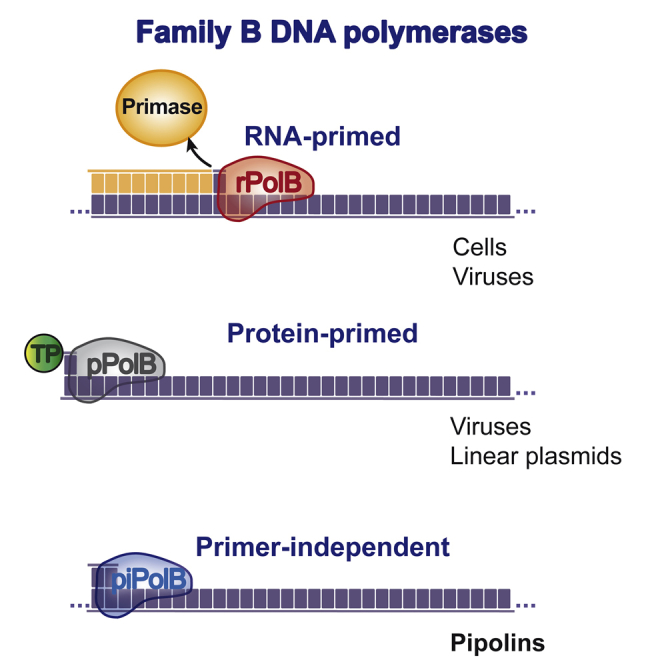
DNA polymerases (DNAPs) are key enzymes essential for genome replication, recombination, and repair across all cellular life forms and their viruses. Family B DNAPs (PolBs) are involved in genome replication in Eukarya and Archaea, but also in viruses from the three domains of life (Koonin, 2006). For the initiation of DNA synthesis, all PolBs characterized thus far depend on the presence of an external primer, a hydroxyl group presented either by a nucleic acid (RNA or DNA) or the so-called terminal protein (TP). Thus, based on the primer requirement for the initiation of genome replication as well as phylogenetic clustering, these enzymes can be broadly divided into two major groups (Filée et al., 2002): RNA primed (rPolBs) and protein primed (pPolBs). The evolutionary relationship between the two groups is unknown, and it is thus unclear whether the ancestral PolB would have employed a protein or an RNA oligonucleotide as a primer. The rPolB group contains mainly replicases devoted to accurate and efficient copying of large cellular and viral genomes. By contrast, pPolBs are exclusive to selfish mobile genetic elements (MGEs) and viruses with moderately sized linear genomes (<50 kb) (Kazlauskas and Venclovas, 2011, Krupovic and Koonin, 2015). The signature of pPolBs is the presence of specific subdomains, named TPR1 and TPR2, which were originally described in Φ29 DNAP. TPR1 is required for the DNAP interaction with the TP, whereas TPR2 endows pPolB with the processivity and strand displacement capacity (Dufour et al., 2000, Rodríguez et al., 2005, Salas et al., 2016), the two properties ensuring the superiority of Φ29DNAP in various molecular biology applications, such as multiple-displacement single-molecule DNA amplification (Hutchison et al., 2005, Sidore et al., 2016).
Protein priming has been described for a number of viruses (Berjón-Otero et al., 2016, Hoeben and Uil, 2013, Salas, 1991) as well as for linear eukaryotic plasmids (Klassen and Meinhardt, 2007). More recently, pPolB-encoding genes were also identified in two superfamilies of MGE integrated into various cellular genomes (Krupovic and Koonin, 2016). The first superfamily comprises eukaryotic virus-like transposable elements, called Polintons (also known as Mavericks), which, besides pPolB, encode retrovirus-like integrases and a set of proteins predicted to be involved in the formation of viral particles (Krupovic et al., 2014a). The second supergroup, denoted casposons, is present in a wide range of archaea and some bacteria (Krupovic et al., 2014b). For integration into the cellular genome, casposons employ endonucleases homologous to Cas1, a signature enzyme of the CRISPR-Cas systems (Béguin et al., 2016, Krupovic et al., 2017). It has been postulated that pPolBs participate in the replication of the casposon and polinton genomes (Kapitonov and Jurka, 2006, Krupovic et al., 2014b); accordingly, these MGEs are referred to as self-synthesizing or self-replicating elements.
Here we uncover a highly diverse superfamily of self-replicating MGEs, dubbed pipolins, which are present in three major bacterial phyla, as well as in mitochondria, and encode divergent PolB carrying TPR1 and TPR2 subdomains. Biochemical characterization of a representative enzyme encoded by a pipolin from Escherichia coli showed that the protein displays a versatile and efficient DNA replication capacity. Strikingly, the protein is also capable of an intrinsic de novo DNA synthesis, i.e., DNA-priming activity, not previously described in members of the PolB family. This group of DNAPs, which we denote primer-independent PolB (piPolB), should be sufficient to initiate and carry out an entire replication cycle of the circular pipolin DNA in vivo. Moreover, enhanced survival of E. coli cells expressing piPolB upon replication blockage by DNA-damaging agents suggests an additional role of piPolB in bacterial DNA damage tolerance.
Results
A Third Major Group of PolBs
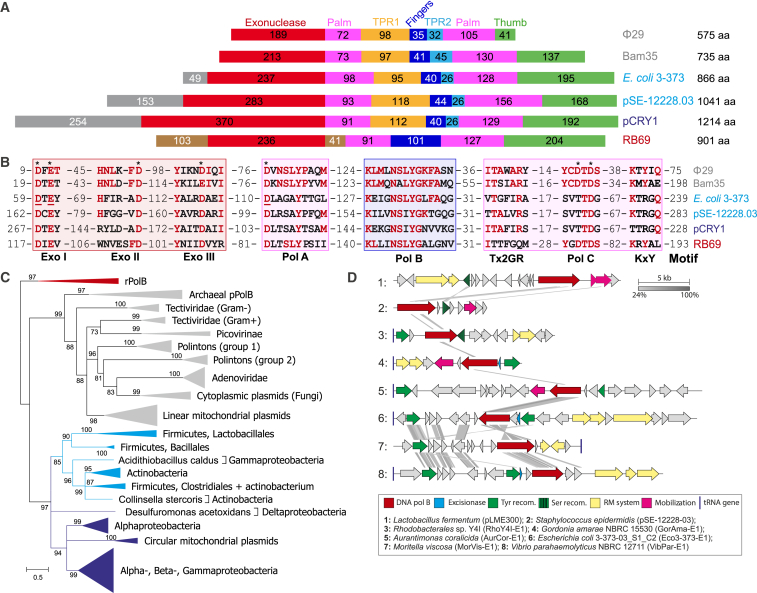
Position-specific iterative (PSI)-BLAST searches against the RefSeq bacterial genome database at NCBI seeded with the sequence of experimentally characterized pPolB from bacteriophage Bam35 (NP_943751) retrieved numerous hits to highly divergent PolBs (∼16%–20% sequence identity to the pPolB of Bam35) encoded within chromosomes and several plasmids from widely diverse bacteria, such as Firmicutes, Actinobacteria, and Proteobacteria. Nevertheless, analysis of multiple sequence alignments of these divergent DNAPs showed that all of them contained the TPR1 and TPR2 subdomains, a hallmark of pPolBs (Figure 1A), and the active site residues of the exonuclease and DNAP domains of PolBs (Braithwaite and Ito, 1993) were conserved, albeit with notable variations within the KxY and PolC motifs (Figure 1B, see also below). The PolC motif (YxDTDS) is almost universally conserved in PolBs (Braithwaite and Ito, 1993) and contains two catalytic aspartic acid residues required for protein activity (Bernad et al., 1990, Copeland and Wang, 1993). We noticed that, in all members of the divergent PolB group, the first of the two aspartates within the PolC motif was substituted for a threonine residue (SxTTDG, see Figure 1B). Notably, some archaeal pPolBs also showed variation within this motif (Bath et al., 2006, Krupovic et al., 2014b, Peng et al., 2007), but none of these proteins has been experimentally characterized.
Figure 1.
A Major Group of PolBs
(A) Schematic representation of representative PolBs. Conserved domains are represented as colored boxes and their length is indicated. Non-conserved N-terminal domains of RB69 and piPolBs are colored in tawny and gray, respectively.
(B) Detail of conserved PolB motifs. Conserved residues D59/E61 and D368 in E. coli 373 piPolB that were mutated to generate exonuclease- and polymerase-deficient variants, respectively, are underlined.
(C) Maximum likelihood phylogeny of piPolBs. The tree is rooted with diverse cellular and viral RNA-primed PolBs (red) and other protein-primed PolBs are also shown (gray). The two groups of piPolBs are colored with light and dark shades of blue (see text for details). The scale bar represents the number of substitutions per site. Branches with support values below 70% were collapsed. The tree in which piPolB clades are expanded is shown in Figure S1.
(D) Genomic organization of representative pipolins (see Figure S2 and Table S2 for details).
Additional searches seeded with representative sequences of the divergent PolB group from proteobacteria, such as Escherichia coli (KDU42669) or Rhodobacterales bacterium Y4I (WP_008555115), yielded significant hits to several homologs encoded by pCRY1-like circular mitochondrial plasmids. Notably, the latter plasmids were distinct from the extensively studied linear mitochondrial plasmids, which encode pPolBs (see below). Sequence analysis of the mitochondrial proteins confirmed their close similarity to the divergent group of bacterial PolBs (Figures 1A and 1B).
Maximum likelihood phylogenetic analysis of representative sequences from all known clades of PolBs revealed that the divergent pPolBs formed a distinct, well-supported clade, which we denote piPolB (see below), separated from all other pPolBs (Figure 1C), suggesting that it diverged early in the evolution of PolBs. Thus, piPolB represents the third major group of PolBs, besides rPolBs and pPolBs. Within the piPolB clade, there are two major groups, which are roughly congruent with the bacterial taxonomy (Figure 1C; Figure S1). The first one predominantly includes sequences from Actinobacteria and several orders of Firmicutes, namely, Bacillales, Lactobacillales, and Clostridiales. The second group contains sequences from different classes of Proteobacteria. Notably, the latter group also includes piPolBs from circular mitochondrial plasmids, which cluster with sequences from alphaproteobacteria (Figure 1C; Figure S1).
piPolBs Are Encoded within Self-Replicating MGEs
Genomic context analysis provided compelling evidence that the majority of piPolBs are encoded within MGEs integrated into bacterial chromosomes. Unlike casposons and polintons, the vast majority of piPolB-carrying MGEs encoded integrases of the tyrosine recombinase superfamily (Y-integrases). Some of the elements carried additional copies of Y-integrases or integrases/invertases of the serine recombinase superfamily (Figure 1D; Figure S2). Nevertheless, several bacterial and all mitochondrial piPolB homologs were encoded by extrachromosomal, rather than integrated, plasmids, and, accordingly, they lacked the integrase genes, suggesting that integration into the chromosome is optional for these MGEs. Hence, we refer to all these bacterial and mitochondrial elements as piPolB-encoding MGEs (pipolins).
The Y-integrases typically catalyze recombination between homologous sites present on the cellular genome and the circular double-stranded DNA (dsDNA) molecules of the MGEs. Thorough analysis of the piPolB-encompassing genomic regions allowed us to define the precise integration sites for many pipolins from diverse bacterial taxa (Table S1; Figure S2), with tRNA genes being the most common integration site. Comparative genomic analysis of pipolins showed that they formed groups that were generally consistent with the phylogeny of the piPolBs (Figure 1D; Figure S2). Besides piPolB and integrases, pipolins often encoded excisionases, components of type I and type II restriction modification systems, and various plasmid mobilization proteins (Figure S2; Table S2). In addition, the less conserved genes found in pipolins encoded different DNA-binding proteins with ribbon-helix-helix, zinc-finger, or helix-turn-helix motifs, as well as histone-like H-NS chromatin proteins, various nucleases, and toxin-antitoxin systems. None of the elements encoded virus-specific proteins. By contrast, the pangenome of pipolins consisted of various genes typical of plasmids (Table S2). Consistent with this assertion, four of the bacterial and five mitochondrial piPolBs were encoded by circular plasmids (Figure S1). Notably, the mitochondrial plasmids carry no other genes than those encoding piPolB (Gobbi et al., 1997, Li and Nargang, 1993, Schulte and Lambowitz, 1991), suggesting that, following the introduction of a mitochondrial ancestor into a proto-eukaryotic host, the MGE underwent reductive evolution.
Although piPolB is the only DNA replication-associated protein conserved in all pipolins, some elements encode putative helicases of superfamilies 1 and 2, 3′-5′ exonucleases, uracil-DNA glycosylases, ribonucleases H, and an Orc1/Cdc6-like AAA+ ATPase. Unlike pPolB-encoding plasmids and viruses, which, as a rule, have linear genomes, pipolins represent circular dsDNA molecules and, thus, the protein-priming mechanism is unlikely to be applicable. The overwhelming majority (94%) of dsDNA viruses encoding RNA-primed DNAPs also encode their own primases (Kazlauskas et al., 2016). By contrast, none of the pipolins possesses genes for recognizable primases, raising questions regarding the priming mechanism.
Collectively, results of the phylogenetic and comparative genomic analyses underscore the uniqueness of piPolBs and pipolins, which may be considered as the third major superfamily of self-replicating MGEs, next to polintons and casposons.
Pipolin DNAP Is a Proficient Replicase
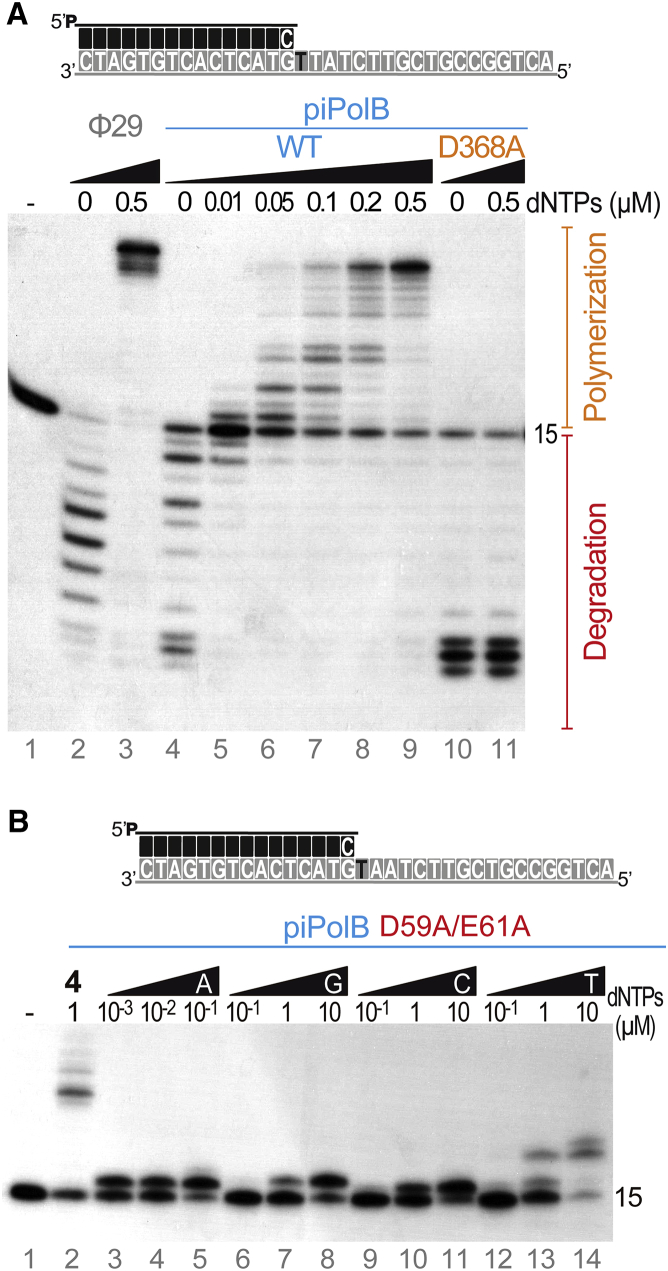
To verify whether piPolBs were indeed active DNAPs, we chose a representative enzyme from E. coli 3-373-03-S1_C2 pipolin, and we purified its recombinant form. We first analyzed the synthetic and degradative activities of this protein in a primer extension assay (Figure 2A, lanes 4–9). As expected, only degradation products could be detected in the absence of dinucleotide triphosphates (dNTPs). However, the addition of dNTPs resulted in a switch from exonucleolysis to polymerization activity, indicating that both activities were coordinated. Protein variants with deficient polymerization (D368A; Figure 2A, lanes 10 and 11; Figure S3A) or exonuclease (D59A/E61A; Figure S3A) activities confirmed that 5′-3′ synthetic and 3′-5′ degradative capacities were intrinsic to the recombinant purified piPolB. The presence of proficient DNA polymerization activity in piPolB confirmed that only the second carboxylate moiety in the PolC motif is required for metal coordination, in agreement with the previous suggestions (Brautigam and Steitz, 1998, Wang et al., 1997).
Figure 2.
Recombinant piPolB from E. coli 3-373-03_S1_C2 Pipolin Is an Active and Faithful DNAP with Intrinsic Proofreading Activity
(A) Primer extension assays with an oligonucleotide template/primer duplex substrate as depicted above the gel. Reactions were incubated for 10 min at 30°C in the presence of either wild-type or D368A polymerase-deficient variant of piPolB and the indicated amount of dNTPs and triggered with 10 mM MgCl2.
(B) Nucleotide insertion preference by the D59A/E61A exonuclease-deficient piPolB variant in the presence of increasing amounts of each dNTP as indicated.
We next analyzed the insertion preference for Watson-Crick base pairs using the piPolB exonuclease-deficient variant D59A/E61A. As shown in Figure 2B, insertion of the correct nucleotide could be detected at approximately 1,000-fold lower dNTP concentration compared with the incorrect dNTP. These results confirm that piPolB of pipolins is an efficient and faithful DNAP.
PiPolB Is Endowed with Intrinsic Translesion Synthesis across DNA Containing Non-bulky Nucleotide Analogs
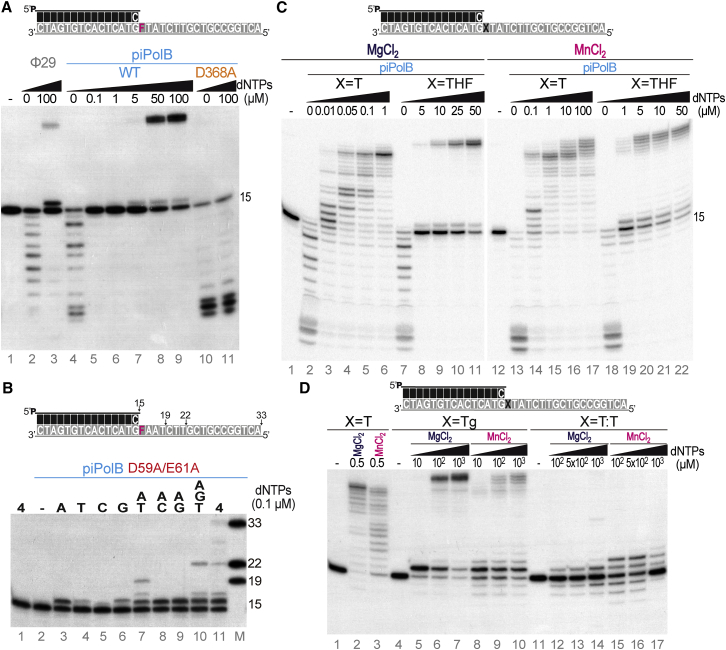
Abasic (AP) sites constitute the most common DNA lesion that may arise from spontaneous depurination, but they also occur as intermediates in base excision repair. A prevailing model is that high-fidelity replicative DNAPs are unable to replicate through such lesions, leading to stalled replication and subsequent triggering of DNA damage tolerance mechanisms, involving specialized DNAPs that can bypass the DNA damage by translesion synthesis (TLS) (Broyde et al., 2008, Vaisman and Woodgate, 2017). However, recent works reported examples of TLS by cellular and viral replicases from families A, B, or C during processive genome replication (Berjón-Otero et al., 2015, Nevin et al., 2017, Sun et al., 2015). As shown in Figure 3A, piPolB was also able to insert the first nucleotide and extend the primer beyond a tetrahydrofuran (THF) moiety, a stable analog of an abasic site (lines 4–9), whereas Φ29DNAP only gave rise to negligible replication, as expected (Berjón-Otero et al., 2015) (lines 2 and 3). The bypass capacity often depends on the sequence context and is counteracted by the proofreading activity (Berjón-Otero et al., 2015, Choi et al., 2010, Tanguy Le Gac et al., 2004, Zhu et al., 2008). However, piPolB TLS capacity did not seem to be affected by the template sequence context (Figure S3A). A minor band of partial product at the lesion site (16-mer) could be detected, suggesting that elongation of the primer beyond the abasic site was a limiting step in the TLS by piPolB, despite the fact that replication of both undamaged and damaged oligonucleotide templates could be processive (Figure S3B).
Figure 3.
Characterization of piPolB TLS Capacity
(A) Primer extension experiment opposite abasic site-containing template. “F” stands for THF abasic site analog.
(B) Step-by-step monitoring of piPolB replication of THF-containing template by the sequential addition of dNTPs (0.1 μM).
(C) Effect of divalent metal cofactors on piPolB polymerization capacity on undamaged and damaged templates. Reactions were triggered either with 1 mM MnCl2 or 10 mM MgCl2, as indicated.
(D) TLS capacity of piPolB on alternative DNA-damaged templates.
We then analyzed the incorporation preference opposite to the THF site. Using the exonuclease-deficient variant D59A/E61A, we found that piPolB preferentially inserted purines over pyrimidines (Figure 3B, lanes 3–6), in the preference order A > G > T > C, in agreement with the so-called “A rule” previously described for many DNAPs (Strauss, 2002). The TLS by DNAPs may occur via a misalignment mechanism, resulting in a 1- or 2-nt deletion and, accordingly, a shorter DNA product (Laverty et al., 2017, Yang, 2014). Thus, we monitored step-by-step polymerization in a primer extension assay in the presence of different dNTP combinations (Figure 3B, lanes 7–11). In particular, we provided deoxyadenosine triphosphate (dATP) in combination with another single dNTP (deoxythymidine triphosphate [dTTP], deoxycytidine triphosphate [dCTP], and deoxyguanosine triphosphate [dGTP], lanes 7–9, respectively). Whereas insertion opposite to the THF was detected in all cases, only the combination dATP and dTTP (AT, lane 7) allowed primer extension beyond the abasic site, giving rise to a product that corresponded with the 19-mer marker (Figure 3B, lane M), indicating accurate replication (see substrate scheme above the gel). Consistently, the presence of dATP, dTTP, and dGTP (ATG, lane 10) allowed the copy of the template up to the 22-mer product length, and only when the four dNTPs were provided could the full-length replication product be detected (lane 11). Taken together, these results indicate that TLS capacity of piPolB preferably inserts an A opposite to the abasic sites and subsequently elongates the primer processively without introducing frameshift mutations.
We next analyzed the abasic site bypass with different metal cofactors and replication-blocking DNA damages. As shown in Figure 3C, abasic site TLS in the presence of manganese ions was more efficient at lower dNTP concentrations than with magnesium (lanes 19–22 versus 8–11), in agreement with previous reports on other PolBs (Tanguy Le Gac et al., 2004, Villani et al., 2002). We noted that replication of undamaged template required a higher dNTP concentration in the presence of manganese ions (lanes 14–17) when compared with the magnesium-triggered reactions (lanes 3–6). We also explored the template specificity of piPolB TLS capacity with substrates containing thymine-glycol (Tg)-oxidized base and cyclobutane thymine dimers (T:T). Our results indicated that piPolB was able to bypass Tg in the presence of magnesium ions (Figure 3D, lanes 5–7). However, primer extension beyond the damage was less efficient, since the 16-mer pause was stronger than in the case of the THF-containing template (lanes 9–11 in Figure 3C versus lanes 5–7 in Figure 3D). In line with the impairment in processive primer extension beyond the damage, manganese ions apparently did not stimulate the TLS. On the contrary, Tg bypass was reduced in the presence of this metal cofactor (lanes 9 and 10). In the case of T:T, insertion of only 1 or 2 nt opposite to the damage could be detected (lanes 12–17). In conclusion, piPolB has an efficient TLS capacity that allows it to bypass abasic sites and oxidative base modifications, but it is unable to overcome bulkier modifications such as T:T, likely because this damage induces major structural changes in the DNA helix that strongly obstruct DNA replication.
Primer-Independent DNA Replication
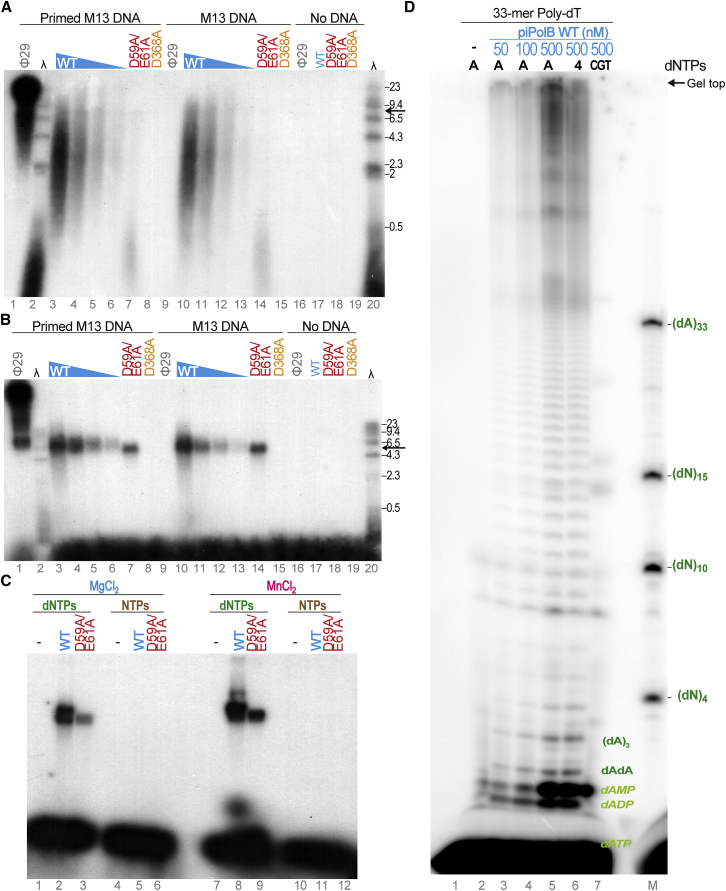
Due to its high processivity, coupled with strand displacement capacity, Φ29DNAP was able to synthesize very large single-stranded DNA (ssDNA) fragments in singly primed M13 DNA rolling circle replication assays (Salas and de Vega, 2016) (Figure 4A, lane 1). By contrast, piPolB gave rise to a smeared signal of replication products spanning 0.5–10 kb, with an apparent peak at ∼3 kb (Figure 4A, lanes 3–6), which indicates that piPolB was not as processive as Φ29DNAP. However, the maximal product length obtained with piPolB remained similar even at 20-fold lower enzyme concentration, suggesting that it is a processive DNA replicase. A considerable portion of replication products was larger than the M13 DNA, suggesting that piPolB DNA replication is coupled with strand displacement. The latter activity was subsequently confirmed using an oligonucleotide template/primer substrate with a 5-nt gap (Figure S4).
Figure 4.
Primer-Independent DNA Synthesis by piPolB
(A and B) Alkaline (A) and non-denaturing TAE (B) agarose electrophoresis of primed (lanes 1 and 3–8) or not-primed (lanes 9–15) M13 ssDNA replication products. Wild-type and D368A and D59A/E61A variants of piPolB were assayed at 500 nM or, when indicated, decreasing concentrations of wild-type piPolB (500, 250, 100, and 50 nM, lanes 3–6 and 10–13). See the Experimental Procedures for details.
(C) Non-denaturing TAE agarose electrophoresis of M13 replication products in the presence of either 100 μM dNTPs (lanes 1–3 and 7–9) or NTPs (lanes 4–6 and 10–12), as indicated. Replication assays were carried out with wild-type or D59A/E61A piPolB variants and triggered with either 10 mM MgCl2 (lanes 1–6) or 1 mM MnCl2 (lanes 7–12).
(D) Primer synthesis and replication of homopolymeric poly-dT DNA template (1 μM) by piPolB. Reactions were triggered with 1 mM MnCl2 and resolved in high-resolution 8 M urea-20% PAGE.
Strikingly, a very similar replication pattern was detected regardless of whether the M13 was primed or not (Figure 4A, lanes 10–13). By contrast, as expected, Φ29DNAP was unable to synthesize any product in the absence of a primer (lane 9). When the same samples were loaded on a non-denaturing agarose gel (Figure 4B), the replication product appeared as a single band that corresponded to the expected M13 unit length, suggesting that the ssDNA products detected in alkaline-denaturing electrophoresis gel are M13 replication products. Consistently, no product could be detected either with the D368A variant deficient for polymerization activity (lanes 8 and 15) or in the absence of input DNA template (lanes 16–19). Notably, the fragments detected with the D59A/E61A mutant were slightly smaller (by <0.5 kb) than with the wild-type enzyme (Figure 4A, lanes 7 and 14), presumably because exonuclease deficiency gives rise to the accumulation of replication mistakes that may result in the impairment of strand displacement or processivity (Soengas et al., 1992). These results further confirmed that M13 DNA replication, with or without the added primer, was intrinsic to piPolB. De novo DNA synthesis on non-primed M13 DNA could be detected using both, magnesium or manganese ions, as cofactors (Figure 4C, lanes 2 and 3 and 8 and 9), albeit with a somewhat higher intensity of total replication product with manganese ions. However, replication was not detected when deoxyribonucleotides were substituted with ribonucleotides (lanes 5 and 6 and 11 and 12), as expected for a PolB that contains the conserved tyrosine steric gate (Bonnín et al., 1999, Brown and Suo, 2011) (Pol B motif in Figure 1B). Smaller DNA fragments were detected with the wild-type polymerase that might be products of the exonucleolytic degradation (lane 8).
To investigate a possible sequence requirement for de novo initiation of DNA replication, we performed assays using a single-stranded homopolymeric poly-dT 33-mer as a template. As shown in Figure 4D, DNA replication in the presence of the complementary dATP gave rise to large DNA products and a laddered pattern, indicating that replication started de novo, with the synthesis of short primers. This laddered pattern could correspond to either a distributive replication or alternative initiation positions throughout the template. Replication products obtained using the exonuclease-deficient piPolB were overall shorter, suggesting a processivity impairment, as found in the case of M13 replication (Figure S5, lanes 8–10 versus 11–13). Using this short, homopolymeric substrate, DNA primer synthesis was negligible with magnesium ions (Figure S5, lanes 2–7 versus 8–13), underlining the higher efficiency of manganese as a cofactor for DNA priming. Interestingly, when all dNTPs were added, generation of large DNA products was somewhat reduced (Figure 4D, lane 6), and, if dATP was reduced to the labeled nucleotide (16 nM compared with 100 μM of the non-labeled, lane 7), replication products were negligible, which suggested that formation of correct Watson-Crick base pairs was required for replication initiation.
Collectively, these results indicate that piPolB from E. coli 3-373-03_S1_C2 pipolin is able to initiate and perform DNA replication of circular and linear templates in the absence of pre-existing primers or additional protein factors. Furthermore, replication of homopolymeric DNA substrates suggests that, contrary to canonical DNA primases (Frick and Richardson, 2001, García-Gómez et al., 2013), piPolB DNA-priming capacity does not rely on a specific template sequence.
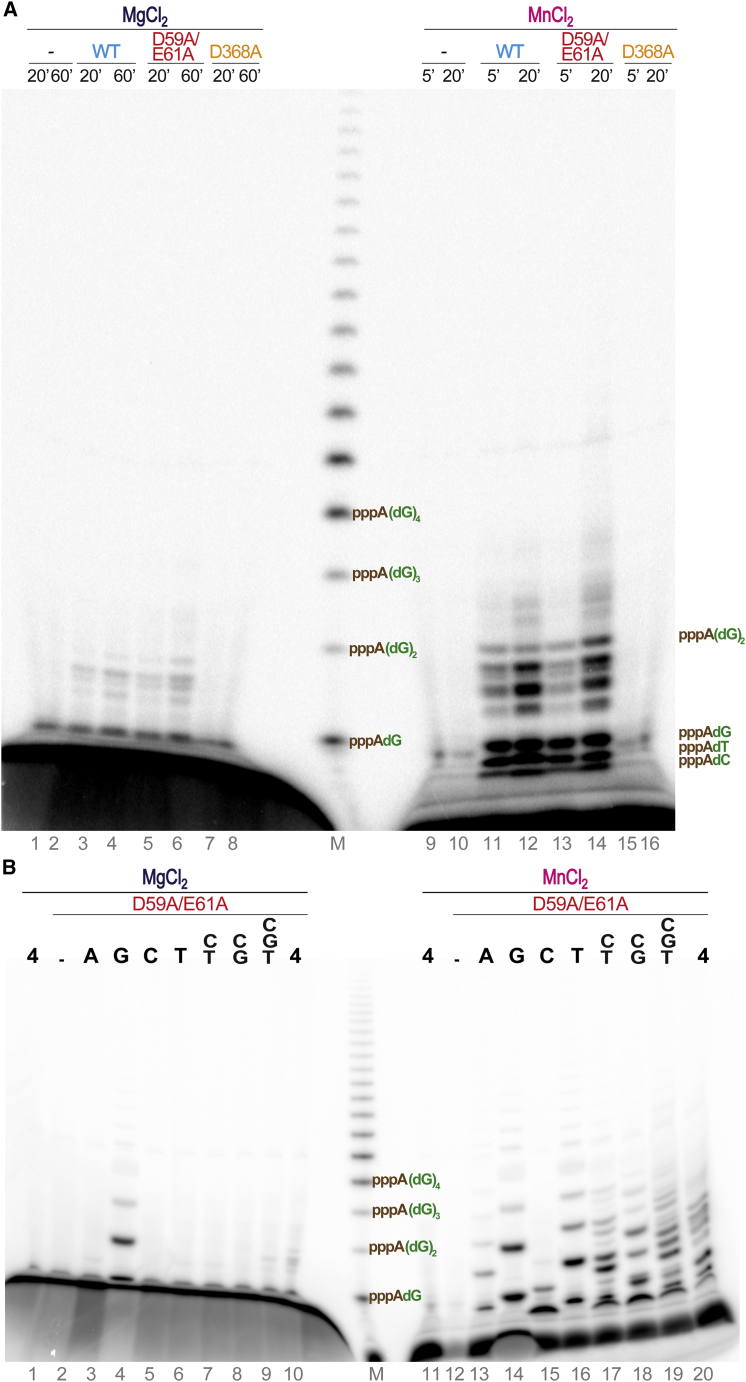
De Novo Synthesis of DNA Primers
To further confirm that piPolB is able to synthesize DNA de novo, we performed M13 ssDNA replication using γ32P-ATP as a labeled nucleotide. Thus, only newly synthesized DNA fragments would be detected. As shown in Figure 5A, small DNA fragments (up to 4–5 nt in length) were generated in a distributive manner by wild-type and exonuclease-deficient piPolBs, but not by the D368A variant. Again, this reaction was considerably more efficient in the presence of manganese ions than with magnesium ions (lanes 1–8 versus 9–16). Furthermore, the products were only detected in the presence of dNTPs, but not with NTPs (not shown). Instead of the large DNA fragments detected in the assays described above (Figure 4A), we observed only di- and trinucleotide primers, which may be abortive initiation products resulting from the incorporation of a ribonucleotide (rather than dNTP) as a terminal 5′ nucleotide.
Figure 5.
De Novo DNA Synthesis by piPolB
(A) Primer synthesis by piPolB. M13 DNA was incubated with either dNTPs or NTPs and wild-type (WT) piPolB or the polymerase- (D368A) or exonuclease- (D59A/E61A) deficient variants. Detected products are labeled with [γ32P]ATP (1 μCi) that only could be incorporated in the 5′ position of the newly synthetized primers. A [γ32P]ATP-(dGMP)n ladder was used a size marker (lane M).
(B) Insertion preference for the first steps of DNA primer synthesis by exonuclease-deficient piPolB. The assay was performed as in (A) but with each dNTP provided independently or in the indicated combinations. Reactions were triggered with either 10 mM MgCl2 or 1 mM MnCl2 and resolved in high-resolution 8 M urea-20% PAGE.
The use of high-resolution PAGE allowed us to identify alternative di- and trinucleotide primers with similar intensity, suggesting that DNA synthesis initiation by piPolB does not require a specific template sequence. In line with this, when each dNTP was provided separately (Figure 5B), the reaction was clearly stimulated by dGTP, in the presence of either magnesium or manganese ions (lanes 4 and 14) and, to a lesser extent, by dCTP and dTTP, either alone or in combination with other deoxyribonucleotides. The fact that A-dG dinucleotide was the most efficiently synthetized initiation product is in agreement with the observation that pyrimidines are the preferential template substrates for the priming reaction by most DNA primases (Frick and Richardson, 2001). In line with these results, single-nucleotide changes in the poly-dT homopolymer substrate did not substantially change the efficiency of de novo DNA synthesis (Figure S6), although short di- and trinucleotides could be detected when one or two Cs were included in the template sequence, even at the 5′ end of the template molecule (lanes 7 and 12). Taken together, these results demonstrate that piPolB was able to initiate de novo DNA primer synthesis without a strong requirement for specific template sequence.
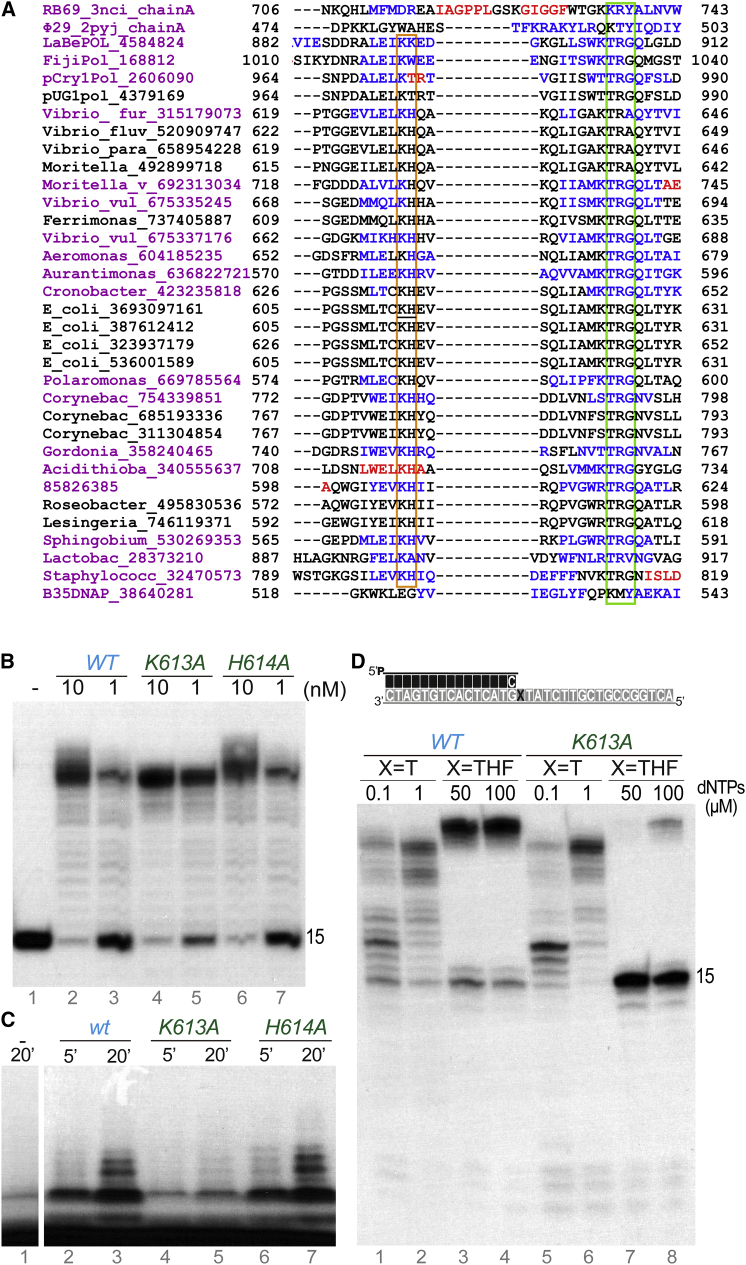
An Invariable Lysine Plays a Role in TLS and Primer Synthesis Activities
PolBs contain a conserved KxY motif within a β strand in the palm domain involved in stabilization of the primer terminus (Berman et al., 2007, Blasco et al., 1995). We hypothesized that structural adaptations of this motif or nearby residues would be required for stable binding of a nucleoside triphosphate at the 5′ side of the nascent primer to allow dinucleotide formation. Indeed, analysis of the multiple sequence alignment showed that the piPolBs lack the canonical KxY motif (Figure 1B) and instead contain an alternative conserved sequence KTRG (Figure 6A). An additional KH pattern within an N-terminal extension of the same β strand is also highly conserved in piPolB homologs, defining an extended KH-X8-KTRG motif. We generated alanine variants of K613, H614, K623, and R625 residues. In agreement with a putative role in primer terminus stabilization, K623A and R625A variants had impaired primer extension capacity (Figure S7A) and primer synthesis beyond the dinucleotide formation (Figure S7B). On the other hand, K613A and H614A proteins had normal primer extension capacity under the tested conditions (Figure 6B). However, whereas H614A was able to synthetize new primers with a similar pattern as the wild-type piPolB (Figure 6C, lanes 6 and 7), K613A priming capacity was strongly reduced (lanes 4 and 5), suggesting a specific role of this residue during the de novo DNA synthesis.
Figure 6.
The piPolBs Invariant K613 Residue Plays a Role in TLS and De Novo Primer Synthesis
(A) Details of a sequence alignment of piPolBs showing a predicted palm β sheet containing the conserved KxY motif. For reference, RB69, Bam35, and Φ29 DNAPs were included. Significantly different sequences are highlighted in magenta, and the predicted secondary structures of these sequence are in blue and red for β sheet and α helix, respectively. The PolB KxY and the piPolB counterpart KTRG motifs are boxed in green, whereas the KH motif is boxed in orange. The mutated K613 and H614 residues in the E. coli 3-373-03_S1_C2 pipolin piPolB (GI 693097161) are underlined.
(B) Primer extension assays of wild-type, K613A, and H614A His-tagged piPolBs. Assays were carried out for 10 min at 30°C in the presence of 1 nM primer/template duplex, 10 μM dNTPs, and the indicated concentration of piPolB variants.
(C) Primer synthesis by wild-type, K613A, and H614A His-tagged piPolBs.
(D) Comparison of primer extension capacity of wild-type and K613A His-tagged piPolBs opposite to undamaged (X = T) or damaged (X = THF) templates. Reactions were triggered with 1 mM MnCl2 and resolved in 8 M urea-20% PAGE.
Furthermore, the TLS capacity of K613A protein was also strongly impaired compared to the wild-type piPolB (Figure 6D, lanes 3 and 4 versus 7 and 8). Thus, although DNA primer synthesis and primer extension opposite to the undamaged and damaged substrates appeared to rely on the same conserved catalytic residues, as shown for the D368A variant (see above), we were able to partially uncouple these activities. This result further confirmed the unique intrinsic TLS and DNA primase capacities of piPolB, and also it unveils the role of the extended primer stabilization motif of the piPolB group that would be required for these activities.
Biological Role of piPolB in De Novo DNA Synthesis
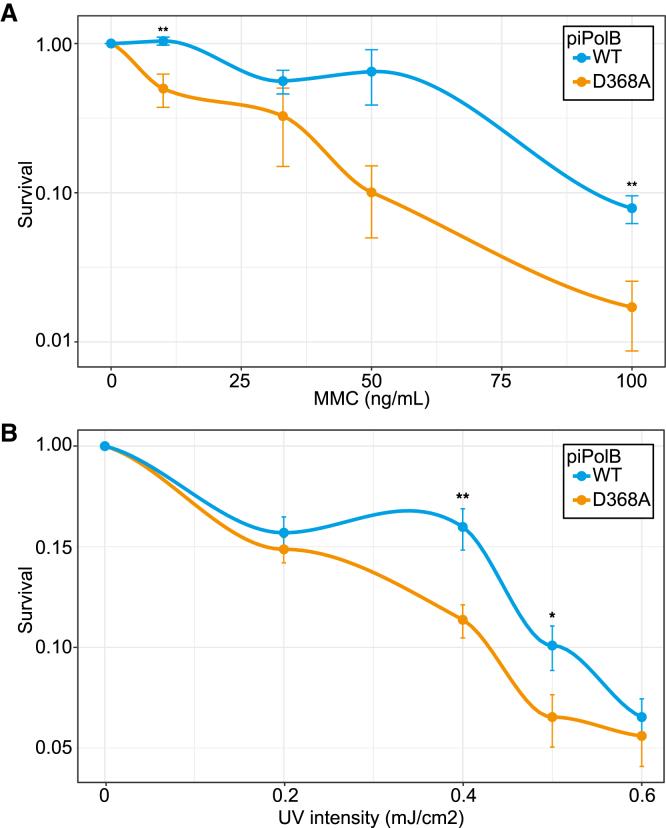
Considering the DNA-priming capacity of piPolB, which is unprecedented in PolB family enzymes, we decided to investigate its biological role in vivo. To this end, we challenged the piPolB-expressing bacteria with Mitomycin C (MMC) and UV irradiation, the two DNA-damaging agents known to block DNA replication by introducing bulky base modifications and interstrand crosslinks. Since piPolB was unable to replicate a T:T-containing template (Figure 3D), it is unlikely that its TLS capacity may allow bypass of DNA damage induced by MMC treatment or UV irradiation. However, given that replication blockage on the leading strand can be circumvented by re-priming events downstream of the UV-generated lesions (Heller and Marians, 2006), we hypothesized that the de novo DNA synthesis by piPolB might contribute to relieving the genotoxic stress generated by DNA-damaging agents. Our results suggested that this is indeed the case, since expression of the wild-type piPolB in E. coli Bl21(DE3) cultures significantly enhanced cell survival upon both MMC treatment and UV irradiation, as compared with bacteria expressing D368A inactive piPolB variant (Figure 7). These results indicate a possible role of piPolB in DNA damage tolerance or repair in the context of E. coli cells.
Figure 7.
Survival of E. coli (DE3) Cells Expressing Wild-Type of Inactive D368A piPolB Variants upon DNA Damage Challenges
(A and B) The graphs show relative survival (mean and SE of four independent experiments) of cells overexpressing wild-type of DNA polymerization-deficient piPolB variants after genotoxic challenge with MMC (A) or UV irradiation (B). The p values are indicated as ∗p ≤ 0.1, ∗∗p ≤ 0.05, and ∗∗∗p ≤ 0.01. See the Supplemental Experimental Procedures for details.
Discussion
Here we report the discovery and biochemical characterization of a previously overlooked major group of replicative PolBs, which we named piPolB due to their unique capacity to perform primer-independent, templated DNA synthesis. Within the global PolB phylogeny, piPolB forms a distinct, ancient clade on par with the two previously described groups, rPolB and pPolB. The piPolB-encoding genes are found in MGEs, dubbed pipolins, most of which are integrated into genomes of bacteria from phyla Firmicutes, Actinobacteria, and Proteobacteria, but also replicating as circular plasmids in mitochondria. The distribution of pipolins is rather patchy, which is typical of integrated MGEs (Forterre, 2012, Makarova et al., 2014). To a large extent, pipolins seem to have co-evolved with their hosts, because piPolB-based phylogeny is congruent with the general bacterial taxonomy. Notably, phylogenetic analysis showed that piPolBs from mitochondrial plasmids cluster with alphaproteobacterial homologs (Figure S1). Given that in all likelihood mitochondria have evolved from an alphaproteobacterial ancestor at the onset of eukaryogenesis (Gray, 2012), it is tempting to speculate that piPolBs were introduced into eukaryotes along with the proto-mitochondrial alphaproteobacterial endosymbiont. According to conservative estimates based on the microfossil record, eukaryotes emerged ∼2 billion years ago (Dyall et al., 2004, López-García and Moreira, 2015). Thus, the piPolB clade should be at least as old if not older, especially if the emergence of pipolins predated the divergence of the major bacterial phyla.
The piPolBs share the conserved active site as well as the TPR1 and TPR2 subdomains with pPolBs (Figure 1). Consistently, we showed that piPolB displays efficient DNA polymerization and strand displacement activities. Furthermore, piPolB also showed intrinsic TLS capacity across non-bulky base damages (Figure 3). Strikingly, unlike all other PolBs, piPolB does not require an externally provided primer for DNA replication. Conversely, we found that piPolB is able to initiate DNA synthesis de novo, a capacity so far exclusive to DNA primases. In the case of Φ29DNAP, the TPR1 motif makes contacts with the template strand and plays a key role in the interaction with the TP during the early steps of protein-primed replication (Dufour et al., 2000, Kamtekar et al., 2006). Given that piPolBs do not interact with a TP, the function of TPR1 region may be limited to the interaction with the DNA or certain cellular cofactors, which would modulate the piPolB activity in vivo.
The use of manganese as divalent cofactor instead of magnesium increased TLS across abasic sites (Figure 3C) as well as de novo DNA synthesis (Figures 4C and 5; Figure S5). Although the roles of divalent metal ions in DNA synthesis have been controversial, a number of recent findings suggest a physiological role of manganese ions as a cofactor in DNA damage tolerance and repair pathways (Andrade et al., 2009, Cannavo and Cejka, 2014, Kent et al., 2016), as in the case of the human PrimPol (García-Gómez et al., 2013). PrimPol domain-containing DNA primases have been shown to possess multiple enzymatic activities in vitro, including primer-dependent and primer-independent DNAP activity, nucleotidyl-transferase, TLS, and even reverse-transcriptase activities (Gill et al., 2014, Guilliam et al., 2015, Iyer et al., 2005, Lipps et al., 2003, Martínez-Jiménez et al., 2015). Although in certain virus- or plasmid-encoded proteins the PrimPol domain is fused to various helicases and can synthesize large DNA products (Zhu et al., 2017), these enzymes lack the exonuclease domain, and their DNA polymerization on longer templates appears to be mainly distributive. Thus, it is generally considered that the role of PrimPol proteins in vivo is largely restricted to the synthesis of short primers, which are extended by the cellular replicative DNAPs (Beck et al., 2010, Gill et al., 2014). By contrast, piPolBs are full-fledged replicative DNAPs endowed with the proofreading and strand displacement capacities. Thus, our results challenge a long-standing dogma in the field, which states that replicative DNAPs are unable to synthesize DNA de novo, without a pre-existing primer providing a hydroxyl moiety to anchor the incoming nucleotide (Kornberg and Baker, 1992, Kuchta and Stengel, 2010).
Our finding that one enzyme acts both as a primase and a DNAP poses evolutionary and mechanistic questions. In particular, why do cellular organisms require two enzymes to fulfill de novo DNA synthesis? The inability of cellular replicases to perform as primases might be dictated by the challenges associated with the binding of the priming 5′ nucleotide, because the active site pocket of a primase has to accommodate the triphosphate moiety, which is substantially more negatively charged compared to the pre-existing primer typically faced by a DNAP. The maintenance of the unstable, short oligonucleotides on the templates is also a considerable challenge that arguably cannot be tackled by processive replicases (Kuchta and Stengel, 2010). We have shown that piPolBs have a unique KTRG motif, alternative to the conserved KxY motif of PolBs, which interacts with the primer terminus (Blasco et al., 1995). Moreover, an invariant lysine nearby the KTRG motif plays a key role both in TLS and de novo primer synthesis (Figure 6). Given the positive charge of this and nearby residues in the piPolB group, it is likely that the extended KH-X8-KTRG motif may induce a highly stable primer terminus-binding mechanism that may favor the binding of the incoming nucleotide and the subsequent stabilization of the ternary complex, which would result in enhanced polymerization capacity. These results establish a structural liaison between TLS and priming capacities of piPolB that should be further explored in the future by structural and biochemical approaches.
As mentioned above, all DNA primases lack proofreading capacity. This seems advantageous for the efficient synthesis of short-lived Okazaki fragments. Conversely, the 3′-5′ exonuclease-proofreading activity, which is necessary for faithful DNA replication, could hinder the primase capacity. Thus, piPolB synthetic and degradative activities must be highly coordinated to allow efficient primer synthesis and faithful DNA replication. Furthermore, the piPolB exonuclease activity is also compatible with TLS of non-bulky base damages, which, as reported previously for pPolB of bacteriophage Bam35 (Berjón-Otero et al., 2015), does not require template strand misalignment but tolerates damage-containing mismatches during processive DNA synthesis. The TLS capacity might be of particular importance for mitochondrial pipolins, which, similar to mitochondrial genomes, are likely to be frequently exposed to reactive oxygen species regularly produced during normal mitochondrial respiration (Valentine et al., 1998). Previous studies have shown that replication of pCRY1-like pipolins from fungal mitochondria (Gobbi et al., 1997, Li and Nargang, 1993) can be initiated from multiple origins rather than from a fixed origin (Baidyaroy et al., 2012). However, this observation remained unexplained. In light of our current results, such a replication pattern is consistent with the possibility that pCRY1-like pipolins are replicated by their cognate piPolBs in a primer-independent manner. Analogously, the circular episomal form of bacterial pipolins could be replicated by piPolBs from multiple origins.
Replication across bulkier DNA lesions that could not be bypassed by piPolB might benefit from possible downstream re-priming. Replication re-start in UV-exposed E. coli chromosome was suggested in the late 60s (Rupp and Howard-Flanders, 1968), and an origin-independent leading strand re-initiation has been demonstrated experimentally (Heller and Marians, 2006). Accordingly, we have shown that expression of the wild-type piPolB promotes survival of E. coli cells exposed to replication-blocking DNA-damaging agents (Figure 7). Hence, we hypothesize that piPolB might have evolved to maintain pipolins’ DNA by providing faithful and processive de novo DNA replication as well as tolerance to DNA damage, which may also increase the fitness of the host bacteria. The latter mechanism resembles the recently proposed roles of human PrimPol in DNA damage tolerance and bypass (Guilliam and Doherty, 2017, Martínez-Jiménez et al., 2015, Mourón et al., 2013), which in the case of piPolB would be provided by an MGE. According to this hypothesis, pipolins may act as bacterial symbionts contributing to maintenance of the host genome upon genotoxic stress.
Importantly, piPolB holds a great promise for developing novel biotechnological applications. For instance, in vitro activities of piPolB, namely, strand displacement and faithful, processive DNA polymerization, can be harnessed for efficient primer-independent whole-genome amplification, whereas the TLS can be useful for amplification of damaged or ancient DNA templates. Given that piPolBs do not display strong sequence requirement for replication initiation, replication origins may be selected in a random manner, a property useful for whole-genome amplification. In this framework it is worth mentioning that a recently developed method, dubbed TruePrime, using a combination of an AEP primase, TthPrimPol, and the Φ29DNAP, has been proposed for whole-genome amplification from single cells (Picher et al., 2016). Similarly, piPolB could become a single-enzyme solution to achieve the same goal in single-cell genomic applications. Further structural and functional characterization of piPolBs and the pipolins that encode them will help to understand the details of this unique replication mechanism and to harness the potential of these enzymes for versatile biotechnological applications.
Experimental Procedures
Primer Extension Assays
Assays were performed in 20 μL final volume containing 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% (v/v) glycerol, 0.1 mg/mL BSA, 0.05% (v/v) Tween 20, and, unless otherwise stated, 1 nM of the indicated 5′-labeled primer/template duplex, 10 nM DNAP, and the indicated dNTP concentration. Reactions were triggered by the addition of either 10 mM MgCl2 or 1 mM MnCl2, as indicated, and, after incubation for the indicated times at 30°C, the reactions were stopped by adding 10 μL formamide loading buffer (98% formamide, 20 mM EDTA, 0.5% [w/v] bromophenol blue, and 0.5% [w/v] xylene cyanol). Samples were analyzed by 8 M urea-20% PAGE (20 × 30 × 0.5 mm) in 1× Tris-borate-EDTA (TBE) buffer. Gel bands were detected either by autoradiography or phosphorimages (Typhoon FLA 7000) and processed with ImageJ software.
Replication of ssDNA
The reaction mixture contained, in a final volume of 25 μL, 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 4% (v/v) glycerol, 0.1 mg/mL BSA, 0.05% (w/v) Tween 20, 20 mM ammonium sulfate, 100 μM dNTPs, 0.5 μCi [α-32P]dATP, 3.2 nM primed or non-primed M13mp18 ssDNA, and the indicated concentrations of each DNAP. Reactions were triggered by the addition of either 10 mM MgCl2 or 1 mM MnCl2 and incubated for 20 min at 30°C. Reactions were then quenched by adding 5 μL 250 mM EDTA and 5% (w/v) SDS, and they were directly loaded in Tris-Acetate-EDTA buffer (40 mM tris, 20 mM acetic acid, and 1 mM EDTA, pH 8.0) (TAE)1x non-denaturing agarose electrophoresis. For alkaline agarose electrophoresis, an aliquot (15 μL) was subjected to gel filtration through Sephadex G-15 spin columns containing 0.1% (w/v) SDS. Lambda DNA ladder used as a size marker was labeled by filling in with Klenow fragment (New England Biolabs) in the presence of [α-32P]dATP (Sambrook and Russell, 2001).
Replication of homopolymeric single-stranded oligonucleotides is described in the Supplemental Experimental Procedures.
Statistical Methods
Data analysis and representation was performed using R and R-Studio (Studio, Boston, MA; http://www.rstudio.com), using packages Dplyr, Stats, and Ggplot2, available from CRAN (the comprehensive R archive network; https://cran.rstudio.com). Based on Shapiro-Wilk normality tests, results were analyzed by either paired t test or Dependent 2-group Wilcoxon signed rank test.
Author Contributions
Conceptualization, M.R.-R.; Investigation, M.R.-R., C.D.O., M.B.-O., J.M.-G., C.A.-M., and M.K.; Writing – Original Draft, M.R.-R., M.S., and M.K.; Writing – Review & Editing, M.R.-R., P.F., M.S., and M.K.; Funding Acquisition, M.R.-R., P.F., and M.S.; Resources, M.S.
Acknowledgments
We thank Laurentino Villar for protein purifications, Dr. Miguel de Vega for valuable suggestions and critical reading of the manuscript, and Professor Luis Blanco for helpful discussions and the [γ-32P]ATP-(dGMP)n ladder. M.S.’s lab is funded by the Spanish Ministry of Economy and Competitiveness (BFU2014-52656P). M.R.-R. was supported by a ComFuturo Grant (NewPols4Biotech) from Fundación General CSIC. C.D.O. was holder of a “Plan de Empleo Juvenil” contract from Madrid Regional Government (funded by YEI program from European Social Fund, EC). An institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa is also acknowledged. P.F. was funded by the European Research Council under the European Union’s Seventh Framework Program (FP/2007-2013)/Project EVOMOBIL - ERC Grant Agreement 340440.
Published: November 7, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.10.039.
Contributor Information
Modesto Redrejo-Rodríguez, Email: modesto.redrejo@csic.es.
Margarita Salas, Email: msalas@cbm.csic.es.
Mart Krupovic, Email: krupovic@pasteur.fr.
Supplemental Information
References
- Andrade P., Martín M.J., Juárez R., López de Saro F., Blanco L. Limited terminal transferase in human DNA polymerase mu defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl. Acad. Sci. USA. 2009;106:16203–16208. doi: 10.1073/pnas.0908492106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidyaroy D., Hausner G., Bertrand H. In vivo conformation and replication intermediates of circular mitochondrial plasmids in Neurospora and Cryphonectria parasitica. Fungal Biol. 2012;116:919–931. doi: 10.1016/j.funbio.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Bath C., Cukalac T., Porter K., Dyall-Smith M.L. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Beck K., Vannini A., Cramer P., Lipps G. The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis. Nucleic Acids Res. 2010;38:6707–6718. doi: 10.1093/nar/gkq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguin P., Charpin N., Koonin E.V., Forterre P., Krupovic M. Casposon integration shows strong target site preference and recapitulates protospacer integration by CRISPR-Cas systems. Nucleic Acids Res. 2016;44:10367–10376. doi: 10.1093/nar/gkw821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjón-Otero M., Villar L., de Vega M., Salas M., Redrejo-Rodríguez M. DNA polymerase from temperate phage Bam35 is endowed with processive polymerization and abasic sites translesion synthesis capacity. Proc. Natl. Acad. Sci. USA. 2015;112:E3476–E3484. doi: 10.1073/pnas.1510280112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berjón-Otero M., Villar L., Salas M., Redrejo-Rodríguez M. Disclosing early steps of protein-primed genome replication of the Gram-positive tectivirus Bam35. Nucleic Acids Res. 2016;44:9733–9744. doi: 10.1093/nar/gkw673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman A.J., Kamtekar S., Goodman J.L., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Lázaro J.M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc. Natl. Acad. Sci. USA. 1990;87:4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A., Méndez J., Lázaro J.M., Blanco L., Salas M. Primer terminus stabilization at the phi 29 DNA polymerase active site. Mutational analysis of conserved motif KXY. J. Biol. Chem. 1995;270:2735–2740. doi: 10.1074/jbc.270.6.2735. [DOI] [PubMed] [Google Scholar]
- Bonnín A., Lázaro J.M., Blanco L., Salas M. A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type phi29 DNA polymerase. J. Mol. Biol. 1999;290:241–251. doi: 10.1006/jmbi.1999.2900. [DOI] [PubMed] [Google Scholar]
- Braithwaite D.K., Ito J. Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res. 1993;21:787–802. doi: 10.1093/nar/21.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam C.A., Steitz T.A. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- Brown J.A., Suo Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry. 2011;50:1135–1142. doi: 10.1021/bi101915z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyde S., Wang L., Rechkoblit O., Geacintov N.E., Patel D.J. Lesion processing: high-fidelity versus lesion-bypass DNA polymerases. Trends Biochem. Sci. 2008;33:209–219. doi: 10.1016/j.tibs.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E., Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Choi J.Y., Lim S., Kim E.J., Jo A., Guengerich F.P. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland W.C., Wang T.S. Mutational analysis of the human DNA polymerase alpha. The most conserved region in alpha-like DNA polymerases is involved in metal-specific catalysis. J. Biol. Chem. 1993;268:11028–11040. [PubMed] [Google Scholar]
- Dufour E., Méndez J., Lázaro J.M., de Vega M., Blanco L., Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- Dyall S.D., Brown M.T., Johnson P.J. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- Filée J., Forterre P., Sen-Lin T., Laurent J. Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 2002;54:763–773. doi: 10.1007/s00239-001-0078-x. [DOI] [PubMed] [Google Scholar]
- Forterre P. Darwin’s goldmine is still open: variation and selection run the world. Front. Cell. Infect. Microbiol. 2012;2:106. doi: 10.3389/fcimb.2012.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D.N., Richardson C.C. DNA primases. Annu. Rev. Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- García-Gómez S., Reyes A., Martínez-Jiménez M.I., Chocrón E.S., Mourón S., Terrados G., Powell C., Salido E., Méndez J., Holt I.J., Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S., Krupovic M., Desnoues N., Béguin P., Sezonov G., Forterre P. A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2. Nucleic Acids Res. 2014;42:3707–3719. doi: 10.1093/nar/gkt1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbi E., Carpanelli A., Firrao G., Locci R. The Cryphonectria parasitica plasmid pUG1 contains a large ORF with motifs characteristic of family B DNA polymerases. Nucleic Acids Res. 1997;25:3275–3280. doi: 10.1093/nar/25.16.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam T.A., Doherty A.J. PrimPol-Prime Time to Reprime. Genes (Basel) 2017;8:E20. doi: 10.3390/genes8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliam T.A., Keen B.A., Brissett N.C., Doherty A.J. Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 2015;43:6651–6664. doi: 10.1093/nar/gkv625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R.C., Marians K.J. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Hoeben R.C., Uil T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a013003. doi: 10.1101/cshperspect.a013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.A., 3rd, Smith H.O., Pfannkoch C., Venter J.C. Cell-free cloning using phi29 DNA polymerase. Proc. Natl. Acad. Sci. USA. 2005;102:17332–17336. doi: 10.1073/pnas.0508809102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L.M., Koonin E.V., Leipe D.D., Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamtekar S., Berman A.J., Wang J., Lázaro J.M., de Vega M., Blanco L., Salas M., Steitz T.A. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci. USA. 2006;103:4540–4545. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas D., Venclovas C. Computational analysis of DNA replicases in double-stranded DNA viruses: relationship with the genome size. Nucleic Acids Res. 2011;39:8291–8305. doi: 10.1093/nar/gkr564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas D., Krupovic M., Venclovas Č. The logic of DNA replication in double-stranded DNA viruses: insights from global analysis of viral genomes. Nucleic Acids Res. 2016;44:4551–4564. doi: 10.1093/nar/gkw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T., Mateos-Gomez P.A., Sfeir A., Pomerantz R.T. Polymerase θ is a robust terminal transferase that oscillates between three different mechanisms during end-joining. eLife. 2016;5:e13740. doi: 10.7554/eLife.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R., Meinhardt F. Microbial Linear Plasmids. Springer; 2007. Linear protein-primed replicating plasmids in eukaryotic microbes; pp. 187–226. [Google Scholar]
- Koonin E.V. Temporal order of evolution of DNA replication systems inferred by comparison of cellular and viral DNA polymerases. Biol. Direct. 2006;1:39. doi: 10.1186/1745-6150-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Baker T.A. W.H. Freeman; 1992. DNA Replication. [Google Scholar]
- Krupovic M., Koonin E.V. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat. Rev. Microbiol. 2015;13:105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Koonin E.V. Self-synthesizing transposons: unexpected key players in the evolution of viruses and defense systems. Curr. Opin. Microbiol. 2016;31:25–33. doi: 10.1016/j.mib.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Bamford D.H., Koonin E.V. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct. 2014;9:6. doi: 10.1186/1745-6150-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Makarova K.S., Forterre P., Prangishvili D., Koonin E.V. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M., Béguin P., Koonin E.V. Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr. Opin. Microbiol. 2017;38:36–43. doi: 10.1016/j.mib.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta R.D., Stengel G. Mechanism and evolution of DNA primases. Biochim. Biophys. Acta. 2010;1804:1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty D.J., Averill A.M., Doublié S., Greenberg M.M. The A-Rule and Deletion Formation During Abasic and Oxidized Abasic Site Bypass by DNA Polymerase θ. ACS Chem. Biol. 2017;12:1584–1592. doi: 10.1021/acschembio.7b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nargang F.E. Two Neurospora mitochondrial plasmids encode DNA polymerases containing motifs characteristic of family B DNA polymerases but lack the sequence Asp-Thr-Asp. Proc. Natl. Acad. Sci. USA. 1993;90:4299–4303. doi: 10.1073/pnas.90.9.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps G., Röther S., Hart C., Krauss G. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 2003;22:2516–2525. doi: 10.1093/emboj/cdg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P., Moreira D. Open Questions on the Origin of Eukaryotes. Trends Ecol. Evol. 2015;30:697–708. doi: 10.1016/j.tree.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K.S., Wolf Y.I., Forterre P., Prangishvili D., Krupovic M., Koonin E.V. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles. 2014;18:877–893. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Jiménez M.I., García-Gómez S., Bebenek K., Sastre-Moreno G., Calvo P.A., Díaz-Talavera A., Kunkel T.A., Blanco L. Alternative solutions and new scenarios for translesion DNA synthesis by human PrimPol. DNA Repair (Amst.) 2015;29:127–138. doi: 10.1016/j.dnarep.2015.02.013. [DOI] [PubMed] [Google Scholar]
- Mourón S., Rodriguez-Acebes S., Martínez-Jiménez M.I., García-Gómez S., Chocrón S., Blanco L., Méndez J. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol. 2013;20:1383–1389. doi: 10.1038/nsmb.2719. [DOI] [PubMed] [Google Scholar]
- Nevin P., Gabbai C.C., Marians K.J. Replisome-mediated translesion synthesis by a cellular replicase. J. Biol. Chem. 2017;292:13833–13842. doi: 10.1074/jbc.M117.800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Basta T., Häring M., Garrett R.A., Prangishvili D. Genome of the Acidianus bottle-shaped virus and insights into the replication and packaging mechanisms. Virology. 2007;364:237–243. doi: 10.1016/j.virol.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Picher A.J., Budeus B., Wafzig O., Krüger C., García-Gómez S., Martínez-Jiménez M.I., Díaz-Talavera A., Weber D., Blanco L., Schneider A. TruePrime is a novel method for whole-genome amplification from single cells based on TthPrimPol. Nat. Commun. 2016;7:13296. doi: 10.1038/ncomms13296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez I., Lázaro J.M., Blanco L., Kamtekar S., Berman A.J., Wang J., Steitz T.A., Salas M., de Vega M. A specific subdomain in phi29 DNA polymerase confers both processivity and strand-displacement capacity. Proc. Natl. Acad. Sci. USA. 2005;102:6407–6412. doi: 10.1073/pnas.0500597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W.D., Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- Salas M., de Vega M. Protein-Primed Replication of Bacteriophage Φ29 DNA. In: Laurie S.K., Marcos Túlio O., editors. The Enzymes. Academic Press; 2016. pp. 137–167. [DOI] [PubMed] [Google Scholar]
- Salas M., Holguera I., Redrejo-Rodríguez M., de Vega M. DNA-binding proteins essential for protein-primed bacteriophage Φ29 DNA replication. Front. Mol. Biosci. 2016;3:37. doi: 10.3389/fmolb.2016.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. Fourth Edition. Cold Spring Harbor Laboratory Press; New York: 2001. Molecular cloning: a laboratory manual. [Google Scholar]
- Schulte U., Lambowitz A.M. The LaBelle mitochondrial plasmid of Neurospora intermedia encodes a novel DNA polymerase that may be derived from a reverse transcriptase. Mol. Cell. Biol. 1991;11:1696–1706. doi: 10.1128/mcb.11.3.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidore A.M., Lan F., Lim S.W., Abate A.R. Enhanced sequencing coverage with digital droplet multiple displacement amplification. Nucleic Acids Res. 2016;44:e66. doi: 10.1093/nar/gkv1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas M.S., Esteban J.A., Lázaro J.M., Bernad A., Blasco M.A., Salas M., Blanco L. Site-directed mutagenesis at the Exo III motif of phi 29 DNA polymerase; overlapping structural domains for the 3′-5′ exonuclease and strand-displacement activities. EMBO J. 1992;11:4227–4237. doi: 10.1002/j.1460-2075.1992.tb05517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss B.S. The “A” rule revisited: polymerases as determinants of mutational specificity. DNA Repair (Amst.) 2002;1:125–135. doi: 10.1016/s1568-7864(01)00014-3. [DOI] [PubMed] [Google Scholar]
- Sun B., Pandey M., Inman J.T., Yang Y., Kashlev M., Patel S.S., Wang M.D. T7 replisome directly overcomes DNA damage. Nat. Commun. 2015;6:10260. doi: 10.1038/ncomms10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguy Le Gac N., Delagoutte E., Germain M., Villani G. Inactivation of the 3′-5′ exonuclease of the replicative T4 DNA polymerase allows translesion DNA synthesis at an abasic site. J. Mol. Biol. 2004;336:1023–1034. doi: 10.1016/j.jmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Vaisman A., Woodgate R. Translesion DNA polymerases in eukaryotes: what makes them tick? Crit. Rev. Biochem. Mol. Biol. 2017;52:274–303. doi: 10.1080/10409238.2017.1291576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine J.S., Wertz D.L., Lyons T.J., Liou L.L., Goto J.J., Gralla E.B. The dark side of dioxygen biochemistry. Curr. Opin. Chem. Biol. 1998;2:253–262. doi: 10.1016/s1367-5931(98)80067-7. [DOI] [PubMed] [Google Scholar]
- Villani G., Tanguy Le Gac N., Wasungu L., Burnouf D., Fuchs R.P., Boehmer P.E. Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucleic Acids Res. 2002;30:3323–3332. doi: 10.1093/nar/gkf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sattar A.K., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- Yang W. An overview of Y-Family DNA polymerases and a case study of human DNA polymerase η. Biochemistry. 2014;53:2793–2803. doi: 10.1021/bi500019s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Song L., Stroud J., Parris D.S. Mechanisms by which herpes simplex virus DNA polymerase limits translesion synthesis through abasic sites. DNA Repair (Amst.) 2008;7:95–107. doi: 10.1016/j.dnarep.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Wang L., Mitsunobu H., Lu X., Hernandez A.J., Yoshida-Takashima Y., Nunoura T., Tabor S., Richardson C.C. Deep-sea vent phage DNA polymerase specifically initiates DNA synthesis in the absence of primers. Proc. Natl. Acad. Sci. USA. 2017;114:E2310–E2318. doi: 10.1073/pnas.1700280114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.