INTRODUCTION
The pathogenesis of asthma is multifactorial, including genetic, environmental and social factors, and the interaction between them (gene-environment interactions).1 A further challenge facing investigations of this condition is that there are likely multiple “phenotypes” or subtypes of what has been clinically diagnosed as “asthma”.2–5 and potential heterogeneity in risk factors for asthma between racial/ethnic groups.
Clinicians have long recognized the increased risk of asthma in certain families and genetic epidemiologic studies have calculated heritabilities between 35 and 95%.6,7 Twin studies provide similar support for strong heritability, with concordance rates among monozygotic twins being about 75% compared to 35% among dizygotic twins.8 Although there are variable results depending on methodology and temporal factors, many studies show that asthma is more prevalent in the American Indian (AI) population when compared to all other races,9–12 but little is known about specific causes or phenotypes. One study found American Indians have the highest asthma rate among single-race groups, with 18.5% of American Indians diagnosed with asthma, while only 11% are diagnosed with asthma among the US adult population.9
There are several, well-replicated genetic variants that have been previously associated with asthma, notably variants at 17q21.7,13–17 While associated SNPs in this region encompass at least a 380Kb span7 including 5 annotated genes,13,18 initial attention has centered primarily on ORMDL3 and GSDMB. Calcium homeostasis, sphingolipid metabolism and lymphocyte function are affected by variants influencing expression of ORMDL3.19–22 whereas GSDMB is highly expressed in T-lymphocytes and thus potentially impacts the inflammatory response, particularly to viral infection.16,23 Calcium sensing and regulation have recently been found to have a profound influence on airway reactivity and suggest therapeutic targets for asthma control.24 Additional genetic variants showing a robust association with asthma phenotypes have been found at 2q12.1,14,17,25 5q22.1,25 5q31.1,14,15,17 7q22.3,14 9p24.1,14,15 and 11q13.5.25 The function of candidate genes at these loci are involved in a myriad of functions,26 including cytokine regulation of inflammatory cells (TSLP, IL1RL1, TMEM182, C11orf30, IL33, DPP10), controlling cellular maturation and differentiation (HLA-DQB1, CDHR3), DNA repair (RAD50); and associated with various inflammatory pathologies, such as eosinophilic esophagitis,27 inflammatory bowel disease28 and allergic rhinitis.25
While variants at these six genetic loci have been replicated across independent studies, the majority have been implicated solely in individuals with European ancestry. Thus, it is unknown if these variants represent universal genetic risk factors for asthma across human populations, and in many instances replication attempts have failed when attempted in a different racial/ethnic group. Furthermore, none of these studies have included American Indian populations, and thus it is unknown whether similar genetic risk factors are present in this population. Our objective was to investigate the contribution of genetic variants at six previously implicated asthma-associated loci in the development of asthma in American Indian children.
METHODS
This analysis derives from a case-control study of the environmental and genetic influences on risk of asthma among an American Indian population in north-central United States. Most primary medical care for this community of predominantly tribal members is provided mainly by federal funding to the Indian Health Service (IHS) and a tribal health department.
The population is located in the north central portion of South Dakota in an area covering 4,266 square miles. The area’s population is approximately 8,500, giving a population density of between 2 and 3 people per square mile. Most live in cluster housing near small towns or in cluster sites far removed from basic services. Beyond federally supported work in health care and education, ranching and farming provide the bulk of employment. Two counties in this area have 33 and 42% of residents with incomes below the poverty line, making them the 11th and 4th lowest per capita income counties in America respectively. In addition, over 20% have less than a high school education.29
Cases were ascertained through automated query of the IHS electronic medical records system, searching for an inclusive array of ICD-9 codes between 493.00 and 493.92, in addition to codes 786.07 (wheezing) and V17.5 (family history of asthma). The search was limited to individuals between and including the ages of 6 and 17 years. Additional cases were sought by contact with local non-IHS providers. This identified over 900 individuals, who were then consented for further review of medical records to determine potential eligibility.
Case definition criteria required both of the following criteria:
a diagnosis of asthma on at least 2 occasions by more than one provider during the past 2 years.
refills of asthma treatment medications on at least 2 occasions during the past 2 years.
Exclusionary criteria were:
Birthweight less than 2500 grams
Neonatal ventilator treatment
Hospitalization at birth greater than 15 days.
Congenital heart anomaly requiring surgery
Diagnosis of cystic fibrosis
Congenital lung, diaphragm, chest wall, or airway anomaly
Diagnosis of pneumonia, pertussis, or tuberculosis within the past year
Congenital muscular disorder
It was found that many of the potential cases initially identified had been assigned an ICD-9 code indicating “asthma” by the pharmacist filling a prescription for a bronchodilator, although the prescribing physician had not indicated a diagnosis of typical “asthma” and was intending to ameliorate the bronchospastic component of a pulmonary infection. These latter children did not meet diagnostic criteria; but did require considerable recruiter effort to contact parents and determine their status via medical record review.
For each case, two controls were initially recruited by identifying the two children born the day after and before the index case and contacting the parents for consent to review medical records for possible inclusion. As the study progressed, this methodology did not yield sufficient controls and many controls were later recruited from previously identified families with children born almost exclusively (>99%) within 6 months of the index case. Initial recruitment was concentrated on cases and later focused on controls. This resulted in a slight bias toward controls being older at the time of exam (even though birth dates were generally within protocol limits). None the less, all but 5 (2.3%) of the pairs were examined within one year of each other. Controls met the same exclusionary criteria as cases in addition to:
No diagnosis of asthma by any provider during the past 2 years.
No prescriptions of any asthma meds during the past 2 years.
Consenting cases and controls were then examined according to study protocol, which included anthropomorphic measures, spirometry, salivary DNA collection and a non-fasting blood draw. Environmental measures of home air quality and dust exposure were made.
A questionnaire collected social, demographic, and medical history from both cases and controls, most of which will be reported in the future. One question: “Has a medical person ever said that your child had hay fever or seasonal allergies?” sought to gauge atopic symptomatology. Total white cell count (WBC), % eosinophils (%eos) and serum measures of high sensitivity C-reactive protein (CRP), total immunoglobulin E (IgE) and specific IgE reactive to 5 airborne antigens (dog and cat dander, dust mite, cockroach and alternia mold) were assessed as covariates for analysis. A specific IgE antibody titer above the detection limit to at least one of the above 5 aeroallergens was defined as “atopy”. Salivary cotinine levels were used to adjust for tobacco smoke exposure, since self-reported exposure is subject to bias, especially considering public recognition of adverse effects for children with asthma, and for some reluctance of minors to admitting of smoking behavior.
Genetic variants previously associated with asthma risk were chosen from the literature as indicated, and referenced in the introduction.
Genomic DNA was collected and extracted from salivary samples using the Oragene (DNA Genotek Inc) system and the manufacturer’s directions. Pre-designed “TaqMan” (Applied Biosystems Inc) genotyping assays and protocols were implemented for SNPs on a real-time, Mini-Opticon (Bio-Rad Laboratories Inc), 4 color thermocycler. Positive controls were identified for each of the three possible genotypes for each SNP and included with no template controls in each genotyping assay. There were 11 samples that consistently failed genotyping attempts, probably due to failed preservation. The number of case-control pairs analyzed varied slightly by SNP since primer reagents were occasionally exhausted and it was not cost efficient to reorder primers for a very small number of additional samples.
Clinical, environmental, and genetic comparisons between asthma cases and controls were performed using a McNemar chi-square test for discrete variables, and a paired t-test for continuous variables. Univariate and multivariate logistic regression was used to explore the effects of genotype and covariates on risk of asthma under an additive model. All models were adjusted for age, as there was a significant difference between cases and controls due to factors noted previously. Descriptive analyses were done using SPSS 13.0.1 (IBM Inc, Armonk, NY). Logistic regression was performed using LogXact-11 (Cytel Inc, Cambridge, MA).
RESULTS
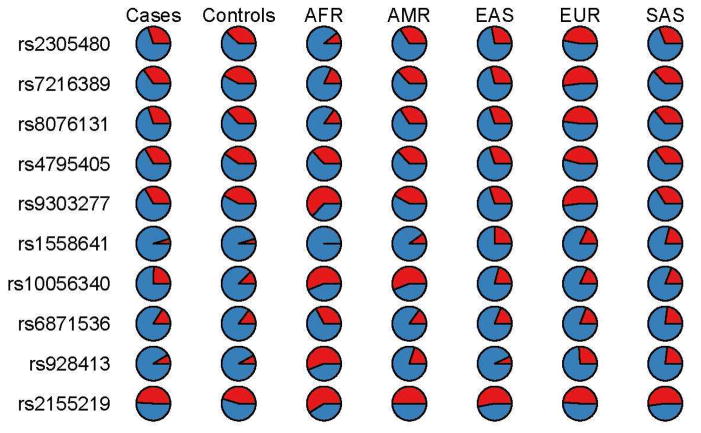
All ten variants previously found to be associated with asthma in populations of European ancestry were polymorphic in this American Indian population in North Central United States. Table 1 provides an orientation to the 10 genetic variants analyzed here, their frequency in the combined cases and controls, and results of tests for Hardy-Weinberg equilibrium. The prevalence of these alleles in other populations can be seen in Figure 1.
Table 1.
Characteristics of SNPs studied and study population prevalences (both cases and controls).
| dbSNP ID | Chromosome Band | Nearest Gene | Annotation | Alleles (major/minor) | Minor allele frequency* | 95% CI | H-W p value |
|---|---|---|---|---|---|---|---|
| rs2305480 | 17q21 | GSDMB | missense | G/A | 35.0% | 31.2 – 38.7 | 0.674 |
| rs7216389 | 17q21 | GSDMB | intronic | T/C | 39.6% | 35.8 – 43.4 | 0.872 |
| rs8076131 | 17q21 | ORMDL3 | intronic | A/G | 34.4% | 30.7 – 38.2 | 0.512 |
| rs4795405 | 17q21 | LRRC3C | intronic | C/T | 37.9% | 34.1 – 41.7 | 0.792 |
| rs9303277 | 17q21 | IKZF3 | intronic | C/T | 39.0% | 35.1 – 42.9 | 0.705 |
| rs1558641 | 2q11.2 | IL1R1 | 5′ upstream | G/A | 4.6% | 3.0 – 6.3 | 0.088 |
| rs10056340 | 5q22.1 | SLC25A46-TSLP | intergenic | T/G | 16.5% | 13.6 – 19.3 | 0.317 |
| rs6871536 | 5q31 | RAD50 | intronic | T/C | 15.2% | 12.4 – 18.0 | 0.596 |
| rs928413 | 9q21 | IL33 | 5′ upstream | A/G | 8.4% | 6.2 – 10.5 | 0.541 |
| rs2155219 | 11q13.4 | EMSY-LRRC32 | intergenic | G/T | 47.3% | 43.4 – 51.2 | 0.781 |
Minor allele frequency among cases and controls combined
Figure 1.
Prevalence of genotyped SNPs in cases and controls of present study, as well as selected global populations. Minor allele (see Table 1) is shown in red. The populations are indicated by standard “1000 Genomes Project” notation, ie AFR: African, AMR: Admixed American, EAS: East Asian, EUR: European, SAS: South Asian.
The characteristics of cases and controls are shown and compared in Table 2. Asthma controls were significantly older than asthma cases, likely due to several factors related to recruitment protocols, as noted in the methods section. The mean difference in age is approximately 4 months older for the controls; and for this reason all logistic regression models were adjusted for age. Asthma cases had significantly higher body mass index (BMI), total serum IgE and white cell count as compared to asthma controls; and atopy was more prevalent among the cases (Table 2). High sensitivity CRP was not significantly different between cases and controls.
Table 2.
Characteristics of matched cases and controls.
| Characteristic | Cases | Controls | p value |
|---|---|---|---|
| Number (N) | 108 | 215 | N/A |
| Male gender, N (%) | 57 (52.8) | 109 (50.7) | 0.773* |
| Age, mean years (SD) | 11.80 (3.22) | 12.14 (3.20) | <0.001** |
| Body-mass index (BMI) Kg/m2 | 25.43 (8.16) | 23.59 (6.60) | 0.005*** |
| White Blood Cells (WBC), X1000 | 7.54 (2.38) | 6.88 (1.82) | 0.017 |
| Eosinophils (%) | 4.99% | 3.79% | 0.931 |
| CRP (mg/L) | 2.17 (2.66) | 1.72 (2.21) | 0.173 |
| IgE**** (total, kU/L) | 486.4 (705.2) | 219.3 (371.3) | <0.001 |
| > one antibody over detection limit | 66 (61%) | 61 (28%) | <0.001 |
| rs2305480, 17q21, A***** | 65/216 =30.1% | 154/410 =37.6% | 0.076****** |
| rs7216389, 17q21, C | 75/216 =34.7% | 176/418 =42.1% | 0.086 |
| rs8076131, 17q21, G | 65/216 =30.1% | 152/414 =36.7% | 0.116 |
| rs4795405, 17q21, T | 72/216 =33.3% | 166/412 =40.3% | 0.105 |
| rs9303277, 17q21, T | 70/210 =33.3% | 168/400 =42.0% | 0.046 |
| rs1558641, 2q11.2, A | 9/214 =4.7% | 19/414 =4.6% | 0.987 |
| rs10056340, 5q22.1, G | 53/216 =24.5% | 51/416 =12.3% | <0.001 |
| rs6871536, 5q31, C | 34/212 =16.0% | 61/414 =14.7% | 0.755 |
| rs928413, 9q21, G | 19/216 =8.8% | 33/406 =8.1% | 0.893 |
| rs2155219, 11q13.4, T | 106/216 =49.1% | 187/412 =45.4% | 0.426 |
Differences between discrete variables evaluated with McNemar’s Chi square test
Differences between means evaluated with paired t test, see Results section for explanation of the significant difference in age
p value for all continuous variables based on analysis of natural log transformed values
Total Immunoglobulin E (IgE)
frequency of designated allele, 65/216 indicates 65 minor alleles (A in this case) and 151 major alleles for a total of 216
Differences in allele frequency evaluated with Chi square test
An analysis of genotypes discordant between case and control pairs is summarized in Table 3. One SNP at 5q22.1, rs10056340, showed a significant association with case-control status following Bonferroni adjustment for ten comparisons (α10=0.005, p<0.0003), considering either the major or minor allele in a dominant model. One SNP at 17q21 was associated at p=0.03 (rs9303277), and three additional SNPs were suggestive at p<0.1 (rs2305480, rs7216389 and rs8076131) considering the major allele in a dominant model.
Table 3.
Genotypes associated with case/control (matched-pair) status.
| Discord. pairs* | Discord. pairs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| dbSNP | allele | Maj Dom | Alter | p value | allele | Min Dom | alter | p value | total pairs |
| rs2305480, 17q21 | G | 15 | 28 | 0.066 | A | 39 | 55 | 0.121 | 205 |
| rs7216389, 17q21 | T | 20 | 34 | 0.076 | C | 39 | 55 | 0.121 | 209 |
| rs8076131, 17q21 | A | 15 | 28 | 0.066 | G | 41 | 53 | 0.256 | 207 |
| rs4795405, 17q21 | C | 18 | 30 | 0.111 | T | 37 | 53 | 0.113 | 206 |
| rs9303277, 17q21 | C | 17 | 33 | 0.033 | T | 37 | 47 | 0.326 | 194 |
| rs1558641, 2q11.2 | G | 0 | 2 | NA | A | 14 | 15 | 1.000 | 205 |
| rs10056340, 5q22.1 | G | 18 | 2 | 0.0008 | T | 67 | 30 | 0.0003 | 208 |
| rs6871536, 5q31 | T | 6 | 3 | 0.508 | C | 43 | 40 | 0.826 | 203 |
| rs928413, 9q21 | A | 2 | 2 | 1.000 | G | 29 | 26 | 0.788 | 203 |
| rs2155219, 11q13.4 | G | 47 | 31 | 0.089 | T | 46 | 37 | 0.380 | 206 |
“Discord. pairs” indicates the number of case/control pairs that are discordant for a particular genotype “Maj Dom”: where the major allele is dominant for the case. “Alter” indicates the alternate situation where the control genotype is dominant for the major allele. Similarly “Min Dom” indicates discordant pairs where the case genotype is dominant for the minor allele. “Total pairs” shows the total number of matched pairs, including both concordant and discordant pairs. “NA” refers to “not applicable”, eg less than 2 groups of discordant pairs.
Results of primarily univariate logistic regression analysis of clinical and environmental covariates are presented in the E-Supplement, Table 1. Since age was shown to be slightly more advanced among controls compared with cases, all univariate analyses were adjusted for age. The demographic and immune measure covariates of BMI, WBC, % eos, total IgE, and atopy were all found to be significantly associated with case status in univariate logistic analysis. C-reactive protein levels did not appear to be associated with asthma in this analysis. While the non-significant estimate of the salivary cotinine level was in the direction of a protective effect, this may be due to parents properly limiting exposure of their children with asthma to tobacco smoke. Self-reported tobacco smoke exposure was not significantly associated with asthma in univariate analysis for any of 4 different questionnaire responses (minimum p value =0.553).
Multivariate logistic regression was then used to assess each SNP in an additive genetic model adjusting for age, BMI and atopy. Although BMI became attenuated below p=0.05 when multivariate models included WBC, the two measures were strongly correlated (Pearson coefficient 0.351, p<0.001) and it was felt that BMI was the primary determinant of the two measures. The results of these analyses are found in Table 4. Rs10056340 was significantly associated with asthma in the multivariate model following Bonferroni correction for ten comparisons, and all five 17q21 SNPs show an association with asthma at p<0.05 after adjustment for these three covariates. No individual 17q21 SNP is statistically significant following a conservative Bonferroni correction for ten comparisons. However, it is only expected to identify one SNP at p<0.05 by chance given ten comparisons, and five are observed, demonstrating an enrichment of true positive associations with asthma at 17q21.
Table 4.
Multivariate logistic regression analysis of factors associated with asthma.
| Characteristic | OR | 95% CI | p Value | |
|---|---|---|---|---|
| Age | 0.909 | 0.833 – 0.992 | 0.033 | |
| Body-mass index (BMI) Kg/m2 | 3.219 | 1.204 – 8.610 | 0.020 | |
| > one specific antibody over detection limit | 3.889 | 2.370 – 6.381 | <0.001 | |
| Above 3 covariates plus genetic variants | ||||
| rs2305480, 17q21, A allele | Additive | 0.635 | 0.434 – 0.927 | 0.019 |
| rs7216389, 17q21, C allele | Additive | 0.681 | 0.472 – 0.982 | 0.040 |
| rs8076131, 17q21, G allele | Additive | 0.668 | 0.459 – 0.971 | 0.035 |
| rs4795405, 17q21, T allele | Additive | 0.680 | 0.469 – 0.985 | 0.041 |
| rs9303277, 17q21, T allele | Additive | 0.648 | 0.447 – 0.940 | 0.022 |
| rs1558641, 2q11.2, A allele | Additive | 1.044 | 0.457 – 2.386 | 0.918 |
| rs10056340, 5q22.1, G allele | Additive | 2.020 | 1.283 – 3.180 | 0.002 |
| rs6871536, 5q31, C allele | Additive | 0.968 | 0.591 – 1.585 | 0.896 |
| rs928413, 9q21, G allele | Additive | 1.187 | 0.634 – 2.222 | 0.592 |
| rs2155219, 11q13.4, T allele | Additive | 1.206 | 0.842 – 1.729 | 0.306 |
To further examine the interaction of atopy and BMI with these SNPs and risk of asthma, we performed stratified analyses according to atopy and BMI. Among atopic individuals (adjusting for age and BMI) the association remained statistically significant for rs10056340 (OR=2.620, 95% CI 1.334 – 5.149, p=0.005), whereas among non-atopic individuals, the results were in the same direction (OR=1.561, 95% CI 0.817 – 2.980, p=0.178). The converse was true for the 5 SNPs at 17q21 which showed an association at p <0.05 among non-atopic individuals; but no association for among atopic individuals. The absolute number of circulating eosinophils (ABS/EOS) was highly correlated to atopy status and cumulative number of specific IgE’s showing sensitization. Substitution of ABS/EOS for atopy in logistic models gave similar results; but when atopy was added to the model, ABS/EOS became non-significant. The same pattern of interaction between atopy as it relates to rs10056340 and the 17q21 SNPs was seen when the ABS/EOS variable was substituted for atopy. Similarly, the leukocyte count (lnWBC) lost significance if included in models with the primary covariates shown in Table 4; and final analyses including representative 5q22.1 and 17q21 SNPs were unchanged.
Stratifying on BMI, rs10056340 showed a similar trend in both those below and above the median lnBMI (p=0.020 and 0.066, respectively, adjusting for age, lnBMI, and atopy). Four of the five 17q21 SNPs also showed nominally significant association; but only in analyses limited to those with lnBMI below the median (maximum p value <0.048).
Fully adjusted models including age, lnBMI, atopy status showed no significant interaction between either rs10056340 or rs2305480 (the prototypic 17q21 SNP) and any of the environmental factors listed in Table 4.
DISCUSSION
We document here that ten common genetic variants previously associated with asthma in other racial/ethnic groups are polymorphic in a Northern Plains American Indian population, and six of those variants are similarly associated with asthma at 5q22.1 and 17q21. Our results demonstrate the presence of shared genetic risk factors for asthma in the Northern Plains American Indian population with other racial/ethnic groups at 5q22.1 and 17q21. The fact that these associations have been confirmed in an American Indian population lends further credence to the likely functional effects of these variants, and/or the existence of an extended haplotype which is shared between American Indian and more distantly related populations.
The relationship between many of these SNPs and asthma risk may depend on the presence of atopy, with associations at 17q21 SNPs stronger in non-atopic individuals, and association with rs10056340 at 5q22.1, stronger among atopic individuals. Similarly, the influence of the 17q21 SNPs on risk of asthma may also show interaction with BMI, with an association seen primarily among those below the median lnBMI. However, these observations are largely qualitative, given the differences observed may reflect the reduction in power from a reduced sample size in a stratified analysis. Nevertheless, without adjustment for atopy or BMI in our logistic model, associations with SNPs at 17q21 become non-significant. Of note, the relationship between asthma and obesity has been seen previously in American Indian30 and other populations.31,32
Our most significant association was with a non-coding variant 3′ downstream of SLC25A46 and 5′ upstream of TSLP (rs10056340 at 5q22.1). SLC25A46 is a mitochondrial solute carrier protein involved in mitochondrial fission,33 however a functional role of this protein in asthma remains to be seen. TSLP promotes cellular response of T helper type 2 (TH2), which contributes to many inflammatory diseases including asthma, allergy, and chronic obstructive pulmonary disease. Variation at 5q22.1 has been associated with asthma in a meta-analysis including African Americans, Hispanic/Latinos, and European Americans, with evidence of association in all three racial/ethnic groups.34 Variants at 5q22.1, including rs10056340 have been previously associated with allergic sensitization in individuals with European ancestry,25,35 and with atopic dermatitis in the Chinese Han population.36 The relationship between asthma and allergic disease has been well established 37,38 and thus the association at rs10056340 and asthma in Northern Plains American Indians suggests atopic asthma is an important sub-phenotype present in this population. We further note the relationship between SNPs at 17q21 and asthma was sensitive to adjustment for the presence of atopy, which further emphasizes the importance of considering asthma sub-phenotypes in genetic analyses.
Variants at 17q21 have consistently been associated with asthma in prior studies, and our results demonstrate the importance of this locus and asthma susceptibility in Northern Plains American Indians. This is arguably the most well-replicated asthma locus including ORMDL3 and GSMDB, with established associations in individuals with European ancestry,39,40 Slovenians,41 Chinese residents of Singapore,42 Pakastani’s,43 Koreans,44 African Americans,45 Puerto Ricans and Mexicans.45 Variants at this locus are most strongly associated with childhood asthma, and given that participants in our study were between the ages of 6–17 we may have expected a stronger signal of genetic association. However, due to the presence of high linkage disequilibrium and the resulting long haplotypes at 17q21, causal variants remain to be identified and may not have been directly genotyped in our study. Thus, additional studies in American Indian populations are necessary to identify causal variants at 17q21, including genotyping a broader spectrum of variants or direct sequencing in a greater number of individuals.
Genetic associations from additional variants queried, including those at 2q12 (IL1R1), 5q31 (RAD50), 9p24, and 11q13 could not be established despite being polymorphic, and sufficiently common in our study population. This may indicate either differences in the genetic architecture of asthma in Northern Plains American Indians at these loci, differences in linkage disequilibrium with the underlying causal variants (assuming the variants typed themselves aren’t causal), or insufficient statistical power to establish a significant association. Furthermore, differences in environmental exposures may also play a role, which can modify the effect of a genetic variant on the risk of asthma (i.e. gene-environment interactions). Environmental factors are known to play an important role in asthma, for example, socioeconomic status, farming, exposure to tobacco smoke and air pollution.
Thus, additional studies are required to fully exclude the possibility that variants at these loci do not contribute to asthma susceptibility in Northern Plains American Indians.
A major strength of our study includes the population-based ascertainment of cases and controls in a Northern Plains American Indian community - a population with high rates of asthma, and yet is notably absent in prior genetic studies of asthma. Unfortunately, for historical reasons, American Indian communities have occasionally taken a skeptical approach to biomedical research and genetic studies in particular. The present investigation benefited from collaboration with a local research organization with a long and deep presence and commitment to the community. Additional strengths include the use of electronic medical records to establish case/control criteria, and the availability of important environmental and clinical covariates, including atopy, for more detailed analyses.
Overall our findings demonstrate the importance of genetic variants at 5q22.1 and 17q21 with risk of asthma in an American Indian population. This is an important step forward to enhance our understanding of the pathophysiology of asthma in a vulnerable population that has been historically absent in genetic studies of asthma. Identifying the spectrum of genes and genetic loci that are relevant to asthma specifically in American Indian children, will facilitate the translation of novel therapeutics and preventative strategies resulting from the genomics era of precision medicine.
Supplementary Material
Acknowledgments
Funding: National Institute on Minority Health and Health Disparities, NIH, Award Number U54MD008164
We thank the study participants, Indian Health Service facilities, and participating tribal communities for their extraordinary cooperation and involvement, which has been critical to the success of this investigation. The views expressed in this paper are those of the authors and do not necessarily reflect those of the Indian Health Service.
Footnotes
Conflicts of interest: None of the authors report any.
Trial registration: not applicable
Authorship:
Concept and design: LGB, MAO, RAO
Data generation: LGB, CA, AS, KJE, SH, DJ, AP, KT
Analysis and interpretation: LGB, MAO, RAO, JMY, DGT
Preparation/revision of manuscript: LGB, MAO, RAO, AP, JMY, DGT
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Colilla S, Nicolae D, Pluzhnikov A, et al. Collaborative Study for the Genetics of Asthma. Evidence for gene-environment interactions in a linkage study of asthma and smoking exposure. J Allergy Clin Immunol. 2003;111:840–6. doi: 10.1067/mai.2003.170. [DOI] [PubMed] [Google Scholar]
- 2.Lang JE, Blake KV. Role of biomarkers in understanding and treating children with asthma: towards personalized care. Pharmgenomics Pers Med. 2013;6:73–84. doi: 10.2147/PGPM.S30626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kudo M, Ishigatsubo Y, Aoki I. Pathology of asthma. Front Microbiol. 2013;4:263. doi: 10.3389/fmicb.2013.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apter AJ. Advances in adult asthma diagnosis and treatment in 2013. J Allergy Clin Immunol. 2014;133:49–56. doi: 10.1016/j.jaci.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Gauthier M, Ray A, Wenzel SE. Evolving Concepts of Asthma. Am J Respir Crit Care Med. 2015;192:660–8. doi: 10.1164/rccm.201504-0763PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Backer V. Estimates of asthma heritability in a large twin sample. Clin Exp Allergy. 2010;40:1054–61. doi: 10.1111/j.1365-2222.2010.03525.x. [DOI] [PubMed] [Google Scholar]
- 9.Center for Disease Control and Prevention. Summary Health Statistics for U.S. Adults: National Health Interview Survey, 2006. 2007;10:112. [Google Scholar]
- 10.Lewis TC, Stout JW, Martinez P, et al. Prevalence of asthma and chronic respiratory symptoms among Alaska Native children. Chest. 2004 May;125(5):1665–73. doi: 10.1378/chest.125.5.1665. [DOI] [PubMed] [Google Scholar]
- 11.Sin DD, Wells H, Svenson LW, Man SF. Asthma and COPD among aboriginals in Alberta, Canada. Chest. 2002;121:1841–6. doi: 10.1378/chest.121.6.1841. [DOI] [PubMed] [Google Scholar]
- 12.Oraka E, Iqbal S, Flanders WD, Brinker K, Garbe P. Racial and ethnic disparities in current asthma and emergency department visits: findings from the National Health Interview Survey, 2001–2010. J Asthma. 2013;50:488–96. doi: 10.3109/02770903.2013.790417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouzigon E, Corda E, Aschard H, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–94. doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 14.Bønnelykke K, Sleiman P, Nielsen K, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46:51–5. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Zhang J, Hu H, Jin Y, Xue M. Polymorphisms of RAD50, IL33 and IL1RL1 are associated with atopic asthma in Chinese population. Tissue Antigens. 2015;86:443–7. doi: 10.1111/tan.12688. [DOI] [PubMed] [Google Scholar]
- 16.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, Romieu I, Shi M, et al. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J Allergy Clin Immunol. 2010;125:321–327. doi: 10.1016/j.jaci.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S, Zhou X, Thibault DM, et al. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol. 2014;134:1153–62. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, et al. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 20.Breslow DK, Collins SR, Bodenmiller B, et al. Orm family proteins mediate sphingolipid homeostasis. Nature. 2010;463:1048–53. doi: 10.1038/nature08787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulenda T, Draber P. The role of ORMDL proteins, guardians of cellular sphingolipids, in asthma. Allergy. 2016;71:918–30. doi: 10.1111/all.12877. [DOI] [PubMed] [Google Scholar]
- 22.Nixon GF. Sphingolipids in inflammation: pathological implications and potential therapeutic targets. Br J Pharmacol. 2009;158:982–93. doi: 10.1111/j.1476-5381.2009.00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calişkan M, Bochkov YA, Kreiner-Möller E, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yarova PL, Stewart AL, Sathish V, et al. Calcium-sensing receptor antagonists abrogate airway hyperresponsiveness and inflammation in allergic asthma. Sci Transl Med. 2015;7:284ra60. doi: 10.1126/scitranslmed.aaa0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bønnelykke K, Matheson MC, Pers TH, et al. Australian Asthma Genetics Consortium (AAGC); EArly Genetics and Lifecourse Epidemiology (EAGLE) Consortium. Meta-analysis of genome-wide association studies identifies ten loci influencing allergic sensitization. Nat Genet. 2013;45:902–6. doi: 10.1038/ng.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. [8/1/16]; http://www.ncbi.nlm.nih.gov/gene.
- 27.Sleiman PM, Wang ML, Cianferoni A, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. doi: 10.1038/ncomms6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latiano A, Palmieri O, Pastorelli L, et al. Associations between genetic polymorphisms in IL-33, IL1R1 and risk for inflammatory bowel disease. PLoS One. 2013;8:e62144. doi: 10.1371/journal.pone.0062144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. [accessed 11/13/15]; http://quickfacts.census.gov/qfd/states/46/46137.html.
- 30.Noonan CW, Brown BD, Bentley B, Conway K, Corcoran M, FourStar K, et al. Variability in childhood asthma and body mass index across Northern Plains American Indian communities. J Asthma. 2010;47:496–500. doi: 10.3109/02770901003759436. [DOI] [PubMed] [Google Scholar]
- 31.Forno E, Han YY, Muzumdar RH, Celedón JC. Insulin resistance, metabolic syndrome, and lung function in US adolescents with and without asthma. J Allergy Clin Immunol. 2015;136:304–11. doi: 10.1016/j.jaci.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon AE, Holguin F, Sood A, Salome CM, Pratley RE, Beuther DA, et al. American Thoracic Society Ad Hoc Subcommittee on Obesity and Lung Disease. An official American Thoracic Society Workshop report: obesity and asthma. Proc Am Thorac Soc. 2010;7:325–35. doi: 10.1513/pats.200903-013ST. [DOI] [PubMed] [Google Scholar]
- 33.Janer A, Prudent J, Paupe V, et al. SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol Med. 2016;8:1019–38. doi: 10.15252/emmm.201506159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torgerson DG, Ampleford EJ, Chiu GY, Gilliland FD, Burchard EG, Martinez FD, Weiss ST, Williams LK, Barnes KC, Ober C, Nicolae DL, et al. Mexico City Childhood Asthma Study (MCAAS)., Children’s Health Study (CHS) and HARBORS study, Genetics of Asthma in Latino Americans (GALA) Study, Study of Genes-Environment and Admixture in Latino Americans (GALA2) and Study of African Americans, Asthma, Genes & Environments (SAGE)., Childhood Asthma Research and Education (CARE) Network., Childhood Asthma Management Program (CAMP)., Study of Asthma Phenotypes and Pharmacogenomic Interactions by Race-Ethnicity (SAPPHIRE)., Genetic Research on Asthma in African Diaspora (GRAAD) Study. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramasamy A, Curjuric I. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J Allergy Clin Immunol. 2011;128:996–1005. doi: 10.1016/j.jaci.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 36.Sun LD, Xiao FL, Li Y, et al. Genome-wide association study identifies two new susceptibility loci for atopic dermatitis in the Chinese Han population. Nat Genet. 2011;43:690–4. doi: 10.1038/ng.851. [DOI] [PubMed] [Google Scholar]
- 37.Stokes JR, Casale TB. Characterization of asthma endotypes: implications for therapy. Ann Allergy Asthma Immunol. 2016;117:121–5. doi: 10.1016/j.anai.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 38.Borish L. The immunology of asthma: Asthma phenotypes and their implications for personalized treatment. Ann Allergy Asthma Immunol. 2016;117:108–14. doi: 10.1016/j.anai.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CN. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–3. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 41.Žavbi M, Korošec P, Fležar M, Škrgat Kristan S, Marc Malovrh M, Rijavec M. Polymorphisms and haplotypes of the chromosome locus 17q12–17q21.1 contribute to adult asthma susceptibility in Slovenian patients. Hum Immunol. 2016;77:527–34. doi: 10.1016/j.humimm.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Andiappan AK, Sio YY, Lee B, et al. Functional variants of 17q12–21 are associated with allergic asthma but not allergic rhinitis. J Allergy Clin Immunol. 2016;137:758–66. e3. doi: 10.1016/j.jaci.2015.08.038. [DOI] [PubMed] [Google Scholar]
- 43.Shahid M, Sabar MF, Bano I, et al. Sequence variants on 17q21 are associated with the susceptibility of asthma in the population of Lahore, Pakistan. J Asthma. 2015;52:777–84. doi: 10.3109/02770903.2015.1012590. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Kang MJ, Kim BJ, et al. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR. Pediatr Pulmonol. 2011;46:701–8. doi: 10.1002/ppul.21424. [DOI] [PubMed] [Google Scholar]
- 45.Galanter J, Choudhry S, Eng C, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.