Abstract
The influenza A virus components of the live, attenuated influenza vaccine (LAIV) encode the HA and NA gene segments from a circulating virus strain and the remaining gene segments from the cold-adapted master donor virus, A/Ann Arbor/6/1960 (H2N2). The master donor virus imparts at least three phenotypes: temperature-sensitivity (ts), attenuation (att), and cold-adaption (ca). The genetic loci responsible for the att and ts phenotypes of LAIV were mapped to PB1, PB2, and NP by reverse genetics experiments using immortalized cell lines. However, some in vivo studies have demonstrated that the M segment, which acquired an alanine (Ala) to serine (Ser) mutation at M2 position 86 during cold adaption – a mutation found in no other influenza A virus strain - contributes to the att phenotype. Prior studies have shown this region of the M2 cytoplasmic tail to be critical for influenza virus replication. Using reverse genetics, we demonstrate that certain amino acid substitutions at M2 positions 83 and 86 alter the replication of influenza A/Udorn/307/72 (H3N2). Importantly, substitution of a Ser at M2 position 86 reduces A/Udorn/307/72 replication in differentiated primary human nasal epithelial cell (hNECs) cultures, but does not considerably affect replication in MDCK cells. When a Ser was substituted for Ala at M2 86 in LAIV, the virus replicated to higher titers and with faster kinetics in hNEC cultures, implicating this amino acid change as contributing to LAIV attenuation. Increased replication also resulted in increased production of IFN-λ. These data indicate the LAIV associated Ser mutation at M2 position 86 contributes to the att phenotype and is associated with a differential regulation of interferon in LAIV infection.
Keywords: influenza, respiratory epithelial cells, LAIV, vaccines
INTRODUCTION
Influenza A virus (IAV) is a member of the Orthomyxoviridae family and contains an 8-segment, negative-sense RNA genome encoding 10 to 14 proteins [1]. The live attenuated influenza vaccine (LAIV) is a 6:2 reassortant vaccine virus containing the PB2, PB1, PA, NP, M, and NS genome segments of a live-attenuated donor virus that was selected for its attenuated (att) and temperature-sensitivity (ts; replication at ≤32°C but not ≥39°C) phenotypes. There are 11 amino acid differences between the LAIV strain and the IAV strain from which it was derived [2]. The att and ts LAIV phenotypes have been mapped to the PB1 (E391, G581, T661), PB2 (S265), and NP (G34) genes by experiments in immortalized cell lines [3–7]. These experiments did not demonstrate a contribution of the M2-A86S mutation to temperature dependent LAIV replication. However, in vivo experiments in hamsters, ferrets and even humans have implicated the LAIV mutation M2-A86S as important for the att phenotype [3,7–9]. Few studies have been performed to understand the molecular mechanisms conferring these phenotypes. During single-step replication in Madin-Darby canine kidney (MDCK) cells at 39°C, viral RNA synthesis and vRNP export from the nucleus are reduced. Furthermore, incorporation of the viral matrix protein M1 into virus particles was reduced, causing heterogeneous and irregular virion morphology [6,7]. LAIV replication is restricted within physiological ranges of temperature (32 to 37°C) during infection of primary, differentiated human nasal epithelial cell (hNEC) cultures [10,11], suggesting that other mutations in the LAIV genome may contribute to reduced virus replication in primary respiratory epithelial cell cultures.
The M2 protein is a 97 amino acid integral membrane protein with a 54 amino acid cytoplasmic tail [12]. It is a highly conserved protein required for virus entry [13], membrane scission [14] and the production of infectious virus particles [15–19]. M2 has been shown to form disulfide-linked homo-tetramers with pH-gated, proton-selective ion channel activity [20,21]. The ion channel activity is critical for virus uncoating - by allowing protons to enter the virion interior, permitting the vRNP to dissociate from M1 [1,22–24] - and preventing the premature cleavage of HA by neutralizing the pH in the late Golgi [25]. M2 has also been shown to be important for inhibition of autophagy and preventing autophagosome fusion with lysosomes [26–28].
The distal region of the M2 cytoplasmic tail is essential for IAV replication and is involved in vRNP incorporation into progeny virions [15–19]. When the M2 cytoplasmic tail is truncated by deleting the last 16 amino acids there is decreased release of infectious virus particles, which is not observed when only the last 8 amino acids are deleted [17]. When the region (M2 82–89) was mutated to alanine residues, infectious particle production was not altered. However, the original alanine residues at M2 positions 83 and 86 were never mutated and their contribution to the M2 protein functions were never investigated. The A83 residue is conserved across a wide range of IAV strains while A86 is highly conserved except for pandemic 2009 H1N1 viruses (Val) and LAIV (Ser).
To investigate the role amino acids at positions 83 and 86 of M2 may play in IAV replication and attenuation, we generated recombinant viruses with substitutions at these amino acids. This allowed us to show that both amino acid positions contributed to efficient virus replication. Both glutamic acid and serine substitutions at position 86 caused an approximately 2-log decrease in virus replication on primary human nasal epithelial cells (hNEC) but not MDCK cells. The magnitude of decrease was dependent on the temperature (greater reduction at 37°C than at the permissive temperature of 32°C) of the cells during infection. Introducing an M2-S86A substitution increased LAIV replication and altered interferon lambda (IFN-λ) production in hNEC cultures, indicating a role for the M2-86S amino acid change in both the att and ts phenotypes of LAIV.
MATERIALS AND METHODS
Plasmids
The plasmid pHH21 expressing full length influenza virus gene segments was used to generate recombinant viruses [16] as described below. All mutations were introduced using the QuikChange Lightning site-directed mutagenesis kit (Agilent). All inserts and mutations were confirmed by sequencing. Primer sequences are available upon request.
Cell culture
MDCK cells and HEK 293T (293T) cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM GlutaMAX (Gibco). Human nasal epithelial cell (hNEC) cultures were isolated from non-diseased tissue after endoscopic sinus surgery for non-infection related conditions and grown at air-liquid interface (ALI) as previously described [11,29–31]. hNEC differentiation medium (DM) and culture conditions have previously been described in detail [11]. The donors for the hNEC cultures were females ages 57 and 61 (Figure 2), males ages 47 and 67 (Figure 3) and males ages 72 and 67 (Figure 4, Figure 5 and Table 1).
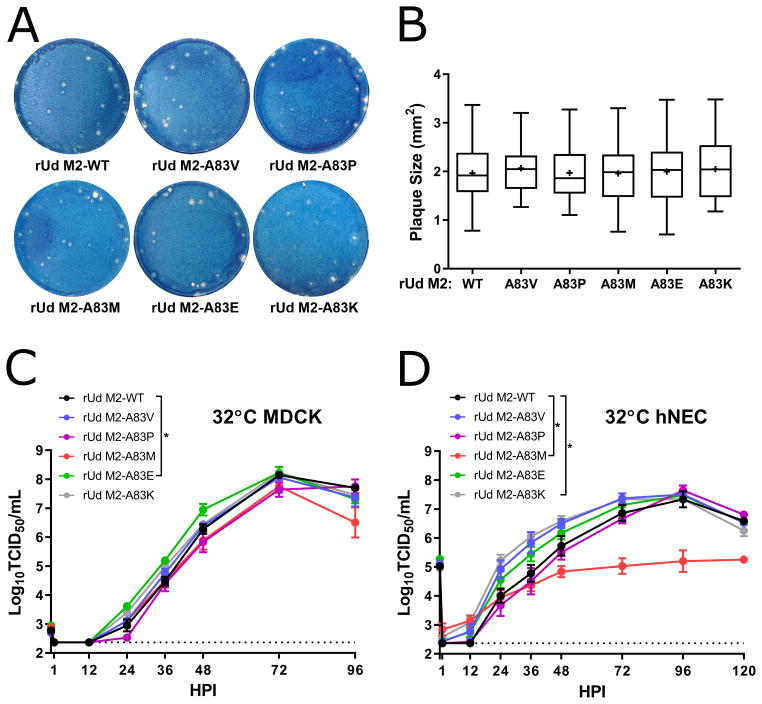
Figure 2. Replication of M2-83 mutants in MDCK cells and hNEC cultures.
(A) Plaque assays performed with indicated virus on MDCK cells. (B) Quantification of plaque diameter from 34–64 individual plaques per virus identified from 2–3 independent experiments. *p < 0.05. No conditions were statistically significant compared to rUd M2-WT. Low MOI multistep growth curves performed on (C) MDCK cells or (D) hNECs with the indicated viruses at 32°C. Data are pooled from two independent replicates each with n = 3 wells per virus (total n = 6 wells per virus). *p < 0.05 compared to rUd M2-WT (two-way ANOVA with Dunnett’s posttest). In (C) no time points were significantly different while in (D) rUd M2-A83M (48, 72, 96, and 120 HPI), rUd M2-A83K (24, 36, and 48 HPI) showed differences. Dotted line indicates limit of detection.
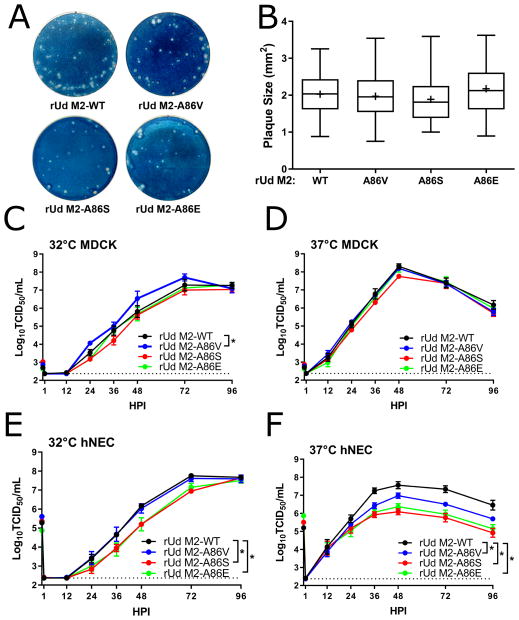
Figure 3. Replication of M2-86 mutants in the rUd background in MDCK cells and hNEC cultures.
(A) Plaque assays performed with indicated virus on MDCK cells. (B) Quantification of plaque diameter from 84–137 individual plaques per virus identified from 2–3 independent experiments. *p < 0.05. No conditions were statistically significant compared to rUd M2-WT. Low MOI multistep growth curves performed on (C and D) MDCK cells or (E and F) hNECs with the indicated viruses at 32°C (C and E) or 37°C (D and F). Data are pooled from two independent replicates each with n = 3 wells per virus (total n = 6 wells per virus). *p < 0.05 compared to rUd M2-WT (two-way ANOVA with Dunnett’s posttest) significant differences (p < 0.05) compared to rUd M2-WT: In (C) no time points were significantly different while in (E) rUd M2-A86S (48 and 72 HPI), rUd M2-A86E (36 and 48 HPI) and (F) rUd M2-A86V (36 and 72 HPI), rUd M2-A86S (36, 48, 72, 96 HPI), rUd M2-A86E (36, 48, 72, 96 HPI) showed differences. Dotted line indicates limit of detection.
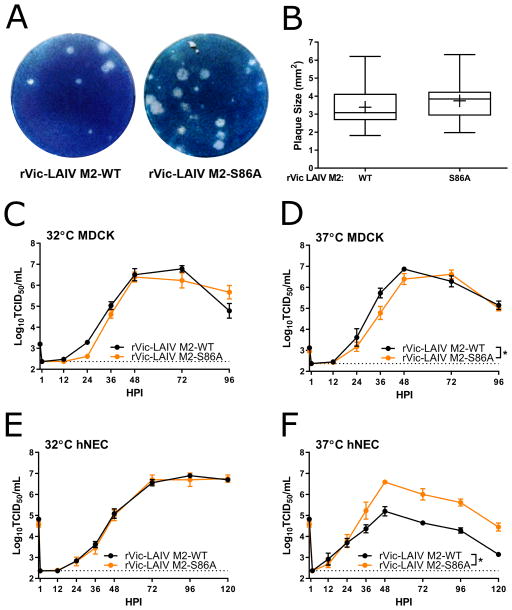
Figure 4. Replication of M2-86 mutants in the rVic-LAIV background in MDCK cells and hNEC cultures.
(A) Plaque assays performed with indicated virus on MDCK cells. (B) Quantification of plaque diameter from 24–47 individual plaques per virus identified from 2–3 independent experiments. *p < 0.05. No mutations were statistically significant compared to rVic-LAIV. Low MOI multistep growth curves performed on (C and D) MDCK cells or (E and F) hNECs with the indicated viruses at 32°C (C and E) or 37°C (D and F). Data are pooled from two independent replicates each with n = 3 wells per virus (total n = 6 wells per virus). *p < 0.05 compared to rVic-LAIV (two-way ANOVA with Dunnett’s posttest). In (D) rVic-LAIV M2-S86A (36 HPI) and (F) rVic-LAIV M2-S86A (36, 48, 72, 96, and 120 HPI) showed differences. Dotted line indicates limit of detection.
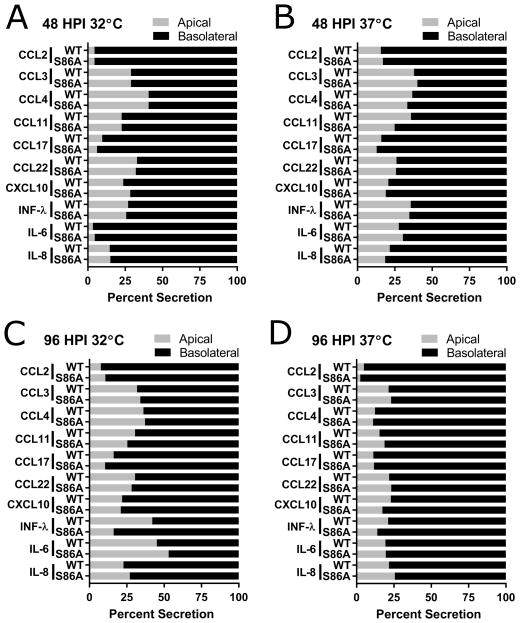
Figure 5. Comparison of apical versus basolateral secretion of interferons, cytokines, and chemokines.
Samples collected 48 (A and B) and 96 (C and D) HPI during low MOI multistep growth curve experiment (Fig. 4E and 4F, Table 1) performed at 32°C (A and C) and 37°C (B and D). WT =rVic LAIV M2-WT; S86A = rVic LAIV M2-S86A. Data are pooled from two independent replicates each with n = 3 wells per virus.
Table 1.
Secretion of chemokines, cytokines and interferon in response to infection of hNEC cultures with LAIV.
| 32°Ca | 37°C | |||||||
|---|---|---|---|---|---|---|---|---|
| 48 HPI | 96 HPI | 48 HPI | 96 HPI | |||||
| M2-WT | M2-S86A | M2-WT | M2-S86A | M2-WT | M2-S86A | M2-WT | M2-S86A | |
| CCL2 | 2.9 ± 0.2 | 3.2 ± 0.5 | 3 ± 0.2 | 4.1 ± 1.5 | 8.5 ± 1.2 | 7.5 ± 0.5 | 26.5 ± 10.1 | 39.3 ± 15 |
| CCL3 | N.D. | N.D. | 13.8 ± 0.4 | 15.5 ± 1.9 | 19.9 ± 2.6 | 16.5 ± 1.8 | 48.9 ± 8.3 | 45.7 ± 9.4 |
| CCL4 | N.D. | N.D. | 5.1 ± 0.5 | 5.8 ± 0.7 | 8.6 ± 2 | 7 ± 1.6 | 34.1 ± 6.3 | 37.7 ± 11.2 |
| CCL11 | N.D. | N.D. | 14.9 ± 1.9 | 13.9 ± 1.5 | 20.1 ± 5.3 | 14.8 ± 2.7 | 54.8 ± 11.1 | 39.5 ± 7.9 |
| CCL13 | 9.9 ± 0.3 | 9.8 ± 0.3 | 9.9 ± 0.3 | 9.8 ± 0.3 | 9.8 ± 0.3 | 9.8 ± 0.3 | 9.9 ± 0.3 | 9.8 ± 0.3 |
| CCL17 | 2.4 ± 0.2 | 3.2 ± 0.2 | 6.1 ± 1.1 | 8.4 ± 0.3 | 15.4 ± 2.6 | 14.4 ± 0.5 | 23 ± 4.9 | 24 ± 1.4 |
| CCL22 | 12.9 ± 0.5 | 13.3 ± 0.6 | 26 ± 2.6 | 22.6 ± 1.7 | 54.4 ± 4* | 35 ± 2.8* | 112.5 ± 3.6 | 110 ± 12.3 |
| CCL26 | 10.8 ± 0.7 | 10.4 ± 0.7 | 10.3 ± 0.8 | 10.4 ± 0.7 | 10.4 ± 0.7 | 10.6 ± 0.7 | 10.4 ± 0.7 | 10.4 ± 0.7 |
| CXCL10 | 15.1 ± 5.1 | 22.3 ± 8.8 | 1000.4 ± 457.3 | 2139.2 ± 1027.1 | 3127.3 ± 1287.1 | 2509.2 ± 1250.9 | 5234.8 ± 543.3 | 5804.8 ± 195.2 |
| IFN-λ | 179.4 ± 7.6 | 184.5 ± 12.3 | 269.9 ± 44.3 | 288.5 ± 53.9 | 920.4 ± 132.9* | 424.6 ± 55* | 3658.8 ± 390.1* | 5888.3 ± 130* |
| IL-6 | 8.1 ± 3.3 | 6.5 ± 2.6 | 7.7 ± 1.3 | 9.6 ± 2.9 | 36.1 ± 6.2 | 20.4 ± 4.4 | 430.7 ± 83.1 | 528.7 ± 85 |
| IL-8 | 1702.7 ± 374.9 | 1655.1 ± 385.8 | 1503.6 ± 87.8 | 1323.2 ± 116.5 | 4557.2 ± 433.1 | 3412.1 ± 620.4 | 27445.7 ± 2865.3 | 25820.9 ± 3920.1 |
Total pg of each protein is listed, which combines the apical and basolateral media.
M2-WT = rVic LAIV M2-WT; M2-S86A = rVic LAIV M2-S86A. Samples were collected from the low MOI multistep growth curve experiment (Fig. 4E and 4F) and are pooled from two independent experiments each with n = 3 wells per virus. None detected (N.D.) indicates levels were below the lower limit of detection. Amounts in bold text are *p < 0.05 for difference between viruses at the indicated time post infection (two-way ANOVA).
Viruses
Recombinant A/Udorn/307/72 (H3N2) (rUd) has been described previously [13,17,32,33]. A recombinant LAIV consisting of the internal gene segments of A/Ann Arbor/6/1960 (H2N2) and the HA and NA segments from A/Victoria/361/2011(H3N2) (rVic-LAIV) was generated using sequences available in Genbank and the NIAID Influenza Research Database [2,34,35]. Recombinant viruses were rescued using the 12-plasmid reverse genetics system [13,32,33,36]. Briefly, 293T cells were transfected with pHH21 plasmids encoding all 8 influenza A virus gene segments along with protein expression plasmids encoding NP, PA, PB1, and PB2. Transfected cells were then co-cultured with MDCK cells, and sampled every 24 hours for the presence of infectious virus. Viruses were then plaque purified and confirmed by sequencing. The entire M segment open reading frame was sequenced in all recombinant virese to ensure the absence of any second site mutations. All rVic-LAIV rescues were performed at 32°C. Stocks of virus were generated by infecting MDCK cells at a low multiplicity of infection (MOI) as described below.
Plaque assays
Plaque assays were performed in 6-well plates of confluent MDCK cells. Serial 10-fold dilutions of the virus inoculum were generated in infection media (IM; DMEM supplemented with 0.3% BSA [Sigma], 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM GlutaMAX [Gibco], and 4 μg/mL N-acetyl trypsin [NAT; Sigma]). Cells were washed twice with PBS containing calcium and magnesium (PBS+) and 250 μL of inoculum was added. The cells were incubated for 1 hour at RT with rocking. The inoculum was removed and replaced with IM containing 1% agarose. Cells were then incubated at 32°C for 4–5 days, fixed in 2% formaldehyde, and stained in a Naphthol Blue Black solution. Individual plaque diameters were determined by ImageJ. For synthetic virus generation, the agar above individual plaques was removed with a sterile pipette, aspirated into 1 mL of IM, and stored at −80°C.
TCID50 assay
MDCK cells were plated in 96-well plates, grown to confluence, and washed twice with PBS+. Tenfold serial dilutions of the virus inoculum were made and 200 μL of dilution was added to each of 6 wells in the plate, followed by incubation for 7 days at 32°C. Cells were then fixed by adding 100 μL of 4% formaldehyde in PBS per well, followed by staining with a Naphthol Blue Black solution. Endpoint calculations were determined by the Reed-Muench method [37].
Low MOI infection
Before all low MOI infections, cells are washed twice with PBS+. Seed stocks (SS) were generated by incubating 250 μL of the plaque pick solution on fully confluent MDCK cells for 1 hour at room temperature. The inoculum was removed, cells were washed twice with PBS+, and then 1 mL of IM was added before returning the cells to the incubator at 32°C. Virus supernatant was harvested when 50% of the cells displayed cytopathic effect upon inspection with light microscopy, usually 24–48 hours post infection (HPI). Seed stocks were then titrated by TCID50 assay, and used to generate working stocks. Working stocks were generated as described for seed stocks, except infections were performed in 75 or 150 cm2 flasks and the inoculum used was SS diluted to an MOI of 0.01 in IM.
Multistep virus growth curves were performed at a MOI of 0.001 in MDCK cells and 0.1 in hNECs. For MDCK cell infections, the inoculum was diluted in IM, added to cells, and allowed to incubate at 32 or 37°C for 1 hour. Cells were then washed 3 times with PBS+ and incubated with fresh IM at 32 or 37°C. At the indicated hours post infection, all media was removed and replaced with fresh IM. The amount of infectious virus in each sample was determined by TCID50 assay on MDCK cells. For hNECs, the apical surface was washed with IM without NAT and the basolateral DM was replaced immediately before infection. Inoculum diluted in IM without NAT was then added and allowed to incubate on cells at the growth curve temperature for 1 hour. The inoculum was then removed, the cells were washed three times with PBS+, left at ALI, and returned to 32°C or 37°C. At the indicated times post infection, IM without NAT was added to the apical chamber of the wells and allowed to incubate at 32°C for 10 minutes, removed, and then the cells were returned to the incubator at ALI. Basolateral DM was removed and replaced every 48 hours for the duration of the growth curves. All samples were stored at −80°C.
Interferon, cytokine and chemokine measurements
Secreted interferons, cytokines, and chemokines were quantified from the apical and basolateral samples from the 48 and 96 hour post infection samples from the hNEC multistep virus growth curves. Measurements were performed using the V-PLEX Human Chemokine Panel 1 (CCL2, CCL3, CCL4, CCL11, CCL13, CCL17, CCL22, CCL26, CXCL10, and IL-8), V-PLEX Human IL-6 kit (Meso Scale Discovery), and the DIY Human IFN Lambda 1/2/3 (IL-29/28A/28B) ELISA (PBL Assay Science) according to manufacturers’ instructions. Of the 12 analytes tested, 10 (CCL2, CCL3, CCL4, CCL11, CCL17, CCL22, CXCL10, IL-8, IL-6, and IFN-λ) were specifically induced in virus-infected cultures. The amount secreted was adjusted by sample volume (either 100 or 150 μl for the apical washes, 500 μl for the basolateral supernatants) to show total pg secreted.
Statistical analyses
Multistep growth curves were analyzed by two-way ANOVA with a Dunnett’s posttest performed for all viruses when significant differences from WT virus were present. Plaque diameters and cytokine and chemokine measurements were analyzed by ANOVA. All statistical analyses were performed in GraphPad Prism 7.0.
RESULTS
Rescue of recombinant viruses with mutations in the M2 cytoplasmic tail
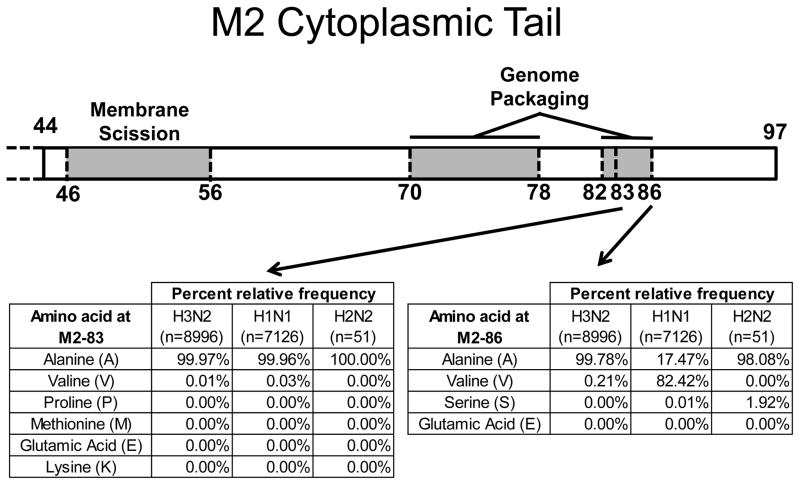
Previous work showed that the region of M2 containing amino acids 82–89 was important for virus replication and efficient genome packaging, but scanning alanine mutagenesis failed to reveal any single residue responsible for the phenotype [16]. However, substitutions at M2 amino acid positions 83 and 86 were not made. Since an Ala residue is present at M2 position 83 in nearly 100% of North American human influenza virus isolates, we replaced Ala83 with a number of amino acids to assess the overall tolerance of that position to mutations (Fig. 1). Approximately 63.5% of IAV strains contain Ala at position 86 (Fig. 1), and Ala is the residue found in most H3N2 influenza A virus isolates. The M2–86Val mutation is present in 36.4% of sequences in the database and is encoded primarily by H1N1 viruses. The A/Ann Arbor/6/1960 (H2N2) LAIV M2 protein contains an Ala to Ser mutation at position 86 of the M2 cytoplasmic tail [2] which was acquired during the cold-adaption of the virus. Given this natural variation in amino acid sequences at M2 position 86, we focused on a limited number of substitutions at this position (Fig. 1). These substitutions were initially introduced into the A/Udorn/307/72 infectious clone, which is an H3N2 virus with highly conserved Ala residues at M2 amino acid positions 83 and 86.
Figure 1. M2 cytoplasmic tail.
M2 cytoplasmic tail schematic depicting amino acid frequency in all H3N2, H1N1, and H2N2 human isolate sequences from North America (complete genomes only) available in the Influenza Research Database. Search was performed 7/2/2017.
Replication of rUd M2-83 mutants in MDCK cells and hNEC cultures
In order to mimic the temperature of the upper respiratory tract, all experiments with the rUd M2-83 viruses were performed at 32°C, the temperature of the upper respiratory tract. Plaque assays performed on MDCK cells showed no difference in plaque size between rUd M2-WT, A83V, A83P, A83M, A83E, and A83K (Fig. 2A and 2B). On MDCK cells, rUd M2-WT, A83V, A83P, A83M, and A83K all replicated with similar kinetics and produced a comparable amount of infectious virus after low MOI infection (Fig. 2C). The rUd M2-A83E showed a modest increase in replication compared to rUd M2-WT (Fig. 2C). In hNEC cultures, rUd M2-WT, A83V, A83P, and A83E all replicated with similar kinetics while rUd M2-A83K replicated slightly faster than rUd M2-WT (Fig 2D). The rUd M2-A83M replicated with significantly reduced kinetics compared to WT (approximately 2 logs lower infectious virus production). The data indicate that rUd M2 position 83 does not have a large effect on virus replication, except in the case of rUd M2-A83M on hNECs, indicating the site is relatively amenable to mutation, despite being highly conserved in natural influenza isolates.
Replication of rUd M2-86 mutants in MDCK cells and hNEC cultures
Plaque assays performed on MDCK cells showed no difference in plaque size between the rUd M2-WT, A86V, A86S, and A86E (Fig. 3A and 3B). In MDCK cells at 32°C, rUd M2-WT, A86S, and A86E all replicated with similar kinetics and produced a comparable amount of infectious virus (Fig 3C). rUd M2-A86V showed a small, but statistically significant, increase in replication compared to rUd M2-WT. Since the M2-86S mutation is associated with LAIV – a virus known to have restricted replication at higher temperatures - the growth curves were also performed at 37°C. All of the viruses replicated with similar kinetics and titers compared to rUd M2-WT (Fig. 3D) at this temperature in MDCK. Virus replication was then assessed at both temperatures in hNEC cultures. At 32°C on hNECs, rUd M2-WT and A86V replicated similarly, while A86S and A86E replicated with decreased kinetics compared to rUd M2-WT (Fig. 3E). Strikingly, on hNECs at 37°C, rUd M2-A86V, A86S, and A86E all replicated with significantly decreased kinetics and to a lower peak titer compared to rUd M2WT (Fig. 3F). Taken together, the data suggest that mutations at rUd M2 position 86 are detrimental to virus replication, and this effect is strongest in hNECs at 37°C.
Replication of rVic-LAIV M2-86 mutants in MDCK cells and hNEC cultures
Since the M2-A86S mutation arose during LAIV cold-adaption and this mutation altered rUd replication in hNEC cultures, we chose to study the effect of this mutation in the LAIV genetic background. Recombinant LAIV expressing the HA and NA proteins from A/Victoria/361/2011 (rVic-LAIV) was rescued along with an rVic-LAIV M2-S86A containing virus. Plaque assays performed on MDCK cells showed no difference in plaque size between rVic-LAIV and rVic-LAIV M2-S86A (Fig. 4A and 4B). Multi-step growth curves performed in MDCK cells at 32 and 37°C (Fig. 4C and 4D) showed consistent virus replication with rVic-LAIV M2-S86A virus showing slightly reduced virus replication kinetics at 37°C. In hNEC cultures, the viruses replicated with identical kinetics at 32°C, but the rVic-LAIV M2-S86A replicated to significantly higher titers at 37°C (Fig. 4E and 4F). Together, the data indicate that the LAIV associated mutation, M2-A86S, contributes to the att and ts on hNEC cultures, but not MDCK cells.
Induction of chemokines, cytokines, and IFN-λ by rVic-LAIV M2-A86S in hNEC cultures
Since LAIV induces a significantly different epithelial cell innate immune response compared to seasonal influenza [10,38], the effect of the M2-S86A mutation on interferon and chemokine secretion in hNEC cultures was determined at 48 and 96 hours post low MOI infection. Expression of ten of the 12 analytes increased with temperature and time in response to infection, irrespective of the infecting virus (Fig. 5 and Table 1). Secretion of the induced factors was directional, with higher amounts detected in the basolateral media (Fig. 5). While IFN-λ secretion is similar between the two viruses at 32°C, differences emerge during infections at a higher temperature. At 37°C, IFN-λ secretion is significantly greater for rVic-LAIV than rVic-LAIV M2-S86A at 48 HPI, but significantly less at 96 HPI (Table 1). Together these data indicate that an M2-S86A mutation increases LAIV virus replication and alters innate immune responses in hNEC cultures at 37°C, identifying it as another mutation responsible for attenuating LAIV replication in a temperature dependent manner.
DISCUSSION
The LAIV associated M segment has been shown to contribute, in part, to LAIV’s attenuation phenotype [3,7,8]. The only M segment mutation between the WT and cold-adapted strains of A/Ann Arbor/6/1960 is a missense mutation at M2 position 86 from an alanine in the WT stain to a serine in the cold-adapted vaccine stain [2]. Our data indicate that the M2-A86S mutation acquired during LAIV cold adaption contributes to reduced virus replication in hNEC cultures but not MDCK cells, in a temperature dependent manner and can alter the production of IFN-λ. This suggests that LAIV attenuation in hNEC cultures is not solely driven by the ts and ca mutations identified in the polymerase complex proteins. In addition to being the location of an LAIV associated mutation, M2 amino acid position 86 is also present in a region of M2 previously identified as critical for IAV replication and budding [15,17]. Scanning alanine mutagenesis of these regions was unable to fully recapitulate the replication defect associated with the M2 truncations used to identify the regions. This could be because alanine is ubiquitous at M2 amino acid position 83 in circulating strains of influenza and common at position 86 (Fig. 1). Mutating the alanine at M2 position 83 did not have a consistent effect on the replication of virus in either MDCK cells or hNECs (Fig. 2C and 2D). However, substituting a methionine at this position led to a defect in replication, indicating that the presence of an alanine at this position is not critical for replication, but the position is not fully malleable.
Since most of the previous studies of LAIV in vitro replication were performed in immortalized cell lines including MDCK cells or embryonated hen’s eggs, it is perhaps not surprising that the contribution of the M2 mutation was not identified previously, as the replication differences we identified were specific to infection of hNEC cultures and not obvious in MDCK cells. Primary, differentiated epithelial cell cultures have been shown to provide greater clinical relevance and phenotype penetrance than immortalized cell lines [10,11,18,39–44]. This is particularly true for studying LAIV [10,11]. These cultures can contain multiple epithelial cell types including ciliated cells, basal cells, goblet cells and club cells [45,46]. Additionally, immortalized cell lines and embryonated chickens’ eggs both lack elements of the epithelial cell innate immune system and virus restriction factors. There is a greater replication restriction of LAIV at both 32 and 37°C in primary, differentiated epithelial cell cultures compared to MDCK cells [11,42]. This difference in replication on primary human respiratory epithelial cells has been associated with a robust, unique innate immune response [10] and an increased ratio of non-infectious to infectious virus particles [11,38]. The M2-A86S mutation attenuated rUdorn virus replication on hNECs at 32 and 37°C (Fig. 3E and 3F). Consistent with this, introducing the M2-S86A mutation intothe LAIV background increased virus replication at 37°C (Fig. 4F) in hNEC cultures. This increase in LAIV replication on hNECs was associated with altered induction of IFN-λ production at 48 HPI (Table 1). IFN-λ receptor expression is restricted to epithelial cells and IFN-λ is critical to protecting epithelial tissues during virus challenge [47,48]. These results are consistent with the robust innate immune response seen with LAIV infections compared to WT viruses [10,38]. The significance of the changes in the kinectics and magnitude of IFN-λ secretion in response to virus infection are not clear at this time, but it will be interesting to investigate the effect of these mutations on LAIV infection in an animal model. Every cytokine and chemokine tested was secreted preferentially into the basolateral media (Supplemental Fig. 1). This agrees with previous reports of a basolateral bias in cytokine secretion during influenza virus infection of primary respiratory epithelial cells [41]. A M2-A86V substitution did show reduced replication in the rUdorn virus background despite it being present in some natural isolates of IAV. This may be due to the fact that rUdorn is a H3N2 virus while the A86V substitution is only found in H1N1 subtype viruses, indicating a strain-specific role for this position. Strain specific effects of mutations in the M2 protein have been documented previously [16–18,33].
Previous work has shown that at the non-permissive temperatures of 38.5 to 40°C, the M1 protein of LAIV viruses is not efficiently packaged into budding particles [49] and particles exhibited a more heterogeneous, enlarged morphology [6]. However, there is no decrease in M1 incorporation into virus particles at the physiologically relevant temperatures of 32 and 37°C [11]. We therefore chose these temperatures to better model the effects of these mutations at the temperatures of the human respiratory tract, 32°C (upper) and 37°C (lower). For the M2-83 mutation experiments, we used 32°C to accurately model infections of the upper respiratory tract. However, since the M2-86 mutation is associated with the temperature-sensitive LAIV vaccine, we used both 32 and 37°C.
Annual influenza vaccination is the primary means of preventing seasonal influenza and decreasing its sociologic and economic impacts. LAIV has been shown to be more efficacious than inactivated influenza in children aged 6–69 months [7,50] and adults [7,51,52]. However, LAIV is not being recommended for use during the 2016–2017 influenza season due to low effectiveness [53]. Few studies have been performed to understand the LAIV attenuation phenotypes at the molecular level, and it remains to be determined if all of the LAIV associated mutations are relevant and necessary for the production of a safe and efficacious vaccine. Our studies show that one can obtain a better understanding of the LAIV attenuation through the use of hNEC cultures and suggest that LAIV attenuation is not solely due to polymerase complex mutations. This implies that LAIV can be re-engineered to replicate either more or less efficiently through the manipulation of the M2 protein sequences including, but not limited to positions 83 and 86. The decreased replication on hNECs and altered innate immune induction associated with the LAIV M2-A86S mutation demonstrates the merit of this strategy and justify the investigation of the other LAIV associated mutations with these methods.
Acknowledgments
We thank the members of the Pekosz laboratory, Sabra Klein, and members of the Klein laboratory for useful and critical discussions of the data. The work was supported by the Shikani/El Hibri Prize for Discovery and Innovation (AP), R01 AI097417 (AP), HHSN272201400007C (AP), R01 AI072502 (APL) and T32 AI007417 (NW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peter Shaw MLP. Orthomyxoviridae. In: Knipe DM, Howley Peter M, editors. Fields Virology. 6. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1151–85. [Google Scholar]
- 2.Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2) Virology. 1988;167:554–67. [PubMed] [Google Scholar]
- 3.Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, Sears SD, Dolin R, Maassab HF, Murphy BR. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. Journal of virology. 1988;62:488–95. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang C-f, Kemble G, Greenberg H. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (flumist) derived from cold-adapted a/ann arbor/6/60. Virology. 2003;306:18–24. doi: 10.1016/s0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 5.Jin H, Zhou H, Lu B, Kemble G. Imparting Temperature Sensitivity and Attenuation in Ferrets to A/Puerto Rico/8/34 Influenza Virus by Transferring the Genetic Signature for Temperature Sensitivity from Cold-Adapted A/Ann Arbor/6/60. Journal of virology. 2004;78:995–8. doi: 10.1128/JVI.78.2.995-998.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan W, Zhou H, Kemble G, Jin H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology. 2008;380:304–11. doi: 10.1016/j.virol.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Jin H, Subbarao K. Live attenuated influenza vaccine. Current topics in microbiology and immunology. 2015;386:181–204. doi: 10.1007/82_2014_410. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Ye Z. Attenuating mutations of the matrix gene of influenza A/WSN/33 virus. Journal of virology. 2005;79:1918–23. doi: 10.1128/JVI.79.3.1918-1923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder MH, Clements ML, De Borde D, Maassab HF, Murphy BR. Attenuation of wild-type human influenza A virus by acquisition of the PA polymerase and matrix protein genes of influenza A/Ann Arbor/6/60 cold-adapted donor virus. Journal of clinical microbiology. 1985;22:719–25. doi: 10.1128/jcm.22.5.719-725.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer WA, 2nd, Chason KD, Brighton M, Jaspers I. Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures. Vaccine. 2014;32:1761–7. doi: 10.1016/j.vaccine.2013.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer WA, 2nd, King LS, Lane AP, Pekosz A. Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells. Vaccine. 2015;33:4495–504. doi: 10.1016/j.vaccine.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb RA, Choppin PW. Identification of a second protein (M2) encoded by RNA segment 7 of influenza virus. Virology. 1981;112:729–37. doi: 10.1016/0042-6822(81)90317-2. [DOI] [PubMed] [Google Scholar]
- 13.Takeda M, Pekosz A, Shuck K, Pinto LH, Lamb RA. Influenza a virus M2 ion channel activity is essential for efficient replication in tissue culture. Journal of virology. 2002;76:1391–9. doi: 10.1128/JVI.76.3.1391-1399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwatsuki-Horimoto K, Horimoto T, Noda T, Kiso M, Maeda J, Watanabe S, Muramoto Y, Fujii K, Kawaoka Y. The Cytoplasmic Tail of the Influenza A Virus M2 Protein Plays a Role in Viral Assembly. Journal of virology. 2006;80:5233–40. doi: 10.1128/JVI.00049-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCown MF, Pekosz A. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. Journal of virology. 2005;79:3595–605. doi: 10.1128/JVI.79.6.3595-3605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. Journal of virology. 2006;80:8178–89. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grantham ML, Stewart SM, Lalime EN, Pekosz A. Tyrosines in the Influenza A Virus M2 Protein Cytoplasmic Tail Are Critical for Production of Infectious Virus Particles. Journal of virology. 2010;84:8765–76. doi: 10.1128/JVI.00853-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. Journal of virology. 2008;82:10059–70. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- 21.Stewart SM, Pekosz A. Mutations in the membrane-proximal region of the influenza A virus M2 protein cytoplasmic tail have modest effects on virus replication. Journal of virology. 2011;85:12179–87. doi: 10.1128/JVI.05970-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–8. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 23.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–28. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 24.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. Journal of virology. 1991;65:232–44. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alvarado-Facundo E, Gao Y, Ribas-Aparicio RM, Jiménez-Alberto A, Weiss CD, Wang W. Influenza Virus M2 Protein Ion Channel Activity Helps To Maintain Pandemic 2009 H1N1 Virus Hemagglutinin Fusion Competence during Transport to the Cell Surface. Journal of virology. 2015;89:1975–85. doi: 10.1128/JVI.03253-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, García-Sastre A, Münz C. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell host & microbe. 6:367–80. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell host & microbe. 2014;15:239–47. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren Y, Li C, Feng L, Pan W, Li L, Wang Q, Li J, Li N, Han L, Zheng X, Niu X, Sun C, Chen L. Proton Channel Activity of Influenza A Virus Matrix Protein 2 Contributes to Autophagy Arrest. Journal of virology. 2015;90:591–8. doi: 10.1128/JVI.00576-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lane AP, Saatian B, Yu XY, Spannhake EW. mRNA for genes associated with antigen presentation are expressed by human middle meatal epithelial cells in culture. The Laryngoscope. 2004;114:1827–32. doi: 10.1097/00005537-200410000-00028. [DOI] [PubMed] [Google Scholar]
- 30.Ramanathan M, Jr, Lane AP. Innate immunity of the sinonasal cavity and its role in chronic rhinosinusitis. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery. 2007;136:348–56. doi: 10.1016/j.otohns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Ramanathan M, Jr, Lee WK, Dubin MG, Lin S, Spannhake EW, Lane AP. Sinonasal epithelial cell expression of toll-like receptor 9 is decreased in chronic rhinosinusitis with polyps. American journal of rhinology. 2007;21:110–6. doi: 10.2500/ajr.2007.21.2997. [DOI] [PubMed] [Google Scholar]
- 32.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9345–50. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grantham ML, Wu WH, Lalime EN, Lorenzo ME, Klein SL, Pekosz A. Palmitoylation of the influenza A virus M2 protein is not required for virus replication in vitro but contributes to virus virulence. Journal of virology. 2009;83:8655–61. doi: 10.1128/JVI.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Research. 2016;44:D67–D72. doi: 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squires RB, Noronha J, Hunt V, García-Sastre A, Macken C, Baumgarth N, Suarez D, Pickett BE, Zhang Y, Larsen CN, Ramsey A, Zhou L, Zaremba S, Kumar S, Deitrich J, Klem E, Scheuermann RH. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza and Other Respiratory Viruses. 2012;6:404–16. doi: 10.1111/j.1750-2659.2011.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neumann G, Ozawa M, Kawaoka Y. Reverse genetics of influenza viruses. Methods in molecular biology (Clifton, NJ) 2012;865:193–206. doi: 10.1007/978-1-61779-621-0_12. [DOI] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. American journal of epidemiology. 1938;27:493–7. [Google Scholar]
- 38.Forero A, Fenstermacher K, Wohlgemuth N, Nishida A, Carter V, Smith EA, Peng X, Hayes M, Francis D, Treanor J, Morrison J, Klein SL, Lane A, Katze MG, Pekosz A. Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells. Vaccine. 2017 doi: 10.1016/j.vaccine.2017.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peretz J, Pekosz A, Lane AP, Klein SL. Estrogenic compounds reduce influenza A virus replication in primary human nasal epithelial cells derived from female, but not male, donors. American journal of physiology Lung cellular and molecular physiology. 2016;310:L415–25. doi: 10.1152/ajplung.00398.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall OJ, Limjunyawong N, Vermillion MS, Robinson DP, Wohlgemuth N, Pekosz A, Mitzner W, Klein SL. Progesterone-Based Therapy Protects Against Influenza by Promoting Lung Repair and Recovery in Females. PLoS pathogens. 2016;12:e1005840. doi: 10.1371/journal.ppat.1005840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newby CM, Sabin L, Pekosz A. The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. Journal of virology. 2007;81:9469–80. doi: 10.1128/JVI.00989-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ilyushina NA, Ikizler MR, Kawaoka Y, Rudenko LG, Treanor JJ, Subbarao K, Wright PF. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. Journal of virology. 2012;86:11725–34. doi: 10.1128/JVI.01477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. Journal of virology. 2006;80:7469–80. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newby CM, Rowe RK, Pekosz A. Influenza A virus infection of primary differentiated airway epithelial cell cultures derived from Syrian golden hamsters. Virology. 2006;354:80–90. doi: 10.1016/j.virol.2006.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adler KB, Cheng PW, Kim KC. Characterization of guinea pig tracheal epithelial cells maintained in biphasic organotypic culture: cellular composition and biochemical analysis of released glycoconjugates. American journal of respiratory cell and molecular biology. 1990;2:145–54. doi: 10.1165/ajrcmb/2.2.145. [DOI] [PubMed] [Google Scholar]
- 46.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. American journal of physiology Lung cellular and molecular physiology. 2002;283:L1315–21. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 47.de Weerd NA, Nguyen T. The interferons and their receptors--distribution and regulation. Immunology and cell biology. 2012;90:483–91. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sommereyns C, Paul S, Staeheli P, Michiels T. IFN-lambda (IFN-lambda) is expressed in a tissue-dependent fashion and primarily acts on epithelial cells in vivo. PLoS pathogens. 2008;4:e1000017. doi: 10.1371/journal.ppat.1000017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odagiri T, Tanaka T, Tobita K. Temperature-sensitive defect of influenza A/Ann Arbor/6/60 cold-adapted variant leads to a blockage of matrix polypeptide incorporation into the plasma membrane of the infected cells. Virus research. 1987;7:203–18. doi: 10.1016/0168-1702(87)90028-1. [DOI] [PubMed] [Google Scholar]
- 50.Belshe RB, Edwards KM, Vesikari T, Black SV, Walker RE, Hultquist M, Kemble G, Connor EM. Live attenuated versus inactivated influenza vaccine in infants and young children. The New England journal of medicine. 2007;356:685–96. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 51.Nichol KL, Mendelman PM, Mallon KP, Jackson LA, Gorse GJ, Belshe RB, Glezen WP, Wittes J. Effectiveness of live, attenuated intranasal influenza virus vaccine in healthy, working adults: a randomized controlled trial. JAMA. 1999;282:137–44. doi: 10.1001/jama.282.2.137. [DOI] [PubMed] [Google Scholar]
- 52.Treanor JJ, Kotloff K, Betts RF, Belshe R, Newman F, Iacuzio D, Wittes J, Bryant M. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine. 1999;18:899–906. doi: 10.1016/s0264-410x(99)00334-5. [DOI] [PubMed] [Google Scholar]
- 53.Grohskopf LA, Sokolow LZ, Broder KR, Olsen SJ, Karron RA, Jernigan DB, Bresee JS. Prevention and Control of Seasonal Influenza with Vaccines. MMWR Recomm Rep. 2016;65:1–54. doi: 10.15585/mmwr.rr6505a1. [DOI] [PubMed] [Google Scholar]