Abstract
Objective
To take a first step towards assembling population based cohorts of bladder cancer patients with longitudinal pathology data, we developed and validated a natural language processing (NLP) engine that abstracts pathology data from full text pathology reports.
Methods
Using 600 bladder pathology reports randomly selected from the Department of Veterans Affairs, we developed and validated an NLP engine to abstract data on histology, invasion (presence versus absence and depth), grade, presence of muscularis propria, and presence of carcinoma in situ. Our gold standard was based on independent review of reports by two urologists, followed by adjudication. We assessed NLP performance by calculating accuracy, positive predictive value (PPV), and sensitivity. We subsequently applied the NLP engine to pathology reports from 10,725 bladder cancer patients.
Results
When comparing the NLP output to the gold standard, NLP achieved the highest accuracy (0.98) for presence versus absence of carcinoma in situ. Accuracy for histology, invasion (presence versus absence), grade, and presence of muscularis propria ranged from 0.83 to 0.96. The most challenging variable was depth of invasion (accuracy 0.68), with acceptable PPV for lamina propria (0.82) and muscularis propria (0.87) invasion. The validated engine was capable of abstracting pathologic characteristics for 99% of bladder cancer patients.
Conclusions
NLP had high accuracy for five of six variables and abstracted data for the vast majority of patients. This now allows for assembly of population based cohorts with longitudinal pathology data.
Keywords: bladder cancer, pathology, natural language processing
Introduction
Bladder cancer is the third most prevalent non-cutaneous cancer in the United States.1 The majority of patients with bladder cancer present with early stage disease, which is rarely lethal.2,3 Thus, median survival for patients with bladder cancer is more than nine years.1,4 During this time, they undergo regular tumor surveillance, including cystoscopy and imaging studies, with the goal of timely detection of tumor recurrences.5 However, despite the high bladder cancer prevalence, current guideline recommendations on surveillance care are based on limited data.6,7 They are largely based on secondary analyses of European clinical trial data from the 1980s and 1990s3,8,9 or on data from institutional observational series.10 However, patients included in the European trials were highly selected and those included in the institutional series were treated at tertiary or quaternary care centers. Thus, surveillance care and outcomes seen in these studies are likely not representative of care in the community.
Population-based studies of bladder cancer care have attempted to address this gap.11 However, the available data sources, primarily Surveillance, Epidemiology and End Results (SEER) and SEER-Medicare data, are limited in the kind of outcomes that can be assessed. This is mainly due to the fact that pathologic details are only abstracted and captured at the time of diagnosis, severely limiting our ability to understand how surveillance care can influence recurrence and progression of bladder cancer. To perform studies addressing these clinically highly relevant outcomes, we need population-based cohorts of bladder cancer patients with longitudinal pathology data.
National Department of Veterans Affairs (VA) data – containing administrative claims as well as full text pathology data – offer the exceptional opportunity to assemble such cohorts. However, in order to develop a population-based cohort of bladder cancer patients with longitudinal bladder pathology data, we needed a method to automate abstraction of important pathological details from tens of thousands of full-text pathology reports. Thus, we embarked on developing and validating a natural language processing (NLP) engine to accomplish this.
Methods
We proceeded in three steps as outlined in detail in the following sections. This included (1) sampling 600 bladder pathology reports and creating a gold standard for NLP development and validation, (2) development and validation of the NLP engine, and (3) application of the NLP engine to all bladder pathology reports for a cohort of patients diagnosed with bladder cancer.
Sampling of Reports
The VA stores national clinical data gathered across the entire network in the Corporate Data Warehouse. Our goal was to sample a nationally representative set of bladder cancer pathology reports from this Corporate Data Warehouse. As described in a previous report focused on the content of these reports,12 we accomplished this via a three step process. First, we identified patients who were diagnosed with bladder cancer between 2005 and 2011. Second, we limited our search to reports with “pathology” in the document title and required one of three keywords to be present in the report text (“bladder”, “urethra”, “ureter”). Third, we randomly selected a sample of 600 reports. The sample size was determined a priori based on sample sizes used in the previous published literature, because there are currently no generally accepted standards of how to determine sample size for validation of natural language processing algorithms.13,14
Human Review of Reports to create a Gold Standard (Annotation)
Development and validation of the NLP engine required a gold standard. This gold standard was created via a rigorous annotation process, during which two human reviewers highlighted full text statements and categorized them according to their meaning as previously described.12 In brief, we first developed an annotation schema, outlining categories of information and content to be abstracted from the reports. Next, two annotators (FRS, a urologic oncologist, and EAP, a urology chief resident) used a specialized annotation tool (ChartReview15) to independently highlight and categorize statements in each report containing information on the following variables: histology, invasion (presence versus absence and depth), grade, and statements regarding presence of carcinoma in situ and of muscularis propria in the specimen. Next, we performed adjudication in two steps: first, we resolved any disagreements between the two annotations by eliminating any obvious errors or omissions. Second, when there was disagreement in the interpretation of a given term between the two annotators, a third independent expert (JDS, a urologic oncologist) functioned as arbiter and decided on the final categorization. The end-result of this process was an annotated dataset of 600 pathology reports that served as the gold standard for development and validation of the NLP engine. Because this analysis focused on urothelial carcinoma, data on grade, invasion, presence of muscularis propria, and presence of carcinoma in situ was only annotated for the 517 reports with urothelial carcinoma.
Development and Validation of the NLP Engine
The 600 pathology reports were randomly split into three groups: a training set (300 reports), a development set (150 reports), and a validation set (150 reports). The NLP engine was built within the VA Corporate Data Warehouse servers using the Apache Unstructured Information Management Architecture Asynchronous Scaleout (UIMA AS)16 and the libraries and tools contained in the Leo framework.17 We developed a rule-based NLP pipeline through an iterative process utilizing the training set for rule design and the development set for error analyses and refinement of rules. The pipeline used regular expressions to find mentions of each concept based on the phrases that were manually annotated in the training set. This set of regular expressions reflected the vocabulary that pathologists use to describe findings. Once a relevant phrase was found in a document, additional heuristics were applied to analyze its context. The heuristics used phrases in the immediate context of the concept mentions to determine if the mentions were negated (e.g. “no invasion was found”), historical (e.g. “history of urothelial carcinoma”), or otherwise irrelevant (e.g. “increased risk of developing adenocarcinoma”, “evaluation for muscle invasion”). The final logic module created output entries for those mentions that were not discarded based on the context. A separate pipeline was developed for each variable except for the invasion variables, so that they can be run separately or together. The pipelines for presence versus absence and depth of invasion were developed together. This was done to differentiate negated statements indicating presence versus absence of invasion from negated statements referencing a certain depth of invasion (e.g., “no lamina propria or muscularis propria invasion” was interpreted as absence of invasion versus “no muscularis propria invasion” was interpreted as absence of muscle invasive disease).
Once we were satisfied with the performance of the NLP engine, we performed a final validation run on the validation set. The validation results reported herein are those obtained from the validation set.
Application of the validated NLP Engine to all Bladder Cancer Pathology Reports
Finally, we applied the validated NLP engine to abstract information from all available bladder pathology reports stored within the Corporate Data Warehouse for a cohort of patients diagnosed with bladder cancer between 2005 and 2011 (10,725 patients, 31,009 non-annotated reports). For each variable and value, we reviewed the 50 most common expressions retrieved by NLP. For a few situations in which NLP captured apparently incorrect information, we applied a post-hoc data cleaning step and removed these instances from the data set. For example, the terms “superior to inferior” and “inferior to superior” were erroneously categorized as “superficial” lamina propria invasion. This erroneous categorization was removed during the post-hoc data cleaning step.
Analyses
To evaluate inter-rater reliability between the two annotators, we calculated Cohen's kappa for each variable prior to any adjudications. We evaluated NLP performance at the pathology report level. For each pathology report and variable, we assigned the highest risk finding mentioned within the report. For example, if statements about both high grade and low grade were captured by the NLP engine within the same report, we assigned a finding of high grade. The full hierarchy is listed in Appendix 1. For each variable, we calculated accuracy, positive predictive value (PPV, also known as precision in information retrieval), and sensitivity (also known as recall in information retrieval) by categorizing the NLP abstracted data as either correct or incorrect based on comparison to the gold standard annotated data.
We determined PPV for no cancer in the biopsy, invasive disease, non-invasive disease, and lamina propria invasion in the non-annotated set of 31,009 bladder pathology reports after applying the post-hoc data cleaning step described above. These four variables were chosen because our plan is to use the pathologic data to study care and outcomes for patients with early stage bladder cancer. In this setting, it will be most important to be certain that included patients truly have non-muscle invasive disease (i.e., non-invasive disease or invasive disease that is limited to lamina propria invasion). In addition, having a biopsy with no cancer in the specimen may be an important marker for potentially avoidable biopsies among patients with a history of low-risk early stage bladder cancer. To determine these PPVs, we sampled a simple random set of 100 reports each that were classified as no cancer in the biopsy, invasive disease, non-invasive disease, or lamina propria invasion by the NLP engine (400 reports in total). These 400 reports were then reviewed by human annotators who confirmed or refuted findings.
Lastly, we obtained some insight into the variability in the language (vocabulary and syntax) used by pathologists to describe findings in the non-annotated reports by assessing the number of unique expressions retrieved by the NLP engine per 1,000 reports for each of the variable values.
Results
Human Review of Reports (Annotation)
Among the 600 annotated reports, 517 represented urothelial carcinoma. Slightly more than half (57.8%) were describing high grade tumors and 43.5% represented non-invasive tumors (Table 1). Other report characteristics as abstracted by the annotators are listed in Table 1. Inter-rater reliability between the two annotators was excellent, ranging from 0.82 to 0.90. It was highest for presence of carcinoma in situ and lowest for presence of muscularis propria in the specimen (Supplemental Table 1).
Table 1.
Information abstracted via human annotation from the national random sample of 600 pathology reports. Because this analysis focused on urothelial carcinoma, data on grade, invasion, presence of muscularis propria, and presence of carcinoma in situ was only annotated for the 517 reports with urothelial carcinoma.
| Variable | Abstracted via Human Annotations |
|---|---|
| Value | N (%) |
| Histology (n=600) | |
| Not from Bladder | 22 (3.7) |
| No Cancer | 36 (6.0) |
| Urothelial Carcinoma | 517 (86.2) |
| PUNLMP | 5 (0.8) |
| Other Histology* | 20 (3.3) |
|
| |
| Grade (n=517) | |
| Low | 159 (30.8) |
| Intermediate | 37 (7.2) |
| High | 299 (57.8) |
| Undifferentiated | 1 (0.2) |
| Not stated / missing | 21 (4.1) |
|
| |
| Carcinoma in situ (n=517) | |
| Present | 62 (12.0) |
| Explicitly absent | 7 (1.4) |
| Not mentioned | 448 (86.7) |
|
| |
| Invasion presence versus absence (n=517) | |
| Non-invasive | 225 (43.5) |
| Suspected invasion | 9 (1.7) |
| Invasive | 230 (44.5) |
| Not stated / missing | 53 (10.3) |
|
| |
| Invasion depth among reports with invasion or suspected invasion (n=239) | |
| Lamina propria | 134 (56.1) |
|
| |
| Muscularis propria | 82 (34.3) |
| Perivesical / Other Organ | 17 (7.1) |
| Not stated / missing | 6 (2.5) |
| Muscularis propria in specimen (n=517) | |
| Present | 358 (69.2) |
| Not present | 103 (19.9) |
| Not stated / missing | 56 (10.8) |
Other histology included squamous cell carcinoma, adenocarcinoma, small cell carcinoma, undifferentiated carcinoma, unspecified carcinoma, among others. PUNLMP = Papillary Urothelial Neoplasm of Low Malignant Potential.
Validation of the NLP Engine
During validation, the NLP engine achieved high accuracy for histology, grade, carcinoma in situ, and presence versus absence of invasion ranging from 0.87 to 0.98. For these variables, PPV and sensitivity were also high with values mostly above 0.85 (Table 2). Figure 1 summarizes results at the pathology report level and shows NLP true positives, false positives, and false negatives. Generally, the vast majority of NLP categorizations were true positives. The most challenging variable to abstract was depth of invasion with sensitivities of 0.71 for lamina propria and 0.57 for muscularis propria invasion. Nevertheless, PPV was acceptable for lamina propria and muscularis propria invasion (0.82 and 0.87, respectively, Table 2). Detailed matrices comparing gold standard to NLP output in the validation sample are provided in Appendix 2.
Table 2.
Positive predictive value, sensitivity, and accuracy of the natural language processing engine at the pathology report level within the final validation data set (n=150 reports, na = not applicable)
| Variable | Number of reports in validation set based on gold standard annotation | Positive Predictive Value | Sensitivity | Accuracy |
|---|---|---|---|---|
| Histology | 0.96 | |||
| No Cancer | 8 | 0.88 | 0.88 | |
| Urothelial Carcinoma | 130 | 0.98 | 0.98 | |
| PUNLMP | 2 | 1.00 | 0.50 | |
| Other Histology* | 3 | 1.00 | 0.67 | |
| Not stated / missing | 7 | na | na | |
|
| ||||
| Grade | 0.93 | |||
| Low | 41 | 0.95 | 0.95 | |
| Intermediate | 5 | 0.80 | 0.80 | |
| High | 75 | 1.00 | 0.92 | |
| Undifferentiated | 1 | 0 | 0 | |
| Not stated / missing | 28 | na | na | |
|
| ||||
| Carcinoma in situ | 0.98 | |||
| Present | 21 | 1.00 | 0.91 | |
| Explicitly absent | 0 | na | na | |
| Not mentioned | 129 | na | na | |
|
| ||||
| Invasion presence versus absence | 0.87 | |||
| Non-invasive | 47 | 0.90 | 0.79 | |
| Suspected invasion | 4 | 1.00 | 0.50 | |
| Invasive | 63 | 0.82 | 0.97 | |
| Not stated / missing | 36 | na | na | |
|
| ||||
| Invasion depth among reports with invasion or suspected invasion by NLP (n=76) | 0.68 | |||
| Lamina propria | 38 | 0.82 | 0.71 | |
| Muscularis propria | 23 | 0.87 | 0.57 | |
| Perivesical / Other Organ | 2 | 0.50 | 0.50 | |
| Not stated / missing | 13 | na | na | |
|
| ||||
| Muscularis propria in specimen | 0.83 | |||
| Present | 92 | 0.91 | 0.80 | |
| Not present | 27 | 0.80 | 0.74 | |
| Not stated | 31 | na | na | |
Other histology included squamous cell carcinoma, adenocarcinoma, small cell carcinoma, undifferentiated carcinoma, unspecified carcinoma, among others. PUNLMP = Papillary Urothelial Neoplasm of Low Malignant Potential.
Figure 1.
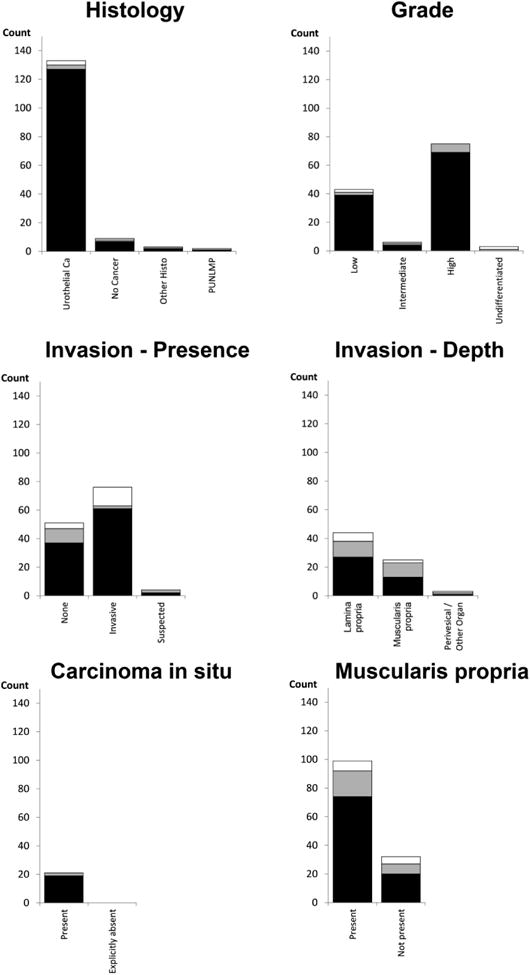
Documents classified correctly as well as false positive and false negatives among the 150 bladder cancer pathology reports included in the validation sample. Black bars indicate correctly identified reports, grey bars are NLP false negatives, and white bars are NLP false positives. Black and grey bars together represent the count based on the gold standard annotation.
Application of the validated NLP Engine to non-annotated reports
Next, we applied the NLP engine to 31,009 non-annotated bladder pathology reports for 10,725 patients. The NLP engine was able to retrieve information for 98% of reports (30,498 of 31,009 reports) and 99% of patients (10,593 of 10,725 patients). The information abstracted is presented in Table 3 and largely mirrors the distribution of pathologic findings across the annotated 600 bladder pathology reports (Table 1). PPV among a random sample of 100 reports each from these full text bladder pathology reports was 0.99 for no cancer in the biopsy, 0.95 for invasive disease, 0.96 for non-invasive disease, and 0.91 for lamina propria invasion.
Table 3.
The NLP engine was able to retrieve information from 30,498 full text bladder pathology reports. Because this analysis focused on urothelial carcinoma, data on grade, invasion, presence of muscularis propria, and presence of carcinoma in situ is only reported for the 20,515 reports with urothelial carcinoma
| Variable | Abstracted by NLP engine |
|---|---|
| Value | N (%) |
| Histology (n=30,498) | |
| Not from Bladder | 1,560 (6.3) |
| No Cancer | 5,577 (18.3) |
| Urothelial Carcinoma | 20,515 (67.3) |
| PUNLMP | 213 (0.7) |
| Other Histology* | 717 (2.4) |
| Missing | 1,916 (6.3) |
|
| |
| Grade (n=20,515) | |
| Low | 6,708 (32.7) |
| Intermediate | 1,271 (6.2) |
| High | 9,548 (46.5) |
| Undifferentiated | 194 (1.0) |
| Not stated / missing | 2,794 (13.6) |
|
| |
| Carcinoma in situ (n=20,515) | |
| Present | 2,630 (12.8) |
| Explicitly absent | 224 (1.1) |
| Not mentioned / missing | 17,661 (86.1) |
|
| |
| Invasion presence vs absence (n=20,515) | |
| Non-invasive | 7,869 (38.4) |
| Suspected invasion | 175 (0.9) |
| Invasive | 9,008 (43.9) |
| Not stated / missing | 3,463 (16.9) |
|
| |
| Invasion depth among reports with invasion or suspected invasion by NLP (n=9,183) | |
| Lamina propria | 3,741 (40.8) |
| Muscularis propria | 2,270 (24.7) |
| Perivesical / Other Organ | 283 (3.1) |
| Not stated / missing | 2,889 (31.5) |
|
| |
| Muscularis propria in specimen (n=20,515) | |
| Present | 10,305 (50.2) |
| Not present | 3,617 (17.6) |
| Not stated / missing | 6,593 (32.1) |
Other histology included squamous cell carcinoma, adenocarcinoma, small cell carcinoma, undifferentiated carcinoma, unspecified carcinoma, among others. PUNLMP = Papillary Urothelial Neoplasm of Low Malignant Potential.
Finally, we evaluated the variability in language used by pathologists to describe findings. We found a high number of unique expressions used to describe depth of invasion ranging from 261 separate expressions for muscularis propria invasion to 863 expressions for perivesical invasion per 1,000 reports. This number of unique expressions was much higher than that for variables with the highest accuracy, PPV, and sensitivity, including urothelial histology (30 per 1,000 reports), grade (40 to 330 per 1,000 reports), and carcinoma in situ (32 per 1,000 reports).
Discussion
We developed and validated an NLP engine to abstract pathology data from full text bladder cancer pathology reports. Accuracy, PPV, and sensitivity were high for most variables. Invasion depth and presence versus absence of muscularis propria in the specimen were harder to abstract, likely because of the substantial variation in the language used by pathologists to describe these types of findings. We successfully used the validated engine to abstract data from a large national set of bladder cancer pathology reports.
Our findings are consistent with prior reports in the literature on the use of NLP to abstract pathology data. For example, an NLP engine developed to classify breast pathology histology achieved a PPV of 0.97 when compared to human annotators as the gold standard.18 Similarly, NLP has been used to abstract histology from colon pathology reports and clinically relevant variables from prostatectomy pathology reports.14,19 These prior results – as well as our own – demonstrate that NLP is highly accurate to retrieve certain components of pathology reports such as histology and grade. However, other components such as tumor stage are more difficult to accurately abstract. In our set of bladder pathology reports, only 20% included an explicitly stated tumor T-stage.12 To derive a T-stage, we had to evaluate statements used by pathologists to describe depth of invasion. The language used in these statements was highly variable, which led to higher levels of text misinterpretation by the NLP engine. NLP would likely be more accurate if pathologists were using more standardized language to describe depth of invasion. The College of American Pathologists suggests use of a synoptic report which includes standardized language,20 but this has only rarely been adopted for bladder pathology reports within VA.12 Wider use of standardized language could facilitate both NLP and clinical care in the future. Nevertheless and in spite of the variability in language, we were able to identify tumors with lamina propria invasion (T1) with a high PPV of 0.91 in the final cleaned data set, and our accuracy is comparable to that of a prior study that was focused on abstracting staging information from full text lung cancer pathology reports (0.68 in the current study versus 0.72 in the prior study).21
Our study has several limitations that warrant discussion. First, we used a national sample of pathology reports from the VA. Thus, our findings are not directly generalizable to settings outside of the VA system. However, our approach enabled us to develop, validate, and apply the NLP engine on a large national sample of bladder pathology reports. While pathologists likely use similar language to describe findings in bladder pathology reports outside of the VA, the structure and format of pathology reports will vary across different healthcare systems. Thus, deploying our NLP engine outside of the VA would warrant further adaptation and validation, and we are currently considering additional studies focused on such external validation. Second, we believe it is important to point out that our goal was to use the data abstracted by NLP in health services research. While there may be a role to use such automatically abstracted data in the clinical setting, for example by incorporating it into automatic risk prediction tools, different uses of the data may warrant different configuration of the NLP engine. Third, while we were able to differentiate between non-invasive and invasive pathology with a high accuracy close to ninety percent, accuracy dropped to 0.68 for depth of invasion, mainly because of a low sensitivity for the identification of muscularis propria invasion (0.57, Table 2). This low sensitivity indicates that the NLP engine had difficulties identifying muscularis propria invasion among the reports that were annotated as muscle-invasive, likely due to the limited number of reports with muscle invasive disease that were available for training (47/300 reports in the training set had muscle invasive disease, 15.7%). This limitation will have to be carefully considered in future studies using the data abstracted with the NLP engine.
These limitations notwithstanding, we believe that our NLP engine is an important step forward in population-based health services research in early stage bladder cancer. Given the high PPV values, our engine can be used to develop population based cohorts of patients that have specific pathology (e.g. identifying a cohort of patients with non-invasive low-grade versus high-grade disease). Pathology data abstracted by the NLP engine can also be used to ascertain the development of invasive disease among patients who were initially diagnosed with non-invasive urothelial carcinoma, because sensitivity for presence of invasion was excellent at 0.97.
In conclusion, we developed and validated an NLP engine that accurately abstracts details from full text bladder pathology reports for a vast majority of patients. We will now use this abstracted data to assemble cohorts of early stage bladder cancer patients with longitudinal pathology information. We will then use these cohorts to examine how different patterns of surveillance care impact patient outcomes, including time to recurrence and progression to invasive disease.
Supplementary Material
Acknowledgments
Funding: FRS is supported by the Department of Veterans Affairs, Veterans Health Administration, VISN1 Career Development Award, by a pilot grant from the American Cancer Society (IRG-82-003-30), by a Conquer Cancer Foundation Career Development Award, and by the Dow-Crichlow Award of the Department of Surgery at the Dartmouth-Hitchcock Medical Center. PPG is supported by a grant from the Food and Drug Administration FDA (U01FD005478-01, Sedrakyan = PI). This project was supported using resources and facilities at the White River Junction VA Medical Center, the VA Salt Lake City Health Care System, and the VA Informatics and Computing Infrastructure (VINCI), VA HSR RES 13-457.
Footnotes
Conflicts of Interest: none
Author contributions: Conceptualization (FRS, DJR, BS, PPG), Methodology (FRS, OVP, PRA, SLD), Software (OVP, PRA, SLD), Validation (FRS, OVP, PRA, SLD), Formal Analysis (FRS, OVP, PRA), Resources (FRS, SLD, BS, PPG), Data Curation (SLD), Drafting (FRS) and Editing (all authors) of Manuscript, Visualization (FRS), Supervision (FRS, SLD, BS, PPG), Project Administration (FRS, SLD), Funding Acquisition (FRS, PPG)
Disclaimer: Opinions expressed in this manuscript are those of the authors and do not constitute official positions of the U.S. Federal Government or the Department of Veterans Affairs.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Howlader N, Noone AM, Krapcho M, et al. Natl Cancer Inst. Bethesda MD: 2014. [accessed August 30, 2014]. SEER Cancer Statistics Review, 1975-2011. Available at: http://seer.cancer.gov/csr/1975_2011/ [Google Scholar]
- 2.Burger M, Catto JWF, Dalbagni G, et al. Epidemiology and Risk Factors of Urothelial Bladder Cancer. Eur Urol. 2013;63:234–241. doi: 10.1016/j.eururo.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Cambier S, Sylvester RJ, Collette L, et al. EORTC Nomograms and Risk Groups for Predicting Recurrence, Progression, and Disease-specific and Overall Survival in Non–Muscle-invasive Stage Ta–T1 Urothelial Bladder Cancer Patients Treated with 1–3 Years of Maintenance Bacillus Calmette-Guérin. Eur Urol. 2016;69:60–69. doi: 10.1016/j.eururo.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 4.Ries LAG, Young JL, Keel GE, et al. SEER Survival Monograph: Cancer Survival Among Adults: US SEER Program, 1988-2001, Patient and Tumor Characteristics Pub No 07-6215. Bethesda, MD: NIH; 2007. [accessed August 12, 2014]. Available at: http://seer.cancer.gov/archive/publications/survival/seer_survival_mono_lowres.pdf. [Google Scholar]
- 5.Holmäng S. Follow-up of patients with noninvasive and superficially invasive bladder cancer. Semin Urol Oncol. 2000;18:273–279. [PubMed] [Google Scholar]
- 6.Babjuk M, Boehle A, Burger M, et al. European Association of Urology (EAU) Guidelines on Non-muscle-invasive Bladder Cancer: Update 2016. Eur Urol. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 7.Chang SS, Boorjian SA, Chou R, et al. [accessed May 3, 2016];Non-Muscle Invasive Bladder Cancer: American Urological Association / SUO Guideline. 2016 Available at: https://www.auanet.org/education/guidelines/non-muscle-invasive-bladder-cancer.cfm.
- 8.Sylvester RJ, van der Meijden APM, Oosterlinck W, et al. Predicting recurrence and progression in individual patients with stage Ta T1 bladder cancer using EORTC risk tables: a combined analysis of 2596 patients from seven EORTC trials. Eur Urol. 2006;49:466–475. doi: 10.1016/j.eururo.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Gomez J, Madero R, Solsona E, et al. Predicting Nonmuscle Invasive Bladder Cancer Recurrence and Progression in Patients Treated With Bacillus Calmette-Guerin: The CUETO Scoring Model. J Urol. 2009;182:2195–2203. doi: 10.1016/j.juro.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Soukup V, Babjuk M, Bellmunt J, et al. Follow-up After Surgical Treatment of Bladder Cancer: A Critical Analysis of the Literature. Eur Urol. 2012;62:290–302. doi: 10.1016/j.eururo.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Chamie K, Litwin MS, Bassett JC, et al. Recurrence of high-risk bladder cancer: A population-based analysis. Cancer. 2013;119:3219–3227. doi: 10.1002/cncr.28147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeck FR, Pattison EA, Denhalter DW, et al. Early Stage Bladder Cancer – Do Pathology Reports Tell Us What We Need to Know? Urology. 2016;98:58–63. doi: 10.1016/j.urology.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harkema H, Chapman WW, Saul M, et al. Developing a natural language processing application for measuring the quality of colonoscopy procedures. J Am Med Inform Assoc. 2011;18:i150–i156. doi: 10.1136/amiajnl-2011-000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imler TD, Morea J, Kahi C, et al. Natural Language Processing Accurately Categorizes Findings From Colonoscopy and Pathology Reports. Clin Gastroenterol Hepatol. 2013;11:689–694. doi: 10.1016/j.cgh.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DuVall S, Forbush T, Cornia RC, et al. Reducing the Manual Burden of Medical Record Review through Informatics. Pharmacoepidemiol Drug Saf. 2014;23:415. [Google Scholar]
- 16.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10:327–348. [Google Scholar]
- 17.Cornia RC, Patterson OV, Ginter T, et al. Rapid NLP Development with Leo. AMIA Annu Symp Proc. 2014 Abstract #1356. [Google Scholar]
- 18.Buckley JM, Coopey SB, Sharko J, et al. The feasibility of using natural language processing to extract clinical information from breast pathology reports. J Pathol Inform. 2012;3:23. doi: 10.4103/2153-3539.97788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim BJ, Merchant M, Zheng C, et al. Second Prize: A Natural Language Processing Program Effectively Extracts Key Pathologic Findings from Radical Prostatectomy Reports. J Endourol. 2014;28:1474–1478. doi: 10.1089/end.2014.0221. [DOI] [PubMed] [Google Scholar]
- 20.Amin MB, Delahunt B, Bochner BH, et al. Protocol for the Examination of Specimens From Patients With Carcinoma of the Urinary Bladder. [accessed November 10, 2015];Coll Am Pathol. 2013 Available at: http://www.cap.org/ShowProperty?nodePath=/UCMCon/Contribution%20Folders/WebContent/pdf/urinary-13protocol-3210.pdf.
- 21.Nguyen AN, Lawley MJ, Hansen DP, et al. Symbolic rule-based classification of lung cancer stages from free-text pathology reports. J Am Med Inform Assoc. 2010;17:440–445. doi: 10.1136/jamia.2010.003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


