Abstract
It is increasingly common for apex predators to face a multitude of complex conservation issues. In Australia, dingoes are the mainland apex predator and play an important role in ecological functioning. Currently, however, they are threatened by hybridization with modern domestic dogs in the wild. As a consequence, we explore how increasing our understanding of the evolutionary history of dingoes can inform management and conservation decisions. Previous research on whole mitochondrial genome and nuclear data from five geographical populations showed evidence of two distinct lineages of dingo. Here, we present data from a broader survey of dingoes around Australia using both mitochondrial and Y chromosome markers and investigate the timing of demographic expansions. Biogeographic data corroborate the presence of at least two geographically subdivided genetic populations, southeastern and northwestern. Demographic modeling suggests that dingoes have undergone population expansion in the last 5,000 years. It is not clear whether this stems from expansion into vacant niches after the extinction of thylacines on the mainland or indicates the arrival date of dingoes. Male dispersal is much more common than female, evidenced by more diffuse Y haplogroup distributions. There is also evidence of likely historical male biased introgression from domestic dogs into dingoes, predominately within southeastern Australia. These findings have critical practical implications for the management and conservation of dingoes in Australia; particularly a focus must be placed upon the threatened southeastern dingo population.
Keywords: Australia, biogeography, conservation, demography, dingoes, hybridization, mitochondrial DNA, mtDNA, population expansion, Y chromosome
1. INTRODUCTION
The effect of removing large socially complex apex consumers such as whales, big cats, bears, wolves, and dingoes from ecosystems is poorly documented (Estes et al., 2011). Apex predators are in decline, globally, which has lead to and threatens continuing impacts to entire ecosystems (Estes et al., 2011; Morris & Letnic, 2017; Ripple et al., 2014, 2016, 2017). Estes et al. (2011) suggest that worldwide large apex consumer declines can cause extensive trophic cascading, exacerbated by agricultural land management, widespread habitat degradation, pollution, and ultimately climate change. On the Australian continent, indigenous apex predators went extinct thousands of years ago, leaving the dingo as the sole remaining apex predator on the mainland. As such, the dingo plays a central ecological role. Today, dingoes are threatened by extensive lethal control programs, habitat fragmentation, and genetic dilution from hybridization with domestic dogs (Stephens, Wilton, Fleming, & Berry, 2015).
In this study, we explore the evolutionary history of Australian dingoes with a goal of informing management and conservation decisions (Figure 1). Since 1788, dingoes have been subject to hybridization pressure from modern domestic dogs brought by Europeans, particularly in regions where human populations are high (Stephens et al., 2015). The observation of hybridization in species and populations is an increasingly common conservation concern; well documented examples include bison, coyotes, wolves, wild cats, and even Galapagos tortoises (Garrick et al., 2012; Halbert & Derr, 2007; Hertwig et al., 2009; vonHoldt et al., 2016; Reich, Wayne, & Goldstein, 1999).
Figure 1.

A pair of typical dingoes, as found across the Australian mainland. Dingoes are commonly observed with ginger, white, sable, and black and tan pelage. © Photograph: L. Watson, Australian Dingo Foundation (2017)
Historically, the dingo is thought to have arrived on mainland Australia approximately 5,000 years before present (BP) (Gollan, 1984; Macintosh, 1975; Savolainen, Leitner, Wilton, Matisoo‐Smith, & Lundeberg, 2004). The minimum arrival time of dingoes is approximately 3,500 years BP based upon the oldest fossil observed in southern Western Australia (Macintosh, 1964). The dingo is frequently blamed for the thylacine extinction on mainland Australia 2,000–3,000 years BP (Fillios, Crowther, & Letnic, 2012; Johnson & Wroe, 2003; Letnic, Fillios, & Crowther, 2012), although new modeling suggests climate change and human population growth may have played a more significant role (Prowse, Johnson, Bradshaw, & Brook, 2014). Dingoes and New Guinea Singing Dog (NGSD) likely came to Oceania with humans. However, there is uncertainty concerning which human colonization event they accompanied and whether dingoes were brought directly to Australia or immigrated via the prehistoric land bridge between Papua New Guinea and Australia prior to 8,000 years BP. It is commonly presumed that dingoes were brought to Australia 5,000 years ago as part of the Neolithic human expansion (Sacks et al., 2013; Savolainen et al., 2004). However, Fillios and Taçon (2016) argue that the Neolithic people are unlikely to be responsible for the arrival of the dingo because Australia lacks key Neolithic cultural indicators.
An alternative hypothesis is that dingoes and NGSD are part of an older dog radiation that immigrated into Australia via the land bridge between Australia and Papua New Guinea, which flooded 6,000–8,000 years BP (Cairns & Wilton, 2016). Some ethnographic evidence supports this hypothesis, for example, the lack of Neolithic cultural items, such as chickens and pigs, in Australia prior to European colonization (Larson et al., 2010; Oskarsson et al., 2011), lack of human genetic signatures indicating contact between South East Asia and Indigenous Australians (Brown, 2013; Haak et al., 2010; van Holst Pellekaan, 2001, 2013; Karafet et al., 2005; McEvoy et al., 2010; Pugach, Delfin, Gunnarsdóttir, Kayser, & Stoneking, 2013), and the finding that dingoes only carry the two ancestral Amylase gene copies, consistent with their having diverged from modern domestic dogs before the agricultural era (Arendt, Cairns, Ballard, Savolainen, & Axelsson, 2016; Freedman et al., 2014). More recent molecular dating efforts based on mitochondrial divergence time suggest that dingoes could have arrived in Australia approximately 8,000–10,000 years BP (Cairns & Wilton, 2016; Oskarsson et al., 2011). Bayesian skyline modeling may help inform this debate by testing when dingoes underwent population expansion and/or contraction.
Knowledge concerning the population biology of dingoes can provide insight into the arrival patterns and origin of this enigmatic canine, which may have important conservation implications. In 2016, Cairns and Wilton identified the presence of at least two dingo lineages, southeastern (SE) and northwestern (NW), in Australia with a pattern of geographical subdivision. Early studies using mitochondrial DNA were unable to elucidate continent‐wide biogeographic patterns, as they restricted genetic sampling to the mitochondrial control region (Oskarsson et al., 2011; Savolainen et al., 2004). Y chromosome studies either had samples from mostly Western Australia or mostly eastern Australia, and differences in genetic marker sampling made it difficult to compare datasets (Ardalan et al., 2012; Sacks et al., 2013).
Ecological theory predicts that shifts in the distribution and abundance of apex predators and herbivores may result in sizable changes in ecosystem dynamics (Fretwell, 1987; Hairston, Smith, & Slobodkin, 1960). In Australia, the distribution and abundance of dingoes is influenced by historical biogeography as well as current hybridization with dogs. This study is the first broad biogeographic survey of dingoes utilizing both mitochondrial and Y chromosome markers that aims to investigate broad patterns of biogeography as a fundamental prerequisite for conservation management. Our data shed light on modern and historical migration and dispersal patterns in male and female dingoes on a continental scale. Bayesian demographic modeling of the distinct dingo lineages suggests the populations may have distinct evolutionary histories, which may impact on conservation. The research has practical implications for dingo conservation and management strategies across Australia, particularly concerning hybridization.
2. MATERIALS AND METHODS
2.1. Canid sampling
In order to investigate biogeography, migration, male and female dispersal patterns, and immigration routes, we sampled 127 dingoes broadly across Australia and five NGSD from the North American captive population (Figure 2, Table 1). Five of the dingoes were sampled from the captive dingo population. We also incorporated a dataset of Y chromosome and mitochondrial control region data from 173 male dogs, including 94 dingoes and 18 NGSD from Sacks et al. (2013).
Figure 2.
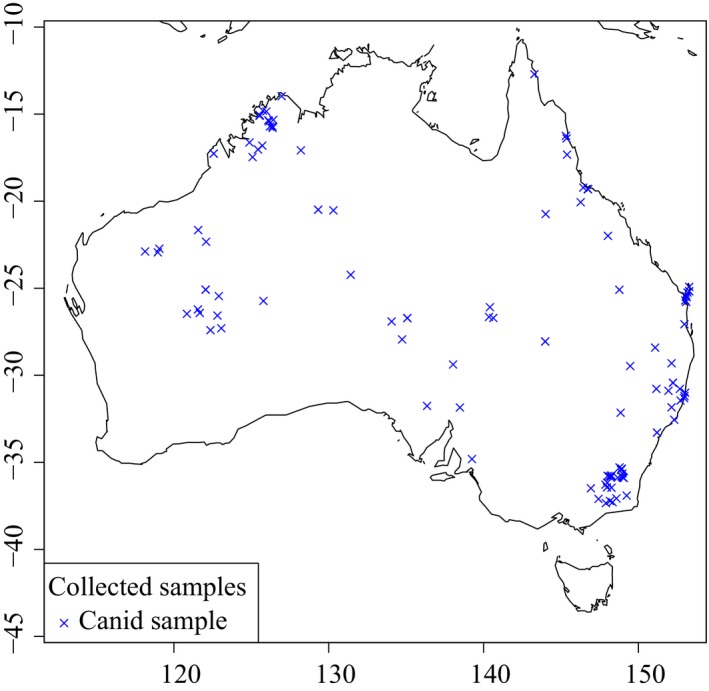
Map depicting geographic sampling of dingoes across Australia. Crosses represent individual samples. New Guinea Singing Dogs are depicted in Papua New Guinea; however, samples were sourced from the North American captive population
Table 1.
Sample data; identifier, geographical locale, latitude, longitude, and genetic identity
| Dingo name | Dingo ID | Gender | State | Longitude | Latitude | CR haplotype (collapsed) | CR haplotype (gaps considered) | MtDNA Clade | MtDNA Type | Y chromosome haplotype | GenBank accession # |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alpine 1 | 96.2 | M | VIC | −37.29 | 148.33 | A209 | A209 | SE | a9 | H60‐k11 | JX088688 |
| Alpine 2 | WD170 | M | NSW | −36.46 | 148.26 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | JX088692 |
| Alpine 3 | 44.5 | M | ACT | −35.84 | 148.98 | A29 | A29 | SE | a3 | H1*/H2*‐6t | JX088692 |
| Alpine 4 | 135.10 | F | VIC | −37.07 | 148.58 | A29 | A179 | SE | a12 | JX088680 | |
| Alpine 5 | 119.1 | M | VIC | −36.17 | 147.99 | A29 | A179 | NW | d23 | H60‐k11 | JX088679 |
| Fraser 1 | 21.3 | F | QLD | −25.25 | 153.17 | A29 | A179 | SE | f1 | JX088676 | |
| Fraser 2 | 21.1 | M | QLD | −25.17 | 153.28 | A29 | A179 | SE | f1 | H60‐n25 | JX088676 |
| Fraser 3 | 184.4 | F | QLD | −25.18 | 153.28 | A29 | A179 | SE | f1 | JX088676 | |
| Fraser 4 | 184.1 | M | QLD | −25.7 | 153.03 | A29 | A179 | SE | f1 | H60‐n25 | JX088676 |
| Fraser 5 | 21.4 | F | QLD | −25.79 | 153.08 | A29 | A179 | SE | f1 | JX088676 | |
| Fraser 6 | 21.2 | F | QLD | −24.915 | 153.281 | A29 | A179 | SE | a1 | KC346413, KC346430 | |
| Gibson 1 | 9.40 | F | WA | −26.22 | 121.55 | A203 | A203 | NW | d5 | JX088685 | |
| Gibson 2 | 19.84 | F | WA | −27.3 | 123.06 | A29 | A29 | NW | d5 | JX088675 | |
| Gibson 3 | DE17 | M | WA | −26.42 | 121.67 | A200 | A200 | NW | d5 | H60‐n24 | JX088673 |
| Gibson 4 | DE13 | F | WA | −25.45 | 122.9 | A29 | A29 | NW | d5 | JX088687 | |
| Gibson 5 | 18.35 | F | WA | −25.08 | 122.05 | A200 | A200 | NW | d5 | JX088672 | |
| Kimberley 1 | 3.45 | F | WA | −15.1 | 125.53 | A29 | A29 | NW | d5 | JX088683 | |
| Kimberley 2 | 7.27 | M | WA | −16.63 | 124.88 | A9 | A9 | NW | d7 | H3‐n4 | JX088691 |
| Kimberley 3 | 19.11 | M | WA | −17.47 | 125.08 | A9 | A9 | NW | d7 | H3‐n20 | JX088681 |
| Kimberley 4 | 4.55 | F | WA | −15.35 | 126.1 | din27 | din27 | NW | d5 | JX088682 | |
| Kimberley 5 | 24.96 | M | WA | −17.27 | 122.57 | A29 | A29 | NW | d5 | H3‐n4 | JX088684 |
| Simpson 1 | 182.6 | F | SA | −26.65 | 140.35 | A29 | A179 | NW | d5 | JX088677 | |
| Simpson 2 | X1777 | M | NT | 25.35 | 133.71 | A200 | din35 | NW | d5 | H60‐9k | JX088671 |
| Simpson 3 | 142.3 | F | SA | −27.94 | 134.74 | A200 | A200 | NW | d4 | JX088678 | |
| Simpson 4 | 217.2 | F | SA | −26.91 | 134.06 | A200 | A200 | NW | d14 | JX088686 | |
| Simpson 5 | X1783 | F | NT | −24.23 | 131.42 | A200 | A200 | NW | d5 | JX088693 | |
| Simpson 6 | 193.5 | M | NT | −26.086056 | 140.411131 | A29 | A29 | NW | d3 | H60‐n25 | KC346412, KC346429 |
| Captive 1 | Gunyah | M | Captive | — | — | A29 | A29 | SE | a3 | H1*/H2*‐k7 | KC346422, KC346439 |
| Captive 2 | N858 | M | Captive | — | — | A29 | A205 | NW | d5 | H60‐k8 | KC346411, KC346428 |
| Captive 3 | N895 | M | Captive | — | — | A29 | A29 | SE | a3 | H1*/H2*‐n7 | KC346422, KC346439 |
| Captive 4 | N834 | M | Captive | — | — | A29 | A29 | SE | a3 | H3‐12d | KC346422, KC346439 |
| Captive 5 | N887 | M | Captive | — | — | A29 | A198 | NW | d5 | H60‐n24 | KC346411, KC346428 |
| Central Australia 1 | X167 | M | QLD | −28.06092 | 143.9805968 | din32 | din32 | NW | d20 | H1*/H2*‐6q | JX090189, JX090195, MF784880 |
| Central Australia 2 | X179 | M | QLD | −28.06092 | 143.9805968 | din32 | din32 | NW | d20 | H1*/H2*‐6q | JX090189, JX090195, MF784880 |
| Central Australia 3 | 182.2 | M | NT | −26.714283 | 140.627761 | A29 | A179 | NW | d5 | H1*/H2*‐6q | KC346411, KC346428 |
| Central Australia 4 | 197.1 | M | SA | −31.854931 | 138.474641 | din32 | din32 | NW | d5 | H1*/H2*‐6q | KC346411, KC346428, MF784880 |
| Central Australia 5 | 193.2 | F | NT | −26.718428 | 135.073047 | A29 | A29 | NW | d1 | KC346416, KC346433 | |
| Central Australia 6 | 193.3 | F | NT | −26.712483 | 135.076839 | — | — | NW | d2 | KC346424, KC346441 | |
| Central Australia 7 | TA101 | M | NT | −20.49 | 129.319 | A200 | A200 | NW | d9 | H60‐0i | MF774083, MF774092 |
| Central Australia 8 | TA91 | M | NT | −20.5335 | 130.29875 | A200 | A200 | NW | d9 | H60‐n24 | MF774083, MF774092 |
| Central Australia 9 | TA94 | M | NT | −20.49 | 129.319 | A29 | A29 | NW | d5 | H60‐n25 | KC346411, KC346428 |
| Central Australia 10 | TA95 | M | NT | −20.509 | 129.462 | A29 | A29 | — | — | H60‐n29 | |
| Central Australia 11 | X2060 | M | NT | −16.791 | 137.526 | A17 | A17 | — | — | H3‐6z | |
| Central Australia 12 | 150.1 | M | SA | −34.814542 | 139.247263 | din32 | din32 | NW | d21 | H60‐k10 | MF774086, MF774095, MF784880 |
| Central Australia 13 | 155.5 | M | SA | −31.759594 | 136.350219 | din31 | din31 | NW | d5 | H60‐n25 | KC346411, KC34642, MF784879 |
| Central Australia 14 | 185.1 | M | SA | −29.38 | 138.03 | A29 | A179 | NW | d22 | H1*/H2*‐6q | MF774087, MF774096 |
| Dubbo | 146.1r | M | NSW | −32.151371 | 148.847676 | A29 | A199 | NW | d8 | H3‐k9 | MF774090, MF774099 |
| Inglewood | X2654 | F | QLD | −28.41 | 151.08 | — | — | NW | d16 | JX094432, JX094433 | |
| Moree | X920 | M | NSW | −29.4735 | 149.4685 | A29 | A208 | SE | a10 | H1*/H2*‐6t | JX090188, JX090194 |
| Northeastern 1 | WD333 | F | QLD | −17.33 | 145.39 | A29 | A199 | NW | d16 | JX094432, JX094433 | |
| Northeastern 2 | WD386 | M | QLD | −12.71 | 143.28 | A29 | A207 | NW | d16 | H60‐n28 | JX094432, JX094433 |
| Northeastern 3 | WD402 | F | QLD | −20.06 | 146.269062 | A201 | din34 | NW | d16 | JX094432, JX094433 | |
| Northeastern 4 | X2159 | M | QLD | −16.2498 | 145.3214 | A29 | A207 | NW | d17 | H60‐n27 | JX090184, JX090190 |
| Northeastern 5 | X987 | F | QLD | −19.3017 | 146.7258 | A29 | A179 | NW | d16 | JX094432, JX094433 | |
| Northeastern 6 | X980 | F | QLD | −19.3117 | 146.7358 | A29 | A179 | NW | d16 | JX094432, JX094433 | |
| Northeastern 7 | X983 | F | QLD | −19.3217 | 146.7438 | A29 | A179 | NW | d16 | JX094432, JX094433 | |
| Northeastern 8 | X985 | M | QLD | −19.2228 | 146.44217 | A29 | A179 | NW | d16 | H60‐n24 | JX094432, JX094433 |
| Northeastern 10 | 157.1 | M | QLD | −20.737969 | 144.007975 | A29 | A199 | NW | d5 | H60‐9k | KC346411, KC346428 |
| Northeastern 11 | 219.2 | M | QLD | −16.40125 | 145.36163 | A29 | A199 | NW | d5 | H60‐n27 | KC346411, KC346428 |
| Northwestern 1 | 1.64 | M | WA | −26.468389 | 120.835219 | A200 | din36 | NW | d5 | H60‐n17 | KC346411, KC346428 |
| Northwestern 2 | 11.55 | M | WA | −27.414 | 122.36 | A29 | A29 | NW | d15 | H60‐n22 | KC346415, KC346432 |
| Northwestern 3 | 3.46 | M | WA | −15.061 | 125.541 | A29 | A29 | NW | d5 | H3‐12d | KC346411, KC346428 |
| Northwestern 4 | 4.53 | M | WA | −15.416 | 126.135 | A29 | A29 | NW | d19 | H3‐n21 | KC346423, KC346440 |
| Northwestern 5 | 4.54 | F | WA | −15.703 | 126.362 | A29 | A29 | NW | d10 | KC346417, KC346434 | |
| Northwestern 6 | 17.87 | M | WA | −16.809 | 125.712 | A202 | A202 | NW | d6 | H3‐n21(?) | KC346425. KC346442 |
| Northwestern 7 | 21.72 | F | WA | −22.33 | 122.0811 | A29 | A179 | NW | d13 | KC346421, KC346438 | |
| Northwestern 8 | 9.39 | F | WA | −25.725917 | 125.780583 | A200 | A200 | NW | d5 | KC346411, KC346428 | |
| Northwestern 9 | 24.94 | F | WA | −17.273 | 122.568 | A29 | A29 | NW | d15 | KC346415, KC346432 | |
| Northwestern 10 | 18.38 | F | WA | −25.08333 | 122.05 | A29 | A29 | NW | d12 | KC346426, KC346443 | |
| Northwestern 11 | 3.47 | M | WA | −14.853 | 125.97 | A29 | A29 | NW | d5 | H3‐n4 | KC346411, KC346428 |
| Northwestern 12 | 4.52 | F | WA | −15.796 | 126.372 | A29 | A29 | NW | d5 | KC346411, KC346428 | |
| Northwestern 13 | 7.36 | F | WA | −15.708944 | 126.203139 | din27 | din27 | NW | d18 | KC346418, KC346435, MF784878 | |
| Northwestern 14 | 21.23 | F | WA | −17.0307 | 125.4269 | A29 | A29 | NW | d5 | KC346411, KC346428 | |
| Northwestern 15 | DE11 | F | WA | −26.56755 | 122.8074 | A200 | A200 | NW | d5 | KC346411, KC346428 | |
| Northwestern 16 | 14.95 | F | WA | −13.96689 | 126.95647 | A29 | A29 | NW | d24 | KC346419, KC346436 | |
| Northwestern 17 | 3.48 | F | WA | −14.798 | 125.753 | A29 | A29 | NW | d5 | KC346411, KC346428 | |
| Northwestern 18 | 4.62 | M | WA | −15.333 | 126.416 | A29 | A29 | NW | d10 | H3‐n21 | KC346417, KC346434 |
| Northwestern 19 | N866 | M | WA | −17.076 | 128.203 | A202 | A202 | NW | d11 | H60‐k11 | MF774084, MF774093 |
| Northwestern 20 | X2581 | M | WA | −22.9366 | 118.9646 | A210 | din33 | NW | d5 | H60‐n25 | KC346411, KC346428 |
| Northwestern 21 | X2583 | M | WA | −22.8917 | 118.141892 | A203 | A203 | NW | d5 | H60‐n22 | KC346411, KC346428 |
| Northwestern 22 | X2601 | M | WA | −22.7264 | 119.06011 | A29 | A29 | NW | d5 | H60‐n25 | KC346411, KC346428 |
| Northwestern 23 | X3291 | M | WA | −21.65537 | 121.57327 | A29 | A29 | NW | d5 | H60‐k3 | KC346411, KC346428 |
| Southeastern 1 | X1267 | M | QLD | −21.99 | 148.03 | A213 | A213 | SE | a10 | H1*/H2*‐1c | JX090188, JX090194 |
| Southeastern 2 | X1273 | F | QLD | 27.0667 | 152.966 | A29 | A179 | SE | a10 | JX090188, JX090194 | |
| Southeastern 3 | X229 | F | NSW | −31.4468 | 152.723 | A29 | A29 | SE | a8 | JX090187, JX090193 | |
| Southeastern 4 | X1020 | M | ACT | −35.871771 | 148.99979 | A29 | A29 | SE | a2 | H1*/H2*‐6t | JX090186, JX090192 |
| Southeastern 5 | 127.1 | F | VIC | −37.1 | 147.417 | — | — | SE | a4 | KC346420, KC346437 | |
| Southeastern 6 | 96.4 | M | VIC | −37.22222 | 148.16168 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 7 | 184.3 | M | QLD | −25.515 | 153.123333 | A29 | A179 | SE | f2 | H60‐n25 | KC346410, KC346427 |
| Southeastern 8 | 144.8 | F | NSW | −35.77729 | 148.24837 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 9 | 144.9 | F | NSW | −35.77729 | 148.24837 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 10 | X874 | F | NSW | −35.745734 | 148.25977 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 11 | X931 | F | NSW | −35.8547 | 148.212694 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 12 | WD192 | F | NSW | −35.29261833 | 148.7790664 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 13 | X791 | F | NSW | −30.42866792 | 152.2332262 | A29 | A29 | SE | a11 | JX090185, JX090191 | |
| Southeastern 14 | 44.2 | F | ACT | −35.797513 | 148.913353 | A29 | A29 | SE | a3 | KC346422, KC346439 | |
| Southeastern 15 | 85.1 | F | VIC | −37.344511 | 147.9035156 | A29 | A29 | SE | a3 | KC346422, KC346439 | |
| Southeastern 16 | X296 | M | QLD | −25.4779854 | 153.0553244 | A29 | A179 | SE | f3 | H60‐n25 | MF774082, MF774091 |
| Southeastern 17 | 21.5 | F | QLD | −25.45 | 153.067 | A29 | A179 | SE | a1 | KC346413, KC346430 | |
| Southeastern 18 | 65.1 | F | VIC | −36.27755288 | 147.8651983 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 19 | 166.4 | F | VIC | −36.43853 | 147.96559 | A29 | A29 | SE | a2 | JX090186, JX090192 | |
| Southeastern 20 | X1006 | M | ACT | −35.365882 | 148.9296677 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 21 | X1012 | M | ACT | −35.885449 | 149.0373287 | A29 | A29 | SE | a3 | H1*/H2*‐6t | KC346422, KC346439 |
| Southeastern 22 | X1049 | M | ACT | −35.427117 | 148.8781602 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 23 | X1050 | M | ACT | −35.427108 | 148.87816 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 24 | X1062 | M | ACT | −35.362466 | 148.920683 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 25 | X2279 | M | ACT | −35.6406 | 148.9629667 | A29 | A29 | SE | a3 | H1*/H2*‐6t | KC346422, KC346439 |
| Southeastern 26 | 156.3 | M | QLD | −25.090277 | 148.767356 | A29 | A29 | SE | a7 | H60‐9k | MF774088, MF774097 |
| Southeastern 27 | W0143 | M | NSW | −33.29289321 | 151.1958311 | A29 | A29 | SE | a3 | H1*/H2*‐6t | KC346422, KC346439 |
| Southeastern 28 | W0144 | M | NSW | −33.29289321 | 151.1958311 | A29 | A29 | SE | a3 | H1*/H2*‐6t | KC346422, KC346439 |
| Southeastern 29 | W0151 | M | NSW | −36.90873 | 149.23903 | A29 | A179 | SE | a3 | H60‐n24 | KC346422, KC346439 |
| Southeastern 30 | WD036 | M | NSW | −35.777 | 148.011 | A29 | A29 | SE | a3 | H3‐k2 | KC346422, KC346439 |
| Southeastern 31 | X1311 | M | NSW | −31.31306346 | 152.9537903 | A29 | A29 | SE | a1 | H3‐k9 | KC346413, KC346430 |
| Southeastern 32 | X2256 | M | NSW | −35.904304 | 149.045024 | A29 | A29 | SE | a3 | H1*/H2*‐k1 | KC346422, KC346439 |
| Southeastern 33 | X2389 | M | NSW | −31.1867476 | 152.9664462 | A29 | A29 | SE | a1 | H3‐k9 | KC346413, KC346430 |
| Southeastern 34 | X2405 | M | NSW | −30.99843908 | 153.0199648 | A29 | A29 | SE | a1 | H3‐k9 | KC346413, KC346430 |
| Southeastern 35 | X2482 | M | NSW | −31.84246908 | 152.1376204 | A29 | A29 | SE | a6 | H1*/H2*‐6t | MF774089, MF774098 |
| Southeastern 36 | X2484 | M | NSW | −30.77870485 | 152.6794424 | A29 | A29 | SE | a1 | H60‐n25 | KC346413, KC346430 |
| Southeastern 37 | X2529 | M | NSW | −30.42666371 | 152.2269422 | A29 | A29 | SE | a1 | H1‐k6 | KC346413, KC346430 |
| Southeastern 38 | X2764 | M | NSW | −30.78488 | 151.15678 | A29 | A208 | SE | a5 | H3‐k9 | MF774085, MF774094 |
| Southeastern 39 | X2792 | M | NSW | −32.54665335 | 152.3076804 | A29 | A29 | SE | a1 | H60‐k4 | KC346413, KC346430 |
| Southeastern 40 | X2931 | M | NSW | −30.8945242 | 151.9407884 | A29 | A29 | SE | a1 | H1*/H2*‐6t | KC346413, KC346430 |
| Southeastern 41 | X3508 | M | NSW | −29.309439 | 152.142707 | A29 | A29 | SE | a1 | H1‐k6 | KC346413, KC346430 |
| Southeastern 42 | X580 | M | NSW | −35.804173 | 148.1267073 | A29 | A29 | SE | a3 | H21*‐7d | KC346422, KC346439 |
| Southeastern 43 | X606 | M | NSW | −35.88248 | 148.6595 | A29 | A29 | SE | a3 | H60‐k11 | KC346422, KC346439 |
| Southeastern 44 | 16.1 | M | VIC | −37.222881 | 147.531358 | A29 | A29 | — | — | H3‐k2 | |
| Southeastern 45 | X2070 | M | VIC | −36.486 | 146.93 | A29 | A29 | SE | a4 | H1*/H2*‐6t | KC346420, KC346437 |
| NGSD 1 | NGSD4 | F | Papua New Guinea | — | — | A79 | A79 | NGSD | ng1 | JX088674 | |
| NGSD 2 | NGSD2 | F | Papua New Guinea | — | — | A79 | A79 | NGSD | ng1 | KC346414, KC346431 | |
| NGSD 3 | NGSD3 | F | Papua New Guinea | — | — | A79 | A79 | NGSD | ng1 | KC346414, KC346431 | |
| NGSD 4 | NGSD6 | M | Papua New Guinea | — | — | A79 | A79 | NGSD | ng1 | H60‐k10 | KC346414, KC346431 |
| NGSD 5 | NGSD5 | M | Papua New Guinea | — | — | A79 | A79 | NGSD | ng1 | H60‐k10 | KC346414, KC346431 |
Blood and/or tissue samples were collected, and all dingoes were screened for genetic purity, using a microsatellite‐based assay for domestic dog introgression (Wilton, 2001; Wilton, Steward, & Zafiris, 1999). Only pure or genetically intact dingoes were allocated to this research project (Stephens, 2011; Wilton, 2001; Wilton et al., 1999).
2.2. Mitochondrial gene analysis
Mitochondrial and nuclear phylogenetic analyses found that there are at least two dingo lineages, with eight diagnostic mitochondrial nucleotide differences between them (SE and NW, Cairns & Wilton, 2016). Two mitochondrial DNA regions harboring diagnostic mutations and the mitochondrial control region were amplified and sequenced (Table 2). The two diagnostic regions were selected as they contained three of the eight differences between the SE and NW mitochondrial lineages (Cairns & Wilton, 2016). Nonrandom genetic sampling has the potential to overestimate a posteriori significance so care must be taken in interpreting results. The regions selected were 676 bp (positions 7,685–8,361 including a region of ATP6 and ATP8) and 1,028 bp (positions 14,098–15,126 including a region of cytochrome b) in length. The mitochondrial control region is 582 bp (incorporating nucleotide positions 15,458–16,039 as in Savolainen et al. (2004)).
Table 2.
PCR amplification primers and conditions for mitochondrial PCR amplification and sequencing of the dingo and NGSD
| Primer name | Sequence | Nucleotide position | Reference | |
|---|---|---|---|---|
| Diagnostic region pair 1 | G8_F | CCAATGATACTGAAGCTATG | 7,340 | Designed by KMC |
| G8_R | ATTTTAGCAGGTTTGGTTAT | 7,915 | ||
| Diagnostic region pair 2 | G13_R | CTAAAAGTCAGAATAGGCATT | 15,150 | Designed by KMC |
| P16_F | TTCAGAACAATCGCACAACC | 13,973 | Designed in Wilton Laba | |
| Control region | H15422 | CTCTTGCTCCACCATCAGC | 15,422 | Savolainen et al. (2004) |
| L16106 | AAACTATATGTCCTGAAACC | 16,106 |
Designed by M. Wong during his 2010 Honours Thesis (unpublished data) and supervised by AN Wilton KMC.
Qiagen DNeasy kits (Qiagen Sciences, Germantown, USA) were used to extract DNA, and mitochondrial loci were amplified using PCR (Table 2). Briefly, PCR reactions were carried out in 25 μl containing water, 5× Crimson polymerase buffer (New England Biolabs Inc., MA, USA), 1.5 mmol/L of MgCl2, 6.25 pmol of each primer, 7.5 mmol/L of dNTPs, 2.5 U of Taq DNA Polymerase (New England Biolabs Inc., MA, USA), and 20–50 ng of DNA template. All PCR reactions were cycled using the following thermal profile: 98°C for 2 min, 95°C for 3 min (add Taq polymerase), then 95°C for 15 s, 52°C for 1 min, 65°C for 1 min for 10 cycles, then 95°C for 15 s, 52°C for 1 min, and 65°C for 1 min (increase time by 5 s each cycle) for 25 cycles followed by 65°C for 10 min.
Prior to sequencing, PCR amplicons underwent purification by ExoSAP‐IT® (USB Amersham, Buckinghamshire, UK). Purified templates underwent Sanger sequencing using standard BigDye terminator v3.1 (Applied Biosystems Inc., Foster City, USA) technology. DNA sequence chromatograms were analyzed and aligned using Sequencher 5.1 (Gene Codes corp., Ann Arbor, USA).
2.3. Y chromosome gene analysis
The iPLEX Sequenom MassARRAY system (Sequenom Inc., San Diego, USA) was used to genotype 29 single nucleotide polymorphisms (SNPs) from the nonrecombining Y chromosome (NRY) region as described in Sacks et al. (2013). These 29 SNPs form a panel of markers enabling differentiation between most observed dog Y chromosome haplogroups (Ardalan et al., 2012; Brown et al., 2011; Ding et al., 2011; Natanaelsson et al., 2006; Sacks et al., 2013). As in Sacks et al. (2013), we use H1 to refer to H1*, H2*, and H1 haplotypes. Five dinucleotide repeat–single tandem repeats (STR) were also genotyped from the NRY region: 650‐79.2b, 650‐79.3b, 990‐35, MS34CA, and MS41B as previously described (Brown et al., 2011; Sacks et al., 2013).
2.4. Neutrality tests
To investigate whether the genetic variation present within the mitochondrial genome departs from the expectations of neutrality, Tajima's D, Fu and Li's F*, and Fu and Li's D* (Fu, 1997; Fu & Li, 1993; Tajima, 1989) statistics were calculated in DnaSP v 5.10.1 (Librado & Rozas, 2009). These statistics can be used to investigate the presence of demographic or selective pressures acting upon the molecular evolution of a DNA sequence. Significantly negative values indicate population expansion and/or purifying selection, whilst significantly positive values indicate balancing selection and/or a decrease in population size. Nonsignificant values indicate that the null hypothesis of neutrality cannot be rejected, that is, no indication of demographic or selective pressures. These neutrality statistics were calculated for all dingoes and then specific dingo populations separately.
2.5. Biogeographic analyses
Median spanning networks were calculated in Networks v4.6 (Bandelt, Forster, & Rohl, 1999; Forster et al., 2000) using the mitochondrial diagnostic region, mitochondrial control region, and Y chromosome datasets. As in Sacks et al. (2013), the median‐joining (MJ) algorithm with default settings was used (r = 2, ε = 0). Mitochondrial networks were created for the concatenated diagnostic region and control region separately. Control region data were analyzed separately to allow incorporation of and comparison to the existing dingo control region dataset (Oskarsson et al., 2011; Sacks et al., 2013; Savolainen et al., 2004). As the control region is not phylogenetically informative in dingoes, it was not included in the mitochondrial diagnostic region analysis (Cairns & Wilton, 2016). Mitochondrial networks are unrooted. Y chromosome networks were calculated using concatenated SNP and STR data. Y chromosome SNPs and STRs were weighted as described by Sacks et al. (2013). Briefly, STRs were weighted as: 650‐79.2b = 5, 650‐79.3b = 2, 990‐35 = 9, MS34CA = 6, MS41B = 1, and SNP loci = 90 (Brown et al., 2011; Sacks et al., 2013). Y networks were drawn using our collected data and an additional dataset including 112 Oceanic samples from Sacks et al. (2013).
To further investigate the relationship between the dingo and NGSD (Cairns & Wilton, 2016), we ran Bayesian phylogenetic analyses in Beast v1.7.5 (Drummond, Suchard, Xie, & Rambaut, 2012), allowing us to estimate the posterior probability value of the inferred relationship. Cairns and Wilton (2016) found that the posterior probability value was low (0.4), suggesting uncertainty regarding the position of the NGSD lineage within dingoes. The Bayesian analysis was conducted on a set of 124 dingoes plus 5 NGSD; three dingoes were excluded due to PCR amplification difficulties. Bayesian analyses were run in Beast v1.7.5 (Drummond, Suchard, Xie, & Rambaut, 2012) under a skyline coalescent model with a strict clock, substitution rate of 7.7027 × 10−8 mutations−1 site−1 year−1 with a standard deviation of 5.4848 × 10−9 (Brown & Yang, 2011; Cairns & Wilton, 2016). All runs were optimized for MCMC chain steps to ensure that the estimated sampling size of all variables was above 200 in Tracer 1.5 (Rambaut & Drummond, 2007). We sampled every 5,000 steps with a 10% burn‐in. The resulting maximum clade credibility tree was midpoint rooted.
The biogeographic distribution of each individual belonging to each mitochondrial or Y chromosome haplogroup was then plotted onto maps using the maps package (Brownrigg, Minka, Becker, & Wilks, 2014) in R, allowing visualization of the distribution of the mitochondrial and Y chromosome lineages across Australia. Simple contingency table analyses, comparing mitochondrial lineage (columns) and Y chromosome haplogroup (rows), were used to evaluate whether the distribution of Y chromosome haplogroups between the mitochondrial lineages was nonrandom.
To investigate the relationship of Y chromosome haplotypes found in dingoes and NGSD with those found in Island Southeast Asia, a network was calculated based upon data from 173 dingoes, 20 NGSD, and 79 Southeast Asian dogs, incorporating our dingo and NGSD dataset as well as the dataset from Sacks et al. (2013). The resulting network was color‐coded relative to geographical region.
2.6. Demographic analyses
To investigate historical patterns of demographic change in the dingo, Bayesian skyline plots were constructed in Tracer 1.5 (Rambaut & Drummond, 2007). Bayesian analyses were carried out in Beast v1.7.4 (Drummond et al., 2012) as detailed above. Skyline plots were constructed based upon the combined mitochondrial DNA dataset and each mitochondrial clade separately.
3. RESULTS
3.1. Neutrality tests
Tajima's D statistics were calculated for all dingoes as grouped by mitochondrial lineage using the mitochondrial diagnostic region (Table 3). Statistics could not be calculated for the NGSD as all individuals carried the same mitochondrial DNA sequence. The NW lineage statistics were found to be significantly negative, indicating the presence of purifying selection and/or population expansion. Statistics calculated for the SE lineage were negative but not significant.
Table 3.
Nucleotide variation and neutrality statistics on mitochondrial DNA (1,706 bp) from 124 dingoes
| π | θ | Hd | Tajima's D | Fu and Li's F* | Fu and Li's D* | |
|---|---|---|---|---|---|---|
| NW lineage | 8.9 × 10−4 | 3.70 × 10−3 | 0.77 | −2.43a | −4.58a | −4.52a |
| SE lineage | 1.08 × 10−3 | 1.40 × 10−3 | 0.79 | −0.65 | −1.91 | −2.10 |
p < .02.
3.2. Biogeographic analyses
When ignoring indels, we observed 12 mitochondrial control region (CR) haplotypes with three novel CR haplotypes in 124 dingoes (three dingoes were excluded due to PCR difficulties) and five NGSD (Table 1). The novel haplotypes (din31, din32, and din33) were found in 1–4 individuals and differed by 1–2 nucleotide substitutions from A29. One dingo carried the A9 haplotype thought to have arisen in dingoes independently from dogs (Savolainen et al., 2004). A single dingo out of 124 carried A17, a CR haplotype hypothesized to be introgressed from domestic dogs (Savolainen et al., 2004). Incorporating all the CR data from previously published studies into our own yielded a star‐shaped genetic network (Figure 3).
Figure 3.
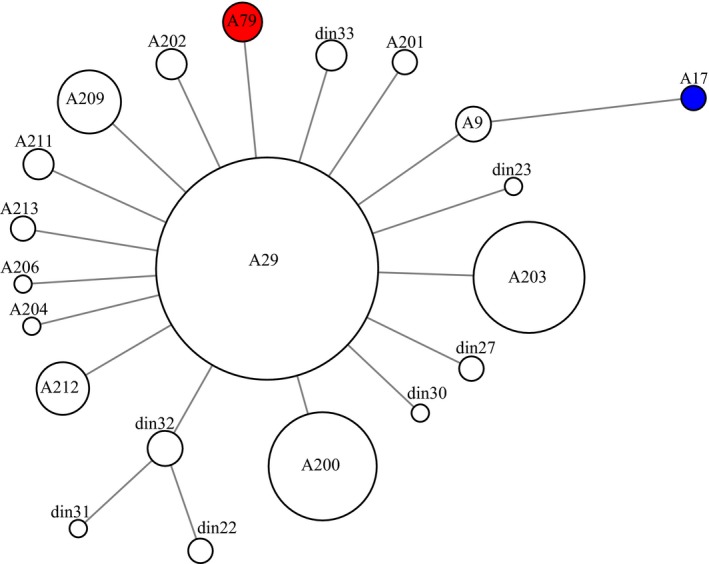
Median spanning network based on mitochondrial control region data from 450 dingoes and 23 NGSD. Red color indicates NGSD samples, whilst blue indicates a haplotype hypothesized to be introgressed from domestic dogs. Circles are proportional to the number of individuals carrying that haplotype. The network was calculated including 94 dingoes and 18 NGSD from Sacks et al. (2013) and 232 dingoes and three NGSD from Oskarsson et al. (2011)
A total of 39 mitochondrial diagnostic region haplotypes were observed in 124 dingoes and 5 NGSD (Table 1). As with the CR data, none was consistent with non‐dingo mitochondrial lineages. The 5 NGSD all carried the same haplotype. The mitochondrial diagnostic region network displays a more interesting pattern consistent with the presence of two mitochondrial haplogroups, SE and NW, in Australia (Figure 4). The genetic network also corroborated the close relationship between the SE lineage and NGSD. The biogeographic distribution of the two mitochondrial haplogroups across Australia was plotted, indicating strong geographic subdivision with limited mixing between the two populations (Figure 5). The SE mitochondrial lineage was restricted to Fraser Island and the southeastern coastal region of Australia (Queensland, New South Wales, Australian Capital Territory and Victoria), whilst the NW mitochondrial lineage was widespread from Western Australia to northern/central Queensland and south into South Australia. A single NW mitochondrial lineage individual was observed within the Australian Alpine region. Within the captive dingo population both NW and SE haplogroups were observed. Captive animals were not plotted on the map.
Figure 4.
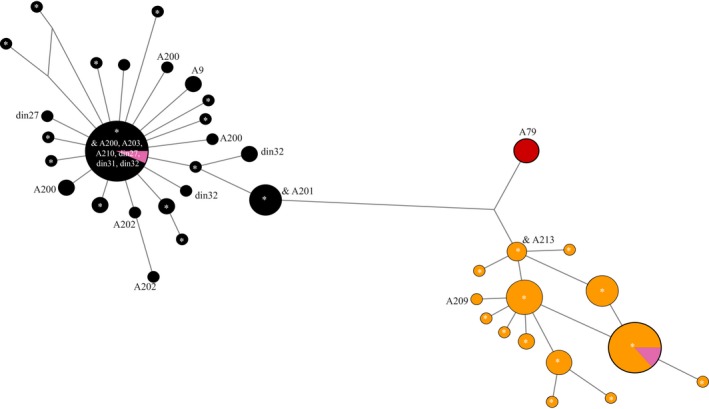
Median spanning network based upon the mitochondrial diagnostic region (1,706 bp) in 124 dingoes and five NGSD. Black coloration indicates NW lineage haplotypes, orange SE lineage haplotypes, red NGSD haplotypes, and pink captive individuals. Branch lengths are relative to the number of mutations separating mitochondrial haplotypes. Mitochondrial control region haplotypes are shown with A29 depicted as * and less common haplotypes as text
Figure 5.
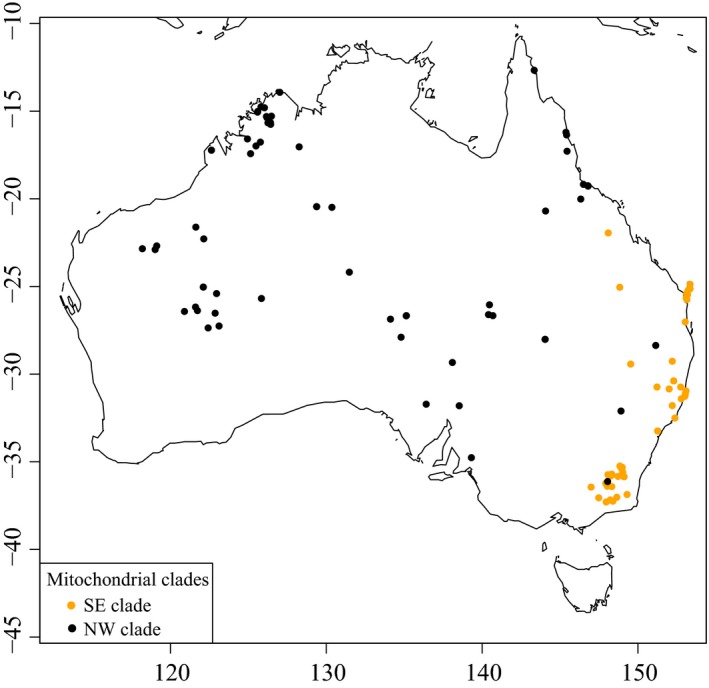
Biogeographical map of 120 dingoes and their mitochondrial lineage designation. Black circles indicate NW lineage haplotypes and orange SE lineage haplotypes. Only wild dingoes were plotted onto the map
To further investigate the relationship between the dingo and NGSD, a Bayesian analysis was conducted on the combined sample of 129 animals. This included 124 dingoes and five NGSD (Table 1). This analysis corroborated the whole mitochondrial genome Bayesian phylogenetic analyses suggesting that the NGSD is more closely related to the SE dingo lineage than the NW lineage (Cairns & Wilton, 2016), with an increased posterior probability node support of 0.84 (Figure 6).
Figure 6.
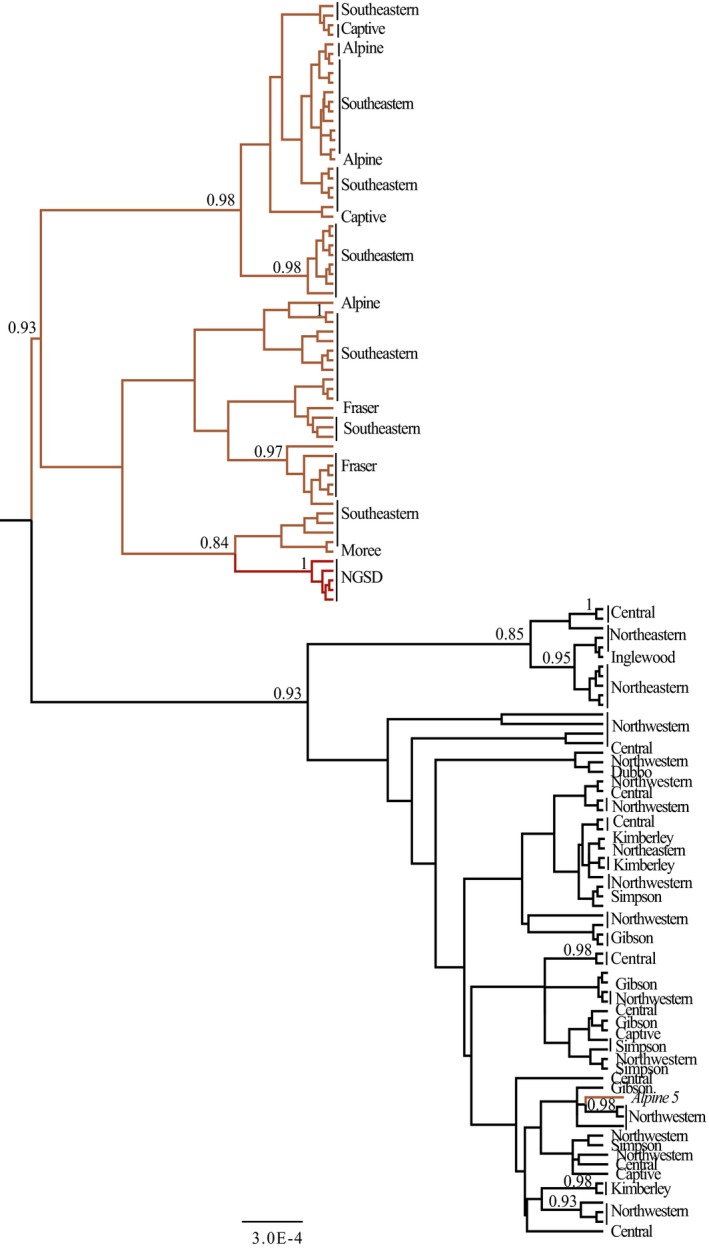
Bayesian analysis of 124 dingo and five NGSD mitochondrial diagnostic region (1,706 bp) sequences. Analyses constructed in BEAST v1.7.5 (Drummond et al., 2012) using a GTR + G + I substitution model and a constant population size coalescent model. Integers below nodes are posterior probability values and values less than 0.6 are not shown. Colors represent geographical sampling population, black for NW, orange for SE, and red for NGSD. The scale bar indicates an estimate of the average per site substitutions between two nodes
We observed 30 Y chromosome haplotypes in our dataset of 79 dingoes and two NGSD (Table 1). Y chromosome network analysis identified three main haplogroups present within dingoes and NGSD, H1, H3, and H60 (Figure 7). A contingency table analysis, with two columns (mitochondrial lineage) and three rows (Y chromosome haplogroup), suggests that the distribution of Y chromosome haplogroups between the mitochondrial lineages was nonrandom in dingoes (χ 2 = 18.1, df = 2, p < .001). To further investigate the distribution of the Y chromosome haplogroups across Australia, we incorporated the dingo and NGSD data from Sacks et al. (2013) resulting in a total of 194 samples (Figure 8). Within the combined dataset representing 173 male dingoes and 20 NGSD, we observed an additional 6 Y chromosome SNP‐STR haplotypes (Figure 8). The 20 NGSD sampled between the two datasets all carried H60 haplotypes.
Figure 7.
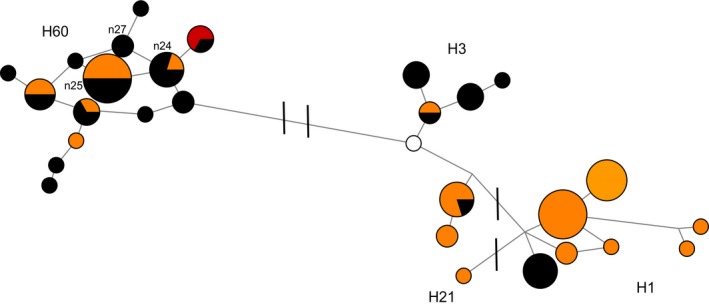
Median spanning network based upon Y chromosome SNP and STR haplotypes for 79 dingoes and two NGSD. Black coloration indicates NW mitochondrial lineage individuals, orange SE mitochondrial lineage individuals, red NGSD individuals, and white unknown mitochondrial lineage. Strokes across branches indicate the presence of Y chromosome SNP mutations differentiating between Y chromosome haplogroups. Branch lengths are relative to the number of STR mutations between Y chromosome haplotypes
Figure 8.
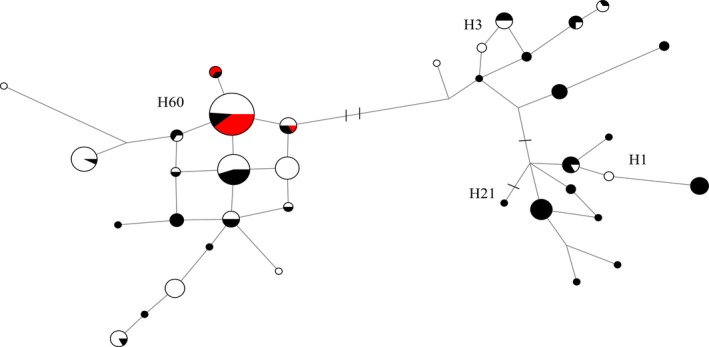
Median spanning network based upon Y chromosome SNP and STR haplotypes for 173 dingoes and 20 NGSD. Black coloration indicates dingoes from this study, red indicates NGSD individuals, and white indicates dingo samples from Sacks et al. (2013). Strokes across branches indicate the presence of Y chromosome SNP mutations differentiating between Y chromosome haplogroups. Branch lengths are relative to the number of STR mutations between Y chromosome haplotypes
When Y chromosome haplogroup information was plotted on a map (Figure 9), we observed that H1 was largely restricted to the southeastern region of Australia, H3 was restricted to the southeastern and Kimberley regions, and H60 was predominantly found throughout northern, Western, and central Australia. Of the four H3 haplogroup alleles observed in the Kimberley region, all were endemic except H3_12d, which was also observed in southeastern Australia.
Figure 9.
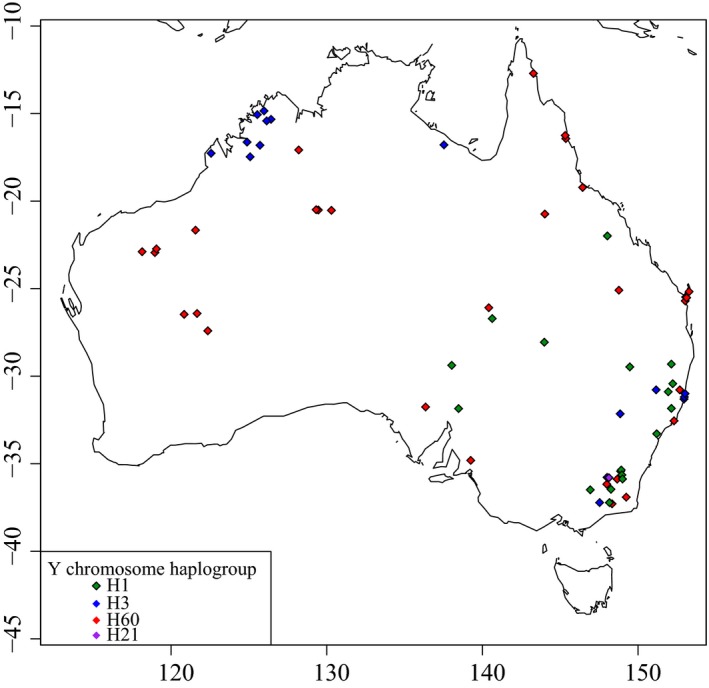
Biogeographical map of 169 dingoes and their Y chromosome haplogroup designation. Coloration indicates Y chromosome haplogroup; red for H60, blue for H3, purple for H21, and green for H1. Only wild dingoes were plotted onto the map
To investigate the relationship between Y chromosome haplotypes observed in dingoes and NGSD and Southeast Asian dogs, a MJ network was calculated based upon a combined dataset incorporating a total of 272 samples. This comprised 173 dingoes, 79 from our dataset and 94 from Sacks et al. (2013); 20 NGSD, two from our dataset and 18 from Sacks et al. (2013); and 79 Southeast Asian dogs from Sacks et al. (2013) (Figure 10). We observed that the H1 and H3 haplotypes found in dingoes were largely unique (not shared with SEA dogs). Further investigation, however, suggests that H1‐1c, H1‐6t, H1‐n7, and H1‐6q were observed in European domestic dog breeds (Brown et al., 2011; Sacks et al., 2013). Three novel H1 haplotypes were observed (H1‐k1, H1‐k7, and H1‐k5) that were unique to dingoes.
Figure 10.
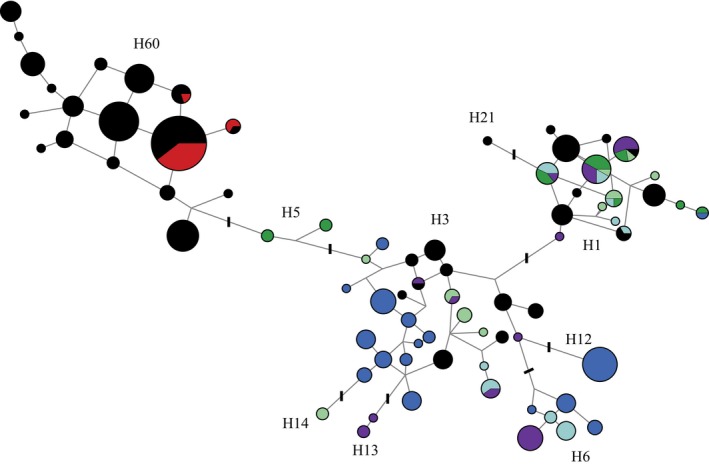
Median spanning network based upon Y chromosome SNP and STR haplotypes for 173 dingoes, 20 NGSD and 79 Southeast Asian dogs. Coloration indicates geographical region/species type: black for dingoes, red for NGSD individuals, dark green for Taiwan, purple for Thailand, light blue for Brunei, light green for Philippines, and dark blue for Bali. Strokes across branches indicate the presence of Y chromosome SNP mutations differentiating between Y chromosome haplogroups. Branch lengths are relative to the number of STR mutations between Y chromosome haplotypes
3.3. Demographic analyses
Bayesian skyline plots constructed on the combined dingo mitochondrial dataset indicate that the population was stable until approximately 5,000 years ago when it began increasing steadily (Figure 11). There was some evidence of a small decline in dingo numbers in the last 200 years. Skyline plots modeled for the individual mitochondrial clades separately suggest differences in demographic histories. The SE clade plot indicates a historically stable population size, which began increasing rapidly in the last 1,000–2,000 years (Figure 12). The NW clade plot depicts a more gradual population increase from about 6,000 years ago stabilizing approximately 3,000–4,000 years ago (Figure 12).
Figure 11.
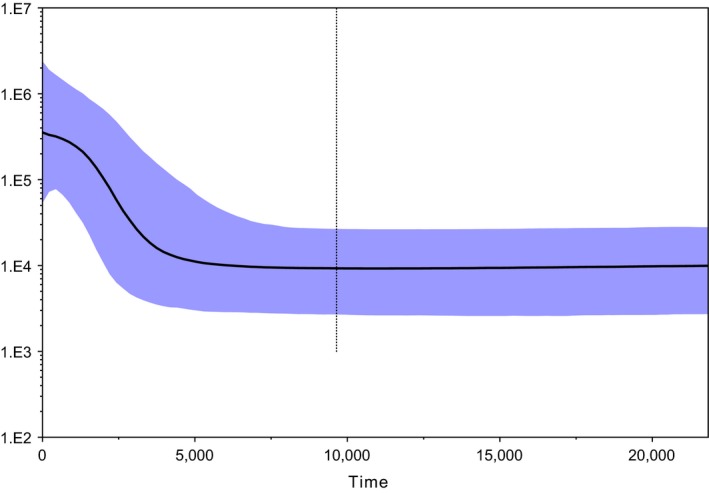
Bayesian skyline plot built using mtDNA diagnostic region (1,706 bp) sequences from 124 dingoes. Analyses constructed using a GTR + G + I substitution model and a skyline coalescent model in BEAST v1.7.5 (Drummond et al., 2012). A strict clock was enforced with a substitution rate of 7.7027 × 10−8 mutations−1 site−1 year−1 with a standard deviation of 5.4848 × 10−9. The skyline plot was constructed in Tracer 1.5 (Rambaut & Drummond, 2007)
Figure 12.
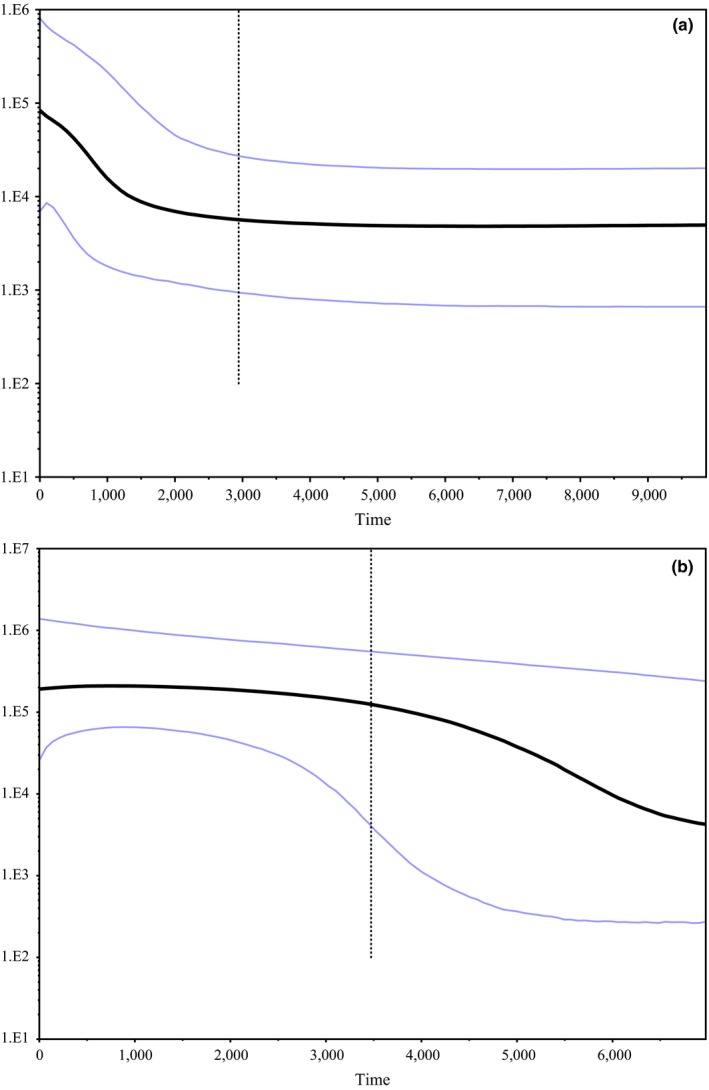
Bayesian skyline plots based on modeling on (a) 57 SE lineage or (b) 67 NW lineage dingoes using mitochondrial diagnostic region (1,706 bp) sequences. Analyses constructed using a GTR + G + I substitution model and a skyline coalescent model in BEAST v1.7.5 (Drummond et al., 2012). A strict clock was enforced with a substitution rate of 7.7027 × 10−8 mutations−1 site−1 year−1 with a standard deviation of 5.4848 × 10−9. The skyline plots were constructed in Tracer 1.5 (Rambaut & Drummond, 2007)
4. DISCUSSION
Understanding the ecological roles of apex predators often comes after their populations have declined to endangered levels, necessitating precautionary management (Estes et al., 2011; Ripple et al., 2014). In the case of the dingo, the findings documented here suggest the potential for sufficiently long‐standing population structure to support management for multiple locally adapted populations. Understanding the population biology, demography, and biogeography of dingoes across Australia is central to the question of how best to manage and conserve them, whilst limiting hybridization.
4.1. Multiple immigrations—different evolutionary lineages
Mitochondrial and Y chromosome data corroborate the presence of at least two discrete populations of dingo, NW (H60/H3), and SE (H3/H1) (Ardalan et al., 2012; Cairns & Wilton, 2016; Sacks et al., 2013). A lack of intermediate haplotypes despite broad geographical sampling suggests that the observed pattern of population structure is due to historical events. Previous studies did not observe the presence of population structure in dingoes, due to restricted sampling of the mitochondrial control region and limited geographic sampling across the Australian continent (Oskarsson et al., 2011; Sacks et al., 2013; Savolainen et al., 2004).
Our data suggest that the two divergent Y chromosome lineages observed in dingoes have different geographical origins and are plausibly the result of multiple immigrations into Australia, as postulated by Cairns and Wilton (2016). Notably, the Y haplogroups H3 and H60, which are both observed in dingoes, are not immediately related (Figure 10; Natanaelsson et al., 2006; Brown et al., 2011; Ardalan et al., 2012; Sacks et al., 2013). The H3 Y chromosome haplogroup is also observed in Southeast Asia. However, seven of the eight haplotypes observed in dingoes were endemic to Australia, indicating shared ancestry with a history of isolation between dingoes and Southeast Asian dogs (Figure 10; Brown et al., 2011; Sacks et al., 2013). On the other hand, the H60 haplogroup is unique to dingoes and NGSD and most closely related to H5, a haplogroup found in Taiwan (Figure 10; Brown et al., 2011; Ardalan et al., 2012; Sacks et al., 2013). The distribution of Y chromosome haplotypes between the two mitochondrial lineages is nonrandom, corroborating the presence of strong geographic subdivision in dingoes (Ardalan et al., 2012; Cairns & Wilton, 2016).
These data have intriguing implications for the movements of canids, and presumably humans, in Australasia. Cairns and Wilton (2016) postulate that dingoes immigrated into Australia via the now flooded land bridge between Papua New Guinea and Australia. Indeed mitochondrial data suggest that SE dingoes and the NGSD are closely related (Figures 4 and 6 and Cairns & Wilton, 2016). Y chromosome data, however, suggest that the NGSD share a closer paternal relationship with the NW lineage (Figures 7, 8, and 10; Sacks et al., 2013). Conflicting maternal and paternal histories mean the exact relationship between NGSD and the two dingo populations is uncertain. However, the close relationship does indicate that dingoes likely arrived in Australia through Papua New Guinea. Furthermore, the disparate geographical origins of the two Y chromosome haplogroups support the hypothesis that dingoes immigrated into Australia twice. A single homogeneous introduction, as suggested by Savolainen et al. (2004), is unlikely given the strong biogeographical subdivision at maternal and paternal markers and the divergent evolutionary relationships between the two populations. This suggests that in Southeast Asia and Oceania, the movements of dogs, and presumably humans, are much more complex than assumed. Indeed, genetic research finds little evidence of Neolithic or Austronesian gene flow into Australia (Bergström et al., 2016; Haak et al., 2010; van Holst Pellekaan, 2013; Karafet et al., 2005; McEvoy et al., 2010; Rasmussen et al., 2011). Intriguingly, human mitochondrial research found a pattern of continuous strong geographic subdivision dating back to approximately 50,000 years BP, after Australians first spread into the continent, with little evidence of migration between populations (Tobler et al., 2017).
4.2. Dating, demography, and dispersal
Demographic modeling and neutrality test results based on mitochondrial data should be treated cautiously but can give insight into modern and historical demographic patterns. Bayesian skyline plots based upon the individual mitochondrial lineages suggest that the SE population size has been stable until about 1,000 years ago, when it underwent rapid expansion (Figure 12). The NW population on the other hand has a history of gradual population expansion from approximately 4,000–6,000 years ago (Figure 12). Possibly Bayesian demographic modeling is reflective of rapid range expansion of dingoes in southeastern Australia following the decline of thylacines on the mainland, which occurred approximately 2,000 years BP (Figure 12; Johnson & Wroe, 2003; Fillios et al., 2012; Letnic et al., 2012; Prowse et al., 2014). It is also possible that the pattern of population expansion in SE dingoes is the result of extensive culling and baiting practices in southeastern Australia within the last 200 years (Fleming, Corbett, Harden, & Thomson, 2001; Wallach, Ritchie, Read, & O'Neill, 2009). The pattern of population expansion observed in the NW dingo population could be the result of long‐term but gradual range expansion after immigration into Australia. Demographic modeling on the entire dingo dataset depicts a population expansion approximately 3,000–8,000 years BP (Figure 11). Ethnographic and molecular dating suggests dingoes arrived in Australia prior to 5,000 years BP (Cairns & Wilton, 2016; Fillios & Taçon, 2016; Oskarsson et al., 2011). It should be noted that uncertainty in the demographic modeling makes it difficult to discern the approximate arrival time of dingoes or pinpoint when range expansions occurred.
Biogeographic patterns within Australia provide insight into the modern dispersal and migration of dingoes. We observed that the geographical distribution of the two mitochondrial lineages, SE and NW, exhibits strong geographical subdivision (Figure 5). Only a single instance of discordance between mitochondrial lineage and geographic origin was observed, indicating that maternal migration is limited. The geographical distribution of three Y chromosome haplogroups, H1, H3, and H60, is similar to that of the mitochondrial lineages, but more diffuse, suggesting higher levels of paternal than maternal migration (Figures 5 and 9). Introgression between the NW and SE populations seems to be west to east biased, with few H3 haplogroup individuals found in northern, Western, or central Australia. Conversely, there are some individuals in southeastern Australia harboring H60 haplogroup types, either the result of male dispersal from the NW population into the SE population or historical distribution patterns. These patterns are likely a factor of male dispersal; male dingoes and dogs range more widely and are more likely to disperse to new areas (Pal, Ghosh, & Roy, 1998; Thomson, Rose, & Kok, 1992). Human‐mediated dispersal may also be a factor in facilitating the movement of dingoes, by breaking apart pack structures through culling/baiting management practices (Corbett, 1988; Fleming et al., 2006; Glen, Dickman, Soulè, & Mackey, 2007; Thomson, 1992; Wallach, Johnson, Ritchie, & O'Neill, 2010; Wallach et al., 2009).
Contrary to demographic modeling, the neutrality test results indicate that the two dingo populations may be experiencing different demographic and/or selective pressures (Table 3 and Cairns & Wilton, 2016). These data are consistent with mitochondrial network analyses depicting a complex pattern in the SE population but a more star‐like pattern in the NW population indicative of population expansion (Figures 3 and 4). There is also evidence of a west to east biased dispersal pattern which might be the result of NW population dingoes moving into vacant niches opened up by extensive culling and baiting practices in southeastern Australia (Figure 5).
4.3. Introgression from modern domestic dogs
The H1 Y chromosome haplogroup is considered to be a European domestic dog haplogroup (Ardalan et al., 2012; Brown et al., 2011; Ding et al., 2011; Sacks et al., 2013), and it is often observed in domestic dog breeds or Southeast Asian dogs thought to have breed ancestry. The observation of H1 haplotypes within dingoes is suggestive of paternal introgression from European domestic dogs into dingoes (Figures 7, 8, and 10). The presence of the H1 haplogroup in genetically tested “pure” dingoes suggests that there has likely been historical, post‐European colonization, introgression from domestic dogs into dingoes. It is unlikely that the introgression is modern because the genetic test is capable of detecting hybridization events on a recent timescale (Cairns, Wilton, & Ballard, 2011; Wilton, 2001; Wilton et al., 1999). The uniparental inheritance of the Y chromosome means that a single hybridization event will be reflected in the paternal lineage of a dingo despite extensive backcrossing. The lack of non‐dingo‐like mitochondrial lineages suggests that the introgression from domestic dogs is predominately due to male domestic dogs mating with female dingoes.
The distribution of the H1 haplogroup in southeastern Australia further suggests it is likely the result of introgression (Figure 9). First, domestic dogs have been present in southeastern Australia for a longer period of time, having arrived with European colonists in 1788, allowing for a longer period of sympatry with dingoes (Corbett, 2001). Second, southeastern Australia has the densest human population, and areas with dense human populations are generally associated with higher incidences of hybridization (Stephens, 2011; Stephens et al., 2015). Thirdly, lethal management strategies such as baiting and culling are widespread in southeastern Australia due to the sheep industry (Fleming et al., 2001). Fatal management strategies are believed to lead to increased levels of hybridization due to the breakdown of pack structure (Fleming et al., 2006; Wallach et al., 2009). This finding is a significant conservation concern in the context of the genetic identity and integrity of the SE dingo population, which is under threat of extinction through introgression and ecological exclusion through lethal management programs.
4.4. Conservation implications
The most important implication of these data is that conservation and management efforts should be focused on maintaining the existing dingo population structure and thus treating the two populations as distinct conservation units. Care should be taken not to deliberately translocate individuals between wild populations. Captive breeding programs may need to ensure that the two dingo populations are maintained separately, with mitochondrial and Y chromosome testing used to identify population ancestry. Fraser Island dingoes appear to share NW paternal lineage ancestry but carry SE mitochondrial types; if genetic rescue is attempted for this inbred island population, then individuals from appropriate genetic lineages should be located.
Conservation groups have long described the presence of multiple morphological varieties of dingo, generally alpine, desert, and tropical; however, it is not clear whether this phenotypic variation is associated with the genetic subdivisions or phenotypic plasticity, although the boundaries do overlap. A future study of morphological and phenotypic variation as well as genetic variation may help answer this question. It is possible that the two dingo populations have different ecological or biological characteristics relevant to the conservation and management of the species or its role in specific ecosystems. Patterns of genetic subdivision in other large carnivores have been linked to ecologically relevant characteristics such as neonatal dispersal (Sacks, Bannasch, Chomel, & Ernest, 2008; Sacks, Brown, & Ernest, 2004), prey specialization (Carmichael, Nagy, Larter, & Strobeck, 2001), environmental climes (Carmichael et al., 2001; Rueness, Jorde et al., 2003; Rueness, Stenseth et al., 2003; Stenseth et al., 2004), and sociality (Randall, Pollinger, Argaw, Macdonald, & Wayne, 2010). This is a key knowledge gap, which needs to be interrogated by future ecological research.
The presence of the H1 haplogroup in southeastern Australia has important implications for conservation and future management strategies; namely, it highlights the importance of inhibiting further hybridization. Neutering male dogs and/or restricting them from reproducing with wild dingoes may help achieve this. Particularly, best practice should dictate that pet, livestock guardian, or working dogs in rural localities should be neutered or chemically castrated to avoid further risk of hybridization. Widespread lethal control measures are shown to also facilitate hybridization by breaking apart pack structures (Wallach et al., 2009). Alternative livestock protection measures should be explored, such as livestock guardians and improved dog‐proof fencing (van Bommel & Johnson, 2012; Fleming et al., 2001). This observation also suggests the need for a higher accuracy “next generation” DNA test for distinguishing dingoes from hybrids; the current method is likely sufficient for monitoring wild populations but not for captive breeding programs.
Knowledge concerning levels of genetic integrity in wild populations is necessary to inform management and conservation programs. The southeastern population of dingoes is under particular extinction pressure from both fatal management strategies and hybridization; steps should be taken to preserve this population before it is too late. A broad genetic survey of dingoes in National Parks and State Forests in southeastern states would be needed to pinpoint high dingo ancestry populations and thus where to focus conservation efforts. Currently state and federal legislation do not protect the dingo sufficiently and allow widespread fatal control measures (Davis, 2001; Downward & Bromell, 1990; Fleming et al., 2001, 2006). Revision of legislation must be achieved to reflect the ecological, cultural, and taxonomic importance of the dingo, balancing the need to conserve this enigmatic canine with any agricultural concerns.
5. CONCLUSIONS
Distinct populations of apex consumers can exhibit different behaviors and prey on disparate trophic niches (Paiva, Fagundes, Romão, Gouveia, & Ramos, 2016). These differences could be due to ecological plasticity or genetically inherited differences. This study corroborates the presence of at least two dingo populations in Australia. It is plausible, given the divergent evolutionary histories of these populations that they are the result of multiple immigrations into Australia via the now flooded land bridge between Papua New Guinea and Australia. The two dingo populations are geographically subdivided, with one restricted to the southeast of Australia and the other widespread across central, northern, and Western Australia. Furthermore, there are differences in male and female dispersal as evidenced by rare matrilineal migration and more diffuse patrilineal subdivision. Demographic modeling suggests that the SE population of dingoes may have expanded either in response to the extinction of thylacines on the mainland or due to widespread lethal control management in the last 1,000 years. In contrast, the NW population appears to have been gradually expanding since the dingoes’ arrival. Further research into historical demographic patterns may help inform hypotheses concerning the arrival and spread of dingoes into Australia. There is evidence of historical, post‐European colonization, paternal introgression from domestic dogs into the SE dingo population. Conservation, management, and legislative practices need to be revised to reflect the presence of two dingo populations and to limit further hybridization particularly in the SE population.
AUTHOR CONTRIBUTIONS
KMC completed experimental design, mitochondrial DNA data collection, analysis, and writing of manuscript. BNS and SKB provided advice concerning experimental design and analysis, collected the Y chromosome data, and edited the manuscript text. JWOB provided advice and comments concerning experimental design and manuscript text.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
ACKNOWLEDGMENTS
Thanks must be directed to Dr Paul Waters (UNSW), Dr Carolina Correa (UNSW), and Dr Barbara Zangerl (UNSW) for providing valuable and constructive comments on the manuscript. Dr Danielle Stephens (UWA) and Dr Alan N Wilton (deceased, UNSW) provided access to dingo samples for this study. As well, appreciation must go to the large number of conservation organizations, dingo trappers, land managers, and government agencies for contributing dingo samples to genetic research projects on the dingo. Janice Koler‐Matznick and other NGSD conservationists in North America supplied the NGSD samples for this project. This work was supported by a Hermon Slade Foundation Research grant (HSF11/6) to KMC, ANW, and JWOB. Australian Research Council grant DP150102038, awarded to JWOB and Claire Wade, supported the publication of this work.
Cairns KM, Brown SK, Sacks BN, Ballard JWO. Conservation implications for dingoes from the maternal and paternal genome: Multiple populations, dog introgression, and demography. Ecol Evol. 2017;7:9787–9807. https://doi.org/10.1002/ece3.3487
REFERENCES
- Ardalan, A. , Oskarsson, M. , Natanaelsson, C. , Wilton, A. , Ahmadian, A. , & Savolainen, P. (2012). Narrow genetic basis for the Australian dingo confirmed through analysis of paternal ancestry. Genetica, 140, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt, M. , Cairns, K. M. , Ballard, J. W. O. , Savolainen, P. , & Axelsson, E. (2016). Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity, 117, 301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt, H. , Forster, P. , & Rohl, A. (1999). Median‐joining phylogenies for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37. [DOI] [PubMed] [Google Scholar]
- Bergström, A. , Nagle, N. , Chen, Y. , McCarthy, S. , Pollard, M. O. , Ayub, Q. , … Tyler‐Smith, C. (2016). Deep roots for aboriginal Australian Y chromosomes. Current Biology, 26, 809–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, P. (2013). Palaeoanthropology: Of humans, dogs and tiny tools. Nature, 494, 316–317. [DOI] [PubMed] [Google Scholar]
- Brown, S. K. , Pedersen, N. C. , Jafarishorijeh, S. , Bannasch, D. L. , Ahrens, K. D. , Wu, J.‐T. , … Sacks, B. N. (2011). Phylogenetic distinctiveness of middle eastern and southeast asian village dog Y chromosomes illuminates dog origins. PLoS ONE, 6, e28496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R. P. , & Yang, Z. (2011). Rate variation and estimation of divergence times using strict and relaxed clocks. BMC Evolutionary Biology, 11, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownrigg, R. , Minka, T. P. , Becker, R. A. , & Wilks, A. R. (2014). Maps: Draw geographical maps. [Google Scholar]
- Cairns, K. M. , & Wilton, A. N. (2016). New insights on the history of canids in Oceania based on mitochondrial and nuclear data. Genetica, 144, 553–565. [DOI] [PubMed] [Google Scholar]
- Cairns, K. M. , Wilton, A. N. , & Ballard, J. W. O. (2011). The identification of dingoes in a background of hybrids In Urbano K. V. (Ed.), Advances in genetics research (pp. 309–327). New York, NY: Nova Science Publishers. [Google Scholar]
- Carmichael, L. E. , Nagy, J. A. , Larter, N. C. , & Strobeck, C. (2001). Prey specialization may influence patterns of gene flow in wolves of the Canadian northwest. Molecular Ecology, 10, 2787–2798. [DOI] [PubMed] [Google Scholar]
- Corbett, L. K. (1988). Social dynamics of a captive dingo pack: Population regulation by dominant female infanticide. Ethology, 78, 177–198. [Google Scholar]
- Corbett, L. K. (2001). The dingo in Australia and Asia. Sydney, NSW: University of NSW Press. [Google Scholar]
- Davis, E. (2001). Legislative issues relating to control of dingoes and other wild dogs in new south wales. 1. Approaches to future management In Dickman C. R., & Lunney D. (Eds.), A symposium on the dingo (pp. 39–41). Sydney, NSW: Royal Zoological Society of New South Wales. [Google Scholar]
- Ding, Z. L. , Oskarsson, M. , Ardalan, A. , Angleby, H. , Dahlgren, L. G. , Tepeli, C. , … Zhang, Y. P. (2011). Origins of domestic dog in southern East Asia is supported by analysis of Y‐chromosome DNA. Heredity, 108, 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward, R. J. , & Bromell, J. E. (1990). The development of a policy for the management of dingo populations in South Australia. Proceedings of the Fourteenth Vertebrate Pest Conference. Paper 23.
- Drummond, A. J. , Suchard, M. A. , Xie, D. , & Rambaut, A. (2012). Bayesian phylogenetics with beauti and the BEAST 1.7. Molecular Biology and Evolution, 29, 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes, J. A. , Terborgh, J. , Brashares, J. S. , Power, M. E. , Berger, J. , Bond, W. J. , … Wardle, D. A. (2011). Trophic downgrading of planet earth. Science, 333, 301. [DOI] [PubMed] [Google Scholar]
- Fillios, M. , Crowther, M. S. , & Letnic, M. (2012). The impact of the dingo on the thylacine in Holocene Australia. World Archaeology, 44, 118–134. [Google Scholar]
- Fillios, M. A. , & Taçon, P. S. C. (2016). Who let the dogs in? A review of the recent genetic evidence for the introduction of the dingo to Australia and implications for the movement of people. Journal of Archaeological Science: Reports, 7, 782–792. [Google Scholar]
- Fleming, P. J. S. , Allen, L. R. , Lapidge, S. J. , Robley, A. , Saunders, G. R. , & Thomson, P. C. (2006). A strategic approach to mitigating the impacts of wild canids: Proposed activities of the invasive animals cooperative research centre. Australian Journal of Experimental Agriculture, 46, 753–762. [Google Scholar]
- Fleming, P. , Corbett, L. K. , Harden, R. , & Thomson, P. (2001). Managing the impacts of dingoes and other wild dogs. Canberra, ACT: Bureau of Rural Sciences. [Google Scholar]
- Forster, P. , Röhl, A. , Lünnemann, P. , Brinkmann, C. , Zerjal, T. , Tyler‐Smith, C. , … Brinkmann, B. (2000). A short tandem repeat–based phylogeny for the human Y chromosome. American Journal of Human Genetics, 67, 182–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, A. H. , Gronau, I. , Schweizer, R. M. , Ortega‐Del Vecchyo, D. , Han, E. , Silva, P. M. , … Novembre, J. (2014). Genome sequencing highlights the dynamic early history of dogs. PLoS Genetics, 10, e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretwell, S. D. (1987). Food chain dynamics: The central theory of ecology? Oikos, 50, 291–301. [Google Scholar]
- Fu, Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, 147, 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X. , & Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics, 133, 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick, R. C. , Benavides, E. , Russello, M. A. , Gibbs, J. P. , Poulakakis, N. , Dion, K. B. , … Caccone, A. (2012). Genetic rediscovery of an ‘extinct’ galápagos giant tortoise species. Current Biology, 22, R10–R11. [DOI] [PubMed] [Google Scholar]
- Glen, A. S. , Dickman, C. R. , Soulè, M. E. , & Mackey, B. G. (2007). Evaluating the role of the dingo as a trophic regulator in Australian ecosystems. Austral Ecology, 32, 492–501. [Google Scholar]
- Gollan, K. (1984). The Australian dingo: In the shadow of man In Archer M., & Clayton G. (Eds.), Vertebrate zoogeography & evolution in Australasia. Carlisle, WA: Hesperian Press. [Google Scholar]
- Haak, W. , Balanovsky, O. , Sanchez, J. J. , Koshel, S. , Zaporozhchenko, V. , Adler, C. J. , … the Genographic Consortium (2010). Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biology, 8, e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hairston, N. G. , Smith, F. E. , & Slobodkin, L. B. (1960). Community structure, population control, and competition. The American Naturalist, 94, 421–425. [Google Scholar]
- Halbert, N. D. , & Derr, J. N. (2007). A comprehensive evaluation of cattle introgression into US federal bison herds. Journal of Heredity, 98, 1–12. [DOI] [PubMed] [Google Scholar]
- Hertwig, S. T. , Schweizer, M. , Stepanow, S. , Jungnickel, A. , Böhle, U. R. , & Fischer, M. S. (2009). Regionally high rates of hybridization and introgression in german wildcat populations (felis silvestris, carnivora, felidae). Journal of Zoological Systematics and Evolutionary Research, 47, 283–297. [Google Scholar]
- Johnson, C. N. , & Wroe, S. (2003). Causes of extinction of vertebrates during the holocene of mainland australia: Arrival of the dingo, or human impact? The Holocene, 13, 941–948. [Google Scholar]
- Karafet, T. M. , Lansing, J. S. , Redd, A. J. , Reznikova, S. , Watkins, J. C. , Surata, S. P. , … Hammer, M. F. (2005). Balinese Y‐chromosome perspective on the peopling of indonesia: Genetic contributions from pre‐neolithic hunter‐gatherers, Austronesian farmers, and Indian traders. Human Biology, 77, 93–114. [DOI] [PubMed] [Google Scholar]
- Larson, G. , Liu, R. , Zhao, X. , Yuan, J. , Fuller, D. , Barton, L. , … Li, N. (2010). Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proceedings of the National Academy of Sciences of the United States of America, 107, 7686–7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letnic, M. , Fillios, M. , & Crowther, M. S. (2012). Could direct killing by larger dingoes have caused the extinction of the thylacine from mainland Australia? PLoS ONE, 7, e34877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado, P. , & Rozas, J. (2009). Dnasp v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25, 1451–1452. [DOI] [PubMed] [Google Scholar]
- Macintosh, N. W. (1964). 4 Thousand year old thylacine tooth (dasyuridae) + 3 thousand year old dingo skeleton from D J Mulvaneys archaeological excavations at fromms landing lower murray river. Journal of Anatomy, 98, 491–495. [Google Scholar]
- Macintosh, N. W. G. (1975). The origin of the dingo: An enigma In Fox M. W. (Ed.), The wild canids – Their systematics, behavioural ecology and evolution (pp. 87–106). New York, NY: Van Nostrand Reinhold Company. [Google Scholar]
- McEvoy, B. P. , Lind, J. M. , Wang, E. T. , Moyzis, R. K. , Visscher, P. M. , van Holst Pellekaan, S. M. , … Wilton, A. N. (2010). Whole‐genome genetic diversity in a sample of Australians with deep aboriginal ancestry. American Journal of Human Genetics, 87, 297–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, T. , & Letnic, M. (2017). Removal of an apex predator initiates a trophic cascade that extends from herbivores to vegetation and the soil nutrient pool. Proceedings of the Royal Society B: Biological Sciences, 284, 20170111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natanaelsson, C. , Oskarsson, M. C. , Angleby, H. , Lundeberg, J. , Kirkness, E. , & Savolainen, P. (2006). Dog Y chromosomal DNA sequence: Identification, sequencing and snp discovery. BMC Genetics, 7, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson, M. C. R. , Klütsch, C. F. C. , Boonyaprakob, U. , Wilton, A. , Tanabe, Y. , & Savolainen, P. (2011). Mitochondrial DNA data indicate an introduction through mainland Southeast Asia for australian dingoes and polynesian domestic dogs. Proceedings of the Royal Society B: Biological Sciences, 279, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva, V. H. , Fagundes, A. I. , Romão, V. , Gouveia, C. , & Ramos, J. A. (2016). Population‐scale foraging segregation in an apex predator of the north Atlantic. PLoS ONE, 11, e0151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, S. K. , Ghosh, B. , & Roy, S. (1998). Dispersal behaviour of free‐ranging dogs (Canis familiaris) in relation to age, sex, season and dispersal distance. Applied Animal Behaviour Science, 61, 123–132. [Google Scholar]
- Prowse, T. A. A. , Johnson, C. N. , Bradshaw, C. J. A. , & Brook, B. W. (2014). An ecological regime shift resulting from disrupted predator–prey interactions in Holocene Australia. Ecology, 95, 693–702. [DOI] [PubMed] [Google Scholar]
- Pugach, I. , Delfin, F. , Gunnarsdóttir, E. , Kayser, M. , & Stoneking, M. (2013). Genome‐wide data substantiate Holocene gene flow from India to Australia. Proceedings of the National Academy of Sciences of the United States of America, 110, 1803–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , & Drummond, A. (2007). Tracer. Retreived from http://beast.bio.ed.ac.uk/Tracer.
- Randall, D. , Pollinger, J. , Argaw, K. , Macdonald, D. , & Wayne, R. (2010). Fine‐scale genetic structure in Ethiopian wolves imposed by sociality, migration, and population bottlenecks. Conservation Genetics, 11, 89–101. [Google Scholar]
- Rasmussen, M. , Guo, X. , Wang, Y. , Lohmueller, K. E. , Rasmussen, S. , Albrechtsen, A. , … Willerslev, E. (2011). An aboriginal Australian genome reveals separate human dispersals into Asia. Science, 334, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, D. E. , Wayne, R. K. , & Goldstein, D. B. (1999). Genetic evidence for a recent origin by hybridization of red wolves. Molecular Ecology, 8, 139–144. [DOI] [PubMed] [Google Scholar]
- Ripple, W. J. , Chapron, G. , López‐Bao, J. V. , Durant, S. M. , Macdonald, D. W. , Lindsey, P. A. , … Zhang, L. (2016). Saving the world's terrestrial megafauna. BioScience, 66, 807–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripple, W. J. , Chapron, G. , López‐Bao, J. V. , Durant, S. M. , Macdonald, D. W. , Lindsey, P. A. , … Zhang, L. (2017). Conserving the world's megafauna and biodiversity: The fierce urgency of now. BioScience, 67, 197–200. [Google Scholar]
- Ripple, W. J. , Estes, J. A. , Beschta, R. L. , Wilmers, C. C. , Ritchie, E. G. , Hebblewhite, M. , … Wirsing, A. J. (2014). Status and ecological effects of the world's largest carnivores. Science, 343, 1241484. [DOI] [PubMed] [Google Scholar]
- Rueness, E. K. , Jorde, P. E. , Hellborg, L. , Stenseth, N. C. , Ellegren, H. , & Jakobsen, K. S. (2003). Cryptic population structure in a large, mobile mammalian predator: The Scandinavian lynx. Molecular Ecology, 12, 2623–2633. [DOI] [PubMed] [Google Scholar]
- Rueness, E. K. , Stenseth, N. C. , O'Donoghue, M. , Boutin, S. , Ellegren, H. , & Jakobsen, K. S. (2003). Ecological and genetic spatial structuring in the Canadian lynx. Nature, 425, 69–72. [DOI] [PubMed] [Google Scholar]
- Sacks, B. N. , Bannasch, D. L. , Chomel, B. B. , & Ernest, H. B. (2008). Coyotes demonstrate how habitat specialization by individuals of a generalist species can diversify populations in a heterogeneous ecoregion. Molecular Biology and Evolution, 25, 1384–1394. [DOI] [PubMed] [Google Scholar]
- Sacks, B. N. , Brown, S. K. , & Ernest, H. B. (2004). Population structure of California coyotes corresponds to habitat‐specific breaks and illuminates species history. Molecular Ecology, 13, 1265–1275. [DOI] [PubMed] [Google Scholar]
- Sacks, B. N. , Brown, S. K. , Stephens, D. , Pedersen, N. C. , Wu, J.‐T. , & Berry, O. (2013). Y chromosome analysis of dingoes and Southeast Asian village dogs suggests a neolithic continental expansion from Southeast Asia followed by multiple Austronesian dispersals. Molecular Biology and Evolution, 13, 1265–1275. [DOI] [PubMed] [Google Scholar]
- Savolainen, P. , Leitner, T. , Wilton, A. N. , Matisoo‐Smith, E. , & Lundeberg, J. (2004). A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proceedings of the National Academy of Sciences of the United States of America, 101, 12387–12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth, N. C. , Shabbar, A. , Chan, K.‐S. , Boutin, S. , Rueness, E. K. , Ehrich, D. , … Jakobsen, K. S. (2004). Snow conditions may create an invisible barrier for lynx. Proceedings of the National Academy of Sciences of the United States of America, 101, 10632–10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, D. (2011). The molecular ecology of australian wild dogs: Hybridisation, gene flow and genetic structure at multiple geographic scales. School of Animal Biology, University of Western Australia. Doctor of Philosophy: 134.
- Stephens, D. , Wilton, A. N. , Fleming, P. J. S. , & Berry, O. (2015). Death by sex in an Australian icon: A continent‐wide survey reveals extensive hybridization between dingoes and domestic dogs. Molecular Ecology, 24, 5643–5656. [DOI] [PubMed] [Google Scholar]
- Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, P. (1992). The behavioural ecology of dingoes in north‐western Australia. Iv. Social and spatial organistaion, and movements. Wildlife Research, 19, 543–563. [Google Scholar]
- Thomson, P. , Rose, K. , & Kok, N. (1992). The behavioural ecology of dingoes in north‐western australia. Vi. Temporary extraterritorial movements and dispersal. Wildlife Research, 19, 585–595. [Google Scholar]
- Tobler, R. , Rohrlach, A. , Soubrier, J. , Bover, P. , Llamas, B. , Tuke, J. , … Cooper, A. (2017). Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia. Nature, 544, 180–184. [DOI] [PubMed] [Google Scholar]
- van Bommel, L. , & Johnson, C. N. (2012). Good dog! Using livestock guardian dogs to protect livestock from predators in Australia's extensive grazing systems. Wildlife Research, 39, 220–229. [Google Scholar]
- van Holst Pellekaan, S. M. (2001). Origins of the Australian and New Guinean aborigines. In Els Chichester, U.K: John Wiley & Sons, Ltd. [Google Scholar]
- van Holst Pellekaan, S. (2013). Genetic evidence for the colonization of Australia. Quaternary International, 285, 44–56. [Google Scholar]
- vonHoldt, B. M. , Cahill, J. A. , Fan, Z. , Gronau, I. , Robinson, J. , Pollinger, J. P. , … Wayne, R. K. (2016). Whole‐genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Science Advances, 2, e1501714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach, A. D. , Johnson, C. N. , Ritchie, E. G. , & O'Neill, A. J. (2010). Predator control promotes invasive dominated ecological states. Ecology Letters, 13, 1008–1018. [DOI] [PubMed] [Google Scholar]
- Wallach, A. D. , Ritchie, E. G. , Read, J. , & O'Neill, A. J. (2009). More than mere numbers: The impact of lethal control on the social stability of a top‐order predator. PLoS ONE, 4, e6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton, A. (2001). DNA methods of assessing australian dingo purity In Dickman C. R., & Lunney D. (Eds.), A symposium on the Australian dingo (pp. 49–55). Sydney, NSW: Royal Zoological Society of New South Wales. [Google Scholar]
- Wilton, A. N. , Steward, D. J. , & Zafiris, K. (1999). Microsatellite variation in the Australian dingo. Journal of Heredity, 90, 108–111. [DOI] [PubMed] [Google Scholar]


