Abstract
Male Tsumura Suzuki obese diabetes (TSOD) mice spontaneously develop obesity and obesity-related metabolic syndrome. Gut dysbiosis, an imbalance of gut microbiota, has been implicated in the pathogenesis of metabolic syndrome, but its mechanisms are unknown. Short-chain fatty acids (SCFAs) are the main fermentation products of gut microbiota and a link between the gut microbiota and the host’s physiology. Here, we investigated a correlation among gut dysbiosis, SCFAs, and metabolic syndrome in TSOD mice. We detected enriched levels of Gram-positive bacteria and corresponding decreases in Gram-negative bacteria in 24-wk-old metabolic syndrome-affected TSOD mice compared with age-matched controls. The abundance of Bacteroidetes species decreased, the abundance of Firmicutes species increased, and nine genera of bacteria were altered in 24-wk-old TSOD mice. The total plasma SCFA level was significantly lower in the TSOD mice than in controls. The major plasma SCFA—acetate—decreased in TSOD mice, whereas propionate and butyrate increased. TSOD mice had no minor SCFAs (valerate and hexanoate) but normal mice did. We thus concluded that gut dysbiosis and consequent disruptions in plasma SCFA profiles occurred in metabolic syndrome-affected TSOD mice. We also propose that the TSOD mouse is a useful model to study gut dysbiosis, SCFAs, and metabolic syndrome.
Introduction
Metabolic syndrome comprises a combination of obesity-related metabolic alterations that increases the risk of type 2 diabetes mellitus and cardiovascular disease1,2. The clinical hallmarks of this syndrome include insulin resistance, hyperglycemia, hyperlipidemia, and nonalcoholic fatty liver disease (NAFLD)3,4. The essential feature of metabolic syndrome is a state of low-grade inflammation5. Gut microbiota has been implicated as a pathogenic factor that affects a host’s metabolism6. Mammals harbor diverse and immensely active gut microbiota that consists of more than 10 trillion microbial cells and more than 1000 microbial strains7. Via dynamic crosstalk with a host, this commensal microbiota can have a number of functions that affect the host’s physiology, from immune responses to energy metabolism8,9. Growing evidence supports the belief that gut microbiota is closely involved in the development of various diseases, including chronic gastrointestinal diseases10, neurological diseases11,12, and systemic diseases13,14. Alterations in gut microbiota composition may play a critical role in the development of metabolic syndrome, which is especially relevant to obesity-associated inflammation (i.e., diabetes mellitus, NAFLD, and nonalcoholic steatohepatitis [NASH])15–27. The mechanism by which gut microbiota affects a host’s physiology may be at least partly mediated by short-chain fatty acids (SCFAs), which contain 1–6 carbons and are the most abundant product of bacterial fermentation of undigested dietary fibers8,28–30. SCFAs can activate G-coupled receptors, inhibit histone deacetylase (HDAC), and be used as an energy substrate, thereby affecting the host’s physiological processes30.
Tsumura Suzuki obese diabetes (TSOD) mice were originally established as a spontaneous model of type 2 diabetes mellitus31,32 and were also shown to spontaneously develop NASH33, a progressive phenotype of NAFLD34. Given that these pathological manifestations in TSOD mice are closely related to low-grade inflammation, we hypothesized that alterations in gut microbiota and plasma SCFA profiles in TSOD mice may affect a host’s immune system and induce inflammation, which would underlie the development of metabolic syndrome in TSOD mice. In this study, we analyzed the gut microbiome and plasma SCFA profiles in 24-wk-old TSOD mice that had already developed insulin resistance and NASH33.
Results
Analysis of the gut microbiome in TSOD mice
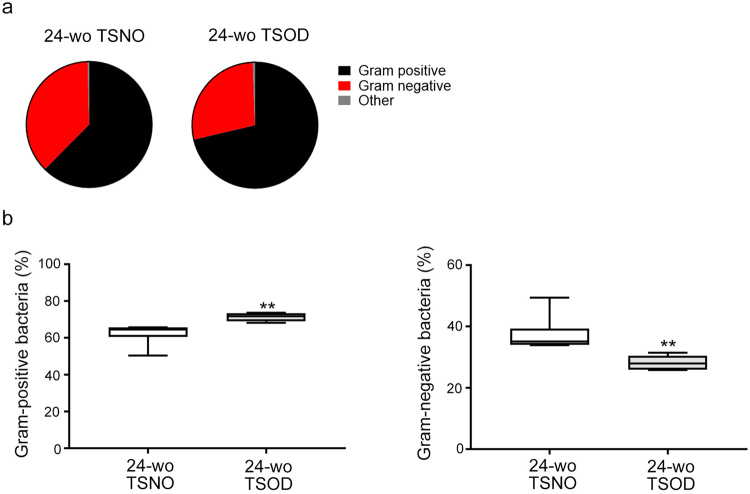
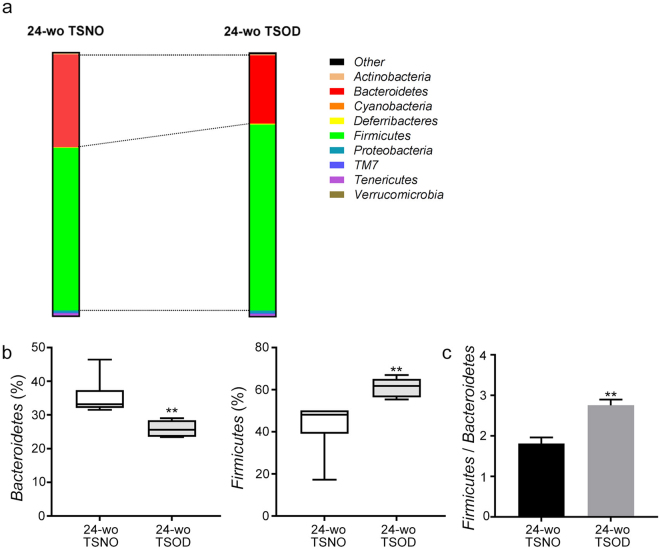
We first confirm that body weights, the ratios of visceral fat or liver to body weight were significantly higher in 24-wk-old TSOD mice (Supplementary Fig. S1a–c). We also scored the liver histology according to our previous report (Supplementary Fig. S1d)33. Because microbiota in fecal samples is widely accepted as a surrogate for gut microbiota, we collected feces from 24-wk-old TSOD male mice and Tsumura Suzuki non-obesity (TSNO) mice (controls), and we analyzed the taxonomic compositions of the microbiota by using 16S ribosomal RNA (rRNA) gene sequencing of DNA extracted from the fecal samples. As Fig. 1a shows, 24-wk-old metabolic syndrome-affected TSOD33 mice and age-matched control mice manifested different compositions of Gram-positive and Gram-negative bacteria. Compared with control mice, TSOD mice had a significantly higher content of Gram-positive bacteria but a significantly lower content of Gram-negative bacteria (Fig. 1b). We also observed marked changes in the composition of intestinal flora at the phylum level in TSOD mice (Fig. 2a). The percentage of the “obese bacteria”, i.e., Firmicutes, was significantly higher and that of the “lean bacteria”, i.e., Bacteroidetes, was significantly lower in TSOD mice compared with controls (Fig. 2b). These findings were consistent with results from a study of obese human subjects and animals18,19. Also, the ratio of Firmicutes to Bacteroidetes was higher in TSOD mice (Fig. 2c). These results reflected the changes in the composition of the populations of Gram-positive and Gram-negative bacteria (Fig. 1), inasmuch as Firmicutes and Bacteroidetes were the most abundant phyla for Gram-positive and Gram-negative bacteria, respectively. These data suggest that the TSOD mouse is a useful model for metabolic syndrome and can manifest the characteristics of the obesity-specific gut microbiome18,19.
Figure 1.
Increased levels of Gram-positive bacteria and decreased levels of Gram-negative bacteria in 24-wk-old (24-wo) TSOD mice. (a) Composition of fecal bacteria in 24-wk-old TSOD mice and age-matched TSNO mice. (b) Comparison of the percentages of Gram-positive bacteria and Gram-negative bacteria in 24-wk-old TSOD mice and age-matched TSNO mice. Boxes indicate the interquartile ranges between the first and third quartiles, and the lines within the boxes indicate the medians. If no error bars appear, the experimental error was smaller than the symbol itself. **P < 0.01 versus TSNO mice.
Figure 2.
Analysis of fecal bacteria at the phylum level. (a) Composition of fecal bacteria at the phylum level in 24-wk-old TSOD mice and age-matched TSNO mice. (b,c) Comparison of the percentages (b) and ratio (c) of the obese microbiota Firmicutes and the lean microbiota Bacteroidetes in 24-wk-old TSOD mice and age-matched TSNO mice. (b) Boxes indicate the interquartile ranges between the first and third quartiles, and the lines within the boxes indicate the medians. If no error bars appear, the experimental error was smaller than the symbol itself. (c) Data are means ± SEM (n = 6). **P < 0.01 versus TSNO mice.
Also, among the major bacterial families, we found two families in the phylum Firmicutes—Clostridiaceae and Erysipelotrichaceae—that showed significant increases in 24-wk-old TSOD mice (Table 1). These bacteria were reportedly involved in a host’s inflammatory response35,36. At the genus level, we detected about 50 bacterial species in both groups of mice. Among these genera, the percentages of nine changed significantly in 24-wk-old TSOD mice (Table 2). As an important finding, these bacteria included Bilophila, Turicibacter, and Lactobacillus, which have been implicated in obesity or NAFLD37–40.
Table 1.
Relative percentages of bacterial groups at the family level that contained more than one genus.
| Phylum | Class | Order | Family | TSNO | TSOD |
|---|---|---|---|---|---|
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | 9.858 | 7.370 |
| 6.925–12.210 | 6.784–9.423 | ||||
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 9.577 | 5.173 |
| 7.100–10.980 | 4.179–7.909 | ||||
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | 2.614 | 9.891 |
| 1.523–5.542 | 5.262–20.110* | ||||
| Firmicutes | Clostridia | Clostridiales | Peptococcaceae | 0.748 | 0.500 |
| 0.474–1.386 | 0.440–0.659 | ||||
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | 0.645 | 3.476 |
| 0.485–0.947 | 3.192–4.629** | ||||
| Firmicutes | Clostridia | Clostridiales | Mogibacteriaceae | 0.551 | 0.512 |
| 0.363–0.718 | 0.454–0.638 | ||||
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | 0.299 | 0.398 |
| 0.227–0.592 | 0.198–0.564 | ||||
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | 0.065 | 0.356 |
| 0.020–0.548 | 0.228–0.419 |
Values are medians and interquartile ranges. *P < 0.05, **P < 0.01 versus 24-wk-old TSNO mice.
Table 2.
List of bacteria whose percentages changed significantly in 24-wk-old TSOD mice.
| Phylum | Class | Order | Family | Genus | TSNO | TSOD |
|---|---|---|---|---|---|---|
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 6.350 | 3.524 |
| 4.374–8.140 | 3.334–4.001* | |||||
| Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae | Lactobacillus | 5.629 | 12.130 |
| 1.817–10.070 | 11.030–13.240** | |||||
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Dorea | 0.965 | 0.434 |
| 0.629–1.316 | 0.403–0.632* | |||||
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Allobaculum | 0.521 | 9.607 |
| 0.078–1.734 | 4.505–19.320** | |||||
| Firmicutes | Erysipelotrichi | Erysipelotrichales | Erysipelotrichaceae | Coprobacillus | 0.437 | 0.130 |
| 0.315–0.650 | 0.068–0.276* | |||||
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Candidatus Arthromitus | 0.254 | 0.059 |
| 0.184–0.363 | 0.011–0.223** | |||||
| Actinobacteria | Actinobacteria | Bifidobacteriales | Bifidobacteriaceae | Bifidobacterium | 0.004 | 0.066 |
| 0–0.024 | 0.013–0.177* | |||||
| Firmicutes | Bacilli | Turicibacterales | Turicibacteraceae | Turicibacter | 0.000 | 5.346 |
| 0.000–0.008 | 4.348–6.740** | |||||
| Proteobacteria | Deltaproteobacteria | Desulfovibrionales | Desulfovibrionaceae | Bilophila | 0.000 | 0.169 |
| 0.000–0.000 | 0.150–0.254* |
Values are medians and interquartile ranges. *P < 0.05, **P < 0.01 versus 24-wk-old TSNO mice.
Analysis of the quantity and type of SCFAs in plasma
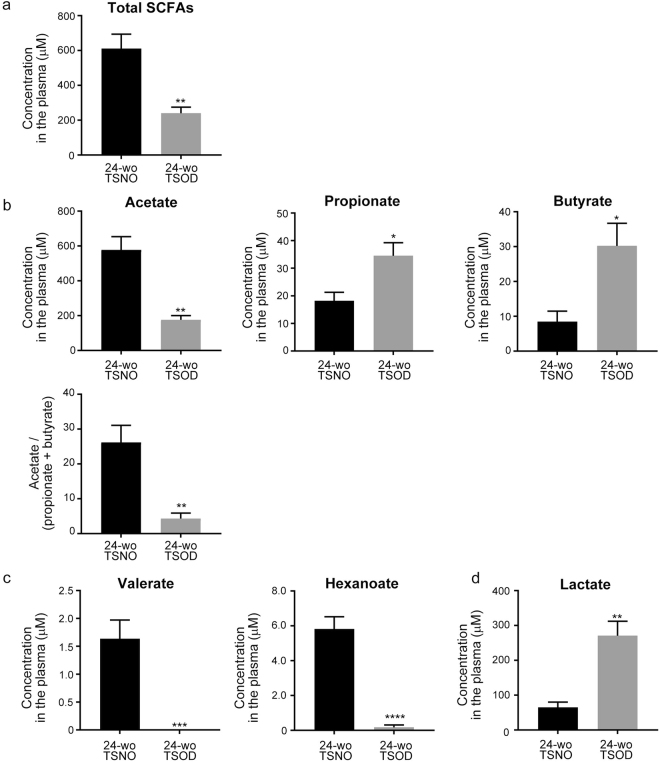
Whereas the homeostatic balance of gut microbiota benefits a host, an imbalance between beneficial and pathogenic bacteria in the presence of dysbiosis would be detrimental to the host. SCFAs are the main fermentation product of gut microbiota and perform diverse functional roles that affect the host’s physiology30. We therefore investigated whether the plasma SCFA profiles changed in TSOD mice. As Fig. 3 shows, we detected acetate, propionate, and butyrate as the major SCFAs, and valerate and hexanoate as the relatively minor SCFAs, in the plasma of TSOD and TSNO mice. The total concentration of plasma SCFAs was significantly lower in 24-wk-old TSOD mice than in the age-matched control mice (Fig. 3a). Acetate, which was the most abundant SCFA, decreased to approximately 30% of the control value in 24-wk-old TSOD mice (Fig. 3b). However, the plasma concentrations of the other major SCFAs—propionate and hexanoate—were 2–3 times higher in 24-wk-old TSOD mice than in controls (Fig. 3b). In agreement with a previous study reporting that leaner people showed a higher ratio of acetate to butyrate plus propionate41, the ratio of acetate to butyrate plus propionate in TSOD mice was significantly lower compared with that in controls (Fig. 3b). However, the minor SCFAs valerate and hexanoate, whose plasma concentrations were in the range of 1.5–6.0 nM in TSNO mice, were almost absent in 24-wk-old TSOD mice (Fig. 3c). Furthermore, the plasma concentration of lactate, which is not only the precursor of SCFAs but also a signaling molecule that reportedly affects a host’s physiology by modulating HDAC and G protein-coupled receptor 81 signaling30, significantly increased in 24-wk-old TSOD mice (Fig. 3d). These results indicate that the dysbiosis in TSOD mice led to a loss of type and quantity of SCFAs, which may have a role in the development of metabolic syndrome in this mouse model.
Figure 3.
Analysis of plasma SCFAs in TSOD mice and age-matched control mice. (a) Total concentration of plasma SCFAs. (b) Concentrations of the major SCFAs acetate, propionate, and butyrate, and the ratio of acetate to propionate plus butyrate. (c) Concentrations of the minor SCFAs valerate and hexanoate and the precursor of SCFAs (lactate). Data are means ± SEM (n = 6). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
To investigate the hypothesis that alterations in the gut microbial community may contribute to the spontaneous development of metabolic syndrome in TSOD mice, we analyzed the gut microbiota and plasma SCFA profiles in 24-wk-old TSOD mice. As expected, we found the following modifications in the microbial community compared with TSNO mice: (1) an increased ratio of Gram-positive bacteria to Gram-negative bacteria, (2) an increased abundance of the obese microbiota Firmicutes and a decreased abundance of the lean microbiota Bacteroidetes, (3) an altered abundance of several bacteria, including Bilophila, Turicibacter, and Lactobacillus, among others (Table 2).
Dysbiosis can generally be categorized into three types: (1) loss of beneficial microbes, (2) excessive growth of harmful microorganisms, and (3) loss of overall microbial diversity42. These categories are not mutually exclusive; in most cases, they can occur simultaneously. On the basis of our results described above, we concluded that dysbiosis occurred in 24-wk-old TSOD mice. Gut dysbiosis is defined as qualitative and quantitative changes in gut microbiota, metabolic activity, and local distribution43. In addition to having a role in the host’s digestive system, gut dysbiosis has been implicated in the development of obesity and metabolic syndrome by activating a host’s immune response and inflammation, disturbing the intestinal barrier integrity, and causing metabolic abnormalities44. The exact role of the gut dysbiosis that we observed here in the etiology or pathology of spontaneous metabolic syndrome in TSOD mice is yet to be elucidated. Colonization experiments such as transfer of microbiota from TSNO mice into TSOD mice or vice versa are required. We previously reported that TSOD mice developed a hepatic tumor that histopathologically resembled human hepatocellular carcinoma (HCC)45. In the present study, the percentage of Gram-positive bacteria was significantly higher in 24-wk-old TSOD mice compared with that in controls. As an interesting result, an obesity-associated increase in Gram-positive bacteria in the gut reportedly produced secondary bile acids that promoted HCC development46. Bile acids also reportedly activated a signaling network in hepatocytes that triggered hepatic inflammation47,48. Given that increased bile acids in the gut are known to favor Gram-positive bacteria49, the increase in Gram-positive bacteria in TSOD mice may at least partly contribute to hepatic carcinogenesis. Thus, TSOD mice may be useful for analyzing the effects of novel therapeutic agents, especially those that modulate gut microbiota, for the prevention and treatment of metabolic syndrome-associated HCC.
The gut microbiota was reportedly significantly affected by obesity in humans and in animal models. Ley et al. reported that the abundance of Bacteroidetes decreased together with a proportional increase in the phylum Firmicutes in obese mice, compared with their control counterparts, independently of diet20. In agreement with this finding, a reduced abundance of intestinal Bacteroidetes associated with an increased abundance of Firmicutes was also observed in obese humans18. Also, patients with NASH had a lower percentage of Bacteroidetes compared with both patients with simple steatosis and healthy controls22. Although other studies did not always reproduce these results50,51, in the present study, the abundance of Firmicutes increased together with a corresponding decrease in Bacteroidetes in the spontaneous mouse model of metabolic syndrome. These alterations in Firmicutes and Bacteroidetes abundance are thought to contribute to the development of obesity by switching the host’s metabolism to increase the adsorption of fatty acids and calories and thereby lead to weight gain19,52. Bacteroidetes reportedly increased the production of SCFAs53. Thus, the disruption in gut microbiota compositions of Firmicutes and Bacteroidetes in the current study may contribute to the development of obesity in TSOD mice. As an interesting finding, several studies reported an increase in the Firmicutes to Bacteroidetes ratio in patients with irritable bowel syndrome, which shares the characteristic of chronic inflammation with metabolic syndrome54–56.
At the microbial family level, we found that the percentages of Erysipelotrichaceae and Clostridiaceae were significantly higher in 24-wk-old TSOD mice than those in controls (Table 1). In other studies, the former increased in patients with type 2 diabetes57 and arthritis58, both of which are associated with severe inflammation. The latter increased in patients with autism, with unknown mechanisms and functions59,60. Bacteria in these families have been implicated in inflammation35,36, which suggests that these bacteria may contribute to the pathology in TSOD mice via exacerbation of inflammation. In addition, the percentages of several microbial genera changed in 24-wk-old TSOD mice (Table 2). In agreement with previous studies of patients with NAFLD39,40, the percentage of Lactobacillus increased in TSOD mice. Inasmuch as this bacterial group can often be used as a probiotic, elucidating the role of Lactobacillus in the etiology and pathology of NAFLD or NASH is a future challenge. We found that the percentage of Turicibacter, which was implicated in the production of butyrate61, was markedly higher in 24-wk-old TSOD mice, in agreement with the increase in plasma butyrate in 24-wk-old TSOD mice. Dimova et al. showed that Turicibacter increased in mice fed a high-fat diet38. However, two studies reported a decrease in Turicibacter in mice in response to high-fat feeding62,63. Although the role of Turicibacter in the development of metabolic syndrome is unknown, it is notable that one study reported an increased abundance of Turicibacter in the gut of patients with rheumatoid arthritis, an immune-mediated disease64. Another important finding is the presence of the pathobiont Bilophila in 24-wk-old TSOD mice, whereas Bilophila was absent in control mice. Bilophila is a sulfite-reducing pathobiont and causes an interleukin-10-mediated immune response, which leads to colitis in mice65. The growth of Bilophila can reportedly be promoted by increased taurine-conjugated bile acids66, which is consistent with the possibility that bile acid-favoring Firmicutes increased in TSOD mice.
Plasma SCFA profiles in TSOD mice also differed from those in control mice. Compositions of SCFAs depend on the microbial community compositions and the type and quantity of fermentation substrates (i.e., dietary fibers)37,67,68. In the present study, because we fed both TSNO and TSOD mice standard chow, the major determinant of plasma SCFAs would supposedly be the composition of gut microbiota. However, because the plasma SCFA profile is an emergent property of the microbial community, making predictions from taxon-based analysis and identifying certain microbes as responsible factors are difficult69. Besides acting as an energy substrate, SCFAs can function as signaling molecules by modulating neuroendocrine and anti-inflammatory responses in various tissues and organs69. Thus, the type and quantity of SCFAs produced by the gut microbiota are also important for the development of obesity and metabolic syndrome. In the present study, the total SCFA concentrations decreased significantly in 24-wk-old TSOD mice compared with concentrations in controls. With regard to specific SCFAs, the plasma concentration of acetate was significantly lower and concentrations of propionate and butyrate were significantly higher in 24-wk-old TSOD mice than in control mice. Inasmuch as the gut microbiome in TSOD mice favored Firmicutes species, which mainly produce butyrate, and did not favor Bacteroidetes species, which primarily produce acetate and propionate70, alterations in the SCFA profiles in TSOD mice seemed to roughly reflect the altered Firmicutes/Bacteroidetes ratio of the microbial community. The mechanism of the increase in propionate in TSOD mice remains to be elucidated, though it may depend on the mouse strains.
Acetate reportedly mediated a Bifidobacterium-induced improvement in the intestinal barrier against bacterial endotoxin, possibly by strengthening tight junctions of epithelial cells71,72. Although whether the reduced abundance of Bifidobacterium in the current study is solely accountable for the decrease in plasma acetate is unknown, that the decrease in plasma acetate may contribute to development of the inflammation-related pathology of metabolic syndrome in 24-wk-old TSOD mice is highly likely. Butyrate has anti-inflammatory activity via modulating HDAC73. Propionate also has an anti-inflammatory property74. However, Schwiertz et al. reported that the propionate level increased in fecal samples of overweight and obese subjects75. In the present study, both plasma butyrate and propionate levels increased in 24-wk-old TSOD mice, which strongly suggests that butyrate and propionate may have pathogenic roles via mechanisms yet to be elucidated. We also observed that the level of lactate, the precursor of SCFAs30, increased in 24-wk-old TSOD mice, a finding that was consistent with results from previous reports showing that the plasma lactate concentration was higher in subjects with type 2 diabetes and obesity than in normal subjects76,77. Given that lactate acts as a mediator of inflammation78, increased lactate in TSOD mice may be a pathogenic factor for induction or maintenance of low-grade inflammation. Acetate, which was decreased in TSOD mice, is mainly produced by the Bacteroidetes phylum70 of which abundance was also decreased in TSOD mice. As lactate can be further metabolized to acetate79, the increase in plasma lactate in TSOD mice might be at least partly due to a decrease in the acetate production by Bacteroidetes. With regard to the minor SCFAs, i.e., valerate and hexanoate, we cannot currently compare our results with others, because of the lack of relevant literature.
In the present study, we observed altered SCFA profiles, with a decrease in total SCFA amounts and diversity, in 24-wk-old TSOD mice. An altered SCFA profile or a disease-specific SCFS profile has been implicated in the pathology of several inflammatory diseases, including Hirschsprung’s-associated enterocolitis80, familial Mediterranean fever81, and celiac diseases82,83. In addition to the respective roles of each SCFA, the total SCFA profile itself, including composition and diversity, may affect a host’s physiology81,84. SCFAs reportedly enhanced secretion of glucagon-like peptide (GLP-1) that enhances glucose tolerance85. Via the SCFA receptors, SCFAs reportedly modulated the release of proinflammatory cytokines such as tumor necrosis factor α and interleukin 6 that may alter insulin sensitivity and contribute to the development of persistent chronic inflammation86,87, which has been implicated in cancer development including that of HCC88. It is also reported that gut microbiota suppress fat accumulation via the SCFA receptor89. These lines of evidence suggest that the altered SCFA profile observed in TSOD mice potentially contribute to the development of metabolic syndrome and HCC via multiple pathways. Although the physiological and pathological roles of the minor SCFAs are not fully understood, our results suggest that the disrupted plasma SCFA profiles may play a role in the development of obesity and metabolic syndrome in TSOD mice. Additional studies are warranted.
In summary, we observed gut dysbiosis and disruptions in plasma SCFA profiles in TSOD mice. Identification of the mechanisms as well as specific bacteria and bacterial metabolites that are responsible for the pathology and etiology of metabolic syndrome in this mouse model will allow for development of precise treatments to prevent or manage chronic inflammatory diseases. We also propose that the TSOD mouse, which demonstrated its own “microbial signature,” is a useful model to study gut dysbiosis, SCFAs, and metabolic syndrome.
Methods
Animals
Six male TSOD mice and six male TSNO mice were purchased from the Institute for Animal Reproduction (Ibaraki, Japan). Two or three mice were reared in plastic cages in a non-barrier-sustained animal room at 23 ± 2 °C in 50 ± 10% relative humidity under a 12/12-h light/dark cycle. All mice were maintained with the basal diet MF (Oriental Yeast Co., Ltd., Tokyo, Japan) and chlorinated water ad libitum. Fecal samples were collected from the colon. The study was performed in accordance with the animal experiment guidelines specified by the University of Tokushima. All experimental protocols were approved by the animal research committee of Tokushima University.
Analysis of the gut microbiome
DNA was extracted from fecal samples by using the isopropanol precipitation technique. Briefly, 30–40 mg of mouse feces was suspended in 19× volume of phosphate-buffered saline and was homogenized by using the FastPrep-24 homogenizer (MP Biomedicals, Santa Ana, CA). A sample consisting of 250 µL of TE buffer (200 mM Tris-HCl, 80 mM ethylenediaminetetraacetic acid, pH 9.0), 50 µL of 10% sodium dodecyl sulfate, 500 µL of TE-saturated phenol (Nippon Gene Co., Ltd., Tokyo, Japan), and 0.3 g of glass beads (diameter 0.1 mm; As-One Co., Ltd., Osaka, Japan, #BZ-01) was added to 200 µL of the ground fecal sample. The fecal samples were further homogenized with a FastPrep-24 homogenizer for 30 s, after which they were centrifuged at 15,000 rpm for 5 min at 4 °C. A sample of 400 µL of a phenol/chloroform/isoamyl alcohol (25:24:1) mixture (Nippon Gene Co., Ltd.) was added to the supernatant, vortexed for 10 s, and centrifuged at 15,000 rpm for 5 min at 4 °C. Isopropanol (250 µL) (Wako Pure Chemical Industries Ltd., Osaka, Japan) was added to 250 µL of the supernatant, mixed by flipping, and kept at room temperature for 10 min followed by centrifugation at 15,000 rpm for 10 min at room temperature. The supernatant was removed, and the resultant pellet was washed with 400 µL of ice-cold ethanol. The extracted DNA was air-dried and then dissolved in 2000 µL of TE buffer (pH 8.0). The V3-V4 region of the bacterial 16S rRNA gene was amplified by using PCR with the TaKaRa Ex Taq HS Kit (TaKaRa Bio, Shiga, Japan) and the primer sets of Tru357F (5ʹ-CGCTCTTCCGATCTCTGTACGGRAGGCAGCAG-3ʹ) and Tru806R (5ʹ-CGCTCTTCCGATCTGACGGACTACHVGGGTWTCTAAT-3ʹ). The DNA was concentrated by amplifying, in triplicate, via PCR: preheating at 94 °C for 3 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s, extension at 72 °C for 30 s, and a final terminal extension at 72 °C for 5 min. The amplicon was prepared for a sequencing instrument with the method described in a previous report90.
Analysis of SCFAs in plasma
Overall, nine analytes were targeted for SCFA analysis. Acetic acid, lactic acid, propionic acid (PA), butyric acid, isobutyric acid, valeric acid (VA), isovaleric acid (iso-VA), pivalic acid (tert-butyl-VA), and caproic acid (CA) were purchased from Wako Pure Chemical Co. For internal standards (IS), PA-d 6, BA-d 5, VA-d 9, and CA-d 11 were obtained from Sigma-Aldrich Co. (St. Louis, MO) and CDN Isotopes Co. (Quebec, Canada). Triphenylphosphine (TPP), 2,2-dipyridyl disulfide (DPDS), and 2-picolylamine were obtained from Tokyo Kasei Co. (Tokyo, Japan). These stock solutions were adjusted by using methanol.
The ultra-performance liquid chromatography (UPLC) system was a Waters Acquity H Class (Waters Co., Milford, MA). A reverse phase analysis was performed via an Acquity UPLC BEH C18 column (1.7 μm, 2.1 × 100 mm) at 40 °C. The injection volume was 5 μL. The mobile phase consisting of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in methanol) was delivered at a flow rate of 0.3 mL/min. The gradient elution was as follows: B% = 2, 2, 35, 45, and 98 (0, 3, 10, 12, and 14 min). A Waters Xevo TQD triple quadrupole mass spectrometer was operated with an electrospray ionization (ESI) source in the positive mode. The ionization source conditions were as follows: capillary voltage, 2.00 kV; cone voltage, 20–70 V; collision energy, 10–40 eV; source temperature, 150 °C; and desolvation temperature, 400 °C. The cone and desolvation gas flows were 50 and 800 L/h, respectively, and were obtained by using a nitrogen source (N2 Supplier Model 24S; Anest Iwata, Yokohama, Japan). On the basis of a previous report for useful derivatization of carboxylic acids91, mixed SCFAs and IS solutions were diluted by adding methanol. These solutions were reacted with 2-picolylamine in DPDS and TPP in acetonitrile at 60 °C for 10 min. The reaction mixtures were removed and re-dissolved in 100 μL of methanol/water (80:20, v/v). Finally, the derivatization solutions (5 μL) were analyzed by means of UPLC-ESI-MS/MS. Plasma samples after thawing were added to IS and mixed with equal volumes of methanol and QuEChERS (Supel QuE PSA (EN) 25 mg), vortexed vigorously, and centrifuged at 15,000 rpm for 5 min. The supernatant was then removed, and the remaining residue was re-dissolved in methanol and derivatized by using the process described above for 2-picolylamine. The sample was then analyzed by means of UPLC-ESI/MS/MS.
Statistical analysis
Values were analyzed via the Mann-Whitney U-test (Figs 1b and 2b, Tables 1 and 2) or unpaired Student’s t-test (Figs 2c and 3), by using Prism software (GraphPad Software, La Jolla, CA). Differences were regarded as significant when P < 0.05.
Electronic supplementary material
Acknowledgements
The authors would like to thank the Support Center for Advanced Medical Sciences, Institute of Biomedical Sciences, Tokushima University Graduate School. This work was supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (B) Number 24390181, (C) Number 25340121 and Grant-in-Aid for challenging Exploratory Research Number 15K15098 to K.T.
Author Contributions
K.N. performed the experiments, interpreted the data, and wrote the paper. J.-z.X. analyzed the gut microbiome and interpreted the data. R.N. and K.I. measured concentration of SCFAs and interpreted the data. H.U. and Y.M. performed the experiments. H.A. contributed reagents, materials, and analytical tools and supervised the entire project. K.T. designed the research, interpreted the data, and wrote the paper and takes full responsibility for the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-16189-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Grundy SM, et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 2.Wilson PW, D’Agostino RB, Parise H, Sullivan L, Meigs JB. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation. 2005;112:3066–3072. doi: 10.1161/CIRCULATIONAHA.105.539528. [DOI] [PubMed] [Google Scholar]
- 3.Kaur J. A comprehensive review on metabolic syndrome. Cardiol Res Pract. 2014;2014:943162. doi: 10.1155/2014/943162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Nathan C. Epidemic inflammation: pondering obesity. Mol Med. 2008;14:485–492. doi: 10.2119/2008-00038.Nathan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassaing B, Gewirtz AT. Gut microbiota, low-grade inflammation, and metabolic syndrome. Toxicol Pathol. 2014;42:49–53. doi: 10.1177/0192623313508481. [DOI] [PubMed] [Google Scholar]
- 6.Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Curr Opin Clin Nutr Metab Care. 2007;10:729–734. doi: 10.1097/MCO.0b013e3282efdebb. [DOI] [PubMed] [Google Scholar]
- 7.Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 8.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 10.Leone V, Chang EB, Devkota S. Diet, microbes, and host genetics: the perfect storm in inflammatory bowel diseases. J Gastroenterol. 2013;48:315–321. doi: 10.1007/s00535-013-0777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borre YE, et al. Microbiota and neurodevelopmental windows: implications for brain disorders. Trends Mol Med. 2014;20:509–518. doi: 10.1016/j.molmed.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Fung TC, Olson CA, Hsiao EY. Interactions between the microbiota, immune and nervous systems in health and disease. Nat Neurosci. 2017;20:145–155. doi: 10.1038/nn.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu H, Tremaroli V, Backhed F. Linking Microbiota to Human Diseases: A Systems Biology Perspective. Trends Endocrinol Metab. 2015;26:758–770. doi: 10.1016/j.tem.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 15.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abu-Shanab A, Quigley EM. The role of the gut microbiota in nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:691–701. doi: 10.1038/nrgastro.2010.172. [DOI] [PubMed] [Google Scholar]
- 17.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 20.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 22.Mouzaki M, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 23.Zhu L, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 24.De Minicis S, et al. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology. 2014;59:1738–1749. doi: 10.1002/hep.26695. [DOI] [PubMed] [Google Scholar]
- 25.Boursier J, Diehl AM. Implication of gut microbiota in nonalcoholic fatty liver disease. PLoS Pathog. 2015;11:e1004559. doi: 10.1371/journal.ppat.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 27.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 29.Neves AL, et al. The microbiome and its pharmacological targets: therapeutic avenues in cardiometabolic diseases. Curr Opin Pharmacol. 2015;25:36–44. doi: 10.1016/j.coph.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 30.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 31.Miura T, et al. Impairment of insulin-stimulated GLUT4 translocation in skeletal muscle and adipose tissue in the Tsumura Suzuki obese diabetic mouse: a new genetic animal model of type 2 diabetes. Eur J Endocrinol. 2001;145:785–790. doi: 10.1530/eje.0.1450785. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi A, et al. Insulin resistance and low sympathetic nerve activity in the Tsumura Suzuki obese diabetic mouse: a new model of spontaneous type 2 diabetes mellitus and obesity. Metabolism. 2006;55:1664–1669. doi: 10.1016/j.metabol.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Nishida T, et al. Spontaneous onset of nonalcoholic steatohepatitis and hepatocellular carcinoma in a mouse model of metabolic syndrome. Lab Invest. 2013;93:230–241. doi: 10.1038/labinvest.2012.155. [DOI] [PubMed] [Google Scholar]
- 34.Matteoni CA, et al. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116:1413–1419. doi: 10.1016/S0016-5085(99)70506-8. [DOI] [PubMed] [Google Scholar]
- 35.Scarpa M, et al. Relationship between mucosa-associated microbiota and inflammatory parameters in the ileal pouch after restorative proctocolectomy for ulcerative colitis. Surgery. 2011;150:56–67. doi: 10.1016/j.surg.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Dinh DM, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dimova LG, Zlatkov N, Verkade HJ, Uhlin BE, Tietge UJF. High-cholesterol diet does not alter gut microbiota composition in mice. Nutr Metab (Lond) 2017;14:15. doi: 10.1186/s12986-017-0170-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raman M, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(868-875):e861–863. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 40.Jiang W, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan SH, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm Bowel Dis. 2016;22:1137–1150. doi: 10.1097/MIB.0000000000000750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holzapfel WH, Haberer P, Snel J, Schillinger U, Huis in’t Veld JH. Overview of gut flora and probiotics. Int J Food Microbiol. 1998;41:85–101. doi: 10.1016/S0168-1605(98)00044-0. [DOI] [PubMed] [Google Scholar]
- 44.Sun M, Wu W, Liu Z, Cong Y. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52:1–8. doi: 10.1007/s00535-016-1242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi T, et al. Histopathological characteristics of glutamine synthetase-positive hepatic tumor lesions in a mouse model of spontaneous metabolic syndrome (TSOD mouse) Mol Clin Oncol. 2016;5:267–270. doi: 10.3892/mco.2016.924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 47.Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175–186. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai SY, et al. Bile acids initiate cholestatic liver injury by triggering a hepatocyte-specific inflammatory response. JCI Insight. 2017;2:e90780. doi: 10.1172/jci.insight.90780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 2010;52:1836–1846. doi: 10.1002/hep.24001. [DOI] [PubMed] [Google Scholar]
- 51.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semova I, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe. 2012;12:277–288. doi: 10.1016/j.chom.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeffery IB, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997–1006. doi: 10.1136/gutjnl-2011-301501. [DOI] [PubMed] [Google Scholar]
- 55.Krogius-Kurikka L, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. doi: 10.1186/1471-230X-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajilic-Stojanovic M, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 57.Larsen N, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Angelis M, et al. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8:e76993. doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finegold SM, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Zhong Y, Nyman M, Fak F. Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Mol Nutr Food Res. 2015;59:2066–2076. doi: 10.1002/mnfr.201500187. [DOI] [PubMed] [Google Scholar]
- 62.Liu W, et al. Diet- and Genetically-induced Obesity Produces Alterations in the Microbiome, Inflammation and Wnt Pathway in the Intestine of Apc + /1638N Mice: Comparisons and Contrasts. J Cancer. 2016;7:1780–1790. doi: 10.7150/jca.15792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Everard A, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forbes JD, Van Domselaar G, Bernstein CN. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Devkota S, Chang EB. Interactions between Diet, Bile Acid Metabolism, Gut Microbiota, and Inflammatory Bowel Diseases. Dig Dis. 2015;33:351–356. doi: 10.1159/000371687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang C, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 68.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ha CW, Lam YY, Holmes AJ. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J Gastroenterol. 2014;20:16498–16517. doi: 10.3748/wjg.v20.i44.16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.den Besten G, et al. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 72.Hsieh, C. Y. et al. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol Rep3 (2015). [DOI] [PMC free article] [PubMed]
- 73.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4:4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 76.Chen YD, Varasteh BB, Reaven GM. Plasma lactate concentration in obesity and type 2 diabetes. Diabete Metab. 1993;19:348–354. [PubMed] [Google Scholar]
- 77.Lovejoy J, Mellen B, Digirolamo M. Lactate generation following glucose ingestion: relation to obesity, carbohydrate tolerance and insulin sensitivity. Int J Obes. 1990;14:843–855. [PubMed] [Google Scholar]
- 78.Wu Y, et al. Lactate, a Neglected Factor for Diabetes and Cancer Interaction. Mediators Inflamm. 2016;2016:6456018. doi: 10.1155/2016/6456018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Demehri FR, et al. Altered fecal short chain fatty acid composition in children with a history of Hirschsprung-associated enterocolitis. J Pediatr Surg. 2016;51:81–86. doi: 10.1016/j.jpedsurg.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ktsoyan ZA, et al. Systemic Concentrations of Short Chain Fatty Acids Are Elevated in Salmonellosis and Exacerbation of Familial Mediterranean Fever. Front Microbiol. 2016;7:776. doi: 10.3389/fmicb.2016.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Di Cagno R, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 2011;11:219. doi: 10.1186/1471-2180-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tjellstrom B, et al. Gut microflora associated characteristics in children with celiac disease. Am J Gastroenterol. 2005;100:2784–2788. doi: 10.1111/j.1572-0241.2005.00313.x. [DOI] [PubMed] [Google Scholar]
- 84.Ktsoyan ZA, et al. Management of familial Mediterranean fever by colchicine does not normalize the altered profile of microbial long chain fatty acids in the human metabolome. Front Cell Infect Microbiol. 2013;3:2. doi: 10.3389/fcimb.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tolhurst G, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puddu A, Sanguineti R, Montecucco F, Viviani GL. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediators Inflamm. 2014;2014:162021. doi: 10.1155/2014/162021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2001;280:E745–751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 88.Berasain C, et al. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci. 2009;1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 89.Kimura I, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Odamaki T, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Higashi T, et al. Simple and practical derivatization procedure for enhanced detection of carboxylic acids in liquid chromatography-electrospray ionization-tandem mass spectrometry. J Pharm Biomed Anal. 2010;52:809–818. doi: 10.1016/j.jpba.2010.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.